Abstract
Models for genome-wide prediction and association studies usually target a single phenotypic trait. However, in animal and plant genetics it is common to record information on multiple phenotypes for each individual that will be genotyped. Modeling traits individually disregards the fact that they are most likely associated due to pleiotropy and shared biological basis, thus providing only a partial, confounded view of genetic effects and phenotypic interactions. In this article we use data from a Multiparent Advanced Generation Inter-Cross (MAGIC) winter wheat population to explore Bayesian networks as a convenient and interpretable framework for the simultaneous modeling of multiple quantitative traits. We show that they are equivalent to multivariate genetic best linear unbiased prediction (GBLUP) and that they are competitive with single-trait elastic net and single-trait GBLUP in predictive performance. Finally, we discuss their relationship with other additive-effects models and their advantages in inference and interpretation. MAGIC populations provide an ideal setting for this kind of investigation because the very low population structure and large sample size result in predictive models with good power and limited confounding due to relatedness.
Keywords: multiple traits, quantitative traits, Bayesian networks, genome-wide predictions, Multiparent Advanced Generation Inter-Cross (MAGIC), multiparental populations, MPP
UNDERSTANDING the behavior of complex traits involves modeling a web of interactions among the effects of genes, environmental conditions, and other covariates. Ignoring one or more of these factors may substantially affect the accuracy and the generality of the conclusions that can be drawn from the model (Li et al. 2006; Hartley et al. 2012; Alimi et al. 2013), both in the context of genome-wide association studies (GWAS) and genomic selection (GS). Indeed a lot of attention has been devoted in recent literature to improving traditional additive genetic models, which were originally defined using only allele counts (e.g., Meuwissen et al. 2001), by supplementing them with additional information. Some examples include marker-based kinship coefficients (Speed et al. 2012), spatial heterogeneity and dominance (Finley et al. 2009), and gene expression data (Druka et al. 2008).
However, most studies in plant and animal genetics still focus on a single phenotypic trait at a time despite the availability of a set of simultaneously measured traits for each genotyped individual. Models for analyzing multiple traits have been available since Henderson and Quaas (1976) introduced the multivariate extension of the genetic best linear unbiased prediction (GBLUP) models, and have been investigated as recently as Stephens (2013) in the context of GWAS. More recent additions include structural equation models (SEM; Li et al. 2006), a Bayesian extension of seemingly unrelated regression (SUR; Banerjee et al. 2008), the MultiPhen ordinal regression (O’Reilly et al. 2012), and spatial models (Banerjee et al. 2012).
In this article we use Bayesian networks (BNs; Pearl, 1988; Koller and Friedman, 2009) to build a multivariate dependency model that accounts for simultaneous associations and interactions among multiple single nucleotide polymorphisms (SNPs) and phenotypic traits. BNs have been applied to the analysis of several kinds of genomic data such as gene expression (Friedman 2004), protein–protein interactions (Jansen et al. 2003; Sachs et al. 2005), pedigree analysis (Lauritzen and Sheehan 2004), and the integration of heterogeneous genetic data (Chang and Mcgeachie 2011). Their modular nature makes them ideal for analyzing large marker profiles. As far as SNPs are concerned, BNs have been used to investigate linkage disequilibrium (LD; Mourad et al. 2011; Morota et al. 2012) and epistasis (Han et al. 2012) and to determine disease susceptibility for anemia (Sebastiani et al. 2005), leukemia (Chang and Mcgeachie, 2011), and hypertension (Malovini et al. 2009). The same BN can simultaneously highlight SNPs potentially involved in determining a trait (e.g., for association purposes) and be used for prediction (e.g., for selection purposes): a network capturing the relationship between genotypes and phenotypes can be used to compute the probability that a new individual with a particular genotype will have the phenotype of interest (Lauritzen and Sheehan 2004; Cowell et al. 2007).
Materials and Methods
A BN is a probabilistic model in which a directed acyclic graph G is used to define the stochastic dependencies quantified by a probability distribution (Pearl 1988; Koller and Friedman 2009). The variables X = {Xi} under investigation in this context include T traits and S SNPs , each of which is associated with a node in G. The arcs between the nodes represent direct stochastic dependencies and determine how the global distribution of X decomposes into a set of local distributions,
| (1) |
one for each variable Xi, depending only on its parents . This modular representation can capture direct and indirect associations between SNPs and phenotypes and associations between SNPs due to linkage and population structure.
In the spirit of commonly used additive genetic models for quantitative traits (e.g., Meuwissen et al. 2001), we make some further assumptions on the BN:
each variable Xi is normally distributed, and X is multivariate normal;
stochastic dependencies are assumed to be linear;
traits can depend on SNPs (i.e., ) but not vice versa (i.e., not ), and they can depend on other traits (i.e., ); and
SNPs can depend on other SNPs (i.e., ).
We also assume that dependencies between traits broadly follow the temporal order in which they are measured; for instance, traits that are measured when a plant variety is harvested can depend on those that are measured while it is still in the field (and obviously on the markers as well), but not vice versa. In other words, assumptions 3 and 4 define BNs that describe the dependencies of phenotypes on genotypes in a prognostic model, as opposed to a diagnostic model in which genotypes depend on phenotypes. The latter is often preferred over the former because it results in simpler models when the Xi are discrete (Sebastiani and Perls 2008); in that setting, the number of parameters grows exponentially with the number of parents of each node. However, this is not the case here due to assumptions 1 and 2. Under these assumptions, the local distribution of each trait is a linear model of the form
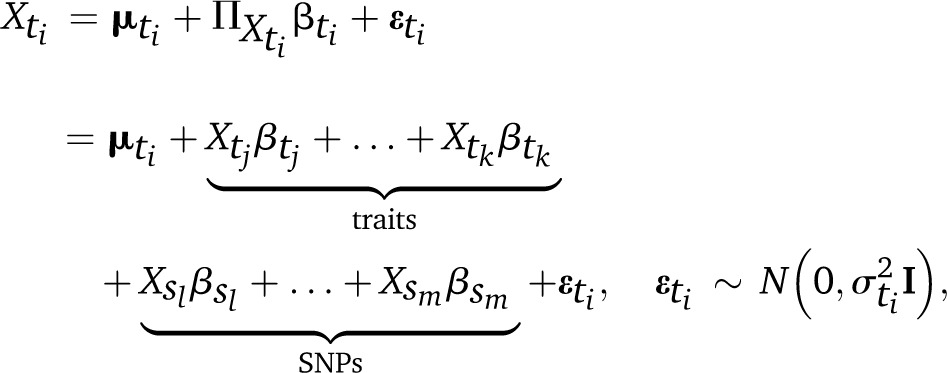 |
(2) |
where I is the identity matrix. SNPs will typically be coded using their allele counts (0, 1, 2), although extensions to multiallelic SNPs and to account for dominance are trivial. Similarly, the local distribution of each SNP is
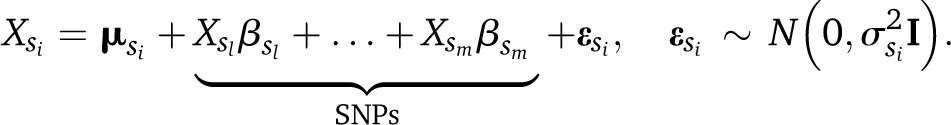 |
(3) |
Therefore, each parent adds only one parameter to a local distribution.
The regression parameters in (2) and (3) can be estimated in different ways. When G is sparse, ordinary least squares (OLS) are often used because each local distribution is estimated independently and contains few regressors. Otherwise, penalized estimators such as ridge regression (RR; Hoerl and Kennard 1970) can be used when G is dense. The resulting BN can then be considered a flexible implementation of multivariate ridge regression, which has a number of of desirable properties over OLS (Brown and Zidek 1980).
Equivalently, we can describe a BN using its global distribution, denoted with P(X) in (1). Following assumption 1, X has a multivariate normal distribution, say X ∼ N(μ, Σ). In addition, by definition graphical separation of two nodes Xi and Xj in G implies the conditional independence of the corresponding variables given the rest. As a result, some elements of the precision matrix Ω = Σ−1 will be equal to zero and some will be strictly positive according to the structure of G. The link with the parameterization based on the local distributions arises from the fact that in each the regression coefficient associated with Xj will be βj = −Ωij/Ωii; so βj = 0 if and only if the (i, j) element of Ω is itself equal to zero (Cox and Wermuth 1996, pp. 68–69).
It is interesting to note that this formulation defines BNs that are equivalent to multivariate GBLUP models (Henderson and Quaas 1976). For simplicity of notation, assume we are modeling only two traits and with a common set of SNP genotypes XS. In this case a multivariate GBLUP model has the form
| (4) |
where are the random effects for the two traits; ZS is the design matrix of the genotypes XS; are the population means; and are the error terms. and are independent of each other and distributed as multivariate normals with zero mean and covariance matrices
| (5) |
The covariance matrix models the pleiotropic effects of the SNPs on traits, potentially increasing the accuracy of multivariate GBLUP compared to a single-trait model.
As was the case in (2), each trait has a population mean and an error term that is normally distributed and independent of the SNP effects. The residual variance is also specific to each trait. The two traits depend directly on each other because of the covariances and indirectly through the covariance structure of the SNP effects . If we denote as G and as R, we can write
| (6) |
which is the covariance matrix of the global distribution. The structure of the BN defined over and corresponding to the multivariate GBLUP in (4) arises from Ω = Σ−1 as discussed above. Finally, it is important to note that even though GBLUP does not model the SNP effects using the allele counts directly as in (2) and (3), when and have the form the linear dependence on can be equivalently expressed as a random regression in the allele counts (Piepho 2009; Piepho et al. 2012). The form of determines how the allele counts are scaled or weighted in the regression. This formulation of GBLUP results in a more natural interpretation of SNP effects, which is in fact analogous to the interpretation they are given in a BN (Scutari et al. 2013).
Another interesting property of the BN defined above is that the covariance matrix of the SNP genotypes, which is a submatrix ΣSS of Σ (the global covariance matrix), is used in computing Ω and determines which arcs are present in G between the SNPs. Furthermore, ΣSS encodes the LD patterns between the SNPs as measured by the squared allelic correlation r2. This has been shown to be useful in exploring complex LD patterns in an inbred Holstein cattle population, albeit with a discrete BN (Morota et al. 2012) and measuring LD in a way that is closer to D and D′ (Falconer and Mackay 1995). Such patterns are reflected in the BN through Ω, providing an intuitive representation of LD as well as of genetic effects on phenotypes as a single, coherent whole.
BNs present two other advantages over classic multivariate regression models such as multivariate GBLUP and ridge regression. First, there is a vast literature on performing causal modeling with BNs from both experimental and observational data (Pearl 2009). Given the lack of a formal distinction between response and explanatory variables in BNs, the same algorithms can be used for inference on the traits based on the genotypes and vice versa. The former includes the estimation of phenotypic EBVs, which is the basis of genomic selection; the latter can be used for association mapping in polygenic traits and when the desired phenotype is a combination of conditions on several traits. Second, the fundamental properties of BNs do not depend on the distributional assumptions of the data. Therefore, accommodating heterogeneous traits (discrete, ordinal,and continuous) in the model requires only specifying the form of the local distributions.
Estimating a BN from data are typically performed as a two-step process. The first step consists in finding the graph G that encodes the conditional independencies present in the data and is called structure learning. This can be achieved using conditional independence tests (constraint-based learning), goodness-of-fit scores (score-based learning), or both (hybrid learning) to identify statistically significant arcs. The second step is called parameter learning and deals with the estimation of the parameters of the local distributions; G is known from the previous step and defines which variables are included in each one. In addition, we propose using structure learning to retain in the BN only those SNPs that are required to make inference on the traits and that make the remaining SNPs redundant. For each trait, such a subset is called the Markov blanket (; Pearl 1988) and includes the parents, the children, and the other nodes that share a child with the trait. Therefore, we can disregard all the SNPs that are not part of any such Markov blanket and reduce drastically the dimension of the model. We have shown in previous work (Scutari et al. 2013) how Markov blankets are effective when used in this setting.
From these considerations, we used the R packages bnlearn (Scutari 2010) and penalized (Goeman 2012) to implement the following hybrid approach to BN learning.
Structure Learning
For each trait , use the SI-HITON-PC algorithm (Aliferis et al. 2010) to learn the parents and the children of the trait; this is sufficient to identify because the only nodes that can share a child with are other traits or SNPs that are parents of other traits due to assumption 3. The choice of SI-HITON-PC is motivated by its similarity to single-SNP analysis, which is improved on with a subsequent backward selection to remove false positives. Dependencies are assessed with Student’s t-test for Pearson’s correlation (Hotelling 1953) and α = 0.01, 0.05, 0.10.
Drop all the markers that are not in any .
Learn the structure of the BN from the nodes selected in the previous step, setting the directions of the arcs according to assumptions 3 and 4. We identify the optimal structure as that which maximizes the Bayesian information criterion (BIC; Schwarz 1978).
Parameter Learning
Learn the parameters of the local distributions using OLS and RR. For comparison, we also fitted an elastic net (ENET) model (Zou and Hastie 2005) and a univariate GBLUP individually on each trait and on all the available SNPs using the glmnet (Friedman et al. 2010) and synbreed (Wimmer et al. 2012) R packages. Since we have shown BNs to be equivalent to a multivariate GBLUP, we did not fit the latter as a separate model. We investigated the properties of the resulting models using, in each case, 10 runs of 10-fold cross-validation. Predictive power was assessed by averaging the cross-validated correlations arising from the 10 runs and computing confidence intervals as in Hooper (1958). In the case of BNs, predictions in the cross-validation folds were performed jointly on all traits and in two different ways: by conditioning only on the SNPs in the BN, to provide a measure of genetic predictive ability (ρG) and a fair comparison with single-trait models, and by conditioning on the parents of each trait, which may in turn be traits themselves, to provide a tentative measure of causal predictive ability (ρC).
To perform inference, we produced an averaged BN using the 100 networks we obtained in the course of cross-validation. First, we created an averaged network structure using their graphs as in Scutari and Nagarajan (2013): we kept only those arcs that appear with a frequency higher than a threshold estimated from the graphs themselves. SNPs that ended up as isolated nodes (i.e., they were not connected to any other SNP or trait) were dropped. We then estimated the parameters of the averaged BN with RR using the whole data set. We used the resulting BN to generate samples of 106 random observations from the conditional distributions of various traits and SNPs with either logic sampling or likelihood weighting (Koller and Friedman 2009) to explore their properties and interplay under different conditions. Statistics estimated from such a big sample are very precise and can capture even small differences reliably.
We based our analysis on a winter wheat population produced by the UK National Institute of Agricultural Botany (NIAB) comprising 15877 SNPs for 720 genotypes. Seven traits were measured: yield (YLD; tonnes per hectare), flowering time (FT; 6–54, aggregate of five scores taken at 3- to 7-day intervals), height (HT; centimeters), yellow rust in the glasshouse (YR.GLASS; 1–9) and in the field (YR.FIELD; 1–9), Fusarium (FUS; 1–9), and mildew (MIL; 1–9). Disease scores from 1 to 9 reflect increasing level of infection, and flowering time scores from 6 to 54 increasing lateness in flowering. The population was created using a multiparent advanced generation inter-cross (MAGIC) scheme. Such a scheme is designed to produce a mapping population from several generations of intercrossing among eight founders and has the potential to improve quantitative trait loci (QTL) mapping precision (for more details and to access the data see Mackay et al. 2014). The use of multiple founder varieties results in a population that is segregating for more QTL and traits than a biparental population, and the balanced crossing used in each generation reduces LD and family structure by ensuring that each founder has an equal opportunity to contribute to each genotype.
SNPs were preprocessed by removing those with minor allele frequencies <1% and those with >20% missing data. Missing data in the remaining SNPs were imputed using the impute R package (Hastie et al. 2013). Other widely used imputation methods in genetics, such as that implemented in MaCH (Li et al. 2010), could not be used because of the lack of precise mapping information at the time of the analysis; a 90K consensus map has just been submitted for publication (Wang et al. 2014). Subsequently, we removed one SNP from each pair whose allele counts have correlation >0.95 to increase the numerical stability of the models. In the end, 3164 SNPs were left for analysis. Phenotypes were adjusted for kinship using a univariate BLUP model for each trait based on pedigree information, thus accounting for population structure. Individuals with missing pedigree information or phenotypes were dropped from the analysis, leaving 600 individuals with complete records.
Results
Table 1 shows genetic predictive correlations (ρG) and causal predictive correlations (ρC) for single-trait ENET, single-trait GBLUP, and BNs fitted with α = 0.01, 0.05, 0.10. Only the results for BNs whose parameters are estimated with RR are reported, because using OLS provides essentially the same performance. The average ρG obtained with RR across all traits is 0.324 for α = 0.01, 0.327 for α = 0.05, and 0.331 for α = 0.10, all with a standard deviation of ±0.004; with OLS we obtain 0.322 for α = 0.01, 0.325 for α = 0.05, and 0.324 for α = 0.10, again with a standard deviation of ±0.004. Similar considerations can be made for ρC.
Table 1. Genetic (ρG) and causal (ρC) predictive correlations for the 7 traits and for single-trait elastic net (ENET), single-trait GBLUP and BNs estimated with α = 0.01, 0.05, 0.10 and RR.
| YLD | FT | HT | YR.FIELD | YR.GLASS | MIL | FUS | ||
|---|---|---|---|---|---|---|---|---|
| ENET | ρG | 0.15 | 0.30 | 0.48 | 0.39 | 0.59 | 0.21 | 0.27 |
| GBLUP | ρG | 0.10 | 0.15 | 0.19 | 0.22 | 0.32 | 0.21 | 0.12 |
| BN,0.01 | ρG | 0.20 | 0.29 | 0.46 | 0.37 | 0.60 | 0.12 | 0.22 |
| ρC | 0.38 | 0.29 | 0.45 | 0.44 | 0.62 | 0.13 | 0.33 | |
| BN,0.05 | ρG | 0.18 | 0.27 | 0.46 | 0.39 | 0.61 | 0.12 | 0.25 |
| ρC | 0.34 | 0.27 | 0.45 | 0.44 | 0.63 | 0.14 | 0.32 | |
| BN,0.10 | ρG | 0.18 | 0.28 | 0.45 | 0.40 | 0.62 | 0.13 | 0.25 |
| ρC | 0.34 | 0.28 | 0.45 | 0.45 | 0.63 | 0.14 | 0.31 |
Standard deviations computed as in Hooper (1958) is 0.01 for all correlations. Traits are yield (YLD), flowering time (FT), height (HT), yellow rust in the field (YR.FIELD) and in the glasshouse (YR.GLASS), mildew (MIL), and Fusarium (FUS).
First, we note that BNs and single-trait ENET have comparable predictive power for ρG: BNs are best for YLD, YR.GLASS, and YR.FIELD, while ENET is best for FT, HT, MIL, and FUS. Overall, the average ρG across all seven traits is 0.343 ± 0.004 for ENET and 0.331 ± 0.004 for BNs with α = 0.10. Therefore, while ENET outperforms BNs on average, BNs still provide the best ρG in three traits of seven. In addition, both ENET and BNs outperform single-trait GBLUP, which has ρG = 0.186 ± 0.005 overall. As expected, the choice of the kinship matrix used in GBLUP does not significantly affect ρG because we accounted for the effect of family structure on the traits as a preliminary step. Using different marker-based estimates of kinship such as allele sharing (Habier et al. 2007) or allelic correlation (Astle and Balding 2009) provides no benefit over not using a kinship matrix at all.
It is also apparent that increasing α does not produce any marked increase in ρG; while larger values of α result in larger BNs, the small increase in predictive power is not worth the longer time required to estimate the model under cross-validation. On average, we learned BNs with 47 nodes (including the 7 traits) in a few seconds for α = 0.01, with 75 nodes in 20 min for α = 0.05, and with 89 nodes in 2.5 hr for α = 0.10. Further increasing α as in Scutari et al. (2013) only exacerbates the problem (24 days for α = 0.15, results not shown). Of all the SNPs included in BNs, few are not parents of any trait and thus appear to be false positives: 1 of 40 (2.5%) for α = 0.01, 2 of 68 (2.9%) for α = 0.05, and 4 of 82 (4.8%) for α = 0.10. The dimension of the BNs is in stark contrast with the average number of nonzero SNP effects in the ENET models: 110 nonzero coefficients for YR.GLASS, 2661 for YLD, 55 for HT, 105 for YR.FIELD, 333 for FUS, 1725 for MIL, and 24 for FT.
As far as causal predictive correlations ρC are concerned, we observe a distinct improvement compared to ρG for three traits: YLD, YR.FIELD, and FUS. As for the other four traits, the difference between ρG and ρC is not as marked, even though it is statistically significant in all cases except flowering time. Overall, ρC = 0.373 ± 0.004, which is higher than both BN’s ρG = 0.331 ± 0.04 for α = 0.10 and the ENET’s ρG = 0.343 ± 0.004.
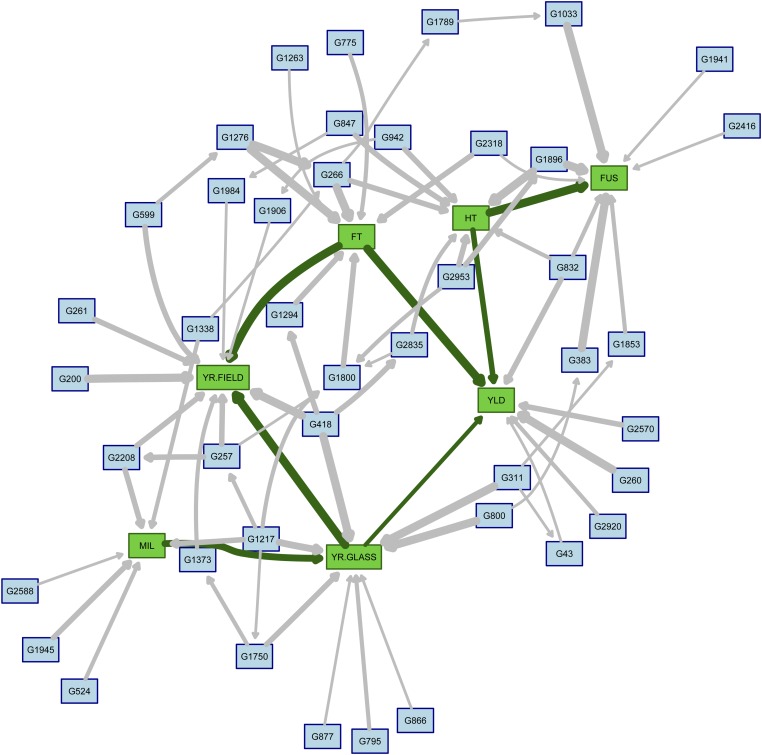
The averaged BN for α = 0.10 is shown in Figure 1; it has 50 nodes and 78 arcs. For ease of plotting, the SNP names corresponding to the labels used in the figure are reported in Table 2. The dimension of the BN is comparable to that obtained for α = 0.01 (30 nodes, 44 arcs) and α = 0.05 (44 nodes, 66 arcs). In all three cases the threshold for arc inclusion estimated as in Scutari and Nagarajan (2013) is 0.49, which is close to the intuitive choice of including in the averaged BN those arcs that appear in more than half of the BNs obtained during cross-validation. All SNPs in the averaged BN are linked with at least one trait, with the exception of G1789 (D_contig28346_467). Their minor allele frequencies range from 0.02 (G2208; IAAV1322) to 0.47 (G1945; Excalibur_c29304_176). Furthermore, the BN is small enough that RR and OLS parameter estimates are practically equivalent.
Figure 1.
Averaged network obtained from the cross-validated BNs for α = 0.10. Green nodes correspond to traits: yield (YLD), flowering time (FT), height (HT), yellow rust in the field (YR.FIELD) and in the glasshouse (YR.GLASS), mildew (MIL), and Fusarium (FUS). Blue nodes correspond to SNPs. The thickness of the arcs represents the strength of the corresponding dependence relationships as measured by their frequency in the BNs produced during cross-validation.
Table 2. SNPs included in the averaged BN.
| LABEL | NAME | LABEL | NAME |
|---|---|---|---|
| G418 | BobWhite_c5756_516 | G311 | BobWhite_c37358_208 |
| G800 | BS00022299_51 | G877 | BS00022830_51 |
| G866 | BS00022703_51 | G795 | BS00022270_51 |
| G2570 | Kukri_c7241_322 | G260 | BobWhite_c29014_241 |
| G832 | BS00022473_51 | G1896 | Excalibur_c19078_210 |
| G2953 | Tdurum_contig64772_417 | G942 | BS00024496_51 |
| G266 | BobWhite_c30043_150 | G847 | BS00022562_51 |
| G2835 | RFL_Contig4790_1091 | G200 | BobWhite_c22728_78 |
| G2208 | IAAV1322 | G257 | BobWhite_c28819_733 |
| G1906 | Excalibur_c20837_868 | G261 | BobWhite_c2905_590 |
| G1984 | Excalibur_c37696_192 | G599 | BS00009575_51 |
| G383 | BobWhite_c47401_491 | G2416 | Kukri_c100613_331 |
| G1033 | BS00035141_51 | G1941 | Excalibur_c27950_459 |
| G1853 | Excalibur_c11795_934 | G1338 | BS00066211_51 |
| G524 | BS00000721_51 | G1945 | Excalibur_c29304_176 |
| G1276 | BS00064538_51 | G1789 | D_contig28346_467 |
| G2318 | IACX11305 | G1800 | D_GBUVHFX01DSLGX_212 |
| G1294 | BS00065110_51 | G775 | BS00022148_51 |
| G1750 | CAP12_c2800_262 | G43 | BobWhite_c11692_148 |
| G1373 | BS00067203_51 | G1217 | BS00062679_51 |
| G2588 | Kukri_rep_c102953_304 | G1263 | BS00064140_51 |
| G2920 | Tdurum_contig42584_1190 |
The labels are those used in Figure 1, while the SNP names are from Mackay et al. (2014) and Wang et al. (2014).
As far as phenotypic traits are concerned, the averaged BN captures several known relationships. YR.FIELD is influenced by FT (FT → YR.FIELD in Figure 1); early flowering genotypes will have their leaves exposed to the pathogens for a longer time than later genotypes, resulting in higher yellow rust scores even if they have the same level of true disease resistance. This is substantiated by the posterior distribution of the disease score conditional on flowering time being in the bottom quartile ([21.0, 29.7]) or in the top quartile ([33.8, 42.0]): it has mean 2.54 in the first case and 2.33 in the second. Standard deviation is 0.47 in both cases. The same is true for YR.GLASS, which has means 2.50 and 2.48 for early and late flowering genotypes; standard deviation is 0.43. The network structure suggests that the YR.GLASS is not influenced directly by FT (i.e., there is no FT → YR.GLASS arc). The two yellow rust scores (YR.GLASS → YR.FIELD) are positively correlated (0.34), likely because of durable resistance. In addition, we note that YR.FIELD summarizes adult resistance to a mixed population of pathotypes, which may include the specific pathotype used to measure juvenile resistance in YR.GLASS.
We can also see from Figure 1 that YLD depends directly on both HT (HT → YLD) and FT (FT → YLD), but it is affected only indirectly by all the disease scores except YR.GLASS. Conditional on the combinations of bottom and top quartiles for FT and HT ([64.3, 74.5] and [79.5, 87.7]), the expected yield is 7.54, 7.71, 7.15, and 7.33, respectively. Standard deviation is 0.47 in all four scenarios. Therefore, we observe a marginal increase in YLD of ∼0.15 when comparing short and tall genotypes, and a marginal decrease of ∼0.4 when comparing early and late flowering genotypes; this is consistent with Flintham et al. (1997) and Snape et al. (2001). The interplay between HT and FT appears to be negligible in determining yield. Conditioning on the bottom and top quartiles of the disease scores, we see a difference in the mean YLD of +0.08 (FUS), −0.02 (MIL), −0.01 (YR.GLASS), and −0.10 (YR.FIELD).
The apparent increase in YLD associated with high FUS scores is the result of the confounding effect of HT, which is directly linked to both variables in the BN (FUS ← HT → YLD). This is expected because susceptibility to Fusarium is known to be positively related to HT (Srinivasachary et al. 2009), which in turn affects YLD. Conditional on each quartile of HT, FUS has a negative effect on YLD ranging from −0.04 to −0.06.
The last interaction between phenotypes in the BN is between MIL and YR.GLASS (MIL → YR.GLASS). This can be explained by the increased susceptibility to one disease in genotypes that are weakened by the onset of the other, by disease resistance being controlled by shared regions in the genome (Spielmeyer et al. 2005; Lillemo et al. 2008), and to a lesser extent by the influence of weather conditions (Beest et al. 2008). The BN in Figure 1 identifies 9 SNPs that are linked to at least one of MIL and YR.GLASS and may be tagging pleiotropic QTL for disease resistance. By contrasting low and high level of both diseases (scores ≤1.5 and ≥3.5, respectively), we can infer which allele may be linked with resistance to both diseases using the conditional expected allele counts, nLOW and nHIGH. For three of the nine genes, the difference between the two is marked: G418 (BobWhite_c5756_516; nLOW = 0.5, nHIGH = 1.9), G311 (BobWhite_c37358_208; nLOW = 1.1, nHIGH = 1.7), and G1217 (BS00062679_51; nLOW = 0.8, nHIGH = 1.7). The 90K consensus map in Wang et al. (2014) locates G418 in chromosome 2D along with other SNPs conferring resistance to YR.GLASS. The same is true also for G311 in chromosome 2B and for G2127 in chromosome 2A. As for the other six SNPs, |nLOW − nHIGH| < 0.5, which suggests that their individual effects are small and that they might work in concert with other genes producing polygenic effects.
Similar analyses on the other traits identify two more SNPs with |nLOW − nHIGH| ≤ 0.5 that may be tagging known genes. G1896 (Excalibur_c19078_210) has nLOW = 0.3, nHIGH = 1.2 when contrasting top and bottom quartiles for HT and has nLOW = 0.2, nHIGH = 1.7 when contrasting the bottom quartile of HT and FUS ≥ 3.5 with the top quartile of HT and FUS ≤ 1.5. The latter pair of scenarios is motivated by the fact that taller plants are less susceptible to Fusarium than shorter plants. The LD analysis in Mackay et al. (2014) suggests that this SNP is located in chromosome 4D in this population and that it may be tagging Rht-D1b, a dwarfing gene that is also closely associated with resistance to Fusarium (Srinivasachary et al. 2009). In addition, G266 (BobWhite_c30043_150) appears to be located in chromosome 2D and to be tagging Ppd-D1, which controls photoperiod response. Contrasting the bottom quartiles of both FT and HT with the top quartiles we have nHIGH = 0 and nLOW = 0.8.
Discussion
Modeling multiple quantitative traits simultaneously has been known to result in better predictive power than targeting one trait at a time in the context of additive genetic models (Henderson and Quaas 1976). BNs provide a general framework in which to estimate and analyze such models. They also provide an accompanying graphical representation that is intuitive yet rigorous; a plot such as that in Figure 1 can be very useful for exploratory analysis, to disseminate results and to motivate further quantitative and qualitative analyses in GWAS and GS studies.
From a theoretical point of view, BNs are more versatile than additive models in common use. By assuming that variables are normally distributed, we have shown that BNs are in fact equivalent to multivariate GBLUP and, by extension, to single-trait GBLUP. Furthermore, the separation between structure and parameter learning makes it possible to accommodate different parametric assumptions with relatively few changes and subsume models such as univariate and multivariate ridge-regression (Hoerl and Kennard 1970; Brown and Zidek 1980). As far as inference is concerned, several established methods from the literature can be used to predict traits from SNPs and vice versa; two examples are logic sampling and likelihood weighting (Koller and Friedman 2009). Both allow exploration of complex scenarios of practical relevance by estimating informative statistics from the corresponding conditional distributions of traits and SNPs. This is made easier by the lack of a formal distinction between response and explanatory variables in the BN, which is central in traditional linear models. As a result, BNs can be used for association studies as well as genomic prediction. In the former, we can condition on some complex combination of traits and predict the expected allele counts of SNPs. Such an approach has the potential of detecting which SNPs tag relevant QTL and which of their alleles are favorable. In the latter, we have shown that BNs are competitive with a state-of-the-art model such as single-trait ENET when predicting traits from SNPs and that they outperform single-trait GBLUP for the population analyzed in this article. As evidenced by the difference between ρG and ρC, using BNs as a multitrait model and performing predictions based on those variables identified as putative causal for each trait outperforms ENET as well by leveraging pleiotropic effects (Hartley et al. 2012). This shows that it is possible to improve genomic selection for traits that are expensive to measure by incorporating cheaper ones in the predictions. Clearly, the impact of correlated phenotypes on the predictive power of BNs depends on the strength of their correlation.
Based on the BN in Figure 1, we can also observe some interesting properties of BNs as genetic models. First, the difference in the number of SNPs included in the BNs compared to the ENET models can be attributed to the limited ability of BNs to capture small epistatic effects (Han et al. 2012). Consider, for instance, a polygenic effect in which two SNPs are jointly associated with a trait but in which each SNP is not significant on its own. Such an effect will not be captured because both SNPs will be discarded by the single-SNP screening performed at the beginning of feature selection. As observed in other studies, this does not have a significant impact on predictive ability if a large enough α threshold is used, as Markov blankets are very effective at feature selection (Chang and McGeachie 2011; Scutari et al. 2013). Second, SNPs with pleiotropic effects are included in the BN even when association with a single phenotype is detected; at that point they can be linked to all relevant phenotypes. This is the case of the SNPs controlling resistance to both mildew and yellow rust discussed above. Furthermore, direct and indirect effects of such SNPs and of traits are correctly separated for the observed traits, as in the case of the Fusarium effect on yield.
MAGIC populations provide an ideal starting point for fitting BNs. On the one hand, the particular pattern of crosses used to produce a MAGIC population results in a very low population structure. This reduces the confounding effect of relatedness on the estimation of SNP effects (Astle and Balding 2009) and on mapping approaches based on LD (Mackay et al. 2014). On the other hand, the size of of the population is large enough to detect weak associations and associations with rare variants. Both are in fact present in the averaged BN, which includes SNPs with minor allele frequencies as low as 0.02 and SNPs that are significant (e.g., for MIL and YR.GLASS) only when considering multiple traits at the same time.
Finally, SNPs of interest can be made to segregate in the population by choosing the founders appropriately, since balanced crosses ensure opportunities for recombination among the founders. This is particularly important in modeling multiple phenotypes, as we need to ensure that as many relevant QTL and genes as possible are tagged to correctly dissect their genetic layout.
Acknowledgments
The work presented in this article forms part of the Molecular Improvement of Disease Resistance in Barley project, which is funded by the UK Technology Strategy Board (TSB) and Biotechnology & Biological Sciences Research Council (BBSRC), grant TS/I002170/1. The MAGIC population was developed within BBSRC Crop Science Initiative project BB/E007201/1. Field trials and SNP genotyping were funded by the NIAB Trust.
Footnotes
Communicating editor: F. van Eeuwijk
Literature Cited
- Aliferis C. F., Statnikov A., Tsamardinos I., Mani S., Xenofon X. D., 2010. Local causal and Markov blanket induction for causal discovery and feature selection for classification part I: algorithms and empirical evaluation. J. Mach. Learn. Res. 11: 171–234 [Google Scholar]
- Alimi N. A., Bink M. C. A. M., Dieleman J. A., Magán J. J., Wubs A. M., et al. , 2013. Multi-trait and multi-environment QTL analyses of yield and a set of physiological traits in pepper. Theor. Appl. Genet. 126(10): 2597–2625 [DOI] [PubMed] [Google Scholar]
- Astle W., Balding D. J., 2009. Population structure and cryptic relatedness in genetic association studies. Stat. Sci. 24(4): 451–471 [Google Scholar]
- Banerjee S., Yandell B. S., Yi N., 2008. Bayesian quantitative trait loci mapping for multiple traits. Genetics 179: 2275–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Finley A. O., Waldmann P., Ericsson T., 2012. Hierarchical spatial process models for multiple traits in large genetic trials. J. Am. Stat. Assoc. 105(490): 506–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beest D. E. T., Paveley N. D., Shaw M. W., van den Bosch F., 2008. Disease–weather relationships for powdery mildew and yellow rust on winter wheat. Phytopatology 98: 609–617 [DOI] [PubMed] [Google Scholar]
- Brown P. J., Zidek J. V., 1980. Adaptive multivariate ridge regression. Ann. Stat. 8(1): 64–74 [Google Scholar]
- Chang, H.-H., and M. McGeachie, 2011 Phenotype prediction by integrative network analysis of SNP and gene expression microarrays, pp. 6849–6852 in Proceedings of the 33rd Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) IEEE Press, New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell R. G., Dawid A. P., Lauritzen S. L., Spiegelhalter D. J., 2007. Probabilistic Networks and Expert Systems. Springer-Verlag, New York [Google Scholar]
- Cox D. R., Wermuth N., 1996. Multivariate Dependencies: Models, Analysis and Interpretation, Chapman & Hall, Boca Raton, FL [Google Scholar]
- Druka A., Druka I., Centeno A., Li H., Sun Z., et al. , 2008. Towards systems genetic analyses in barley: integration of phenotypic, expression and genotype data into GeneNetwork. BMC Genet. 9(1): 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1995. Introduction to Quantitative Genetics, Ed 4 Prentice Hall, Harlow, UK [Google Scholar]
- Finley A. O., Banerjee S., Waldmann P., Ericsonn T., 2009. Hierarchical spatial modeling of additive and dominance genetic variance for large spatial trial datasets. Biometrics 61(2): 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flintham J. E., Börner A., Worland A. J., Gale M. D., 1997. Optimizing wheat grain yield: effects of Rht (Gibberellin-insensitive) dwarfing genes. J. Agric. Sci. 128(1): 11–25 [Google Scholar]
- Friedman J. H., Hastie T., Tibshirani R., 2010. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33(1): 1–22 [PMC free article] [PubMed] [Google Scholar]
- Friedman N., 2004. Inferring cellular networks using probabilistic graphical models. Science 303(5659): 799–805 [DOI] [PubMed] [Google Scholar]
- Goeman, J. J., 2012 penalized R package R package version 0.9–41.
- Habier D., Fernando R. L., Dekkers J. C. M., 2007. The impact of genetic relationship information on genome-assisted breeding values. Genetics 177: 2389–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B., Chen X., Talebizadeh Z., Xu H., 2012. Genetic studies of complex human diseases: characterizing SNP-disease associations using Bayesian networks. BMC Syst. Biol. 6(Suppl. 3): S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley S. W., Monti S., Liu C.-T., Steinberg M. H., Sebastiani P., 2012. Bayesian methods for multivariate modeling of pleiotropic SNP associations and genetic risk prediction. Front. Genet. 3(176): 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie, T., R. Tibshirani, B. Narasimhan, and G. Chu, 2013 impute: imputation for microarray data R package version 1.36.0.
- Henderson C. R., Quaas R. L., 1976. Multiple trait evaluation using relatives’ records. J. Anim. Sci. 43: 1188–1197 [Google Scholar]
- Hoerl A. E., Kennard R. W., 1970. Ridge regression: biased estimation for nonorthogonal problems. Technometrics 12(1): 55–67 [Google Scholar]
- Hooper J. W., 1958. The sampling variance of correlation coefficients under assumptions of fixed and mixed variates. Biometrika 45(3/4): 471–477 [Google Scholar]
- Hotelling H., 1953. New light on the correlation coefficient and its transforms. J. R. Stat. Soc., B 15(2): 193–232 [Google Scholar]
- Jansen R., Yu H., Greenbaum D., Kluger Y., Krogan N. J., et al. , 2003. A Bayesian networks approach for predicting protein–protein interactions from genomic data. Science 302(5644): 449–453 [DOI] [PubMed] [Google Scholar]
- Koller D., Friedman N., 2009. Probabilistic Graphical Models: Principles and Techniques. MIT Press, Cambridge, MA [Google Scholar]
- Lauritzen S. L., Sheehan N. A., 2004. Graphical models for genetic analysis. Stat. Sci. 18: 489–514 [Google Scholar]
- Li R., Tsaih S.-W., Shockley K., Stylianou I. M., Wergedal J., et al. , 2006. Structural model analysis of multiple quantitative traits. PLoS Genet. 2(7): e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Willer C. J., Ding J., Scheet P., Abecasis G. R., 2010. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 34: 816–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillemo M., Asalf B., Singh R. P., Huerta-Espino J., Chen X. M., et al. , 2008. The adult plant rust resistance loci Lr34/Yr18 and Lr46/Yr29 are important determinants of partial resistance to powdery mildew in bread wheat line Saar. Theor. Appl. Genet. 116: 1155–1166 [DOI] [PubMed] [Google Scholar]
- Mackay I., Bansept-Basler P., Barber T., Bentley A., Cockram J. et al, 2014. An eight-parent multiparent advanced generation intercross population for winter-sown wheat: creation, properties and first results. G3 (Bethesda) 4: 1603–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovini A., Nuzzo A., Ferrazzi F., Puca A., Bellazzi R., 2009. Phenotype forecasting with SNPs data through gene-based Bayesian networks. BMC Bioinformatics 10(Suppl. 2): S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morota G., Valente B. D., Rosa G. J. M., Weigel K. A., Gianola D., 2012. An assessment of linkage disequilibrium in holstein cattle using a Bayesian network. J. Anim. Breed. Genet. 129(6): 474–487 [DOI] [PubMed] [Google Scholar]
- Mourad R., Sinoquet C., Leray P., 2011. A hierarchical Bayesian network approach for linkage disequilibrium modeling and data-dimensionality reduction prior to genome-wide association studies. BMC Bioinformatics 12(1): 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly P. F., Hoggart C. J., Pomyen Y., Calboli F. C. F., Elliott P., et al. , 2012. MultiPhen: joint model of multiple phenotypes can increase discovery in GWAS. PLoS ONE 7(5): e34861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl J., 1988. Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference. Morgan Kaufmann, San Francisco [Google Scholar]
- Pearl J., 2009. Causality: Models, Reasoning and Inference, Ed 2 Cambridge University Press, Cambridge, UK [Google Scholar]
- Piepho H.-P., 2009. Ridge regression and extensions for genomewide selection in maize. Crop Sci. 49(4): 1165–1176 [Google Scholar]
- Piepho H.-P., Ogutu J. O., Schulz-Streeck T., Estaghvirou B., Gordillo A., et al. , 2012. Efficient computation of ridge-regression best linear unbiased prediction in genomic selection in plant breeding. Crop Sci. 52(3): 1093–1104 [Google Scholar]
- Sachs K., Perez O., Pe’er D., Lauffenburger D. A., Nolan G. P., 2005. Causal protein-signaling networks derived from multiparameter single-cell data. Science 308(5721): 523–529 [DOI] [PubMed] [Google Scholar]
- Schwarz G. E., 1978. Estimating the dimension of a model. Ann. Stat. 6(2): 461–464 [Google Scholar]
- Scutari M., 2010. Learning Bayesian networks with the bnlearn R package. J. Stat. Softw. 35(3): 1–2221603108 [Google Scholar]
- Scutari M., Nagarajan R., 2013. On identifying significant edges in graphical models of molecular networks. Artif. Intell. Med. 57(3): 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scutari M., Mackay I., Balding D. J., 2013. Improving the efficiency of genomic selection. Stat. Appl. Genet. Mol. Biol. 12(4): 517–527 [DOI] [PubMed] [Google Scholar]
- Sebastiani P., Perls T. T., 2008. Complex genetic models, pp. 53–72 in Bayesian Networks: a Practical Guide to Applications, edited by Pourret O., Naïm P., Marcot B. Wiley, Hoboken, NJ [Google Scholar]
- Sebastiani P., Ramoni M. F., Nolan V., Baldwin C. T., Steinberg M., 2005. Genetic dissection and prognostic modeling of overt stroke in sickle cell anemia. Nat. Genet. 37(4): 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snape J. W., Butterworth K., Whitechurch E., Worland A. J., 2001. Waiting for fine times: genetics of flowering time in wheat. Euphytica 119(1–2): 185–190 [Google Scholar]
- Speed D., Hermani G., Johnson M. R., Balding D. J., 2012. Improved heritability estimation from genome-wide SNPs. Am. J. Hum. Genet. 91(6): 1011–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmeyer W., McIntosh R. A., Kolmer J., Lagudah E. S., 2005. Powdery mildew resistance and Lr34/Yr18 genes for durable resistance to leaf and stripe rust cosegregate at a locus on the short arm of chromosome 7D of wheat. Theor. Appl. Genet. 111: 731–735 [DOI] [PubMed] [Google Scholar]
- Srinivasachary N. Gosman, A. Steed, T. W. Hollins, R. Bayleset al, 2009. Semi-dwarfing Rht-B1 and Rht-D1 loci of wheat differ significantly in their influence or resistance to fusarium head blight. Theor. Appl. Genet. 118: 695–702 [DOI] [PubMed] [Google Scholar]
- Stephens M., 2013. A unified framework for association analysis with multiple related phenotypes. PLoS ONE 8(7): e65245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., D. Wong, K. Forrest, A. Allen, S. Chao et al, 2014 Characterization of polyploid wheat genomic diversity using a high-density 90,000 SNP array. Plant Biotech. J. 12: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer V., Albrecht T., Auinger H.-J., Schön C.-C., 2012. synbreed: framework for the analysis of genomic prediction data using R. Bioinformatics 18(15): 2086–2087 [DOI] [PubMed] [Google Scholar]
- Zou H., Hastie T., 2005. Regularization and variable selection via the elastic net. J. R. Stat. Soc. B 67(2): 301–320 [Google Scholar]