Abstract
Environmental stresses induce several plant pathogenic bacteria into a viable but nonculturable (VBNC) state, but the basis for VBNC is largely uncharacterized. We investigated the physiology and morphology ofthe copper-induced VBNC state in the plant pathogen Ralstonia solanacearum in liquid microcosm. Supplementation of 200 μM copper sulfate to the liquid microcosm completely suppressed bacterial colony formation on culture media; however, LIVE/DEAD BacLight bacterial viability staining showed that the bacterial cells maintained viability, and that the viable cells contain higher level of DNA. Based on electron microscopic observations, the bacterial cells in the VBNC state were unchanged in size, but heavily aggregated and surrounded by an unknown extracellular material. Cellular ribosome contents, however, were less, resulting in a reduction of the total RNA in VBNC cells. Proteome comparison and reverse transcription PCR analysis showed that the Dps protein production was up-regulated at the transcriptional level and that 2 catalases/peroxidases were present at lower level in VBNC cells. Cell aggregation and elevated levels of Dps protein are typical oxidative stress responses. H2O2 levels also increased in VBNC cells, which could result if catalase/peroxidase levels are reduced. Some of phenotypic changes in VBNC cells of R. solanacearum could be an oxidative stress response due to H2O2 accumulation. This report is the first of the distinct phenotypic changes in cells of R. solanacearum in the VBNC state.
Keywords: copper, dps, oxidative stress, Ralstonia solanacearum, viable but nonculturable (VBNC) state
Ralstonia solanacearum (Smith) (Yabuuchi et al., 1995), a causative agent of bacterial wilt, is a member of the β-proteobacteria infecting over 200 plant species belonging to more than 50 botanical families. Bacterial wilt by soilborne R. solanacearum causes significant yield losses of solanaceous crops worldwide (Hayward, 1991). Although economically important, there are few management options for the bacterial wilt. The most effective control relies on resistant cultivars, although breeding of wilt resistance cultivars is difficult because the genetics of resistance is complex (Saddler, 2005). No effective chemical control is available and persistence of the organism in soils and waters complicates disease management.
Persistence of R. solanacearum has been studied in soils and waters, where the bacterium R. solanacearum can survive for long periods (Kelman, 1956). Extended survival of R. solanacearum in water is a major cause of contamination in irrigation water, which introduces the pathogen into agricultural soils and causes disease outbreaks (Van Elsas et al., 2001). Many environmental factors influence bacterial survival in aquatic habitats (Van Elsas et al., 2001). In contrary to the rather consistent reports on bacterial survival in water, there are conflicting reports on the survival of R. solanacearum in soil (Coutinho, 2005). It has been suggested that R. solanacearum can survive for long periods in a nutrient-depleted bulk soil environment (Grey and Steck, 2001). Detection of bacterial survivors in soil and water is largely dependent on standard cultivation of live bacterial cells. However, standard bacterial cultivation methods cannot detect viable but nonculturable (VBNC) cells in many environments (Xu et al., 1982).
Since the discovery of the VBNC state (Xu et al., 1982), it has been considered a bacterial survival mechanism for non-spore-forming bacteria. The conditions that induce the VBNC state vary by bacterial species and include nutrient starvation, low temperature, osmotic stress, oxygen tension, and the presence of heavy metals (Oliver, 2010). The VBNC state is frequently confirmed by restoring bacterial culturability, i.e., “resuscitation,” and in many cases resuscitation may occur in response to relief of the stress condition (Oliver, 2010), while in some cases, resuscitation is more complex (Kell et al., 1998). Cell division capability, metabolic activity, gene expression, and intact cell membrane assays have served as measures of VBNC status (Boulos et al., 1999; Kogure et al., 1979; Rodriguez et al., 1992).
VBNC induction has been reported in many soil bacteria, plant and animal pathogenic bacteria (Oliver, 2010). R. solanacearum enters the VBNC state under low temperature, starvation, and the presence of copper sulfate (Álvarez et al., 2008; Caruso et al., 2005; Van Elsas et al., 2000; Van Elsas et al., 2001). Van Elsas et al. (2000, 2001) showed that low temperature induced the VBNC state in R. solanacearum biovar 2 strains in unsterile soils and irrigation water. These bacteria are a potential threat to secure crop production as they are not easily detected. The VBNC state of R. solanacearum can be also induced by low concentrations of copper in soil, while infectivity and culturability are maintained in the presence of host plants (Grey and Steck, 2001). Resuscitation and some ecology of the VBNC state of R. solanacearum have been documented by several groups (Grey and Steck, 2001; Imazaki and Nakaho, 2009; Van Elsas et al., 2000; Van Elsas et al., 2001), however, physiological and morphological studies are lacking. Understanding the VBNC state of R. solanacearum may provide insights into effective management strategies for bacterial wilt since detection and eradication of VBNC R. solanacearum in soil and in water is an essential part of any management program.
Our objectives in this study were to evaluate bacterial cell morphology, intracellular changes, and gene expression in the VBNC state of R. solanacearum cells maintained in a liquid microcosm. We hypothesized that the VBNC state of R. solanacearum induced by copper in a water microcosm represents the distinct physiological and morphological states that differs from both the culturable and the dead states, and that some of the unique VBNC phenotypes are important for maintenance of the unculturable state. Our results suggest that the copper-induced VBNC state of R. solanacearum might be distinct from other bacterial physiological states and some of the changes associated with the R. solanacearum VBNC state may be induced by the oxidative stress response to H2O2 accumulation.
Materials and Methods
Bacterial strains, growth conditions, and virulence assays
Ralstonia solanacearum strain SL341 (Jeong et al., 2007) was routinely grown at 30 °C in casamino acid peptone glucose (CPG) broth (Schaad et al., 2001) or on CPG plus 0.005% 2,3,5-triphenyl tetrazolium chloride (TZC) agar medium (Kelman, 1954). To induce the VBNC state in a liquid microcosm, R. solanacearum SL341 was grown in mannitol-glutamate (MG) medium. The virulence of R. solanacearum SL341 in the culturable and VBNC states was assessed by using tomato plants and pepper plants as previously described (Lee et al., 2012).
Preparation of liquid microcosm and VBNC induction
Liquid microcosm used in this study was routinely prepared in 0.5× phosphate-buffered saline (0.5× PBS, pH 7.0) as follows; R. solanacearum SL341 cells were grown in MG broth at 30°C for 24 h with 150 rpm shaking, and then harvested by centrifugation (7,610 × g for 10 min at 4 °C). The bacterial cells were washed 3 times by repeated centrifugation (7,610 × g for 10 min at 4 °C) to remove extracellular polysaccharide (EPS), and finally suspended in 0.5× PBS to a cell density of approximately 108 cfu/ml. Whenever a bacterial suspension was prepared, the initial bacterial density was determined using a direct viable count on CPG medium. The cell suspension was divided into 2 liquid microcosms containing 25 ml of the cell suspension in 125-ml Erlenmeyer flasks. One of the liquid microcosms was supplemented with 50 or 200 μM of copper sulfate to induce the VBNC state. The liquid microcosms were incubated at 25°C. To determine bacterial culturability, cells were harvested using centrifugation (16,100 × g for 10 min at room temperature), resuspended in 0.5× PBS, diluted as appropriate, and spread on CPG medium. Bacterial colonies were counted after incubation at 30 °C until 5 days. When no bacterial colonies were formed, the bacterial suspension was concentrated 10 to 100 fold and spreaded on CPG medium to confirm the lack of colony formation.
To measure H2O2 production by bacterial cells in liquid microcosm, R. solanacearum SL341 cells maintained for 24 h in liquid microcosm with or without copper (each 2 ml solution) were harvested by centrifugation (9,300 × g for 15 min at room temperature) to separate bacterial cells and the supernatant. H2O2 contents in both bacterial cells and the supernatant were measured at 390 nm by following the previously described method (Loreto and Velikova, 2001). H2O2 content was evaluated by comparing the absorbance at 390 nm with a standard calibration curve.
Viability of bacterial cells
Bacterial viability was determined by using the LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Oregon, USA) by following the instructions provided by the manufacturer. Briefly, cells in liquid microcosm were harvested by centrifugation (16,100 × g for 10 min at room temperature) and resuspended in saline (0.85% NaCl). The bacterial suspension was mixed with an equal volume of 2× staining solution (6 μM SYTO-9 and 30 μM propodium iodide (PI)) and incubated at room temperature for 15 min in the dark. When necessary, live cell control suspension was prepared by suspending SL341 cells at late exponential growth stage in saline. Dead cell control suspension was prepared by killing SL341 cells with 70% isopropanol for 1 h, harvesting by centrifugation (16,100 × g for 10 min at room temperature), and re-suspending in saline. Each control suspension was prepared to approximately 108 cells/ml.
VBNC and culturable cells stained with SYTO-9 and PI were investigated using a Cytomics FC500 flow cytometer (Beckman Coulter, Brea, USA) equipped with an argon-ion laser with excitation at 488 nm. Green fluorescence was measured at 525 nm and red fluorescence was measured at 620 nm. Ten thousand events were analyzed for each sample and the experiments were repeated at least 3 times. Green SYTO-9 fluorescence was measured by excitation at 480 nm and emission at 500 nm, and red PI fluorescence was measured by excitation at 490 nm and emission at 635 nm.
Microscopy
Fluorescence-labeled cells were also visualized using a model LSM510 confocal laser scanning microscope (CLSM, Carl Zeiss, Germany). The light source was a laser that provided an excitation wavelength of 488 nm (Argonion laser). Fluorescence signals were detected with the filter sets for fluorescein isothiocyanate (FITC; BP 505–530 green) and for red fluorescence (LP560). Image acquisition was carried out using a 10× objective and processed using the standard LSM510 software package.
Transmission electron microscopy was performed to investigate the cellular changes associated with the VBNC state of R. solanacearum SL341. Both negative staining and ultrastructure investigations were conducted using cells maintained for 5 days in liquid microcosm (Kim et al., 2012). Negative staining to investigate cell size and general morphology was carried out by staining the cell suspension with 2% uranyl acetate and briefly dried on carbon-coated copper grids for 1 min. The stained cells were examined under a JEM-2010 transmission electron microscope (TEM) (JEOL Co., Tokyo, Japan) operated at an accelerating voltage of 80 kV.
The cellular ultrastructure of VBNC cells was also examined. Cells were harvested by centrifugation (7,610 × g for 15 min at 4 °C) and fixed in 2.5% paraformaldehyde-glutaraldehyde mixture buffered with 0.1 M sodium phosphate (pH 7.2) for 2 h. The samples were post-fixed in 1% (w/v) osmium tetroxide in the same buffer for 1 h. The post-fixed samples were dehydrated in graded ethanol and propylene oxide, and embedded in Epon-812. Ultra-thin sections, made with an Ultracut-E ultramicrotome (Leica, Wein, Austria), were stained with uranyl acetate and lead citrate and examined under CM 20 TEM (Philips, Eindhoven, The Netherlands) at 100 kV.
Total protein analysis
Bacterial cells maintained for 3 days in liquid microcosm were harvested by centrifugation (7,610 × g for 15 min at 4 °C) and extracted with Mg/NP-40 buffer (0.5 M Tris-HCl (pH 8.3), 2% (v/v) NP-40, 20 mM MgCl2, 5% β-mercaptoethanol, 1 mM phenyl methyl sulfonyl fluoride and 1% (w/v) polyvinyl polypyrrolidone) in a sonicator, followed by phenol extraction. Water-saturated phenol (50 ml) was added to the extracted supernatant solution and thoroughly mixed, then centrifuged at 12,000 × g for 15 min at room temperature. The phenol extraction was repeated twice. The combined organic phase was carefully collected and extracted again with 1 M sucrose to collect the supernatant phenol phase. Four volumes of methanol containing 100 mM ammonium acetate were added to precipitate proteins at −20 °C overnight. The precipitated proteins were recovered by centrifugation (12,000 × g for 10 min at 4 °C) and washing with 80% methanol containing 100 mM ammonium acetate, repeated 3 times. Finally, the precipitated proteins were washed with 80% acetone and stored at −20 °C. Two-dimensional gel electrophoresis (2-DE) and downstream 2-D gel analyses were performed as follows. Briefly, 250 μg protein dissolved in rehydration buffer (8 M (w/v) urea, 2% (w/v) CHAPS, 0.002% (w/v) bromophenol G, 20 mM DTT, 0.5% (w/v) pharmalyte (pH 5–8)) was loaded on 18-cm IPG strips (pH 4–7). The IPG strips were equilibrated with rehydration buffer for 12 h and focused at 50 V for 8 h, 100 V for 1 h, 500 V for 1 h, 1,000 V for 1 h, 2,000 V for 1 h, 4,000 V for 2 h, 8,000 V for 5 h, 8,000 V for 3 h, and 20 V for 2 h with an IPGphore3 platform (GE Healthcare, Little Chalfont, UK). SDS-PAGE in the second dimension was conducted as described by Laemmli by using 12.5% polyacrylamide gels. The 2-D gels were stained by colloidal coomassie brilliant blue G-250 (colloidal CBB).
By comparing two 2-D gels stained with colloidal CBB, the changed protein spots were selected for further identification by mass spectroscopy. The protein spots were excised from the gel, cut into small pieces, washed with 50% acetonitrile in 0.1 M NH4HCO3, and dried under vacuum. Protein digestion and all procedures for MALDI-TOF-MS (Voyager STR, PerSeptive Biosystems, Framingham, USA) analysis and database searching were performed as described (Kim et al., 2008). Protein analysis by MALDI-TOF mass spectrometry was conducted with the following parameters. Parent ion masses were measured in the reflection/delayed extraction mode with an accelerating voltage of 20 kV, a grid voltage of 76%, a guide wire voltage of 0.010%, and a delay time of 150 ns. For data processing, the MoverZ (http://bioinformatics.genomicsolutions.com) software was used. The mass spectra were compared to the database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) by using a peptide mass fingerprint (PMF) search program, MASCOT (http://www.matrixscience.com). A peptide mass accuracy of below 50 ppm was applied. The results with MOWSE scores higher than 65 (p < 0.05) were considered valuable. Only the best matches with high MOWSE scores were selected.
Reverse transcription PCR (RT-PCR) and quantitative RT-PCR (RT-qPCR)
Bacterial cells maintained in liquid microcosm were harvested by centrifugation (7,610 × g for 15 min at 4 °C) and total RNA was isolated using Hybrid-R™ RNA extraction kit (GeneAll Biotechnology Inc., Seoul, Korea). Any trace amount of contaminating DNA was removed by DNase treatment and a regular RT-PCR was performed with 1 μg of total RNA for VBNC cells and culturable cells using a one-step RT-PCR kit (Quiagen, Hilden,Germany). All primers were synthesized by GenoTech Corp. (Daejeon, Korea) and are listed in the Table 1. The PCR amplification was carried out under the following conditions; 95 °C for 15 min of initial denaturation; 30 cycles of denaturation at 94°C for 1 min, annealing at variable temperature (Table 1) for 30 s, extension at 72 °C for 30 s, and a final extension at 72 °C for 10 min. The amplified products were analyzed by agarose gel electrophoresis.
Table 1.
Primers used for RT-PCR and RT-qPCR analyses
| Gene (2-DE spot) | Forward primer (5′-3′) Reverse primer (5′-3′) |
aTa (°C) | Amplicon size (bp) |
|---|---|---|---|
| omp (spot 14) | F: ACTCGTGGCTCTACTACG | 50 | 381 |
| R: ATGCGTTGTTATACAGG | |||
|
| |||
| omp (spot 15) | F: ATCACGGCACAAGACGC | 60 | 255 |
| R: AGCGTTCTGAGCAGCGAACG | |||
|
|
|||
| for RT-qPCR | F: GCGCTGGCAACCTGCGTTCG | 60 | 166 |
| R: GTGCGCTTGGACAGCGCGTA | |||
|
| |||
| rps (spot 16) | F: AAGCGCCAGTCGCGTGAGATCC | 60 | 407 |
| R: TTGCTCGAGTCGTCGTTACC | |||
|
| |||
| dps (spot 17) | F: TCAAGACCCACAACTTCC | 54 | 249 |
| R: TGACGGACTCGTGACCG | |||
|
|
|||
| for RT-qPCR | F: GGACCGCCGTGGATTCGGTGG | 60 | 189 |
| R: CCGGGAACAGGCTGCGTGCG | |||
|
| |||
| epsD (spot 18) | F: TGCTGGAATCCACTTCG | 50 | 360 |
| R: TCTCATCACAGATCATCG | |||
Annealing temperature for PCR amplification
The RNA expression levels of R. solanacearum SL341 in liquid microcosm were determined by RT-qPCR. Bacterial total RNAs were directly used as the template for cDNA synthesis by a Prime Script™ RT Master Mix kit (TaKaRa Biotech. Co., Japan), following the manufacturer’s instructions. Quantitative PCR using the cDNAs synthesized from the above-mentioned method was conducted by a CFX384 Real Time System (Bio-Rad, Hercules, CA, USA). The qPCR reaction components contained SYBR® Premix Ex Taq™ mix (TaKaRa), cDNA template, and 10 μM of primers (Table 1). Thermal cycling included the following steps: an initial preheat for 3 min at 95°C, followed by 40 cycles at 95 °C for 5 sec, 60 °C for 10 sec, 72 °C for 35 sec. The qPCR data were analyzed to compare relative RNA expression of dps using the CFX manager software version 1.6 (Bio-Rad). Student’s t-test was conducted to determine statistically significant difference between VBNC cells and culturable cells. Relative expression levels of dps were calibrated and normalized against the expression levels of outer membrane porin gene.
Results
Determination of viable but nonculturable Ralstonia solanacearum
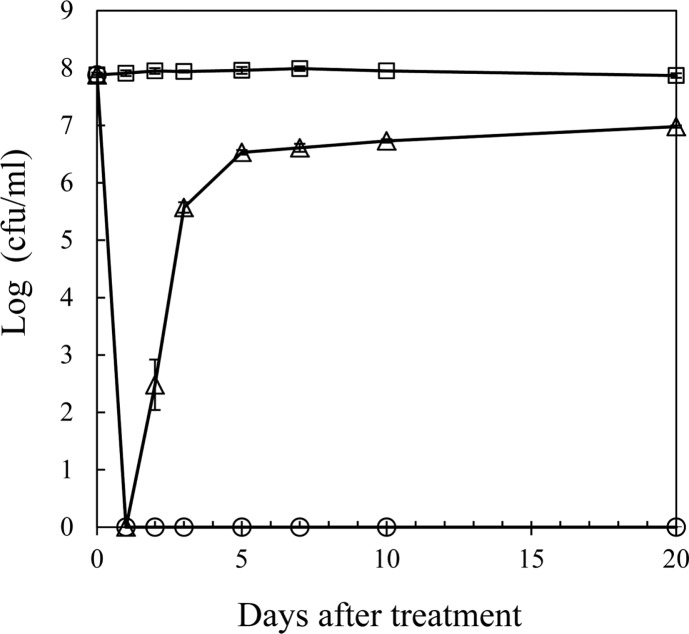
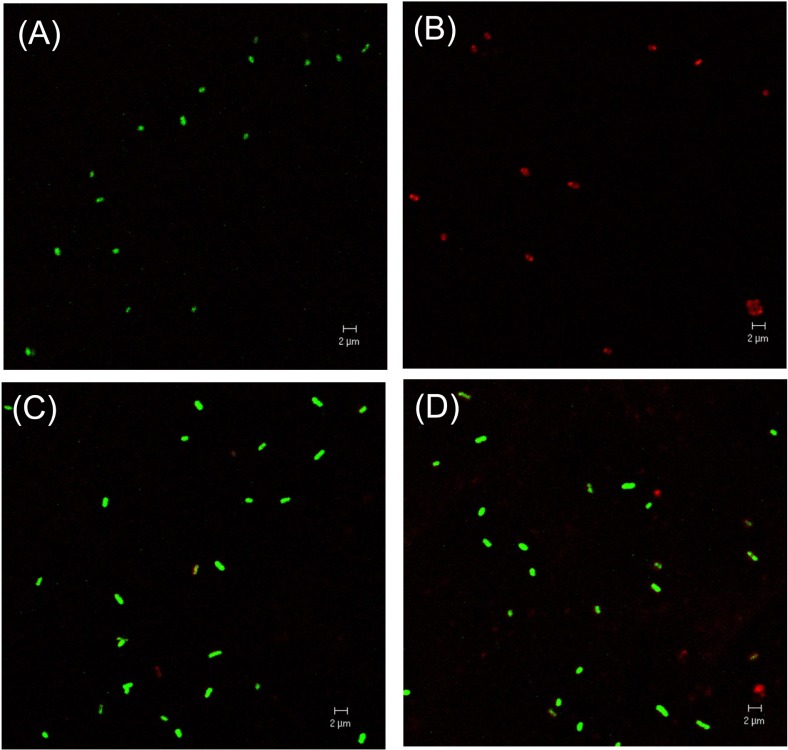
R. solanacearum isolate SL341 was originally isolated from tomato plants in Korea (Jeong et al., 2007). This isolate is a member of race 1 (phylotype I) and stably maintains its viability and culturability when stored in pure water or 0.5× PBS (pH 7.0). However, treatment with 200 μM copper sulfate in 0.5× PBS resulted in the rapid loss of bacterial culturability in most culture media (Fig. 1). This phenomenon was previously documented in R. solanacearum by using liquid microcosm and sterile soil amended with copper sulfate (Grey and Steck, 2001). At 50 μM copper sulfate in liquid microcosm bacterial culturability was temporarily lost for the first day. However, culturability was slowly restored, beginning 3 days after treatment. This result may suggest that the bacteria, which were in a nonculturable state on the first day, may not be in a death stage. Therefore, to determine if the bacterial cells in liquid microcosm supplemented with copper sulfate were alive or not, we stained the bacterial cells by using the LIVE/DEAD BacLight bacterial viability kit and observed using CLSM. Most of the cells in copper sulfate solution appeared to be green, similar to live cell controls but different from the dead cells, which appear red (Fig. 2). Therefore, we conclude that SL341 entered into a VBNC state induced by copper sulfate treatment in liquid microcosm. Viability in the VBNC state was maintained for over 60 days, as determined by Live/Dead staining (data not shown).
Fig. 1.
Culturable cells of R. solanacearum in liquid microcosm over time. Culturable cells of R solanacearum in microcosm containing 0 (square), 50 (triangle), and 200 μM (circle) copper sulfate. The culturable cell numbers were detected by formation of visible colonies on CPG medium. Vertical bars represent standard deviation from 3 replicates.
Fig. 2.
Confocal laser scanning micrograph of culturable and VBNC R. solanacearum. Images of SYTO-9 and PI were merged. (A), live cell control; (B), dead cells killed by isopropanol; (C), VBNC cells maintained for 1 day in microcosm; (D), VBNC cells maintained for 2 days in microcosm. Green cells and red cells represent viable and dead cells, respectively. Magnification was 1,600× and the bar represents 2 μm.
Visualization of viable but nonculturable state
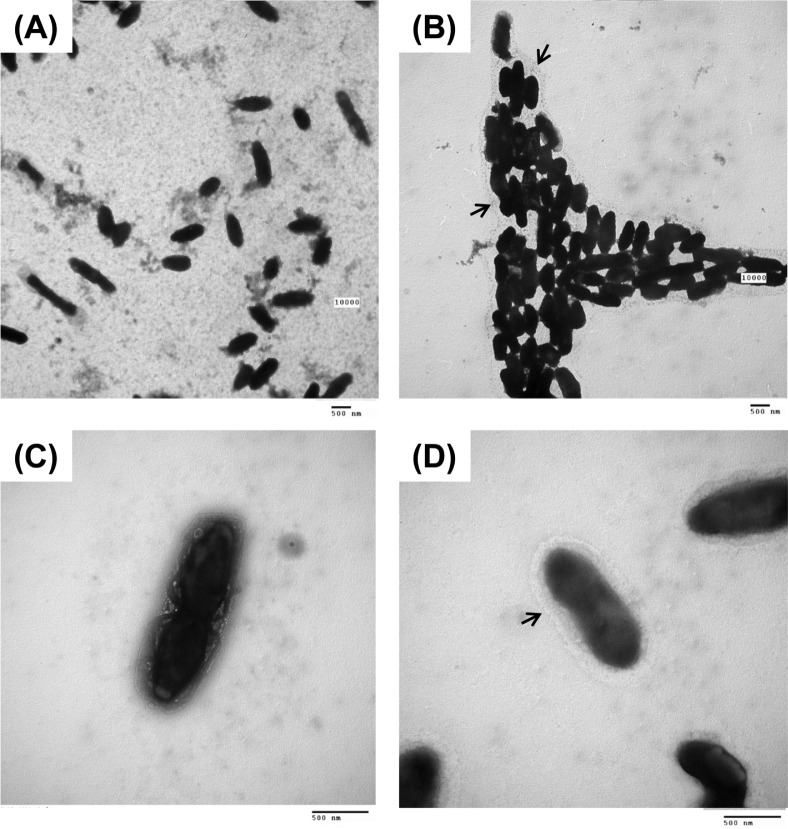
While observing VBNC cells using CLSM, we noticed that the cells were enlarged in comparison to dead or live cell controls (Fig. 2). Thus, we carefully compared bacterial cell size between VBNC cells and culturable cells by using TEM. However, no difference in cell size of the different states was apparent (Fig. 3 and Fig. 4). Interestingly, TEM with negative staining revealed that VBNC cells of R. solanacearum aggregated each other and were surrounded by an unidentified extracellular matrix (Fig. 3B, 3D). Bacterial cells in a culturable state were not highly aggregated and did not produce the same extracellular matrix (Fig. 3A, 3C). We quantified EPS production by culturable and VBNC R. solanacearum SL341 using a method described by Brumbley and Denny (1990). However, we found no significant difference in the amount of EPS production by these different bacterial states (data not shown).
Fig. 3.
Transmission electron micrograph of culturable and VBNC R. solanacearum shown by negative staining. Culturable R. solanacearum in liquid microcosm (A and C), VBNC R. solanacearum in microcosm with 200 μM copper sulfate (B and D). Arrows in (B) and (D) indicate unknown extracellular material produced by VBNC cells. Magnifications for (A and B) and (C and D) were 10,000× and 30,000×, respectively. Bars represent 500 nm.
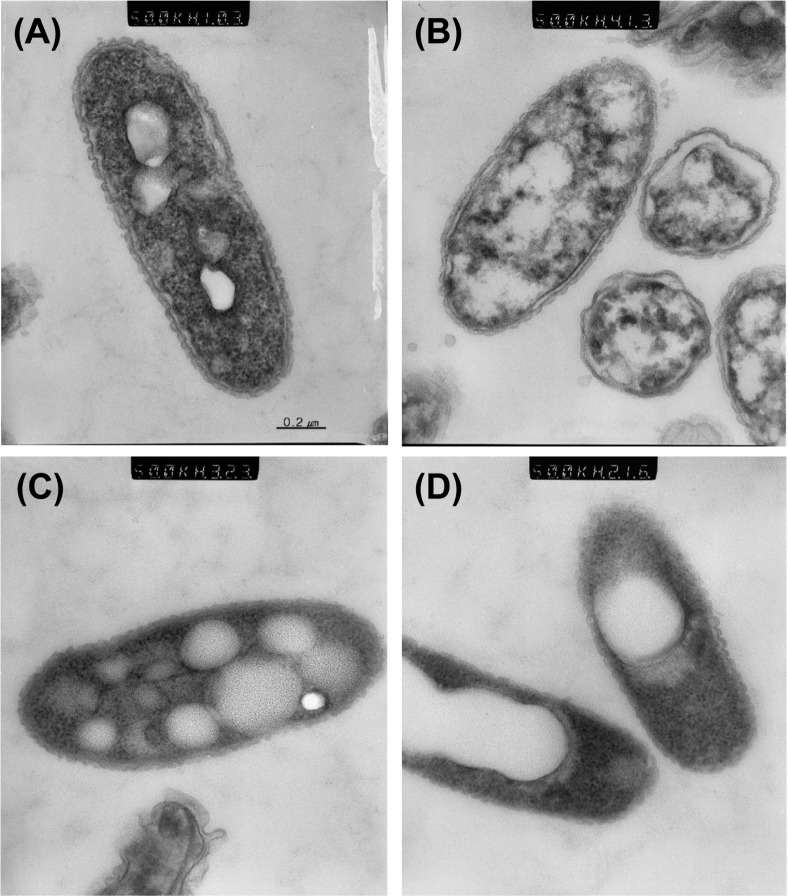
Fig. 4.
Ultrastructure of actively growing cells. (A), VBNC cells in microcosm maintained for 5 days with 200 μM copper sulfate (B), culturable cells in microcosm maintained for 5 days without copper (C), and dead cells (D) observed after ultra-thin section. Note the reduction of electron-dense granules and PHB disintegration in VBNC cells in panel B. Magnification for all 4 figures was 50,000×.
Ultrastructure of the bacterial cells under different conditions was observed by TEM. Bacterial cells grown in CPG broth (Fig. 4A) and those maintained in copper-free water for 5 days (Fig. 4C) showed the presence of uniform intracellular inclusion bodies. The inclusion bodies are composed of poly-β-hydroxybutyrate (PHB), a common poly-β-hydroxyalkanoate (PHA),which accumulates as a cellular reserve in Ralstonia species (Potter et al., 2004). The culturable bacterial cells maintained in liquid microcosm exhibited clear and relatively uniform PHB inclusions in the protoplasm. The inclusion bodies were apparent in dead cells (isopropanol treated) but were conglomerated (Fig. 4D). However, bacterial cells in the VBNC state exhibited significant disintegration of the PHB inclusions, which were dispersed in the protoplasm (Fig. 4B). Another noticeable change in the VBNC cells was the remarkable decrease of electron-dense granules in the protoplasm, in comparison to the live or dead cells. The electron-dense granules are probably ribosomes, which are abundant in the protoplasm. Therefore, we postulated that the cellular contents of ribosomes and ribosomal RNA (rRNA) might be dramatically reduced in the copper-treated VBNC cells in comparison to the culturable or live cells.
Altered nucleic acid contents
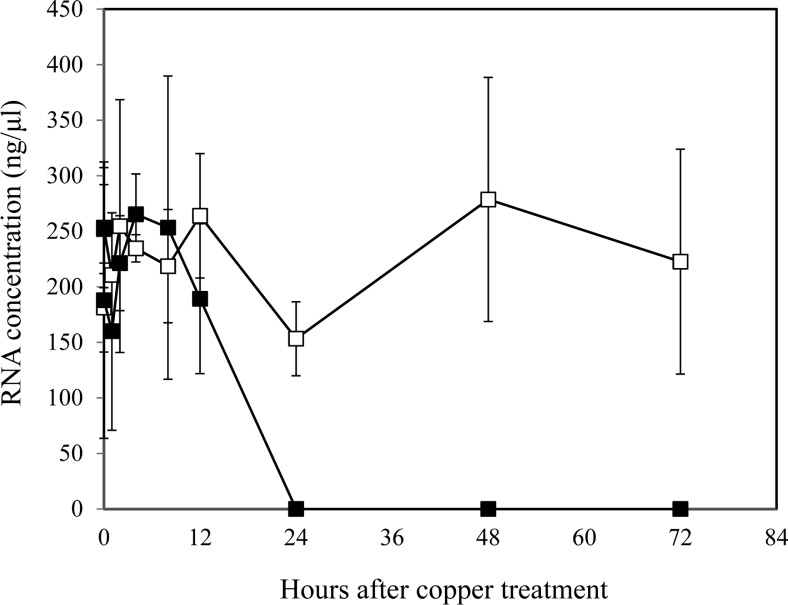
While CLSM observation of VBNC cells suggested that the cells were enlarged (Fig. 2), this was not evident in the TEM analysis (Fig. 3 and Fig. 4). Therefore, we investigated the possibility that VBNC cells take up more SYTO-9 dye during the viability staining process, which may be caused by an increased concentration of nucleic acids. Flow cytometry of culturable and VBNC cells showed that SYTO-9 staining was significantly increased 2.7- to 4.8-fold in comparison to culturable cells (Table 2). In contrast, PI staining of VBNC cells was only increased 1.6- to 2.4-fold (Table 2). When we measured the total RNA content of culturable cells and VBNC cells, VBNC cells exhibited dramatically reduced amounts of RNA within 12 h after copper treatment and RNA was not detectable after 24 h (Fig. 5). However, the total RNA content of culturable cells was stably maintained. Therefore, flow cytometry after viability staining indicated an increase of total DNA content in the VBNC state of R. solanacearum SL341.
Table 2.
Flow cytometry of culturable and VBNC R. solanacearum SL341 for SYTO-9 and PI staining.
| Days | Mean fluorescence intensity (SYTO-9) | Mean fluorescence intensity (PI) | ||
|---|---|---|---|---|
|
|
|
|||
| Culturable cells | VBNC cells | Culturable cells | VBNC cells | |
| 0 | 2.77 ± 0.029 | 2.77 ± 0.029 | 0.317 ± 0.029 | 0.317 ± 0.029 |
| 3 | 0.92 ± 0.029 | 2.50 ± 0.229 (2.7×) | 0.217 ± 0.029 | 0.350 ± 0.000 (1.6×) |
| 5 | 0.82 ± 0.104 | 3.42 ± 0.104 (4.2×) | 0.233 ± 0.058 | 0.567 ± 0.029 (2.4×) |
| 7 | 0.97 ± 0.058 | 2.75 ± 0.132 (2.8×) | 0.267 ± 0.029 | 0.433 ± 0.029 (1.6×) |
Fluorescence intensity of SYT-O9 and PI were counted up to 10,000 cells to generate mean intensity using live/dead stained cells of R. solanacearum in microcosm. Three independent sets from the same bacterial suspension were used to generate average intensities and standard deviations. The numbers in parenthesis under VBNC cells is the fold increase in mean intensity compared to culturable cells.
Fig. 5.
Total RNA contents in culturable (open square) and VBNC (closed square) R. solanacearum measured by spectrophotometry. Vertical bars represent the standard deviation from 3 replicates.
Proteome profile and gene expression
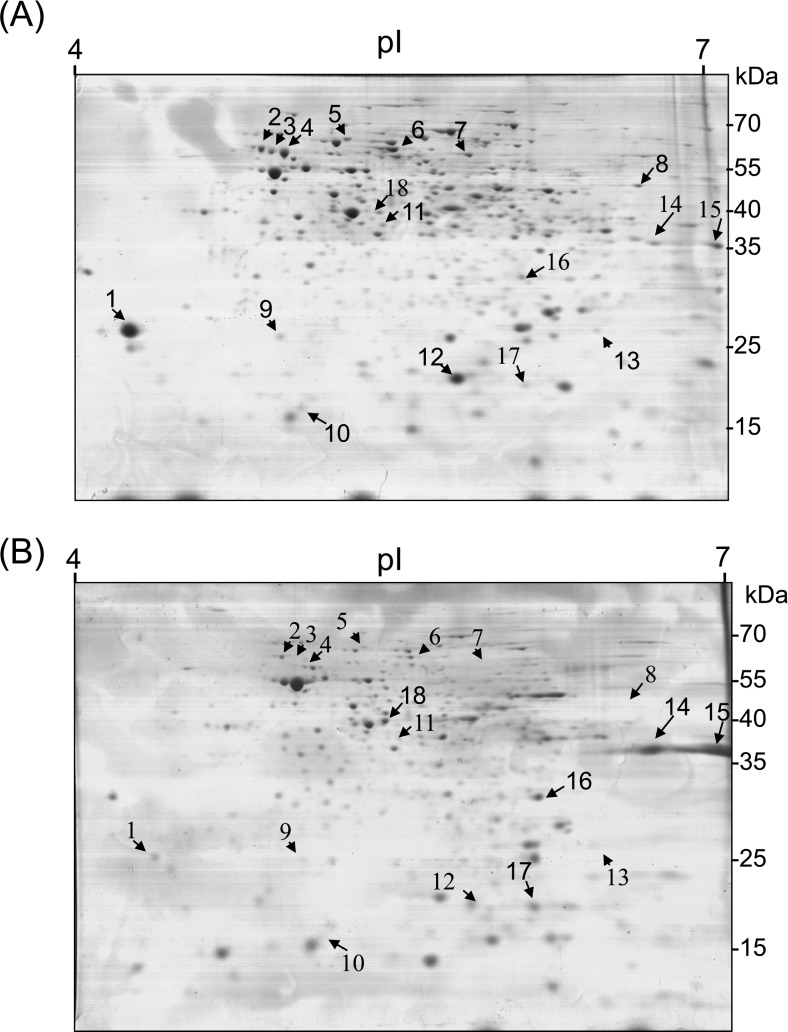
To compare protein production between culturable and VBNC cells of R. solanacearum SL341, total proteins were extracted and analyzed by 2-DE (Fig. 6). Twenty-eight proteins were differentially expressed in the VBNC cells and 18 of these were identified (Table 3). The remaining 10 proteins were not identified. Thirteen proteins were down-regulated and 5 proteins were up-regulated in the VBNC state of R. solanacearum SL341 (Table 3). Most of the identified proteins were highly similar to proteins from R. solanacearum GMI1000. Specifically, the induced proteins in VBNC cells were identified as Dps (spot 17), outer membrane porin proteins (spot 14, 15), small subunit ribosomal protein S2P (spot 16), and NDP-N-acetyl-galactosaminuronic acid dehydrogenase oxidoreductase (EpsD) (spot 18). Dps is a DNA-binding protein involved in DNA protection during starvation or oxidative stress, which was first described in starved Escherichia coli (Almirón et al., 1992). Many other proteins were repressed in VBNC cells, including catalase/peroxidase(spot 6 and 8) and several oxidoreduction enzymes (Table 3).
Fig. 6.
Proteome analysis of culturable (A) and VBNC (B) R. solanacearum. Proteins extracted from cells maintained for 3 days in liquid microcosm were separated by 2-DE. Differentially expressed proteins were marked with arrows. Numbered arrows are identified proteins and arrows without numbers indicate unidentified proteins.
Table 3.
MALDI-TOF MS identification of proteins repressed or induced by the VBNC state in Ralstonia solanacearum SL341
| Spot | Protein name (organism)a | Accession no. | MOWSE Score | MP | SC (%) | MW (KDa)/pI | Reg.b |
|---|---|---|---|---|---|---|---|
| 1 | SMF protein (Sulfurimonas denitrificans) | 78777855 | 12658 | 7 | 27.9 | 28.5/6.6 | Down |
| 2 | Hypothetical protein | 17428510 | 6.4E+05 | 13 | 23.2 | 63.2/5.2 | Down |
| 3 | Hypothetical protein | 17428510 | 1.7E+07 | 18 | 31.7 | 63.2/5.2 | Down |
| 4 | Hypothetical protein | 17428510 | 4089 | 7 | 13.9 | 63.2/5.2 | Down |
| 5 | Probable elongation factor G | 17431276 | 2.29E+08 | 15 | 28 | 77.6/5.3 | Down |
| 6 | Peroxidase/catalase, oxidoreductase | 17427787 | 4E+07 | 14 | 34.5 | 49.6/5.3 | Down |
| 7 | Probable transketolase | 17429772 | 5.62E+08 | 17 | 23.3 | 72.4/6.0 | Down |
| 8 | Peroxidase/catalase, oxidoreductase | 17427787 | 1986 | 6 | 13.8 | 49.6/5.3 | Down |
| 9 | Inosine-5’-monophosphate dehydrogenase | 83749992 | 1.7E+08 | 20 | 55.4 | 52.0/7.2 | Down |
| 10 | Putative glyoxalase/bleomycin resistance protein/dioxygenase | 17431071 | 19338 | 8 | 57.3 | 15.6/5.2 | Down |
| 11 | N-acetyl-γ-glutamyl-phosphate reductase, oxidoreductase protein | 17427150 | 672 | 5 | 16 | 36.4/5.5 | Down |
| 12 | Hypothetical Toxin corregulated pilus biosynthesis protein T (Cupriavidus metallidurans) | 56410289 | 141648 | 12 | 19.8 | 66.8/6.4 | Down |
| 13 | Probable transcription antitermination protein | 17430063 | 1124 | 5 | 26.8 | 21.8/6.9 | Down |
|
| |||||||
| 14 | Probable outer membrane porin signal peptide (Omp) | 17430115 | 67044 | 10 | 28.2 | 39.5/8.8 | Up |
| 15 | Putative porin signal peptide protein (Omp) | 17429956 | 3E+06 | 14 | 40.1 | 39.6/9.3 | Up |
| 16 | SSU ribosomal protein S2P (Rps) | 83748807 | 2962 | 10 | 35.6 | 27.2/6.6 | Up |
| 17 | Putative DNA protection protein during starvation or oxidative stress (Dps) | 17429709 | 1450 | 5 | 33.1 | 18.4/6.2 | Up |
| 18 | NDP-N-acetyl-galactosaminuronic acid dehydrogenase oxidoreductase (EpsD) | 17431489 | 1.42E+08 | 14 | 39 | 46.7/5.5 | Up |
Homolog protein from other bacterial species but not in R. solanacearum
Proteins repressed (Down) or induced (Up) by copper treatment MP, matched peptides; SC, sequence coverage; MW, molecular weight; Reg, Regulation
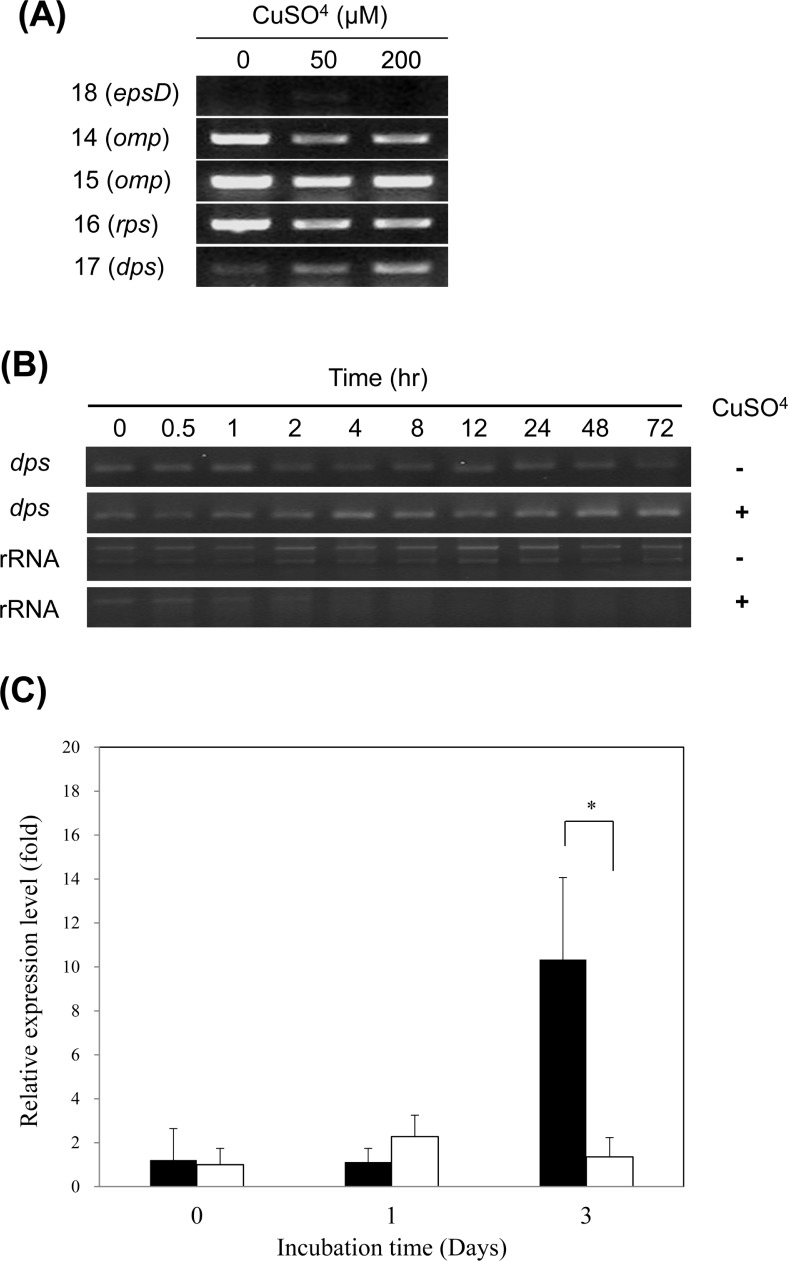
In order to examine if the induced proteins were regulated at the transcriptional level, RT-PCR was performed using total RNA extracted from culturable and VBNC cells, which were maintained for 3 days in liquid microcosm. Transcription of epsD was slightly induced by 50 μM copper sulfate, while its expression was not evident at 200 μM copper sulfate. Two genes for outer membrane porin proteins and a gene for ribosomal protein S2P were transcribed constitutively or down-regulated, while the proteins were increased by the VBNC state. The dps gene was transcriptionally induced by copper treatment in a concentration-dependent manner (Fig. 7A). Therefore, it is likely that Dps production in VBNC R. solanacearum SL341 might be transcriptionally regulated, while the other 4 proteins may be post-transcriptionally or translationally regulated. Furthermore, transcription of dps in the VBNC state increased over time in comparison to the culturable cell control (Fig. 7B). Up-regulation dps expression was also apparent in VBNC cells compared to culturable cells of SL341, when analyzed by RT-qPCR. Especially, expression of dps in VBNC cells was significantly increased in 3 days incubation compared to culturable cells (Fig. 7C). Dps is involved in DNA protection from starvation or oxidative stress (Almirón et al., 1992; Martinez and Kolter, 1997; Saumaa et al., 2007).
Fig. 7.
RT-PCR analysis of genes encoding induced proteins in VBNC R. solanacearum. (A) Total RNAs were extracted from bacterial cells maintained for 3 days in liquid microcosm containing various amount of copper sulfate. The numbers on the left represent the corresponding protein spot number from the 2-DE gel. (B) Detection of dps transcript over time in culturable (Cu−) and VBNC (Cu+) R. solanacearum. (C) Relative expression level of dps over time in VBNC cells (black) and culturable cells (white) of R. solanacearum. Y-axis represents the relative amount of dps transcript over omp transcript. The vertical bar represents the standard deviation of 3 replicates. Asterisk represents significant difference between VBNC cells and culturable cells, P < 0.05.
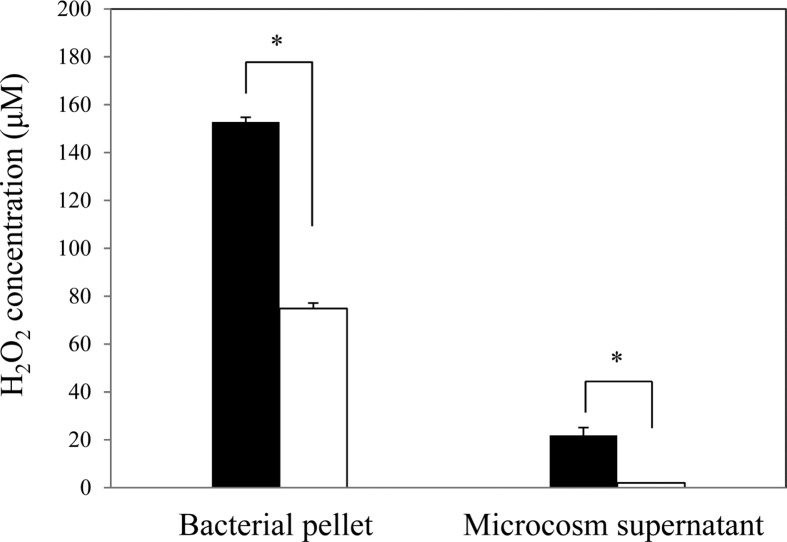
H2O2 accumulation in the VBNC bacterial cell
Twenty-four hours after copper sulfate treatment, H2O2 was significantly increased in the VBNC cells compared to culturable cells, both in the cells and the liquid microcosm supernatant (Fig. 8). H2O2 accumulation in the VBNC bacterial pellet from a 2-ml microcosm (ca. 2 × 108 cells/ml) was 152.8 μM, while that in the culturable state was 74.8 μM. H2O2 concentration in the microcosm supernatant (2 ml) was 21.9 μM in the VBNC state but was undetectable in the culturable state. The increase of H2O2 production was coincident with down regulation of catalase/peroxidase and induction of Dps in the VBNC state of R. solanacearum SL341. Gene expression using RT-PCR showed that no gene expression was detected for the above-mentioned 4 genes in isopropanol-treated R. solanacearum SL341 cells (data not shown). Exogenous application of H2O2 into bacterial cell suspension (approximately 5 × 108 cells/ml) did not induce unculturability up to 1 mM, while supplementation with 150 μM H2O2 in bacterial culture medium CPG completely inhibited bacterial growth (data not shown).
Fig. 8.
H2O2 production by culturable (white) and VBNC (black) R. solanacearum maintained for 24 h in liquid microcosm. H2O2 production was quantified in cells and supernatants in liquid microcosm. The vertical bar represents the standard deviation of 3 replicates. Asterisk represents significant difference between VBNC cells and culturable cells, P < 0.05.
Discussion
Copper-induced VBNC was previously documented in R. solanacearum in liquid microcosm and sterile soil (Grey and Steck, 2001) and in several plant pathogenic bacteria (Alexander et al., 1999; Ghezzi and Steck, 1999; Ordax et al., 2006). Induction of VBNC by 200 μM copper sulfate in R. solanacearum SL341 was similar to previous reports. The unculturability of R. solanacearum by copper treatment was noted in other strains such as GMI1000 (Boucher et al., 1985) and SL2029 (Jeong et al., 2007) (data not shown). The concentration of copper sulfate in liquid microcosm was lower than the minimal inhibitory concentration (850 μM) for SL341 and the average amount of copper in soil in the United States (Holmgren et al., 1993). Although copper is not the sole inducer of the VBNC state in R. solanacearum (Álvarez et al., 2008; Grey and Steck, 2001; Van Elsas et al., 2000), little is known about the detailed physiological and morphological changes in the copper-induced VBNC state.
Morphological analysis of VBNC R. solanacearum by using TEM revealed interesting phenotypes such as bacterial cell aggregation surrounded by extracellular material (Fig. 3B and 3D) and PHB disintegration (Fig. 4B). Since EPS production did not differ between culturable and VBNC cells, we do not know the nature of the extracellular material in VBNC state. The extracellular material must be sticky, leading to aggregation. In fact, cell pellets from liquid microcosm containing copper sulfate were sticky compared to culturable bacterial pellets. Another noticeable change in VBNC cells was PHB disintegration. PHB is a common PHA, the cellular reserve in Ralstonia species (Potter et al., 2004). PHB in the VBNC state was severely disrupted and was dispersed irregularly in the bacterial cytoplasm (Fig. 4B). Disintegration of PHB by environmental stress has not been previously reported. Divalent copper ion may disrupt the chemical linkage of PHB granules in the bacterial cytoplasm.
Although we do not know the nature of the extracellular material produced by the VBNC R. solanacearum, it may protect the cells in harsh environments (Blat and Eisenbach, 1995). Bacterial cell aggregation was previously described after prolonged starvation of R. solanacearum (phylotype II) in water microcosm (Álvarez et al., 2008). The phylotype II strain changed to cocci after prolonged starvation and aggregated. It has been frequently reported that cell aggregation and pattern formation might be triggered by oxidative stress, such as that triggered by H2O2 (Blat and Eisenbach, 1995; Budrene and Berg, 1991) and may provide the living cells in the cluster with nutrients released from lysing or dead cells (Shapiro, 1988). Cell aggregation of R. solanacearum might also be triggered by H2O2 accumulation. In fact, H2O2 production was much higher in VBNC cells than in culturable cells (Fig. 8).
One of the remarkable phenotype changes in R. solanacearum SL341 induced by copper treatment was the dramatic reduction of ribosomes and the rapid decrease in total RNA content (Fig. 4 and Fig. 5). Reduction of total RNA in VBNC cells is probably due to the dramatic reduction of ribosomes, because ribosomal RNAs comprise the major components of total RNA (over 80%). While total RNAs were severely reduced, mRNA must be stably retained in VBNC cells because we successfully detected transcripts by using RT-PCR. Because there are fewer ribosomes in the VBNC state, de novo protein synthesis will be much reduced and metabolic activity might be significantly affected. Nonetheless, proteome (Fig. 6) and RT-PCR analyses (Fig. 7) showed that many genes are actively expressed to produce certain proteins in VBNC cells. The gene expression detected by RT-PCR in the VBNC state would be an excellent indicator to prove that SL341 maintains its viability. Our trials to recover colony-forming VBNC cells all failed in various culture media and under various culture conditions (data not shown). In addition, inoculation of VBNC cells into a plant host, such as tomato and pepper, seldom caused bacterial wilt (data not shown). Grey and Steck (2001) reported the recovery of bacterial culturability “resuscitation” and bacterial virulence by using copper-induced VBNC R. solanacearum. Our failure to resuscitate SL341 may be due to different experimental conditions; for example, we used only liquid microcosm applied to the pots of adult plants, while Grey and Steck (2001) performed their recovery studies in a soil environment. The severely reduced ribosomes in the VBNC state could be a barrier for recovery due to reduced metabolic activity. Reduction of bacterial ribosomes in VBNC cells has not been described previously. In fact, this result is in contrast to the VBNC state of several probiotic Bifidobacterium spp., which retain high levels of rRNA, despite losing culturability in fermented food (Lahtinen et al., 2008).
Increases SYTO-9 intensity was another intriguing change in VBNC cells of R. solanacearum (Table 1). Because SYTO-9 selectively binds to DNA, the increase of SYTO-9 intensity in the VBNC state could be a consequence of increased DNA content, possibly due to genome multiplication. It is not clear whether the entire bacterial genome was multiplicated or a particular region in the genome was amplified in VBNC R. solanacearum. Further investigation of the increased DNA content by comparative genome hybridization is necessary to compare bacterial genomes in culturable and VBNC cells (Mergaert et al., 2006). Genome duplication or multiplication in Lactococcus lactis strains (Michelsen et al., 2010) and in human pathogenic Neisseria spp. (Tobiason and Seifert, 2010) has been described, although the cases were not related with VBNC. Endoreduplication of the genome has been also reported in legume symbiotic bacteroids under the control of symbiotic plant cells (Mergaert et al., 2006).
Gene expression investigated by proteomic approaches and RT-PCR was a good indicator of the viable state of the unculturable R. solanacearum. One of the VBNC-induced proteins was Dps, which is involved in DNA protection during starvation or oxidative stress (Grant et al., 1998). Dps in R. solanacearum also plays a role in host plant colonization and bacterial wilt (Colburn-Clifford et al., 2010), in addition to oxidative stress tolerance and stationary phase survival. Bacterial cells in liquid microcosm with copper sulfate might be confronted with both starvation and oxidative stress. Therefore, it is likely that Dps induction in the VBNC state is correlated with increased H2O2 production and DNA content. Since expression of catalase/peroxidase in R. solanacearum was suppressed (Table 3), H2O2 may steadily accumulate in the VBNC state. Some of the physiological and morphological changes shown in this study, such as cell aggregation with extracellular material and Dps induction, could be consequences of copper-induced H2O2 accumulation. Several studies have suggested that H2O2 might play a central role in inducing the VBNC state in a variety of bacteria (Imazaki and Nakaho, 2009; Kong et al., 2004; Mizunoe et al., 1999; Oliver, 2010). Since amendment with 150 μM H2O2 in bacterial growth media suppresses growth of R. solanacearum (data not shown), the amount of H2O2 (152.8 μM) accumulated after 24 h in the VBNC state may be enough to suppress colony formation. This speculation is coincident with the idea that H2O2 may play a central role in inducing the VBNC state in various bacteria (Mizunoe et al., 1999). While VBNC cells contained a high concentration of H2O2, treatment of SL341 cell suspension (approximate density of ca. 108 cells/ml) with H2O2 did not cause bacterial unculturability. This is probably because of the indigenous catalase activity in R. solanacearum. In fact, there are at least 3 different genes encoding catalase/peroxidase in the R. solanacearum GMI 1000 genome (Salanoubat et al., 2002). Thus, H2O2 applied exogenously might be rapidly eliminated by the catalase activity of the cell suspension before inducing any VBNC-related phenotypes in SL341 cells.
The VBNC state induced by copper in liquid microcosm is somewhat artificial and thus the VBNC state in natural watercourses or soils may differ from the phenotypes described in this study, such as reduction of ribosome content, gene expression profiles, ultrastructure, and cell aggregation. It will be important to investigate the VBNC state of R. solanacearum in natural environments, although it will not be easy to distinguish VBNC cells, dead cells, and culturable cells in situ. Nonetheless, we conclude that the copper-induced VBNC state of R. solanacearum might be distinct from other bacterial physiological states. The VBNC state is accompanied by morphological and physiological changes with accumulation of hydrogen peroxide.
Acknowledgments
This research was supported by Mid-career Researcher Program through the National Research Foundation of Korea (NRF) grant funded by the Ministry of Education, Science and Technology (NRF-2012R1A2A2A01045039), by a grant from the Next-Generation BioGreen 21 Program (No. PJ0082012013), Rural Development Administration and by a grant (Project No. 609002-5) from the Screening Center for Disease Resistant Vegetable Crops of TDPAF funded by MIFAFF, Republic of Korea.
References
- Alexander E, Pham D, Steck TR. The viable-but-nonculturable conditions is induced by copper in Agrobacterium tumefaciens and Rhizobium leguminosarum. Appl Environ Microbiol. 1999;65:3754–3756. doi: 10.1128/aem.65.8.3754-3756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almirón M, Link A, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- Álvarez B, López MM, Biosca EG. Survival strategies and pathogenicity of Ralstonia solanacearum phylotype II subjected to prolonged starvation in environmental water microcosms. Microbiology. 2008;154:3590–3598. doi: 10.1099/mic.0.2008/019448-0. [DOI] [PubMed] [Google Scholar]
- Blat Y, Eisenbach M. Tar-dependent and -independent pattern formation by Salmonella typhimurium. J Bacteriol. 1995;177:1683–1691. doi: 10.1128/jb.177.7.1683-1691.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher CA, Barberis P, Trigalet AP, Demery DA. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J Gen Microbiol. 1985;131:2449–2457. [Google Scholar]
- Boulos L, Prévost M, Barbeau B, Coallier J, Desjardins R. LIVE/DEAD®Bac LightTM: application of a new rapid staining method for direct enumeration of viable ad total bacteria in drinking water. J Microbiol Meth. 1999;37:77–86. doi: 10.1016/s0167-7012(99)00048-2. [DOI] [PubMed] [Google Scholar]
- Brumbley SM, Denny TP. Cloning of wild-type Pseudomonas solanacearum phcA, a gene that when mutated alters expression of multiple traits that contribute to virulence. J Bacteriol. 1990;172:5677–5685. doi: 10.1128/jb.172.10.5677-5685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budrene EO, Berg HC. Complex patterns formed by motile cells of Escherichia coli. Nature. 1991;349:630–633. doi: 10.1038/349630a0. [DOI] [PubMed] [Google Scholar]
- Caruso P, Palomo JL, Bertolini E, Álvarez B, López MM, Biosca EG. Seasonal variation of Ralstonia solanacearum biovar 2 populations in a Spanish river; recovery of stressed cells at low temperature. Appl Environ Microbiol. 2005;71:140–148. doi: 10.1128/AEM.71.1.140-148.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colburn-Clifford JM, Scherf JM, Allen C. Ralstonia solanacearum Dps contributes to oxidative stress tolerance and to colonization of and virulence on tomato plants. Appl Environ Microbiol. 2010;76:7392–7399. doi: 10.1128/AEM.01742-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho TA. Introduction and prospectus on the survival of R solanacearum. In: Allen C, Prior P, Hayward AC, editors. Bacterial wilt disease and the Ralstonia solanacearum species complex. APS press; St. Paul, MN: 2005. pp. 29–38. [Google Scholar]
- Ghezzi JI, Steck JM. Induction of the viable but non-culturable condition in Xanthomonas campestris pv. campestris in liquid microcosms and sterile soil. FEMS Microbiol Ecol. 1999;30:203–208. doi: 10.1111/j.1574-6941.1999.tb00648.x. [DOI] [PubMed] [Google Scholar]
- Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nature Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- Grey B, Steck TR. The viable but nonculturable state of Ralstonia solanacearum may be involved in long-term survival and plant infection. Appl Environ Microbiol. 2001;67:3866–3872. doi: 10.1128/AEM.67.9.3866-3872.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward AC. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol. 1991;29:65–87. doi: 10.1146/annurev.py.29.090191.000433. [DOI] [PubMed] [Google Scholar]
- Holmgren GGS, Meyer M, Chaney R, Daniels R. Cadmium, lead, zinc, copper, and nickel in agricultural soils of the United States of America. J Environ Qual. 1993;22:335–348. [Google Scholar]
- Imazaki I, Nakaho K. Temperature-upshift-mediated revival from the sodium-pyruvate-recoverable viable but non-culturable state induced by low temperature in Ralstonia solanacearum: linear regression analysis. J Gen Plant Pathol. 2009;75:213–226. [Google Scholar]
- Jeong Y, Kim J, Kang Y, Lee S, Hwang I. Genetic diversity and distribution of Korean isolates of Ralstonia solanacearum. Plant Dis. 2007;91:1277–1287. doi: 10.1094/PDIS-91-10-1277. [DOI] [PubMed] [Google Scholar]
- Kell D, Kaprelyants A, Weichart D, Harwood C, Barer M. Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie Leeuwenhoek. 1998;73:169–187. doi: 10.1023/a:1000664013047. [DOI] [PubMed] [Google Scholar]
- Kelman A. The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium medium. Phytopathology. 1954;44:693–695. [Google Scholar]
- Kelman A. Survival of Pseudomonas solanacearum in water. Phytopathology. 1956;46:16–17. [Google Scholar]
- Kim ST, Kim SG, Kang YH, Wang Y, Kim J-Y, Yi N, Kim J-K, Rakwal R, Koh J-K, Kang KY. Proteomics analysis of rice lesion mimic mutant (spl1) reveals tightly localized probenazole-induced protein (PBZ1) in cells undergoing programmed cell death. J Proteome Res. 2008;7:1750–1760. doi: 10.1021/pr700878t. [DOI] [PubMed] [Google Scholar]
- Kim YH, Kim KS, Riggs RD. Initial subcellular responses of susceptible and resistant soybeans infected with the soybean cyst nematode. Plant Pathol J. 2012;28:401–408. [Google Scholar]
- Kogure K, Simidu U, Taga N. A tentative direct microscopic method for counting living marine bacteria. Can J Microbiol. 1979;25:415–420. doi: 10.1139/m79-063. [DOI] [PubMed] [Google Scholar]
- Kong I-S, Bates TC, Hülsmann A, Hassan H, Oliver JD. Role of catalase and oxyR in the viable but nonculturable state of Vibrio vulnificus. FEMS Microbiol Ecol. 2004;50:133–142. doi: 10.1016/j.femsec.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Lahtinen SJ, Ahokoski H, Reinikainen JP, Gueimonde M, Nurmi J, Ouwehand AC, Salminen SJ. Degradation of 16S rRNA and attributes of viability of viable but nonculturable probiotic bacteria. Lett Appl Microbiol. 2008;46:693–698. doi: 10.1111/j.1472-765X.2008.02374.x. [DOI] [PubMed] [Google Scholar]
- Lee YH, Choi CW, Kim SH, Yun JG, Chang SW, Kim YS, Hong JK. Chemical pesticides and plant essential oils for disease control of tomato bacterial wilt. Plant Pathol J. 2012;28:32–39. [Google Scholar]
- Loreto F, Velikova V. Isoprene produced by leaves protects the photosynthetic apparatus against ozone damage, quences ozone products, and reduces lipid peroxidation of cellular membranes. Plant Physiol. 2001;127:1781–1787. [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P, Uchiumi T, Alunni B, Evanno G, Cheron A, Catrice O, Mausset AE, Barloy-Hubler F, Galibert F, Kondorosi A, Kondorosi E. Eukaryotic control of bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. USA. 2006;103:5230–5235. doi: 10.1073/pnas.0600912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelsen O, Hansen FG, Albrechtsen B, Jensen PR. The MG1363 and IL1403 laboratory strains of Lactococcus lactis and several dairy strains are diploid. J Bacteriol. 2010;192:1058–1065. doi: 10.1128/JB.00900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizunoe Y, Wai SN, Takade A, Yoshida S. Restoration of culturability of starvation-stressed Escherichia coli O157 cells by using H2O2-degrading compounds. Arch Microbiol. 1999;172:63–67. doi: 10.1007/s002030050741. [DOI] [PubMed] [Google Scholar]
- Oliver JD. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev. 2010;34:415–425. doi: 10.1111/j.1574-6976.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- Ordax M, Marco-Noales E, López MM, Biosca EG. Survival strategy of Erwinia amylovora against copper: induction of the viable-but-nonculturable state. Appl Environ Microbiol. 2006;72:3482–3488. doi: 10.1128/AEM.72.5.3482-3488.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter M, Müller H, Reinecke F, Wieczorek R, Fricke F, Bowien B, Friedrich B, Steinbüchel A. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology. 2004;150:2301–2311. doi: 10.1099/mic.0.26970-0. [DOI] [PubMed] [Google Scholar]
- Rodriguez GG, Phipps D, Ishiguro K, Ridgway HF. Use of a fluorescent redox probe for direct visualization of actively respiring bacteria. Appl Environ Microbiol. 1992;58:1801–1808. doi: 10.1128/aem.58.6.1801-1808.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddler GS. Management of bacterial wilt disease. In: Allen C, Prior P, Hayward AC, editors. Bacterial wilt disease and the Ralstonia solanacearum species complex. APS press; St. Paul, MN: 2005. pp. 121–132. [Google Scholar]
- Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, Arlat M, Billault A, Brottier P, Camus JC, Cattolico L, Chandler M, Choisne N, Claudel-Renard C, Cunnac S, Demange N, Gaspin C, Lavie M, Moisan A, Robert C, Saurin W, Schiex T, Siguier P, Thébault P, Whalen M, Wincker P, Levy M, Weissenbach J, Boucher CA. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature. 2002;415:497–502. doi: 10.1038/415497a. [DOI] [PubMed] [Google Scholar]
- Saumaa S, Tover A, Tark M, Tegova R, Kivisaar M. Oxidative DNA damage defense systems in avoidance of stationary-phase mutagenesis in Pseudomonas putida. J Bacteriol. 2007;189:5504–5514. doi: 10.1128/JB.00518-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad NW, Jones JB, Chun W. Laboratory guide for identification of plant pathogenic bacteria. 3rd ed. APS press; St. Paul, MN: 2001. [Google Scholar]
- Shapiro JA. Bacteria as multicellular organisms. Sci Am. 1988;258:62–69. [Google Scholar]
- Tobiason DM, Seifert HS. Genomic content of Neisseria species. J Bacteriol. 2010;192:2160–2168. doi: 10.1128/JB.01593-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elsas JD, Kastelein P, de Vries PM, van Overbeek LS. Effects of ecological factors on the survival and physiology of Ralstonia solanacearum bv. 2 in irrigation water. Can J Microbiol. 2001;47:842–854. doi: 10.1139/w01-084. [DOI] [PubMed] [Google Scholar]
- van Elsas JD, Kastelein P, van Bekkum P, van der Wolf JM, de Vries PM, van Overbeek LS. Survival of Ralstonia solanacearum biovar 2, the causative agent of potato brown rot, in field and microcosm soils in temperate climates. Phytopathology. 2000;90:1358–1366. doi: 10.1094/PHYTO.2000.90.12.1358. [DOI] [PubMed] [Google Scholar]
- Xu H, Roberts N, Singleton FL, Attwell RW, Grimes DJ, Colwell RR. Survival and viability of nonculturable Escherichia coli and Vibrio cholera in the estuarine and marine environment. Microb Ecol. 1982;8:313–323. doi: 10.1007/BF02010671. [DOI] [PubMed] [Google Scholar]
- Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi YY. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: Proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]