Abstract

We discuss the description of water and hydration effects that employs an approximate density functional theory, DFTB3, in either a full QM or QM/MM framework. The goal is to explore, with the current formulation of DFTB3, the performance of this method for treating water in different chemical environments, the magnitude and nature of changes required to improve its performance, and factors that dictate its applicability to reactions in the condensed phase in a QM/MM framework. A relatively minor change (on the scale of kBT) in the O–H repulsive potential is observed to substantially improve the structural properties of bulk water under ambient conditions; modest improvements are also seen in dynamic properties of bulk water. This simple change also improves the description of protonated water clusters, a solvated proton, and to a more limited degree, a solvated hydroxide. By comparing results from DFTB3 models that differ in the description of water, we confirm that proton transfer energetics are adequately described by the standard DFTB3/3OB model for meaningful mechanistic analyses. For QM/MM applications, a robust parametrization of QM-MM interactions requires an explicit consideration of condensed phase properties, for which an efficient sampling technique was developed recently and is reviewed here. The discussions help make clear the value and limitations of DFTB3 based simulations, as well as the developments needed to further improve the accuracy and transferability of the methodology.
Introduction
Water is arguably the most important molecule in life and physical sciences.1,2 The importance of water to the structure, stability, dynamics, and function of biomolecules has been well documented for decades. For example, since hydrophobic interaction is a major driving force for the stability of proteins, the structure and dynamics of water near protein are essential to the folding/association of proteins3,4 and dictate, at least in part, how small solutes influence biomolecular processes.5,6 Being a highly polar and polarizable molecule, water is often found in the active site of enzymes for the stabilization of otherwise high-energy intermediates;7,8 solvation of metal ions in the protein interior has been shown to make a significant contribution to the transport mechanism of these ions through ion channels and transporters,9,10 and interfacial water molecules between proteins have also been demonstrated to facilitate interprotein electron transfers.11 Another unique feature of the water molecule is its ability to relay the transfer of proton or “proton hole”12 (hydroxide) through the celebrated Grotthus mechanism;13,14 thus, water networks are commonly observed in proteins that mediate long-range proton transports.1,15−18 These unique physical and chemical properties of water manifest themselves in processes that occur in nonbiological systems as well, such as reactions in aqueous solutions and at various interfaces. As a recent example, the water layer at the surface of a Pt electrode was suggested to exhibit hydrophobic characteristics,19,20 which are expected to influence the composition and reactivity at the electrode/water interface.
This very incomplete list of properties and roles of water highlights why it is the most studied molecule by both experiments and theoretical means. For example, sophisticated spectroscopic techniques have been developed in recent years to analyze the structure and dynamics of water at various interfaces, the active site/surface of proteins, and also bulk water under different conditions (e.g., various phases of ice).21−24 From a theoretical and simulation point of view, water, without any doubt, is the molecule for which the largest number of models have been developed;25,26 these include quantum mechanical models,27−31 classical force fields with various levels of treatment for many-body effects,25,32−40 and statistical mechanical models in the framework of liquid state theories.41,42 Some of the models have been developed for general purpose simulations, while others have been motivated with more specific applications, such as investigation of nonlinear spectroscopies.43,44
In our own work, we have been focusing on developing methods for simulating water in different environments at two distinct levels and resolutions. One level is a semiempirical QM model45 referred to as density-functional-tight-binding (DFTB),46−48 which allows one to routinely carry out the QM/MM type of simulations for condensed phase systems at a 1–10 ns time scale, making it a valuable complement to ab initio QM/MM methods49−52 for problems that require extensive sampling. The other level is a coarse-grained (CG) model referred to as the big multipole water (BMW) model,53 based on which we also developed CG models for amino acids and common lipids in the general framework of the MARTINI force field,54 leading to the BMW-MARTINI model.55 Clearly, CG models are required to extend the length and time scales of simulations to address questions of increasing complexity.56−60
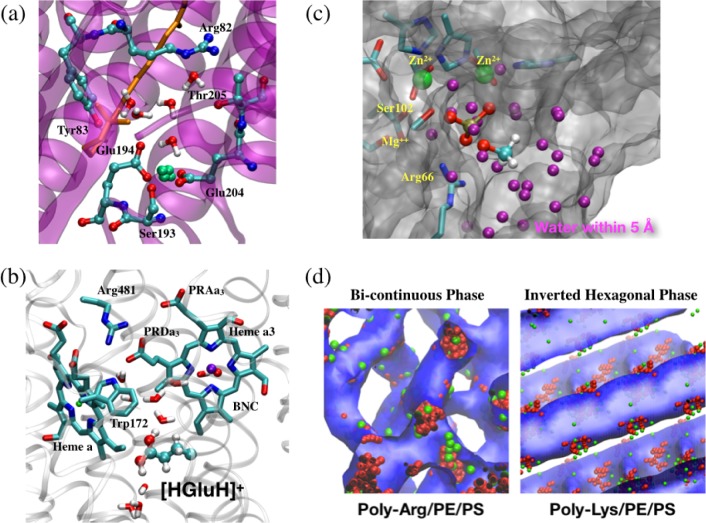
In Figure 1, we use several recent applications from our research to illustrate that calibrated semiempirical QM and CG models are uniquely valuable for a broad range of applications. For example, DFTB/MM simulations47,48,61−63 were used to probe the chemical nature of the proton storage group in bacteriorhodopsin (bR, Figure 1a)64−67 and the proton pumping mechanism in cytochrome c oxidase (CcO, Figure 1b).68,69 In those applications, calibration of proton affinity for the key groups involved and conducting adequate sampling of local protein/water motions proved to be essential. More importantly, both examples implicated rather unusual chemical species: a pair of glutamates interacting strongly with nearby water in bR65−67 and a transiently doubly protonated glutamate in CcO.69 Therefore, the use of a QM description for the region of interest was indispensable. Active site water molecules were found to play a major role in stabilizing these unusual species and therefore also needed to be treated at a QM level. The example of alkaline phosphatase (AP)70,71 (Figure 1c) highlighted that some enzyme active sites are rather solvent accessible, especially those with a significant degree of catalytic promiscuity.72 A proper description of the level of hydration requires a careful treatment of QM/MM interactions; otherwise, spurious stabilization of certain intermediates can be observed.73 Finally, the example in Figure 1d illustrated the need of CG simulations in the analysis of the phase behavior of complex materials (e.g., peptide/lipid mixtures),74 which remain difficult to study using atomistic simulations.
Figure 1.
Examples that illustrate recent applications of DFTB3/MM and CG (BMW-MARTINI) models where the proper description of water proved essential: (a) the proton storage site in bacteriorhodopsin involves a proton delocalized between amino acid side chains that are solvated by active site water;65−67 (b) proton transfers in cytochrome c oxidase proceed through water wire and a neutral glutamate near a hydrophobic cavity;69,75 (c) catalytic specificity and promiscuity in alkaline phosphatase are modulated in part by solvent molecules accessible to the active site;70,71 (d) capturing the distinct phase behaviors of cationic peptide/lipid mixtures with coarse-grained simulations relies on a careful treatment of electrostatics and water.74
Another topic we will not discuss extensively in this article but hope to highlight, nevertheless, concerns the sampling of hydration state, particularly the change of the level of hydration, when studying various water mediated processes in proteins and at interfaces. The importance of sampling water penetration coupled to events in the protein interior, such as deprotonation of a buried titratable group, has been emphasized by several authors.76,77 A notable example concerns an internal cavity in the protein cytochrome c oxidase, for which our recent simulation study suggested that the change of the hydration state of this cavity is likely of major functional significance.75 The importance of sampling hydration change at solid/liquid interfaces, however, has not been broadly emphasized. A recent example discussed by us78 is the binding of small molecules at the interface between water and metal oxide (e.g., TiO2). For the adsorbate to bind, the interfacial waters that occupy the potential binding site need to desorb. Due to the highly ionic nature of the metal oxide, however, the interfacial layer of water is strongly bound, making the desolvation of the binding site a rare event and therefore difficult to sample when it is not explicitly considered as part of the binding process. Both multidimensional PMF calculations and preliminary committor analysis78 underscored the importance of explicitly including surface desolvation as part of the binding reaction coordinate.
Considering the rich literature on water, including in particular excellent recent reviews on the computational models for water,25−29 the goal of this work is not to review the current status of water simulations in general. Rather, motivated by the discussion above, we focus on several remaining challenges for modeling water or hydration effects in different environments using DFTB based QM or QM/MM simulations; this is worthwhile considering the promise of these methods for biological and materials applications.47,48,62,63,79 To avoid distraction, we will not discuss our ongoing developments of the BMW model, although it is tempting to imagine a QM/MM/BMW framework for multiscale simulations, since BMW treats electrostatic interactions carefully and therefore is uniquely suited for integration with QM/MM models. In the following, we first discuss the status, limitations, and ongoing improvements of DFTB in the context of modeling water in different protonation states. Next, we discuss recent developments related to the DFTB-MM interactions; these are essential to the establishment of a reliable QM/MM treatment of solvation effects in solution and protein active sites. Finally, we end with concluding remarks.
DFTB3: Toward a QM Model That Bridges Clusters and Bulk
General Background
One promising semiempirical method that has emerged in recent years is the density-functional-tight-binding (DFTB) method,46 which has been thoroughly reviewed recently,79,80 including by some of us.47,48,81 Briefly, it is based on a Taylor expansion of the total electronic energy in the framework of density functional theory around a reference density, ρ0. The reference density is usually taken to be the sum of (slightly modified) atomic densities; the original method truncates at the second order, and recent developments81−84 extended the expansion to include terms up to the third order. The energy in DFTB is generally written in the following form:
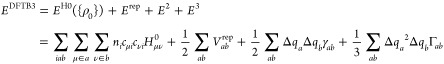 |
1 |
The first two terms contain only first-order contributions in the density fluctuations and constitute the DFTB1 model, which is not appropriate for biological applications.46,81 Including the third term, E2, leads to the DFTB2 model (originally referred to as SCC-DFTB46) which has been successfully applied to many biological problems.62,63 DFTB2 has been tested extensively for reaction energies, geometries, and vibrational frequencies of large sets of organic molecules.62,85,86 However, two rather systematic failures indicated a shortcoming of the computational model: the systematic underestimation of hydrogen bonding interactions and the accuracy for proton affinities. As discussed in detail previously,81−83 the function γab approximates the electron–electron interaction in the second-order terms in a way that lacks transferability across the periodic table. In particular, for interactions where hydrogen is involved, this approximation needs to be refined. A modified interaction term γabh was proposed,81 which seems to resolve these issues. Furthermore, for molecules where a net charge is highly localized, the description of the density within a minimal basis in the second-order terms is deficient. An extension to third order has led to a systematic improvement. In the first step, only the diagonal contributions to the third-order terms E3 were implemented,81,82 resulting in the DFTB3-diag model which also uses the γab interaction. Recently, the full third-order contributions were implemented,83 and the model is called DFTB3. Note that DFTB has been augmented with a damped empirical 1/R6 term to address the dispersion interactions early on,87 which are not included in the usual DFT-GGA functionals.
Most of the electronic contributions to the DFTB models are computed rather than fitted from DFT calculations,47 while there are only several adjustable parameters that determine the reference density, basis sets, and thus accuracy of the results.84,88Erep in eq 1 is a sum of two-body potentials that are fitted with respect to reference data. Since Erep depends only on the reference density, its form is in principle the same for DFTB1, DFTB2, or DFTB3. Indeed, the performance of DFTB3 using the repulsive potentials determined for DFTB2, referred to as “mio”, is surprisingly favorable.83 However, a fitting at each level can improve the performance of the model; therefore, a new set of parameters has been developed especially for the DFTB3 model based on a list of organic molecules of general biological interest; this parameter set is called “3OB”. Thus, a full specification of a DFTB method has to indicate both the model and the parametrization, e.g., DFTB2/mio or DFTB3/3OB. For reference, we summarize the DFTB models discussed here in Table 1, although we will focus primarily on the DFTB3 models.
Table 1. DFTB Models Discussed in This Work.
| nomenclature | third-order terms | parameterization | SRP?b | reference | applicability |
|---|---|---|---|---|---|
| DFTB2a | no | mio | no | (46) | general |
| DFTB3-diag | diagonal | mio | no | (81, 82) | improved proton affinity and H-bonding |
| DFTB3-diag+gaus | diagonal | mio | yes | (12) | improved proton transfer barrier |
| SCC-DFTBPR | diagonal | mio | yes | (89) | phosphate hydrolysis reaction |
| DFTB3 | full | 3OB | no | (84) | improved atomization energies for organic/biomolecules |
| DFTB3 | full | 3OBw | yes | this work | improved bulk water properties |
Referred to as SCC-DFTB in previous literature.
Whether the model is designed as a specific reaction parameter (SRP) approach.
The DFTB models involve several approximations; therefore, not all molecular properties can be determined using one model/parametrization with high accuracy. Following the strategy of semiempirical models, we have introduced “specific reaction parameters” (SRPs) for specific purposes.90 For example, with DFTB3-diag and “mio” parameters, proton transfer barriers and phosphate reaction energies were not well described. Accordingly, we have introduced SRPs that improved the performance for these special purposes, referred to as DFTB3-diag+gaus12,82 and SCC-DFTBPR,89 respectively (see Table 1). It was encouraging that, with the development of DFTB3/3OB,84,88 a SRP is no longer necessary for many of those properties, although there remain several limitations for complex reactions such as phosphate hydrolysis,88 indicating the necessity of continuing development of the DFTB model.
In the specific context of describing water mediated processes, DFTB3 and its earlier variants have been applied to many biological systems in which water participates in proton or proton hole transfers.12,91−95 In these problems, proton (hole) transfer generally takes place via small clusters of water molecules in protein interiors, which are very different from bulk water. Hence, to reiterate our statement in ref (96), for most biological applications, benchmark calculations using small clusters are much more relevant than setting the behavior of an excess proton in bulk water as the reference; even more importantly, it is essential to calibrate the electrostatics of the active site by, for example, microscopic pKa calculations of proton donor/acceptor groups.75,77,92,97,98 Nevertheless, considering that water is ubiquitous as a solvent, it is worthwhile to benchmark and improve the DFTB method for the description of bulk behaviors.96,99−101 Ultimately, the power of a reliable QM model lies in its transferability among typical, if not all, environments of potential interest. Along this line, we mention here that there are several specifically parametrized semiempirical methods that have been tuned to work well for either water clusters102 or bulk under ambient conditions,103,104 although their broad applicability to reactions in water remains to be tested.
For small neutral and protonated water clusters, DFTB3/3OB has been shown84 to give generally adequate results in terms of the magnitude of total binding energy and relative energies of different conformers. However, several recent studies96,99−101 have indicated the oversolvation tendency of all DFTB3 variants in bulk aqueous environments (e.g., see Table 2 and Figure 2), for neutral water as well as for a hydrated proton/hydroxide, which also likely leads to a higher density than 1 g/cm3 under ambient conditions. In these studies, detailed analyses96,100 revealed that the oversolvation behavior is largely associated with water population in the “interstitial” sites around the central species, such as occupation of the faces of the coordination tetrahedron around a neutral water molecule or presence of water molecules in between the three H-bond-accepting water molecules around a central hydronium.
Table 2. Integrated First Peak, Denoted by nO and nH, of gO–O and gO–H, Respectively, of Neutral Bulk Water with Different DFTB Models.
Figure 2.
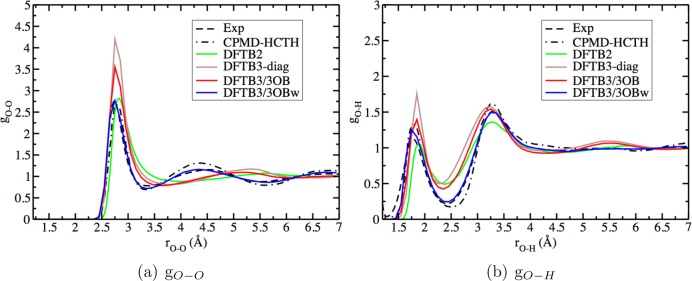
Comparison of neutral bulk water radial distribution functions from computations and experimental data.105 The CPMD and DFTB2 results are from ref (100), while DFTB3-diag data is from ref (96).
There are several origins for the limited accuracy of DFTB3 for the description of water, especially for bulk properties. First of all, the charge fluctuations in DFTB3 are treated at the monopole level, while multipole terms are known to be essential to electrostatic interactions, such as the angular dependence of hydrogen bonding interactions;106,107 due to electronic elements in DFTB3, however, the angular dependence of hydrogen bonding interactions appears to be properly described.108 Second, with a minimal basis set, DFTB3 has limited electronic polarizability109 and underestimated Pauli repulsion. For example, the dipole moment of water changes from ∼1.9 D in the gas phase to ∼2.4 D in the bulk (see the Supporting Information); the latter is comparable to the values for nonpolarizable water models, although larger values (∼3 D) were reported from DFT simulations of bulk water.110 The underestimation of Pauli repulsion is also reflected by the observation that internal rotational barriers are systematically too low with DFTB methods.47 Third, the Pauli repulsion is treated with the reference density and explicit charge dependence of short-range interactions111 is not included. In fact, considering these limitations, it is rather remarkable that DFTB3 works generally well for small to medium neutral and protonated water clusters; apparently, error cancellation is involved and not transferable to the bulk. Along this line, it is worth noting that York and co-workers developed a fragment based DFTB model that features multipolar interactions and Lennard-Jones potentials among water sites;102,108 the model was found to give reliable results for water clusters and bulk water under ambient conditions, although a systematic analysis of its performance for water in different protonation states and environments remains to be carried out.
Reverse Monte Carlo (RMC) Optimization of the O–H Repulsive Potential in DFTB3/3OB for Neutral Bulk Water Solvation Structure
To improve the transferability of DFTB3 for water in different environments, one possible avenue is to systematically evaluate its performance for water clusters of different sizes and structures and compare various components of intermolecular interactions (e.g., two-body and three-body contributions) to those from high-level ab initio calculations.38−40 The three issues mentioned above can then be improved to minimize the difference between DFTB3 and ab initio components. Since improving these terms will have a major impact on the parameters in DFTB3, systematic reparamerization is required,112 which may not be the most productive avenue for applications in the near future. Therefore, here we explore an alternative approach: since DFTB3 with the 3OB parametrization84 already provides a rather encouraging description of hydrogen-bonding interactions, we hypothesize that it is possible to derive relatively minor modifications to the repulsive potential to improve the description of bulk water structure under ambient conditions; we focus on the repulsive potential because its computation is decoupled from the self-consistent solution of the charge fluctuations; thus, modification of the repulsive potential does not require reparameterization of the electronic parameters. The questions worth investigating are as follows: (i) At what intermolecular distance range are modifications necessary and what is the magnitude of the required changes? (ii) To what degree are modifications developed based on neutral bulk water structure transferable to other properties and water in other scenarios, such as water clusters of different protonation states and solvated proton/hydroxide?
Specifically, we adopt a reverse Monte Carlo (RMC)113 scheme (also known as iterative Boltzmann inversion114) based on eq 2 to tune the O–H repulsive potential for the 3OB83,84 parametrization of DFTB3. (As the current manuscript was in preparation, Rothlisberger and co-workers reported an empirically adjusted DFTB2 (SCC-DFTB) model based on similar ideas,115 although many details differ from the current work.) In eq 2, gOHexp(r) denotes the target, experimental (although new experimental data became available recently,116 we have used the “standard” experimental result of Soper105), OH radial distribution function (RDF), while VOH(r) and gOH(i)(r) indicate the repulsive potential and the calculated RDF in the ith iteration of RMC calculations; the calculated RDF is from NVT simulation following the same setup as in ref (96).
| 2 |
A comparison of the “3OB” and RMC-generated “3OBw” O–H repulsive potentials in Figure 3 shows that, upon RMC optimization, this potential becomes longer-ranged and more repulsive at O–H distances in the range 1.6–3.3 Å, thus affecting both the first and second solvation shells of a central water molecule. The magnitude of the modification is fairly modest and on the scale of kBT for most ranges. Nevertheless, the collective effect of the change produces a very significant improvement in the bulk solvation structure. As shown in Figure 2, although only the O–H repulsive potential is modified, leaving the H–H and O–O repulsive potentials untouched, both O–O and O–H RDFs are substantially improved; the H–H RDF remains somewhat different from the experimental data (see the Supporting Information).
Figure 3.

Comparison of the original 3OB and RMC-generated (3OBw) O–H repulsive potentials.
Table 2 shows that, compared to other DFTB variants, DFTB3/3OBw leads to a much lower oxygen coordination number around a central water oxygen of 4.3, which is close to the experimental105 and CPMD-HCTH100 values of 4.7 and 4.1, respectively. Also, the spatial distribution function (SDF) plots of oxygen atoms about a central water O in Figure 4 show that the new parameters lead to a much sharper distribution of water molecules around another in the bulk, more closely resembling a tetrahedral coordination environment rather than an indiscriminate distribution.
Figure 4.
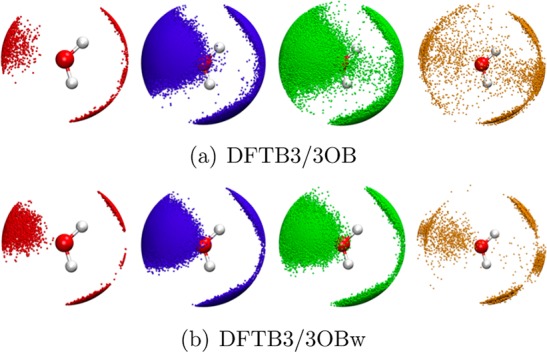
SDF of water O atoms about a central O in neutral bulk water with the different colors denoting different O–O distance ranges. Only SDF > 3 is shown. Red, 0.0 Å < rO–O < 2.6 Å; blue, 2.6 Å < rO–O < 2.8 Å; green, 2.8 Å < rO–O < 3.0 Å; orange, 3.0 Å < rO–O < 3.3 Å.
Transferability Test of the 3OBw Model
Other Properties of Bulk Water
The 3OBw model is established by considering only the structural properties of bulk water at ambient density. It is of interest to explore how other properties of bulk water are affected by the change of the repulsive potential. Although it is clear that the RMC scheme is state-dependent, it is also of interest to probe to what degree the modification applies to other thermodynamic conditions. In the following, we focus on the dynamic and spectroscopic observables of bulk water with 3OB and 3OBw models. In the Supporting Information, we discuss the temperature dependence of water with DFTB3 models and NPT simulations.
In Table 3, the diffusion coefficient of water calculated with different DFTB models is compared to CPMD and experimental values. The diffusion of water is rather similar for all three third-order variants, with the diffusion coefficient being almost twice larger than the experimental value. Compared to the second-order methods, the DFTB3 approaches represent a major improvement.
Table 3. Diffusion Coefficient (Å2/ps) of the Oxygen Atom in Neutral Bulk Water (DH2O).
We describe the orientational dynamics in liquid water by the second rank rotational time-correlation function of the O–H bond vector:
| 3 |
where û is a unit vector along the O–H bond and θ the angle between û at time “t” and at time 0. Figure 5 shows that DFTB3/3OB predicts C(t) for H2O to decay faster than DFTB3/3OBw; the C(t)’s are computed by averaging independent NVE trajectories that sum to ∼300 ps. Table 4 shows the DFTB3/3OBw relaxation times to be lower than those from experiments and from the classical SPC/E water model. The inset in Figure 5 shows that, like the SPC/E water model, the DFTB3 models predict a very short librational response followed by an initial fast decay and then a long time exponential decay (the DFTB3 relaxation times are obtained by explicit integration of the correlation function until t = 2.5 ps and integration from 2.5 to 30 ps of the exponential fit to the part of the correlation function from 2.5 to 5 ps).
Figure 5.
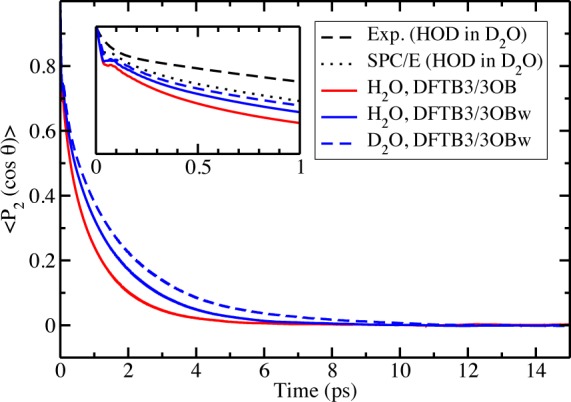
P2 correlation function of the OH(/D) bond vector in light water (H2O) and heavy water (D2O). The experimental and SPC/E data for HOD in D2O are from ref (119).
Table 4. Orientational Relaxation Time in ps from the P2 Correlation Function (eq 3).
Figure 6 shows a comparison of calculated and experimental infrared spectra for bulk H2O and D2O, with the same sets of trajectories used to compute the P2 correlation function. The calculated spectra are obtained by a Fourier transformation of the dipole autocorrelation function, followed by application of the quantum harmonic correction.127 The peak at around 3500 cm–1 for H2O and that at ∼2500 cm–1 for D2O, which correspond to O–H and O–D stretching vibrations, respectively, agree well with the experimental sets (although a splitting of the peak, especially for D2O, is observed). As expected, the frequencies are lower compared to corresponding gas-phase normal-mode frequencies (denoted by vertical lines). The calculated peaks corresponding to angle bending vibrations (in the 1000–2000 cm–1 region) are red-shifted compared to the experimental peaks; the red-shift, however, is also present in the gas phase, as we noted earlier.128 The peaks below 1000 cm–1 correspond to librational motions and agree quite well with experiment. The shoulder at low (∼300–500 cm–1) frequencies, which has been postulated to be seen in calculated spectra when polarization is accounted for in the simulation model,129 is also seen in the calculated spectra. The relative intensities of the different peaks are also predicted quite well by DFTB3/3OB(w), especially for D2O.
Figure 6.
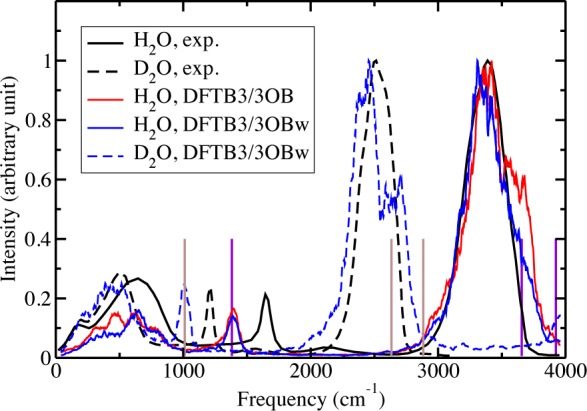
Infrared spectrum of light water (H2O) and heavy water (D2O). The experimental results for H2O and D2O are taken from refs (130) and (131), respectively. The vertical violet and brown lines denote the O–H/D bending and stretching frequencies in an isolated H2O and D2O molecule, respectively, with DFTB3/3OB (the frequencies are the same with DFTB3/3OBw) and are assigned arbitrary heights. All liquid-phase spectra are scaled so as to have a maximum intensity of 1.
Gas Phase Water Clusters with a Proton or Hydroxide
Concerning the structure and energetics of small protonated water clusters, we note that the RMC optimization above does not affect the O–H repulsive potential at O–H distances less than 1.6 Å (the distance range in which proton transfer barriers generally occur). Hence, proton transfer barriers with DFTB3/3OB and DFTB3/3OBw are very similar, and both show improvement over the previously tested DFTB3-diag+gaus variant96 (see the Supporting Information for a discussion based on H+(H2O)2). In H+(H2O)6 and H+(H2O)22, DFTB3/3OB is able to provide a description of the Eigen–Zundel balance in low-energy isomers comparable to DFTB3-diag+gaus.96 Compared to high-level MP2 calculations, DFTB3/3OB and DFTB3/3OBw give similar results for the relative energies of different conformers for H+(H2O)6, with the errors actually smaller with 3OB (see the Supporting Information). However, for the large cluster H+(H2O)22, where the oversolvation problem can start to affect the relative energies of the isomers, DFTB3/3OBw is seen to yield much lower errors in the relative energies (see Tables 5 and 6). For one of the two isomers (B and G) of H+(H2O)22 which have the excess proton in the interior of the cluster rather than on the surface, Figure 7 illustrates that DFTB3/3OBw removes “crowding” of water molecules around the hydronium. These results reflect a considerable degree of transferability of the 3OBw parameters optimized for the neutral bulk water solvation structure to systems with an excess proton; this is further confirmed by tests for a proton in bulk water (see below).
Table 5. Energies Relative to Isomer A (kcal/mol) and Zundel/Eigen Character of Low-Energy Isomers of H+(H2O)22a.
| isomer | RIMP2b | DFTB2 | DFTB3-diag+gaus | DFTB3/3OB | DFTB3/3OBw | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 0.0 | (E) | (E) | 0.0 | (Z) | (E-Z) | 0.0 | (E) | (E) | 0.0 | (E) | (E) | 0.0 | (E) | (E) |
| B | 9.5 | (E) | (E) | 8.8 | (Z) | (E-Z) | 6.2 | (E) | (E) | 7.0 | (E) | (E) | 9.1 | (E) | (E) |
| C | 2.9 | (E) | (E) | 0.9 | (Z) | (E-Z) | 2.1 | (E) | (E) | 2.5 | (E-Z) | (E) | 2.5 | (E-Z) | (E) |
| D | 7.4 | (E) | (E) | 1.9 | (Z) | (E-Z) | 0.0 | (E) | (E) | 1.8 | (E) | (E) | 5.2 | (E) | (E) |
| E | 6.5 | (E) | (E) | 4.1 | (Z) | (Z) | 1.2 | (E) | (E) | 2.2 | (E) | (E) | 5.5 | (E-Z) | (E) |
| F | 5.3 | (E) | (E) | 2.6 | (Z) | (Z) | 3.3 | (E) | (E) | 1.5 | (E) | (E) | 1.8 | (E) | (E) |
| G | 9.8 | (E-Z) | (E) | 6.9 | (Z) | (Z) | 5.3 | (E) | (E) | 7.0 | (E) | (E) | 10.5 | (E) | (E) |
| H | 2.7 | (E) | (E) | 1.8 | (E-Z) | (E) | –1.2 | (E) | (E) | 0.5 | (E) | (E) | 3.8 | (E) | (E) |
| I | 2.4 | (E) | (E) | 1.7 | (Z) | (E) | –1.4 | (E) | (E) | 0.4 | (E) | (E) | 4.5 | (E) | (E) |
| J | 0.5 | (E) | (E) | 1.6 | (E-Z) | (E) | 1.0 | (E) | (E) | 1.1 | (E) | (E) | 0.9 | (E) | (E) |
| K | 12.0 | (E) | (E) | 8.0 | (Z) | (E-Z) | 6.6 | (E) | (E) | 7.8 | (E) | (E) | 9.8 | (E) | (E) |
| L | 3.4 | (E) | (E) | 4.5 | (Z) | (E-Z) | 3.3 | (E) | (E) | 3.3 | (E) | (E) | 4.0 | (E) | (E) |
| M | 6.8 | (E) | (E) | 3.9 | (Z) | (Z) | 4.5 | (E) | (E) | 5.2 | (E-Z) | (E) | 6.3 | (E-Z) | (E) |
In the column for each method, the three subcolumns indicate the energy relative to isomer A, the Z/E/E-Z classification according to the criterion based on ROO,132 and the Z/E/E-Z classification according to the criterion based on δ.100
The RIMP2 relative energies are from ref (133). The RIMP2 results therein were obtained by single-point calculations with the aug-cc-pVTZ basis set at B3LYP/6-31+G(d) optimized geometries. The Z/E/E-Z classification is based on the B3LYP/6-31+G(d) geometries.
Table 6. Summary of Results for H+(H2O)22a.
Figure 7.
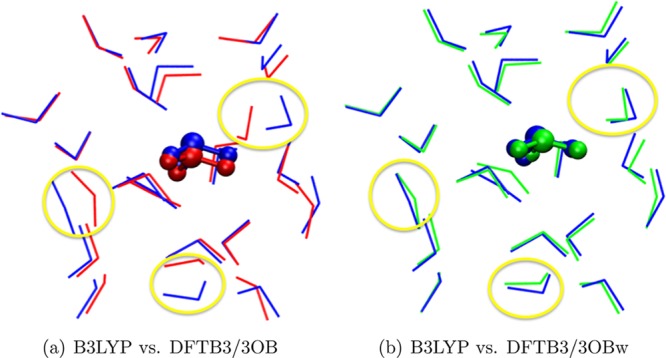
Isomer G of H+(H2O)22 optimized by different methods. Blue, B3LYP/6-31+G(d); red, DFTB3/3OB; green, DFTB3/3OBw. The yellow circles highlight the water molecules “crowding” around the central hydronium in the DFTB3/3OB optimized structure.
When finite temperature effects are considered, the Eigen–Zundel balance in protonated water clusters remains well described with the 3OBw parameters. An example studied in previous work is the “magic” cluster, H+(H2O)21. As shown in Figure S6 (Supporting Information), the results using DFTB3/3OBw compare well with DFTB3-diag+gaus, which was found96 to be consistent with CPMD-BLYP studies.134 By contrast, DFTB2 is qualitatively different, especially at temperatures higher than 150 K.
For the hydroxide–water clusters, both DFTB3/3OB and DFTB3/3OBw predict somewhat shorter O–O and O–H distances around the hydroxide ion in the small gas-phase clusters (H2O)2OH– and (H2O)3OH–, in addition to a very symmetric (H2O)OH– when compared to MP2/aug-cc-pVD(/T)Z (see the Supporting Information). For an isomer of (H2O)4OH– with three water molecules H-bonded to the hydroxide ion and one water molecule in the “second solvation shell” of OH–, while DFTB3/3OB optimization results in a collapse to an isomer with a tetra-coordinated OH–, the DFTB3/3OBw optimized structure remains close to the MP2/aug-cc-pVDZ structure (Figure 8b). For both (H2O)4OH– and (H2O)5OH–, the relative energies of the two isomers investigated here are close at the levels of MP2/aug-cc-pVDZ and DFTB3/3OBw (see Table 7). By comparison, DFTB3/3OB overestimates the stabilization of isomer I of (H2O)5OH– (see the Supporting Information for structures), which has a more highly coordinated OH–. Therefore, the DFTB3/3OBw variant also improves the oversolvation of a hydroxide.
Figure 8.

Optimized structures of two isomers of (H2O)4OH– with MP2/aug-cc-pVDZ (colored by atom type), DFTB3/3OB (colored in blue), and DFTB3/3OBw (colored in green). The starting structures are based on ref (135). The black dotted circle depicts the collapse of a tricoordinated OH– structure to a tetra-coordinated OH– structure with DFTB3/3OB.
Table 7. EIsomerI – EIsomerII for (H2O)4OH– (Figure 8) and (H2O)5OH– (See the Supporting Information for Structures).
| ΔE (kcal/mol) |
||
|---|---|---|
| (H2O)4OH– | (H2O)5OH– | |
| MP2/aug-cc-pVDZ | –4.4 | 0.1 |
| DFTB3/3OB | –1.2 | –1.8 |
| DFTB3/3OBw | –4.5 | –0.6 |
Excess Proton in the Bulk
The DFTB3/3OBw variant leads to an improved solvation structure around a proton in bulk water, compared to earlier variants of DFTB.96 This is evident from Table 8 which shows that the integration of the first peak of different kinds of RDFs from DFTB3/3OBw leads to coordination numbers in good agreement with available EPSR, CPMD-HCTH, and MS-EVB3 data. Figure 9 further helps to illustrate the loss of oversolvation of the hydrated proton, with the first peak of the O0–O RDF from DFTB3/3OBw almost completely comprised of the three nearest neighbors of the hydronium, in contrast to the situation with DFTB3/3OB. Parts a and b of Figure 10 show that, with the new parameters, the RDFs of water O atoms about the “hydronium” oxygen and hydrogen atoms, represented by gO0–O and gH0–O, respectively, have a better position for the first peak and a very distinct first minimum, in addition to much more pronounced second and third solvation shells for the O0–O RDF, compared to other DFTB3 variants. The improved position of the first peak is also reflected in the better agreement between MP2/aug-cc-pVDZ and DFTB3/3OB for the position of the minima in the H5O2+ potential curves (see the Supporting Information), and in the absence of a split first peak in gH0–O (unlike DFTB3-diag+gaus96). The large improvement in the solvation structure is also reflected in other RDFs like gO0–H, gO1x–O, and gO1yz–O, as illustrated in the Supporting Information.
Table 8. Integrated First Peak of Different RDFs Associated with an Excess Proton in Bulk Water.
Figure 9.
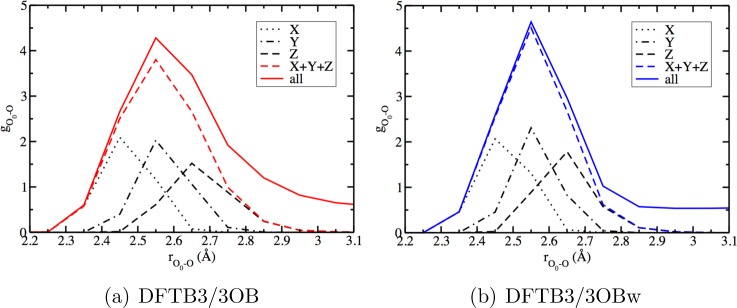
Decomposition of the first solvation peak of the O0–O radial distribution function for bulk water with an excess proton (O0 is the hydronium oxygen). The curves labeled X, Y, and Z represent the contributions of O1x, O1y, and O1z, respectively. The curve labeled X+Y+Z represents the total contribution from these three atoms, and that labeled “all” represents the g(r) for all the water O atoms.
Figure 10.
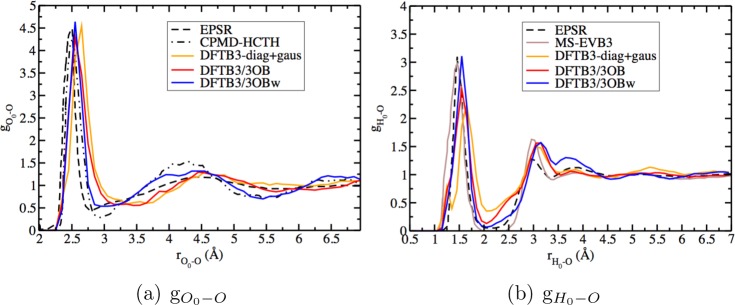
RDF of water O atoms around the hydronium O, denoted O0, and around the hydronium hydrogen, denoted H0. The EPSR, CPMD-HCTH, MS-EVB3, and DFTB3-diag+gaus data are from refs (136) and (137), ref (100), ref (138), and ref (96), respectively. For additional plots, see the Supporting Information.
In previous work,96 we found that water molecules around the “hydronium” occupy so-called interstitial sites when DFTB3-diag+gaus is used. The SDFs from DFTB3/3OBw plotted in Figure 11 (see also the Supporting Information for the SDFs of water O atoms around the hydronium O) and compared to those from DFTB3/3OB reveal elimination of water population in the interstitial sites and hence a correct water distribution around the central “hydronium”.
Figure 11.

SDF of water O atoms around the hydronium H atoms (left, side view; right, top view) for a system comprised of an excess proton in bulk water. The different colors denote different H–O distance ranges. Only SDF > 3 is shown. Green, 0.0 Å < rH0–O < 2.0 Å; blue, 2.0 Å < rH0–O < 2.5 Å.
Compared to DFTB3-diag+gaus,96 both DFTB3/3OB and DFTB3/3OBw provide a better description of the energetics of proton transfer in bulk water (Figure 12). Both of the 3OB variants predict the Zundel configuration to be the “transition state” and the Eigen configuration to be the “resting state” for the proton transfer, in agreement with CPMD-HCTH100 and MS-EVB3.138 While the proton transfer barrier is somewhat higher than that predicted by CPMD-HCTH100 (more so with DFTB3/3OBw), previous (classical) MS-EVB2 and MS-EVB3138 models also predict this barrier to be close to ∼1 kcal/mol.
Figure 12.
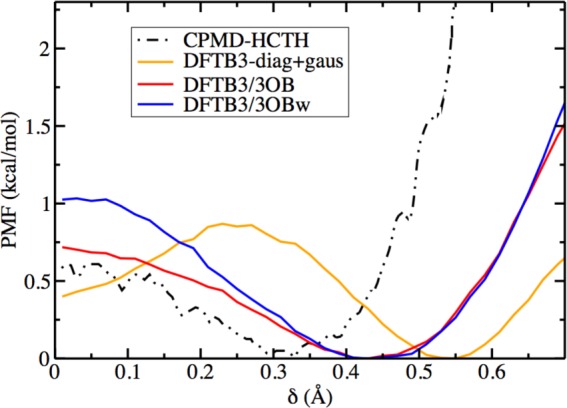
Potential of mean force for the transfer of an excess proton in bulk water. The reaction coordinate is δ = |r⃗O0H – r⃗O1xH|, where O0 is the hydronium oxygen and O1x is the “special pair” partner of O0. δ ≤ 0.1 corresponds to the Zundel form, and δ ≥ 0.2 corresponds to the Eigen form. The CPMD-HCTH and DFTB3-diag+gaus data are from refs (100) and (96), respectively.
Hence, overall, the results in this section illustrate that the RMC-optimized 3OBw parameters, developed on the basis of the solvation structure of neutral bulk water, are fairly transferable to a system with an excess proton in the bulk, providing an encouraging description of both the solvation structure and the proton transfer energetics.
Excess Hydroxide in the Bulk
Choi et al.101 recently reported the performance of several variants of DFTB for the description of the solvation structure and dynamical properties of a hydroxide ion in bulk water. All investigated variants, including DFTB3/3OB, were found to predict OH– to be oversolvated in the bulk. We find that DFTB3/3OBw can provide appreciable improvement in this oversolvation behavior. In the discussions below, to gain insight into the dynamical behavior of the hydroxide ion, we make use of a coordinate δ = min|rO0H – rOaH| (also employed in the relevant literature;101,139−141 note that this is different from δ defined in the section on proton in bulk water). Here, Oa is any water O atom which shares a hydrogen (H) with the hydroxide oxygen, O0, such that the associated displacement coordinate δ is the smallest. Large values of δ imply configurations far from a proton transfer event, while low values are associated with configurations likely involved in proton transfer. Below, to be consistent with the literature,139,140 we choose δ ≥ 0.5 and δ ≤ 0.1 to classify configurations far from and near a proton transfer event, respectively. The hydroxide hydrogen is designated H′.
As Figure 13 illustrates, compared to DFTB3/3OB, DFTB3/3OBw leads to a narrowing of the first peak of both gO0–O and gO0–H. Integration of the first peak of gO0–O from DFTB3/3OBw leads to a value of 5.8 (Table 9), which, while still greater by 1 compared to CPMD-BLYP simulations,139,140 is an improvement over the value of 6.9 from DFTB3/3OB. The first peak of gO0−H also integrates to a value (4.9) higher by 1 compared to the CPMD-BLYP value of 3.9, but providing improvement over the DFTB3/3OB value of 5.6. Compared to results published by Choi et al.,101 DFTB3/3OBw performs better than all other variants (including DFTB2(-γh) which Choi et al. found to be better than/on par with DFTB3/3OB for these properties).
Figure 13.
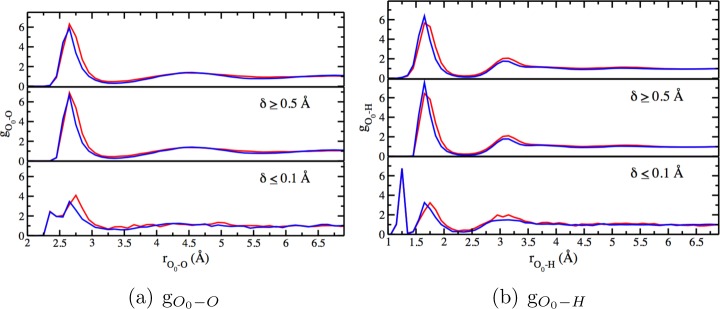
RDF (a) of water O atoms around the hydroxide O, denoted O0, and (b) of water H atoms around O0. The topmost panel in both parts a and b compares the RDFs obtained with DFTB3/3OB (in red) and DFTB3/3OBw (in blue), using all simulation data. The middle panels show the same comparison for configurations far from proton transfer events (large δ), while the bottom panels do so for configurations corresponding to the “transition state” of proton transfer (small δ).
Table 9. Integrated First Peak of Different RDFs Associated with a Hydroxide Ion in Bulk Watera.
|
gO0–O |
gO0–H |
gH′–O |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| all | δ ≥ 0.5 | δ ≤ 0.1 | all | δ ≥ 0.5 | δ ≤ 0.1 | all | δ ≥ 0.5 | δ ≤ 0.1 | |
| DFTB3/3OB | 6.9 | 7.0 | 6.4 | 5.6 | 5.7 | 4.5 | 0.8 | 0.8 | 0.8 |
| DFTB3/3OBw | 5.8 | 5.9 | 5.4 | 4.9 | 5.0 | 4.0 | 0.7 | 0.7 | 0.7 |
| CPMD-BLYP139,140 | 4.8 | 3.9 | 0.67 | ||||||
The hydroxide oxygen is denoted as O0, and its hydrogen, as H′.
The SDF of water O atoms about O0 in Figure 14a (note that the SDF does not show the oxygen population H-bonding to the hydroxide hydrogen) reveals the presence of a H-bond donor population below the “expected” square planar arrangement of H-bond donors around O0, thus overestimating the H-bond donor population by 1. Figure 14b shows greater oversolvation of hydroxide with DFTB3/3OB compared to DFTB3/3OBw (higher density of green spheres) in “interstitial” sites.
Figure 14.
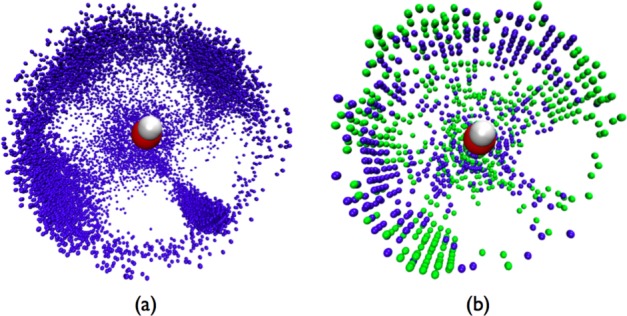
SDF of water O atoms around a hydroxide O solvated in bulk water: (a) SDF from DFTB3/3OBw; (b) a difference plot of the SDF from DFTB3/3OBw (in blue) and DFTB3/3OB (in green). In both cases, points with SDF > 3 and within 3 Å from the hydroxide O are shown (but, for clarity, excluding those within 2.5 Å from the hydroxide H). The reference coordinate frame is defined such that the hydroxide O is at the origin, the hydroxide ion is aligned with the z axis, and the hydroxide ion and its nearest H-bonding water H atom constitute the xz plane. For part a, a 0.1 Å bin-width along r and a 1° bin-width each along θ and ϕ are used. For part b, for clarity, a 0.2 Å bin-width along r and a 5° bin-width each along θ and ϕ are used.
The H-bond acceptor population around the hydroxide ion is also improved with DFTB3/3OBw, as reflected in the lowering of the first peak height (Figure S11, Supporting Information (top panel)) and a lower integrated first peak value compared to DFTB3/3OB (Table 9).
Concerning the dynamical behavior of the hydroxide ion, while it is clear from Table 9 that for both DFTB3/3OBw and DFTB3/3OB, in concert with proton transfer, a reduction of the H-bond donor population around the hydroxide ion takes place, it is less clear whether proton transfer is accompanied by a change in the H-bond acceptor population. While the middle and bottom panels of Figure S11 (Supporting Information) indicate a sharper first peak in the H′–O RDF accompanying proton transfer, the degeneration of the first peak into a broad plateau for large δ values observed in CPMD-BLYP simulations139,140 is not observed even with DFTB3/3OBw. Figure S12 (Supporting Information) shows the proton transfer barrier with DFTB3/3OBw to be similar to that with DFTB3/3OB (although with a slight improvement in the position of the minimum), implying that the lowering in H-bond donor population by 0.5–0.7 with DFTB3/3OBw compared to DFTB3/3OB does not have an appreciable impact on the proton transfer barrier. Thus, the dynamical properties of solvated hydroxide described by DFTB3/3OBw are similar to those reported by Choi et al.101 using DFTB3/3OB.
Hence, while the 3OBw parameters yield appreciable improvement in the solvation structure around the hydroxide ion in bulk water compared to previous DFTB variants (besides improving the structure and energetics of gas-phase hydroxide clusters), the dynamical properties of the hydroxide ion and remaining deficiencies in the description of solvation structure still present a challenge which can guide further development of DFTB. This is a somewhat expected result in that treating hydroxide solvation likely requires a careful description of the charge dependence of Pauli repulsion, which is missing in the current formulation of DFTB and also not captured empirically in the RMC procedure based on neutral bulk water (the modification of the O–H repulsive potential occurs mainly at long range).
Proton Transfer Barrier in a Model Channel
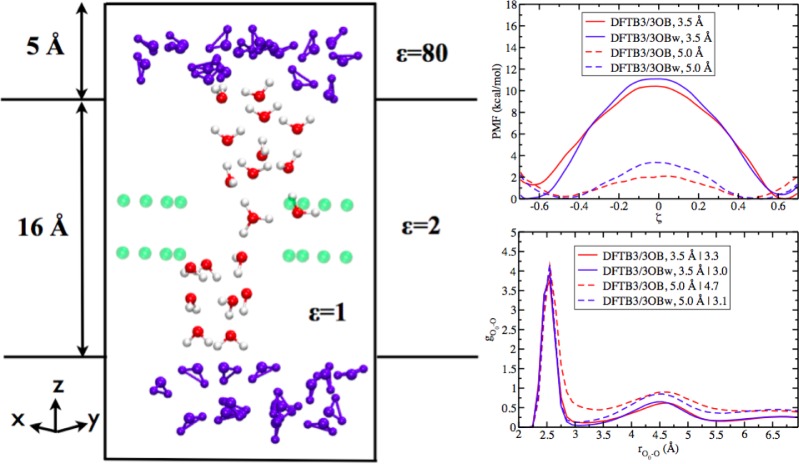
Having observed the difference between DFTB3/3OB and DFTB3/3OBw results for the hydration of excess proton, it is important to ask whether such a difference leads to any significant impact on the computed proton transfer energetics in biomolecules. To answer this question, we study the proton transfer in a simplified ion channel model that we established in previous work142,143 (Figure 15, left); the advantage of using such a model system over more realistic biomolecular systems is that issues with QM/MM interactions73,144 and sampling can be largely avoided, allowing us to directly assess the impact of different proton hydration in a confined environment. We study two model channels with a radius of 3.5 and 5 Å, respectively, to explore the magnitude of the effect with different degrees of confinement; these correspond to typical sizes of proton conducting channels found in biomolecules. The computational setup and simulation protocols for the proton transfer PMF along the collective ζ coordinate follow those reported in refs (142) and (143). Umbrella sampling involves 13 windows for each setup; the production run for each window is 500 ps for the wide channel and 800 ps for the narrow channel.
Figure 15.
Left: An illustration of the model channel. The “bulk” MM water molecules are shown in blue, while the QM water molecules are colored by atom type. The eight dipoles are shown in green. For the channel of radius 3.5 (5.0) Å, the (inner region) box edge-length along X and Y is 17.2 (20.4) Å. Each dipole is comprised of charges of +0.5e (+0.6e) and −0.5e (−0.6e) separated by 1.5 Å for the narrower (wider) channel, with the positive end pointing toward the channel. Top right: Proton transfer free energy profiles along ζ (two MM oxygen atoms fixed at (0, 0, 10) and (0, 0, −10) are used as the donor and acceptor for defining ζ. Bottom right: RDF of water O atoms about the “hydronium” O when it is near the center of the channel along z. The number after “|” in the legend results from integration of the first peak of the RDF.
As shown in Figure 15 (right top panel), the description of proton hydration has an impact on the computed barrier for the proton transport, and the magnitude of the effect varies depending on the degree of confinement. The levels of hydration of the proton when it is at the center of the channel (along ζ, see Figure 15, right bottom panel) differ with the 3OB and 3OBw models by 0.3 water molecules in the first solvation shell in the narrower channel model; the difference goes up to 1.6 water molecules in the wider channel model. Nevertheless, the difference in the PMF barriers between DFTB3/3OB and DFTB3/3OBw is on the order of 1–2 kcal/mol. Since this level of error in the energetics does not lead to any qualitative difference in most mechanistic analysis, we confirm that DFTB3/3OB is well suited for mechanistic analysis of proton transport in biomolecules.
Along this line, a recent study145 analyzed the issue of proton oversolvation in a synthetic ion channel and concluded that the findings “called into question the applicability of the DFTB3 approach for proton transport in biomolecules”. Although the issue of proton oversolvation should certainly be resolved, our study here highlights that the key energetic properties for proton transport in biomolecules from DFTB3/MM simulations are meaningful for most purposes. The work of ref (145) compared proton dynamics in the channel at the time scale of 100 ps and observed a qualitative difference between DFTB3/MM and B(3)LYP/MM simulations. What ref (145) failed to point out, however, is that the local free energy minimum for the proton in the middle of the channel is separated from the mouth of the channel by a small barrier of merely 2 kcal/mol according to MS-EVB calculations.146 Therefore, small differences in barrier heights from different calculations will lead to distinct proton dynamics at the 10–100 ps time scale. A more relevant comparison is the PMF profile, as we have done here for the model channel, and the results indicate that proton oversolvation likely has a limited impact on the proton transfer energetics for most purposes.
QM/MM Simulations Involving Water: Importance of the QM-MM Hamiltonian and Methods of Calibration
In most biological and many chemical applications, the bulk of the water environment can be treated at the classical level with only the site (solute) of interest treated at the QM level. The interface between the QM region and nearby MM atoms is expected to be important to the quantitative aspect of the simulation. In typical QM/MM implementations for biological applications,147 the QM-MM interaction includes electrostatic and van der Waals contributions (we will not discuss QM/MM partition across covalent bonds here147−150). The electrostatic component is usually described with one-electron Hamiltonians that involve the interaction between QM electrons and MM charge distributions, which, depending on the force field, may include point charges, permanent multipoles, and polarizabilities. The van der Waals interactions are usually taken to be decoupled from the determination of the QM wave function/density and described by the empirical Lennard-Jones expression,151 although more physical expressions like the Buckingham potential have been used;111 a formulation of QM-MM van der Waals interactions that depend on the charge distribution of the QM region has also been developed.111 In terms of empirical parameters, the simplest QM/MM implementation involves only the Lennard-Jones parameters for the QM atoms; several previous studies144,151 have shown that the QM-MM van der Waals interactions may substantially perturb the structural properties of the MM atoms around the QM region (e.g., solvent distribution around the QM solute); the effect on energetic properties of typical interest, such as reduction potential, pKa, and reaction energies, is, however, more modest (1–2 kcal/mol). For quantitative calculations, nevertheless, a careful parametrization of the QM van der Waals parameters is worthwhile.
Regarding the QM-MM electrostatics, there is no additional parameter in the most straightforward implementation. It has been recognized, however, that, when point charge MM models are used, it is beneficial to smear the MM charges to partially take the effect of charge penetration into consideration.152,153 This protocol involves additional parameters such as the width of the smeared MM charges, which are usually described with Gaussian functions. Specifically for DFTB, we also recognized the importance of treating the QM-MM electrostatics more carefully, especially when the QM region is substantially charged. In the following sections, we first briefly review our recent development along this line and then discuss ways that QM-MM interactions can be calibrated, including a novel sampling protocol that can be particularly useful in this context.
QM-MM Electrostatics in DFTB/MM Simulations
The most physical way to describe QM-MM electrostatics should consider the finite size of the corresponding charge distributions, as in the Gaussian-blur approach.152,153 Semiempirical methods45 do not need to evaluate the additional core integrals between the electron density and the external point charges, but the idea of charge “blurring” was implemented already in their QM/MM extensions. For example, the pioneering QM/MM work by Field and co-workers evaluated the QM-MM electrostatics with the ⟨ss|ss⟩ electron integral154 that takes the form of the Klopman–Ohno (KO) expression45 for the two-center two-electron integral, ⟨μaνa|λbσb⟩,
| 4 |
where the summations are over the multipole charges (Qi’s) used to mimic atomic orbitals and di’s are parameters determined on the basis of the condition that as the distance Rab approaches zero the expression reduces to known one-center two-electron integrals. For the interactions between two spherical charges (s orbitals), the di’s are nothing but the corresponding inverse chemical hardness values (1/Ui), which are fitted in semiempirical methods to reproduce certain molecular properties. Note that these inverse chemical hardness values represent on the one hand the electron–electron interaction and on the other hand the atomic size.46,81 Also, the KO integral is very similar to the γab function in the DFTB total energy expression, although the KO expression has a simpler functional form.
In semiempirical theory, ξ in eq 4 is set to equal unity, but to model the atomic size in QM/MM interactions, which is an effective blurring of the point charges, a scaling of ξ < 1 seems appropriate. In practice, several semiempirical QM/MM implementations have found different optimal values of ξ. In ref (154), ξ = 1 was chosen and Uj was set to zero when j indicates a MM atom. In other approaches, ξ was either optimized to roughly 0.1155 or was set to zero.156 In an early DFTB2/MM implementation,157 we found that ξ = 0 leads to the best DFTB2/MM hydrogen bonding energies, and this value was also adapted in a subsequent DFTB2/MM implementation in CHARMM.61 This choice is related to the fact that DFTB2 underestimates hydrogen bonding energies systematically, and therefore an undamped 1/R scaling leads to improved interaction energies. However, the QM-QM, QM-MM, and MM-MM hydrogen bonding energies are then not well balanced. With DFTB3/3OB, hydrogen bonding energies are better treated,84 and thus, a simple 1/R scaling is no longer appropriate.
Therefore, we recently explored using the KO expression to compute DFTB3-MM electrostatic interactions as follows:73
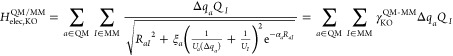 |
5 |
in which ξa and αa are element dependent parameters; together with the van der Waals parameters in the QM-MM Hamiltonian, there are four QM-MM parameters for each element type. For the Hubbard parameter of a MM atom (UI), we simply took the computed value for an atom.84 We note that, by including the charge dependence of the Hubbard parameters in DFTB3, the effective atomic size information is directly integrated into the SCF determination of the QM wave function/density. Due to the difference in physical origin, this does not replace the charge dependence of the QM-MM van der Waals interaction,111 although it is an important step toward the reliable description of chemical reactions that involve a significant charge redistribution.
Calibration of QM-MM Hamiltonian: From Clusters to the Condensed Phase
To ensure transferability, we take the parameters to be element-dependent; thus, there are four parameters (ξa, αa in eq 5 and the QM Lennard-Jones parameters) per element. Alternatively, we could make ξ element-independent as in previous semiempirical approaches,155 and instead fit the Hubbard parameters for the MM atoms (UI). What is the best approach to determine these parameters? In most previous studies,144,151 parameters in the QM-MM Hamiltonian (e.g., the QM Lennard-Jones parameters) were empirically fitted on the basis of small molecule models, following the way that nonbonded parameters are fitted in empirical force fields.158 For example, a MM water molecule is used as a probe to interact with a “solute” in different geometries, the QM-MM Hamiltonian is adjusted such that QM/MM results best match those from full QM calculations.
In our recent work,73 we were concerned that these small solute–water systems were not representative of the solution environment. Therefore, an alternative strategy was followed in which larger clusters that included solutes interacting with multiple water molecules were studied; the structures were collected on the basis of DFTB/MM simulations of the solute in water. Test calculations indicated that including multiple water molecules was essential to a successful parametrization and including only pairwise models as in previous studies144 did not capture the complexity of interactions in the condensed phase and therefore did not lead to as transferrable parameters. Calculations in ref (73) showed that this strategy led to a set of KO parameters that appeared to be rather transferrable for the SCC-DFTBPR89 variant of DFTB; e.g., for 16 stable states and 24 transition states involved in 10 model phosphate hydrolysis reactions in RNA from the QCRNA database established by the York group,159 which were not included in the fitting of the KO parameters, the mean unsigned errors (MUEs) were 3.5 and 4.8 kcal/mol for the stable and transition states, respectively, for the comparison of solute–water interaction between SCC-DFTBPR/MM and full SCC-DFTBPR calculations. We note that, due to the significant charges of these molecules, the total solute–water interactions are on the order of 100–200 kcal/mol; thus, a MUE of ∼4–5 kcal/mol is very satisfactory. For instance, by comparison, the MUEs for calculations that used the original (Coulombic) QM-MM model were 14.2 and 17.6 kcal/mol for the stable and transition states, respectively. The use of γKOQM-MM in SCC-DFTBPR/MM simulations was essential in the study of several phosphate hydrolysis reactions in solution73 and enzymes.71 For example, in our study71 of mono ester hydrolysis in alkaline phosphatase (AP), using the Coulombic QM-MM Hamiltonian led to significant overpolarization and thus instability in the simulations. With γKO, SCC-DFTBPR/MM simulations were much more stable; both structural and energetic properties of mono ester hydrolysis in two enzymes in the AP superfamily were consistent with available experimental data.
Despite these encouraging initial results, we note that the preliminary QM/MM-KO parameters were fitted on the basis of gas phase clusters rather than an authentic solution environment; moreover, the reference results were full SCC-DFTBPR calculations rather than high quality QM or experimental data. Therefore, a more robust protocol is to parametrize the QM-MM Hamiltonian based explicitly on condensed phase properties and experimental data; as an initial step, we use solvation free energies as the reference data, similar to force field developments.106,158 To illustrate the promise of this protocol, we show the solvation free energies computed using several DFTB models with different QM-MM Hamiltonians for several neutral and charged small molecules. These calculations take advantage of the thermodynamic cycle in Figure 16, which has been adopted by several authors in the past,160 most notably Gao161 and Warshel.162 The advantage of following this thermodynamic cycle is that the steps that require the most sampling are carried out with MM potential functions, while the most expensive QM/MM calculations are involved in the vertical processes only, which converge much faster due to the relatively small perturbation in solute–solvent interactions.
Figure 16.
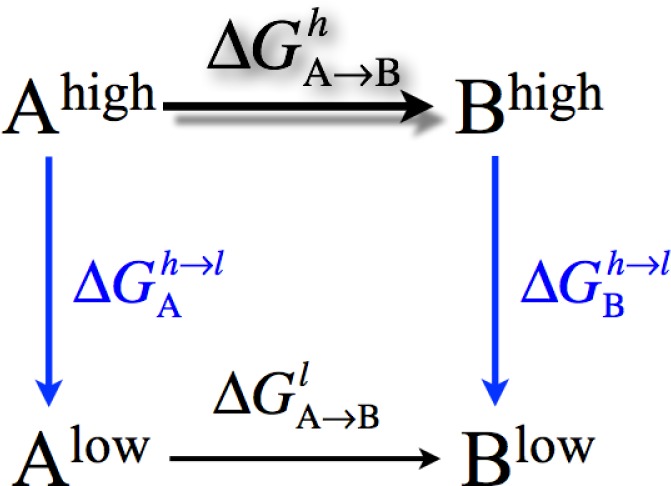
A general thermodynamic cycle used to compute the free energy change of a process (A → B) at different (high, low) levels of theory. For this work, the process corresponds to solvation and the high (low) level of theory corresponds to the treatment with a QM (MM) potential function. Thus, the high-level potential function is only used in simulations that involve the vertical processes: conversion of the solute from a MM to a QM description once in the gas phase and once in the solution phase.171
As shown in Table 10, the solvation free energies are modestly underestimated with the original DFTB2/MM model, when the QM atoms use van der Waals parameters from the CHARMM27 force field.158 With DFTB3 as the QM model and otherwise the identical QM-MM Hamiltonian, the solvation free energies are typically overestimated, in agreement with the expectation based on the larger magnitudes of DFTB3 charges as discussed above. With the γKOQM-MM parametrized in ref (73), SCC-DFTBPR/MM simulations lead to underestimated solvation free energies for charged solutes. Although the qualitative trend is expected since the KO expression (eq 5) damps QM-MM interactions, the magnitude of the effect is surprisingly large. As seen in Figure 17, the number of solvent molecules in the first solvation shell is smaller by ∼1 in the γKO based SCC-DFTBPR/MM simulations compared to either MM or DFTB/MM simulations using the Coulombic (1/R) QM-MM Hamiltonian.
Table 10. Relative Solvation Free Energy ΔΔGgas→aq(MM → QM) (in kcal/mol) with Different QM and QM-MM Electrostatic Hamiltoniansa.
| DFTB2 | SCC-DFTBPR | DFTB3/3OB | ||
|---|---|---|---|---|
| solute | MM ref. (Exp.)b | Coulomb | Klopman–Ohno (eq 5)c | Coulomb |
| H2O | –6.6 (−6.3) | +4.4 | +1.7 | |
| CH3COOH | –4.5 (−6.7) | –0.1 | –0.0 (+0.0) | –3.4 |
| CH3COO– | –81.9 (−77.6/–80.7163) | +3.1 | 13.1 (15.3) | –5.4 |
| CH3O– | –102.2 (−95.0) | +6.8 | 23.8 (24.9) | –3.9 |
| H3PO4 | –15.2 (−26.0) | –1.4 | –3.9 (−3.8) | –8.8 |
| H2PO4– | –80.8 (−76.0) | –1.7 | 10.1 (11.1) | –17.2 |
The MM here is the CHARMM22 force field. The water molecules are treated with TIP3P. Unless specified otherwise, the QM atoms use the standard CHARMM van der Waals parameters.
The “MM ref.” values are absolute solvation free energies computed following the standard protocol163,164 with periodic boundary conditions; no correction related to the gas/liquid interface has been included.165−167 Values in parentheses are experimental values from ref (168) for the first four solutes and from ref (169) for the phosphate species.
The values in parentheses use the CHARMM van der Waals parameters for the QM atoms; those without parentheses use the van der Waals parameters optimized in ref (73).
Figure 17.

Oxygen–oxygen radial distribution functions (solid lines) for water near a charged QM solute (acetate or methoxide) in DFTB/MM simulations with different DFTB models and QM-MM electrostatic Hamiltonians; the integrated distribution functions are also shown as dotted lines.
Calculations of solute–solvent interactions using clusters collected from different windows confirm our previous observation73 that γKOQM-MM based SCC-DFTBPR/MM gives results very consistent with full SCC-DFTBPR and DFTB3, which give systematically weaker solute–solvent interactions than both full MM and 1/R based DFTB3/MM calculations (see Table S3 in the Supporting Information). Since the nonpolarizable TIP3P water model is overpolarized for the purpose of condensed phase applications, this observed trend for gas-phase clusters is not surprising. We defer a more detailed discussion to a separate work that reports the systematic parametrization of QM-MM Hamiltonian (including QM van der Waals parameters) for DFTB3. The purpose here is to illustrate that parametrizing the QM-MM Hamiltonian explicitly based on condensed phase properties is potentially very important. We note that, since the change of local solvation during a typical chemical reaction is unlikely very large, the errors in computed reaction free energies71,73 due to the QM-MM Hamiltonian are not as large as those in the solvation free energies shown in Table 10. Nevertheless, adequately reproducing solvation free energies is clearly much more preferable than relying on error cancellation. Along this line, an equally important benchmark that also benefits from the thermodynamic cycle in Figure 16 is the calculation of QM/MM binding free energies of ligands to protein (enzyme) active sites. These will be reported elsewhere.
To facilitate the use of solvation/binding free energies in the parametrization and calibration of the QM-MM Hamiltonian, it is essential to maximize the efficiency of the MM ↔ QM/MM free energy perturbation simulations. When the MM and QM/MM potential functions have a good overlap in distribution, it is possible to use reweighing techniques based on, for example, a novel application of the Bennett acceptance ratio approach.170 In general, however, it is difficult to predict the degree of overlap between the MM and QM/MM distributions, especially when the solute is flexible. Therefore, it is valuable to develop efficient free energy protocols that involve explicit sampling using the QM/MM potential function but without overwhelming computational cost.
Motivated by this consideration and other applications,78 we have recently developed a novel sampling protocol referred to as the integrated Hamiltonian sampling (IHS).171 As explained in more detail in ref (171), in IHS, we introduce an effective potential (Ub(R)) whose canonical distribution is the integrated distributions of the intermediate and two-end states
| 6 |
where Ω(λ) is a weight function to be determined and Uλ takes the usual form (although other forms are also possible, like in free energy perturbations in general),
| 7 |
In the specific case of MM ↔ QM/MM free energy perturbation, U0 and U1 correspond to MM and QM/MM potential functions, respectively. In the more general applications, U0 can be the potential function of a physical system while U1 the potential function of a fictitious system introduced to enhance the sampling of U0; e.g., specific torsional barriers or nonbonded interactions are reduced.
The key to the efficiency of IHS lies in the choice of the weight function Ω(λ). The aim is to weight the contributions from different intermediate Hamiltonians (potential functions) such that transition among them, including the end states, is facile. To this end, one criterion is to set the expectation value of the probability for the weighted intermediate states to be uniform
| 8 |
where ⟨···⟩b is the ensemble average over the configurations sampled with the potential Ub (eq 6). Guided by this criterion, we have developed an efficient protocol to optimize the weight function (discretized as Ω(λi)) on the fly using a combination of the histogram flattening172 and weighted histogram173 approaches. This automated protocol makes it rather straightforward to employ multiple end-state Hamiltonians to facilitate sampling along different chemical and conformational degrees of freedom. The IHS approach is closely related to several enhanced sampling techniques in the literature; its development was inspired by the integrated tempering approach of Gao,174,175 and it has similar motivations to Hamiltonian replica exchange molecular dynamics (HREMD),176,177 enveloping distribution sampling,178 and λ-dynamics.179 For more detailed discussions, see the original reference.171 We only point out here that IHS requires only one trajectory, and the computational cost is essentially independent of the number of intermediate Hamiltonians if simple interpolation between the end-state Hamiltonians (eq 7) is used. Therefore, the computational cost of IHS can be substantially reduced compared to HREMD, especially when the distributions of the end-state Hamiltonians do not overlap well and many intermediate states are required for efficient sampling. This feature is particularly desirable when expensive Hamiltonians such as (ab initio) QM/MM potentials are needed.
As an illustration of the convergence behavior of IHS, we show the convergence of the weight function for the MM ↔ QM conversion for an acetate ion in solution. As Figure 18 indicates, satisfactory convergence is reached essentially after about 50 ps. Simulations in ref (171) also indicate that the standard deviation of computed free energies is substantially smaller in IHS simulations when compared to regular free energy perturbation simulations of similar length (per λ window). It is also straightforward to compute properties for the end-states using trajectories from IHS simulations with proper reweighting.171 Therefore, we anticipate that, by combining efficient IHS and multistate reweighting techniques,180 it is straightforward to automate optimization of QM-MM Hamiltonians based on solution properties.
Figure 18.

Convergence of weight functions (β–1 ln Ω(λi)) for MM ↔ QM (DFTB2) perturbation of an acetate ion in the gas phase (top) and water (bottom), using the integrated Hamiltonian sampling (IHS).171 Solvent molecules are described by TIP3P. Cross points describe when the weights were updated, and blue, green, red, cyan, purple, and ochre lines represent λ = 0.0, 0.2, 0.4, 0.6, 0.8, and 1.0, with U0 and U1 being QM/MM and MM Hamiltonians, respectively.
Concluding Remarks
The role of water in many chemical and biological processes is clearly beyond being merely a spectator solvent. Depending on the relevant length scale and specific role(s) of water in the problem of interest, different computational models are required. As a bulk solvent under ambient conditions, water is well treated by standard classical force fields because they were parametrized for this purpose. As the condition deviates from ambient conditions, however, the applicability of standard force field models is no longer obvious and careful tests need to be carried out to establish the appropriate model.181 In problems of biological relevance, examples include concentrated solutions, water in the interior of proteins and near the surface of nucleic acids. When water molecules are explicitly involved in the chemistry, such as proton transfers or proton-coupled electron transfers, they clearly need to be treated at a quantum mechanical level. In fact, a quantum mechanical treatment of many-body effects in water appears to be important even for nonreactive events such as ion diffusion; so far, it appears that only a quantum mechanical simulation is able to properly capture the temperature dependence of ion diffusion in water.182 This recent study again highlights the importance of and subtleties associated with many-body effects in condensed phase systems.
In this article, we have focused on our own studies of water using an approximate density functional theory, DFTB3, which is of interest because many applications require striking the proper balance between accuracy of the potential function and the degree of sampling. Our aim is not to argue that DFTB3 is already a reliable model for water that works under different conditions. Rather, our goal is to explore, with the current formulation of DFTB3, the performance of this method for treating water in different chemical environments, the magnitude and nature of changes required to improve its performance, and factors that dictate its applicability to reactions in the condensed phase with a QM/MM framework. This type of study helps establish both the value and limitations of the DFTB3 based simulations, and helps make clear the developments needed to further improve the accuracy and transferability of the methodology.
In previous studies, we and others found that DFTB3 generally gives encouraging results (e.g., low energy structures) for small water clusters, in both neutral and protonated forms. However, DFTB3 overpredicts the level of solvation of water and an excess proton in the bulk;96,99,100,145 there is also a significant tendency to oversolvate hydroxide even in gas phase clusters.101 Likely physical explanations for these limitations have been recognized47 and are discussed in this article, which together with recent work of York and co-workers102,108 highlight the potential importance of including multipole terms and a better treatment of Pauli repulsion in the next generation of DFTB models; the effect of dispersion, whose importance has been discussed for large water clusters183,184 and bulk water simulations with DFT,30,185 also needs to be analyzed with the empirical D3 model recently parametrized for DFTB3.184 What we demonstrate here is that a relatively minor change (on the scale of kBT) in the 3OB/O–H repulsive potential can substantially improve the description of bulk water structure under ambient conditions; several relevant dynamic properties and the infrared spectrum also show improvements. Moreover, this simple change has a considerable degree of transferability to protonated water clusters and solvated protons; it also improves the solvation of hydroxide, although further improvement of Pauli repulsion for charged species is needed to completely remove the oversolvation of hydroxide.
This simple but ad hoc correction based on the reverse Monte Carlo (RMC) scheme is by no means a satisfactory solution to the description of water in the DFTB framework. In fact, removal of oversolvation with the current RMC scheme is accomplished at the cost of reducing water–water interactions and therefore notably deteriorated heat of vaporization (see the Supporting Information), emphasizing the importance of further extending the DFTB3 model in a more systematic fashion as discussed above. Therefore, the 3OBw model should be applied with caution, especially in applications where there is a significant variation in water density (e.g., a liquid/vapor interface).
By comparing results using DFTB3 models that differ in the description of water, we are able to confirm that proton transfer energetics are adequately described by the standard DFTB3/3OB model for meaningful mechanistic analyses. This finding is consistent with the approach that we have been employing to calibrate the DFTB models over the years for proton transfer studies,12,63,92,186,187 i.e., focusing on proton affinities, pKa values, and relative solvation free energies of different protonation states (including hydroxide12). Along this line, it is clear from the discussions here that a careful consideration of QM-MM interaction is also essential, and a robust parametrization requires an explicit consideration of condensed phase properties rather than gas phase clusters only. This requirement demands the development of efficient sampling algorithms, another technical issue that we have briefly touched upon in this article. The issue of sampling is particularly relevant in processes where the change of local hydration level plays a significant role, since such a change may be gated or be part of the kinetic bottleneck.75,78
As a final reflection, we emphasize again that the accuracy and thus the applicability of any approximate method should be judged in the context of the problem of interest. For the analysis of complex problems, a major part of the challenge for any computational study is indeed to establish the required level of accuracy to properly answer the question. For instance, although an explicit consideration of nuclear quantum effects is not critical to many mechanistic studies at room temperature, it is essential for kinetic isotope effects188,189 and potentially subtle spectroscopic features.66,67 Therefore, it is essential to establish the most relevant benchmark calculations, such as pKa calculations and solvation free energies for the discussion of proton transfers, while short-time behaviors sensitive to barriers of 1–2 kcal/mol145 are often of more limited relevance. Another useful strategy is to cross-validate the computational results using methods with very different approximations, such as comparing QM/MM and continuum electrostatic models for pKa predictions.75 In the end, the ultimate goal is to establish a conceptual framework to guide the development of novel mechanistic hypotheses and to stimulate new experiments to evaluate them.
Acknowledgments
The developments reviewed here have been made possible by generous support over the past years from the National Science Foundation grants CHE-0957285 and CHE-1300209 to Q.C. Some of the applications to biological systems have been supported by grants from the National Institutes of Health to Q.C. (R01GM084028 and R01GM106443). Computational resources from the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by NSF grant number OCI-1053575, are greatly appreciated; computations are also supported in part by NSF through a major instrumentation grant (CHE-0840494) to the Chemistry department. We also thank all the co-workers who have contributed to the work reviewed here.
Biographies
Puja Goyal received a B.Sc. (2006) in Chemistry from Presidency College, Kolkata, India, an M.Sc. (2008) in Chemistry from the Indian Institute of Technology, Kharagpur, and a Ph.D. (2013) in Chemistry from the University of Wisconsin—Madison. Currently, she is a postdoctoral researcher at the University of Illinois at Urbana–Champaign.
Hu-Jun Qian received a B.S. (2002) in Polymer Materials and Engineering and a Ph.D (2007) in Physical Chemistry from Jilin University. He was a postdoctoral researcher at Technische Universität Darmstadt (2007–2009) and Nagoya University (2009–2011). He joined the faculty of Jilin University in 2011 as an Associate Professor in the State Key Laboratory of Theoretical and Computational Chemistry.
Stephan Irle received a Diploma in Chemistry (1992) from the University of Siegen in Germany and a Ph.D. (1997) in Chemistry from the University of Vienna. In 1998, he became Associate Scientist at the Cherry L. Emerson Center for Scientific Computation at Emory University in Atlanta, GA, USA. In 2006, he moved to Japan as an Associate Professor at Nagoya University, where he became a full Professor of Chemistry in 2011. Since 2013, he also holds an appointment as PI at the WPI-Institute for Transformative Bio-Molecules (ITbM).
Xiya Lu received her B.S. (2010) in Chemistry from Nankai University in China. Currently, she is a Ph.D. candidate in Chemistry at University of Wisconsin—Madison under the supervision of Professor Qiang Cui. Her thesis research focuses on the parameterization of DFTB3 and QM/MM free energy simulations in metalloenzymes.
Daniel Roston received a B.A. (2007) in Chemistry from Hamilton College and a Ph.D. (2013) in Chemistry from the University of Iowa. He is currently a post-doctoral researcher at the University of Wisconsin.
Toshifumi Mori received a B.S. (2005) from Kyoto University and an M.S. (2007) and a Ph.D. (2010) in Chemistry from the Graduate School of Science, Kyoto University. After being a postdoctoral researcher at Stanford University and University of Wisconsin—Madison, he became an Assistant Professor at the Institute of Molecular Science in 2013.
Marcus Elstner received his Diploma in Physics (1993) from Technical University of Berlin and Ph.D. from the University of Paderborn (1998). Following postdoctoral studies at Harvard University with Prof. Kaxiras, he was a Junior Professor in Paderborn. In 2006, he joined TU Braunschweig, and in 2009, he moved to Karlsruhe Institute of Technology as a Professor of Theoretical Chemical Biology.
Qiang Cui received a B.S. (1993) in Chemical Physics from the University of Science and Technology of China and a Ph.D. (1997) in Chemical Physics from Emory University, working with Professor Keiji Morokuma. Following postdoctoral studies at Harvard University with Professor Martin Karplus, in 2001, he joined University of Wisconsin—Madison, where he remained since as a Professor of Chemistry.
Supporting Information Available
Additional results for the performance of DFTB3/3OB vs DFTB3/3OBw for different gas phase and condensed phase systems are included. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
○ Department of Chemistry, University of Illinois at Urbana–Champaign, 505 South Mathews Avenue, Urbana, IL 61801.
Author Present Address
∇ Institute for Molecular Science, Myodaiji, Okazaki, Aichi, 444-8585, Japan.
Author Contributions
# Contributed equally.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Ball P. Water as an active constituent in cell biology. Chem. Rev. 2008, 108, 74–108. [DOI] [PubMed] [Google Scholar]
- Eisenberg D.; Kauzmann W.. The structure and properties of water; Oxford University Press: Oxford, U.K., 2005. [Google Scholar]
- Dill K. A. Dominant forces in protein folding. Biochemistry 1990, 29, 7133–7155. [DOI] [PubMed] [Google Scholar]
- Levy Y.; Onuchic J. N.Water mediation in protein folding and molecular recognition. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 389–415. [DOI] [PubMed] [Google Scholar]
- Record M. T.; Zhang W. T.; Anderson C. F. Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: A practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects, and osmotic effects of salts. Adv. Protein Chem. 1998, 51, 281–353. [DOI] [PubMed] [Google Scholar]
- Xie W. J.; Gao Y. Q. A simple theory for the Hofmeister series. J. Phys. Chem. Lett. 2013, 4, 4247–4252. [DOI] [PubMed] [Google Scholar]
- Fersht A.Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding; W.H. Freeman and Company: New York, 1999. [Google Scholar]
- Warshel A.; Åqvist J. Electrostatic energy and macromolecular function. Annu. Rev. Biophys. Biophys. Chem. 1991, 20, 267–298. [DOI] [PubMed] [Google Scholar]
- Roux B.; Allen T. W.; Berneche S.; Im W. Theoretical and computational models of biological channels. Q. Rev. Biophys. 2004, 37, 15–103. [DOI] [PubMed] [Google Scholar]
- Zhu F. Q.; Hummer G. Pore opening and closing of a pentameric ligand-gated ion channel. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 19814–19819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. P.; Balabin I. A.; Beratan D. N. The nature of aqueous tunneling pathways between electron-transfer proteins. Science 2005, 310, 1311–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardi D.; König P.; Prat-Resina X.; Yu H.; Elstner M.; Frauenheim T.; Cui Q. “Proton holes” in long-range proton transfer reactions in solution and enzymes: A theoretical analysis. J. Am. Chem. Soc. 2006, 128, 16302–16311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth G. A. Computer simulation of proton solvation and transport in aqueous and biomolecular systems. Acc. Chem. Res. 2006, 39, 143–150. [DOI] [PubMed] [Google Scholar]
- Marx D. Proton transfer 200 years after von Grotthuss: Insights from ab initio simulations. ChemPhysChem 2006, 7, 1848–1870. [DOI] [PubMed] [Google Scholar]
- Decoursey T. E. Voltage-gated proton channels and other proton transfer pathways. Physiol. Rev. 2002, 83, 475–579. [DOI] [PubMed] [Google Scholar]
- Silverman D. N.; McKenna R. Solvent-mediated proton transfer in catalysis by carbonic anhydrase. Acc. Chem. Res. 2007, 40, 669–675. [DOI] [PubMed] [Google Scholar]
- Sham Y. Y.; Muegge I.; Warshel A. Simulating proton translocations in proteins: Probing proton transfer pathways in the Rhodobacter sphaeroides reaction center. Proteins: Struct., Funct., Bioinf. 1999, 36, 484–500. [PubMed] [Google Scholar]
- Swanson J. M. J.; Maupin C. M.; Chen H. N.; Petersen M. K.; Xu J. C.; Wu Y. J.; Voth G. A. Proton solvation and transport in aqueous and biomolecular systems: Insights from computer simulations. J. Phys. Chem. B 2007, 111, 4300–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmer D. T.; Willard A. P.; Madden P.; Chandler D. Hydration of metal surfaces can be dynamically heterogeneous and hydrophobic. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 4200–4205. [Google Scholar]
- Golze D.; Iannuzzi M.; Nguyen M.; Passerone D.; Hutter J. Simulation of adsorption processes at metallic interfaces: an image charge augmented QM/MM approach. J. Chem. Theory Comput. 2013, 9, 5086–5097. [DOI] [PubMed] [Google Scholar]
- Asbury J. B.; Steinel T.; Stromberg C.; Corcelli S. A.; Lawrence C. P.; Skinner J. L.; Fayer M. D. Water dynamics: Vibrational echo correlation spectroscopy and comparison to molecular dynamics simulations. J. Phys. Chem. A 2004, 108, 1107–1119. [Google Scholar]
- Pal S. K.; Zewail A. H. Dynamics of water in biological recognition. Chem. Rev. 2004, 104, 2099–2123. [DOI] [PubMed] [Google Scholar]
- Shen Y. R.; Waychunas G. A. In Vibrational spectroscopy at Electrified Interfaces; Wieckowski A., Korzeniewski C., Braunschweig B., Eds.; J. Wiley & Sons, Inc.: New York, 2013; Chapter SFG studies of oxide-water interfaces: protonation states, water polar orientations, and comparison with structure results from X-ray scattering. [Google Scholar]
- Bakker H. J.; Skinner J. L. Vibrational spectroscopy as a probe of structure and dynamics in liquid water. Chem. Rev. 2010, 110, 1498–1517. [DOI] [PubMed] [Google Scholar]
- Vega C.; Abascal J. L. F. Simulating water with rigid non-polarizable models: a general perspective. Phys. Chem. Chem. Phys. 2011, 13, 19663–19688. [DOI] [PubMed] [Google Scholar]
- Darre L.; Machado M. R.; Pantano S. Coarse-grained models of water. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2012, 2, 921–930. [Google Scholar]
- Grossman J. C.; Schwegler E.; Draeger E. W.; Gygi F.; Galli G. Towards an assessment of the accuracy of density functional theory for first principles simulations of water. J. Chem. Phys. 2004, 120, 300–311. [DOI] [PubMed] [Google Scholar]
- Ma Z. H.; Zhang Y. L.; Tuckerman M. E. Ab initio molecular dynamics study of water at constant pressure using converged basis sets and empirical dispersion corrections. J. Chem. Phys. 2012, 137, 044506. [DOI] [PubMed] [Google Scholar]
- Gordon M. S.; Smith Q. A.; Xu P.; Slipchenko L. V. Accurate first principles model potentials for intermolecular interactions. Annu. Rev. Phys. Chem. 2013, 64, 553–578. [DOI] [PubMed] [Google Scholar]
- Schmidt J.; VandeVondele J.; Kuo I. F. W.; Sebastiani D.; Siepmann J. I.; Hutter J.; Mundy C. J. Isobaric-isothermal molecular dynamics simulations utilizing density functional theory: an assessment of the structure and density of water at near-ambient conditions. J. Phys. Chem. B 2009, 113, 11959–11964. [DOI] [PubMed] [Google Scholar]
- Ben M. D.; Schonherr M.; Hutter J.; VandeVondele J. Bulk liquid water at ambient temperature and pressure from MP2 theory. J. Phys. Chem. Lett. 2013, 4, 3753–3759. [DOI] [PubMed] [Google Scholar]
- Stern H. A.; Rittner F.; Berne B. J.; Friesner R. A. Combined fluctuating charge and polarizable dipole models: Application to a five-site water potential function. J. Chem. Phys. 2001, 115, 2237–2251. [Google Scholar]
- Lamoureux G.; MacKerell A. D. Jr.; Roux B. A simple polarizable model of water based on classical Drude oscillators. J. Chem. Phys. 2003, 119, 5185–5197. [Google Scholar]
- Fanourgakis G. S.; Xantheas S. S. Development of transferable interaction potentials for water. V. Extension of the flexible, polarizable, Thole-type model potential (TTM3-F, v. 3.0) to describe the vibrational spectra of water clusters and liquid water. J. Chem. Phys. 2008, 128, 074506. [DOI] [PubMed] [Google Scholar]
- Bukowski R.; Szalewicz K.; Groenenboom G. C. A. van der Avoird, Predictions of the properties of water from first principles. Science 2007, 315, 1249–1252. [DOI] [PubMed] [Google Scholar]
- Wang Y. M.; Huang X. C.; Shepler B. C.; Braams B. J.; Bowman J. M. Flexible, ab initio potential, and dipole moment surfaces for water. I. Tests and applications for clusters up to the 22-mer. J. Chem. Phys. 2011, 134, 094509. [DOI] [PubMed] [Google Scholar]
- Bartok A. P.; Gillan M. J.; Manby F. R.; Csanyi G. Machine-learning approach for one- and two-body corrections to density functional theory: Applications to molecular and condensed water. Phys. Rev. B 2013, 88, 054104. [Google Scholar]
- Gillan M. J.; Alfe D.; Bartok A. P.; Csanyi G. First-principles energetics of water clusters and ice: A many-body analysis. Phys. Rev. B 2013, 139, 244504. [DOI] [PubMed] [Google Scholar]
- Medders G. R.; Babin V.; Paesani F. A critical assessment of two-body and three-body interactions in water. J. Chem. Theory Comput. 2013, 9, 1103–1114. [DOI] [PubMed] [Google Scholar]
- Babin V.; Medders G. R.; Paesani F. Development of a “first principles” water potential with flexible monomers. II: trimer potential energy surface, third virial coefficient, and small clusters. J. Chem. Theory Comput. 2014, 10, 1599–1607. [DOI] [PubMed] [Google Scholar]
- Dill K. A.; Truskett T. M.; Vlachy V.; Hribar-Lee B. Modeling water, the hydrophobic effect, and ion solvation. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 173–199. [DOI] [PubMed] [Google Scholar]
- Reddy G.; Lawrence C. P.; Skinner J. L.; Yethiraj A. Liquid state theories for the structure of water. J. Chem. Phys. 2003, 119, 13012–13016. [Google Scholar]
- Tainter C. J.; Pieniazek P. A.; Lin Y. S.; Skinner J. L. Robust three-body water simulation model. J. Chem. Phys. 2011, 134, 184501. [DOI] [PubMed] [Google Scholar]
- Ni Y. C.; Gruenbaum S. M.; Skinner J. L. Slow hydrogen-bond switching dynamics at the water surface revealed by theoretical two-dimensional sum-frequency spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 1992–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel W. Perspectives on semiempirical molecular orbital theory. Adv. Chem. Phys. 1996, 93, 703–757. [Google Scholar]
- Elstner M.; Porezag D.; Jungnickel G.; Elsner J.; Haugk M.; Frauenheim T.; Suhai S.; Seifert G. Self-consistent-charge density-functional tight-binding method for simulations of complex materials properties. Phys. Rev. B 1998, 58, 7260–7268. [Google Scholar]
- Gaus M.; Cui Q.; Elstner M. Density Functional Tight Binding (DFTB): Application to organic and biological molecules. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2014, 4, 49–61. [Google Scholar]
- Cui Q.; Elstner M. Density Functional Tight Binding: values of semi-empirical methods in an ab initio era. Phys. Chem. Chem. Phys. 2014, 16, 14368–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Pseudobond ab initio QM/MM approach and its applications to enzyme reactions. Theor. Chem. Acc. 2006, 116, 43–50. [Google Scholar]
- Hu H.; Yang W. T. Free energies of chemical reactions in solution and in enzymes with ab initio quantum mechanics/molecular mechanics methods. Annu. Rev. Phys. Chem. 2008, 59, 573–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerlin S. C. L.; Haranczyk M.; Warshel A. Progress in ab initio QM/MM free-energy simulations of electrostatic energies in proteins: accelerated QM/MM studies of pKa, redox reactions and solvation free energies. J. Phys. Chem. B 2009, 113, 1253–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laio A.; VandeVondele J.; Rothlisberger U. A Hamiltonian electrostatic coupling scheme for hybrid Car-Parrinello molecular dynamics simulations. J. Chem. Phys. 2002, 116, 6941–6947. [Google Scholar]
- Wu Z.; Cui Q.; Yethiraj A. A new coarse-grained model for water: the importance of electrostatic interactions. J. Phys. Chem. B 2010, 114, 10524–10529. [DOI] [PubMed] [Google Scholar]
- Marrink S. J.; Risselada H. J.; Yefimov S.; Tieleman D. P.; de Vries A. H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [DOI] [PubMed] [Google Scholar]
- Wu Z.; Cui Q.; Yethiraj A. A new coarse-grained force field for membrane-peptide simulations. J. Chem. Theory Comput. 2011, 11, 3793–3802. [DOI] [PubMed] [Google Scholar]
- Praprotnik M.; Delle Site L.; Kremer K. Multiscale simulation of soft matter: From scale bridging to adaptive resolution. Annu. Rev. Phys. Chem. 2008, 59, 545–571. [DOI] [PubMed] [Google Scholar]
- Marrink S. J.; de Vries A. H.; Tieleman D. P. Lipids on the move: Simulations of membrane pores, domains, stalks and curves. Biochim. Biophys. Acta, Biomembr. 2009, 1788, 149–168. [DOI] [PubMed] [Google Scholar]
- Nielsen S. O.; Bulo R. E.; Moore P. B.; Ensing B. Recent progress in adaptive multi-scale molecular dynamics simulations of soft matter. Phys. Chem. Chem. Phys. 2010, 12, 12401–12414. [DOI] [PubMed] [Google Scholar]
- Saunders M. G.; Voth G. A. Coarse-graining of multiprotein assemblies. Curr. Opin. Struct. Biol. 2012, 22, 144–150. [DOI] [PubMed] [Google Scholar]
- Shinoda W.; DeVane R.; Klein M. L. Computer simulation studies of self-assembling macromolecules. Curr. Opin. Struct. Biol. 2012, 22, 175–186. [DOI] [PubMed] [Google Scholar]
- Cui Q.; Elstner M.; Kaxiras E.; Frauenheim T.; Karplus M. A QM/MM implementation of the self consistent charge density functional tight binding (SCC-DFTB) method. J. Phys. Chem. B 2001, 105, 569–585. [Google Scholar]
- Elstner M.; Frauenheim T.; Suhai S. An approximate DFT method for QM/MM simulations of biological structures and processes. THEOCHEM 2003, 632, 29. [Google Scholar]
- Riccardi D.; et al. Development of effective quantum mechanical/molecular mechanical (QM/MM) methods for complex biological processes. J. Phys. Chem. B 2006, 110, 6458–6469. [DOI] [PubMed] [Google Scholar]
- Garczarek F.; Gerwert K. Functional waters in intraprotein proton transfer monitored by FTIR difference spectroscopy. Nature 2006, 439, 109–112. [DOI] [PubMed] [Google Scholar]
- Phatak P.; Ghosh N.; Cui Q.; Elstner M. Amino acids with an intermolecular proton bond as the proton storage site in bacteriorhodopsin. Proc. Acad. Natl. Sci. U.S.A. 2008, 105, 19672–19677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal P.; Ghosh N.; Phatak P.; Clemens M.; Gaus M.; Elstner M.; Cui Q. Proton storage site in bacteriorhodopsin: new insights from QM/MM simulations of microscopic pKa and infrared spectra. J. Am. Chem. Soc. 2011, 133, 14981–14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S.; Freier E.; Gerwert K. A delocalized proton-binding site within a membrane protein. Biophys. J. 2014, 107, 174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaila V. R.; Verkhovsky M. V.; Wikstr̈om M. Proton-coupled electron transfer in cytochrome c oxidase. Chem. Rev. 2010, 110, 7062–7081. [DOI] [PubMed] [Google Scholar]
- Goyal P.; Yang S.; Cui Q.. QM/MM analysis provides the microscopic basis for kinetic gating in Cytochrome c oxidase. Chem. Sci. 2014, submitted for publication [DOI] [PMC free article] [PubMed]
- Hou G. H.; Cui Q. QM/MM analysis suggests that Alkaline Phosphatase (AP) and Nucleotide pyrophosphatase/phosphodiesterase (NPP) slightly tighten the transition state for phosphate diester hydrolysis relative to solution: implication for catalytic promiscuity in the AP superfamily. J. Am. Chem. Soc. 2012, 134, 229–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou G. H.; Cui Q. Stabilization of different types of transition states in a single enzyme active site: qm/mm analysis of enzymes in the alkaline phosphatase superfamily. J. Am. Chem. Soc. 2013, 135, 10457–10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien P. J.; Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 1999, 4, R91–R105. [DOI] [PubMed] [Google Scholar]
- Hou G.; Zhu X.; Elstner M.; Cui Q. A modified QM/MM Hamiltonian with the Self-Consistent-Charge Density-Functional-Tight-Binding Theory for highly charged QM regions. J. Chem. Theory Comput. 2012, 8, 4293–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.; Cui Q.; Yethiraj A. Why do Arginine and Lysine organize lipids differently? Insights from coarse-grained and atomistic simulations. J. Phys. Chem. B 2013, 117, 12145–12156. [DOI] [PubMed] [Google Scholar]
- Goyal P.; Lu J.; Yang S.; Gunner M. R.; Cui Q. Changing hydration level in an internal cavity modulates the proton affinity of a key glutamate in Cytochrome c Oxidase. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 18886–18891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom D. G.; Castaneda C. A.; Velu P. D.; Garcia-Moreno B.; Charges E. in the hydrophobic interior of proteins. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 16096–16100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty S.; Warshel A. Capturing the energetics of water insertion in biological systems: The water flooding approach. Proteins: Struct., Funct., Bioinf. 2013, 81, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T.; Hamers R. J.; Pedersen J. A.; Cui Q. An explicit consideration of de-solvation is critical to binding free energy calculations of charged molecules at ionic surfaces. J. Chem. Theory Comput. 2013, 9, 5059–5069. [DOI] [PubMed] [Google Scholar]
- Frauenheim T.; Seifert G.; Elstner M.; Niehaus T.; Kohler C.; Amkreutz M.; Sternberg M.; Hajnal Z.; Di Carlo A.; Suhai S. Atomistic simulations of complex materials: ground-state and excited-state properties. J. Phys.: Condens. Matter 2002, 14, 3015–3047. [Google Scholar]
- Seifert G.; Joswig J. O. Density-functional tight binding - an approximate density-functional theory method. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2012, 2, 456–465. [Google Scholar]
- Elstner M. SCC-DFTB: What is the proper degree of self-consistency?. J. Phys. Chem. A 2007, 111, 5614–5621. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Yu H.; York D.; Cui Q.; Elstner M. Extension of the Self-Consistent Charge Density-Functional Tight-Binding method: Third-order expansion of the Density Functional Theory total energy and introduction of a modified effective Coulomb interaction. J. Phys. Chem. A 2007, 111, 10861–10873. [DOI] [PubMed] [Google Scholar]
- Gaus M.; Cui Q.; Elstner M. DFTB-3rd: Extension of the self-consistent-charge density-functional tight-binding method SCC-DFTB. J. Chem. Theory Comput. 2011, 7, 931–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaus M.; Goez A.; Elstner M. Parametrization and benchmark of DFTB3 for organic molecules. J. Chem. Theory Comput. 2013, 9, 338–354. [DOI] [PubMed] [Google Scholar]
- Sattelmeyer K. W.; Tirado-Rives J.; Jorgensen W. Comparison of SCC-DFTB and NDDO-based semiempirical molecular orbital methods for organic molecules. J. Phys. Chem. A 2006, 110, 13551–13559. [DOI] [PubMed] [Google Scholar]
- Otte N.; Scholten M.; Thiel W. Looking at Self-Consistent-Charge Density Functional Tight Binding from a semiempirical perspective. J. Phys. Chem. A 2007, 111, 5751–5755. [DOI] [PubMed] [Google Scholar]
- Elstner M.; Hobza P.; Frauenheim T.; Suhai S.; Kaxiras E. Hydrogen bonding and stacking interactions of nucleic acid base pairs: A density-functional-theory based treatment. J. Chem. Phys. 2001, 114, 5149–5155. [Google Scholar]
- Gaus M.; Lu X.; Elstner M.; Cui Q. Parameterization of DFTB3/3OB for Sulfur and Phosphorus for chemical and biological applications. J. Chem. Theory Comput. 2014, 10, 1517–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Yu H.; York D.; Elstner M.; Cui Q. Description of phosphate hydrolysis reactions with the Self-Consistent-Charge Density-Functional-Tight-Binding (SCCDFTB) theory 1. Parametrization. J. Chem. Theory Comput. 2008, 4, 2067–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi I.; Truhlar D. G. Parameterization of NDDO wave functions using genetic algorithms - an evolutionary approach to parameterizing potential energy surfaces and direct dynamics calculations for organic reactions. Chem. Phys. Lett. 1995, 233, 231–236. [Google Scholar]
- Riccardi D.; Koenig P.; Guo H.; Cui Q. Proton transfer in carbonic anhydrase is controlled by electrostatics rather than the orientation of the acceptor. Biochemistry 2008, 47, 2369–2378. [DOI] [PubMed] [Google Scholar]
- Riccardi D.; Yang S.; Cui Q. Proton transfer function of Carbonic Anhydrase: Insights from QM/MM simulations. Biochim. Biophys. Acta 2010, 1804, 342–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Yu H.; Cui Q. Extensive conformational changes are required to turn on ATP hydrolysis in myosin. J. Mol. Biol. 2008, 381, 1407–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Cui Q. Does water relayed proton transfer play a role in phosphoryl transfer reactions? A theoretical analysis of uridine 3′-m-nitrobenzyl phosphate isomerization in water and tert-butanol. J. Phys. Chem. B 2009, 113, 4930–4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar A. N.; Smith J. C.; Elstner M. Mechanism of a proton pump analyzed with computer simulations. Theor. Chem. Acc. 2010, 125, 353–363. [Google Scholar]
- Goyal P.; Elstner M.; Cui Q. Application of the SCC-DFTB method to neutral and protonated water clusters and bulk water. J. Phys. Chem. B 2011, 115, 6790–6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M.; Pisliakov A. V.; Warshel A. The barrier for proton transport in Aquaporins as a challenge for electrostatic models: The role of protein relaxation in mutational calculations. Proteins: Struct., Funct., Bioinf. 2006, 64, 829–844. [DOI] [PubMed] [Google Scholar]
- Ghosh N.; Prat-Resina X.; Cui Q. Towards a reliable molecular model of cytochrome c oxdiase: insights from microscopic pKa calculations. Biochemistry 2009, 48, 2468–2485. [DOI] [PubMed] [Google Scholar]
- Hu H.; Lu Z.; Elstner M.; Hermans J.; Yang W. Simulating water with the Self-Consistent-Charge Density Functional Tight Binding method: From molecular clusters to the liquid state. J. Phys. Chem. A 2007, 111, 5685–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin C.; Aradi B.; Voth G. The Self-Consistent Charge Density Functional Tight Binding method applied to liquid water and the hydrated excess proton: benchmark simulations. J. Phys. Chem. B 2010, 114, 6922–6931. [DOI] [PubMed] [Google Scholar]
- Choi T. H.; Liang R.; Maupin C. M.; Voth G. A. Application of the SCC-DFTB method to hydroxide water clusters and aqueous hydroxide solutions. J. Phys. Chem. B 2013, 117, 5165–5179. [DOI] [PubMed] [Google Scholar]
- Giese T. J.; Chen H. Y.; Dissanayake T.; Giambasu G. M.; Heldenbrand H.; Huang M.; Kuechler E. R.; Lee T. S.; Panteva M. T.; Radak B. K.; et al. A variational linear-scaling framework to build practical, efficient next-generation orbitalbased quantum force fields. J. Chem. Theory Comput. 2013, 9, 1417–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.; Thiel W.; Pezeshki S.; Lin H. Specific reaction path hamiltonian for proton transfer in water: reparameterized semiempirical models. J. Chem. Theory Comput. 2013, 9, 2672–2686. [DOI] [PubMed] [Google Scholar]
- Han J.; Mazack M. J. M.; Zhang P.; Truhlar D. G.; Gao J. L. Quantum mechanical force field for water with explicit electronic polarization. J. Chem. Phys. 2013, 139, 054503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper A. K.; Benmore C. J. Quantum differences between heavy and light water. Phys. Rev. Lett. 2008, 101, 065502. [DOI] [PubMed] [Google Scholar]
- Ponder J. W.; et al. Current status of the AMOEBA polarizable force field. J. Phys. Chem. B 2010, 114, 2549–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z. Y.; Zhou N. J.; Wu Q.; Zhang Y. K. Directional dependence of hydrogen bonds: a density-based energy decomposition analysis and its implications on force field development. J. Chem. Theory Comput. 2011, 7, 4038–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese T. J.; Chen H. Y.; Huang M.; York D. M. Parametrization of an orbital-based linear-scaling quantum force field for noncovalent interactions. J. Chem. Theory Comput. 2014, 10, 1086–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski S.; Giese T. J.; Gaus M.; York D. M.; Elstner M. Extended polarization in third-order SCC-DFTB from chemical-potential equalization. J. Phys. Chem. A 2012, 116, 9131–9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu S.; Tan M.-L.; Ichiye T. The large quadrupole of water molecules. J. Chem. Phys. 2006, 124, 074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese T. J.; York D. M. Charge-dependent model for many-body polarization, exchange, and dispersion interactions in hybrid quantum mechanical/molecular mechanical calculations. J. Chem. Phys. 2007, 127, 194101. [DOI] [PubMed] [Google Scholar]
- Bodrog Z.; Aradi B. Possible improvements to the self-consistent-charges density-functional tight-binding method within the second order. Phys. Status Solidi B 2012, 249, 259–269. [Google Scholar]
- Lyubartsev A. P.; Laaksonen A. Calculation of effective interaction potentials from radial distribution functions: a reverse Monte Carlo approach. Phys. Rev. E 1995, 52, 3730–3737. [DOI] [PubMed] [Google Scholar]
- Reith D.; Pütz M.; Müller-Plathe F. Deriving effective mesoscale potentials from atomistic simulations. J. Comput. Chem. 2003, 24, 1624–1636. [DOI] [PubMed] [Google Scholar]
- Doemer M.; Liberatore E.; Kanup J. M.; Tavernelli I.; Rothlisberger U. In situ parameterisation of SCC-DFTB repulsive potentials by iterative Boltzmann inversion. Mol. Simul. 2014, 111, 3595–3607. [Google Scholar]
- Skinner L. B.; Huang C.; Schlesinger D.; Pettersson L. G. M.; Nilsson A.; Benmore C. J. Benchmark oxygen-oxygen pair-distribution function of ambient water from x-ray diffraction measurements with a wide Q-range. J. Chem. Phys. 2013, 138, 074506. [DOI] [PubMed] [Google Scholar]
- Mills R. Self-Diffusion in normal and heavy water in the range 1–450. J. Phys. Chem. 1973, 77, 685–688. [Google Scholar]
- Price W. S.; Ide H.; Arata Y. Self-Diffusion of supercooled water to 238 K using PGSE NMR diffusion measurements. J. Phys. Chem. A 1999, 103, 448–450. [Google Scholar]
- Fecko C. J.; Loparo J. J.; Roberts S. T.; Tokmakoff A. Local hydrogen bonding dynamics and collective reorganization in water: Ultrafast infrared spectroscopy of HOD/D2O. J. Chem. Phys. 2005, 122, 054506. [DOI] [PubMed] [Google Scholar]
- Lawrence C. P.; Skinner J. L. Vibrational spectroscopy of HOD in liquid D2O. III. Spectral diffusion, and hydrogen-bonding and rotational dynamics. J. Chem. Phys. 2003, 118, 264–272. [Google Scholar]
- Tan H. S.; Piletic I. R.; Fayer M. D. Orientational dynamics of water confined on a nanometer length scale in reverse micelles. J. Chem. Phys. 2005, 122, 174501. [DOI] [PubMed] [Google Scholar]
- Rezus Y. L. A.; Bakker H. J. On the orientational relaxation of HDO in liquid water. J. Chem. Phys. 2005, 123, 114502. [DOI] [PubMed] [Google Scholar]
- Winkler K.; Lindner J.; Bursing H.; Vohringer P. Ultrafast Raman-induced Kerr effect of water: Single molecule versus collective motions. J. Chem. Phys. 2000, 113, 4674. [Google Scholar]
- Ropp J.; Lawrence C.; Farrar T. C.; Skinner J. L. Rotational motion in liquid water is anisotropic: A nuclear magnetic resonance and molecular dynamics simulation study. J. Am. Chem. Soc. 2001, 123, 8047–8052. [DOI] [PubMed] [Google Scholar]
- van der Spoel D.; van Maaren P. J.; Berendsen H. J. C. A systematic study of water models for molecular simulation: Derivation of water models optimized for use with a reaction field. J. Chem. Phys. 1998, 108, 10220. [Google Scholar]
- Laage D.; Hynes J. T. A Molecular Jump Mechanism ofWater Reorientation. Science 2006, 311, 832–835. [DOI] [PubMed] [Google Scholar]
- Ramirez R.; Lopez-Ciudad T.; Kumar P.; Marx D. Quantum corrections to classical time-correlation functions: Hydrogen bonding and anharmonic floppy modes. J. Chem. Phys. 2004, 121, 3973. [DOI] [PubMed] [Google Scholar]
- Yu H.; Cui Q. The vibrational spectra of protonated water clusters: A benchmark for SCC-DFTB. J. Chem. Phys. 2007, 127, 234504. [DOI] [PubMed] [Google Scholar]
- Wang L.-P.; Head-Gordon T.; Ponder J. W.; Ren P.; Chodera J. D.; Eastman P. K.; Martinez T. J.; Pande V. S. Systematic improvement of a classical molecular model of water. J. Phys. Chem. B 2013, 117, 9956–9972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Miller W. H.; Fanourgakis G. S.; Xantheas S. S.; Imoto S.; Saito S. Insights in quantum dynamical effects in the infrared spectroscopy of liquid water from a semi-classical study with an ab initio-based flexible and polarizable force field. J. Chem. Phys. 2011, 135, 244503. [DOI] [PubMed] [Google Scholar]
- Max J.-J.; Chapados C. Isotope effects in liquid water by infrared spectroscopy. III. H2O and D2O spectra from 6000 to 0 cm(−1). J. Chem. Phys. 2009, 131, 184505. [DOI] [PubMed] [Google Scholar]
- Choi T.; Jordan K. Application of the SCC-DFTB method to H+(H2O)6, H+(H2O)21, and H+(H2O)22. J. Phys. Chem. B 2010, 114, 6932–6936. [DOI] [PubMed] [Google Scholar]
- Kumar R.; Christie R.; Jordan K. A modified MS-EVB force field for protonated water clusters. J. Phys. Chem. B 2009, 113, 4111–4118. [DOI] [PubMed] [Google Scholar]
- Singh N. J.; Park M.; Min S. K.; Suh S. B.; Kim K. S. Magic and antimagic protonated water clusters: Exotic structures with unusual dynamic effects. Angew. Chem., Int. Ed. 2006, 45, 3795–3800. [DOI] [PubMed] [Google Scholar]
- Masamura M. Ab initio MO study on the structures of OH- (H2O)(n) in the gas phase. J. Mol. Struct.: THEOCHEM 2000, 498, 87–91. [Google Scholar]
- Botti A.; Bruni F.; Imberti S.; Ricci M. A.; Soper A. K. Ions in water: The microscopic structure of a concentrated HCl solution. J. Chem. Phys. 2004, 121, 7840–7848. [DOI] [PubMed] [Google Scholar]
- Botti A.; Bruni F.; Ricci M. A.; Soper A. K. Eigen versus Zundel complexes in HCl-water mixtures. J. Chem. Phys. 2006, 125, 014508. [DOI] [PubMed] [Google Scholar]
- Wu Y.; Chen H.; Wang F.; Paesani F.; Voth G. An improved multistate empirical valence bond model for aqueous proton solvation and transport. J. Phys. Chem. B 2008, 112, 467–482. [DOI] [PubMed] [Google Scholar]
- Tuckerman M. E. Structure and dynamics of OH-(aq). Acc. Chem. Res. 2006, 39, 151–158. [DOI] [PubMed] [Google Scholar]
- Marx D.; Chandra A.; Tuckerman M. E. Aqueous basic solutions: hydroxide solvation, structural diffusion, and comparison to the hydrated proton. Chem. Rev. 2010, 110, 2174–2216. [DOI] [PubMed] [Google Scholar]
- Roberts S. T.; Petersen P. B.; Ramasesha K.; Tokmakoff A.; Ufimtsev I. S.; Martinez T. J. Observation of a Zundel-like transition state during proton transfer in aqueous hydroxide solutions. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 15154–15159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig P.; Ghosh N.; Hoffman M.; Elstner M.; Tajkhorshid E.; Frauenheim T.; Cui Q. Towards theoretical analysis of long-range proton transfer kinetics in bio-molecular pumps. J. Phys. Chem. A 2006, 110, 548–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zienau J.; Cui Q. Implementation of the solvent macromolecule boundary potential and application to model and realistic enzyme systems. J. Phys. Chem. B 2012, 116, 12522–12534. [DOI] [PubMed] [Google Scholar]
- Riccardi D.; Li G.; Cui Q. The importance of van der Waals interactions in QM/MM simulations. J. Phys. Chem. B 2004, 108, 6467–6478. [DOI] [PubMed] [Google Scholar]
- Liang R. B.; Swanson J. M.; Voth G. A. Benchmark study of the SCC-DFTB approach for a biomolecular proton channel. J. Chem. Theory Comput. 2014, 10, 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Ilan B.; Voth G. A. Charge delocalization in proton channels II: the synthetic LS2 channel and proton selectivity. Biophys. J. 2007, 92, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn H. M.; Thiel W. QM/MM methods for biomolecular systems. Angew. Chem., Int. Ed. 2009, 48, 1198–1229. [DOI] [PubMed] [Google Scholar]
- Konig P.; Hoffmann M.; Frauenheim T.; Cui Q. A critical evaluation of different QM/MM frontier treatments with SCC-DFTB as the QM method. J. Phys. Chem. B 2005, 109, 9082–9095. [DOI] [PubMed] [Google Scholar]
- Reuter N.; Dejaegere A.; Maigret B.; Karplus M. Frontier bonds in QM/MM methods: A comparison of different approaches. J. Phys. Chem. A 2000, 104, 1720–1735. [Google Scholar]
- Gao J. L.; Amara P.; Alhambra C.; Field M. J. A generalized hybrid orbital (GHO) method for the treatment of boundary atoms in combined QM/MM calculations. J. Phys. Chem. A 1998, 102, 4714–4721. [Google Scholar]
- Freindorf M.; Gao J. L. Optimization of the Lennard-Jones parameters for a combined ab initio quantum mechanical and molecular mechanical potential using the 3-21G basis set. J. Comput. Chem. 1996, 17, 386–395. [Google Scholar]
- Das D.; Eurenius K. P.; Billings E. M.; Sherwood P.; Chatfield D. C.; Hodoscek M.; Brooks B. R. Optimization of quantum mechanical molecular mechanical partitioning schemes: Gaussian delocalization of molecular mechanical charges and the double link atom method. J. Chem. Phys. 2002, 117, 10534–10547. [Google Scholar]
- Wang B.; Truhlar D. G. Including charge penetration effects in molecular modeling. J. Chem. Theory Comput. 2010, 6, 3330–3342. [DOI] [PubMed] [Google Scholar]
- Field M. J.; Bash P. A.; Karplus M. A combined quantum-mechanical and molecular mechanical potential for molecular-dynamics simulations. J. Comput. Chem. 1990, 11, 700–733. [Google Scholar]
- Vasilyev V. V.; Bliznyuk A. A.; Voityuk A. A. A combined quantum chemical molecular mechanical study of hydrogen-bonded systems. Int. J. Quantum Chem. 1992, 44, 897–930. [Google Scholar]
- Cummins P. L.; Gready J. E. Coupled semiempirical molecular orbital and molecular mechanics model (QM/MM) for organic molecules in aqueous solution. J. Comput. Chem. 1997, 18, 1496–1512. [Google Scholar]
- Han W. G.; Elstner M.; Jalkanen K. J.; Frauenheim T.; Suhai S. Hybrid SCCDFTB/molecular mechanical studies of H-bonded systems and of N-acetyl-(L-Ala)(n) N ′-methylamide helices in water solution. Int. J. Quantum Chem. 2000, 78, 459–479. [Google Scholar]
- MacKerell A. D. Jr.; Bashford D.; Bellott M.; Dunbrack R. L. Jr.; Evenseck J. D.; Field M. J.; Fischer S.; Gao J.; Guo H.; Ha S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- Giese T. J.; York D. M.; et al. QCRNA 1.0: A database of quantum calculations for RNA catalysis. J. Mol. Graphics Modell. 2006, 25, 423–433. [DOI] [PubMed] [Google Scholar]
- Woods C. J.; Manby F. R.; Mulholland A. J. An efficient method for the calculation of quantum mechanics/molecular mechanics free energies. J. Chem. Phys. 2008, 128, 014109. [DOI] [PubMed] [Google Scholar]
- Gao J. In Reviews in Computational Chemistry VII; Lipkowitz K. B., Boyd D. B., Eds.; VCH: New York, 1995; p 119. [Google Scholar]
- Warshel A.Computer Modeling of Chemical Reactions in Enzymes and Solution; Wiley: New York, 1991. [Google Scholar]
- Deng Y. Q.; Roux B. Hydration of amino acid side chains: Nonpolar and electrostatic contributions calculated from staged molecular dynamics free energy simulations with explicit water molecules. J. Phys. Chem. B 2004, 108, 16567–16576. [Google Scholar]
- Shivakumar D.; Deng Y. Q.; Roux B. Computations of absolute solvation free energies of small molecules using explicit and implicit solvent model. J. Chem. Theory Comput. 2009, 5, 919–930. [DOI] [PubMed] [Google Scholar]
- Kastenholz M. A.; Hünenberger P. H. Computation of methodology-independent ionic solvation free energies from molecular simulations. II. the hydration free energy of the sodium cation. J. Chem. Phys. 2006, 124, 224501-1–224501-20. [DOI] [PubMed] [Google Scholar]
- Lu X.; Cui Q. Charging free energy calculations using the Generalized Solvent Boundary Potential (GSBP) and periodic boundary condition: a comparative analysis using ion solvation and reduction potential in proteins. J. Phys. Chem. B 2013, 117, 2005–2018. [DOI] [PubMed] [Google Scholar]
- Lin Y.; Aleksandrov A.; Simonson T.; Roux B. An overview of electrostatic free energy computations for solutions and proteins systems. J. Chem. Theory Comput. 2014, 10, 2690–2709. [DOI] [PubMed] [Google Scholar]
- Marenich A. V.; Kelly C. P.; Thompson J. D.; Hawkins G. D.; Chambers C. C.; Giesen D. J.; Winget P.; Cramer C. J.; Truhlar D. G.. Minnesota Solvation Database; 2012.
- George P.; Witonsky R. J.; Trachtman M.; Wu C.; Dorwart W.; Richman L.; Richman W.; Shurayh F.; Lentz B. Squiggle-H2O - An enquiry into importance of solvation effects in phosphate ester and anhydride reactions. Biochim. Biophys. Acta 1970, 223, 1–15. [DOI] [PubMed] [Google Scholar]
- König G.; Hudson P. S.; Boresch S.; Woodcock H. L. Multiscale free energy simulations: An efficient method for connecting classical MD simulations to QM or QM/MM free energies using Non-Boltzmann Bennett reweighting schemes. J. Chem. Theory Comput. 2014, 10, 1406–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T.; Hamers R. J.; Pedersen J. A.; Cui Q. Integrated Hamiltonian Sampling: a simple and versatile method for free energy simulations and conformational sampling. J. Phys. Chem. B 2014, 118, 8210–8220. [DOI] [PubMed] [Google Scholar]
- Wang F.; Landau D. Efficient, multiple-range random walk algorithm to calculate the density of states. Phys. Rev. Lett. 2001, 86, 2050–2053. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Rosenberg J. M.; Bouzida D.; Swendsen R. H.; Kollman P. A. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992, 13, 1011–1021. [Google Scholar]
- Gao Y. Q. An integrate-over-temperature approach for enhanced sampling. J. Chem. Phys. 2008, 128, 064105. [DOI] [PubMed] [Google Scholar]
- Yang L. J.; Gao Y. Q. A selective integrated tempering method. J. Chem. Phys. 2009, 131, 214109. [DOI] [PubMed] [Google Scholar]
- Mitsutake A.; Okamoto Y. Replica-exchange simulated tempering method for simulations of frustrated systems. Chem. Phys. Lett. 2000, 332, 131–138. [Google Scholar]
- Fukunishi H.; Watanabe O.; Takada S. On the Hamiltonian replica exchange method for efficient sampling of biomolecular systems: Application to protein structure prediction. J. Chem. Phys. 2002, 116, 9058–9067. [Google Scholar]
- Christ C. D.; van Gunsteren W. F. Enveloping distribution sampling: A method to calculate free energy differences from a single simulation. J. Chem. Phys. 2007, 126, 184110. [DOI] [PubMed] [Google Scholar]
- Kong X.; Brooks C. L. λ-dynamics: A new approach to free energy calculations. J. Chem. Phys. 1996, 105, 2414. [Google Scholar]
- Paliwal H.; Shirts M. R. Using multistate reweighting to rapidly and efficiently explore molecular simulation parameters space for nonbonded interactions. J. Chem. Theory Comput. 2013, 9, 4700–4717. [DOI] [PubMed] [Google Scholar]
- Li Y. P.; Li J. C.; Wang F. Liquid-liquid transition in supercooled water suggested by microsecond simulations. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 12209–12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y.; Hassanali A. A.; Parrinello M. Anomalous water diffusion in salt solutions. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 3310–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lilienfeld O. A.; Tkatchenko A. Two- and three-body interatomic dispersion energy contributions to binding in molecules and solids. J. Chem. Phys. 2010, 132, 234109. [DOI] [PubMed] [Google Scholar]
- Risthaus T.; Grimme S. Benchmarking of London dispersion-accounting density functional theory methods on very large molecular complexes. J. Chem. Theory Comput. 2013, 9, 1580–1591. [DOI] [PubMed] [Google Scholar]
- Yoo S.; Xantheas S. S. Communication: The effect of dispersion corrections on the melting temperature of liquid water. J. Chem. Phys. 2011, 134, 121105. [DOI] [PubMed] [Google Scholar]
- Riccardi D.; Schaefer P.; Cui Q. pKa calculations in solution and proteins with QM/MM free energy perturbation simulations. J. Phys. Chem. B 2005, 109, 17715–17733. [DOI] [PubMed] [Google Scholar]
- Riccardi D.; Zhu X.; Goyal P.; Yang S.; Hou G.; Cui Q. Toward molecular models of proton pumping: challenges, methods and relevant applications. Sci. China: Chem. 2012, 55, 3–18. [Google Scholar]
- Gao J.; Truhlar D. G. Quantum mechanical methods for enzyme kinetics. Annu. Rev. Phys. Chem. 2002, 53, 467. [DOI] [PubMed] [Google Scholar]
- Cui Q.; Karplus M. QM/MM studies of the Triosephosphate isomerase (TIM) catalyzed reactions: The effect of geometry and tunneling on proton transfer rate constants. J. Am. Chem. Soc. 2002, 124, 3093–3124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.