Summary
The past twenty years have seen tremendous advances in our understanding of the mechanisms underlying bacterial cytokinesis, particularly the composition of the division machinery and the factors controlling its assembly [1]. At the same time, we understand very little about the relationship between cell division and other cell cycle events in bacteria. Here we report that inhibiting division in Bacillus subtilis and Staphylococcus aureus quickly leads to an arrest in the initiation of new rounds of DNA replication followed by a complete arrest in cell growth. Arrested cells are metabolically active but unable to initiate new rounds of either DNA replication or division when shifted to permissive conditions. Inhibiting DNA replication results in entry into a similar quiescent state, in which cells are unable to resume growth or division when returned to permissive conditions. Our data suggest the presence of two failsafe mechanisms: one linking division to the initiation of DNA replication and another linking the initiation of DNA replication to division. These findings contradict the prevailing view of the bacterial cell cycle as a series of coordinated but uncoupled events. Importantly, the terminal nature of the cell cycle arrest validates the bacterial cell cycle machinery as an effective target for antimicrobial development.
Results
An extended block in division leads to a metabolically active but unrecoverable state
To determine the impact of an extended block in cell division on cell physiology, we took advantage of genetic and chemical methods to conditionally inhibit division. We utilized three B. subtilis strains: one in which synthesis of the essential tubulin-like cell division protein FtsZ is dependent on a xylose inducible promoter, Pxyl-ftsZ (ftsZ::spc, thrC::Pxyl-ftsZ, xylA::tet) [2], one encoding a heat-sensitive allele of ftsZ, ftsZts (ftsZ::ftsZ-gfp) [3], and one encoding a heat-sensitive allele of a protein required for septum formation, divICts (divIC::divICts) [4]. FtsZ assembles into a ring structure that serves as a scaffold for the division machinery at nascent septal sites [1]. DivIC is a membrane-anchored protein that contributes to the integrity of the cytokinetic ring [5]. Upon removal of inducer in Pxyl-ftsZ, FtsZ levels quickly decreased and were reduced to 3% at 5.2 MDP (Figure S1A). In contrast, both the ftsZts and divICts strains retained near wild-type levels of FtsZ following a division block (Figure S1A). In addition, we inhibited division in wild-type S. aureus (Newman), using PC190723, a derivative of 3-methoxybenzamide that inhibits FtsZ assembly [6, 7]. All four strains form FtsZ rings and septa under permissive conditions, although the divICts strain is somewhat impaired for division even at 30°C (Figure S2).
Following a shift to nonpermissive conditions, cells continued to increase in mass at normal rates for several generations until growth abruptly arrested (Figure 1A-D). For comparative purposes, we normalized growth rates between strains and culture conditions using exponential mass doubling period(s) (MDP) (Table S1). Pxyl-ftsZ and ftsZts increased in mass for ~5 MDP while divICts and PC190723 treated S. aureus grew for ~3 MDP after the shift to nonpermissive conditions.
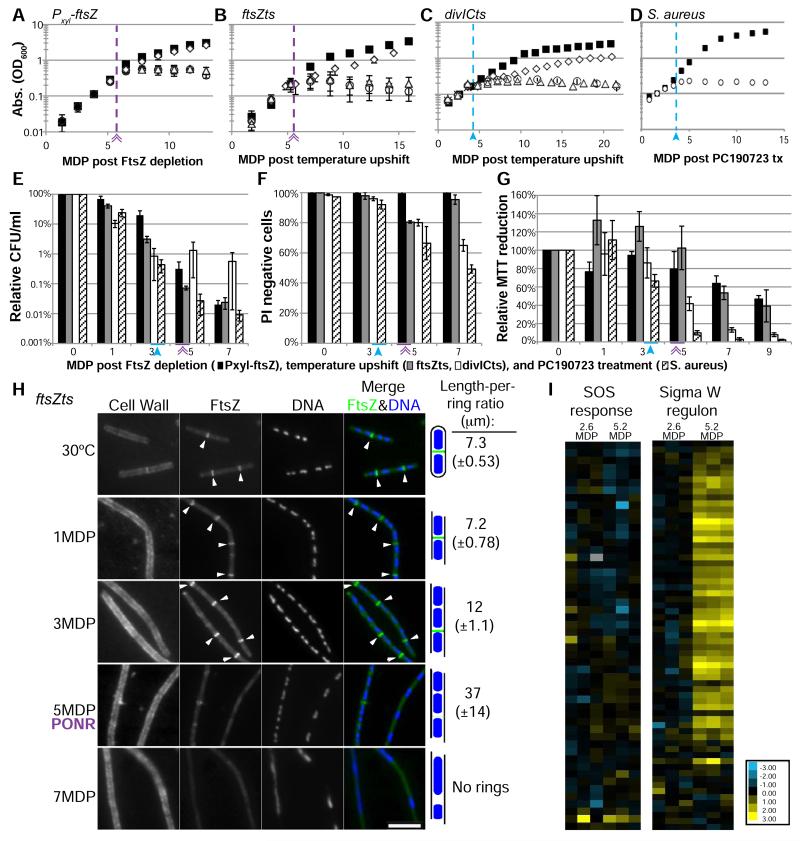
Figure 1. An extended block in cell division leads to a growth arrest and entry into a permanent, quiescent state at the Point of No Return (PONR).
(A-D) Cell growth as measured by OD600 of Pxyl-ftsZ (A), ftsZts (B), divICts (C) and PC190723 treated S. aureus (D). (A-G) Purple lines/double arrow (Pxyl-ftsZ, ftsZts) and blue lines/closed arrow (divICts) delineate the PONR boundary. Growth under permissive conditions (black squares), nonpermissive conditions (open circles), shifted to permissive conditions ~ 1-2 Mass Doubling Periods (MDP) before the PONR (3.9 MDP – Pxyl-ftsZ, 4.4 MDP – ftsZts, 3 MDP – divICts) (open diamonds), or shifted to permissive conditions ~1 generation after the PONR (6.5 MDP – Pxyl-ftsZ, 7 MDP – ftsZts, 5 MDP – divICts) (open triangles). (E-G) Cells are metabolically active but unrecoverable following the PONR. Pxyl-ftsZ (black bars), ftsZts (gray bars), divICts (white bars) and S. aureus (dashed bars). (E) Colony forming ability (plating efficiency) was determined after culturing cells at nonpermissive conditions prior to plating cells under permissive conditions. (F) Propidium iodide (PI) is a marker for loss of membrane integrity. PI negative (live) cells were quantified after cells were cultured for the indicated number of generations under nonpermissive conditions. (G) Percent MTT reduction serves as a direct readout of metabolic activity. Permissive levels were set to 100%. (A-G) Error bars = SD, n=3-5. (H) Cells are refractile to FtsZ assembly after the PONR. ftsZts cells were cultured under nonpermissive conditions for 1, 3, 5 and 7 MDP prior to a shift to permissive conditions for 1 MDP. Length-per-ring ratios were calculated by dividing total cell length by the number of FtsZ rings. ~200 wild-type cell lengths scored per replicate. n=3-4, SD in parentheses. Arrows indicate FtsZ rings. Bar = 5 μm. (I) Relative expression of genes in the SOS response or the Sigma W regulon. Transcript levels for 3 replicates are represented as the fold change for FtsZ depleted cells compared to FtsZ+ cells. See also Figure S1, S2 and Table S1.
The growth arrest was irreversible in all strains. Shifting cultures to permissive conditions following the growth arrest failed to yield any significant increase in optical density (Figure 1A-C). In addition, colony-forming ability under permissive condition was reduced at least 100-fold in cells sampled at the growth arrest and ~1000 fold 7 MDP post division inhibition (Figure 1E).
Based on the inability of cells blocked for division to recover, we have termed the time of growth arrest as the Point of No Return (PONR). The inability to recover following the shift to permissive conditions differentiates these cells from persisters, which are dormant variants that occur stochastically in wild-type populations and can divide and grow following the removal of an antibiotic [8].
Cells retain membrane integrity and metabolic activity after the PONR
To determine if cells remain alive subsequent to the PONR, we assessed membrane integrity and metabolic levels in cells sampled before and after the growth arrest. We assessed membrane integrity using propidium iodide (PI), a nucleic acid binding agent that cannot pass through intact membranes [9]. Prior to the growth arrest, Pxyl-ftsZ and ftsZts cells were refractory to PI staining (Figure 1F). At and even after the PONR, the majority of cells (~60-100%, depending on strain) remained impermeable to PI staining (Figure 1F). We observed 100% PI labeling for heat killed and ethanol killed cells (Figure S1B).
To assess metabolic activity, we employed 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), which measures NAD(P)H-dependent oxidoreductase activity [10]. MTT is reduced from its yellow, water-soluble form to purple insoluble formazan crystals in metabolically active cells. Resuspending these crystals in DMSO and OD550 provides a direct measurement of cell metabolism [10].
MTT reduction indicates that cells inhibited for division are alive and metabolically active at the growth arrest. At the PONR, MTT reduction rates in were 65-100% of cells cultured under permissive conditions (Figure 1G). Pxyl-ftsZ and ftsZts exhibited a gradual decline in metabolic activity the PONR, exhibiting ~40% of the activity of cells at permissive conditions at 9 MDP (Figure 1G). divICts and S. aureus rapidly declined in MTT reduction after the PONR, with MTT reduction rates falling to <20% of cells at permissive conditions (Figure 1G). Heat killed cells had 0-4% of permissive activity (Figure S1C). Consistent with high levels of metabolic activity, 35S-methionine labeling indicated that protein synthesis rates remain near wild type in Pxyl-ftsZ cells up to 9 MDP following FtsZ depletion (Figure S1D).
We speculate the increased sensitivity of divICts and S. aureus strains following a division block is due to physiological differences between these bacteria and Pxyl-ftsZ and ftsZts strains, specifically with regard to membrane integrity. divICts mutants grow slowly and are somewhat filamentous even under permissive conditions while S. aureus exhibits an extremely rapid increase in diameter following inhibition of division (Figure S2C,G, Table S1).
Inhibiting cell division triggers an arrest in DNA replication
Although cells remained intact and metabolically active, DNA staining revealed a marked impact on chromosome number and morphology following an extended block in division. As previously reported, nucleoids (discrete DNA-containing units) appear normal with regard to morphology and segregation up to the initial arrest in growth [11-13] (Figure 2A, S2). After the arrest, however, we observed a clear deterioration in both the size and position of nucleoids, culminating in bright discontinuous structures (Fig 2A, S2). The nucleoid morphology differences following the PONR most likely result from cells at different stages of the cell cycle at the onset of the division block.
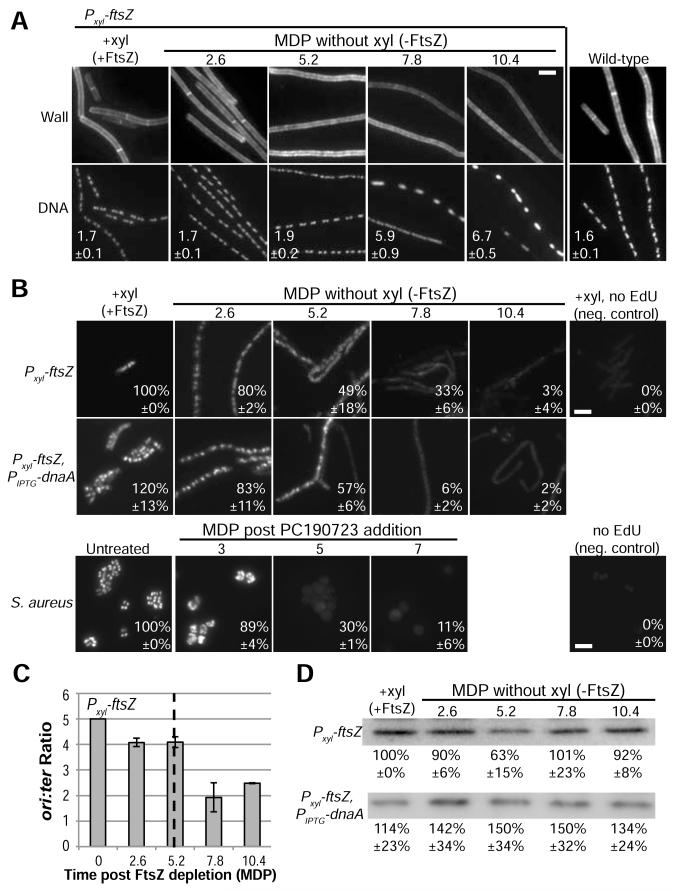
Figure 2. Inhibiting cell division triggers an arrest in DNA replication.
(A) WGA labeled cell wall and dapi-labeled DNA in wild-type and Pxyl-ftsZ cells. The average length per nucleoid ratio ±SD in μm are shown. n=3, ~200 cells scored per replicate. (B) Newly replicated DNA labeled with EdU (Methods). The fluorescence of induced Pxyl-ftsZ or untreated S. aureus was set to 100%. Average ±SD of fluorescence is shown. n=3, ~100 cells per replicate. (C) Marker frequency analysis of the origin to terminus (ori:ter) ratios of Pxyl-ftsZ after depleting FtsZ. Error bars = SD, n=3. (D) Quantitative immunoblots of DnaA in Pxyl-ftsZ and Pxyl-ftsZ, PIPTG-dnaA cells depleted for FtsZ. The average ±SD is shown below a representative blot. Induced Pxyl-ftsZ (+xyl) was set to 100%, n=3. (A-B) Bar = 5 μm. See also Figure S3.
Consistent with the abnormal nucleoid morphology, we find that DNA replication is inhibited in cells subjected to a prolonged block in division (Figure 2B). Labeling Pxyl-ftsZ cells with the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU) [14, 15], revealed a ~20% reduction in DNA replication at 2.6 MDP post FtsZ depletion, a ~60% reduction at 5.2 MDP and replication was essentially non-existent after 10.4 MDP (Figure 2B). S. aureus treated with PC190723 also exhibited a decline in EdU labeling that is detectable before the PONR (Figure 2B).
DNA replication is blocked at initiation following division inhibition
The gradual reduction in EdU incorporation following division inhibition suggested cells were blocked at initiation, the earliest stage in DNA replication. Defects in initiation allow ongoing replication forks to complete but block new rounds of replication [11]. Fast growing B. subtilis cultured in nutrient rich medium undergo multifork replication, with up to 16 replication forks proceeding simultaneously and can complete ~3 rounds of chromosome segregation in the absence of new initiation events [16].
To confirm that DNA replication was blocked at initiation, we employed marker frequency analysis using qPCR to amplify origin and terminus proximal sequences and calculate the ratio of the origin to terminus DNA (ori:ter) [17]. Inhibiting replication at initiation but permitting completion of ongoing replication forks thus leads to a reduction in the ori:ter ratio.
In support of the idea that replication initiation is coupled to the successful completion of cytokinesis [18], we observed a steady decline in the ori:ter ratio in Pxyl-ftsZ cells cultured in the absence of inducer. Marker frequency analysis revealed a reduction in the ori:ter ratio from 5:1 to 4:1 at the PONR (5.2 MDP) (Figure 2C). The ori:ter ratio continued to decline after the PONR approaching 2:1 at 7.8 MDP (Figure 2C). The failure to reach a 1:1 ori:ter ratio (the expected outcome of a complete initiation arrest) suggests the presence of a secondary arrest in elongation after the PONR. Localization of a CFP fusion to Spo0J, which binds near the origin of replication, further supports a block at initiation in FtsZ depleted cells (Figure S3C).
We hypothesized the defect in replication initiation might be due to a reduction in the highly conserved initiation regulator DnaA [19]. However, while we observed a transient drop in DnaA concentration to ~60% of wild type levels at the PONR, modest overproduction of DnaA (~1.3 to 1.5 times wild-type) was insufficient to bypass the replication arrest in FtsZ depleted cells (Figure 2B, D). The dip in DnaA levels is most likely the result of autorepression in response to a transient increase in the ratio of DnaA to oriC DNA at the growth arrest [17, 20].
FtsZ rings are unable to form after the PONR
The block in DNA replication together with the unrecoverable nature of cells subsequent to the PONR suggested the onset of a terminal cell cycle arrest. To confirm this possibility, we examined the ability of cells cultured past the PONR to support assembly of FtsZ rings at nascent division sites following a shift to permissive temperatures.
Consistent with a terminal cell cycle arrest, the PONR served as a sharp boundary in the ability to form FtsZ rings when shifted to permissive conditions (Figure 1H, S2). In ftsZts cells, Extant FtsZ rings were stable under restrictive conditions and new FtsZ rings readily assembled in cells downshifted to permissive conditions prior to the PONR (1 or 3 MDP) (Figure S2A, 1H). However, FtsZ rings failed to form in ftsZts cells downshifted after the PONR (7 MDP) (Figure 1H, S2B). The timing of the division arrest rules out delocalization of the MinCD division inhibitor as FtsZ rings form readily in cells downshifted prior to the PONR, when MinCD should already be delocalized [21].
Pxyl-ftsZ and divICts cells cultured past the PONR were similarly incapable of forming new FtsZ rings (Figure S2C-F). DivIC is required for steps in division subsequent to FtsZ assembly, reinforcing the idea that a general block in division is sufficient to trigger the PONR.
An extended block in DNA replication initiation leads to entry into a quiescent, but unrecoverable state
The onset of the replication arrest ~3 MDP prior to the PONR suggested that inhibition of DNA replication, rather than the division block, might be the proximal trigger for entry into the quiescent state. To test this possibility, we employed a strain encoding a conditional mutation in dnaB, the replicative helicase co-loader [22]. dnaB134ts (dnaBts) mutants can initiate new rounds of DNA replication at the permissive temperature of 30°C but are unable to do so upon a shift to the nonpermissive temperature of 45°C [22].
As observed in cells subjected to a prolonged block in division, inhibiting initiation resulted in a growth arrest and entry into a non-recoverable state. dnaBts cells increased in mass for ~3 MDP after a shift to 45°C, before growth abruptly arrested (Figure 3A). Downshifting dnaBts cells to 30°C prior to the growth arrest resulted in almost full recovery (as measured by OD600), however mutants were unable to recover when shifted to permissive conditions after 5 MDP at 45°C (Figure 3A). Colony-forming ability was similarly impaired after the growth arrest in dnaBts cells cultured at 45°C (Figure 3B).
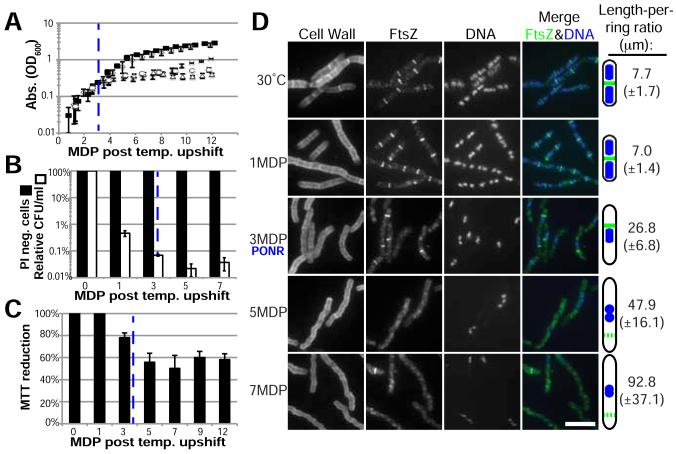
Figure 3. An extended block in DNA replication initiation results in a terminal cell cycle arrest.
(A) Cell growth of dnaB134ts under permissive conditions (black squares), nonpermissive conditions (open circles), shifted to permissive conditions at 1 MDP (open diamonds), shifted to permissive conditions approximately at 3 MDP (X’s), or shifted to permissive conditions at 5 MDP (open triangles). (B) PI negative (live) cells and colony forming ability (plating efficiency) was determined after culturing cells under nonpermissive conditions for the indicated MDP (PI) and prior to plating cells at permissive conditions (CFU). (C) MTT reduction levels indicate metabolic activity of cells blocked for initiation. (A-C) Error bars = SD, n=3, dashed blue line indicates the PONR. (D) Cells were cultured under nonpermissive conditions for 1, 3, 5 and 7 MDP. Length-per-ring ratios were calculated as in Figure 1. n=3, SD in parentheses. Bar = 5 μm.
Despite their unrecoverable nature, ~97-100% of dnaBts cells cultured at 45°C retained intact membranes well past the growth arrest (Figure 3B). dnaBts cells also were metabolically active after the growth arrest, retaining MTT reduction rates of 60% after 12 MDP at non-permissive conditions (Figure 3C).
A prolonged block in replication initiation triggers a terminal arrest in division
Consistent with a terminal cell cycle arrest, dnaBts cells subjected to a prolonged block in replication were unable to form FtsZ rings (Figure 3D, S4A). As has been observed in other DNA replication mutants, dnaBts cells were elongated and contained single FtsZ rings adjacent to the medially positioned nucleoids 3 MDP after the shift to nonpermissive conditions, (Figure 3D) [23-26]. While dnaBts cells cultured at 45°C were able to form FtsZ rings at the growth arrest (3 MDP), they were unable to do so 2 MDP later, even if shifted to permissive conditions (Figure 3D, S4A). FtsZ levels remained unchanged over the course of the experiment (Figure S4B). A loss of function mutation in the gene encoding Noc –implicated in coordinating chromosome segregation and division [27]– had no impact on the onset of the growth arrest or the ability of cells to recover after the PONR (Data not shown).
Microarray analysis of FtsZ-depleted cells suggests division is coordinated with DNA replication via a post-transcriptional mechanism
Microarray analysis of Pxyl-ftsZ cells prior to and at the PONR suggests the both the replication arrest and entry into the quiescent state are controlled via a post-transcriptional mechanism. Comparison of gene expression in FtsZ-depleted cells and mock-depleted cells revealed the overwhelming majority of genes maintained constant expression levels, similar to what has been reported in E. coli [28]. We did not observe induction of genes known to play a role in cell cycle regulation, nor did we see induction of the SOS response, which controls the cell cycle arrest associated with DNA damage (Figure 1I) [29]. Importantly, we did not observe significant induction of stationary-phase specific gene expression, consistent with the PONR being a distinct physiological state.
We observed upregulation of the Sigma W regulon, a set of genes involved in the response to a variety of cell envelope stresses including antibiotic challenge and osmotic shock [30]. However, deletion of the transcription factor controlling this regulon, Sigma W, had only a modest impact on the growth and plating efficiency of Pxyl-ftsZ cells cultured past the PONR (Figure 1I; S1E-F; Table S2). Additional upregulated transcripts at 5.2 MDP included the flagellar component motA; two genes involved in mannitol phosphorylation, mtlA and mtlD; two putative phage genes (ydjG and yqaL) and several putative and hypothetical proteins (Table S2).
Discussion
Our findings support a model in which inhibiting division for an extended period of time (~5 MDP in rich medium), triggers entry into a quiescent state characterized by a terminal cell cycle arrest. Although a link between division and DNA replication in bacteria has been proposed [18], our data are the first to demonstrate a direct connection between these two cell cycle processes. Importantly, we observed a significant reduction in DNA replication prior to the PONR, with a complete arrest occurring several generations after the PONR.
Failsafe mechanisms couple cell division and DNA replication in bacteria
Based on these data, we propose there is a failsafe mechanism that inhibits DNA replication in the absence of division (Figure 4). Coupling division to replication ensures that bacterial cells initiate only one round of replication per mass doubling period, regardless of whether or not they are undergoing multifork replication. Our finding that an extended block in DNA replication leads to an arrest in new rounds of division supports the presence of a second failsafe mechanism that prevents division in the absence of DNA replication. In function, if not in form, these mechanisms are akin to the cell cycle checkpoints that ensure the orderly progression through the eukaryotic cell cycle. Such failsafe mechanisms ensure that cells defective in cell division or DNA replication are removed from the growing population.
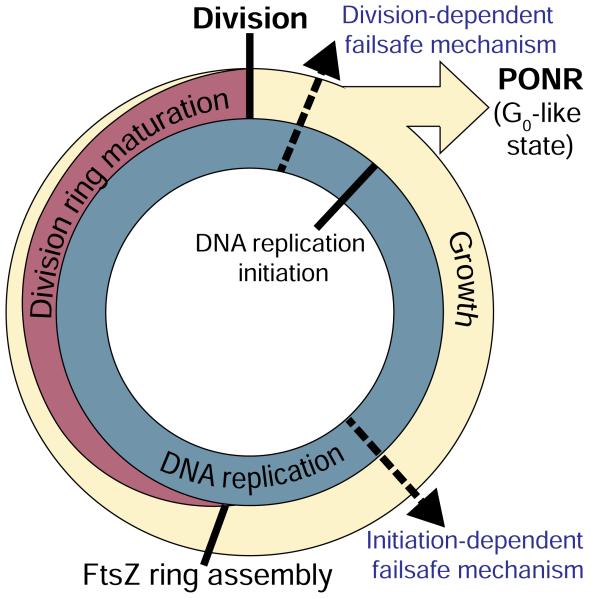
Fig 4. Failsafe mechanisms couple division and DNA replication in the bacterial cell cycle.
DNA replication (blue) and growth (beige) are ongoing and the division ring is present for approximately the second half of each cell cycle. A division-dependent failsafe mechanism ensures one DNA replication initiation occurs per each division. Failure to divide leads to a block in replication initiation followed by entry into a terminal cell cycle arrest at the PONR, equivalent to a eukaryotic G0 state. A second initiation-dependent failsafe mechanism ensures that cells impaired in the initiation of DNA replication do not assemble division rings. The dashed arrows indicate hypothetical localizations of the failsafe mechanisms.
We favor the idea that the terminal cell cycle arrest observed in cells subjected to an extended block in cell division or DNA replication is a consequence of an infinite loop (a “vicious cell cycle”) in which cells that cannot divide cannot initiate DNA replication and cells that cannot initiate replication cannot divide (Figure 4). The earlier onset of the growth arrest in dnaBts cells suggests the block in DNA replication might be the proximal cause of the PONR following the division block.
The cell division machinery is an ideal target for antibiotic development
In addition to providing fundamental and unforeseen insights into our understanding of the bacterial cell cycle, our findings validate the bacterial cell division machinery as an ideal target for the development of new antibiotics. Not only does inhibiting division prevent the production and proliferation of viable daughter cells, the concurrent arrest in DNA replication serves as a barrier to the acquisition of antibiotic resistant mutations. Most importantly, the terminal cell cycle arrest at the PONR ensures that treated cells cannot recover after the drug has been metabolized.
Supplementary Material
Highlights.
Inhibiting cell division or DNA replication results in a terminal growth arrest.
Cells remain alive and metabolically active following the growth arrest.
Impeding division leads to an arrest in DNA replication.
Impeding DNA replication leads to a terminal arrest in cell division.
Acknowledgements
We thank Blaise Boles, John Helmann, David Rudner and Shu Ishikawa (Nara Institute of Science and Technology) for the gifts of S. aureus Newman, RNpFtsZ-1, HB0042, BNS1762, and NIS2020 respectively. We are grateful to the technical assistance of Wandy Beatty (Imaging Facility, Washington University School of Medicine) and Shalon Ledbetter. We thank members of the Levin and Margolin labs, Joe Jez, Daniel Haeusser, Jim Skeath, Kelsey Tinkum, Elizabeth Tuck, Heath Murray and Dhruba Chattoraj for critical analysis of the manuscript and/or helpful discussions. The following supported this work: National Institutes of Health grants GM64671 to PAL, T32 GM007067 to HAA, GM084003 to JDW, and NIAID/R01AI08093 to JTS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental information includes four figures, 2 tables and Supplemental Experimental Procedures.
Contributions:
HAA and AK performed research. JDS and NAS contributed reagents (PC190723). HAA, AK, JDW, and PAL designed research and analyzed data. HAA and PAL wrote the paper.
References
- 1.Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 2.Weart RB, Levin PA. Growth rate-dependent regulation of medial FtsZ ring formation. J Bacteriol. 2003;185:2826–2834. doi: 10.1128/JB.185.9.2826-2834.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levin P, Kurtser I, Grossman A. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc Natl Acad Sci USA. 1999;96:9642–9647. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levin PA, Losick R. Characterization of a cell division gene from Bacillus subtilis that is required for vegetative and sporulation septum formation. J Bacteriol. 1994;176:1451–1459. doi: 10.1128/jb.176.5.1451-1459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katis VL, Wake RG, Harry EJ. Septal Localization of the Membrane-Bound Division Proteins of Bacillus subtilis DivIB and DivIC Is Codependent Only at High Temperatures and Requires FtsZ. J Bacteriol. 2000;182:3607–3611. doi: 10.1128/jb.182.12.3607-3611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haydon DJ, Stokes NR, Ure R, Galbraith G, Bennett JM, Brown DR, Baker PJ, Barynin VV, Rice DW, Sedelnikova SE, et al. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science. 2008;321:1673–1675. doi: 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- 7.Tan C, Therien A, Lu J, Lee SH, Caron A, Gill CJ, Lebeau-Jacob C, Benton-Perdomo L, Monteiro JM, Pereira PM, et al. Restoring methicillin-resistant Staphylococcus aureus susceptibility to β-lactam antibiotics. Sci Transl Med. 2012;4:1–11. doi: 10.1126/scitranslmed.3003592. [DOI] [PubMed] [Google Scholar]
- 8.Lewis K. Persister Cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 9.Breeuwer P, Abee T. Assessment of viability of microorganisms employing fluorescence techniques. International Journal of Food Microbiology. 2000;55:193–200. doi: 10.1016/s0168-1605(00)00163-x. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Cheng H, Wang F, Wei D, Wang X. An improved 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) reduction assay for evaluating the viability of Escherichia coli cells. J Microbiol Methods. 2010;82:330–333. doi: 10.1016/j.mimet.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Hirota Y, Ryter A, Jacob F. Thermosensitive mutants of E. coli affected in the processes of DNA synthesis and cellular division. Cold Spring Harbor Symposia on Quantitative Biology. 1968;33:677–693. doi: 10.1101/sqb.1968.033.01.077. [DOI] [PubMed] [Google Scholar]
- 12.Dai K, Lutkenhaus J. ftsZ is an essential cell division gene in Escherichia coli. J Bacteriol. 1991;173:3500–3506. doi: 10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Callister H, Wake RG. Characterization and mapping of temperature-sensitive division initiation mutations of Bacillus subtilis. J Bacteriol. 1981;145:1042–1051. doi: 10.1128/jb.145.2.1042-1051.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferullo D, Cooper D, Moore H, Lovett S. Cell cycle synchronization of Escherichia coli using the stringent response, with fluorescence labeling assays for DNA content and replication. Methods. 2009;48:8–13. doi: 10.1016/j.ymeth.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaechter M, Ingraham JL, Neidhardt FC. The cell division cycle. Microbe. 2006:169–182. [Google Scholar]
- 17.Hill NS, Kadoya R, Chattoraj DK, Levin PA. Cell size and the initiation of DNA replication in bacteria. PLoS Genet. 2012;8:e1002549. doi: 10.1371/journal.pgen.1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bates D, Kleckner N. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol. 2010;8:163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 20.Ogura Y, Imai Y, Ogasawara N, Moriya S. Autoregulation of the dnaA-dnaNOperon and Effects of DnaA Protein Levels on Replication Initiation in Bacillus subtilis. J Bacteriol. 2001;183:3833–3841. doi: 10.1128/JB.183.13.3833-3841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 1998;12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rokop ME, Auchtung JM, Grossman AD. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Molecular Microbiology. 2004;52:1757–1767. doi: 10.1111/j.1365-2958.2004.04091.x. [DOI] [PubMed] [Google Scholar]
- 23.Rowland SL, Katis VL, Partridge SR, Wake RG. DivIB, FtsZ and cell division in Bacillus subtilis. Molecular Microbiology. 1997;23:295–302. doi: 10.1046/j.1365-2958.1997.2141580.x. [DOI] [PubMed] [Google Scholar]
- 24.Harry EJ, Rodwell J, Wake RG. Co-ordinating DNA replication with cell division in bacteria: a link between the early stages of a round of replication and mid-cell Z ring assembly. Molecular Microbiology. 1999;33:33–40. doi: 10.1046/j.1365-2958.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- 25.Moriya S, Rashid R, Rodrigues C, Harry E. Influence of the nucleoid and the early stages of DNA replication on positioning the division site in Bacillus subtilis. Mol Microbiol. 2010;76:634–647. doi: 10.1111/j.1365-2958.2010.07102.x. [DOI] [PubMed] [Google Scholar]
- 26.Bernard R, Marquis KA, Rudner DZ. Nucleoid occlusion prevents cell division during replication fork arrest in Bacillus subtilis. Molecular Microbiology. 2010;78:866–882. doi: 10.1111/j.1365-2958.2010.07369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu LJ, Errington J. Coordination of Cell Division and Chromosome Segregation by a Nucleoid Occlusion Protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Arends S, Weiss D. Inhibiting cell division in Escherichia coli has little if any effect on gene expression. J Bacteriol. 2004 doi: 10.1128/JB.186.3.880-884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Au N, Kuester-Schoeck E, Mandava V. Genetic composition of the Bacillus subtilis SOS system. J Bacteriol. 2005;187:7655–7666. doi: 10.1128/JB.187.22.7655-7666.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho TD, Ellermeier CD. Extra cytoplasmic function σ factor activation. Curr Opin Microbiol. 2012;15:182–188. doi: 10.1016/j.mib.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.