Abstract
Raman spectroscopy provides many advantages compared to other common analytical techniques due to its ability of rapid and accurate identification of unknown specimens as well as simple sample preparation. Here, we described potential of Raman spectroscopic technique as an efficient and high throughput method to detect plants infected by economically important viruses. To enhance the detection sensitivity of Raman measurement, surface enhanced Raman scattering (SERS) was employed. Spectra of extracts from healthy and Turnip yellow mosaic virus (TYMV) infected Chinese cabbage leaves were collected by mixing with gold (Au) nanoparticles. Our result showed that TYMV infected plants could be discriminated from non-infected healthy plants, suggesting the current method described here would be an alternative potential tool to screen virus-infection of plants in fields although it needs more studies to generalize the technique.
Keywords: discrimination, Raman spectroscopy, surface enhanced Raman scattering (SERS), Turnip yellow mosaic virus (TYMV), virus detection
Virus infection of vegetable plants is a serious problem in farming industry since it substantially retards growth of plants and finally leads to produce vegetables with no commercial value. Moreover, an infected plant could transmit viruses to neighboring plants by diverse courses such as contact by insects or growers as well as ground water, so all plants in a cultivation field could be infected in a worst case. Therefore, a virus-infected plant should be identified as soon as possible and removed to prevent further spread. Conventionally, enzyme-linked immunosorbent assay (ELISA) and polymerase chain reaction (PCR) have been used for diagnosis of virus infection. However, the length of time required for their analysis is long and also both methods need complicated sample preparation steps. Especially, when large numbers of plants are subject to simultaneous diagnosis for the purpose of large scale omnidirectional screening, an analytical method needs to be fast with ability of high throughput analysis.
One of candidates to meet the above-mentioned analytical requirement could be Raman spectroscopy since it provides rich structural and chemical information. It relies on inelastic scattering, or Raman scattering, of monochromatic light such as laser. Raman scattering, resulted from the shift in energy (or wavewlength), usually provides information about the vibrational modes of a measured sample. Also, the measurement is fast and non-destructive (El-Abassy et al., 2009; Mozharov et al., 2010; Park et al., 2007). Recently, Raman spectroscopy has also been employed for rapid identification of bacteria (Beier et al., 2012; Prucek et al., 2012). In this study, we have exploited the possibility of Raman spectroscopy for detection of Turnip yellow mosaic virus (TYMV) (Chalcroft and Matthews, 1966; Matthews, 1958; Reid and Matthews, 1966; Turano et al., 1976) infection on Chinese cabbage plants. We artificially infected the plants with TYMV, and both extracts from TYMV-infected as well as healthy (non-infected) leaves were measured. If TYMV concentration is low in the extracts, its Raman feature would not be recognizable since Raman spectroscopy itself is not a sensitive analytical method. To enhance the sensitivity of measurement, SERS (surface enhanced Raman scattering) spectra were collected by mixing the extracts with gold (Au) nanoparticles. The enhancement of Raman signal arises from the generation of strong electromagnetic fields that are induced by the localized surface plasmon resonance of noble metal nanostructures (Moskovits, 1978; Moskovits, 2005; Schatz and Van Duyne, 2002), so providing ~106 to 1010 fold increase over normal Raman signal when a molecule is adsorbed on the metal surface.
To examine Raman spectral feature of TYMV, the purification of TYMV was initially performed (Zaihlin, 1975). Leaves (40 g) were ground with a warming blender for 60 seconds in 4 vol. of 0.1 M sodium phosphate buffer (pH 7.0) containing 0.01 M Na-DIECA and 0.01% thioglycolic acid. The homogenate was stirred for 30 min with adding 1:1 mixture of 8% chloroform and n-butanol, and then followed by centrifugation at 8000 rpm for 25 min. The supernatant obtained was stirred for 60 min and left in refrigerator. Pellets were suspended in 0.01 M sodium phosphate buffer (pH 7.0). After low centrifugation, virus in the supernatant was pelleted by ultracentrifugation at 130,000 rpm for 2 h. After 2 repeat of low- and ultracentrifugation, the partially purified virus was purified further by 10–40% sucrose density gradient centrifugation at 24,000 rpm for 2 h (SRP28SA). Milkish virus band formed on medium of sucrose gradients was removed with hypodermic syringe, and the virions were concentrated by ultracentrifugation.
For the spectral collection, 20 μl of 740 nM TYMV solution was dropped on a quartz window and then dried. This procedure was repeated 3 times to increase the population of TYMV to obtain reasonable Raman intensity. Raman spectra were collected using a dispersive Raman microscope (Kaiser Optical Inc., laser wavelength: 785 nm). Laser beam (approximately 10 μm in diameter) was focused using an objective lens (10 ×/0.25 numerical aperture) for the data acquisition (spectral resolution: 4 cm−1).
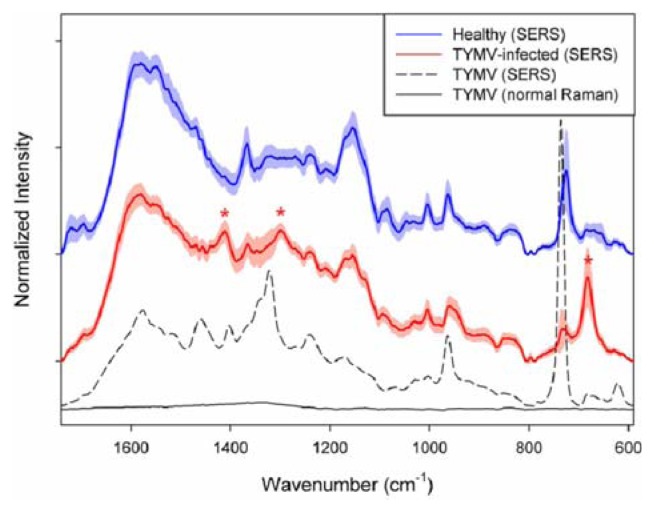
The obtained Raman spectrum (black solid line in the bottom) in the 1,740–590 cm−1 range is shown in Figure 1. The spectrum is a result of averaging 20 spectra collected at different spots on the sample. Before the averaging, the baseline of each spectrum was corrected at 1,740, 802, 790, 697 and 590 cm−1. As shown, no distinct spectral feature corresponding to TYMV is observed due to inherent low sensitivity of normal Raman measurement.
Fig. 1.
Normal Raman spectrum (black) and SERS spectrum (dashed) of pure turnip yellow mosaic virus (TYMV). SERS spectra of the extracts (1 μl) from healthy (blue) and TYMV-infected (red) leaves. The shades with the same color indicate the corresponding standard deviation (1σ). The spectra of samples were collected with an exposure time of 40 seconds (excitation wavelength: 785 nm). The laser power at the sample was approximately 45 mW.
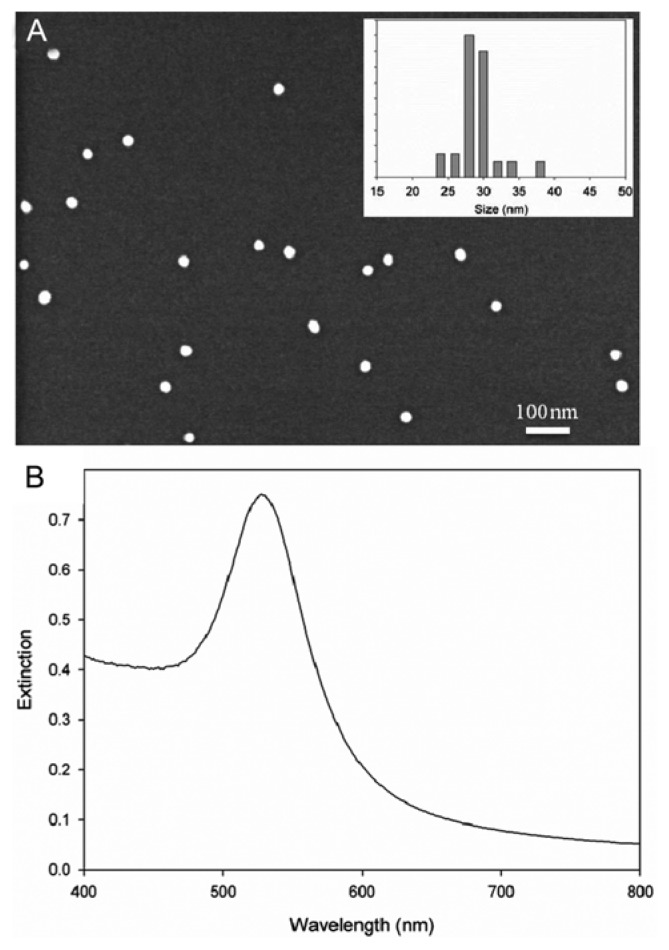
For the collection of SERS spectra, a sample is usually mixed with a solution of Au nanoparticles to allow molecules to adsorb on the Au surface. In this study, citrate reduction method (Chi et al., 2010; Turkevich et al., 1951) was used to synthesize Au nanoparticles. Fig. 2A shows a scanning electron microscope (SEM) image of the synthesized Au nanoparticles and the corresponding size distribution. The average size was 29.15 ± 2.91 nm with maximum extinction at 525 nm (refer to Fig. 2B). As shown, the size of Au nanoparticles is fairly uniform. A visible spectrum was collected using a UV/VIS spectrometer (Optizen 3220UV), equipped with a silicone photodiode detector (spectral resolution < 1.0 nm).
Fig. 2.
SEM image of the synthesized Au nanoparticles and their size distribution (A) and the corresponding extinction spectrum (B).
On the contrary, distinct and strong TYMV bands are observable in the SERS spectrum as shown in Fig. 1 (dashed line), although the concentration is only 67.3 nM. Two separate samples, mixtures of pure TYMV solution with Au nanoparticles were prepared for SERS measurement and 10 SERS spectra were collected for each sample. The spectrum in the figure is a result of averaging these 20 spectra. As shown, the enhancement of Raman intensity due to SERS effect is tremendous.
Table 1 shows band assignment for both normal Raman as well as SERS bands of TYMV. The positions of normal Raman bands are based the reports from previous publications (Chen and Lord, 1974; Kaper, 1975; Turano et al., 1976; Yu et al., 1975). As shown, there is no significant difference in the band position between normal and SERS spectra of TYMV. The SERS bands shown in Fig. 1 are related with amino acid residue and RNA in TYMV. First of all, the 734 cm−1 peak with the strongest intensity corresponds to vibration of C-S bond, mostly from coat protein of TYMV as found in a previous study (Turano et al., 1976). Sulfur can strongly adsorb on the Au surface as generally known, so the intensity of C-S vibration is greatly enhanced, so resulting in the strongest band intensity. The 961 cm−1 and 1240 cm−1 band are from C-C stretching mode and amide III, respectively. In addition, there are many peaks corresponding to C-H deformation, mostly from the backbone structure.
Table 1.
Band assignment for both normal Raman as well as SERS peaks of Turnip yellow mosaic virus (TYMV). Standard three-letter symbols for amino acids (Phe: Phenylalanine, Tyr: Tyrosine, Trp: Tryptophan) and one-letter symbols for RNA bases (A: Adenine, G: Guanine, U: Uracil, C: Cytosine) are used
| Band position | ||
|---|---|---|
|
|
||
| Normal Raman | SERS | |
| Phe, U, C | 626 cm−1 | 620 cm−1 |
| C-S stretching, A | 726 cm−1 | 734 cm−1 |
| C-C stretching | 962 cm−1 | 961 cm−1 |
| C-N stretching | 1159 cm−1 | 1168 cm−1 |
| Tyr, Phe | 1178 cm−1 | 1184 cm−1 |
| Amide III, C, U | 1248 cm−1 | 1240 cm−1 |
| G, CH deformation | 1319 cm−1 | 1320 cm−1 |
| Trp, A, CH deformation | 1339 cm−1 | 1342 cm−1 |
| Trp, CH deformation | 1362 cm−1 | 1366 cm−1 |
| U, A, G, CO2− symmetric stretching | 1395 cm−1 | 1401 cm−1 |
| CH deformation | 1458 cm−1 | 1460 cm−1 |
| Trp, A, G, Phe, Tyr | 1577 cm−1 | 1574 cm−1 |
A cultivation of Chinese cabbage carried out in an infect-free greenhouse, in National Academy of Agricultural Science, Suwon, Korea, in 2012. The plants at 3 – 5 stage were mechanically inoculated using sap prepared by homogenizing infected leaf samples in 0.01 M phosphate buffer (pH 7.0). Inoculated plants were maintained in an infect-free greenhouse at 20 – 25 °C. The extracts were prepared by homogenizing in 0.01 M phosphate buffer (pH 7.0). The infection of TYMV was double-checked with the use of PCR.
Since there are so many different metabolites as well as TYMV in the extracts from the infected leaves and Raman spectra of these extracts are directly measured without further separation steps, direct identification of TYMV Raman peak would not be easy since it would be obscured by complexly overlapped Raman bands of the extracted metabolites. If TYMV infection alters the plant metabolism, the extracted metabolites from healthy and infected leaves would be different each other, and the difference in metabolite composition could be indirectly used for the detection of infection rather than direct identification of TYMV peaks.
To examine the spectral features of the extracts, normal Raman spectra were initially collected directly for the dried extracts on a quartz window. Although there should be hundreds of extracted metabolites as well as TYMV in the extracts, no clear Raman bands were observed due to the overlapping with strong fluorescence background (the corresponding spectrum is not shown). It is evident that some extracted metabolites containing chromophores in their molecular structure, which are easy to generate the fluorescence by the absorption of laser wavelength.
Fig. 1 also shows average SERS spectra of the extracts from healthy (blue) and TYMV-infected (red) leaves. The shades with the same color indicate the corresponding standard deviation (1 σ). Before the averaging, each raw spectrum was baseline corrected at 1,740, 805, 790 and 592 cm−1, and normalized by dividing each baseline-corrected spectrum by the corresponding peak area in the 1740 – 590 cm−1 range. As shown, the spectral features between two average spectra are quite similar each other; however, the distinguishable bands at 1,411, 1,300 and 682 cm−1 (indicated by red asterisks) are observed in the spectrum of extract from TYMV-infected leaves. These three bands are not dominant in the spectrum of pure TYMV, so it could be concluded that the observed spectral difference between two average spectra of healthy and TYMV-infected leaves may not be originated from the presence of TYMV. Other than TYMV itself, widely different metabolites should be present in the extracts, these compete with TYMV to adsorb on the surface of Au nanoparticles and the corresponding Raman peaks appear as shown in the figure. Consequently, the spectral difference between the average spectra is originated from the compositional difference in the extracted metabolites. Full chemical analysis of extracted metabolites (components) to explain the spectral differences should require highly extensive analytical effort as well as sophisticated instrumentations such as LC-MS and LC-NMR; therefore, it would be beyond the scope of this study. More importantly, different metabolite compositions in the extracts from healthy and TYMV-infected leaves could be effectively utilized for the identification of infection without using either separation or specific tagging methods.
Principal component analysis combined with linear discriminate analysis (PCA-LDA) was used for the identification of TYMV-infection using SERS spectra collected from the extracts. PCA is a widely accepted method to represent complex spectral data in much fewer dimensions using principal components (a.k.a. eigenvectors) and scores (Beebe and Kowalski, 1987; Beebe et al., 1998; Lee et al., 2010). Linear discriminant analysis (LDA) is a well-known method in pattern recognition (Coomans et al., 1979; Lee et al., 2010; Todeschini and Marengo, 1992; Wu et al., 1996). It intends to find a linear combination of features which separates two or more classes of samples. PCA was initially performed using all of the spectral collected from healthy and TYMV-infected leaves, and then the resulting scores were used for the LDA. A combination of two scores was employed since it was easy to visualize the discrimination performance in the two-dimensional domain. The optimal two-score combination was determined by leave-one-out cross-validation. PCA and LDA were conducted using Matlab version 7.0. (The Math-Works Inc., MA, USA).
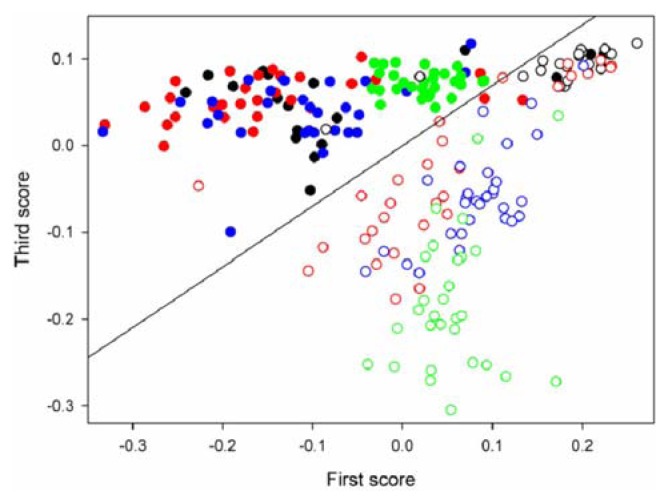
Figure 3 shows the score scatter plot (the first vs. third scores) that resulted from the use of PCA-LDA for the discrimination between healthy (filled circles) and TYMV-infected (open circles) samples. Four colors are used to indicate 4 independent extract samples in each case. The boundary line between these two groups that was determined by LDA is also displayed. As shown, two groups of healthy and virus-infected samples are identifiable with minor overlap along the boundary line and the obtained identification error is 5.5%.
Fig. 3.
Score scatter plot (the first vs. third scores) that resulted from the use of PCA-LDA for the discrimination between healthy (filled circles) and TYMV-infected (open circles) samples. Four colors are used to indicate 4 independent extract samples in each case.
In conclusion, the spectral difference due to dissimilar metabolite composition between healthy and TYMV-infected leaves was the origin for possible identification of infection, although Raman spectral feature of TYMV itself was hard to observe directly in the spectra of extracts. It is obvious that the specificity and accuracy of the proposed method couldn’t be better than the conventional diagnostic methods such as PCR and ELISA. It, however, has a potential for an efficient and high throughput screening method. Obviously, in future, full compositional analysis of extracted metabolites and plant physiology are necessary to combine to explain the different metabolic pathway in the case of virus infection. In addition, the investigation with the larger numbers of samples would be beneficial to impose statistical significance on the achieved accuracy.
Acknowledgments
This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ007511)” Rural Development Administration, Republic of Korea.
References
- Beebe KR, Kowalski BR. An introduction to multivariate calibration and analysis. Anal Chem. 1987;59:1007A–1017A. [Google Scholar]
- Beebe KR, Pell RJ, Seasholtz MB. Chemometrics: A Practical Guide. Wiley-Interscience; New York, USA: 1998. [Google Scholar]
- Beier BD, Quivey RG, Berger AJ. Raman microspectroscopy for species identification and mapping within bacterial biofilms. AMB Express. 2012;2:1–7. doi: 10.1186/2191-0855-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalcroft J, Matthews REF. Cytological changes induced by turnip yellow mosaic virus in Chinese cabbage leaves. Virology. 1966;28:555–562. doi: 10.1016/0042-6822(66)90240-6. [DOI] [PubMed] [Google Scholar]
- Chen MC, Lord RC. Laser-excited Raman spectroscopy of biomolecules. V. Conformational changes associated with the chemical denaturation of lysozyme. J Am Chem SOC. 1974;96:3038–3042. doi: 10.1021/ja00817a003. [DOI] [PubMed] [Google Scholar]
- Chi H, Liu B, Guan G, Zhang Z, Han M-Y. A simple, reliable and sensitive colorimetric visualization of melamine in milk by unmodified gold nanoparticles. Analyst. 2010;135:1070–1075. doi: 10.1039/c000285b. [DOI] [PubMed] [Google Scholar]
- Coomans D, Massart DL, Kaufman L. Optimization statistical linear discriminant analysis in analytical chemistry. Anal. Chim. Acta. 1979;112:97–122. [Google Scholar]
- El-Abassy RM, Donfack P, Materny A. Visible Raman spectroscopy for the discrimination of olive oils from different vegetable oils and the detection of adulteration. J Raman Spectrosc. 2009;40:1284–1289. [Google Scholar]
- Kaper JM. The Chemical Basis of Virus Structure Dissociation and Reassembly. North Holland/American Elsevier; New York, USA: 1975. [PubMed] [Google Scholar]
- Kim JS, Cho JD, Choi HS, Lee SH, Choi GS, Lee SY, Kim HJ, Yoon MK. Ribgrass mosaic tobamovirus occurred on Chinese cabbage in Korea. Plant Pathol J. 2012;26:328–339. [Google Scholar]
- Lee SU, Chung H-I, Choi HS, Cha KJ. Using combinations of principal component scores from different spectral ranges in near-infrared region to improve discrimination for samples of complex composition. Microchemical J. 2010;95:96–101. [Google Scholar]
- Matthews REF. Studies on the relation between protein and nucleoprotein particles in turnip yellow mosaic virus infections. Virology. 1958;5:192–205. doi: 10.1016/0042-6822(58)90018-7. [DOI] [PubMed] [Google Scholar]
- Moskovits M. Surface roughness and the enhanced intensity of Raman scattering by molecules adsorbed on metals. J Chem Phys. 1978;69:4159–4161. [Google Scholar]
- Moskovits M. Surface-enhanced Raman spectroscopy: A brief retrospective. J Raman Spectrosc. 2005;36:485–496. [Google Scholar]
- Mozharov S, Nordon A, Girkin JM, Littlejohn D. Non-invasive analysis in micro-reactors using Raman spectrometry with a specially designed probe. Lab Chip - Miniat Chem Biol. 2010;10:2101–2107. doi: 10.1039/c004248j. [DOI] [PubMed] [Google Scholar]
- Park SC, Kim MJ, Noh JG, Chung HI, Woo YA, Lee JH, Kemper MS. Reliable and fast quantitative analysis of active ingredient in pharmaceutical suspension using Raman spectroscopy. Anal. Chim. Acta. 2007;593:46–53. doi: 10.1016/j.aca.2007.04.056. [DOI] [PubMed] [Google Scholar]
- Prucek R, Ranc V, Kvítek L, Panáèek A, Zboøil R, Koláø M. Reproducible discrimination between Gram-positive and Gram-negative bacteria using surface enhanced Raman spectroscopy with infrared excitation. Analyst. 2012;137:2866–2870. doi: 10.1039/c2an16310a. [DOI] [PubMed] [Google Scholar]
- Reid MS, Matthews REF. On the origin of the mosaic induced by turnip yellow mosaic virus. Virology. 1966;28:563–570. doi: 10.1016/0042-6822(66)90241-8. [DOI] [PubMed] [Google Scholar]
- Ryu KT, Haes AJ, Park HY, Nah SH, Kim JM, Chung HI, Yoon MY, Han SH. Use of peptide for selective and sensitive detection of an anthrax biomarker via peptide recognition and surface-enhanced Raman scattering. J Raman Spectrosc. 2009;41:121–124. [Google Scholar]
- Schatz GC, Van Duyne RP. Electromagnetic mechanism of surface-enhanced spectroscopy. In: Chalmers JM, Griffiths PR, editors. Handbook of Vibrational Spectroscopy. Vol. 1. Wiley; New York, USA: 2002. pp. 759–774. [Google Scholar]
- Todeschini R, Marengo E. Linear discriminant classification tree: a user-driven multicriteria classification method. Chemom Intell Lab Syst. 1992;16:25–35. [Google Scholar]
- Turano TA, Hartman KA, Thomas GJ., Jr Studies of Virus Structure by Laser-Raman Spectroscopy. 3. Turnip Yellow Mosaic Virus. J Phys Chem. 1976;80:1157–1163. [Google Scholar]
- Turkevich J, Stevenson PC, Hilliery J. A study of the nucleation and growth processes in the synthesis of colloidal gold. Discuss Faraday Soc. 1951;11:55. [Google Scholar]
- Wu W, Mallet Y, Walczak B, Penninckx W, Massart DL, Heuerding S, Erni F. Comparison of regularized discriminant analysis, linear discriminant analysis and quadratic discriminant analysis applied to NIR data. Anal. Chim. Acta. 1996;329:257–265. [Google Scholar]
- Yu NT, Lin TS, Tu AT. Laser Raman scattering of neurotoxins isolated from the venoms of sea snakes Lapemis hardwickii and Enhydrina schistosa. J Biol Chem. 1975;250:1782–1785. [PubMed] [Google Scholar]
- Zaitlin M. Tobacco mosaic virus (Type strain) GMI/AAB Descriptions of plant viruses. 1975;(151) [Google Scholar]