Abstract
Among imported plants, seeds are the items that have many latent pathogens and are difficult to inspect. Also, they are the import and export items whose market is expected to expand. The biggest problem with seeds is viruses. Prune dwarf virus (PDV) is the virus that is commonly inspected in Prunus cerasifera, P. persica, P. armeniaca, P. mandshurica, P. cerasus, P. avium or P. serotina seeds. In this study, two RT-PCR primer sets, which can promptly and specifically diagnose plant quarantine seed-transmitted PDV, were developed; and nested PCR primers, where products amplify 739 and 673 nucleotides (nt), and an nested PCR-product, 305 nt, can be obtained as these products are amplified again, were developed. Also, a modified-positive control plasmid was developed, where the restriction enzyme XhoI, which can identify the contamination of samples from the control, was inserted. The method developed in this study has detected PDV in 18 cases since 2007, and is expected to continuously contribute to the plant quarantine in Korea.
Keywords: nested PCR, Prune dwarf virus, quarantine, RT-PCR, seed-transmitted
With the recent trade liberalization, various plants have been imported into Korea, and accordingly, the possibility of the introduction of plant pathogens that are not distributed in Korea has increased (Lee et al., 2013a). Among imported plants, seeds are the items that have many latent pathogens and are difficult to inspect (Min, 2009). Also, the domestic import and export of seeds are expected to increase depending on the global market expansion in the future (Shin, 2009). If seed-transmitted pathogens are introduced into Korea, they could extensively infect healthy plants during the distribution and cultivation processes, and bring about tremendous economic loss (Lee et al., 2013a). The biggest problem with seeds is viruses. In Korea, viruses with high quarantine risk are managed by classifying them into ‘prohibited,’ ‘controlled,’ ‘regulated non-quarantine,’ ‘non-quarantine’ and ‘provisionally regulated,’ based on risk assessment (www.qia.go.kr). PDV is a plant pathogenic virus that belongs to Bromoviridae Ilarvirus. In Korea, it is designated as ‘controlled’ and inspected in Prunus cerasifera, P. persica, P. armeniaca, P. mandshurica, P. avium or P. serotina seeds (including P. cerasus seeds), which are imported into the country (Animal, Plant and Fisheries Quarantine and Inspection Agency, 2013).
For a long time, the inspection of plant quarantine viruses has been performed using enzyme-linked immunosorbent assay (Stein, 1979), but this method has the disadvantages of false positive reaction and low sensitivity (Caruso et al., 2003; Priou et al., 2006). Therefore, RT-PCR methods and nested PCR methods, which are simpler and highly sensitive, have recently been studied and established in Korea (Kim et al., 2000; Lee et al., 2011a; Lee et al., 2011b; Park and Kim, 2004). In other countries, the genome variability (Fonseca et al., 2005) and coat protein expression (Raquel et al., 2008) of PDV during the infection of crops were studied; and for the diagnosis field, the duplex RT-PCR method, including the detection of PDV (Massart et al., 2008), and the multiplex one-step RT-PCR method for eight stone fruit viruses (Peiró et al., 2011) were studied. In Korea, considering the particularity of seed samples, studies using the RT-PCR and nested PCR inspection methods, which have high detection sensitivity, have been conducted, along with the use of a single condition inspection system that meets the demands of quarantine sites, inspection method dualization, alternative primer securement, and the development of a genetically modified-positive control (Lee et al., 2013a; Lee et al., 2013b; Lee et al., 2013c). However, a PDV detection method that meets the demands of domestic quarantine sites has yet to be reported. Therefore, in this study, RT-PCR, nested PCR, and a genetically modified-positive control, which meet the inspection conditions of the domestic quarantine sites of quarantine seed-transmitted PDV, were developed, and the results of the inspection, which had been performed from 2007 to 2013 using the PCR module developed in this study, were reported.
As for the PDV, which is the target for the development of the RT-PCR and nested PCR inspection methods, the leaf tissue of infected plums was imported from England (ADGEN, England) through the approval of prohibited goods import by the Animal and Plant Quarantine Agency. The cDNAs of the Alfalfa mosaic virus (AMV), Tobacco streak virus (TSV), Cucumber mosaic virus (CMV), Arabis mosaic virus (ArMV), Cherry leaf roll virus (CLRV) and Tomato ringspot virus (ToRSV), which were used in this experiment, were purchased from domestic enterprises (Bione, Korea and Plutos, Korea), and all the samples were stored in a deep freezer at −70°C.
For the PDV primer design, from the National Center for Biotechnology Information, 36 kinds of isolates, including PDV RNA-3 (NC_008038) (about 2.13 kb), and 13 kinds of Ilarvirus, including the Apple mosaic virus RNA-3 (NC_0035480), were used (Supplementary Table 1). The obtained sequences of the viruses were aligned using BioEdit (Hall, 1999), and PDV species-specific sequences were searched at the 51–59°C annealing condition using DNAMAN software package version 6.0 (Pan et al., 2000; Lee et al., 2013c). The searched primers were 21 forward primers and 11 reverse primers (total of 33 primers), which were 18–24 nt, respectively (Supplementary Table 2). Using these primers, 167 primer sets that are capable of PCR amplification were combined (Supplementary Table 3).
The RNA extraction from the samples, RT-PCR and nested PCR were carried out using the same methods as those described in previous papers (Lee et al., 2013a; Lee et al., 2013b; Lee et al., 2013c). After the reaction was completed, the electrophoresis of the PCR products was performed for 100 minutes using 1.2% agarose gel (150 ml) by adding 4 μl of TopRed nucleic acid gel stain (Biopure, United Kingdom). Then it was observed under UV light.
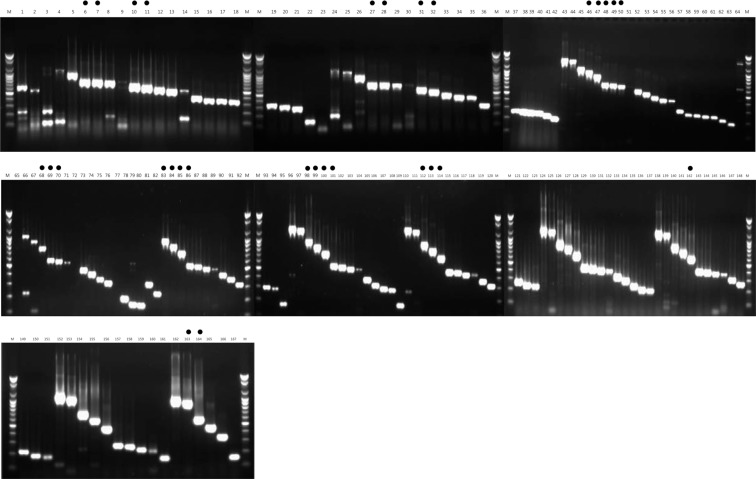
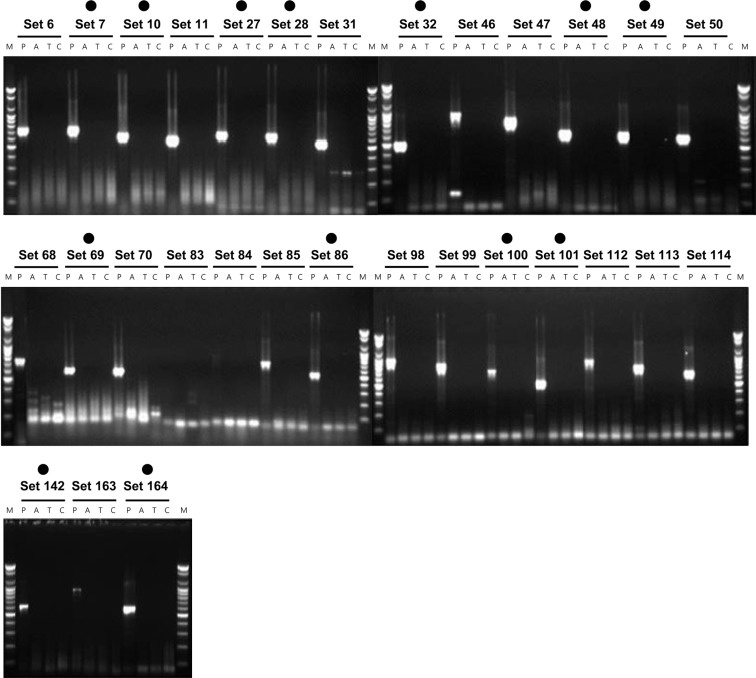
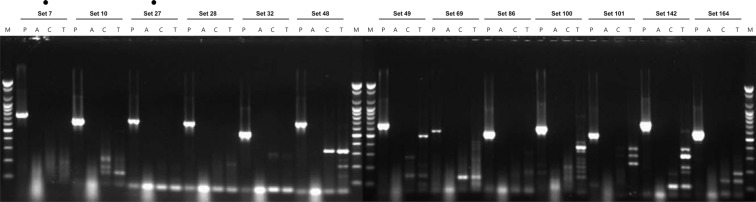
In the first selection, one-step RT-PCR for the primer sets of 167 combinations was performed using the total RNA, including the PDV (i.e., the target virus) as the template. Among these, 30 sets (sets 6, 7, 10, 27, 28, 32, 48, 49, 69, 86, 100, 101, 142 and 164) were selected, which had high reaction intensity and did not show a non-specific reaction (non-specific reaction sets 1, 2, 3, 4, 8, 9, 14, 24, 25, 26, 30, 43, 44, 66, 79, 96, 110, 138, 139, 140, 141, 154 and 155) that interfered with the diagnosis (Fig. 1). In the second selection, the non-specificity to AMV, TSV and CMV, which belong to Bromoviridae, was examined for the selected 30 primer sets. The 17 primer sets, which formed bands for the three kinds of viruses (AMV, TSV and CMV), were excluded, and sets 7, 10, 27, 28, 32, 48, 49, 69, 86, 100, 101, 142 and 164 (total of 13 sets) were selected (Fig. 2). In the third selection, the non-specificity to the three kinds of viruses (ArMV, CLRV and ToRSV), which can infect a host that can be infected with PDV, was examined. Among the 13 primer sets, only two primer set (sets 7 and 27), which did not form non-specific bands, were selected (Fig. 3). Thus, primer set 7 (PDV_F20/C10 and 739 nt) and primer set 27 (PDV_F10/C20 and 673 nt), which can specifically detect PDV, were finally selected (Table 1). Nested PCR primers, which can again amplify the product of the selected primer sets, were also selected. The selected primers were PDV_F60/PDV_C80, and the end-product was 305 bp (data not shown).
Fig. 1.
First selection of species-specific primers for the PDV detection.
Lane M, 100 bp step DNA Ladder maker (Genepia, Korea); lanes 1–167, PDV-specific primer sets 1–167; dot, selected primer set.
Fig. 2.
Second selection of species-specific primers for the PDV detection.
Lane M, 100 bp step DNA Ladder maker (Genepia, Korea); lane P, PDV; lane A, AMV; lane T, TSV; lane C, CMV; dot, selected primer set.
Fig. 3.
Third selection of species-specific primers for the PDV detection.
Lane M, 100 bp step DNA Ladder maker (Genepia, Korea); lane P, PDV; lane A, ArMV; lane C, CLRV; lane T, ToRSV; dot, selected primer set.
Table 1.
Selected primer sets for RT-PCR and nested PCR of PDV
| Primer
|
Sequence | Length (bp) | Product size (bp) | ||
|---|---|---|---|---|---|
| Set | PCR | Name | |||
| 7 | RT | PDV-F20 | ACGATTGGTTAACTCACTTTGT | 22 | 739 |
| PDV-C10 | ATCCACTGACTATTYTATCCATYG | 24 | |||
| Nested | PDV-F60 | AGCTCGGAAGAATAATAATACTAC | 24 | 305 | |
| PDV-C80 | CGAAACCTTTAATGAGTCCGTAGA | 24 | |||
|
| |||||
| 27 | RT | PDV-F10 | GAAGCTTTTGGTGTAACGATTGGT | 24 | 673 |
| PDV-C20 | TCTTTGGCATCGAGTGTTGG | 20 | |||
| Nested | PDV-F60 | AGCTCGGAAGAATAATAATACTAC | 24 | 305 | |
| PDV-C80 | CGAAACCTTTAATGAGTCCGTAGA | 24 | |||
PDV is a ‘controlled’ quarantine virus, and is not distributed in Korea. Accordingly, there were difficulties in using a sample as a control, and thus, the control was produced using a plasmid. Using the pGEM®-T easy vector (Promega, USA), the cloning was performed using the PCR amplification product, which includes all the selected RT-PCR primer sets, as the insert DNA. Using a site-directed mutagenesis kit (Nelson and McClelland, 1992), CTCGAG, which is the sequence with which the restriction enzyme XhoI can react, was inserted into 305 nt, which is the nested PCR amplification product (Supplementary Fig. S1). If the nested PCR product is cut into two bands with the reaction with the restriction enzyme XhoI (Supplementary Fig. S2) or if the inserted six nucleotides are found during the sequencing, it would be judged that the product has been contaminated from the genetically modified-positive control developed in this study.
Using the PCR module (RT-PCR, nested PCR and modified-positive control plasmid) developed in this study, PDV was inspected in Prunus cerasifera, P. persica, P. armeniaca, P. mandshurica, P. cerasus and P. avium and P. serotina seeds, which were imported into Korea from 2007 to September 2013. According to the Disease and Insect Pest Information System (http://10.110.128.100), which is the intranet system of the Animal and Plant Quarantine Agency, PDV has been detected in a total of 18 cases since 2007, in the quarantine inspection for PDV over all the import routes, which included those for cargo (air, ship, railroad and land), mail and carry-on baggage. PDV was detected in Prunus persica seeds (13 cases), P. armeniaca seeds (two cases), P. cerasus seeds (one case), P. avium or P. serotina seeds (one case), and Prunus mandshurica seeds (one case). PDV was detected most frequently in 2009 (12 out of 18 cases).
The RT-PCR and nested PCR inspection methods and the genetically modified-positive control of the plant quarantine seed-transmitted PDV that was developed in this study will be continuously used for plant quarantine in a prompt and accurate manner, and are expected to contribute to the plant quarantine in the future.
Supplementary Material
References
- Animal, Plant and Fisheries Quarantine and Inspection Agency. List of Plant Quarantine Viruses in Korea Newly Revised in 2013. Res Plant Dis. 2013;19:67–75. [Google Scholar]
- Caruso P, Bertolini E, Cambra M, López MM. A new and sensitive co-operational polymerase chain reaction for rapid detection of Ralstonia solanacearum in water. J. Microbiol. Methods. 2003;55:257–272. doi: 10.1016/s0167-7012(03)00161-1. [DOI] [PubMed] [Google Scholar]
- Fonseca F, Neto JD, Martins V, Nolasco G. Genomic variability of Prune dwarf virus as affected by agricultural practice. Arch Virol. 2005;150:1607–1619. doi: 10.1007/s00705-005-0507-z. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Kim D, Hyun J, Hwang H, Lee S. RT-PCR detection of Citrus Tristeza Virus from early satsuma mandarin and Cheju Island. Plant Pathol J. 2000;16:48–51. [Google Scholar]
- Lee JS, Cho WK, Choi HS, Kim KH. RT-PCR detection of five quarantine plant RNA viruses belonging to Poty and Tospoviruses. Plant Pathol J. 2011b;27:291–296. [Google Scholar]
- Lee JS, Cho WK, Lee SH, Choi HS, Kim KH. Development of RT-PCR based method for detecting five non-reported quarantine plant viruses infecting the family Cucurbitaceae or Solanaceae. Plant Pathol J. 2011a;27:93–97. [Google Scholar]
- Lee S, Kang EH, Chu YM, Shin YG, Ahn TY. Development of PCR diagnosis system for plant quarantine seed-borne Wheat streak mosaic virus. Korean J Microbiol. 2013a;49:112–117. [Google Scholar]
- Lee S, Kang EH, Heo NY, Kim SM, Kim YJ, Shin YG. Detection of Carnation necrotic fleck vrus and Carnation ringspot virus using RT-PCR. Res Plant Dis. 2013b;19:1–9. [Google Scholar]
- Lee S, Kang EH, Shin YG, Lee SH. Development of RT-PCR and nested PCR for detection of four quarantine plant viruses belonging to Nepovirus. Res Plant Dis. 2013c;19:220–225. [Google Scholar]
- Massart S, Brostaux Y, Barbarossa L, César V, Cieslinska M, Dutrecq O, Fonseca F, Guillem R, Laviña A, Olmos A, Steyer S, Wetzel T, Kummert J, Jijakli MH. Inter-laboratory evaluation of a duplex RT-PCR method using crude extracts for the simultaneous detection of Prune dwarf virus and Prunus necrotic ringspot virus. Eur J Plant Pathol. 2008;122:539–547. [Google Scholar]
- Min JS. Import and export for seed quarantine. J Korea Soc Seed Sci Ind. 2009;6:8–13. [Google Scholar]
- Pan YB, Burner DM, Legendre BL. An assessment of the phylogenetic relationship among sugarcane and related taxa based on the nucleotide sequence of 5S rRNA intergenic spacers. Genetica. 2000;108:285–295. doi: 10.1023/a:1004191625603. [DOI] [PubMed] [Google Scholar]
- Park MR, Kim KH. RT-PCR detection of three non-reported fruit tree viruses useful for quarantine purpose in Korea. Plant Pathol J. 2004;20:147–154. [Google Scholar]
- Peiró A, Pallás V, Sánchez-Navarro JA. Simultaneous detection of eight viruses and two viroids affecting stone fruit trees by using a unique polyprobe. Eur J Plant Pathol. 2011;132:469–475. [Google Scholar]
- Priou S, Gutarra L, Aley P. An improved enrichment broth for the sensitive detection of Ralstonia solanacearum(biovars 1 and 2A) in soil using DAS-ELISA. Plant Pathol J. 2006;55:36–45. [Google Scholar]
- Raquel H, Lourenço T, Moita C, Oliveira MM. Expression of prune dwarf Ilarvirus coat protein sequences in Nicotiana benthamiana plants interferes with PDV systemic proliferation. Plant Biotechnol Rep. 2008;2:75–85. [Google Scholar]
- Shin YG. An advanced quarantine system for the inspection of seed-borne viruses in Korea. 2009. Ph. D. thesis. Kyungpook National University, Daegu, Gyeongsangbuk-do, Korea.
- Stein A, Loebensten G, Koenic R. Detection of Cucumber mosaic virus and Bean yellow mosaic virus in gladiolus by enzyme-linked immunosobent assay (ELISA) Plant Dis Rep. 1979;63:185–188. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.