Abstract
Multiparental populations are of considerable interest in high-density genetic mapping due to their increased levels of polymorphism and recombination relative to biparental populations. However, errors in map construction can have significant impact on QTL discovery in later stages of analysis, and few methods have been developed to quantify the uncertainty attached to the reported order of markers or intermarker distances. Current methods are computationally intensive or limited to assessing uncertainty only for order or distance, but not both simultaneously. We derive the asymptotic joint distribution of maximum composite likelihood estimators for intermarker distances. This approach allows us to construct hypothesis tests and confidence intervals for simultaneously assessing marker-order instability and distance uncertainty. We investigate the effects of marker density, population size, and founder distribution patterns on map confidence in multiparental populations through simulations. Using these data, we provide guidelines on sample sizes necessary to map markers at sub-centimorgan densities with high certainty. We apply these approaches to data from a bread wheat Multiparent Advanced Generation Inter-Cross (MAGIC) population genotyped using the Illumina 9K SNP chip to assess regions of uncertainty and validate them against the recently released pseudomolecule for the wheat chromosome 3B.
Keywords: linkage map, multiparental populations, recombinant inbred line, recombination fraction, variability, high-density genotyping, Multiparent Advanced Generation Inter-Cross (MAGIC), single nucleotide polymorphism (SNP), multiparental populations, MPP
LINKAGE maps have been fundamental to genetic analysis for many years, both for gaining a better understanding of genomic structure and for utilizing that structure to gain power in mapping gene–trait associations. For humans and many other species, high-density consensus maps have been published and used across multiple mapping studies (Murray et al. 1994; Dietrich et al. 1996; Chowdhary and Raudsepp 2006; Bult et al. 2008; Cox et al. 2009; Wong et al. 2010). However, efforts to increase the saturation of genetic maps with high-throughput genotyping are still being made in many plant species (Poland et al. 2012; Ward et al. 2013; Wang et al. 2014).
Many approaches to genetic map estimation have been proposed and are reviewed along with common challenges in Cheema and Dicks (2009). Perhaps the most challenging step in map construction is ordering markers within a linkage group. Methods for ordering markers in biparental populations have been well studied and include techniques such as seriation (Buetow and Chakravarti 1987), ant colony optimization (Iwata and Ninomiya 2006), minimum spanning trees (Wu et al. 2008), rapid chain delineation (Nascimento et al. 2010), and simulated annealing (Van Ooijen 2011). These in turn form the basis of numerous map-construction software packages. These can be roughly divided up into those relying on multipoint approaches, which incorporate information across the genome to maximize the likelihood of the map (MAPMAKER, Lander et al. 1987; CRI-MAP, Green et al. 1990; JoinMap, Stam 1993; R/qtl, Broman et al. 2003; CARTHAGENE, deGivry et al. 2005), and those relying on two-point approaches, which achieve much greater speed by using only pairwise recombination estimates (RECORD, van Os et al. 2005; OneMap, Margarido et al. 2007; MSTmap, Wu et al. 2008; Lep-MAP, Rastas et al. 2013; HighMap, Liu et al. 2014). The gain in accuracy from multipoint approaches must therefore be balanced against the accompanying computational burden.
The recent increases in genotyping throughput have made high-density genetic maps increasingly valuable, both in fine-mapping trait associations and as anchors for physical maps in progress toward full sequence assembly. However, this increase has also resulted in a number of problems for map construction and ensuing analyses. First, the resolution and coverage of the map are limited by the number of individuals and design of the population. Second, the computational burden of mapping thousands of markers per chromosome limits analysis to the fast two-point methods (Wu et al. 2007; Speed and Zhao 2008; Rastas et al. 2013). Hence, although we typically have low power to distinguish between marker locations when performing high-density mapping, we also rarely have information about uncertainty attached to the resulting map. Indeed, although simulations have shown that map misspecification can bias estimates or reduce power in subsequent analyses such as QTL mapping (Daw et al. 2000), the uncertainty in their estimation is rarely taken into account (Matise et al. 2007). Identification of weak areas of a genetic map could help in avoiding spurious results in further analysis or draw attention to regions that should be analyzed more thoroughly. Since regions of high marker density are likely to correspond to those with greater uncertainty, accounting for the map uncertainty is crucial for appropriate use of high-density maps.
For the first of these issues, multiparental crosses offer a solution by increasing the genetic diversity and opportunities for recombination in the population. In particular, multiparent advanced generation inter-cross (MAGIC) results in a large population of inbred lines with a large number of recombination events accumulated throughout the generations of the pedigree. These populations were proposed as a compromise between advanced inter-crosses (Darvasi and Soller 1995) and heterogeneous stock populations (Mott et al. 2000), combining the creation of highly recombinant inbred lines from the first design with the larger and more diverse set of founders from the second.
MAGIC populations have been created in several plants, including model plant Arabidopsis thaliana (Kover et al. 2009), model crop rice (Bandillo et al. 2013), and unsequenced crops such as wheat (Huang et al. 2012). They have been successfully used as mapping populations (Huang et al. 2012) and in conjunction with biparental populations to produce a high-density consensus map in bread wheat (Cavanagh et al. 2013). In these studies, the authors found that not only could more markers be mapped than in individual biparental populations, but the multiparental map often allowed linkage groups to be joined together in the consensus map since the increased diversity enabled greater coverage of markers across the genome.
Regarding the second issue of statistical confidence in genetic maps, two classes of methods have been considered. This again reflects the difference between more accurate multipoint methods and methods based on fewer markers, which are more practical for high-throughput data. Several packages for linkage map construction include functions to compare orders for a set of markers, either based on the multipoint likelihood or the number of crossovers (Broman et al. 2003; Margarido et al. 2007). Other confidence measures have considered analysis of the whole map through Bayesian or bootstrap approaches (Servin et al. 2010; Ronin et al. 2010). However, these can be very computationally demanding, and in practice are used to refine the order rather to indicate regions of low confidence. As the computational burden typically limits comparison of likelihoods to smaller subregions, DeWan et al. (2002) proposed an order support score examining the likelihood ratio of the two most likely orders within a triplet of markers. More recently Gilks et al. (2012) proposed a Bayesian method for estimating the uncertainty in triplets of markers, but this can be applied only to populations of biparental inbred lines where each marker segregates with equal probability. In particular, it is not applicable to multiparental populations.
In light of the potential of multiparental populations for high-density map construction and the limits of current approaches, we developed a novel sliding window approach to assessing map uncertainty in such populations. For a triplet of markers, we derive the asymptotic distribution of the pairwise recombination fraction matrix between the markers. Given an estimated map, we can then use this distribution to construct an uncertainty measure based on a test of whether the data agree with the current order and marker distances. We compare this to the support score method for triplets through simulation to demonstrate the ability of each method to highlight regions where markers may be misordered during the map construction process. Finally, we apply our approach to a map constructed from a four-parent wheat MAGIC population genotyped at high density and validate the resulting uncertain areas against genome sequence.
Materials and Methods
Preliminaries
Mapping:
We can establish a general statistical framework for genetic mapping. Assume that we have a set of K arbitrarily ordered genetic markers {M1, M2, … MK}. Let θ represent the matrix of true recombination fractions between each pair of markers. Element θjk gives the true recombination fraction between markers j and k. Instead of specifying a map via a matrix of recombination fractions, a common alternative is to specify the order of the markers and the recombination fractions between the adjacent markers (George 2005). We let δ = (δ1, …, δK)T represent the true order of the markers, which is a permutation of (1, …, K). Let θadjacent be a vector (of length K − 1) of the adjacent recombination fractions, where the kth element corresponds to . Then our true genetic map M can be represented by the set of parameters {δ, θadjacent}, and the mapping task can be framed as estimating these two quantities.
We assume that crossovers occur as a homogeneous Poisson process leading to the use of Haldane’s map function (Haldane 1919) to convert between recombination fractions and genetic distance (Zhao and Speed 1996).
MAGIC populations:
While a variety of designs can be broadly categorized as MAGIC, we initially focus here on a simple version of the MAGIC design. We define a general h-parent MAGIC population, where h can be written as 2j, as follows. Starting with h inbred founder lines, in each of the first j generations, we form crosses by pairing off individuals under the restriction that each of the h founders can appear at most once in the ancestry of the progeny at any generation. The individuals resulting from the jth generation hence contain equal contributions from each of the h founders. These individuals are then selfed until essentially inbred, typically for six or more generations.
Founder distribution patterns (FDP):
Let X be an N × K-matrix of marker genotype data, where represents the marker genotype at locus j for individual i of the N progeny. While some markers may be multiallelic in practice, we assume that we are dealing with the most common scenario, where we have genotyped biallelic SNPs. We write the two alleles as 0 and 1; hence for all i and j. We also define the haplotype at two markers j and k for the ith individual as . Let G be the corresponding unobserved matrix of the originating founders for each of the progeny alleles. In the case of a four-parent cross, and the set of true founder haplotypes is , where . Knowing the founder genotypes and the founder origin of each progeny genotype is sufficient to construct the progeny genotypes.
We refer to the pattern (vector) of observed alleles in the h founders at marker j as the founder distribution pattern (FDP), denoted by fj. Then the complete matrix of FDPs for all markers is denoted by F, where the columns of F are the K FDPs. For biallelic markers in a four-parent population, all FDPs will match one of the seven listed in Table 1, swapping allele labels if necessary. Similarly, for an eight-parent population there are 127 possible FDPs, and for a general h-parent MAGIC cross, there would be possibilities.
Table 1. Possible founder distribution patterns (FDP) for biallelic markers in a four-parent MAGIC population.
| Founder | f1 | f2 | f3 | f4 | f5 | f6 | f7 |
|---|---|---|---|---|---|---|---|
| A | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| B | 1 | 1 | 0 | 1 | 1 | 0 | 0 |
| C | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| D | 0 | 1 | 1 | 1 | 0 | 0 | 1 |
| % | 13.7 | 17.5 | 15.2 | 26.1 | 5.3 | 8.9 | 13.2 |
The last row (%) indicates the observed percentages of each FDP among 4606 biallelic SNPs mapped in the wheat four-parent MAGIC population.
As we consider pairs of markers in map construction, we define two important quantities based on pairs of FDPs. When observing biallelic markers in a multiparental population, different originating founder haplotypes can result in the same observed biallelic haplotype. Let , which is the set of all biallelic haplotypes for two markers j and k. Then and , respectively, can be defined as the number of nonrecombinant and recombinant elements of T, which result in a specific value of a. For example, if the FDP at marker j is (0 1 1 1)T, and the FDP at marker k is (1 0 1 0)T, then and , while and . Although there are 21 possible pairwise combinations of the seven FDPs for a four-parent MAGIC, the unordered values of and for each pair reduce to five classes, enumerated in Table 2.
Table 2. Possible coefficient classes in a four-parent MAGIC for two biallelic loci j and k.
| Class 1 | Class 2 | Class 3 | Class 4 | Class 5 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| a | ||||||||||
| 00 | 2 | 2 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 3 |
| 01 | 0 | 4 | 0 | 3 | 0 | 2 | 1 | 2 | 1 | 3 |
| 10 | 0 | 4 | 0 | 3 | 1 | 5 | 1 | 2 | 1 | 3 |
| 11 | 2 | 2 | 3 | 6 | 2 | 4 | 2 | 7 | 1 | 3 |
| % | 2.8 | 2.8 | 14.1 | 14.1 | 39.9 | 39.9 | 38.5 | 38.5 | 4.7 | 4.7 |
and are the number of nonrecombinant and recombinant phase-known two-marker haplotypes (respectively) that are represented by an observed haplotype of a. Note that the labels for a may be permuted and result in the same variance class for recombination fraction estimates between the two loci. The last row (%) indicates the percentage of pairwise combinations of 4606 SNPs mapped in wheat four-parent MAGIC populations that fall into each class.
Asymptotic Distribution
Likelihood:
For high-density data, map estimation is typically carried out on the basis of the two-point estimates of the recombination fractions (Cheema and Dicks 2009), primarily due to computational issues. Fundamentally, map uncertainty is due to the sampling error attached to these estimates, which will propagate through to the final map estimate. While the marginal distribution of the recombination fraction estimators have been studied (Neumann 1990; Martin and Hospital 2006; Wu et al. 2007), the joint distribution has received little attention in the literature. Given the pedigree and the marker data, we can specify a likelihood function . For two loci, we can write this in terms of quantities previously defined, using probabilities derived by Broman (2005). For four-parent RILs,
Then the two-locus log-likelihood function can be written as
| (1) |
The fifth class of FDP pairs is a special case that results in a likelihood that does not depend on θjk. In this case, all terms are equal to 1 and all terms are equal to 3 (Table 2). Substituting these values into Equation 1 yields
This no longer depends on and hence cannot be used to estimate the recombination fraction between these pairs. We thus remove the minimal set of markers to avoid including any such pairs in our data prior to map construction. After maximizing the likelihood for all other pairs of markers, we estimate θadjacent by taking a subset from the matrix and derive an order for the markers by minimizing the sum of adjacent recombination fractions (SARF) in θadjacent (Falk 1989).
Composite likelihood:
To derive the asymptotic joint distribution of the pairwise recombination fraction estimates, we approximate the log-likelihood in Equation 1 via a composite log-likelihood in
| (2) |
Composite likelihoods are a special case of misspecified likelihoods, where the full likelihood is approximated by the product of a series of marginal or conditional likelihoods (Varin and Vidoni 2005). Pairwise composite likelihoods have been used previously in statistical genetics to avoid computational issues with high-dimensional data (McVean et al. 2004; Larribe and Fearnhead 2011). However, they have been used primarily for population genetic data in human populations, which differ greatly in structure from inbred line populations. The primary benefit of using this form of the likelihood is that the asymptotic distribution (as sample size goes to infinity) of the maximum composite likelihood estimators is well characterized. Specifically,
| (3) |
where N is the sample size, and
Full asymptotic joint distribution of pairwise recombination fraction estimators:
Assuming that the necessary regularity conditions hold, we derive the asymptotic joint distribution of the pairwise recombination fraction estimate using Equation 3. As each parameter will appear in a single term of the summation in Equation 2, the partial derivatives can be expressed as
| (4), |
where is an indicator variable that takes value one if and zero otherwise. For ease of later reading, let
We begin by computing the covariances between the elements of the score vector to determine V(θ). Suppose we have four markers j, k, l, and m, where some of the markers may overlap (e.g., when we consider the covariance of recombination fraction between j and k with that between j and l)
Note that all the expectations can be replaced by the probabilities of the event occurring, since the product of two indicator functions is also an indicator function. Hence we can write the covariance as
| (5) |
For disjoint intervals, where no markers overlap, this expression contains four-locus probabilities, which are dependent on the order of the four markers. However, V(θ) can be expressed for triplets using only two- and three-locus probabilities, as derived in Broman (2005).
Deriving the Hessian matrix J(θ) is straightforward in comparison. Taking the derivative of Equation 1, we see that
since the expression in brackets depends only on . Hence the off-diagonal elements of the Hessian are 0, and the diagonal elements can be derived from
by taking the negative expectation to get
| (6) |
Equations 5 and 6 are sufficient to calculate V(θ) and J(θ) for an arbitrary number of markers. To consider groups of four or more markers, four-locus probabilities are required, which are difficult to obtain in closed form. By limiting our attention to triplets we can fully specify the covariance matrix using previously published results for these quantities.
We note that in addition to a dependence on sample size and the value of the recombination fraction estimator (and hence the distance between markers), the variance also depends on the FDPs in multiparental populations. The asymptotic variance of the pairwise recombination fraction estimator of is the reciprocal of the Fisher information, given in Equation 6. The and terms result in the dependency of variance of the estimator on the FDPs at markers j and k. We can characterize the classes of FDP pairs in Table 2 according to the variance of the resulting recombination fraction estimator. Pairs of markers contained in the classes described in Table 2 have the same variance for the recombination fraction estimate between the markers, with variance increasing from class 1 to class 4.
Although we focus on a simple version of the MAGIC design here, extensions to more complex designs incorporating additional generations of intercrossing will rely only on the use of appropriate two-loci and three-loci probabilities, many of which have been derived in Broman (2005) and Teuscher and Broman (2007).
Uncertainty Measures
We consider two uncertainty measures derived from the joint asymptotic distribution of the recombination fraction estimates. The first is the probability that the map produced by minimizing the SARF criterion will produce the correct marker order. In addition to providing an estimate of the correctness of the map, we show how to use this probability to estimate the required number of lines to map markers with high certainty under different conditions of marker density and FDPs. The second is a hypothesis test of whether the estimated marker order is correct and as such provides a direct indicator of regions of uncertainty in a map. We compare this in simulation to one other measure of uncertainty, which is briefly described at the end of this section.
Probability of correct order:
We derive an expression for the probability that minimizing SARF in an inbred line genetic mapping experiment will result in a correct estimate of marker order. As a simple example, consider a triplet of markers {X, Y, Z} for which there are three possible orders. Each of these orders results in a possible SARF given by the sum of two entries in the matrix of recombination fraction estimates . For example, the SARF corresponding to the order X–Y–Z is . Let S be a vector containing all SARF corresponding to the possible orders of markers in the map. The key to deriving the probability of correct order (PCO) is to note that S is expressible as an affine transformation of the recombination fractions, written as . Hence an approximation to the distribution of S, based on the asymptotic distribution of given in Equation 4, is
(Wackerly et al. 2008). Now suppose the marker orders are listed in order of increasing SARF, so that the first one is that which minimizes the criterion. The PCO is the probability that Z = S1 − S−1 < 0, where S−1 denotes the vector S with the first entry omitted. However, we note that Z = QS is in turn an affine transformation of S. If s is the number of potential orders, Q can be written as [1s−1 –Is−1], where the two entries denote the vector of ones of length s − 1 and the identity matrix of size s − 1 respectively. Hence , and we define the
| , |
where FZ(z) is the cumulative distribution function of Z. We note that this probability depends on factors such as sample size and marker spacing, as well as FDPs in multiparental populations. While the cumulative distribution function cannot be evaluated analytically, it is straightforward to compute numerically for triplets of markers. However, the dimension of Z grows exponentially with the number of markers, so for larger sets of markers it may be necessary to consider other dimension reduction techniques to reduce the computational burden.
Hypothesis test (MMU):
We define our primary measure of map uncertainty on the basis of the hypothesis test for marker order in triplets of markers X, Y, and Z. We assume that the correct order of the markers is X–Y–Z and write the vector of recombination fractions θ as {θXY, θYZ, θXZ}. Under the assumption that crossovers occur as a Poisson process, crossovers in disjoint intervals are independent, so we can write the third recombination fraction as a function of the other two:
Similarly, each possible order of the three markers gives rise to a different constraint of this form, so we can represent the order of the markers as a nonlinear restriction on the recombination fractions. As the asymptotic distribution of θ is normal, we can construct nonlinear Wald tests based on these formulas (Phillips and Park 1988).
We define our measure of map uncertainty (MMU) as the –log10(P-value) for the test of the null hypothesis that the true order is X–Y–Z. Hence larger values indicate higher uncertainty. This statement is mathematically equivalent to Equation 3; hence our test of the null hypothesis is a Wald test given by
Here and is the estimated variance–covariance matrix of . As we are testing only a single restriction, asymptotically . Here we investigate triplets of markers, but since the estimate of V(θ) based on two- and three-locus probabilities is consistent (Molenberghs and Verbeke 2005, Chap. 9), it could be used to produce confidence intervals and perform hypothesis testing for sets of four or more markers.
Order support score:
We compare our measure to the order support score (OSS) previously proposed by DeWan et al. (2002). They compare the most likely order of a triplet against the second most likely order. The OSS is defined by Keats et al. (1991) as the log-ratio of the likelihood under the two proposed orders. Dewan et al. (2002) define as uncertain those regions where the OSS is less than three and propose removing these markers from the map. We convert the OSS to the same scale as our measures by extending its definition using the Vuong closeness test, which is a likelihood-ratio-based test for comparing two models (Vuong 1989).
For a triplet of arbitrarily ordered markers j, k, and l, we can formulate the test as follows. Let be the maximum-likelihood estimates of the recombination fractions under the first marker order , and let be the maximum-likelihood estimates of the recombination fractions under the second marker order . Let LR be the ratio of the likelihoods under the two models. The Vuong test statistic is given by, whereis the sample variance of the pointwise log-likelihood ratios under each model. To determine a P-value for this test, the statistic is compared to the standard normal distribution.
Data
Simulation studies:
To assess the accuracy of the approximate asymptotic distribution and the performance of our estimation approach we conducted 2000 simulations of four-parent MAGIC populations. In each simulation we generated lines from a four-parent MAGIC pedigree selfed to fixation, using a genetic map with a triplet of equally spaced markers. We considered the effects of varying marker density, sample size, and founder distribution patterns. We varied marker density from 0.5 to 5 cM; population sizes from 500 to 1500; and selected combinations of FDPs with low, medium, and high variances (Supporting Information, Table S1). These variance categories depend on the FDPs for each of the three pairs of markers contained in the triplet and are relative to other possible combinations of the FDP classes categorized in Table 2. The overall variance matrix for the three markers will depend on all of the factors simulated (e.g., marker density, population size, and FDP). For each of these combinations we computed the PCO to determine how much precision was achievable in mapping for given scenarios of markers and population sizes.
For our second proposed measure, the hypothesis test, we undertook a simulation study to estimate its power. As above, we examined the impact of marker density, population size, and founder distribution pattern. In each replicate we calculated the proposed test statistic, using a null hypothesis while generating data under the alternative order X–Z–Y. The power estimate was then the proportion of times that the null hypothesis was rejected.
All simulations were performed using functions from R/qtl (Broman et al. 2003) and scripts written in the R language (R Core Team 2013). Code to generate all simulations can be found in File S1.
Wheat MAGIC:
We estimate uncertainty attached to a map constructed from a four-parent wheat MAGIC population described by Huang et al. (2012). Genotypes from 1088 lines derived from the four parents were collected using the 9K SNP chip (Cavanagh et al. 2013), with 4606 biallelic markers mapped. We focus on chromosome 3B, as the availability of its assembled sequence provides a standard for validation. Genotypes for these 207 markers can be found in File S2 and the analysis script in File S3. For triplet analysis, a representative marker from each set of markers with the same map position was used, resulting in 111 triplets. We estimate uncertainty using the MMU and the OSS and compare regions identified as uncertain to those where the genetic map is inconsistent with the physical map.
To identify regions of inconsistency between the maps, we aligned 362 putative marker sequences from the 9K consensus map for chromosome 3B (Cavanagh et al. 2013, http://www.pnas.org/content/suppl/2013/04/29/1217133110.DCSupplemental/sd03.xls) to the targeted pseudomolecule traes3bPseudomoleculeV1 (http://wheat-urgi.versailles.inra.fr/projects/3bseq, released January 2014, Choulet et al. 2014). Alignment was performed using Biokanga (release 2.97.1, http://sourceforge.net/projects/biokanga/files/) at two levels of stringency. For each marker, two sequences containing, respectively, one variation of the allelic SNP locus were generated, and these sequences were then independently aligned to the targeted 3B pseudomolecule. First, we allowed maximal stringency with zero substitutions; alignments reported at this level of stringency are to loci that match one allele of the target SNP sequence. We then performed a further alignment, relaxing the stringency and allowing at most one substitution per 100 bp of the individual marker sequence. In this case, reported accepted alignments are those where there is an assembly specific variant relative to the marker sequence, which may be at the known allelic base site or at a novel site(s) additional to the known allelic site.
Results and Discussion
Results
Simulation studies:
We performed simulation studies to assess the efficacy of the uncertainty measures in predicting regions of the genome, which are mapped incorrectly. This additionally provides guidelines for FDPs and marker densities that are likely to prove most difficult for map construction.
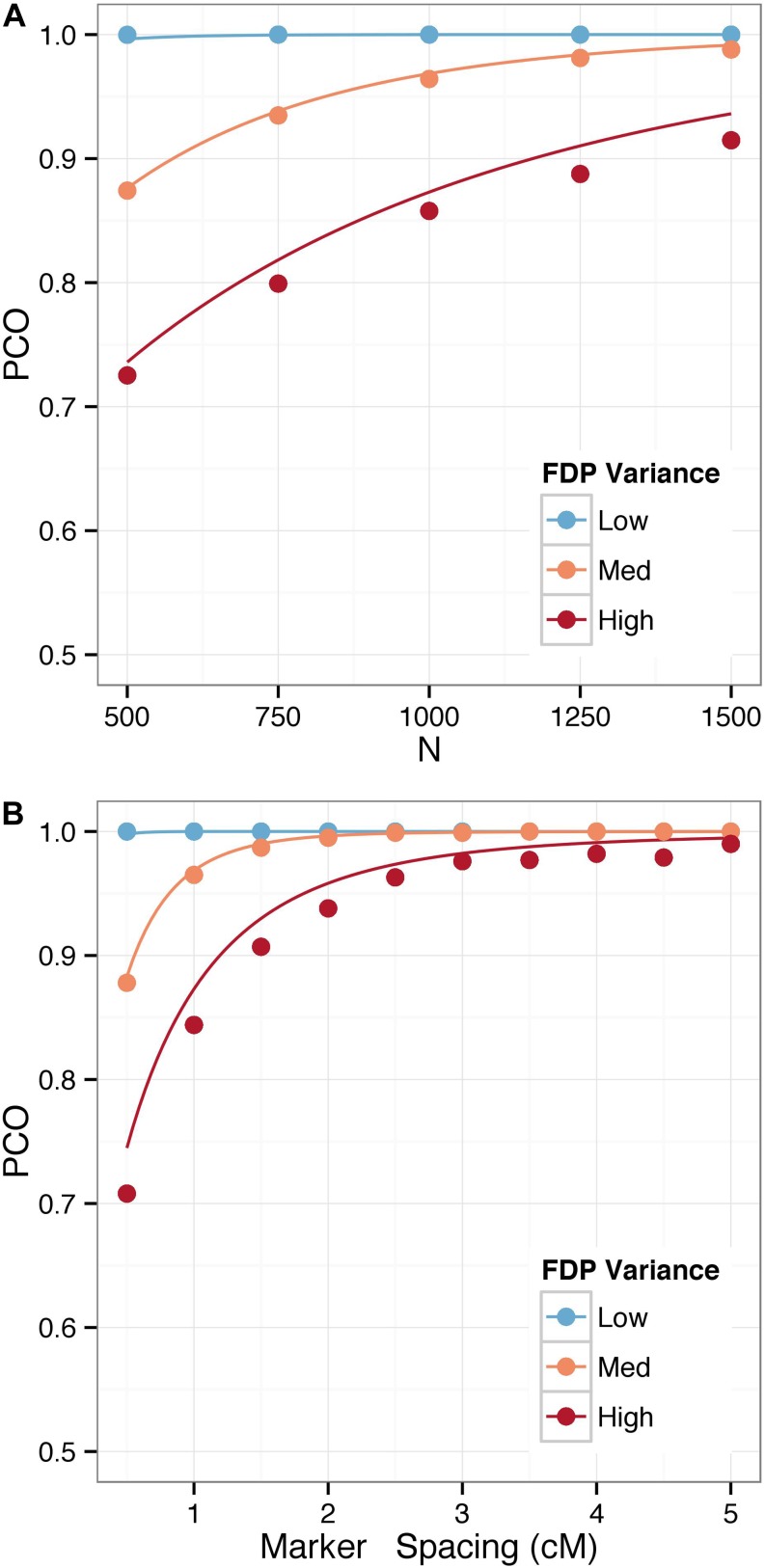
We consider the performance of our approach in typical map construction scenarios in Figure 1 and Figure 2. We vary marker density, sample size, and FDPs for a triplet of markers and estimate the PCO when constructing a map by minimizing the SARF criterion. We see that the empirical estimates match very closely with the theoretical values derived above and, further, that except in cases of high variance FDPs, small sample sizes, and very dense markers, we expect that markers will typically be ordered correctly. In particular, the probability of correctly ordering a triplet of markers when the marker spacing is >2 cM is high for almost any FDP. For sample sizes >1000 lines, the probability is well above 0.9 for all scenarios.
Figure 1.
Probability of correct order (PCO) for a triplet of markers generated from a four-way population with (A) varying sample size for fixed marker spacing of 1 cM and (B) varying marker spacing for fixed sample size of 1000. Lines denote the theoretical values of the PCO, while circles denote the empirical values.
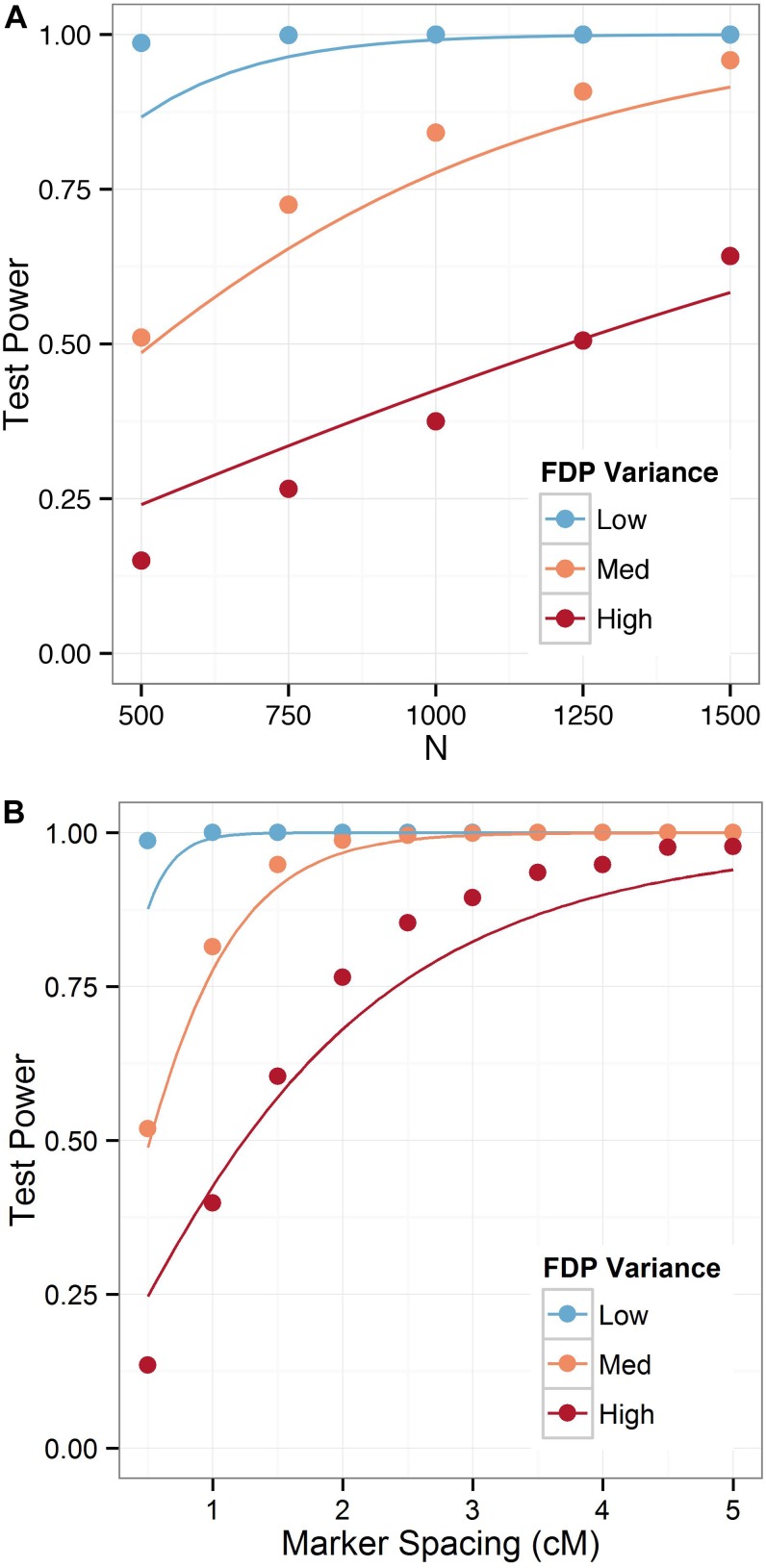
Figure 2.
Power for the hypothesis test that the estimated map is superior to the other two triplet orders for a four-way population with (A) varying sample size for fixed marker spacing of 1 cM and (B) varying marker spacing for a fixed population size of 1000. Lines denote the theoretical values of the power, while circles denote the empirical values.
We also consider the power of the test across various population sizes with a fixed intermarker spacing of 1 cM, and under varying marker density, with a fixed population size of 1000 lines. These two situations are illustrated in Figure 2, A and B, respectively. For each, we plot the theoretical power of the test (solid line), along with the power estimated from simulations (points) for triplets of markers with low, medium, and high variance FDPs. There are a number of factors that influence the power. Since the PCO rises with increasing intermarker separation and with sample size, the number of markers that are misordered falls, and hence so does the importance of statistical power. However, the power also increases with increasing intermarker separation and with sample size. The theoretical curves converge toward 1 with a sigmoid shape. A majority of triplets will resemble the medium or high FDP variance cases (Table 2). The power of the test is >80% when N = 1000 and the intermarker spacing is at least 2.5 cM. The power for the worst (high-variance) FDP improves almost linearly with increased sample size for N near 1000.
Wheat MAGIC:
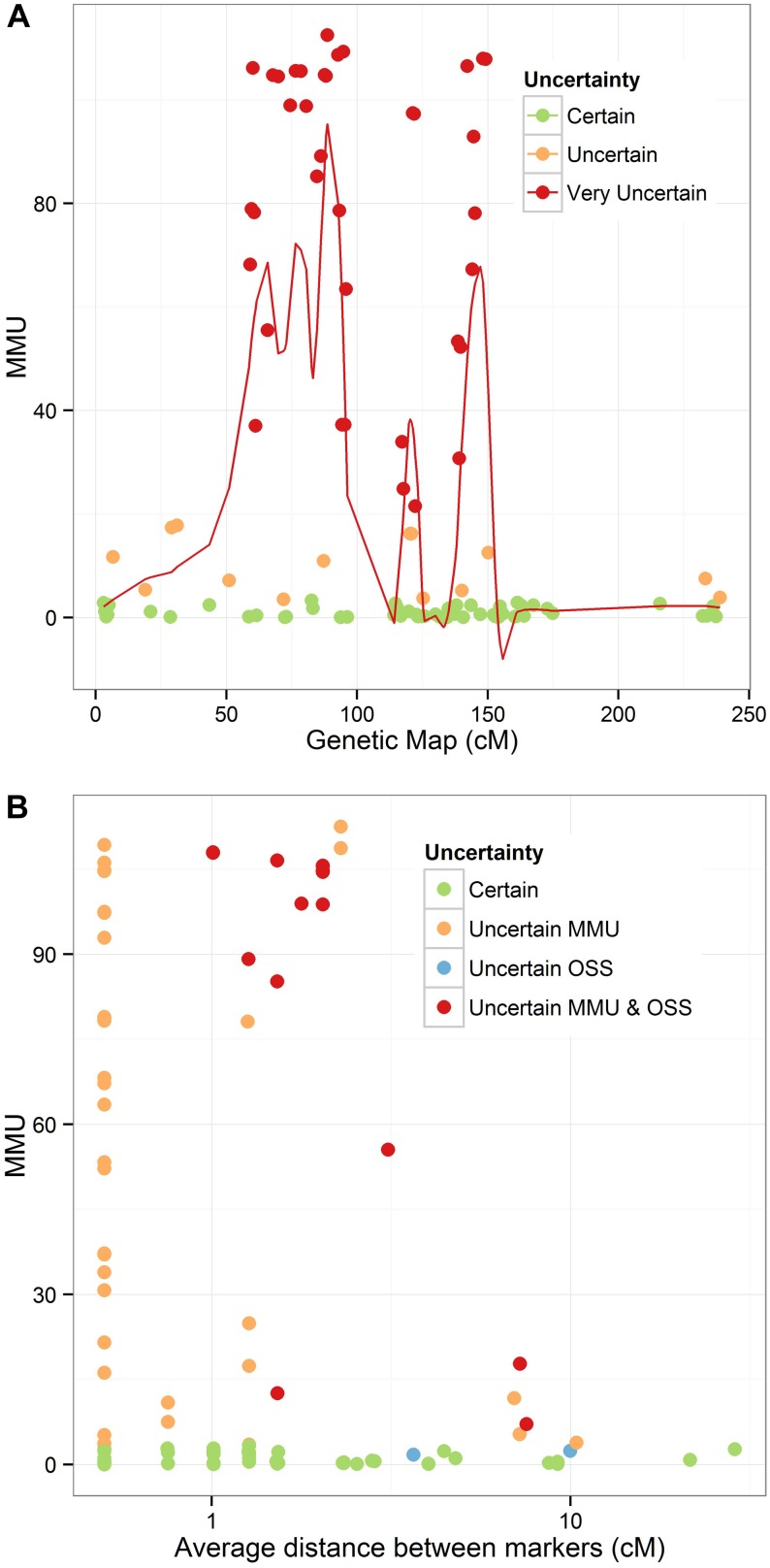
We calculated measures of uncertainty for the map based on the 9K SNP chip in wheat (Cavanagh et al. 2013). In particular, we focused on chromosome 3B (Figure 3A), as the assembled sequence was recently publicly released, and it is the only chromosome for which this is currently available. In Figure 3B, we compare the P-values from our proposed hypothesis test with those from the OSS for 111 triplets. We note that for most triplets labeled as uncertain by our test, the markers are very dense. Of those triplets where the average spacing between markers was <1 cM, 52% are labeled uncertain, indicating the difficulty in ordering these markers unambiguously. The OSS, in contrast, does not label these triplets as uncertain. Only two markers are indicated to be uncertain by the OSS, which do not have high MMU; given that these markers are spaced at >5 cM from the nearest marker and the population size is quite large, our simulations indicate that it is unlikely that these were actually misordered.
Figure 3.
Measures of uncertainty for chromosome 3B. (A) Measure of map uncertainty (MMU), calculated as –log10(P-value) for hypothesis test that the true order is the given map order, for all triplets on the chromosome. Red line denotes a LOESS fit to the MMU across the chromosome, with span of 0.15. (B) Dependency of MMU and –log10(P) for order support score (OSS) on distance to closest marker (log-scale). Points are denoted uncertain if Bonferroni-adjusted P-value from respective tests is <0.05. Points are denoted as very uncertain if the MMU > 20.
To assess our ability to identify uncertain regions in a genetic map, we compared the genetic order of markers in the wheat MAGIC map to physical order. While we do not expect the genetic and physical positions to be linearly related, we would expect the orders to correspond in the case that marker sequence for a location on the genetic map could be uniquely and accurately mapped to a physical position. Fewer than half of 362 markers mapped to chromosome 3B in previous genetic maps (Cavanagh et al. 2013) could be aligned to the pseudomolecule sequence. At the highest level of stringency (zero mismatches allowed), only 56 markers could be aligned; this increased to 147 upon allowing a single mismatch per 100 bp (Table S2). For the MAGIC wheat population, this resulted in 30 triplets being omitted due to lack of information on physical position.
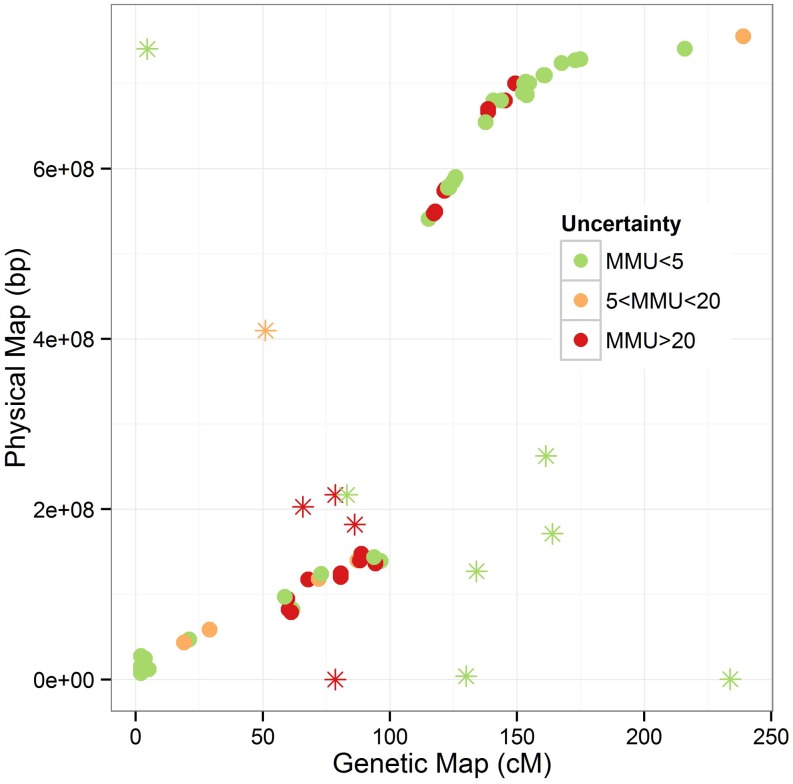
When we compare the genetic map with the physical positions, we identify a number of regions worthy of further investigation (Figure 4). First, we note a large region of markers with high MMU; these are likely centromeric, where very low recombination rates may result in small genetic map distances corresponding to large physical distances. In this region, two markers are identified as having high uncertainty that does not align well with the physical map; the general level of uncertainty in this region indicates that these may have been misordered. Second, we note several markers whose genetic map positions are inconsistent with physical positions. Apart from the two regions already mentioned, none of these are identified as uncertain (high MMU) by our method.
Figure 4.
Genetic and physical positions of 81 markers aligned to chromosome 3B. Points are denoted uncertain if the test that the true order matches the given map order yields a Bonferroni-adjusted P-value <0.05. Points are denoted as very uncertain if the MMU, calculated as –log10(P-value) for the test, >20. Markers whose genetic map position is inconsistent with physical position are denoted by asterisks.
Where possible, we compared the genetic map positions of these markers in the MAGIC map to those in two other biparental mapping populations that had >80 markers aligned to the pseudomolecule (Synthetic × Opata and Gladius × Drysdale; Cavanagh et al. 2013) to determine whether this was likely to be an issue with the MAGIC map. Of the 12 markers indicated to have alignment issues, 4 were not mapped in the other populations; 4 were relatively consistent with physical positions in at least one of the other populations; and 4 showed the same inconsistencies as in the MAGIC map. These last 4 likely indicate issues either with the alignment of the marker sequence to the pseudomolecule or with the physical positions themselves; all were labeled as low uncertainty by our approach. They include the markers with the most drastic differences from physical position (genetic positions at opposite ends of the chromosome). In contrast the middle four markers, which may indicate issues with the MAGIC map, are all located in the centromeric region; 3 of 4 were labeled as highly uncertain (P < 1e-20) in our analysis.
Discussion
We have used the asymptotic distribution of recombination fraction estimators to derive two measures of map uncertainty in the context of multiparental populations. This has not been explored even in biparental populations, as previous attempts to characterize this relationship have been based on simulation studies (Wu et al. 2003; Mollinari et al. 2009) rather than on an analytical approach. We focus on multiparental populations as they offer great promise for high-density mapping and as such uncertainty measures are crucial. However, the measures we define could be used equally well on biparental populations.
Simulations of maps and accompanying uncertainty measures can provide guidelines for map construction in multiparental populations. We note that for populations of size 500 or less, which have been generated in several plants, the only markers that can be reliably ordered when very tightly linked are those whose FDPs fall into the low variance category. In general, the high-variance category of FDP combinations will have the worst performance for ordering and hence highest uncertainty. For marker spacings >2 cM, however, most markers can be confidently ordered using the SARF criterion. Once populations have size >1000, the recombination fractions can be estimated with sufficient precision to order most markers spaced 1 cM apart, and almost all which have >3- to 5-cM spacing.
The FDPs have a major influence on map construction in multiparental populations. This represents the fact that the use of primarily biallelic SNPs in a multiallelic system may introduce significant uncertainty (Weir et al. 2006) and lower marker informativeness, both of which affect the ability to order markers (Wu et al. 2011). Leal (2003) performed a simulation study comparing the variance of recombination fraction estimators under biallelic markers and multiallelic markers in a population genetics scenario. Leal found that the recombination fraction estimates based on biallelic markers had a higher variance than the estimates from the multiallelic markers, which affected the effectiveness of subsequent QTL detection.
As our method relies on the recombination fractions between markers, it is subject to the fallacies of the MAGIC design that result in certain values being nonidentifiable. To simplify our computation, we removed markers for which this would be an issue prior to map construction. In practice this results in a loss of ∼15% of markers, although the exact value will depend on founder distribution patterns. These markers could be added to the map by methods imputing recombination fractions or relying on multipoint probabilities, but to assess the uncertainty in their vicinity will require further investigation. However, any method utilizing pairwise recombination fractions would be subject to this same issue.
One of the measures we derive estimates the probability of ordering markers correctly by minimizing the SARF criterion. While many algorithms determine marker order, minimizing SARF has been shown to perform well as a heuristic (Olson and Boehnke 1990; Wu et al. 2003; Mollinari et al. 2009). Since many methods have similar performance, we do not expect the probability derived for SARF to vary greatly if an alternative is used for map construction. If anything, the PCO should increase for more sophisticated approaches. The other measure is a hypothesis test comparing the currently estimated map order against alternate orders. As this will be calculated across the genome, there is a need to correct for multiple testing when interpreting which regions are uncertain in a map. A Bonferroni correction will suffice to identify the most problematic markers, but given that adjacent triplets contain overlapping markers, this will be overly stringent.
While there are a number of limitations to focusing on triplets of markers, this is a practical division of the genome for high-density maps. Bootstrap approaches such as those in Mester et al. (2003) and Matise et al. (2007) would be computationally prohibitive for maps containing hundreds of thousands of markers, constructed from thousands of individuals. Other groups, such as Gilks et al. (2012), have also investigated the use of triplets. However, this has been derived solely for biparental inbred line populations and requires more investigation to generalize fully to multiparental populations in an efficient manner. In practice, the use of triplets will most easily identify pairwise flips of markers, and markers with larger-scale rearrangements will show a complex pattern of uncertainty. However, from our simulations we have seen that markers spaced over larger distances can be ordered with high accuracy, so considering subsets of markers in triplets may provide additional information about this type of misordering. Further, we plan to extend our approach to an arbitrary number of markers by computing four-locus probabilities. As noted in the composite likelihood section, the formulation of the asymptotic distribution depends only on these probabilities even for arbitrary numbers of markers, so the only limitation once this has been done will be due to computational power. In contrast, extending Gilks’ Bayesian approach, which is based on posterior probabilities, to windows of more than three markers would be very difficult, since its efficacy is dependent on a closed-form expression for the likelihood.
Ideally, measures of uncertainty could be incorporated into downstream analyses directly, but even without doing so they provide useful information for verification of QTL mapping. At a basic level, identifying regions of uncertainty in the map may assist in improving the overall order and hence mapping accuracy. However, it is quite likely that errors in marker ordering will occur, particularly at sub-centimorgan distances. Rather than removing the markers with uncertainty and losing information for downstream analyses it is important to consider the measure of uncertainty (Figure 4) alongside QTL profiles. This will be particularly important in situations where accuracy of the map is vital, such as in targeting regions for fine mapping or utilizing markers in selection. Further, the uncertainty may provide an extra measure of verification in comparing QTL across multiple traits and/or environments to determine whether they represent pleiotropy or multiple effects in a small interval.
Supplementary Material
Acknowledgments
Many thanks to Andrew George and Hien Nguyen for helpful discussions and comments. Daniel Ahfock was supported by an Australian Grains Research and Development Corporation Grains Industry Undergraduate Honours Scholarship (UHS10485). Dr. Huang is the recipient of an Australian Research Council Discovery Early Career Research Award (DE120101127).
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.167577/-/DC1
Communicating editor: R. W. Doerge
Literature cited
- Bandillo N., Raghavan C., Muyco P. A., Sevilla M. A. L., Lobina I. T., et al. , 2013. Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K., 2005. The genomes of recombinant inbred lines. Genetics 169: 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Buetow K., Chakravarti A., 1987. Multipoint gene mapping using seriation. Am. J. Hum. Genet. 41(2): 189–201 [PMC free article] [PubMed] [Google Scholar]
- Bult C. J., Eppig J. T., Kadin J. A., Richardson J. E., Blake J. A., 2008. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 36: D724–D728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh C., Chao S., Wang S., Huang B. E., Stephen S., et al. , 2013. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. USA 110: 8057–8062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheema J., Dicks J., 2009. Computational approaches and software tools for genetic linkage map estimation in plants. Brief. Bioinform. 10(6): 595–608 [DOI] [PubMed] [Google Scholar]
- Choulet F., Alberti A., Theil S., Glover N., Barbe V., et al. , 2014. Structural and functional partitioning of bread wheat chromosome 3B. Science 345: 1249721. [DOI] [PubMed] [Google Scholar]
- Chowdhary B. P., Raudsepp T., 2006. The horse genome. Genome Dyn. 2: 97–110 [DOI] [PubMed] [Google Scholar]
- Cox A., Ackert-Bicknell C. L., Dumont B. L., Ding Y., Bell J. T., et al. , 2009. A new standard genetic map for the laboratory mouse. Genetics 182: 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi A., Soller M., 1995. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141: 1199–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daw E. W., Thompson E. A., Wijsman E. M., 2000. Bias in multipoint linkage analysis arising from map misspecification. Genet. Epidemiol. 19: 366–380 [DOI] [PubMed] [Google Scholar]
- de Givry S., Bouchez M., Chabrier P., Milan D., Schiex T., 2005. CARTHAGENE: multipopulation integrated genetic and radiated hybrid mapping. Bioinformatics 21: 1703–1704 [DOI] [PubMed] [Google Scholar]
- DeWan A. T., Parrado A. R., Matise T. C., Leal S. M., 2002. Map error reduction: using genetic and sequence-based physical maps to order closely linked markers. Hum. Hered. 54(1): 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich W. F., Miller J. C., Steen R., Merchant M. A., Damron-Boles D., et al. , 1996. A comprehensive genetic map of the mouse genome. Nature 380: 149–152 [DOI] [PubMed] [Google Scholar]
- Falk C., 1989. A simple scheme for preliminary ordering of multiple loci: application to 45 CF families. Prog. Clin. Biol. Res. 329: 17–22 [PubMed] [Google Scholar]
- George A., 2005. A novel Markov chain Monte Carlo approach for constructing accurate meiotic maps. Genetics 171: 791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks W., Welham S. J., Wang J., Clark S., King G. J., 2012. Three-point appraisal of genetic linkage maps. Theor. Appl. Genet. 125(7): 1393–1402 [DOI] [PubMed] [Google Scholar]
- Green P., Falls K., Crooks S., 1990. Documentation for CRI-MAP, version 2.4. Washington University School of Medicine, St. Louis, MO [Google Scholar]
- Haldane J., 1919. The combination of linkage values, and the calculation of distances between the loci of linked factors. J. Genet. 8: 299–309 [Google Scholar]
- Huang B. E., George A. W., Forrest K. L., Kilian A., Hayden M. J., et al. , 2012. A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol. J. 10: 826–839 [DOI] [PubMed] [Google Scholar]
- Iwata H., Ninomiya S., 2006. AntMap:constructing genetic linkage maps using an ant colony optimization algorithm. Breed. Sci. 56(4): 371–377 [Google Scholar]
- Keats B. J., Sherman S. L., Morton N. E., Robson E. B., Buetow K. H., et al. , 1991. Guidelines for human linkage maps an international system for human linkage maps. Ann. Hum. Genet. 55(1): 1–6 [DOI] [PubMed] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., et al. , 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5(7): e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Green P., Abrahamson J., Barlow A., Daly M. J., et al. , 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181 [DOI] [PubMed] [Google Scholar]
- Larribe F., Fearnhead P., 2011. On composite likelihoods in statistical genetics. Stat. Sin. 21: 43–69 [Google Scholar]
- Leal S., 2003. Genetic maps of microsatellite and single-nucleotide polymorphism markers: Are the distances accurate? Genet. Epidemiol. 24(4): 243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Ma C., Hong W., Huang L., Liu M., et al. , 2014. Construction and analysis of high-density linkage map using high-throughput sequencing data. PLoS ONE 9(6): e98855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margarido G., Souva S., Garcia A., 2007. OneMap: software for genetic mapping in outcrossing species. Hereditas 144: 78–79 [DOI] [PubMed] [Google Scholar]
- Martin O. C., Hospital F., 2006. Two-and three-locus tests for linkage analysis using recombinant inbred lines. Genetics 173(1): 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise T., Chen F., Chen W., Francisco M., Hansen M., et al. , 2007. A second generation combined linkage–physical map of the human genome. Genome Res. 17(12): 1783–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. A., Myers S. R., Hunt S., Deloukas P., Bentley D. R., et al. , 2004. The finescale structure of recombination rate variation in the human genome. Science 304: 581–584 [DOI] [PubMed] [Google Scholar]
- Mester D., Ronin Y., Minkov D., Nevo E., Korol A., 2003. Constructing large-scale genetic maps using an evolutionary strategy algorithm. Genetics 165: 2269–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenberghs G., Verbeke G., 2005. Models for Discrete Longitudinal Data. Springer, New York [Google Scholar]
- Mollinari M., Margarido G., Vencovsky R., Garcia A., 2009. Evaluation of algorithms used to order markers on genetic maps. Heredity 103(6): 494–502 [DOI] [PubMed] [Google Scholar]
- Mott R., Talbot C. J., Turri M. G., Collins A. C., Flint J., 2000. A new method for fine-mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. USA 97: 12649–12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. C., Buetow K. H., Weber J. L., Ludwigsen S., Scherpbier-Heddema T., et al. , 1994. A comprehensive human linkage map with centimorgan density: Cooperative Human Linkage Center (CHLC). Science 265: 2049–2054 [DOI] [PubMed] [Google Scholar]
- Nascimento M., Cruz C., Peternelli L., Campana A., 2010. Comparison between simulated annealing algorithms and rapid chain delineation in the construction of genetic maps. Genet. Mol. Biol. 33(2): 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P., 1990. Two-locus linkage analysis using recombinant inbred strains and Bayes’ theorem. Genetics 126: 277–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson J. M., Boehnke M., 1990. Monte Carlo comparison of preliminary methods for ordering multiple genetic loci. Am. J. Hum. Genet. 47(3): 470. [PMC free article] [PubMed] [Google Scholar]
- Phillips P. C., Park J. Y., 1988. On the formulation of wald tests of nonlinear restrictions. Econometrica 56(5): 1065–1083 [Google Scholar]
- Poland J. A., Brown P. J., Sorrells M. E., Jannink J.-L., 2012. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 7(2): e32253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2013. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Rastas P., Paulin L., Hanski I., Lehtonen R., Auvinen P., 2013. Lep-MAP: fast and accurate linkage map construction for large SNP datasets. Bioinformatics 29: 3128–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronin Y., Mester D., Minkov D., Korol A., 2010. Building reliable genetic maps: different mapping strategies may result in different maps. Natl. Sci. 2: 576–589 [Google Scholar]
- Servin B., de Givry S., Faraut T., 2010. Statistical confidence measures for genome maps: application to the validation of genome assemblies. Bioinformatics 26: 3035–3042 [DOI] [PubMed] [Google Scholar]
- Speed T., Zhao H., 2008. Chromosome maps, pp. 1–39 in Handbook of Statistical Genetics, Ed. 3, edited by Balding D., Bishop M., Cannings C. Wiley, Hoboken, NJ [Google Scholar]
- Stam P., 1993. Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J. 3: 739–744 [Google Scholar]
- Teuscher F., Broman K. W., 2007. Haplotype probabilities for multiple-strain recombinant inbred lines. Genetics 175: 1267–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen J., 2011. Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet. Res. 93: 343–349 [DOI] [PubMed] [Google Scholar]
- Van Os H., Stam P., Visser R. G. F., van Eeck H. J., 2005. RECORD: a novel method for ordering loci on a genetic linkage map. Theor. Appl. Genet. 112: 30–40 [DOI] [PubMed] [Google Scholar]
- Varin C., Vidoni P., 2005. A note on composite likelihood inference and model selection. Biometrika 92: 519–528 [Google Scholar]
- Vuong Q. H., 1989. Likelihood ratio tests for model selection and non-nested hypotheses. Econometrica 57: 307–333 [Google Scholar]
- Wackerly D., Mendenhall W., Scheaffer R., 2008. Mathematical Statistics with Applications, Ed. 7 Cengage Learning, Stamford, CA [Google Scholar]
- Wang S., Wong D., Forrest K., Allen A., Chao S., et al. , 2014. Characterization of polyploidy wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. DOI: .10.1111/pbi.12183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. A., Bhangoo J., Fernandez-Fernandez F., Moore P., Swanson J. D., et al. , 2013. Saturated linkage map construction in Rubus idaeus using genotyping by sequencing and genome-independent imputation. BMC Genomics 14: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B., Anderson A., Hepler A., 2006. Genetic relatedness analysis: modern data and new challenges. Nat. Rev. Genet. 7: 771–780 [DOI] [PubMed] [Google Scholar]
- Wong A. K., Ruhe A. L., Dumont B. L., Robertson K. R., Guerrero G., et al. , 2010. A comprehensive linkage map of the dog genome. Genetics 184: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Jenkins J., Zhu J., McCarty J., Jr, Watson C., 2003. Monte Carlo simulations on marker grouping and ordering. Theor. Appl. Genet. 107: 568–573 [DOI] [PubMed] [Google Scholar]
- Wu J., Jenkins J. N., McCarty J. C., Lou X.-Y., 2011. Comparisons of four approximation algorithms for large-scale linkage map construction. Theor. Appl. Genet. 123: 649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Ma C., Casella G., 2007. Statistical Genetics of Quantitative Traits: Linkage, Maps, and QTL: Statistics for Biology and Health. Springer, New York [Google Scholar]
- Wu Y., Bhat P. R., Close T. J., Lonardi S., 2008. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 4: e1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Speed T. P., 1996. On genetic map functions. Genetics 142: 1369–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.