Abstract
The incidence of Broad bean wilt virus 2 (BBWV2) on red pepper was investigated using the samples obtained from 24 areas of 8 provinces in Korea. Two hundred and five samples (79%) out of 260 collected samples were found to be infected with BBWV2. While the single infection rate of BBWV2 was 21.5%, the co-infection rate of BBWV2 with Cucumber mosaic virus, Pepper mottle virus, Pepper mild mottle virus and/or Potato virus Y was 78.5%. To characterize the genetic diversity of BBWV2 Korean isolates, 7 isolates were fully sequenced and analyzed. Phylogenetic analyses revealed that BBWV2 isolates could be divided largely into two groups as Group I and Group II. Based on the partial sequence analyses, 153 selected BBWV2 isolates were subgrouped into GS-I (21.6%), GS-II (3.9%) and GS-III (56.9%). BBWV2 GS-III, which was predominant in Korea, appears to be a new combination between Group I RNA-1 and Group II RNA-2. Viral disease incidence of BBWV2 on red pepper was under 2% before 2004. However, the incidence was increased abruptly to 41.3% in 2005, 58.2% in 2006 and 79% in 2007. These rapid increases might be related with the emergence of new combinations between BBWV2 groups.
Keywords: BBWV2, genetic diversity, red pepper
Broad bean wilt virus (BBWV) is the type species of the genus Fabavirus in the family Comoviridae (Taylor et al., 1972). It is transmitted by aphids, mostly Aphis gossypii and Myzus persicae, with infection rate of 60 – 90% in a nonpersistent manner and has a wide host range (Benner et al., 1985; Brunt et al., 1996). By serological and molecular studies, BBWV isolates are classified into two groups, Broad bean wilt virus 1 (BBWV1) and Broad bean wilt virus 2 (BBWV2) (Kobayashi et al., 2003; Uyemoto et al., 1974). Although they show the similar genome structures and functions, the nucleotide (nt) sequence identity between them was limited (39% –67%). The genome is composed of two single stranded positive-sense RNA molecules, RNA-1 and RNA-2, that are encapsidated separately into icosahedral virions (Lisa et al., 1996). Although BBWV1 has not been detected yet, disease incidences caused by BBWV2 have been reported in Korea (Lee et al., 1979).
BBWV2 RNA-1 and RNA-2 are about 6 kb and 4 kb nucleotides in length, respectively, contain a single open reading frame (ORF), and thus translated into a single polyprotein precursor from which functional proteins are produced by proteolytic cleavage. RNA-1 encodes five proteins containing cofactor protease (Co-pro), NTP-binding motif (NTBM), genome-linked viral protein (VPg), protease (Pro) and RNA-dependent RNA polymerase (RdRp) while RNA2 encodes three proteins containing movement protein (MP), large coat protein (LCP) and small coat protein (SCP) (Goldbach et al., 1995). Up to date, the complete nucleotide sequences of eight BBWV2 isolates (Ikegami et al., 1998; 2001; Kobayashi et al., 1999; Koh et al., 2001; Kuroda et al., 2000; Lee et al., 2000; Nakamura et al., 1998; Qi et al., 2000a; 2000b) and partial sequences of twenty-five BBWV2 isolates (Ferrer et al., 2011; Kobayashi et al., 1999; 2004; Kondo et al., 2005; Sui et al., 2009) from various host plants have been reported.
BBWV2 is a destructive pathogen in many economically important horticultural and ornamental crops, and has a worldwide distribution. In Korea, it has been reported that BBWV2 naturally infects red pepper (Lee et al., 2000), broad bean (unpublished), pea (Choi et al., 2001), spinach (Lee et al., 1979), perilla (Choi et al., 2001), lily (Jang et al., 1987), Gentiana spp. (Roh et al., 1998), gladiolus (Park et al., 1998), celery (Hahm et al., 1998), etc. Especially, occurrence of BBWV2 in red pepper has increased by single and mixed infection with Cucumber mosaic virus (CMV) and other viruses (Cho et al., 2007).
Peppers are the economically important vegetable giving rise to many phenotypic diversities depending on their originate countries where there are cultivated and their intended uses, such as fresh or cooked vegetable, ornamental, spice, or extraction of either its natural colorant for food dyes or capsaicin for use in medicine. Peppers are easily infected with fungi, bacteria, and viruses which cause severe damages. Especially, viral diseases are difficult to control directly, so they cause significant economic losses due to yield reduction and low ornamental quality. The majority of viral diseases on red peppers led to overall stunting, mosaic, necrosis, and fruit malformation and reduced fruit. Over 66 viruses belonging to 9 families, namely Partitiviridae, Bromoviridae, Comoviridae, Tombusviridae, Luteoviridae, Bunyaviridae, Potyviridae, Geminiviridae, and Rhabdoviridae, are known to cause damage on red pepper in the world, and twenty-nine viruses are distributed worldwide but the remains are occurred on confined areas at certain geographical countries (Kim et al., 2002; Watterson, 1993).
In Korea, red pepper was cultivated in 44,584 ha and its production was 95,391 tons in 2010 (www.kostate.go.kr). It has been grown continuously during May to September at every year. Since climatic conditions during the red pepper production season are also favorable for aphid populations, the dissemination of vector transmissible viruses are tend to increase dramatically throughout the pepper cultivation season. Although many viruses have been detected in pepper, only 16 viruses were reported in Korea (The List of Plant Diseases in Korea, 2010), i.e. Alfalfa mosaic virus (AMV), BBWV2, CMV, Pepper mottle virus (PepMoV), Pepper mild mottle virus (PMMoV), Pepper severe mosaic virus (PSMV), Potato virus X (PVX), Potato virus Y (PVY), Satellite Tobacco mosaic virus (sTMV), Tobacco mild green mosaic virus (TMGMV), Tobacco mosaic virus (TMV), Tobacco mild green mild mottle virus (TMGMMV), Bell pepper mottle virus (BPMV), Pepper vein mottle virus (PVMV), Pepper vein chlorosis virus (PVCV), and Tomato spotted wilt virus (TSWV).
Major viruses occurred recently in Korean red peppers are CMV, BBWV2, PepMoV and PMMoV. Their average disease incidences from 2002 to 2006 were 29.4%, 25.6%, 13.6%, 14.3%, respectively (Cho et al., 2007). Among them, CMV occurred most frequently and caused significant damage in pepper because of the widest host-range, nonpersistent transmission by a large number of aphid species and the ability of synergistic effects with unrelated plant viruses (Palukaitis et al., 2003). BBWV2 had low incidence under 2% before 2004, however, it was increased abruptly to 41.3% in 2005 and 58.2% in 2006 has steadily increased since. For the types of mixed infections of pepper viruses, the incidence of CMV+BBWV2 was 2.1% in 2004, but increased suddenly to 33.2% in 2005 and to 83.2% in 2006. Especially, mixed infections with BBWV2 in pepper resulted in enhanced symptoms with synergistic symptom expression (Cho et al., 2007).
The purpose of this study was to investigate occurrence of BBWV2 in main cultivation areas of red pepper in 2007 and to understand the genetic diversity of Korean BBWV2 isolates by conducting sequence and phylogenetic analyses using complete nt sequences of seven BBWV2 isolates collected from red pepper from 2007 to 2011.
Materials and Methods
Survey and sample collection.
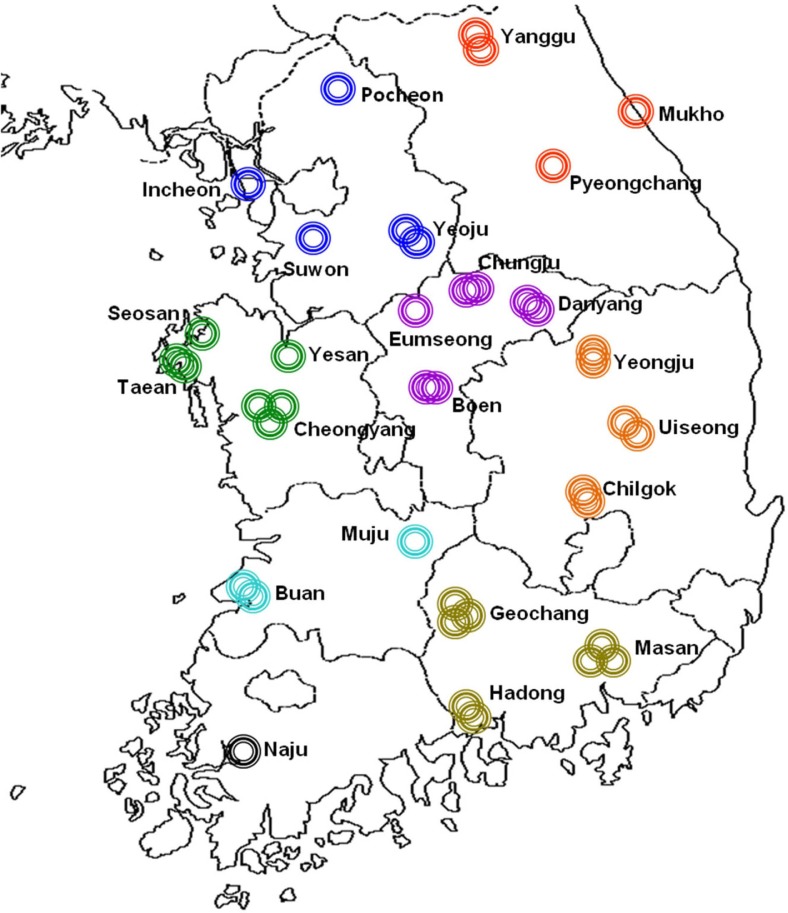
A survey of red pepper virus was carried out in 41 cultivated fields at 24 areas of 8 provinces of Korea from July to October 2007 (Fig. 1). Two hundred and sixty samples of red pepper (Capsicum annuum L.) leaves showing the general virus symptom were collected as follows; 44 samples at 4 areas of Yeoju, Suwon, Pocheon and Incheon in Gyeonggi-do province; 45 samples at 4 areas of Cheongyang, Yesan, Seosan and Taean in Chungcheongnam-do province; 42 samples at 4 areas of Boeun, Chungju, Danyang and Eumseong in Chungcheong-buk-do; 15 samples at 2 areas of Buan and Muju in Jeollabuk-do; 7 samples at Naju area in Jeollanam-do; 27 samples at Yanggu, Pyeongchang and Mukho in Gangwon-do; 32 samples at Chilgok, Uiseong and Yeongju in Gyeongsangbuk-do; 48 samples at Hadong, Geochang Masan in Gyeongsangnam-do (Table 1). Samples collected were maintained at −70 °C and used for the genetic diagnosis by reverse transcription-polymerase chain reaction (RT-PCR) and enzyme linked immuno-sorbent assay (ELISA)
Fig. 1.
Geographical distribution of collection areas for the redpepper specimens without symptoms expression of virus diseasein Korea. Two hundred and sixty red pepper specimens were sampled from 41 cultivated fields in 24 local areas of 8 provinces as indicated.
Table 1.
Legend of red pepper specimens used in this paper
| Province | Areaa | No. of fields Investigated | No. of samplesb collected |
|---|---|---|---|
| Gyeonggi-do | Yeoju | 2 | 16 |
| Suwon | 1 | 11 | |
| Pocheon | 1 | 16 | |
| Incheon | 1 | 1 | |
|
| |||
| Chungcheongnam-do | Cheongyang | 3 | 28 |
| Yesan | 1 | 3 | |
| Seosan | 1 | 6 | |
| Taean | 2 | 8 | |
|
| |||
| Chungcheongbuk-do | Boeun | 2 | 16 |
| Chungju | 2 | 14 | |
| Danyang | 2 | 11 | |
| Eumseog | 1 | 1 | |
|
| |||
| Jeollabuk-do | Buan | 2 | 10 |
| Muju | 1 | 5 | |
|
| |||
| Jeollanam-do | Naju | 1 | 7 |
|
| |||
| Gangwon-do | Yanggu | 2 | 12 |
| Pyeongchang | 1 | 1 | |
| Mukho | 1 | 1 | |
|
| |||
| Gyeongsang-buk-do | Chilgok | 2 | 10 |
| Yeongju | 2 | 9 | |
| Uiseong | 2 | 13 | |
|
| |||
| Gyeongsangnam-do | Hadong | 2 | 14 |
| Geochang | 3 | 19 | |
| Masan | 3 | 15 | |
|
| |||
| Total | 24 | 41 | 260 |
Areas and fields for sample collection were selected by the major control was considered as viral infection. production areas of red pepper in Korea.
Samples of leaves red pepper were collected randomly without regard to symptom expression of virus disease.
Virus isolation.
Seven isolates of BBWV2, RP1 to RP7, collected from red pepper (Capsicum annuum L.) showing vein clearing, mosaic and virus-like symptoms. RP1 and RP2 isolates collected from Cheongyang in 2007, RP3 isolate from Yesan in 2007, RP4 isolate from Suwon in Chungcheong-2007, RP5 isolate from Yeongyang in 2009, RP6 isolate from Imsil in 2010 and RP7 isolate from Suwon in 2011 (Table 2). Virus isolates were inoculated to local lesion at least three times in succession, and propagated in Nicotiana benthamiana.
Table 2.
Database of the complete nucleotide sequence of BBWV2 genome
| Isolates | Host plant | Origin | NCBI accession No.
|
Remark | |
|---|---|---|---|---|---|
| RNA-1 | RNA-2 | ||||
| IP | Capsicum annuum | Japan | AB023484 | AB018698 | |
| ME | Megaskepasma erythrochlamys | Singarpore | NC003003 | NC003004 | |
| IA | Gentiana spp | Japan | AB051386 | AB032403 | |
| K | Capsicum annuum | Korea | AF144234 | AF104335 | |
| B935 | Vicia faba | China | AF149425 | AJ132844 | |
| MB7 | Vicia faba | Japan | AB013615 | AB013616 | |
| XJ14-3 | Lycopersicon esculentum MILL | China | FN985164 | HQ283389 | |
| PatMMVa | Pogostemon patchouli | Japan | NC003975 | NC003974 | |
|
| |||||
| RP1b | Korea-Cheongyang | JX183221 | JX183222 | 2007 | |
| RP2 | Korea-Cheongyang | JX183223 | JX183224 | 2007 | |
| RP3 | Korea-Yesan | JX183225 | JX183226 | 2007 | |
| RP4 | Capsicum annuum | Korea-Suwon | JX183227 | JX183228 | 2007 |
| RP5 | Korea-Yeongyang | JX183229 | JX183230 | 2009 | |
| RP6 | Korea-Imsil | JX183231 | JX183232 | 2010 | |
| RP7 | Korea-Suwon | JX183233 | JX183234 | 2011 | |
Patchouli mild mosaic virus; an isolate of BBWV2
Korean isolates of RP1 to RP7 used in this study.
ELISA
Double antibody sandwich (DAS)-ELISA test kits for ELISA were purchased from Agdia (USA) and JPPA (Japan). The leaf samples were homogenized with a mortar and a pestle with equal volumes (v/w) of 0.01 M phosphate buffer, pH 7.0. The crude sap of 100 μl per microplate well was used respectively. Quantitative measurements with p-nitrophenol were made by absorbance at 405 nm spectrophotometer (EL312e EIA, Bio-Tek Instruments Inc., USA). Absorbance value of 2 times higher than that of negative control was considered as viral infection.
Electron microscopy.
Virus particles were observed in leaf samples by using negatively stained leaf dips. Dip preparations were prepared by grinding a small piece of 1 × 1 mm collected sample with 2–3 drops of 2% phosphotungstic acid, pH 7.0. The extract was mounted on a carbonstabilized and Formvar-coated grid.
Total RNA extraction and RT-PCR.
Total RNAs were extracted from infected leaf samples by easy-spinTM total RNA extraction kit (iNtRON, Korea) according to the manufacturer’s instructions. RT reaction was carried out at 42 °C for 30 min in a final 5 μl volume obtained by adding 1 μl total RNA (2.5 ng/μl), 10 pmole (1 μl) of the down-stream primer, 1 × RT buffer, 2 mM dNTP, and 0.5 U AMV reverse transcriptase (Promega, USA) and made up to volume with dH2O, and was terminated by heating at 95 °C for 5 min. When RT was completed, total 25 μl of 10 pmole (1 μl) of the upstream primer, 1 U Go-Taq DNA polymerase (Promega, USA), 1 × PCR buffer and 1.5 mM MgCl2, were added. Mixtures were then amplified in a thermal Cycler (Bio-Rad, USA) for a total 35 cycles. Each cycle included a denaturing step at 94°C for 30 sec, an annealing step at 55 °C for 30 sec, and extension step at 72 °C for 90 sec, and finally kept at 72 °C for 10 min. PCR products were analyzed by electrophoresis in 1.0% agarose gel, staining with ethidium bromide, and DNA bands were visualized using a UV transilluminator.
Primer design.
Primers for detection of red pepper infecting viruses were designed based on nucleotide sequence of reported red pepper virus isolates registered in GenBank of the National Center for Biotechnology Information (Table 3). Primer pairs for full-length nucleotide sequences were designed group-specifically for RNA-1 and universally for RNA-2 (Table 4). End sequences of the genomes were obtained with the 5′ and 3′ rapid amplification of cDNA ends (RACE) protocol (Boehringer Mannheim, Germany). cDNA clones containing the 5′ end of the genomes were obtained using Xec primers (5′-AAAGAATTCCCCCCCC CCCCC-3′), and BBWV2 5′-RNA1 (5′-GCCATYTCATT GGCATGGAAAAAGC-3′) and BBWV2 5′-RNA2 (5′-GAAGGGTGCAACGATATATGCAAATTC-3′) primers complementary to nucleotides 384–360 and 424–395, respectively, of BBWV2 stain. In addition, cDNA clones containing the 3′ end of the genomes were obtained using BBWV2 3′-RNA1 (5′-AGGCATATGAGAGAGGAGAG-3′) and BBWV2 3′-RNA2 (5′-GAAATGGGGAATGTTC ATGAGTA-3′) primer complementary to nucleotides 5646–5665 and 3269–3291, respectively, and anchor primers (5′-GACCACGCGTATCGATGTCGACTTTTTTTTTTTT TTTTV-3′). Besides, the group-specific primers for strain analysis were selected from Table 4 for RNA-1 genome and designed newly for RNA-2 genome of BBWV2 based on the previously reported complete sequences (Table 5).
Table 3.
Primers used in genetic diagnosis for the specimens of red pepper infecting viruses majored in Korea
| Virusa | Primerb | Sequences | Size (nt) |
|---|---|---|---|
| BBWV2 | BBWV2 1u | 5′-AAACAAACAGCTTTCGTTCCG-3′ | 380 |
| BBWV2 1R | 5′-GCCATCTCATTGGCATGGA-3′ | ||
|
| |||
| CMV | CMV 3-3u | 5′-AAGAARCTTGTTTCGCGCATT-3′ | 629 |
| CMV 3-3d | 5′-TGGTCTCCTTTTRAGGCCCCCA-3′ | ||
|
| |||
| PMMoV | PMMoV u6 | 5′-TAAATGGCGTTAGTAGTCAAGGA-3′ | 1451 |
| PMMoV d6 | 5′-TGGGCCGCTACCCGCGGTTCGGGG-3′ | ||
|
| |||
| PepMoV | PepMoV u1 | 5′-AATGGCACGTCCCCAAA-3′ | 705 |
| PepMoV d1 | 5′-TCTCTCTCATGCCAACTACGA-3′ | ||
|
| |||
| PVY | PVY u1 | 5′-TGGTGCATTGARAATGGAACCTC-3′ | 755 |
| PVY d1 | 5′-GTCTCCTGATTGAAGTTTACAAG-3′ | ||
BBWV2, Broad bean wilt virus 2; CMV, Cucumber mosaic virus; PMMoV, Pepper mild mottle virus; PepMoV, Pepper mottle virus; PVY, Potato virus Y.
Primers were designed based on the nucleotide sequences of pepper viruses registered in GenBank.
Table 4.
Primer pairs for full-lengh sequencing of BBWV2a genomes
| Group | Primer | Sequence (5′ → 3′) | Loci | Size(nt) |
|---|---|---|---|---|
| I of RNA-1 | BBWV2 1-1u | AAACAAACAGCTTTCGTTCCG | 19–915 | 896 |
| BBWV2 1-1R | TTCCATGCATCGTGTTGGCC | |||
|
| ||||
| BBWV2 1-2F | ATTTCAGGCTCCATTAAAAAGAG | 753–1701 | 948 | |
| BBWV2 1-2R | TCTATCAACTTCTCCAAAGCC | |||
| BBWV2 1-3F | GAATGTGATTGCATGGTGGCT | 1515–2400 | 885 | |
|
| ||||
| BBWV2 1-3R | CAAATGAAGTGGCTATGACCGT | |||
| BBWV2 1-4F | GTACACAAGGTTTAAGGAGCA | 2275–3203 | 928 | |
| BBWV2 1-4R | CCAAGCACACTTGTGTTCTCA | |||
|
| ||||
| BBWV2 1-3u | TGGGAG(A)TACAAGCAA(G)TGG C | 2867–3888 | 1021 | |
| BBWV2 1-5R | AAATGCCATTCCTCTGGGG | |||
|
| ||||
| BBWV2 1-6F | GAAGTGCCAGTTTTATGCCGT | 3690–4626 | 936 | |
| BBWV2 1-6R | TCATTCATGACACACTTGGGC | |||
|
| ||||
| BBWV2 1-7F | GGAGGGATGTTTTACCAGCT | 4476–5252 | 776 | |
| BBWV2 1-7R | CTATTGCGGAATTCCTTGGC | |||
|
| ||||
| BBWV2 1-8F | TGAATCCAGATGGCACATGG | 5078–5958 | 880 | |
| BBWV2 1-8R | CCCTCACTACTGAAATTTACTTG | |||
|
| ||||
| II of RNA-1 | BBWV2 1-1u | AAACAAACAGCTTTCGTTCCG | 19–1173 | 1154 |
| BBWV2 1-1r | CTGAAATAGCTGCCATAGCAG | |||
|
| ||||
| BBWV2 1-2f | TTTGCAGCGTGGATGCC | 883–1981 | 1098 | |
| BBWV2 1-2r | CTTGTAGCCTGACCAAAAAGG | |||
|
| ||||
| BBWV2 1-3f | GCTGATAGAGTTGCACAATTCAGT | 1774–2850 | 1076 | |
| BBWV2 1-3r | TACAAATACGCCCTTTGCCA | |||
|
| ||||
| BBWV2 1-4f | GTGGAGCAGATGCAATTGAG | 2603–3664 | 1061 | |
| BBWV2 1-4r | GAAATACTTTGGGGAATTTCCC | |||
|
| ||||
| BBWV2 1-5f | GATGTGGAAACTTTGCCCAA | 3495–4390 | 895 | |
| BBWV2 1-5r | TTGCTCACTTCTTCAAACCAAG | |||
|
| ||||
| BBWV2 1-6f | CTTGATCGTGGTCCTGGTGA | 4272–5355 | 1083 | |
| BBWV2 1-6r | TCCATCTCAAATTTCTCAAGACTT | |||
|
| ||||
| BBWV2 1-7f | TTGGAAATGGCAGAGGCAT | 5239–5958 | 719 | |
| BBWV2 1-7R | CCCTCACTACTGAAATTTACTTG | |||
|
| ||||
| Universal of RNA-2 | BBWV2 2-1F | CAAACAGCTTTCGTTCCGAA | 22–949 | 927 |
| BBWV2 2-1R | AAGCCATCTGCCACCAATTC | |||
|
| ||||
| BBWV2 2-2F | GCCATGTCAGGTGCTGGA | 801–1702 | 900 | |
| BBWV2 2-1d | CGCATA(G)GTC(T)TCT(C)GTA(G)GCA(T)GA | |||
|
| ||||
| BBWV2 2-2u | TACAAT(C)TTG(A)CCTGGG(A)CCTAGC | 1377–2496 | 1119 | |
| BBWV2 2-2r | CATGTTCCCAACCTTTTCAATGT | |||
|
| ||||
| BBWV2 2-3u | ATTT(C)TA(G)GATTTTACT(C)CTC(T)ATGAG | 2348–3511 | 1163 | |
| BBWV2 2-3d | AGGTCATGGAACCCATTTTAATGG | |||
BBWV2, Broad bean wilt virus 2.
Table 5.
Group-specific primer pairs for BBWV2 RNA-1 and RNA-2 RNA
| RNA | Group | Primer | Sequences(5′ → 3′)b | Position | Size (nt) |
|---|---|---|---|---|---|
| 1 | I | BBWV2 1-7F | GGAGGGATGTTTTACCAGCT | 4476–5252 | 776 |
| BBWV2 1-7R | CTATTGCGGAATTCCTTGGC | ||||
|
| |||||
| II | BBWV2 1-5f | GATGTGGAAACTTTGCCCAA | 3495–4390 | 895 | |
| BBWV2 1-5r | TTGCTCACTTCTTCAAACCAAG | ||||
|
| |||||
| 2 | I | BBWV2-R2-F | ARTTGGYAAAGGTAARGCGCA | 2731–3226 | 495 |
| BBWV2-R2-R | GCCATCMACTGGRCCTACAACC | ||||
|
| |||||
| II | BBWV2-r2-f | CATAGGGAAGGYAARACTATA | |||
| BBWV2-r2-r | ACCATCCACGGGACCAACCACT | ||||
BBWV2, Broad bean wilt virus 2.
The primers were selected after alignment of the nucleotide sequences of BBWV2 isolates registered in GenBank.
Cloning and sequencing.
RT-PCR amplified DNA fragments were purified using MEGA quick-spin™ Kit (iNtRON, Korea) and cloned into pGEM-T easy vector (Promega, USA) according to the manufacturer’s instructions followed by transformation into Escherichia coli DH5α strain. Sequencing was done by a commercial company (Genotech, Korea). Complete nucleotide sequences of each genomic RNA segment were determined and submitted to GenBank (Accession Nos. JX183221 – JX183234, Table 2).
Sequence analysis and phylogenetic analysis.
The complete sequences were analyzed using the DNASTAR, Genetyx, Geneious pro 5.13 and Mega 5.10 software. Multiple nucleotide sequence alignments were performed by using ClustalW in Geneious pro 5.13. Phylogenetic analyses were performed by maximum likelihood method in Mega 5.10 software and 1,000 bootstrap replicates and compared with previously reported isolates, e.g. IP (Japan, Kobayashi et al., 1999) and ME (Singarpore, Koh et al., 2001) isolates for Group I and IA (Japan, Kuroda et al., 2000), K (Korea, Lee et al., 2000), B935 (China, Qi et al., 2000a; 2000b), MB7 (Japan, Nakamura et al., 1998), XJ14-3 (China) isolates and PatMMV (Japan, Ikegami et al., 1998; Ikegami et al., 2001) for Group II (Table 1) and with BBWV1 as an outgroup.
Results
Isolates collections and detection of BBWV2.
The representative symptoms observed on the infected red pepper samples were vein clearing, mosaic, malformation and a combination of chlorosis local and yellow mosaic (Fig. 2). The most common symptom was mosaic that is similar to the symptom induced by BBWV2, CMV, PepMoV, and PMMoV reported in Korea (Kim et al., 1990).
Fig. 2.
Representative symptoms observed on red pepper in cultivated fields. (A) Vein chlorosis and (B) mosaic induced by single infection of BBWV2. (C) Mottle and chlorosis induced by BBWV2+CMV. (D) Yellowing mosaic and malformation induced by BBWV2+CMV+PepMoV.
A total 260 leaf specimens of red pepper collected from 41 fields at 24 areas in 8 provinces were analyzed BBWV2 using DAS-ELISA, RT-PCR and electron microscopy. The total infection rate of BBWV2 was 79% of 205 samples as 30 out of 44 samples in Gyeonggi-do, 35 out of 45 samples in Chungcheongnam-do, 34 out of 42 samples in Chungcheongbuk-do, 6 out of 15 samples in Jeollabuk-do, 4 out of 7 samples in Jeollanam-do, 20 out of 27 samples in Gangwon-do, 29 out of 32 samples in Gyeongsangbuk-do, and 47 out of 48 samples in Gyeongsangnam-do (Table 6). BBWV2 was not detected from 55 samples (21%) even though they had viral symptoms. These samples must have been infected with other pepper infecting viruses.
Table 6.
Detection of viruses for red pepper from fields
| Infection type | Virus speciesa | No. of samplesb from | No. of total samples (%)d | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GGc | CN | CB | JN | GW | JB | GB | GN | |||
| Single | BB2 | 7 | 2 | 8 | 0 | 0 | 5 | 1 | 21 | 44(21.5) |
|
| ||||||||||
| Double | BB2+CMV | 19 | 13 | 6 | 2 | 4 | 6 | 1 | 7 | 58(28.3) |
| BB2+PMM | 0 | 1 | 1 | 0 | 0 | 2 | 0 | 0 | 4(1.9) | |
| BB2+Pep | 0 | 0 | 3 | 0 | 0 | 1 | 5 | 2 | 11(5.4) | |
| BB2+PVY | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 2 | 6(2.9) | |
|
| ||||||||||
| Triple | BB2+CMV+Pep | 4 | 13 | 6 | 4 | 0 | 2 | 19 | 12 | 60(29.3) |
| BB2+CMV+PMM | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 3(1.5) | |
| BB2+CMV+PVY | 0 | 3 | 1 | 0 | 0 | 0 | 0 | 2 | 6(2.9) | |
| BB2+Pep+PVY | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1(0.5) | |
|
| ||||||||||
| Quadruple | BB2+CMV+Pep+PMM | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 1 | 5(2.4) |
| BB2+CMV+Pep+PVY | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 3(1.5) | |
|
| ||||||||||
| Quintuple | BB2+CMV+Pep+PMM+PVY | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | 4(1.9) |
|
| ||||||||||
| Total | 30 | 35 | 34 | 6 | 4 | 19 | 30 | 47 | 205(100) | |
BB2: Broad bean wilt virus 2, CMV: Cucumber mosaic virus, Pep: Pepper mottle virus, PVY : Potato virus Y, PMM: Pepper mild mottle virus.
Virus infection was judged by DAS-ELISA, RT-PCR and electron microscopy.
The abbreviation represented as follows; GG: Gyeonggi-do, CN: Chungcheongnam-do, CB: Chungcheongbuk-do, JB: Jeollabuk-do, JN: Jeollanam-do, GW: Gangwon-do, GB: Gyeongsangbuk-do and GN: Gyeongsangnam-do
% in total
BBWV2 infected singly on red peppers with the infection rate of 21.5%. The BBWV2 single infection was detected highly on red pepper from Gyeongsangnam-do province as 21 out of 44 total specimens. The mixed infection with BBWV2 and other viruses including CMV was 78.5%. The total infection rate of double, triple, quadruple and quintuple was 38.5%, 34.2%, 3.9% and 1.9%, orderly. The most prevalent infection type was the double infection of BBWV2+CMV that occurred whole country of 8 provinces and the triple infection of BBWV2+CMV+PepMoV that occurred most provinces excluding Jeonnam province (Table 6). The combined incidence rate of these double (Fig. 2C) and triple (Fig. 2D) infections was 57.6% and thus was the most important virus species infecting red pepper in Korea which causing severe economic damages by synergistic symptom expressions.
Phylogenetic analyses of BBWV2 Korean isolates.
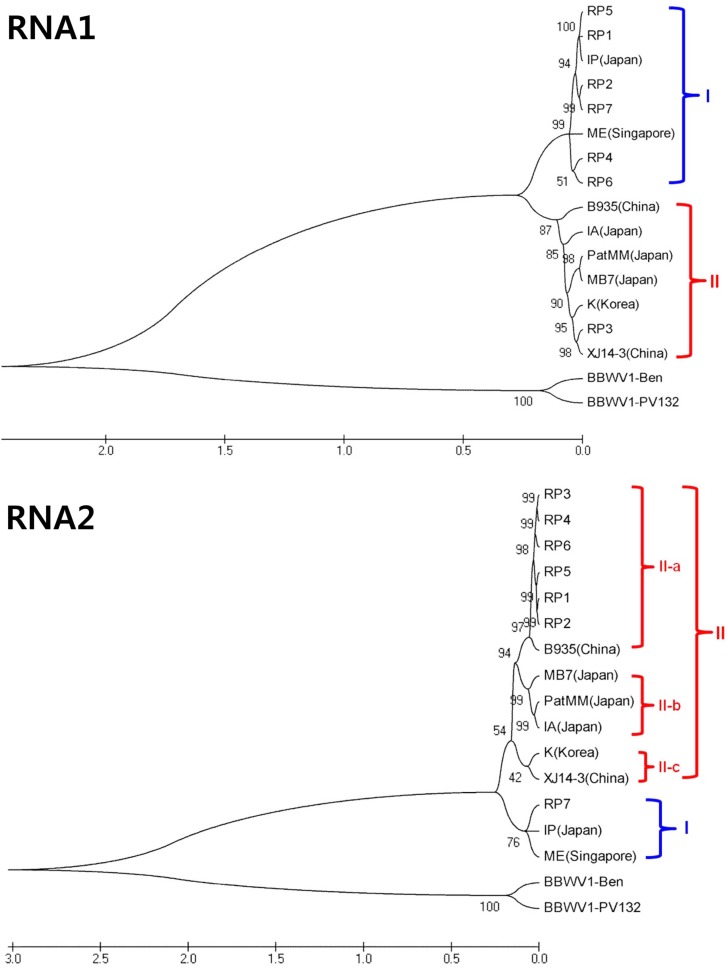
In order to confirm the classification and genetic diversity of BBWV2, the complete nucleotide sequences of RNA-1 and RNA-2 genomes of 7 Korean isolates were determined. Complete nucleotide sequences of 7 Korean BBWV2 isolates (RP1-RP7) were compared with the reported sequences of eight BBWV2 isolates (IP, IA, ME, MB7, K, PatMMV, B935, and XJ14-3) along with two BBWV1 isolates (PV132 and Ben) as outgroups. RNA-1 of BBWV2 Korean pepper isolates consisted of 5950 to 5955 nt encoding 5 proteins, Co-pro (353 aa or 354 aa), NTBM (582 aa), VPg (26 aa), Pro (209 aa) and RdRp (699 aa). RNA-2 contained 3571 to 3596 nt encoding 3 proteins, MP (465 aa), LCP (402 aa) and SCP (197 aa). Based on phylogenetic analyses, BBWV2 isolates could be divided largely into two groups, Group I and Group II in RNA-1 and RNA-2 genomes, respectively (Fig. 3). While nucleotide sequence identity between two groups was about 78% – 79%, nucleotide sequence identity within group was over 84% – 90%. Group I included two isolates (IP and ME) while Group II included six isolates (B935, IA, K, MB7, PatMMV, and XJ114) for RNA-1 and RNA-2 genomes, respectively. In case of RNA-1 genome of Korean isolates, six isolates excluding RP3 were classified as group I and only RP3 was classified as group II. Mean-while, the phylogenetic tree of RNA-2 genome showed that only RP7 was classified as group I and the remaining six isolates were classified as group II. Furthermore, group II of RNA-2 could be divided into 3 subgroups of II-a, II-b and II-c. Six Korean BBWV2 isolates, excluding RP7, belonged to subgroup II-a along with Chinese isolate B935. Group I was predominant in RNA-1 and Group II was predominant in RNA-2 genome in BBWV2 Korean pepper isolates. According to group-combination between RNA-1 and RNA-2 genomes, BBWV2 could be designated as three genetic strains of GS-I (Group I in both RNA-1 and RNA-2), GS-II (Goup II in both RNA-1 and RNA-2) and GS-III (Group I in RNA-1 and Group II in RNA-2). In case of 7 BBWV2 Korean isolates, RP7 belonged to GS-I, RP3 to GS-II and other 5 isolates (RP1, RP2, RP4, RP5 and RP6) to GS-III. GS-III has been detected newly and predominantly in Korean red pepper fields in 2007.
Fig. 3.
Phylogenetic trees reconstructed using the complete nucleotide sequences of RNA-1or RNA-2 of BBWV2 isolates. The maximum likehood method implemented in MEGA 5 was used to reconstruct the phylogenetic trees.
Genetic diversity of Korean BBWV2 isolates.
To investigate the genetic diversity of BBWV2 isolates from naturally infected red pepper plants, 153 isolates out of 205 specimens were selected according to the occurrence areas and analyzed by RT-PCR using group I and II-specific primers for RNA-1 and RNA-2 genome segments, respectively (Table 7). As a result, GS-I and GS-III were detected from 33 (21.6%) and 87 (56.9%) samples, respectively. Mixed infection of GS-I and GS-III was detected from 27 samples (17.6%). In addition, mixed infection of GS-II with either GS-I or GS-III was detected from 6 samples (3.9%). While GS-I and GS-III isolates were detected throughout the investigated areas, GS-II isolates were detected from only Gyeongsangbuk-do with a low incidence rate. Above all, GS-III of BBWV2, which was predominant (78.4%) in Korean red pepper appears to be a new combination between Group I RNA-1 segment and Group II RNA-2 segment.
Table 7.
Genetic diversity of BBWV2a isolates from red peppers
| Provinceb | No. of red peppers | Genome combination of BBWV2 (%)c | |||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| investigated | infected | selected | GS-Id | GS-IIIe | GS-I+GS-IIIf | GS-I+GS-II+GS-IIIg | |
| GG | 44 | 30 | 25 | 10 | 15 | 0 | 0 |
| CN | 45 | 35 | 30 | 1 | 27 | 2 | 0 |
| CB | 42 | 34 | 24 | 11 | 8 | 5 | 0 |
| JB | 15 | 6 | 4 | 2 | 1 | 1 | 0 |
| JN | 7 | 4 | 1 | 1 | 0 | 0 | 0 |
| GW | 27 | 20 | 15 | 2 | 11 | 2 | 0 |
| GB | 32 | 29 | 24 | 2 | 7 | 9 | 6 |
| GN | 48 | 47 | 30 | 4 | 18 | 8 | 0 |
|
| |||||||
| Total | 260 | 205 | 153 | 33(21.6)g | 87(56.9) | 27(17.6) | 6(3.9) |
BBWV2: Broad bean wilt virus 2
The abbreviation represented as follows; GG: Gyeonggi-do, CN: Chungcheongnam-do, CB: Chungcheongbuk-do, JB: Jeollabuk-do, JN: Jeollanam-do, GW: Gangwon-do, GB: Gyeongsangbuk-do and GN: Gyeongsangnam-do
% in total
GS-I strain: Group I of both RNA-1 and RNA-2
GS-III strain: Group I of RNA-1 and Group II of RNA-2
GS-I+GS-III strain: Group I of RNA-1 and Group I & II of RNA-2
GS-I+GS-II+GS-III strain: Group I & II of RNA-1 and Group I & II of RNA-2
% in total.
Discussion
The survey of the pepper virus disease incidence in 2007 was conducted and the genetic diversity of BBWV2 pepper isolates from 24 areas of 8 provinces in Korea was analyzed. A total 205 samples of 260 collected samples were detected positively (79%) with BBWV2 by DAS-ELISA and RT-PCR, which was increased largely in comparison with 41.3% in 2005 and 58.1% in 2006 (Cho et al., 2007). While the single infection rate with BBWV2 was 21.5%, the co-infection with BBWV2 and red pepper infecting viruses including CMV, PepMoV, PMMoV and PVY was 78.5%. Compared to 37.1% of single infection and 54.1% of mixed infection in 2006, the increase of mixed infection is thought to have caused greater damages on red pepper in Korea. The most prevalent co-infection type was the double infection of BBWV2 and CMV and the triple infection of BBWV2+CMV+PepMoV that had the incidence rates of 28.3% and 29.3%, respectively. As the mixed infection of BBWV2 and CMV with other viruses was 67.8% in total, BBWV2 and CMV are still very important viruses on red pepper in Korea.
To characterize the genetic diversity of BBWV2 Korean isolates, 7 representative isolates were fully sequenced and analyzed (Table 2). So far, complete genome sequences of 8 isolates and partial sequences of 25 isolates have been reported and registered in the GeneBank database. In this study, phylogenetic analyses of 7 BBWV2 Korean isolates with the previously reported 8 isolates based on complete genome sequences showed that BBWV2 could be divided largely into two groups, Group I and Group II in RNA-1 and RNA-2 genomes, respectively (Fig. 3). While the nucleotide sequence identity between two groups was approximately 78% in RNA-1 and 79% in RNA-2, the intragroup nucleotide homology was more than 86% – 90% in RNA-1 and 84% – 89% in RNA-2. The nucleotide sequence indentities between BBWV1, which was used as an outgroup, and BBWV2 were approximately 61% and 58% for RNA-1 and RNA-2, respectively (Kobayashi et al., 2003). In case of RNA-2, Group II could be subdivided into 3 subgroups, II-a, II-b and II-c. BBWV2 Korean red pepper isolates except RP7 were subgrouped into II-a. Rosa et al. (2011) reported that 30 BBWV2 isolates could be divided into six subgroups based on the phylogenetic tree of the SCP nucleotide sequences. RNA-2 compared with RNA-1 showed more extensive genetic variation. As mentioned earlier, RNA-2 encodes MP and two coat proteins (LCP and SCP). Plant viral MPs are invovled in the viral cell-to-cell movement by modifying the structure and function of plasmodesmata (Hofmann et al., 2007 and Liu et al., 2009). Some MPs can bind specifically to heterologous CPs to form complexes that function in movement (Carvalho et al., 2003 and Sánchez-Navarro et al., 2006). Therefore, genetic variations in RNA-2 genome can affect virus infection characteristics. According to group-combination between RNA-1 and RNA-2 genomes, BBWV2 could be designated as three genetic strains of GS-I (Group I in both RNA-1 and RNA-2), GS-II (Goup II in both RNA-1 and RNA-2) and GS-III (Group I in RNA-1 and Group II in RNA-2). Eight isolates reported previously could be grouped to GS-I (IP and ME) and GS-II (B935, IA, K, MB7, PatMMV and XJ114), while BBWV2 Korean isolates could be grouped GS-I (RP7), GS-II (RP3) and GS-III (RP1, RP2, RP4, RP5 and RP6). GS-III was a new combination detected only in Korean isolates. Especially, when compared with BBWV-K isolated in Korea in 2000, only RP3 belonged to the same group (i.e. Group II) and all the other Korean isolates (RP1, RP2, RP4, RP5, RP6 and RP7) included in Group I in the phylogenetic tree of RNA-1 (Fig. 3). In addition, RP3 showed typical mosaic symptom while BBWV-K had severe necrotic symptom on C. annuum (Lee et al., 2000). Thus, it is likely that the BBWV2 isolates emerged in Korea in 2007 might not be originated from BBWV-K.
Based on RT-PCR using Group I and II-specific primers (Table 5), 153 BBWV2 isolates out of 205 isolates were further analyzed and group-specified (Table 7). As a result, GS-I was detected from 33 isolates (21.6%) out of 153 isolates, GS-III from 87 isolates (56.9%), the mixed infection of GS-I and GS-III from 27 isolates (17.6%). The mixed infextion of GS-II with either GS-I or GS-III was detected from 6 isolates (3.9%). While GS-I and GS-III was detected almost all over the provinces investigated, GS-II was detected only from Gyeongsangbuk-do with low incidence rate. Above all, GS-III of BBWV2 was predo-minant at 78.4% in Korean red pepper, as new combination with Group I RNA-1 and Group II RNA-2. Incidence of BBWV2 on red pepper was under 2% before 2004. The incidence was increased abruptly, however, to 41.3% in 2005, 58.2% in 2006 (Cho et al., 2007), 79% in 2007 in this study. These rapid increases of viral disease might be related with the emergence of new combination of BBWV2 groups at high incidence. We are currently constructing full-length infectious clones of some BBWV isolates to better understand the relationship between the genetic variation and the pathogenicity in the BBWV2 population.
Acknowledgments
This work was support by a grant from the Agenda Program (PJ00789802), Rural Development Administration, Republic of Korea.
References
- Benner CP, Kuhn CW, Demski JW, Dobson JW, Colditz P, Nutter FW., Jr Identification and incidence of pepper viruses in Northeastern Georgia. Plant Dis. 1985;69:999–1001. [Google Scholar]
- Brunt AA, Crabtree K, Dallwitz MJ, Gibbs AJ, Watson L. Viruses of Plants. Descriptions and Lists from the VIDE Database. CAB International; Wallingford, UK: 1996. [Google Scholar]
- Carvalho CM, Wellink J, Ribeiro SG, Goldbach RW, van Lent JWM. The C-terminal region of themovement protein of cowpea mosaic virus is involved in binding to the large but not to the small coat protein. J. Gen. Virol. 2003;84:2271–2277. doi: 10.1099/vir.0.19101-0. [DOI] [PubMed] [Google Scholar]
- Chang MU, Chung JD. Studies on viruses isolated from Lilium spp. in Korea. Plant Pathol. J. 1987;3:223–235. [Google Scholar]
- Cho JD, Kim JS, Lee SH, Choi GS, Chung BN. Viruses and symptoms on peppers and their infection types in Korea. Res. Plant Dis. (in Korean) 2007;13:75–81. [Google Scholar]
- Choi GS, Kim JH, Lee DH, Kim JS, Ryu KH. Occurrence and distribution of viruses infecting pepper in Korea. Plant Pathol. J. 2005;21:258–261. [Google Scholar]
- Choi HS, Cho JD, Lee KH, Kim JS. Broad bean wilt fabaviruses and their specific ultrastructures. Korean J. Electron Microscopy. 2001;31:215–222. [Google Scholar]
- Ferrer R, Ferriol I, Gueri J, Moreno P, Rubio L. Genetic variation and evolutionary analysis of Broad bean wilt virus 2. Arch. Virol. 2011;156:1445–1450. doi: 10.1007/s00705-011-0990-3. [DOI] [PubMed] [Google Scholar]
- Goldbach R, Martelli GP, Milne RG. Family Comoviridae. In: Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Martelli GP, Mayo MA, Summers MD, editors. Virus taxonomy. Classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. Springer; Wien New York: 1995. pp. 341–347. (Arch Virol [Suppl] 10). [Google Scholar]
- Hahm YI, Kwon M, Kim JS, Seo HW, Ahn JH. Serveys on disease occurrence in major horticultural crops in Kangwon Alpine areas. Plant Pathol. J. 1998;14:668–675. [Google Scholar]
- Hofmann C, Sambade A, Heinlein M. Plasmodesmata and intercellular transport of viral RNA. Biochem. Soc. Transact. 2007;35:142–145. doi: 10.1042/BST0350142. [DOI] [PubMed] [Google Scholar]
- Ikegami M, Kawashima H, Natsuaki T, Sugimura N. Complete nucleotide sequence of the genome organization of RNA2 of Patchouli mild mosaic virus, a new fabavirus. Arch. Virol. 1998;143:2431–2434. doi: 10.1007/s007050050472. [DOI] [PubMed] [Google Scholar]
- Ikegami M, Onobori Y, Sugimura N, Natsuaki T. Complete nucleotide sequence and the genome organization of Patchouli mild mosaic virus RNA1. Intervirology. 2001;44:355–358. doi: 10.1159/000050071. [DOI] [PubMed] [Google Scholar]
- Kim JH, Choi GS, Choi JK. Characterization of Cucumber mosaic virus subgroup II isolated from paprika (Capsicum annuum var. grossum) in Korea. Plant Pathol. J. 2002;18:6–11. [Google Scholar]
- Kim JS, Kim SK, Choi GS, Lee MW. Virus disease incidence and symptom appearance in red pepper. Korean J. Plant Pathol. 1990;6:125–132. [Google Scholar]
- Kobayashi YO, Mikoshiba Y, Honda Y, Omura T. Detection of Broad bean wilt virus 1 and Broad bean wilt virus 2 in Japan. Ann. Rept. Kanto Plant Prot. Soc. 2004;51:43–48. [Google Scholar]
- Kobayashi YO, Nakano M, Kashiwazaki S, Naito T, Mikoshiba Y, Shiota A, Kameya-Iwaki M, Honda Y. Sequence analysis of RNA-2 of different isolates of broad bean wilt virus confirms the existence of two distinct species. Arch. Virol. 1999;144:1429–1438. doi: 10.1007/s007050050599. [DOI] [PubMed] [Google Scholar]
- Kobayashi YO, Kobayashi A, Nakano M, Hagiwara K, Honda Y, Omura T. Analysis of genetic relations between Broad bean wilt virus 1 and Broad bean wilt virus 2. J. Gen. Plant Pathol. 2003;69:320–326. [Google Scholar]
- Kobayashi YO, Nakano M, Kashiwazaki S, Naito T, Mokoshiba Y, Shiota A, Kameya-Iwaki M, Honda Y. Sequence analysis of RNA-2 of different isolates of broad bean wilt virus confirms the existence of two distinct species. Arch. Virol. 1999;144:1429–1438. doi: 10.1007/s007050050599. [DOI] [PubMed] [Google Scholar]
- Koh LH, Cooper JI, Wong SM. Complete sequences and phylogenetic analyses of a Singapore isolate of Broad bean wilt fabavirus. Arch. Virol. 2001;146:135–147. doi: 10.1007/s007050170197. [DOI] [PubMed] [Google Scholar]
- Kondo T, Fuji S, Yamashita K, Kang DK, Chang MU. Broad bean wilt virus 2 in yams. J. Gen. Plant Pathol. 2005;71:441–443. [Google Scholar]
- Kuroda T, Okumura A, Takeda L, Miura Y, Suzuki K. Nucleotide sequence and synthesis of infectious RNA from cloned cDNA of Broad bean wilt virus 2 RNA 2. Arch. Virol. 2000;145:787–793. doi: 10.1007/s007050050671. [DOI] [PubMed] [Google Scholar]
- Lee U, Hong JS, Choi JK, Kim KC, Kim YS, Curtis IS, Nam HG, Lim PO. Broad bean wilt virus causes necrotic symptoms and generates defective RNAs in Capsicum annuum. Phytopathology. 2000;90:1390–1395. doi: 10.1094/PHYTO.2000.90.12.1390. [DOI] [PubMed] [Google Scholar]
- Lee SH, Lee KW, Chung BJ. Investigations on the virus diseases in spinach (Spinacia oleracea L.) 2. Identification of Broad bean wilt virus occuring spinach. Korean J. Plant Port. 1979;18:11–14. [Google Scholar]
- Lisa V, Boccardo G. Fabaviruses: broad bean wilt and allied viruses. In: Harrison BD, Murant AF, editors. The plant viruses, vol. 5: Polyhedral virions and bipartite RNA genomes. Plenum Press; New York: 1996. pp. 229–250. [Google Scholar]
- Liu C, Meng C, Xie L, Hong J, Zhoua X. Cell-to-cell trafficking, subcellular distribution, and binding to coat protein of Broad bean wilt virus 2 VP37 protein. Virus Res. 2009;143:86–93. doi: 10.1016/j.virusres.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Iwai T, Honkura R. Complete nucleotide sequence and genome organization of Broad bean wilt virus 2. Ann. Phytopathol. Soc. Jpn. 1998;64:565–568. [Google Scholar]
- Palukaitis P, Garcia-Arenal F. Cucumoviruses. Adv Virus Res. 2003;62:241–323. doi: 10.1016/s0065-3527(03)62005-1. [DOI] [PubMed] [Google Scholar]
- Park IS, Kim KW, Kyun HJ, Chang MU. The viruses in gladiolus hybridus cultivated in Korea 2. Broad bean wilt virus, Cucumber mosaic virus and Tobacco rattle virus. Plant Pathol. J. 1998;14:83–91. [Google Scholar]
- Qi Y, Zhou X, Li D. Complete nucleotide sequence and infectious cDNA clone of the RNA1 of a Chinese isolate of Broad bean wilt virus 2. Virus Genes. 2000a;20:201–207. doi: 10.1023/a:1008132310111. [DOI] [PubMed] [Google Scholar]
- Qi Y, Zhou X, Xue C, Li D. Nucleotide sequence of RNA2 and polyprotein processing sites of a Chinese isolate of Broad bean wilt virus 2. Prog. Nat. Sci. 2000b;10:680–686. [Google Scholar]
- Roh SH, Chang MU. Three viruses isolated from Gentiana spp. in Korea. Plant Pathol. J. 1998;14:425–432. [Google Scholar]
- Sanchez-Navarro JA, Herranz MC, Pallas V. Cell-to-cell movement of alfalfa mosaic virus can be mediated by the movement proteins of ilar-, bromo-, cucumo-, tobamo- and comoviruses and does not require virion formation. Virology. 2006;346:66–73. doi: 10.1016/j.virol.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Sui C, Wei JH, Zhan QQ, Zhang J. First report of Broad bean wilt virus 2 infecting Bupleurum chinense in China. Plant Dis. 2009;93:844. doi: 10.1094/PDIS-93-8-0844B. [DOI] [PubMed] [Google Scholar]
- Taylor RH, Stubbs LL. Commonw. Mycol. Inst./ Assoc. Appl. Biol. Warwick, U.K: 1972. Broad bean wilt virus. Page 81 in: Description of Plant Viruses. [Google Scholar]
- Uyemoto KJ, Provvidenti R. Isolation and identification of two serotypes of Broad bean wilt virus. Phytopathology. 1974;64:1547–1548. [Google Scholar]
- Watterson JC. Development and breeding of resistance to pepper and tomato viruses. In: Kyle MM, editor. Resistance to Viral Diseases of Vegetables. Timber Press; Portland OR: 1993. pp. 80–101. [Google Scholar]