Abstract
Although research effort is being expended into determining the importance of epistasis and epistatic variance for complex traits, there is considerable controversy about their importance. Here we undertake an analysis for quantitative traits utilizing a range of multilocus quantitative genetic models and gene frequency distributions, focusing on the potential magnitude of the epistatic variance. All the epistatic terms involving a particular locus appear in its average effect, with the number of two-locus interaction terms increasing in proportion to the square of the number of loci and that of third order as the cube and so on. Hence multilocus epistasis makes substantial contributions to the additive variance and does not, per se, lead to large increases in the nonadditive part of the genotypic variance. Even though this proportion can be high where epistasis is antagonistic to direct effects, it reduces with multiple loci. As the magnitude of the epistatic variance depends critically on the heterozygosity, for models where frequencies are widely dispersed, such as for selectively neutral mutations, contributions of epistatic variance are always small. Epistasis may be important in understanding the genetic architecture, for example, of function or human disease, but that does not imply that loci exhibiting it will contribute much genetic variance. Overall we conclude that theoretical predictions and experimental observations of low amounts of epistatic variance in outbred populations are concordant. It is not a likely source of missing heritability, for example, or major influence on predictions of rates of evolution.
Keywords: quantitative genetics, epistasis, additive variance, multilocus models, selection
EPISTATIC variance in quantitative traits arises from the interaction effects or epistasis between segregating genes at two or more loci that affect these complex traits. Such gene interaction is a common phenomenon because many factors have, for example, a regulatory role in a hierarchical system (Phillips 2008). The statistical theory of quantitative genetics following Fisher (1918) is based on a partition between average effects across loci, which contribute to the additive genetic variance, and to interactions within loci and between loci, which contribute to the dominance and epistatic variance, respectively (Cockerham 1954; Kempthorne 1954). The magnitudes of these components of the genotypic variance each depend on the frequencies, the effects, and the interactions among the contributing genes (see also Falconer and Mackay 1996; Lynch and Walsh 1998). The actual causal genetic factors are usually not known, but many quantitative genetic analyses, including selection on metric traits, have been applied successfully without such knowledge.
Among quantitative geneticists, interest in epistasis continues, both to understand the genetic architecture and as a potential way to improve the genomic predictions of disease and quantitative traits, utilizing some of the unexplained parts of the genetic variation (e.g., Carlborg and Haley 2004; Nelson et al. 2013; Mackay 2014). Despite the obvious interactions in the biological system, it has, however, been argued that the proportion of the genotypic variance contributed by epistatic variance expected in outbred populations is small and that data generally support this prediction (Hill et al. 2008). The theory is based mainly on population and statistical genetics arguments (the data of a more indirect form): for many traits the covariances among relatives could largely be explained by additive models, albeit estimation of epistatic variance per se with much precision is difficult in most outbred population structures. Hemani et al. (2013) in contrast, taking an evolutionary perspective, have argued that much of the variation that will remain segregating long term in populations under natural section will be epistatic. It is notable, however, that the loci in the models they simulated that were still segregating all showed overdominance, for which direct data are limited (e.g., Charlesworth and Charlesworth 2009). Using models with multiple loci and two-locus interactions in populations evolving with bottlenecks, Ávila et al. (2014) found only small contributions of epistatic variance. Further, it has been argued that the magnitude of epistatic variation is essentially irrelevant in evolution (and in animal breeding) (Crow 2010), because rates of evolutionary change depend only on the additive variance, even in epistatic and tightly linked systems (Kimura 1965; Nagylaki 1993). Nevertheless Nelson et al. (2013) have recently argued that epistasis has been ignored in quantitative genetics and in its evolutionary studies because of convenience, and consequently statements about its insignificance are misleading.
Genomics offers tools that go much deeper, to identify the genes involved and their interactions within the system, perhaps thereby providing an understanding of the causes of specific complex diseases or traits and a route to their improvement. There has, therefore, been a surge of interest in estimating the magnitude of epistasis and understanding its source. Despite the challenges in estimation, thousands of QTL have been found and many findings of interaction effects reported in crosses of divergent lines of chicken (Pettersson et al. 2011), yeast (Bloom et al. 2013), and Drosophila (Huang et al. 2012). Further, there is recent evidence of detection of epistatic loci for levels of gene expression in human populations (Hemani et al. 2014; Brown et al. 2014).
Nevertheless the variation contributed by these epistatic loci detected in segregating populations is generally found to be small (Huang et al. 2012; Brown et al. 2014; Hemani et al. 2014), which fits with the theoretical predictions (Hill et al. 2008) that only at high heterozygosity levels do loci contribute much epistatic variance; Bloom et al. (2013) note that the epistatic variance they found would have been much lower had they not been using an F1-based population.
The genetic variation accounted for by significant SNPs in genome-wide association studies (GWAS) in humans for both metric traits such as height and for multifactorial disease traits has typically been substantially less than the estimates of the additive genetic variation found in conventional pedigree-based quantitative genetic analyses. For example, the top 150 loci identified by SNPs account for only 10% of the variance in human height (Lango-Allen et al. 2010). If all SNPs are fitted without constraint as to statistical significance, the amount explained rises to ∼40% (Yang et al. 2011) but still falls well short of the estimates of ∼80% based on covariances among relatives. Several hypotheses for the so-called “missing heritability” (Manolio et al. 2009) have been put forward, among these that epistasis is inflating the pedigree-based estimates (Zuk et al. 2012).
We therefore have some dichotomy between, on the one hand, those who argue that, although there may be epistasis, the epistatic variance contributed is likely to be small and generally we can ignore its contribution because it is unimportant, and on the other those who argue that this is a blinkered view. We cannot resolve this here, but we do attempt to provide a stronger theoretical base to the arguments. The analysis and inferences of Hill et al. (2008) took into account the size and direction of interaction effects and the allele frequencies or heterozygosity at all contributing loci. Models were, however, based on pairs of loci, yet as quantitative geneticists have long assumed, have subsequently inferred from the continuing responses to selection for many traits in experiments and breeding programs, and now inferred more directly from GWAS, very many loci influence each trait. For multiple loci (n), however, the numbers of pairs with potential two-way interaction among loci increases as n2 [strictly 1/2n(n – 1)], three-way interactions as n3, etc., and so it is possible that merely accounting for interactions among pairs of loci gives a biased impression.
In this article we therefore consider the magnitude of epistatic variance expected in outbred populations for multilocus models to check on the robustness of the argument that most variance is likely to be additive genetic. We concentrate initially on models without dominance or interactions involving dominance, partly for simplicity of exposition and partly because we showed previously that additive variance is also expected to contribute much more than the dominance variance (Hill et al. 2008). We consider alternative distributions of the gene frequency within the populations at segregating loci affecting the trait because these have an important impact on the magnitude of the genotypic variance and its components. As these distributions are not static in populations evolving under selection, we also consider the impact of consequent gene frequency changes.
Analysis
Genetic model with no dominance
We assume that there are n loci, each of which has two alleles Ai or ai, i = 1, …, n, with frequencies pi and 1 – pi, respectively. We further assume that there is Hardy–Weinberg and linkage equilibrium among all loci. Genotypic values are given in Table 1 (second column) for examples of alternative genotypes, and those for other genotypes are analogous, e.g., that for A1A1a2a2A3a3 is 2a1 + a3 + 2[aa]13 (cf. fourth row). The model readily extends to more loci and to dominance (see below and third column of Table 1) and is fully parameterized.
Table 1. Genotypic values for representative genotypic combinations for a three-locus diploid model including two- and three-locus interaction effects.
| Genotype | Genotypic values without dominance | Additional terms under dominance |
|---|---|---|
| a1a1 a2a2 a3a3 | 0 | 0 |
| a1a1 a2a2 A3a3 | a3 | d3 |
| a1a1 a2a2 A3A3 | 2a3 | 0 |
| a1a1 A2a2 A3a3 | a2 + a3 + [aa]23 | d2 + d3+ [ad]23 + [da]23+ [dd]23 |
| a1a1 A2a2 A3A3 | a2 + 2a3 + 2[aa]23 | d2 + 2[da]23 |
| a1a1 A2A2 A3A3 | 2a2 + 2a3 + 4[aa]23 | 0 |
| A1a1 A2a2 A3a3 | a1 + a2 + a3 + [aa]12 + [aa]13 + [aa]23 + [aaa]123 | d1 + d2+ d3 + [ad]12 +[ad]13 + [ad]23 + [da]12 + [da]13 + [da]23 + [dd]12 + [dd]13 + [dd]23 + [aad]123 + [ada]123 + [daa]123 + [add]123 + [dad]123 + [dda]123 + [ddd]123 |
| A1a1 A2a2 A3A3 | a1 + a2 + 2a3 + [aa]12 + 2[aa]13 + 2[aa]23 + 2[aaa]123 | d1 + d2 + [ad]12 + [da]12 + 2[da]13 + 2[da]23 + [dd]12 + 2[daa]123 + 2[ada]123 + 2[dda]123 |
| A1a1 A2A2 A3A3 | a1 +2a2 + 2a3 + 2[aa]12 + 2[aa]13 + 4[aa]23 + 4[aaa]123 | d1 + 2[da]12 + 2[da]13 + 4[daa]123 |
| A1A1 A2A2 A3A3 | 2a1 + 2a2 + 2a3 + 4[aa]12 + 4[aa]13 + 4[aa]23 + 8[aaa]123 | 0 |
The population mean without dominance is
With H–W and linkage equilibrium, average effects (also called “additive effects”) and the additive variance can be found by averaging over genotypes, by regression of phenotypes on the number of increasing alleles within loci (Fisher 1918), or from the first derivative of the mean with respect to the frequency of the increasing allele (Kojima, 1959, 1961). We adopt Kojima’s approach, which extends simply to epistatic effects using higher derivatives involving the corresponding loci. (In the absence of dominance higher derivatives at individual loci are zero.)
For locus i, the average effect including up to third-order interaction terms is
| (1) |
The additive variance is obtained by summing over the n loci
where Hi = 2pi(1 – pi) is the heterozygosity at locus i. Similarly additive × additive interaction effects and variances are
| (2) |
and so on, with the nth such partial derivatives giving n-locus epistatic effects and variances.
For three loci, for example,
and therefore
| (3a) |
| (3b) |
| (3c) |
The key to subsequent results is that, because two- and three-locus interactions enter the average effects and additive variance, they can make a major contribution to VA, depending on their allele frequencies and sign. Similarly k-locus interaction effects contribute to the epistatic variances of order <k, whereas single-locus contributions and interaction effects of order lower than k do not.
Numbers and magnitude of terms in variance components
While it is assumed that many loci determine most quantitative traits, the number (n) is not known. To obtain some understanding of how the relative magnitudes of additive and epistatic variance depend on n, we undertake some simple calculations on the basis that the partition will depend at least to some extent on the numbers of terms contributing to each. For the additive variance there are contributions from n loci, and expansions such as in Equations 1 and 3a show that for each locus there are
terms in the expression for the average effects and thus n22(n − 1) = n4n − 1 in all when effects are squared (counting terms such as ai[aa]jk and [aa]jkai separately). There are more pairs, trios etc. of loci contributing epistatic variance, but the numbers of terms they each comprise are reduced. For VAA there are = 1/2n(n − 1) pairs of loci but 4n − 2 terms in each pair when squared (Equation 3b). In general, there are a total of 4n − k terms for the kth-order epistatic variance among n loci. While terms such as [aa][aaa] that comprise products of different interaction effects may be negative, 2n − k are squared terms and therefore nonnegative.
In the formulae for the average and other effects, terms in L loci have a coefficient of 2L − k − 1, e.g., 4 for pjpk[aaa]ijk in Equation 1. But coefficients involving products of gene frequencies also appear in these same terms, each of which take values of 0.5 when gene frequencies are one-half. Thus if we assume gene frequencies are 0.5, which is when epistatic variance is typically at a maximum, this cancels the coefficients of 2 alluded to above. Hence while there are arguments for including other terms in these simple models, as a basis we restrict them just to assuming these factors of two cancel. Hence the sum of weighted terms is 4n − k and of squared terms is 2n − k. Examples of these figures are given in Table 2, which shows how the number of contributions to the variances, particularly the additive variance, can be large indeed.
Table 2. Number of variance components of order k among n loci (k = 1 denotes additive variance), numbers of terms in the expansion of the variance from effects, and number of such terms that are squared terms, 2n − k.
| No. of loci n | k | 4n−k | 2n−k | 2n−2k | 2n−4k | |
|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 1 | 0.5 | 0.125 |
| 2 | 1 | 2 | 8 | 4 | 2 | 0.5 |
| 2 | 1 | 1 | 1 | 0.25 | 0.0156 | |
| 3 | 1 | 3 | 48 | 12 | 6 | 1.5 |
| 2 | 3 | 12 | 6 | 1.5 | 0.0938 | |
| 3 | 1 | 1 | 1 | 0.125 | 0.0020 | |
| 4 | 1 | 4 | 256 | 32 | 16 | 4.0 |
| 2 | 6 | 96 | 24 | 6 | 0.375 | |
| 3 | 4 | 16 | 8 | 1 | 0.0156 | |
| 4 | 1 | 1 | 1 | 0.0625 | 0.0002 | |
| 5 | 1 | 5 | 1280 | 80 | 40 | 10 |
| 2 | 10 | 640 | 80 | 20 | 1.25 | |
| 3 | 10 | 160 | 40 | 10 | 0.0781 | |
| 4 | 5 | 20 | 10 | 0.625 | 0.0024 | |
| 5 | 1 | 1 | 1 | 0.0312 | 0.0000 | |
| 6 | 1 | 6 | 6144 | 192 | 96 | 24 |
| 2 | 15 | 3840 | 240 | 60 | 3.75 | |
| 3 | 20 | 1280 | 160 | 20 | 0.313 | |
| 4 | 15 | 240 | 60 | 7.5 | 0.0145 | |
| 5 | 6 | 24 | 12 | 0.375 | 0.0004 | |
| 6 | 1 | 1 | 1 | 0.0156 | 0.0000 |
The number of squared terms are also shown multiplied by the maximum heterozygosity (1/2)k for the k order term, i.e., 2n − 2k, and also by a more typical mean heterozygosity in a long standing population (1/8)k, i.e., 2n − 4k.
Heterozygosity has a maximum of 0.5 at P = 0.5, when, in the absence of epistasis, the additive variance is then at a maximum. As the epistatic variances among k loci depend on products of k heterozygosity terms, the epistatic variances are likely to contribute correspondingly less to the genotypic variance VG than does VA, and most of the epistatic variance is likely to derive from second-order components. Thus if we include heterozygosity in the calculations, letting H be an “average” or representative figure, the kth component of variance is of order Hk4n − k. If H = 0.5, its maximum, the weighted number of terms, becomes (1/2)k2n − k = 4n − k, of which 2n − 2k are squared terms. Further, in all but populations derived from F1 crosses, mean heterozygosity will be less than one-half, and we subsequently consider the impact of different allele frequency distributions on the expected proportion of epistatic variance. Accordingly, the weighted numbers of terms become ∼4n − 4k and 2n − 4k, respectively, i.e., very much smaller indeed as k increases.
Examples of the computed values are given in Table 2. These calculations for multiple loci are intended to serve only as a guide, but illustrate why a high proportion of epistatic variance is unlikely, because even very strong multilocus gene–gene interactions do not automatically lead to a high proportion of epistatic components in the genetic variation (cf. Cheverud and Routman 1995).
Examples
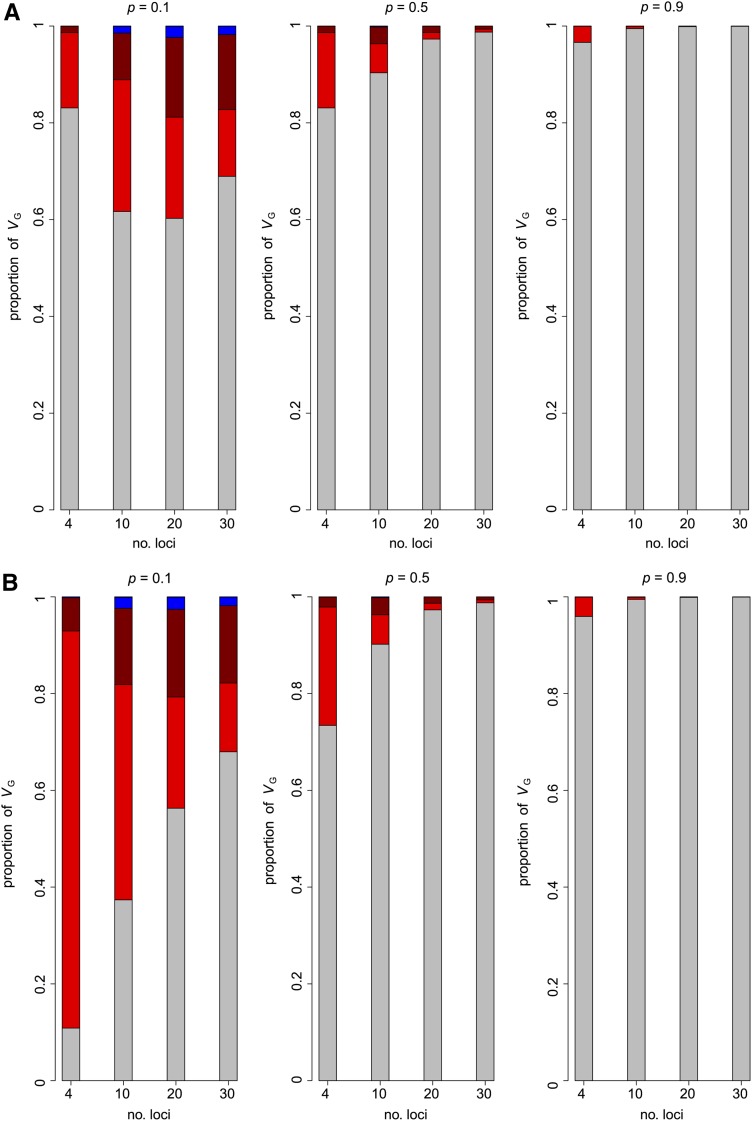
The relative magnitude of additive and epistatic variances for two to five loci including all possible gene–gene interactions is summarized in Figure 1 for two models with the frequency of the increasing allele the same at all loci. In both, the magnitude of gene and (absolute value of) interaction effects are also assumed to be the same at all loci, and are either all synergistic ([aa] = [aaa] = … = a) or antagonistic ([aa] = [aaa] = … = −a). As expected from the above arguments, the genetic variance in many of these examples is seen to be mainly additive and the remainder mainly additive × additive, such that higher-order epistatic variances can generally be ignored. Gene interactions generally make the most substantial contributions to VA when the frequency of the increasing allele is very low, and only negative (antagonistic) interactions cause relatively large ratios of epistatic to additive variance because terms such as ai[aa]jk in VA are then negative.
Figure 1.
Multilocus models with gene interaction involving four to 30 loci. The proportion of additive variance (gray), two-locus (red), three-locus (dark red) and four-locus (blue) epistatic variance in the total genotypic variance for models with different numbers of loci with interactions of all orders among them. The loci have equal effects and allele frequencies (p), and either (A) positive (synergistic) or (B) negative (antagonistic) interaction effects, all of the same absolute value as the single locus effect. (A) [aaaaa] = [aaaa] = [aaa] = [aa] = a. (B) [aaaaa] = [aaaa] = [aaa] = [aa] = −a.
Expected variances for different allele frequency distributions
The components of genetic variance and their relative magnitudes depend on the allele frequencies in addition to the individual gene effects and their interactions. Although there is little information on allele frequencies for quantitative trait genes, it is possible to predict their frequency distribution under different assumptions about relevant evolutionary factors, such as mutation, selection, finite population size (drift), and migration. Allele frequencies of 0.5, i.e., from a recent inbred cross, serve as one reference point and, as evidence suggests that many genes influence metric traits and so there is presumably very mild natural selection on them, theory for neutral alleles in finite populations serves as another.
Two allele frequency distributions are considered, each with mean 0.5. One is a closed finite population under drift without mutation, when the steady-state distribution is uniform over (0, 1) (e.g., Crow and Kimura 1970; Falconer and Mackay 1996). The other is for mutation–drift balance, when the frequency density of mutants is proportional to 1/p (Wright 1931); but, if increasing and decreasing mutants for the trait are assumed equally likely, the frequency distribution of segregating increasing mutants for the trait is U shaped, with f(p)∝1/[p(1 – p)] (Hill et al. 2008). The distributions and expected heterozygosities E(H) are
For the U-shaped distribution and effective population sizes N = 10, 20, 100, 1000, and 100,000, E(H) ∼0.167, 0.136, 0.0944, 0.0658, and 0.0410, respectively. Examples are given in Figure 2 for the partition of variance under different epistatic and frequency distribution models for three and more loci. The largest proportion of epistatic variance is expected when P = 0.5 while uniform and U-shaped allele frequency distributions yield mostly additive variance, even with antagonistic gene–gene interaction. In all cases the epistatic variance is almost entirely contributed by the two-locus epistatic variance.
Figure 2.
Expected components of genetic variance under gene–gene interaction. As Figure 1, but for fewer loci, assumed allele frequency (p) distributions of P = 0.5, uniform and U-shaped (N = 100) distribution (respective expected heterozygosities E(H)) with two- and three-locus interaction effects with (A) constant or (B) declining gene and negative interaction effects with increasing number of loci. (A) [aaa] = [aa] = −a. (B)
When n loci contribute nearly equally to the variation, for given total genetic variance, their individual effects a will tend to decline approximately in proportion to as n increases to and, similarly, epistatic interactions between pairs of loci [aa] are likely to decline roughly in proportion to 1/n and between trios of loci [aaa] by . The diminishing relative size of the interaction effects with increasing number of loci shown above for those with equal effect is therefore exacerbated as the interaction terms become individually smaller (Figure 2A vs. Figure 2B). Hence, if there is a very large number of interacting loci (say >50) almost all the variation is expected to be additive. If their effects differ greatly, then the results become like those for fewer loci.
To illustrate the effect of increasing the number of loci, we include interactions only up to second or third order with positive (or negative) sign consistent over loci. Again, unless interaction terms are such that, for example, [aa] is negative and its products in the expressions for variance balance the positive a2 and [aa]2 term (Figure 2), the general conclusion is that, with a very large number of loci, only in special cases are the positive and negative terms likely to counter each other and the epistatic variance to be larger than the additive variance. Similarly, as the number of loci become very large, epistatic variance of higher order also become trivial compared to VAA.
Dominance
The diploid models can easily be extended to incorporate dominance effects and Kojima’s (1959) method can be used to compute the different components of genetic variance. This has been done to illustrate the close correspondence with additive models (Appendix 1). The inclusion of dominance introduces three additional two-locus interaction terms [ad], [da], and [dd]. Similar to the way in which the additive interactions contribute to the additive variance, the dominance interactions contribute to both the additive and dominance variance. The number of additional terms in the formulae for the variances increases rapidly, so for clarity, results are given only for two loci with positive interactions (Figure 3).
Figure 3.
Components of genetic variance under dominance. Influence of positive two-locus additive and dominance interaction on (A) the expected values of additive, dominance and epistatic components of genetic variation with allele frequency distributions P = 0.5, uniform and U-shaped distribution, model i: a = d and all interaction terms = 0, j: a = d = [aa] = [ad] = [da] = [dd], and (B) the partition of genetic variation with a varying number of loci with equal allele frequency of 0.5 (VA gray, VD dark gray, VAA red, VAD+VDA dark red, VDD black) and effects like in the model j.
As in the case of additive interactions, the proportion of epistatic variance is small, in particular when allele frequencies are not concentrated at intermediate values. The proportion of dominance variance stays the same or is slightly increased when adding the dominance interaction while the ratio between additive and dominance variance is not changed, implying that the interaction terms contribute to these variances in the same way. When the allele frequencies are 0.5 there is most epistatic variance, but its contribution rapidly decreases for a higher number of loci, with the same pattern as with only additive effects and interactions.
Effect of selection on the expected variances
For traits under stabilizing selection with intermediate optimum, frequencies of increasing and decreasing alleles will also tend to be distributed around 0.5 as in the neutral case. In a population newly under directional selection, the mean frequency of increasing alleles will increase >0.5, but under long continued selection stabilize as new mutants come in. Hence we consider the results for neutral distributions to be still relevant. We examine some limited examples, however.
In a population under selection the contribution of epistatic to total variance changes as frequencies change. Thus as Hansen (2013), for example, emphasizes, although selection response at any given time depends almost entirely on the additive variance (Kimura 1965; Crow 2010), the long-term trajectory depends on the nature of the interactions among the loci. Further, allele frequency distributions can no longer remain symmetric around P = 0.5 under directional selection. In a small investigation of the robustness of some of our conclusions under selection, for example, that a high proportion of VG is contributed by VA, we used transition probability matrices to obtain the distributions for pairs of unlinked loci, modeling with quite small population size values. For simplicity a haploid model with population size Nh was assumed. Details of the model are given in Appendix 2.
Several simple situations were modeled using two loci with synergistic or antagonistic epistatic effects ([aa] = +a or –a). These included cases where (i) the increasing alleles at each locus were mutants at initial frequency 1/Nh, (ii) the frequency at one locus was at steady state under neutrality (i.e., U shaped) and at the other locus a mutant allele of either increasing or decreasing effect, and (iii) initial frequencies at the two loci were set as either 1/Nh or 1 – 1/Nh with equal probability. Components of variance were obtained by averaging over 100 generations.
Detailed findings are given in Appendix 2. Although limited in scope, the results are clear: the asymmetry in the allele frequency distributions caused by selection does not seem to influence the relative proportions of additive and nonadditive variance and thus substantially alter the general expectation that the proportion of epistatic variance is likely to be small.
Discussion
Main findings
When the genetic variation is caused by segregation at many loci, the magnitude of the epistatic variance is almost certain to be trivial relative to the additive variance. This result is nearly invariant to the order of the interaction, allele frequencies, and type and magnitude of interaction effects. The models show how the gene-interaction effects appear in the average effects and contribute to the additive variance and produce more positive terms there than in the epistatic components. The epistatic components depend on the magnitude of heterozygosity and are generally small for low heterozygosity. For the same reasons, under most of the assumptions the majority of the epistatic variance is due to the two-locus epistatic component.
When the variation is due to only a few interacting loci or there are major genes with substantial gene–gene interaction effects segregating in the population, there are no general patterns in the proportion of epistatic variance, but the following are the main observations: high heterozygosity (or allele frequency 0.5) and/or negative interaction can generate high proportions of epistatic variance; the proportion of the variance that is epistatic variance is lowest at low heterozygosity, notably for the U-shaped allele frequency distribution but even for the uniform distribution compared to P = 0.5; and deviations from symmetry of the distribution caused by mild selection do not increase the low proportion of epistatic variance.
Models and assumptions
Only biallelic loci were analyzed but we cannot see that the general results would be fundamentally affected if loci are multiallelic. Each of the m two-way interactions for m alleles at each locus with no dominance appear in the average effects for the m alleles at the m loci, giving m3 terms after squaring effects, whereas the number of additive × additive effects and variance components increases only in proportion to the number of allelic pairs, m2, as each has only one two-way interaction. Although we focused on models without dominance, as the interaction effects containing dominance contribute to the dominance variance in the same way as do the additive × additive effects to the additive variance, the qualitative finding remains that the proportion of epistatic variance in the total genotypic variance is small.
Epistasis also arises, even with infinitesimal additive effects on some underlying variable, if there is a nonlinear relationship between genotypic and observed phenotypic values, such as with a multiplicative, optimum, or threshold model (e.g., Wright 1931; Dempster and Lerner 1950; Cockerham 1959). Kojima’s (1959, 1961) methodology provides a simple way to partition the genotypic variance and can also be applied to these models. Basically, the proportion of epistatic variance is high only when the transformation departs far from linearity: a high coefficient of variation in the multiplicative model, the population mean near the optimum value in the optimum model, and with a proportion truncated near 0 or 1 in the threshold model (see also Mäki-Tanila and Hill 2014).
In an attempt to explain some of the missing heritability, especially that estimated from human twin studies, Zuk et al. (2012) suggested that the additive component was overestimated, biased inter alia by epistatic variance. They proposed an extreme type of threshold model, with the all-or-none output dependent on whether, in some underlying system of multiple identically independently (i.i.d.) normally distributed variables, the lowest exceeded the cutoff. This model undoubtedly generates substantial epistatic variance, but has been queried both because it is sensitive to assumptions, such as i.i.d. and is biologically implausible (Stringer et al. 2013). Indeed there is little evidence of such limiting pathways, and models based on flux through systems without “limiting steps” have firmer foundation (Kacser and Burns 1973). Analysis of genetic models of flux in pathways shows that largely additive variance is to be expected (Keightley 1989; Hill et al. 2008). Much of the missing heritability can be accounted for by fitting all SNPs without regard to statistical significance (Purcell et al. 2009; Yang et al. 2011), showing that multiple sites are involved. This observation that multiple loci influence the traits coupled with our demonstration that this lends support to additive rather than nonepistatic models argues against the belief that much of the missing heritability is due to epistatic variation.
In the examples studied, we set the gene–gene interactions [aa], [aaa], …, to be of the same magnitude as the single-locus effects a. This is important in assessing the proportion of epistatic variance for models with very few loci but ceases to do so as the number of loci increase. It seems biologically reasonable to assume that the interaction effects are smaller, however, in which case a higher proportion of additive variance is seen with even a few loci and strong antagonistic pleiotropy. In the models studied we assumed that all loci were interacting with each other, but most interactions would not involve them all and consequently there would be relatively more additive variance.
Linkage disequilibrium
An important assumption made is that there is linkage equilibrium (no LD) among all pairs of loci comprising the analysis, indeed contributing to the trait. In higher organisms most pairs of loci are on different chromosomes or far apart on the same chromosome. Further, even for closely linked loci, LD is likely to be small in populations of large effective size, such as for humans and Drosophila melanogaster, although not so for closely linked loci in domestic livestock (e.g., de Roos et al. 2008, Veroneze et al. 2013) and laboratory populations of experimental species. A theoretical analysis of variance partition in the presence of LD is problematic because trait effects of different loci are not orthogonal; i.e., fitting locus B after A removes less variance than fitting B alone, and epistatic variance may be confounded with that of average effects. An alternative approach is to be pragmatic and consider just the quantities that can readily be estimated from the covariances among relatives such as half-sibs, full-sibs, parent–offspring, and grandparent–offspring. We have analyzed this complete LD scenario for pairs of epistatic loci including only additive effects and interactions. We made a further assumption that, as substantial LD occurs only among very tightly linked loci, recombination between close relatives can be ignored. Our preliminary analysis shows that the magnitude of epistatic variance, as judged say by the sire × dam interaction component or the difference between the full-sib and offspring–parent covariance, remains small relative to the ‘additive’ component (e.g., 4 × covariance of half sibs), as in similar models examined here for linkage equilibrium (Mäki-Tanila and Hill 2014).
Prediction of selection responses due to epistasis
The success of a selection scheme within populations depends on utilizing the additive variance. Griffing (1960) showed with theoretical analyses how the genetic changes obtained with mass selection that derive from the epistatic variation are only temporary, as selected haplotypes break up. Similar predictions were made by Bulmer (1980) by comparing the offspring–parent and grandoffspring–grandparent regressions. Estimates of VA based on four times the half-sib covariance, which has expectation 1/4VA + (1/16)VAA + …, are, however, unlikely to be substantially biased by epistatic variance because, for typical quantitative VAA, they are expected to be small compared to VA. While the rate of recombination loss is lower with linkage, as shown by Kimura (1965) and Crow (2010), only the additive variance contributes to the steady-state response even in the presence of linkage.
Hansen (2013) has argued that epistatic effects must be included in predicting long-term responses to natural selection. While short-term response to selection can be predicted from the additive variance, changes over many generations depend on genetic architecture, including the distribution of gene effects and frequencies at loci that do not exhibit epistasis and therefore on many unknowns. Thus, under most circumstances, Hansen’s objective is not achievable in practice, and adding another level of complexity would only exacerbate the uncertainty. Further, the changes observed in plant and animal breeding have continued for very many generations, often with little or no sign of slowing down even in very intensive selection schemes (Dudley and Lambert 2004; Hill 2010). The achieved changes have been far beyond what could be foreseen in the early stage of selection.
Nelson et al. (2013, p. 671) find classical quantitative genetics incapable of giving “insight to infer the underlying, functional architecture of the traits required to predict phenotypes of individuals in other populations, as desired in medical diagnostics, or long-term changes in populations studied in evolutionary biology.” The incorporation of epistasis in predicting breeding values or individual risks would, for example, also depend on the distribution of gene effects across loci and the genome. Although they consider the existing research insufficient for characterizing epistatic effects needed in predicting individual phenotypes, their objective, like that of Hansen (2013), is laudable. The incorporation of epistasis in predicting breeding values or individual risks would depend on distribution of gene effects across loci and genome. The prediction of response to directional selection over multiple generations, however, already contains many unknown even without epistasis, not least the consequences of mutations. Adding another level of complexity would only exacerbate problems.
Epistatic variance and findings on functional epistasis
Experiments from crosses among lines have revealed many clear cases of epistatic QTL, by Bloom et al. (2013) in yeast, Huang et al. (2012) in Drosophila, and Pettersson et al. (2011) in chickens. These contributed a sizable proportion of the variance. As we have shown, however, the contribution of epistasis is likely to be largest with genes at frequency one-half, and so these findings do not imply substantial contributions of epistatic variance in natural or domesticated populations maintained segregating with large effective population size. In contrast to such studies using defined crosses e.g., in which gene frequencies are all 0.5, in (outbred) human and livestock populations minor allele frequencies are likely to be diverse and typically low. There are hundreds of loci affecting both continuous and disease traits, hence vast numbers of potential locus combinations, most of which are not present in the populations analyzed. This is in line with the findings by Huang et al. (2012) and Hemani et al. (2014) of several epistatic QTL but nevertheless these made very little overall contribution to the genetic variances. In an analysis of gene expression, for loci found to be interacting by variance heterogeneity, Brown et al. (2014) detected on average 4.3% of the variance to be epistatic, with the highest up to 16%. This represented two successive selection processes in the analysis, however.
Mackay (2014) argues for the importance of epistasis, and while acknowledging the fact that substantial amounts of epistasis do not in themselves produce much epistatic variance, provides experimental data on behavioral traits in D. melanogaster. For Drosophila inbred lines that were derived from an outbred population the variance among lines was much more than twice the additive variance within populations expected under an additive model. This comparison is, however, subject to potential biases due to nonadditive gene action within loci: notably low-frequency recessives contribute little variance within lines but a lot between lines. Her group also concluded from comparisons of QTL detected from GWAS and from selection within a population that there was little correspondence in the estimates of effects (regions of large effect) between them, interpreting this as epistasis. In such an experiment, although lack of power in either analysis leads to noncorrespondence and apparent interaction, this was minimized by focusing on variants with highly significant marginal effects.
The best-known biochemical networks of up to a few dozen nodes in laboratory organism exhibit much interaction (Phillips 2008). Therefore it is important that research on functional epistasis of individual genes is pursued to gain more knowledge about the nature of quantitative trait variation. The new findings can be then incorporated in the statistical models elucidating the state and potential in the variation. Quantitative geneticists would like to exploit all the information underlying genotypic values. The challenges in detecting interaction effects are great for a number of reasons: multilocus genotypic classes have lower frequency, epistatic effects are likely to be smaller than main effects, power of detection in GWAS depends on there being LD between two pairs of trait genes and markers, and their detection has to pass more stringent statistical thresholds because so many tests are undertaken. Therefore it is not yet feasible to obtain information on epistasis among individual loci for quantitative traits pending as-yet-unforeseen advances in technology or ideas are achieved.
Conclusions
We show that substantial epistasis among multiple loci is likely to lead to mostly additive genetic variance in outbred populations. The common error is, however, to assume a lot of one implies a lot of the other. Perhaps the controversies arise because it is not fully appreciated or scientists choose not to acknowledge this rather mundane theoretical finding. An understanding of the biology, however, requires that the epistasis be identified. A pragmatic approach is likely to be more successful in areas such as livestock improvement or understanding the variation of most traits in humans. Only when at the individual gene or gene pair do we need to concern ourselves greatly with epistasis.
Acknowledgments
We thank Nick Barton for helpful comments on the manuscript. A.M.T. acknowledges the receipt of a fellowship from the OECD (The Organisation for Economic Co-operation and Development) Co-operative Research Programme: Biological Resource Management for Sustainable Agricultural Systems in 2013 for the visit to Edinburgh.
Appendix
Appendix A: Partition of Variance for Loci Showing Dominance
Inclusion of dominance yields an additional term such that heterozygotes differ from the homozygous mean at locus i by di, with corresponding interaction terms A × D, D × A, and D × D for two loci, defined in Table 1 for three loci and extending naturally to more.
Table A1. The transition matrix approach used to study the influence of genetic background on the impact of a new mutation (either with increasing or decreasing effect) on the proportion of additive variance (VA) in the genotypic variance (VG).
| Mutation b → B | Mutation B → b | |||
|---|---|---|---|---|
| No selection VA/VG | Selection VA/VG | No selection VA/VG | Selection VA/VG | |
| Positive interaction [aa] = a | 0.965 | 0.968 | 0.981 | 0.988 |
| Negative interaction [aa] = −a | 0.923 | 0.904 | 0.793 | 0.742 |
The variation is caused by two interacting loci. A haploid population of size 40 is first iterated for 100 generations with or without selection to reach a steady state on one locus and the impact of mutation (either to decreasing b or increasing B allele) at the other locus is estimated after another 100 generations with or without selection, respectively. Selection intensity applied was 0.29.
Assuming Hardy–Weinberg and linkage equilibrium, the mean is
The average effect at locus i, including only terms up to two loci for illustration, is
and the dominance deviation at the locus, obtained by differentiating again with respect to pi (Kojima 1959), is
To simplify these expressions and those for epistatic effects, we define the following symbolic expressions
and the outcomes of symbolic products, all of which are commutative, among them. For example,
and similarly for higher-order terms.
Hence, from the above and equivalent expressions it can be seen that
giving symbolic form of similar structure to that for μ and ∂μ/∂pi in the additive case (cf. Equation 2). Then
and so on for higher-order terms. This form shows results in a similar pattern to that of the additive model. For the dominance deviation we have
The variances VA and VD are the sums over loci of Hi(1/2∂μ/∂pi)2 and Hi2(1/4∂2μ/∂pi2)2, respectively. The total for VAAD, for example, comprises sums over locus trios
and similarly for higher-order terms.
Appendix B: Method for Analysis of Effect of Selection on the Expected Variances
Based on diffusion equation approximations (e.g., Crow and Kimura 1970) the results should approximate those for populations with larger N values but similar values of N × gene effects (or selection coefficients) for such loci, on a time scale proportional to N. We use a haploid model but approximate a diploid population of (approximate) effective size Nh/2. The vector of gene frequency values has dimension (Nh + 1)2, where each element specifies the probability P(i1, i2) that there are i1 copies of the increasing allele at the first locus and i2 at the second at some generation t. The transition matrix of dimension (Nh + 1)2 × (Nh + 1)2 defines conditional probabilities of joint frequency change between states. Selective values are computed assuming truncation selection on the trait being modelled, with specified selection intensity S and selective values Sα1 and Sα2, where α1 and α2 are the average effects (i.e., including interaction terms) in units of the phenotypic standard deviation SD (based on Robertson 1960).
Mild selection (with S = 0.29 and values of a1 and a2 of 0.1 SD) was applied to represent the selection intensities when there is both genetic variation from many loci and environmental variation. The (haploid) population size was set at Nh = 40. Initial allele frequencies were set at 0.05 (2/Nh) for both loci and iteration was continued for 100 (i.e., 1.5N) generations, i.e., close to fixation. Iteration was also undertaken with the same starting frequencies and random mating, but no selection (i.e., S = 0). The results were computed as the average of the components of variance over the 100 generations. With positive interaction ([aa] = a), 96.8% of the genetic variation is additive with random mating vs. 96.5% with selection. For negative interaction ([aa] = – a), the proportions are 94.6 and 93.3%, respectively. In the results for this two-locus case the epistatic variance has a higher proportion with negative interaction while selection seems to alter this proportion very little. Checks undertaken with Nh = 20, the same starting frequency (1/20), higher intensity (S = 0.58), and fewer generations (50) gave proportions 92.9 vs. 92.2 for positive and 96.3 vs. 96.2% for negative interaction, respectively. The difference due to N is small, so the results with this order of population size serve as good indicators for the effect of selection.
In the second case, the initial frequency at locus j was set to 1/Nh (or 1 –1/Nh) (i.e., mutant in a population of size Nh = 40) and that for locus i at the frequency distribution expected at steady state of mutation and drift without selection (i.e., U shaped) after 100 generations of random mating or selection with the selection intensity as above. With no selection VAA is the same in all the runs, as is the additive variance in the mutated locus j while that for locus i changes greatly depending on the sign of the interaction [aa] of mutation due to the term pjaj[aa]ij in the formula for VA. The epistatic variance is again highest for negative interaction, while the results with and without selection differ very little and, in general, the genetic variation is mainly additive.
In the third example the starting allele frequencies for each of the two loci were set as either 1/Nh or 1 – 1/Nh and the results are presented as weighted averages over these four different initial conditions. Two relative mutation rates at locus B were considered: equal, (B → b) = (b → B), and unequal, (B → b) = 3(b → B), with Nh = 40. With positive interaction ([aa] = a) VA/VG averages 98.2% for the model with equal probability and 98.5% if unequal, when there was no selection and with selection (S = 0.29) 98.3 and 98.8%, respectively. When the gene interaction is negative, these values are slightly smaller, with no selection 90.4 and 84.2%, and with selection 90.3 and 84.2%, respectively. Hence, selection produced negligible difference. In conclusion, the asymmetry in the allele frequency distributions caused by selection does not seem to alter the general picture about the proportion of epistatic variance.
Footnotes
Communicating editor: S. Sen
Literature cited
- Ávila V., Pérez-Figueroa A., Caballero A., Hill W. G., García-Dorado A., et al. , 2014. The action of stabilizing selection, mutation and drift on epistatic quantitative traits. Evolution 68: 1974–1987 [DOI] [PubMed] [Google Scholar]
- Bloom J. S., Ehrenreich I. M., Loo W. T., Lite T. L. V., Kruglyak L., 2013. Finding the sources of missing heritability in a yeast cross. Nature 494: 234–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. A., Buil A., Viñuela A., Lappalainen T., Zheng H. F., et al. , 2014. Genetic interactions affecting human gene expression identified by variance association mapping. eLife 3: e01381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M., 1980. The Mathematical Theory of Quantitative Genetics. Oxford Unversity Press, Oxford [Google Scholar]
- Carlborg Ö., Haley C. S., 2004. Epistasis: Too often neglected in complex trait studies? Nat. Rev. Genet. 5: 618–625 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 2009. Elements of Evolutionary Genetics. Roberts Publishers, Greenwood Village, CO [Google Scholar]
- Cheverud J. M., Routman E. J., 1995. Epistasis and its contribution to genetic variance components. Genetics 139: 1455–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham C. C., 1954. An extension of the concept of partitioning hereditary variance for analysis of covariances among relatives when epistasis is present. Genetics 39: 859–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham C. C., 1959. Partitions of hereditary variance for various genetic models. Genetics 44: 1141–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J. F., 2010. On epistasis: Why it is unimportant in polygenic directional selection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1241–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J. F., Kimura M., 1970. An Introduction to Population Genetics Theory. Harper & Row, New York [Google Scholar]
- Dempster E. R., Lerner I. M., 1950. Heritability of threshold characters. Genetics 35: 212–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Roos A. P., Hayes B. J., Spelman R. J., Goddard M. E., 2008. Linkage disequilibrium and persistence of phase in Holstein–Friesian, Jersey, and Angus cattle. Genetics 179: 1503–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J. W., Lambert R. J., 2004. 100 generations of selection for oil and protein in corn. Plant Breed. Rev. 24: 79–110 [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics, Ed. 4th Longmans Green, Harlow, Essex [Google Scholar]
- Fisher R. A., 1918. The correlation between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. 52: 399–433 [Google Scholar]
- Griffing B., 1960. Theoretical consequences of truncation selection based on the individual phenotype. Aust. J. Biol. Sci. 13: 307–343 [Google Scholar]
- Hansen T. F., 2013. Why epistasis is important for selection and adaptation. Evolution 67: 3501–3511 [DOI] [PubMed] [Google Scholar]
- Hemani G., Knott S., Haley C. S., 2013. An evolutionary perspective on epistasis and the missing heritability. PLoS Genet. 9: e1003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G., Shakhbazov K., Westra H. J., Esko T., Henders A. K., et al. , 2014. Detection and replication of epistasis influencing transcription in humans. Nature 508: 249–253 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hill W. G., 2010. Understanding and using quantitative genetic variation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Goddard M. E., Visscher P. M., 2008. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 4(2): e1000008 10.1371/journal.pgen.1000008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Richards S., Carbone M. A., Zhu D., Anholt R. R., et al. , 2012. Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc. Natl. Acad. Sci. USA 109: 15553–15559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacser H., Burns J. A., 1973. The control of flux. Symp. Soc. Exp. Biol. 27: 65–104 [PubMed] [Google Scholar]
- Keightley P. D., 1989. Models of quantitative variation of flux in metabolic pathways. Genetics 121: 869–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempthorne O., 1954. The correlation between relatives in a random mating population. Proc. R. Soc. Lond. B Biol. Sci. 143: 102–113 [PubMed] [Google Scholar]
- Kimura M., 1965. Attainment of quasi linkage equilibrium when gene frequencies are changing by natural selection. Genetics 52: 875–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K., 1959. Role of epistasis and overdominance in stability of equilibria with selection. Proc. Natl. Acad. Sci. USA 45: 984–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K., 1961. Effects of dominance and size of population on response to mass selection. Genet. Res. 2: 177–188 [Google Scholar]
- Lango Allen H., Estrada K., Lettre G., Berndt S. I., Weedon M. N., et al. , 2010. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467: 832–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA [Google Scholar]
- Mackay T. F., 2014. Epistasis and quantitative traits: using model organisms to study gene–gene interactions. Nat. Rev. Genet. 15: 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäki-Tanila, A., and W. G. Hill, 2014 Contribution of gene–gene interaction to genetic variation and its utilisation by selection. Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, in press. [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagylaki T., 1993. The evolution of multilocus systems under weak selection. Genetics 134: 627–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R. M., Pettersson M. E., Carlborg Ö., 2013. A century after Fisher: time for a new paradigm in quantitative genetics. Trends Genet. 29: 669–676 [DOI] [PubMed] [Google Scholar]
- Pettersson M., Besnier F., Siegel P. B., Carlborg Ö., 2011. Replication and explorations of high-order epistasis using a large advanced intercross line pedigree. PLoS Genet. 7(7): e1002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips P. C., 2008. Epistasis: the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 9: 855–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. M., Wray N. R., Stone J. L., Visscher P. M., O’Donovan M. C., et al. , 2009. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460: 748–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A., 1960. A theory of limits in artificial selection. Proc. R. Soc. Lond. B Biol. Sci. 153: 236–249 [Google Scholar]
- Stringer S., Derks E. M., Kahn R. S., Hill W. G., Wray N. R., 2013. Assumptions and properties of limiting pathway models for analysis of epistasis in complex traits. PLoS ONE 8: e68913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veroneze R., Lopes P.S., S.E. Guimarães, F.F. Silva, M.S. Lopes et al, 2013. Linkage disequilibrium and haplotype block structure in six commercial pig lines. J. Anim. Sci. 91: 3493–3501 [DOI] [PubMed] [Google Scholar]
- Wright S., 1931. Evolution in Mendelian populations. Genetics 16: 97–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Manolio T. A., Pasquale L. R., Boerwinkle E., Caporaso N., et al. , 2011. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 43: 519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O., Hechter E., Sunyaev S. R., Lander E. S., 2012. The mystery of missing heritability: genetic interactions create phantom heritability. Proc. Natl. Acad. Sci. USA 109: 1193–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]