Abstract
Genetic influences on anxiety disorders are well documented; however, the specific genes underlying these disorders remain largely unknown. To identify quantitative trait loci (QTL) for conditioned fear and open field behavior, we used an F2 intercross (n = 490) and a 34th-generation advanced intercross line (AIL) (n = 687) from the LG/J and SM/J inbred mouse strains. The F2 provided strong support for several QTL, but within wide chromosomal regions. The AIL yielded much narrower QTL, but the results were less statistically significant, despite the larger number of mice. Simultaneous analysis of the F2 and AIL provided strong support for QTL and within much narrower regions. We used a linear mixed-model approach, implemented in the program QTLRel, to correct for possible confounding due to familial relatedness. Because we recorded the full pedigree, we were able to empirically compare two ways of accounting for relatedness: using the pedigree to estimate kinship coefficients and using genetic marker estimates of “realized relatedness.” QTL mapping using the marker-based estimates yielded more support for QTL, but only when we excluded the chromosome being scanned from the marker-based relatedness estimates. We used a forward model selection procedure to assess evidence for multiple QTL on the same chromosome. Overall, we identified 12 significant loci for behaviors in the open field and 12 significant loci for conditioned fear behaviors. Our approach implements multiple advances to integrated analysis of F2 and AILs that provide both power and precision, while maintaining the advantages of using only two inbred strains to map QTL.
Keywords: conditioned fear, anxiety, complex traits, QTL mapping, advanced intercross line, genome-wide association study (GWAS), GWAS, mixed models, quantitative genetics, single nucleotide polymorphism (SNP), Multiparent Advanced Generation Inter-Cross (MAGIC), multiparental populations, MPP
ANXIETY disorders are among the most prevalent psychiatric disorders in the world; in the United States, they affect the lives of ∼18% of the adult population (Demyttenaere et al. 2004; Kessler et al. 2005a,b). Many of these debilitating illnesses can be characterized by exaggerations of normal and adaptive emotional response to fearful or stressful events (Mahan and Ressler 2012). Twin and family studies support a genetic basis for anxiety disorders, but attempts to identify the underlying genetic substrates have been disappointing—to date, genome-wide association studies (GWAS) have not reliably replicated candidate genes associated with anxiety disorders (Hettema et al. 2011). As a result, we have little knowledge of the specific genes that underlie these disorders. Genetic loci relevant to these disorders may be difficult to map via GWAS because anxiety disorders are only modestly heritable and, like many psychiatric conditions, are expected to be highly polygenic—that is, modulated by a large number of genetic factors with individually small effects (Sullivan et al. 2012). Therefore, genetic contributions to anxiety are likely to be difficult to distinguish from correlations that occur by chance alone (Flint 2011; Parker and Palmer 2011; Flint and Eskin 2012).
While the full spectrum of any human psychiatric disorder can never be fully recapitulated in a single mouse model, there is substantial behavioral, genetic, and neuroanatomical conservation between humans and mice. When broken down into individual components, many of the symptoms of anxiety disorders can be modeled in mice. Thus, translational mouse models can provide a powerful strategy for understanding the genetic and biological underpinnings of the acquisition of fear, as well as the etiologic processes related to anxiety (Kalueff et al. 2007; Flint and Shifman 2008; Hovatta and Barlow 2008). Animal models provide similarly strong models for comorbidity of traits, like evidence for a shared genetic substrate in anxiety and fear. For example, selective breeding paradigms in mice and rats have shown that selection of anxiety-like behavior also selects for differences in fear and vice versa (Ponder et al. 2007a; López-Aumatell et al. 2009).
Reverse genetic approaches using genetically modified mice have been important for testing hypotheses about specific genes relevant to fear and anxiety, but they have tended to focus on the “usual suspects” underlying anxiety disorders. Alternatively, forward genetic approaches in mice have been developed to measure phenotypes and identify the underlying sources of standing genetic variability in these phenotypes without prior hypotheses. However, forward genetic approaches have been less successful at identifying relevant genes. This may be because forward genetic studies in mice have traditionally used recombinant inbred (RI) lines, backcrosses (BC), or F2 intercrosses to identify quantitative trait loci (QTL)—in these experimental crosses, we can have high statistical power to detect genetic loci, but poor mapping resolution due to limited recombination (Cheng et al. 2010; Flint 2011; Parker and Palmer 2011).
To better pinpoint candidate genes and genetic loci that might influence anxiety, we mapped QTL in a combined F2 intercross and an F34 advanced intercross line (AIL). An AIL is created by successive generations of pseudorandom mating after the F2 generation. Each additional generation leads to the accumulation of new recombinations, which allows for more precise mapping due to a breakdown in linkage disequilibrium. We show that our analysis not only yields strong support for several QTL in anxiety-related phenotypes, but also highlights a narrower set of candidate genes than previous studies of these phenotypes.
AILs have been employed in several previous studies to successfully map QTL for complex traits in mice, including locomotor activity (Cheng et al. 2010), muscle weight (Lionikas et al. 2010), red blood cell characteristics (Bartnikas et al. 2012), body weight (Parker et al. 2011), methamphetamine sensitivity (Parker et al. 2012a), prepulse inhibition (Samocha et al. 2010), and the conditioned fear phenotypes studied in this article (Parker et al. 2012b). This study makes several key contributions over previous work in this area. First, we demonstrate that the combination of an F2 intercross and a 34th-generation AIL allows us to map more QTL for conditioned fear and anxiety-related phenotypes and at a greater precision than our earlier study of an 8th-generation AIL (Parker et al. 2012b). Second, unlike previous studies, we use SNP data to account for the varying levels of genetic sharing that may confound detection of QTL. This is particularly relevant because assessing evidence for QTL in an AIL population is more challenging than in traditional designs such as F2 intercrosses because varying levels of genetic sharing among individuals in the AIL can confound tests for association (Cheng et al. 2010). Third, we demonstrate the benefits of using marker data in place of a pedigree to infer familial relationships in an AIL. We find that the linear mixed model with marker-based relatedness estimates yields greater support for QTL compared to using the pedigree to estimate genetic sharing, a finding that is consistent with a recent comparison of these approaches in simulated populations (Cheng and Palmer 2013). Finally, we show that a simple permutation test that assumes independence of the samples—that is, the permutation test ignores the varying levels of genetic sharing—provides an adequate way to assess significance of QTL in our AIL population.
Materials and Methods
Animals and housing
Subjects consisted of 487 F2 mice (249 males and 241 females) derived from a cross between LG/J and SM/J inbred strains obtained from The Jackson Laboratories (Bar Harbor, ME) and 687 F34 LG/J × SM/J AIL mice (353 males and 334 females) derived from F33 breeders obtained from the laboratory of James Cheverud (Washington University, St. Louis). Three mice were later removed from the F2 cohort because a high proportion of their markers were not reliably genotyped. The colony was maintained on a 12:12-hr light:dark cycle with lights on at 0630 hr. Mice were housed in clear acrylic cages with corn-cob bedding in same-sex groups of 2–5 mice with food and water available ad libitum. The full pedigree of all AILs was recorded so that the ancestry of each mouse could be traced back to the inbred founders.
To produce F34 mice from the F33 breeders, we mated the F33 mice such that no mating pairs shared a common grandparent. (In subsequent generations, which are not described in this article, we selected pairs of mice for breeding that produced the smallest possible inbreeding coefficients in the offspring. This was achieved using R code written by Andrew Skol, which we have made available at http://github.com/pcarbo/breedail.)
Behavioral testing
Behavioral testing in all mice was always conducted during the light phase, between 0800 and 1700 hr. We waited for the mice to acclimatize to the testing room for at least 30 min before beginning the tests. Mice were ∼2–3 months of age on the first day of behavioral testing (F2 range was 53–71 days; AIL range was 50–76 days). All mice went through an identical testing sequence: first, we measured open field (OF) behavior as part of a locomotor testing paradigm (Bryant et al. 2012); 1 week later, we began the conditioned fear (CF) paradigm. All experiments were performed in accordance with the National Institutes of Health guidelines for care and use of laboratory animals. Experimental procedures were approved by the University of Chicago’s Institutional Animal Care and Use Committee.
OF
Our procedures for OF testing have been explained in detail in previous publications (Palmer et al. 2005; Sokoloff et al. 2011). Briefly, after 30 min of acclimation to the testing room, mice were removed from the home cage and placed into individual holding cages for 5 min, and then they were weighed individually, injected i.p. with physiological saline (0.01 ml/g body weight), and immediately placed in the center of the OF (AccuScan Instruments, Columbus, OH). Each OF was housed inside a sound-attenuating environmental chamber (AccuScan Instruments) with overhead lighting providing illumination (∼80 lux) and a fan in the rear wall providing ventilation. Mice were tested in the OF arena (40 × 40 × 30 cm) for 30 min during the locomotor test. The first 10 min were used to measure OF behavior. We measured three phenotypes (Versamax, AccuScan Instruments): (1) distance traveled (centimeters) in the periphery (width: 10 cm), (2) distance traveled (centimeters) in the center (20 × 20 cm), and (3) proportion of time spent in the center of the arena. We mapped QTL for these three phenotypes. After testing, mice were returned to their home cage, and the field was cleaned with 10% isopropanol before the next mouse was tested. At the end of testing, mice were returned to the vivarium. Distance traveled from this same session was also analyzed in a previous article (Cheng et al. 2010).
CF
CF procedures were identical to those described previously (Ponder et al. 2007a). Briefly, mice were tested in standard CF chambers (29 × 19 × 25 cm) housed within sound-attenuating chambers (Med Associates, St. Albans, VT). Lights in each chamber provided dim illumination (∼3 lux), and fans provided a low level of masking background noise. Chambers were cleaned with 10% isopropanol between animals. Behavior was digitally recorded by a computer and subsequently analyzed with FreezeFrame software (Actimetrics, Evanston, IL).
Testing for CF consisted of a 5-min test that occurred three times over three consecutive days. After habituating to the procedure room for 30 min in their home cages, mice were transferred to the CF chambers in individual holding cages. On day 1, baseline freezing (“pretraining freezing”) was measured beginning 30 sec after mice were placed in the test chambers and ending 150 sec later. After the pretraining period, mice were exposed twice to the conditioned stimulus (CS), a 30-sec tone (85 dB, 3 kHz) that coterminated with the unconditioned stimulus (US), a 2-sec, 0.5-mA foot shock delivered through the stainless steel floor grid. After each CS–US pairing, there was a 30-sec period in which no stimuli were delivered to the subject.
Test day 2 began exactly 24 hr after the start of test day 1. The testing environment was identical to that of day 1, except that neither tones nor shocks were presented. Proportion of time freezing in response to the test chamber (“% freezing to context”) was measured over the same time period as pretraining freezing (30–180 sec). We chose this time period to permit immediate comparison to the pretraining freezing scores on day 1 and to avoid measuring freezing behavior during the latter part of the trial in which the mice might have anticipated shocks based on tests from previous days.
Test day 3 began exactly 24 hr after the start of test day 2. On day 3, the context was altered in several ways: (1) a different experimenter conducted the testing and wore a different style of gloves; (2) the transfer cages had no bedding; (3) the metal shock grid, chamber door, and one wall were covered with hard white plastic; (4) yellow film was placed over the chamber lights; (5) chambers and plastic surfaces were cleaned with 0.1% acetic acid solution; and (6) the vent fan was partially obstructed to alter background noise. On day 3, the tone was presented at the same times as on day 1, but there was no shock. We measured “freezing to cue,” which was defined as the average percentage of time spent freezing during the two 30-sec tone presentations (180–210 sec and 240–270 sec).
Genotyping
DNA from the F2 mice was extracted by LGC Genomics (Hoddesdon, Hertfordshire, UK; formerly KBiosciences). F2 genotypes were called using KASPar, a fluorescence-based PCR assay (LGC Genomics), at 162 evenly spaced markers on autosomal and X chromosomes. These markers are a subset of the 1638 SNPs suggested by Petkov et al. (2004) for QTL mapping in mouse strains. AIL genotypes at 4601 SNPs on autosomal and X chromosomes were ascertained using an Illumina Infinium Platform (iSelect) custom genotyping array (http://www.illumina.com), as described previously (Cheng et al. 2010). To avoid having to impute unascertained genotypes in the AIL mice, we discarded 7 of the 162 SNPs genotyped in the F2 cohort that were not genotyped in the F34 samples. Of the final set of 4601 candidate SNPs, 4535 (98.6%) correspond to SNPs in the dbSNP reference database (Sherry et al. 2001). A list of SNPs used in the present study is available at the Mouse Phenome Database (http://phenome.jax.org) under project name “Chicago1” (http://phenome.jax.org/db/q?rtn=projects/projdet&reqprojid=316). The Chicago1 data set includes ∼8200 SNPs; of these, 4601 are polymorphic between LG/J and SM/J mice. All SNP identifiers and locations of the SNPs reported in this article are based on release 37 (July 2007) of the NCBI Mouse Genome Assembly.
Average SNP allele frequencies in the F2 and F34 cohorts were 0.50 and 0.51, respectively, indicating little unintended selection or genetic drift. As expected, we observed greater variation in allele frequencies in the F34 cross; SNP allele frequencies in F2 mice ranged from 0.45 to 0.54, whereas allele frequencies in F34 mice ranged from 0.12 to 0.90. (See Cheng et al. 2010 for further discussion of genetic drift in the F34 AIL.)
Genotypes called with lower confidence were removed and treated as missing. In the F2 mice, <1% of the genotypes were treated as missing. At most 9% of genotypes were missing for any single SNP and at most 26% in any F2 mouse (after removing the three mice with a large fraction of poor-quality genotype calls). In the F34 mice, 1% of the genotypes were treated as missing overall; at most 1.2% of genotypes were missing for any single SNP and at most 5% in any F34 mouse.
Genotype imputation
To analyze the combined data, we estimated the probabilities of missing genotypes and the genotypes of SNPs that were called in the F34 mice and not called in the F2 mice. Probabilities of missing or unavailable genotypes in the F2 and F34 crosses were estimated based on correlation patterns with available genotypes, using recombination models for autosomal chromosomes in advanced intercrosses (Darvasi and Soller 1995). Genotype probabilities were calculated separately in F2 and F34 crosses as they exhibited different patterns of recombination. Even though only a small fraction of the full set of SNPs were genotyped in the F2 mice (155 of 4601), this small panel of SNPs was mostly sufficient to accurately estimate genotypes at other SNPs, since F2 chromosomes experience little recombination, and most alleles occur at high frequencies. In the F2 mice, 85% of genotypes were imputed with high confidence (maximum genotype probability >0.9). Only a small fraction of SNPs were imputed less accurately, presumably because they were not in sufficient linkage disequilibrium with a SNP genotyped in the F2 mice; for 109 SNPs (2.4%), <20% of the genotypes in the F2 mice were estimated with high confidence (maximum genotype probability >0.9). Since we accounted for genotype uncertainty in the QTL mapping, the contribution of each sample to the association signal was appropriately adjusted to reflect the confidence of the genotype estimates. We used the Haley–Knott approximation (Broman and Sen 2009) to account for uncertainty in missing genotypes. Note that we did not perform interval mapping to assess support for QTL between markers.
Estimation of missing genotypes requires genetic distance estimates at all markers. We used the genetic distance estimates from the reference panel described in Cox et al. (2009). These estimates were retrieved from the Mouse Map Converter hosted at Jackson Laboratories (http://cgd.jax.org/mousemapconverter). We checked these genetic distances against intermarker distances estimated from our F2 genotypes. Estimates were obtained using the Lander–Green method (Lander and Green 1987), as implemented in R/qtl (Broman and Sen 2009). Intermarker distances on the X chromosome were estimated using females only. Comparison of the two intermarker genetic distance estimates on X and autosomal chromosomes (Supporting Information, Figure S1) shows that they are well correlated (r = 0.85), and they do not exhibit any bias—that is, neither estimate is consistently larger or smaller than the other.
QTL mapping
Linear mixed model for QTL mapping:
Individuals in an F2 cross are full sibs that share roughly the same amount of the genome, so it is common to map QTL using a simple linear regression approach that ignores familial relatedness. By contrast, genetic sharing can vary considerably in an AIL, so it is important to correct for possible confounding due to varied levels of relatedness (Abney et al. 2000; Cheng et al. 2010). We used the QTLRel mixed-model framework, which was developed for QTL mapping in AILs (Cheng et al. 2010, 2011). What makes QTLRel different from a standard linear regression is the inclusion of a “polygenic effect” that captures how correlations in the phenotype are explained by genome-wide genetic sharing. QTLRel models the phenotype as a linear combination of these variables: additive genotype (allele count), dominance genotype (0 = heterozygous, 1 = homozygous), additional covariates such as sex and coat color, the polygenic effect, and the residual. (Note that the X chromosome requires a separate treatment, as we explain below.) Fitting this model to the data involves estimating the covariance matrix of the polygenic effect, where n is the number of samples. While the general expression for this covariance matrix is derived in Abney et al. (2000) for analyzing quantitative traits in an inbred population, that article suggests using a simpler expression in which only the additive and dominance terms are retained. Following this suggestion, the covariance matrix entry corresponding to pairs of individuals is where is the conditional probability of identity state k for pair otherwise known as the “condensed coefficient of identity”; is the kinship coefficient for pair —i.e., the probability that a pair of randomly chosen alleles from individuals i and j at the same autosomal locus are identical by descent (IBD); and and are parameters to be fitted to the data.
QTLRel uses a two-step procedure to fit the mixed model to the data: first, the model parameters are estimated assuming that no markers have an effect on the phenotype (the null hypothesis); second, for a given marker, the additive and dominance effects are estimated while the other model components are fixed to their values obtained from the first step, up to a scaling factor that is also estimated in this step. This is similar to the strategy used in EMMAX (Kang et al. 2010). [More recently, efficient methods have been developed to integrate model fitting into a single step, which can sometimes lead to improved power to detect QTL (see Lippert et al. 2011; Zhou and Stephens 2012)]. Once the model-fitting steps are completed, QTLRel uses the parameter estimates to compute the log-likelihood-ratio test statistic. For each SNP, we reported support for a genotype–phenotype association using the (base 10) logarithm of the likelihood ratio, commonly called the LOD score.
Since F2 crosses are approximately equally related to each other, QTL mapping that does not account for familial relationships should yield similar results to those of QTLRel. To verify this, we compared our results in the F2 sample using QTLRel against results from a linear regression model that did not include the polygenic component, implemented in R/qtl (Broman and Sen 2009). Conversely, ignoring relatedness in the F34 and combined cohorts is expected to yield very different QTL mapping results (Cheng et al. 2010).
Pedigree- and marker-based estimates of genetic sharing:
To estimate the covariance matrix of the polygenic effect, we must supply matrices of identity coefficients (Abney et al. 2000; Cheng et al. 2010). Specifically, the additive and dominance variance components of this covariance matrix require estimates of the kinship coefficient and identity coefficient for each pair of individuals We derived estimates of identity coefficients in two different ways, using the pedigree, and using the marker data. Algorithms to compute identity coefficients from the pedigree are far too complex for large pedigrees such as the one used here, so we applied the scalable approximation developed by Cheng et al. (2010), building on Karigl (1981). Even this approximate algorithm was not sufficiently scalable for computation of coefficients for F2–F34 pairs, so for each of these pairs we assigned the identity coefficients to their expected values in an inbred cross, and
One advantage of using the markers to estimate relatedness was that it was much easier to compute identity coefficient estimates for all pairs of individuals. In keeping with other methods that use marker-based estimates of relatedness to correct for confounding due to population stratification or familial relationships (Yu et al. 2006; Kang et al. 2008; Lippert et al. 2011; Listgarten et al. 2012; Zhou et al. 2013), we included only the additive portion of the polygenic effect. The rationale is that dominance effects typically make a much smaller contribution to the variance of a complex trait. Therefore, we needed to calculate only kinship coefficients. At a single locus, the estimate of is simply equal to the number of alleles that share the same state between individuals (2 if both genotypes are homozygous and the same, 0 if both genotypes are homozygous and different, and 1 in all other cases). This is equivalent to the number of alleles IBD, since the mice are crosses of inbred founders. Note that other marker-based estimates of the kinship coefficients have been used, and these are based on different derivations of the polygenic covariance matrix. To account for uncertainty in missing genotypes at a given marker, we calculated the expected number of shared alleles.
The final marker-based kinship coefficient estimate was obtained by averaging over markers across the genome, excluding the X chromosome. To avoid “proximal contamination” when assessing evidence for a QTL at a given SNP, this SNP, and all nearby SNPs, should be excluded from estimation of the kinship coefficient (Listgarten et al. 2012; Cheng et al. 2013; Yang et al. 2014). We accomplished this by omitting markers on the same chromosome to estimate relatedness. Therefore, we fitted the mixed model to the data separately for each chromosome and used this mixed model only to quantify support for QTL on that chromosome.
Covariates:
In all analyses, we included four covariates in the regression model of the phenotype: age (in days) and three binary traits, sex (1 = male, 0 = female), albino (1 = white coat color, 0 = non-white), and agouti (1 = agouti coat color, 0 = nonagouti). We found that coat color confounded some of the phenotype measurements obtained from video tracking; if we did not account for the effect of coat color, for some phenotypes we obtained strong evidence for a QTL that mapped to a region overlapping the Tyr gene on chromosome 7, which is the gene variant for the albino trait (Jackson 1994, 1997). Some phenotype and coat color observations were not recorded for some mice; at most 75 samples in any one phenotype were excluded from QTL mapping due to missing phenotype or covariate values.
A larger proportion of F34 mice showed less freezing to context and freezing to cue than F2 mice. However, this difference disappeared if we excluded albino mice, reinforcing the importance of including coat color traits as covariates in the QTL mapping. We observed no systematic differences in distributions of other phenotypes from the F2 and F34 crosses.
Transformation of quantitative traits:
We mapped QTL for six phenotypes: the three measurements recorded in open field testing (distance traveled in the periphery, distance traveled in the center, and percentage of time in the center of the arena) and the three measurements from our conditioned fear tests (pretraining freezing, freezing to context, and freezing to cue). Four of these phenotypes (percentage of time in center, pretraining freezing, freezing to context, and freezing to cue) are proportions between 0 and 1. To obtain numbers on the real line, and to admit a normal model for these phenotypes, we transformed the proportions to the log-odds scale using the logit function, To avoid extreme values after the transformation, any proportions <0.01 or >0.99 were fixed to 0.01 and 0.99, respectively. Observed quantiles of the transformed phenotypes, separately in the F2 and F34 crosses, after removing linear effects of the covariates, closely matched expected quantiles under the normal distribution (results not shown), suggesting that the normal distribution was a good fit for these phenotypes.
Determining significance of genotype–phenotype associations:
To calculate significance thresholds for the LOD scores, we must first obtain the distribution of this test statistic under the null distribution. This distribution is commonly estimated by permuting the phenotype values relative to the genotypes (Broman and Sen 2009). However, this approach will not preserve the covariance structure in the samples that is due to unequal relatedness of the individuals and therefore may lead to inadequately stringent significance thresholds and inflated type 1 error rates (Abney et al. 2002; Zou et al. 2005; Aulchenko et al. 2007; Cheng et al. 2010; Cheng and Palmer 2013). A simple alternative to this approach would be to use a Bonferroni correction of the P-values calculated from the test statistics. But this would lead to overly stringent significance thresholds because it would ignore correlations between the markers—that is, it would ignore the fact that the association tests are not independent.
Despite these concerns with the standard permutation test (that assumes independence of the samples), we have reason to expect that this permutation test will still provide reasonably accurate significance thresholds because (1) we do not observe systematic population stratification in the AIL (e.g., clusters of samples that are more related to each other than they are to other mice) and (2) no individuals are closely related to each other (e.g., no mice from the same litter). Careful inspection of our marker-based estimates of kinship coefficients confirms that expectations 1 and 2 do not apply to our AIL. Therefore, we performed permutation-based tests using R/qtl, ignoring unequal relatedness of the individuals (note that the tests for association still used the full-relatedness model). To obtain an estimate for the significance threshold at the 100(1 − α)th percentile of the test statistic, with α = 0.05, we recorded the maximum LOD score for each permutation replicate and defined the significance threshold to be the empirical percentile obtained from 1000 maximum LOD scores, following the recommendation of Churchill and Doerge (1994).
To provide an independent validation of this significance threshold, which is based on the assumption that the samples are exchangeable, we compared it against a more computationally intensive permutation test that accounts for relatedness among individuals (Cheng et al. 2010; Cheng and Palmer 2013), similar to the permutation test proposed by Aulchenko et al. (2007). This permutation procedure differs from the standard method in two key ways: (1) it estimates parameters of a “null” model that includes the polygenic effect, in which kinship coefficients are calculated using the markers, and (2) instead of permuting the phenotypes, it permutes the genotypes, which preserves the relationship between phenotype and polygenic effect. (Note that the accuracy of this permutation procedure hinges on how well the polygenic covariance matrix captures the true covariance structure in the phenotype, which is, of course, unknown.) Following our procedure described above, we fitted a separate mixed model for each chromosome during the permutation tests to avoid the problem of proximal contamination. In Results, we show empirically that this more sophisticated permutation-based test produced significance thresholds that closely corresponded to the thresholds obtained from the much simpler permutation procedure using R/qtl.
QTL regions:
We used a 1.5-LOD support interval to approximate the confidence interval for the location of the QTL. This is a slightly smaller interval than the suggested 1.8-LOD interval based on simulations in two intercrosses, although in practice the best interval for each QTL depends on a number of factors, including the QTL effect size (Manichaikul et al. 2006). The main reason we chose this interval was to be consistent with our previous studies using AIL mice (e.g., Parker et al. 2011; Bartnikas et al. 2012).
Assessment of multiple QTL:
For several phenotypes, the initial genome-wide scan indicated appreciable support for QTL at multiple locations on the same chromosome. However, testing each marker one at a time cannot indicate whether there is support for multiple QTL on a chromosome. To address this question, we included the SNP with the highest LOD score as a covariate, and then we recalculated the LOD scores for all other SNPs on the same chromosome. We repeated this procedure for each of the 30 QTL regions identified in the initial genome-wide scans for the six phenotypes.
X chromosome:
QTL mapping must be performed differently in the X chromosome due to differences in males and females (Broman and Sen 2009; Wise et al. 2013) and due to recombination frequencies that are specific to the X chromosome (Broman 2012). Therefore, a proper analysis of the X chromosome that corrects for confounding due to familial relationships is beyond the capabilities of QTLRel. Nonetheless, we used QTLRel to obtain rough estimates of support for QTL to investigate whether there was a suggestion that markers might warrant further investigation. To adapt the mixed model to analyze the X chromosome, we included sex as an interactive covariate—that is, we included separate additive and dominance effects for males and females. We then estimated the null distribution of the test statistic using the same model (without marker effects), using 1000 permutation replicates as before. Note that this model has one too many degrees of freedom (there is no need to include both additive and dominance terms in hemizygous males since there are only two possible genotypes), so our analysis was overly conservative, and one could improve on our X chromosome analysis, for example, following the approach of Pan et al. (2007). On the X chromosome, we computed genotype probabilities for missing genotypes separately for females and hemizygous males.
Bioinformatics databases:
To identify genes containing nonsynonymous coding SNPs within 1.5-LOD support intervals, we used sequence data for LG/J and SM/J inbred strains generously provided by James Cheverud and Heather Lawson from the Genome Sequencing Center at Washington University [http://genome.wustl.edu (Norgard et al. 2011)].
The full code and data reproducing the steps of the analyses are available for download at http://github.com/pcarbo/lgsmfear.
Results
QTL mapping for open field and conditioned fear phenotypes
We performed a genome-wide association analysis for six traits: (1) distance traveled in the periphery of the open field, (2) distance traveled in the center of the open field, (3) percentage of time spent in the center of the open field, (4) pretraining freezing, (5) freezing to context, and (6) freezing to cue. To assess support for genotype–phenotype correlations at 4601 candidate SNPs, we used a linear mixed model that accounted for possible confounding due to differences in genetic sharing, in which levels of sharing were estimated from the genotype data. We assessed evidence for QTL separately in the LG/J × SM/J F2 intercross, in the F34 advanced intercross, and in the combined sample (Figure 1, Figure 2, and Table 1).
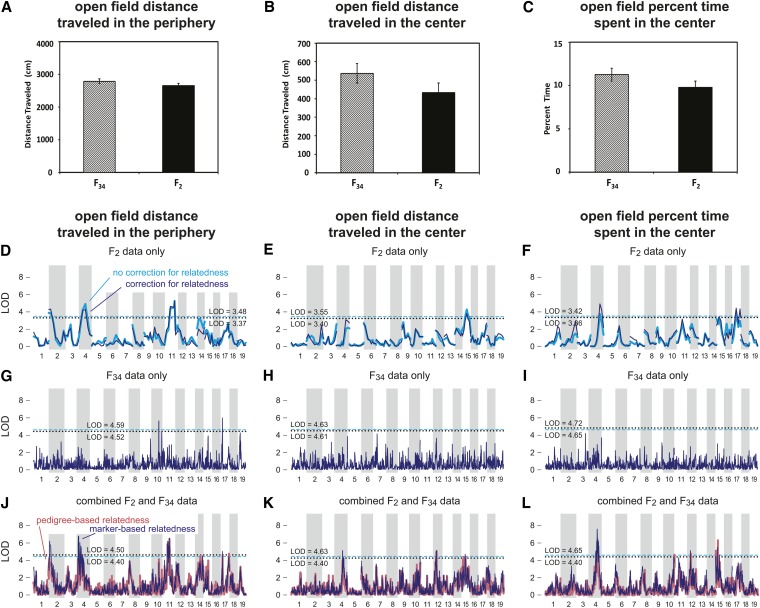
Figure 1.
Open field behavior and genome-wide map of open field behavioral traits in LG/J × SM/J F2, F34, and combined cohorts. A–C summarize open field (OF) testing. Mice were placed in the center of the open field arena, and the following measurements were made in the first 10 min of testing: (A) distance traveled (centimeters) in the periphery, (B) distance traveled (centimeters) in the center, and (C) percentage of time spent in the center. Error bars represent standard error. D–L show genome-wide association signal from F2 samples alone (D–F), from F34 samples alone (G–I), and from combined F2 + F34 cohort (J–L). In all panels, dark blue lines show LOD scores (log10 likelihood ratio) at each genotyped marker. These LOD scores are calculated using marker-based estimates of pairwise relatedness. In D–F, the light blue line shows LOD scores obtained when ignoring hidden relatedness in F2 mice. In J–L, light red lines give LOD scores obtained using pedigree-based estimates of relatedness. For each genome-wide scan, we define the significance threshold as the 95th percentile of the test statistic under the null (α = 0.05). We show significance thresholds obtained from null distributions estimated in two different ways: using a naive permutation test in which samples are assumed to be exchangeable (dashed, light blue line) and using a permutation test that preserves the covariance structure of the samples, which is estimated from the marker data (dotted black line). See Materials and Methods for a detailed explanation of these permutation tests. Autosomal chromosomes 1–19 are shown in alternating shades of gray.
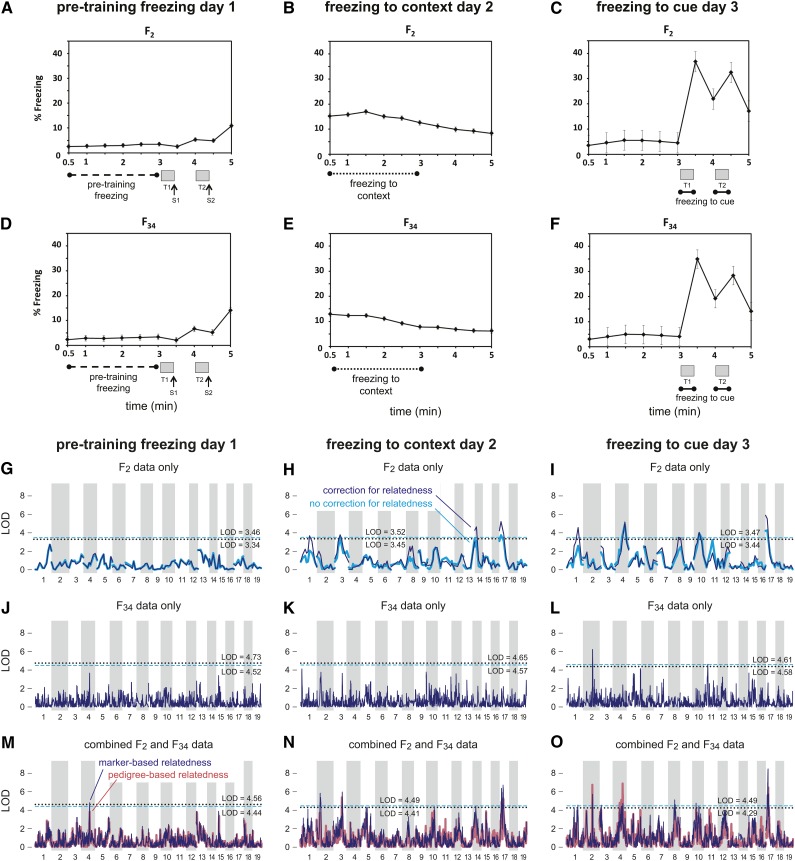
Figure 2.
Conditioned fear and QTL genome-wide map of conditioned fear traits in LG/J × SM/J F2, F34, and combined cohorts. A–F summarize conditioned fear (CF) testing in mice. A 3-day procedure was used to phenotype each subject. On day 1, a baseline indicator of freezing tendency, “pretraining freezing,” was measured in F2 (A) and F34 (D) mice from 30 to 180 sec, after which mice were exposed to two 30-sec tones (indicated by gray bars, labeled T1 and T2) that coterminated with a 2-sec, 0.5-mA foot shock (indicated by arrows, labeled S1 and S2). On day 2, freezing to context was measured in F2 (B) and F34 (E) mice from 30 to 180 sec. On day 3, freezing to cue was measured in F2 (C) and F34 (F); time spent freezing to each tone was averaged to obtain the freezing to cue variable (180–210 sec + 240–270 sec). Each data point represents average percentage of freezing calculated across the 30-sec time bin; error bars represent standard error. G–O show the genome-wide association signal from F2 samples alone (G–I), from the F34 samples alone (J–L), and from the combined F2 + F34 cohort (M–O). See Figure 1 legend for a detailed explanation of the panels.
Table 1. QTL for open field and conditioned fear phenotypes.
| Chr | Peak LOD | Peak SNP | 1.5-LOD interval (Mb) | QTL width | rs start | rs end | No. genes | Genes with NS SNPs | LG/J homozygote mean | Heterozygote mean | SM/J homozygote mean | PVE | Source for QTL previously implicated in fear/anxiety |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Open field activity QTL: distance traveled in the periphery (cm) | |||||||||||||
| 2 | 6.20 | rs27100797 | 9.594–26.988 | 17.39 | rs33088454 | rs13476394 | 177 | 6 | 2614.77 | 2843.57 | 2780.70 | 0.024 | |
| 4 | 6.75 | rs32242095 | 30.271–33.911 | 3.64 | rs27817374 | rs27790884 | 15 | 4 | 2626.90 | 2789.62 | 2939.04 | 0.026 | |
| 11 | 6.54 | rs29391262 | 73.660–81.992 | 8.33 | rs29435763 | rs29431896 | 150 | 42 | 2908.44 | 2836.54 | 2746.93 | 0.025 | Sokoloff et al. (2011) |
| 14 | 4.51 | rs6289260 | 111.373–118.862 | 7.49 | rs3670736 | rs30259301 | 10 | 1 | 2818.45 | 2836.09 | 2634.62 | 0.017 | |
| 17 | 5.01 | rs29501002 | 25.869–29.205 | 3.34 | rs6303335 | rs49110321 | 78 | 23 | 2548.33 | 2783.25 | 3002.24 | 0.019 | Sokoloff et al. (2011) |
| Open field activity QTL: distance traveled in the center (cm) | |||||||||||||
| 4 | 5.11 | rs32263942 | 118.880–121.182 | 2.30 | rs32320402 | rs31878303 | 40 | 8 | 596.55 | 527.09 | 442.72 | 0.019 | Nakamura et al. (2003) |
| 10 | 4.44 | rs6236374 | 127.348–129.068 | 1.72 | rs6157058 | rs6236374 | 78 | 33 | 585.54 | 513.47 | 508.95 | 0.017 | |
| 12 | 5.12 | rs6176279 | 55.962–58.925 | 2.96 | rs29212678 | rs29221863 | 14 | 4 | 456.63 | 520.83 | 618.88 | 0.019 | Jones et al. (1999); Wilson et al. (2011) |
| Open field activity QTL: percentage of time spent in the center | |||||||||||||
| 4 | 5.06 | rs6318014 | 104.577–114.064 | 9.49 | rs32431061 | NA | 83 | 15 | 11.93 | 11.05 | 10.77 | 0.019 | Nakamura et al. (2003) |
| 10 | 4.42 | rs29369956 | 125.235–129.068 | 3.83 | rs29334025 | rs6236374 | 104 | 48 | 12.15 | 10.80 | 10.64 | 0.016 | |
| 12 | 5.12 | rs6176279 | 54.210–58.331 | 4.12 | rs29174777 | rs29134430 | 38 | 3 | 10.15 | 11.00 | 12.40 | 0.019 | Wilson et al. (2011) |
| 15 | 5.46 | rs32422569 | 34.289–38.377 | 4.09 | rs32100330 | rs32514574 | 31 | 8 | 10.45 | 10.80 | 11.96 | 0.020 | Thifault et al. (2008) |
| Conditioned fear QTL: pretraining freezing day 1 | |||||||||||||
| 4 | 4.83 | rs27528211 | 116.993–119.923 | 2.93 | rs27498062 | rs28308541 | 69 | 16 | 3.34 | 3.82 | 4.69 | 0.013 | Nakamura et al. (2003); Owen et al. (1997) |
| Conditioned fear QTL: freezing to context day 2 | |||||||||||||
| 2 | 5.86 | rs33188452 | 26.988–56.359 | 29.37 | rs13476394 | rs32872527 | 236 | 6 | 8.35 | 9.92 | 11.89 | 0.022 | |
| 3 | 5.99 | rs31289463 | 122.098–127.129 | 5.03 | rs30843783 | rs46551270 | 25 | 11 | 8.61 | 11.28 | 11.93 | 0.022 | Thifault et al. (2008) |
| 17a | 6.39 | rs33425954 | 23.684–31.495 | 7.81 | rs33664000 | rs33593781 | 219 | 71 | 8.97 | 9.85 | 10.56 | 0.024 | |
| 17a | 6.74 | rs29506726 | 44.843–49.199 | 4.36 | rs33693466 | rs33454963 | 95 | 17 | 8.66 | 10.36 | 10.79 | 0.025 | Sokoloff et al. (2011) |
| Conditioned fear QTL: freezing to cue day 3 | |||||||||||||
| 1 | 4.57 | rs31862254 | 152.578–161.410 | 8.83 | rs33860680 | rs30802730 | 71 | 6 | 29.85 | 35.89 | 37.81 | 0.018 | Sokoloff et al. (2011); Wilson et al. (2011); Radcliffe et al. (2000); Caldarone et al. (1997) |
| 2b | 5.18 | rs27143196 | 31.169–44.474 | 13.31 | rs6168198 | rs13476468 | 132 | 6 | 34.98 | 33.97 | 35.73 | 0.020 | |
| 2 | 5.75 | rs27453770 | 118.352–125.033 | 6.68 | rs3149909 | rs33186999 | 110 | 59 | 42.11 | 30.63 | 29.29 | 0.023 | Sokoloff et al. (2011) |
| 3 | 4.57 | rs29599008 | 24.613–31.177 | 6.56 | rs13477022 | rs31004134 | 30 | 4 | 28.17 | 35.62 | 34.87 | 0.018 | Owen et al. (1997) |
| 4 | 4.76 | rs27538669 | 129.820–133.601 | 3.78 | rs27517761 | rs27560257 | 65 | 26 | 34.68 | 30.97 | 39.77 | 0.019 | Sokoloff et al. (2011) |
| 8 | 5.14 | rs3710389 | 82.591–91.538 | 8.95 | rs6382412 | rs33287905 | 109 | 37 | 43.58 | 34.12 | 32.20 | 0.020 | |
| 10 | 4.78 | rs6247407 | 56.324–60.568 | 4.24 | rs6314360 | rs6372589 | 34 | 12 | 37.84 | 31.30 | 37.17 | 0.019 | |
| 17 | 8.49 | rs33357930 | 26.747–30.583 | 3.84 | rs3679791 | rs13482944 | 71 | 16 | 32.16 | 32.13 | 39.57 | 0.034 | Sokoloff et al. (2011) |
Shown are regions containing one or more SNPs with LOD scores exceeding the significance threshold. Significance thresholds are defined as the 95th percentile of the test statistic under the null (α = 0.05). LOD scores were obtained by fitting a model to the combined (F2 + F34) cohort, using marker-based estimates of pairwise relatedness. Columns from left to right are: (1) chromosome, (2) maximum LOD score, (3) SNP with maximum LOD score, (4 and 5) region most likely containing causal variant(s) and size of region in megabases (Mb) based on a 1.5-LOD support interval, (6 and 7) SNPs bounding region, (8) number of genes within region annotated to mouse reference sequence, (9) number of genes within region for which nonsynonymous coding polymorphisms are identified (see Table S1), (10) mean phenotypic value for LG/J homozygotes at peak SNP, (11) LG/J × SM/J heterozygotes, (12) SM/J homozygotes, (13) proportion of variance in phenotype explained by peak SNP, and (14) published findings on fear and anxiety QTL that overlap this region. All SNP information and genomic positions are based on Mouse Genome Assembly 37 (July 2007) in the NCBI Assembly Database.
Support for the other QTL on chromosome 17 falls short of our criteria for significance once we condition on this QTL; our data support at most one QTL on chromosome 17.
SNPs in this region exceed the significance threshold only after conditioning on the effect of SNP rs27453770 on the same chromosome.
In the analysis of the combined sample, we identified a total of 24 QTL exceeding significance thresholds (we discuss the significance thresholds in greater detail below). Specifically, we identified 12 QTL for OF phenotypes and 12 QTL for CF phenotypes, and we identified at least 1 QTL for each of the six behavioral traits.
Our analysis did not yield any compelling QTL on the X chromosome; the largest LOD score, 4.6, for the phenotype “percentage of time spent in center,” was considerably below the significance threshold, 5.7 (at α = 0.05). Since sex-linked chromosomes require a separate treatment from that of autosomal chromosomes, X chromosome results are not shown in Table 1, Figure 1, and Figure 2. (Note that our analysis of the X chromosome was probably overly conservative, and an improved analysis of the X chromosome correcting for relatedness could yield stronger support for QTL; see Materials and Methods for more details.)
The initial genome-wide scan (Figure 1 and Figure 2) identified multiple distinct regions located on the same chromosome containing significant QTL (Table 1). In all cases except one—chromosome 2 for freezing to cue—support for a second QTL fell short of our significance thresholds once we conditioned on the estimated effect of the peak SNP from the same chromosome. Table 1 gives two regions on chromosome 17 that each show strong evidence for containing a QTL for freezing to context, although we have sufficient support for a QTL in only one of the two loci.
Several phenotypes were associated with regions that appear to overlap each other (Table 1). However, the QTL mapping did not yield sufficient resolution to determine whether the overlapping QTL regions highlight the same gene or genes. Below, we examine some of the more interesting overlapping QTL.
Most of the QTL with the strongest support from the combined sample were also identified using the F2 sample alone. One exception was the QTL for freezing to cue on chromosome 2. This locus showed no appreciable association signal in the F2 sample, but strong support from the F34 data.
In the F2 analyses, each of the QTL regions (based on 1.5-LOD support intervals) covered a large portion of the chromosome, owing to limited recombination in F2 crosses. Compared to the QTL regions in the F2 cross, the QTL were much narrower in the F34 alone. However, few SNPs reached the threshold for significance at α = 0.05, owing to a loss of power due to many more candidate haplotypes. By comparison, in the combined analysis, we identified 24 QTL with narrow 1.5-LOD support intervals, ranging in size from 1.7 Mb to 29.4 Mb and with a median interval length of 4.3 Mb. Of these, 13 QTL had support intervals <5 Mb in length. The number of annotated genes within these intervals ranged from 10 to 236, with a median of 71 genes.
As expected, support for QTL in the F2 sample for the most part did not change appreciably when we accounted for possible confounding due to relatedness; F2 crosses are full sibs with respect to one another and hence are expected to exhibit roughly the same amount of genetic sharing. Nonetheless, there were several cases where support for a QTL increased notably in the F2 cross after accounting for varying relatedness and occasionally yielded a significant QTL only after correcting for relatedness. This comparison was not systematic, nor did we experience a uniform increase in support across the genome after correcting for relatedness. Nonetheless, this hints at the potential gains in applying mixed models to conventional intercrosses—a topic that we plan to explore in greater depth in subsequent work.
Since we had kept track of the complete pedigree for the F34 mice, we were able to compare the QTL mapping results, correcting for relatedness using (1) marker-based estimates of the kinship coefficients and (2) pedigree-based estimates. Both analyses showed broad agreement; compare the dark blue and light red lines in Figure 1, J–L, and Figure 2, M–O. In several regions, the association signal was considerably stronger in the analysis using marker-based estimates. A notable exception to this tendency was the QTL on chromosome 4 for freezing to cue. These results are consistent with a more systematic comparison of pedigree- and marker-based estimates of relatedness for QTL mapping in simulated populations (Cheng et al. 2013).
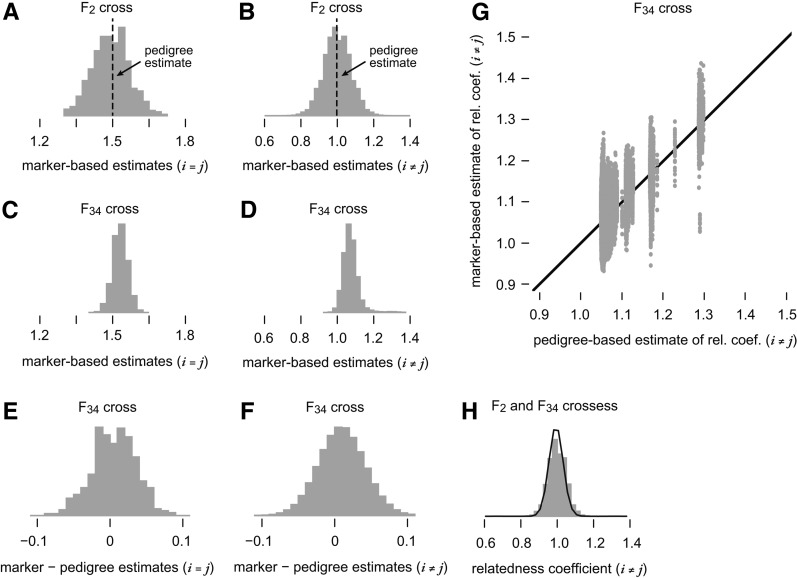
Comparison of the pedigree- and marker-based estimates of the kinship coefficients also offers the opportunity to point out some features of the marker-based estimates. Figure 3 shows the distribution of pairwise relatedness coefficients in the F2 and F34 crosses. First, using the marker data we were able to predict that some F2 mice share a greater proportion of their genome than others (Figure 3B), whereas pedigree-based estimates of sharing are all identical, as the relationship to the inbred founders is the same for all F2 crosses. (For any F2 cross, pairwise relatedness is and for see Lange 2002.) By comparison, using the marker data we predict much less variation in genetic sharing between pairs of F34-generation mice (compare Figure 3B with Figure 3D). This was expected because of the greater number of accumulated crossovers in the F34 mice. Second, we observed that the marker-based estimates were for the most part unbiased; differences in the marker- and pedigree-based estimates were centered near zero (see Figure 3, A, B, E, and F). Even though the genetic sharing estimates in F34 AIL mice agreed on average, the estimates did not agree in a large proportion of the samples—the scatterplot (Figure 3G) illustrates this point in detail. The larger discrepancies in the sharing estimates may explain the differences observed in the QTL mapping (Figure 1 and Figure 2). Finally, the marker data allowed us to estimate sharing for F2–F34 pairs of mice (Figure 3H), whereas these calculations are very complex using the pedigree data (and would not have been meaningful in this case because the F2 are not the direct progenitors of the F34).
Figure 3.
Comparison of marker- and pedigree-based estimates of pairwise relatedness. A–D and H show the extent to which relatedness, or genetic sharing, varies among the AIL mice. More precisely, A–D and H give the observed distributions of estimated pairwise relatedness coefficients, or two times the kinship coefficients, for all pairs of mice (i, j) in which i and j are different individuals () or when i = j (in which case the coefficients correspond to inbreeding levels). These are “realized relatedness” estimates obtained by calculating average allele sharing over all SNPs genotyped in the F2 and F34 crosses. Diagonal entries of the relatedness matrix, are shown in A and C, and off-diagonal entries are shown in B, D, G, and H. To compare the marker-based estimates to the estimates of allele sharing obtained from the AIL pedigree data, in E and F we show the distribution of differences between the marker-based and pedigree estimates of pairwise relatedness. In G we show a scatterplot providing a more detailed comparison of the marker- and pedigree-based pairwise relatedness estimates in which each point in the scatterplot corresponds to a pair of F34 mice. In the F2 cross, all pedigree-based pairwise relatedness estimates are and as indicated in A and B. Finally, H shows the distribution of marker-based pairwise relatedness estimates for all F2–F34 pairs, using only markers available in the F2 mice (shaded bars) and using all markers (solid line).
To assess significance of the LOD scores in our genome-wide scans, we applied a simple permutation test (see Materials and Methods for details). Although this simple permutation test can lead to inadequately stringent significance thresholds when phenotype samples are correlated (Abney et al. 2002; Zou et al. 2005; Aulchenko et al. 2007; Cheng et al. 2010; Cheng and Palmer 2013), as they are here, we have assessed empirically in our AIL that the distribution of the test statistic under the null is similar regardless of whether the permutations account for relatedness (Figure 1 and Figure 2). One possible explanation why the standard permutation test seems to adequately control for type 1 error in our AIL sample is that we have carefully avoided including highly related mice—in particular, no mice share a common grandparent—unlike our previous AIL studies (e.g., Cheng et al. 2010; Cheng and Palmer 2013).
In summary, the combined analysis identified more QTL than either the F2 or the F34 samples alone (due in part to a larger sample size). Importantly, the combined analysis located the QTL within narrower regions compared to those identified from the F2 mice alone.
Sequence analysis
We examined our 1.5-LOD support intervals for the presence of “consequential” SNPs that had the potential to directly alter proteins (i.e., nonsynonymous coding, stop-gain, stop-lost, frameshift, and splice sites). The number of genes with consequential SNPs in significant QTL intervals ranged from 1 to 59, with a median of 12 genes (Table S1).
Discussion
We investigated the genetic basis of anxiety-like behavior, based on open field and conditioned fear tests, in a combined F2 and F34 AIL mouse population. To map QTL across the genome, we analyzed phenotypes using a linear mixed model that accounts for differences in genetic sharing, an important confounding factor in tests for association. Taking this mixed-model approach, we identified a total of 24 QTL affecting six traits related to fear and anxiety-like behavior. Because our study maintained uniform environmental conditions and ensured alleles with high frequency, we expect a limited contribution of gene–environment interactions to variation in these traits.
The integration of the F2 and F34 populations provided good power to detect QTL, presumably because of the F2 cross; and good resolution, presumably because of the F34 advanced intercross. The median size of a QTL region was 4.3 Mb. The QTL were slightly narrower than QTL intervals we identified in this population for other traits, such as red blood cell parameters [median 1.5-LOD support interval = 4.7 Mb (Bartnikas et al. 2012)] and body weight [median 1.5-LOD support interval = 5.5 Mb (Parker et al. 2011)].
In the 1.5-LOD intervals that contained ≤15 genes with coding SNPs, we searched for candidate genes that could plausibly explain the variation in the traits we measured. Some of the genes within these regions had coding SNPs known to be involved in anxiety-like behavior and/or conditioned fear; examples include the nuclear receptor subfamily 6, group A, member 1 gene [Nr6a1 (Heydendael et al. 2013)], the phospholipase D1 gene [Pld1 (Sun et al. 2013)], the cadherin 23 gene [Cdh23 (Terracciano et al. 2010, but see Schwander et al. 2009)], the prosaposin gene [Psap (Hovatta et al. 2005; Donner et al. 2008)], and the SLIT and NTRK-like family, member 5 gene [Slitrk5 (Shmelkov et al. 2010)]. However, we are cautious about interpreting the functional relevance of nonsynonymous coding SNPs; we do not currently have expression QTL (eQTL) data that could be used to detect heritable regulatory polymorphisms. Thus, the current results are not sufficient to identify specific causal genes.
In two instances, contextual and cued fear mapped to overlapping chromosomal regions on chromosomes 2 and 17 (Figure S2 and Figure S3) despite known differences in their neuroanatomical substrates (Fanselow and Ledoux 1999; Jovanovic and Ressler 2010). This may indicate the presence of alleles that influence both traits; alternatively, it could be due to different alleles that are located close to each other in the genome. We (Ponder et al. 2007a,b; Sokoloff et al. 2011; Parker et al. 2012b) and others have reported similar results; Talbot et al. (2003) reported that contextual and cued fear were highly correlated (r = 0.63) in heterogeneous stock mice, and selection for freezing to context has been shown to cause coincident changes in freezing to cue (Radcliffe et al. 2000; Ponder et al. 2007a). Thus, it is likely that contextual and cued fear are modulated by some of the same alleles, but further studies are needed to definitively identify the alleles that give rise to such correlations.
One of the underlying motivations for this study was the belief that a subset of alleles would pleiotropically influence both conditioned fear and anxiety-like behavior in the open field. Previous studies by our laboratory (Ponder et al. 2007a,b; Sokoloff et al. 2011) and other groups (Lopez-Aumatell et al. 2009) have indicated a shared genetic control of these two traits. The overlap between the QTL on chromosome 17 for conditioned fear (freezing to context and freezing to cue) and distance traveled in the periphery (Figure S3) provides some additional support for a shared genetic basis. Interestingly, we observed QTL on chromosome 2 for freezing to context and freezing to cue that were immediately adjacent to, but did not overlap with, the chromosome 2 QTL for distance traveled in the periphery (Figure S2). And in one case, the QTL on chromosome 4 for pretraining freezing showed modest overlap with the chromosome 4 QTL for distance traveled in the center of the arena (Figure S4). However, pretraining freezing (freezing before administration of any tones or shocks) is more indicative of unlearned anxiety rather than conditioned fear. The absence of regions common to several traits may indicate that in this population the genetic origin of fear and anxiety-like behavior is relatively distinct, or it may reflect a lack of power to detect QTL, so we cannot interpret this result as implying that genes in these regions do not jointly affect these traits. An integrated analysis of these traits [e.g., building on multivariate mapping approaches (Stephens 2013)] may yield an improved understanding of how QTL modulate these anxiety-like behaviors and represents an important research direction.
Traditionally, F2 intercrosses are used to identify QTL underlying phenotypic variation, and fine-mapping is carried out as a second step, using congenic strains. This time- and labor-intensive effort to identify specific genes is often derailed by the discovery that a single QTL of large effect is in fact caused by multiple loci of small effect located in the same chromosomal region (Legare et al. 2000; Mott et al. 2000; Cheng et al. 2010; Shao et al. 2010; Parker et al. 2013). An AIL is an improvement over these traditional methods because it merges identification and fine-mapping into a single step, which can often discriminate between loci that are due to single vs. multiple alleles (Darvasi and Soller 1995). The trade-off is that the power to detect QTL in AILs is often lower than in F2 populations. This is because the AIL mice experience greater numbers of crossover events than the F2 mice, so more tests are performed, and a corresponding higher threshold is needed to control for false positives. For example, a QTL was observed on chromosome 1 for freezing to context in the F2 intercross, but in the F34 AIL it appeared to split into two smaller regions, neither of which had a LOD score exceeding significance. On the other hand, we observed a highly significant QTL peak in the F34 AIL for freezing to cue that was not originally seen in the F2 intercross and was subsequently supported in the integrated analysis. While the advantages of an integrated analysis of an AIL have been shown in previous work, this study provides further support for the benefits of this approach and can serve as a prototype for how to identify QTL in AILs using a mixed-model approach that accounts for relatedness, using marker data. (We have made available the code and data used to implement the steps of our analysis at http://github.com/pcarbo/lgsmfear.)
Our main methodological contribution was to show that we can use marker data in the place of a pedigree to infer familial relationships in an AIL. We found that mixed models using either pedigree- or marker-based estimates of relatedness showed broad agreement in the QTL mapping. While there has been a considerable amount of work demonstrating the benefits of marker-based estimates of “realized relatedness” to control for confounding due to population structure or due to familial relationships (Yu et al. 2006; Kang et al. 2008; Lippert et al. 2011; Listgarten et al. 2012; Zhou et al. 2013), there has been little work on demonstrating these benefits in AILs. In this article, we did not aim for a systematic comparison of approaches to correct for relatedness (for an empirical comparison in simulated data sets, see Cheng et al. 2013). Our results nonetheless suggest that it is better to use marker data to correct for relatedness, even when the full pedigree is available, provided genetic variation is ascertained at sufficient resolution throughout the genome. The marker-based estimates often yield more precise estimates of genetic sharing and are usually less costly to obtain.
Our study has several limitations. First, because we have used a cross between two inbred strains, we are studying the alleles that segregate between them and not the total number of alleles that segregate among other laboratory strains or wild mice. For example, we observed little overlap of QTL for conditioned fear between an F8 AIL derived from C57BL/6J × DBA/2J mice (Parker et al. 2012b) and the LG/J × SM/J AIL in the present study. Nonetheless, we did observe some overlap in the QTL we identified in our population with anxiety and fear-related QTL identified in other populations of mice (Table 1), consistent with the possibility that different two-strain combinations may segregate the same alleles. It is possible that some of the QTL identified in our study are the same as those identified by other researchers; one advantage of our study is that we have mapped QTL with greater resolution than in previous studies.
Our approach has dramatically increased the mapping resolution compared to that in more conventional mapping populations. To further reduce the number of candidate genes within our intervals, we focused on genes with coding polymorphisms. However, it is important to note that the polymorphisms underlying the observed trait variance may be due to differences in gene expression, rather than alterations in gene function. For example, some QTL may be explained by SNPs that modulate the recruitment of proteins involved in regulation of gene transcription; studies of complex human traits have shown that a large fraction of the variants underlying these traits coincide with DNA sequences related to gene regulation (Nicolae et al. 2010; Schaub et al. 2012). Availability of genome-wide eQTL data in the LG/J and SM/J strains may help pinpoint the genes underlying these QTL.
Supplementary Material
Acknowledgments
We thank James Cheverud and Heather Lawson for generously providing the F33 mice used to create the F34 and the sequence data in the LG/J and SM/J mice. We also acknowledge Michaelanne Munoz, Tanya Cebollero, and Jackie Lim for help with husbandry and behavioral testing; Kaitlin Samocha for pedigree reconstruction; Karl Broman for advice on dealing with QTL mapping on the X chromosome; Xiang Zhou for technical help on the QTL mapping; Pei-Ting Wu for data entry; and Riyan Cheng and Andrew Skol for helping to develop QTLRel and calling of genotypes used in this study. Finally, we thank the reviewers for their helpful feedback. This work was supported by National Institutes of Health grants 5R01MH079103 (to A.A.P.) and R01HG002899 (to M.A.). P.C. was supported by a cross-disciplinary postdoctoral fellowship from the Human Frontiers Science Program.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.167056/-/DC1.
Communicating editor: F. van Eeuwijk
Literature Cited
- Abney M., McPeek M. S., Ober C., 2000. Estimation of variance components of quantitative traits in inbred populations. Am. J. Hum. Genet. 66: 629–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abney M., Ober C., McPeek M. S., 2002. Quantitative-trait homozygosity and association mapping and empirical genomewide significance in large, complex pedigrees: fasting serum-insulin level in the Hutterites. Am. J. Hum. Genet. 70: 920–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko Y. S., de Koning D.-J., Haley C., 2007. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics 177: 577–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnikas T. B., Parker C. C., Cheng R., Campagna D. R., Lim J. E., et al. , 2012. QTLs for murine red blood cell parameters in LG/J and SM/J F2 and advanced intercross lines. Mamm. Genome 23: 356–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., 2012. Haplotype probabilities in advanced intercross populations. G3 2: 199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman, K. W., and S. Sen, 2009 A Guide to QTL Mapping with R-qtl Springer-Verlag, Berlin/Heidelberg, Germany/New York. [Google Scholar]
- Bryant C. D., Parker C. C., Zhou L., Olker C., Chandrasekaran R. Y., et al. , 2012. Csnk1e is a genetic regulator of sensitivity to psychostimulants and opioids. Neuropsychopharmacology 37: 1026–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldarone B., Saavedra C., Tartaglia K., Wehner J. M., et al. , 1997. Quantitative trait loci analysis affecting contextual conditioning in mice. Nat. Genet 17: 335–337 [DOI] [PubMed] [Google Scholar]
- Cheng R., Palmer A. A., 2013. A simulation study of permutation, bootstrap, and gene dropping for assessing statistical significance in the case of unequal relatedness. Genetics 193: 1015–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R., Lim J. E., Samocha K. E., Sokoloff G., Abney M., et al. , 2010. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics 185: 1033–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R., Abney M., Palmer A. A., Skol A. D., 2011. QTLRel: an R package for genome-wide association studies in which relatedness is a concern. BMC Genet. 12: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R., Parker C. C., Abney M., Palmer A. A., 2013. Practical considerations regarding the use of genotype and pedigree data to model relatedness in the context of genome-wide association studies. G3 3: 1861–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A., Ackert-Bicknell C. L., Dumont B. L., Ding Y., Bell J. T., et al. , 2009. A new standard genetic map for the laboratory mouse. Genetics 182: 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi A., Soller M., 1995. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141: 1199–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demyttenaere K., Bruffaerts R., Posada-Villa J., Gasquet I., Kovess V., et al. , 2004. Prevalence, severity, and unmet need for treatment of mental disorders in the World Health Organization World Mental Health Surveys. JAMA 291: 2581–2590 [DOI] [PubMed] [Google Scholar]
- Donner J., Pirkola S., Silander K., Kananen L., Terwilliger J. D., et al. , 2008. An association analysis of murine anxiety genes in humans implicates novel candidate genes for anxiety disorders. Biol. Psychiatry 64: 672–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanselow M. S., LeDoux J. E., 1999. Why we think plasticity underlying Pavlovian fear conditioning occurs in the basolateral amygdala. Neuron 23: 229–232 [DOI] [PubMed] [Google Scholar]
- Flint J., 2011. Mapping quantitative traits and strategies to find quantitative trait genes. Methods 53: 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., Eskin E., 2012. Genome-wide association studies in mice. Nat. Rev. Genet. 13: 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., Shifman S., 2008. Animal models of psychiatric disease. Curr. Opin. Genet. Dev. 18: 235–240 [DOI] [PubMed] [Google Scholar]
- Hettema J. M., Webb B. T., Guo A.-Y., Zhao Z., Maher B. S., et al. , 2011. Prioritization and association analysis of murine-derived candidate genes in anxiety-spectrum disorders. Biol. Psychiatry 70: 888–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydendael W., Sengupta A., Beck S., Bhatnagar S., 2013. Optogenetic examination identifies a context-specific role for orexins/hypocretins in anxiety-related behavior. Physiol. Behav. 130: 182–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovatta I., Barlow C., 2008. Molecular genetics of anxiety in mice and men. Ann. Med. 40: 92–109 [DOI] [PubMed] [Google Scholar]
- Hovatta I., Tennant R. S., Helton R., Marr R. A., Singer O., et al. , 2005. Glyoxalase 1 and glutathione reductase 1 regulate anxiety in mice. Nature 438: 662–666 [DOI] [PubMed] [Google Scholar]
- Jackson I. J., 1994. Molecular and developmental genetics of mouse coat color. Annu. Rev. Genet. 28: 189–217 [DOI] [PubMed] [Google Scholar]
- Jackson I. J., 1997. Homologous pigmentation mutations in human, mouse and other model organisms. Hum. Mol. Genet. 6: 1613–1624 [DOI] [PubMed] [Google Scholar]
- Jones B. C., Tarantino L. M., Rodriguez L. A., Reed C. L., McClearn G. E., et al. , 1999. Quantitative-trait loci analysis of cocaine-related behaviors and neurochemistry. Pharmacogenetics 9: 607–17 [PubMed] [Google Scholar]
- Jovanovic T., Ressler K. J., 2010. How the neurocircuitry and genetics of fear inhibition may inform our understanding of PTSD. Am. J. Psychiatry 167: 648–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalueff A. V., Wheaton M., Murphy D. L., 2007. What’s wrong with my mouse model? Advances and strategies in animal modeling of anxiety and depression. Behav. Brain Res. 179: 1–18 [DOI] [PubMed] [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S.-Y., et al. , 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karigl G., 1981. A recursive algorithm for the calculation of identity coefficients. Ann. Hum. Genet. 45: 299–305 [DOI] [PubMed] [Google Scholar]
- Kessler R. C., Berglund P., Demler O., Jin R., Merikangas K. R., et al. , 2005a Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch. Gen. Psychiatry 62: 593–602 [DOI] [PubMed] [Google Scholar]
- Kessler R. C., Chiu W. T., Demler O., Merikangas K. R., Walters E. E., 2005b Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch. Gen. Psychiatry 62: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Green P., 1987. Construction of multilocus genetic linkage maps in humans. Proc. Natl. Acad. Sci. USA 84: 2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, K., 2002 Mathematical and Statistical Methods for Genetic Analysis Springer-Verlag, Berlin/Heidelberg, Germany/New York. [Google Scholar]
- Legare M. E., Bartlett F. S., Frankel W. N., 2000. A major effect QTL determined by multiple genes in epileptic EL mice. Genome Res. 10: 42–48 [PMC free article] [PubMed] [Google Scholar]
- Lionikas A., Cheng R., Lim J. E., Palmer A. A., Blizard D. A., 2010. Fine-mapping of muscle weight QTL in LG/J and SM/J intercrosses. Physiol. Genomics 42A: 33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C., Listgarten J., Liu Y., Kadie C. M., Davidson R. I., et al. , 2011. FaST linear mixed models for genome-wide association studies. Nat. Methods 8: 833–835 [DOI] [PubMed] [Google Scholar]
- Listgarten J., Lippert C., Kadie C. M., Davidson R. I., Eskin E., et al. , 2012. Improved linear mixed models for genome-wide association studies. Nat. Methods 9: 525–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Aumatell R., Vicens-Costa E., Guitart-Masip M., Martínez-Membrives E., Valdar W., et al. , 2009. Unlearned anxiety predicts learned fear: a comparison among heterogeneous rats and the Roman rat strains. Behav. Brain Res. 202: 92–101 [DOI] [PubMed] [Google Scholar]
- Mahan A. L., Ressler K. J., 2012. Fear conditioning, synaptic plasticity and the amygdala: implications for posttraumatic stress disorder. Trends Neurosci. 35: 24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A., Dupuis J., Sen S., Broman K. W., 2006. Poor performance of bootstrap confidence intervals for the location of a quantitative trait locus. Genetics 174: 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R., Talbot C. J., Turri M. G., Collins A. C., Flint J., 2000. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. USA 97: 12649–12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Xiu Y., Ohtsuji M., Sugita G., Abe M., et al. , 2003. Genetic dissection of anxiety in autoimmune disease. Hum. Mol. Genet. 12: 1079–86 [DOI] [PubMed] [Google Scholar]
- Nicolae D. L., Gamazon E., Zhang W., Duan S., Dolan M. E., et al. , 2010. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6: e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgard E. A., Lawson H. A., Pletscher L. S., Wang B., Brooks V. R., et al. , 2011. Genetic factors and diet affect long-bone length in the F34 LG,SM advanced intercross. Mamm. Genome 22: 178–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A. A., Verbitsky M., Suresh R., Kamens H. M., Reed C. L., et al. , 2005. Gene expression differences in mice divergently selected for methamphetamine sensitivity. Mamm. Genome 16: 291–305 [DOI] [PubMed] [Google Scholar]
- Pan L., Ober C., Abney M., 2007. Heritability estimation of sex-specific effects on human quantitative traits. Genet. Epidemiol. 31: 338–347 [DOI] [PubMed] [Google Scholar]
- Parker C. C., Palmer A. A., 2011. Dark matter: Are mice the solution to missing heritability? Front. Genet. 2: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. C., Cheng R., Sokoloff G., Lim J. E., Skol A. D., et al. , 2011. Fine-mapping alleles for body weight in LG/J × SM/J F2 and F34 advanced intercross lines. Mamm. Genome 22: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. C., Cheng R., Sokoloff G., Palmer A. A., 2012a Genome-wide association for methamphetamine sensitivity in an advanced intercross mouse line. Genes Brain Behav. 11: 52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. C., Sokoloff G., Cheng R., Palmer A. A., 2012b Genome-wide association for fear conditioning in an advanced intercross mouse line. Behav. Genet. 42: 437–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. C., Sokoloff G., Leung E., Kirkpatrick S. L., Palmer A. A., 2013. A large QTL for fear and anxiety mapped using an F2 cross can be dissected into multiple smaller QTLs. Genes Brain Behav. 12: 714–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov P. M., Ding Y., Cassell M. A., Zhang W., Wagner G., et al. , 2004. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 14: 1806–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder C. A., Kliethermes C. L., Drew M. R., Muller J., Das K., et al. , 2007a Selection for contextual fear conditioning affects anxiety-like behaviors and gene expression. Genes Brain Behav. 6: 736–749 [DOI] [PubMed] [Google Scholar]
- Ponder C. A., Munoz M., Gilliam T. C., Palmer A. A., 2007b Genetic architecture of fear conditioning in chromosome substitution strains: relationship to measures of innate (unlearned) anxiety-like behavior. Mamm. Genome 18: 221–228 [DOI] [PubMed] [Google Scholar]
- Radcliffe R. A., Lowe M. V., Wehner J. M., 2000. Confirmation of contextual fear conditioning QTLs by short-term selection. Behav. Genet. 30: 183–191 [DOI] [PubMed] [Google Scholar]
- Samocha K. E., Lim J. E., Cheng R., Sokoloff G., Palmer A. A., 2010. Fine mapping of QTL for prepulse inhibition in LG/J and SM/J mice using F2 and advanced intercross lines. Genes Brain Behav. 9: 759–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub M. A., Boyle A. P., Kundaje A., Batzoglou S., Snyder M., 2012. Linking disease associations with regulatory information in the human genome. Genome Res. 22: 1748–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander M., Xiong W., Tokita J., Lelli A., Elledge H. M., et al. , 2009. A mouse model for nonsyndromic deafness (DFNB12) links hearing loss to defects in tip links of mechanosensory hair cells. Proc. Natl. Acad. Sci. USA 106: 5252–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H., Sinasac D. S., Burrage L. C., Hodges C. A., Supelak P. J., et al. , 2010. Analyzing complex traits with congenic strains. Mamm. Genome 21: 276–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry S. T., Ward M. H., Kholodov M., Baker J., Phan L., et al. , 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29: 308–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmelkov S. V., Hormigo A., Jing D., Proenca C. C., Bath K. G., et al. , 2010. Slitrk5 deficiency impairs corticostriatal circuitry and leads to obsessive-compulsive-like behaviors in mice. Nat. Med. 16: 598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloff G., Parker C. C., Lim J. E., Palmer A. A., 2011. Anxiety and fear in a cross of C57BL/6J and DBA/2J mice: mapping overlapping and independent QTL for related traits. Genes Brain Behav. 10: 604–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., 2013. A unified framework for association analysis with multiple related phenotypes. PLoS ONE 8: e65245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan P. F., Daly M. J., O’Donovan M., 2012. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat. Rev. Genet. 13: 537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Gooding H. L., Brunton P. J., Russell J. A., Mitchell R., et al. , 2013. Phospholipase D-mediated hypersensitivity at central synapses is associated with abnormal behaviours and pain sensitivity in rats exposed to prenatal stress. Int. J. Biochem. Cell Biol. 45: 2706–2712 [DOI] [PubMed] [Google Scholar]
- Talbot C. J., Radcliffe R. A., Fullerton J., Hitzemann R., Wehner J. M., et al. , 2003. Fine scale mapping of a genetic locus for conditioned fear. Mamm. Genome 14: 223–230 [DOI] [PubMed] [Google Scholar]
- Terracciano A., Sanna S., Uda M., Deiana B., Usala G., et al. , 2010. Genome-wide association scan for five major dimensions of personality. Mol. Psychiatry 15: 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thifault S., Ondrej S., Sun Y., Fortin A., Skamene E., et al. , 2008. Genetic determinants of emotionality and stress response in AcB/BcA recombinant congenic mice and in silico evidence of convergence with cardiovascular candidate genes. Hum. Mol. Genet. 17: 331–44 [DOI] [PubMed] [Google Scholar]
- Wilson Y. M., Brodnicki T. C., Lawrence A. J., Murphy M., 2011. Congenic mouse strains enable discrimination of genetic determinants contributing to fear and fear memory. Behav. Genet. 41: 278–287 [DOI] [PubMed] [Google Scholar]
- Wise A. L., Gyi L., Manolio T. A., 2013. eXclusion: toward integrating the X chromosome in genome-wide association analyses. Am. J. Hum. Genet. 92: 643–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zaitlen N. A., Goddard M. E., Visscher P. M., Price A. L., 2014. Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 46: 100–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W. H., Bi I. V., Yamasaki M., et al. , 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208 [DOI] [PubMed] [Google Scholar]
- Zhou X., Stephens M., 2012. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44: 821–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Carbonetto P., Stephens M., 2013. Polygenic modeling with Bayesian sparse linear mixed models. PLoS Genet. 9: e1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F., Gelfond J. A. L., Airey D. C., Lu L., Manly K. F., et al. , 2005. Quantitative trait locus analysis using recombinant inbred intercrosses: theoretical and empirical considerations. Genetics 170: 1299–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.