Abstract
Whole exome and whole genome sequencing are applications of next generation sequencing transforming clinical care, but there is little evidence whether these tests improve patient outcomes or if they are cost effective compared to current standard of care. These gaps in knowledge can be addressed by comparative effectiveness and patient-centered outcomes research. We designed a randomized controlled trial that incorporates these research methods to evaluate whole exome sequencing compared to usual care in patients being evaluated for hereditary colorectal cancer and polyposis syndromes. Approximately 220 patients will be randomized and followed for 12 months after return of genomic findings. Patients will receive findings associated with colorectal cancer in a first return of result visit, and findings not associated with colorectal cancer (incidental findings) during a second return of result visit. The primary outcome is efficacy to detect mutations associated with these syndromes; secondary outcomes include psychosocial impact, cost-effectiveness and comparative costs. The secondary outcomes will be obtained via surveys before and after each return visit. The expected challenges in conducting this randomized controlled trial include the relatively low prevalence of genetic disease, difficult interpretation of some genetic variants, and uncertainty about which incidental findings should be returned to patients. The approaches utilized in this study may help guide other investigators in clinical genomics to identify useful outcome measures and strategies to address comparative effectiveness questions about the clinical implementation of genomic sequencing in clinical care.
Keywords: Comparative effectiveness research, Genomics, Next generation sequencing, Randomized clinical trial, Outcomes research, Whole exome sequencing
1. Introduction
Next generation sequencing is a transformative technology that is changing the practice of genetics in medical care [1]. Whole exome sequencing (WES) and whole genome sequencing are applications of this technology that can now be implemented in a clinical setting, mainly because of rapidly decreasing technology costs and improved quality control. The potential benefits and complications from applying next generation sequencing in clinical practice should be evaluated before indications for its widespread use are implemented [2]. Part of this evaluation includes measuring outcomes such as morbidity, mortality, and mutation detection rates in genes not routinely investigated by current clinical genetic testing.
However, evaluating the impact of next generation sequencing, specifically of WES, extends beyond the assessment of genetic variants related to the patient's original clinical presentation due to the comprehensive nature of these tests, i.e. the systematic assessment of nearly all genes at one time. One of the potential outcomes is the identification of clinically relevant variants in genes with no apparent relationship to the primary indication for obtaining the test. These results have been called incidental findings (IFs), and are particularly challenging when taken as a group because the clinical implications of IFs are heterogeneous [3,4]. The observance of IFs raises many questions such as which IFs should be reported back to patients, the method of reporting, and when to disclose these results [4,5].
Evaluation of the impact of next generation sequencing, and the return of IFs to patients, will require innovative methodologies [2]. The Institute of Medicine has defined comparative effectiveness research (CER) as “the generation and synthesis of evidence that compares the benefits and harms of alternative methods to prevent, diagnose, treat, and monitor a clinical condition or to improve the delivery of care. The purpose of CER is to assist consumers, clinicians, purchasers, and policy makers to make informed decisions that will improve healthcare at both the individual and population levels” [6]. Patient-centered outcomes research (PCOR), a related concept, is intended to help people and their caregivers communicate and make informed healthcare decisions, allowing their voices to be heard in assessing the value of healthcare options [7].
The objective of this paper is to describe our experience with the incorporation of CER and PCOR methods in the evaluation of next generation sequencing in the adult genetics clinic, and to that end we discuss the design of a randomized controlled trial (RCT) comparing WES to usual care for evaluation of hereditary colorectal cancer and polyposis syndromes (CRCP), a common reason for referral to the University of Washington, Genetic Medicine Clinic. The goals of the RCT are twofold: to evaluate the use of WES to improve CRCP syndrome diagnosis, and to evaluate the implications of using WES unrelated to CRCP using comparative effectiveness and patient-centered outcomes research methods. This study, the NEXT (New Exome Technology) Medicine Study, is part of the CSER (Clinical Sequencing Exploratory Research) consortium, a National Human Genome Research Institute funded initiative [8].
1.1. Clinical context
Colorectal cancer often occurs sporadically, yet familial cancer syndromes occur, especially if the cancers arise at an early age [9]. Lynch syndrome, formerly referred to as hereditary non-polyposis colorectal cancer, is caused mainly by mutations in mismatch repair (MMR) genes and is the most common form of genetically determined colon cancer predisposition, accounting for 2–4% of CRC cases [10–12]. The diagnostic approach to inherited colorectal cancer is to screen those at high risk according to clinical criteria, based on a strong family history or an early age of presentation [13,14]. In patients who meet criteria for further evaluation, this is followed by tumor tissue testing for MMR protein expression with immunohistochemistry and/or microsatellite instability, a consequence of MMR protein deficiency in the tumor. If these functional assays suggest deficiency or a hypo-functional MMR protein, then a conclusive germline sequencing test determines if there is a mutation in one of these genes [15,16]. Thus, this testing approach can involve a sequence of two or more intermediate steps with several visits to the medical geneticist or other healthcare providers depending on the protocol, at a potential cost of thousands of dollars and several weeks to months with no established diagnosis. At the end of this process, considerably less than half of the patients with clinical suspicion of Lynch undergoing testing will have a mutation detected [17].
The use of next generation sequencing to address questions related to hereditary colorectal cancer is promising because the multistep screening process for Lynch can be bypassed by sequencing all MMR genes as well as other genes responsible for less common genetic causes of colorectal cancer such as APC (responsible for familial adenomatous polyposis syndrome), MUTYH (responsible for MUTYH-polyposis) or PTEN (responsible for PTEN-hamartoma syndrome) [18]. This group of conditions, that includes but is not limited to Lynch syndrome, has been described as colorectal cancer and polyposis syndromes (CRCP).
We chose patients with CRCP for the study because this is a common referral indication in our clinic population, there are multiple genes associated with a similar clinical picture, which makes this condition amenable to be tested by WES, and the standard of care has limited clinical sensitivity. Furthermore, Lynch syndrome's usual care testing (i.e., immunohistochemistry and microsatellite instability tissue testing followed by genetic testing) is relatively costly and has modest effectiveness, despite the fact that it is supported by evidence-based recommendations [19] and is considered a good economic value compared to no testing [20].
2. Experimental design
We selected an RCT design in order to evaluate the efficacy and effectiveness of WES compared to usual care in patients with CRCP. A randomized trial is an inherently comparative design because it enables estimation of the attributable effects of the testing intervention by including a comparison group. Another major advantage of an RCT design over an observational study design, often used in the evaluation of diagnostic approaches, is the reduction in confounding from both known and unknown factors. For example, an observational study comparing individuals who received WES with those who did not might show worse anxiety outcomes in this group just because anxious individuals are prone to request more comprehensive tests, and not because WES itself or an incidental finding has generated that level of anxiety.
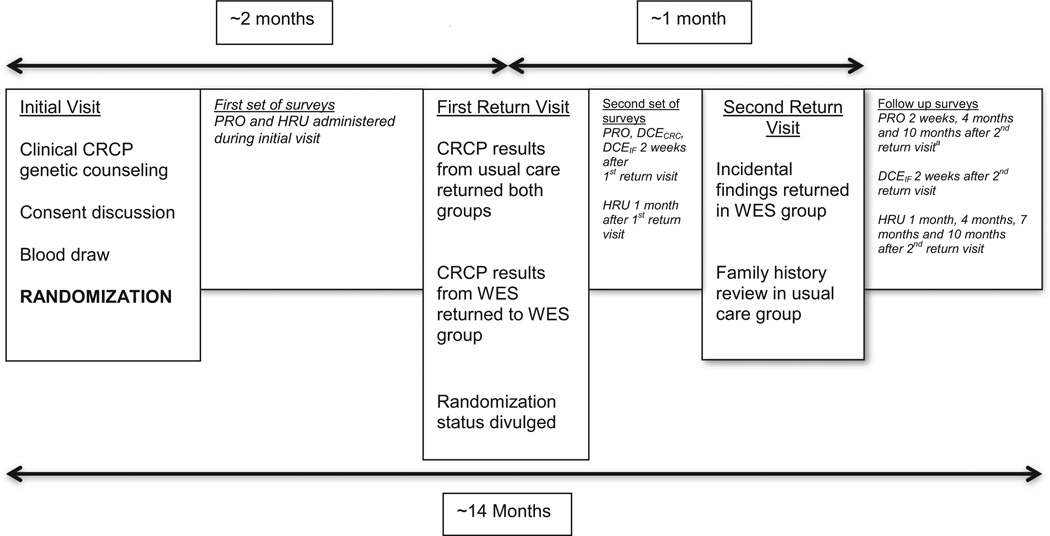
Patients are randomized to a usual care arm or a WES plus usual care arm (Fig. 1). Adding WES to usual care, rather than eliminating any clinical test yields parity to the non-WES experience of patients in both arms and may assure those in the WES arm that their care is not compromised. Patients are evaluated during an initial clinic visit in which genetic testing is ordered and informed consent is obtained followed by randomization. A key aspect of the trial design is that test results are delivered in two separate return visits (Fig. 1). The first return of results visit is scheduled approximately two months after the initial visit, and the results from the usual care genetic testing pertinent to CRCP are returned to both groups, while those in the WES plus usual care arm are also returned CRCP related findings from WES. The second return visit takes place at least one month after the first return visit, and IFs found by WES are returned in the WES plus usual care arm, while the usual care alone group receives the CRCP information about disease risks based on family history, but no IFs. We included two return visits to differentiate the impact of receiving disease diagnosis information from IF information. The second return visit during which IF results are communicated occurs in the context of a research visit with a genetic counselor and a medical geneticist. Known pharmacogenetic variants, which are found in approximately two-thirds of the participants, are returned at this visit. For actionable high penetrance Mendelian conditions that affect adults, we consider a list of approximately 117 genes meeting these criteria, and include most of the adult-relevant 56 genes recommended for return of results by the ACMG [21]. Only pathogenic variants are returned. While the gene list is dynamic, we recently published this list and our criteria for pathogenicity [22]. Of note, neither the patients nor the clinicians are blinded after the first return visit, when the randomization status is divulged. This is done to capture any changes in outcomes from the baseline surveys.
Fig. 1.
Study timeline. Patients are assessed at their initial clinic visit for inclusion/exclusion criteria, and if eligible, asked if they wish to enroll in the study and consented. Blood volumes are drawn simultaneously for usual care and the research protocol, followed by randomization to one of two groups: WES plus usual care or usual care alone. Results related to colorectal cancer/polyposis are returned in the first return visit, two months after the initial visit. During a second return visit that takes place one month after the first, incidental findings from WES are discussed in this group, while further review of family history takes place for the usual care alone participants. Patient reported outcomes (PRO), health resource utilization (HRU) and discrete choice experiment instruments (DCECRC for findings associated to colorectal cancer and polyposis and DCEIF for incidental findings) are given to the patient starting during the initial visit until 10 months after the second return visit. aThe PRO instrument sent 4 months after the second visit contains only the FACToR instrument.
The primary outcome of the trial is the diagnostic effectiveness of WES: the proportion of subjects who receive a result identifying the mutation predicted to be etiological of CRCP in their family. The key secondary outcomes include WES vs. usual care comparative costs, cost-effectiveness and psychosocial impacts. The clinical or analytical sensitivity and specificity of WES compared to Sanger sequencing has been previously established [23].
2.1. Inclusion/exclusion criteria
Participants eligible for randomization have a personal and/or family history of colon cancer and/or polyposis that has resulted in the initiation of clinical genetic testing to identify a causative mutation in the patient. Participants are excluded if they have a history of genetic testing for colon cancer or polyps or if they felt to have a high probability of a single mutated gene, such as hundreds of polyps indicating familial polyposis due to APC gene mutations.
2.2. Sample size/power analysis
Power calculations for the primary outcome assume the following: a usual care rate of identifying a causal variant of 50% vs. 70% in the WES plus usual care group; 2.75 years of enrollment with a rate of 80 subjects/year yielding a total of 220 subjects; a 10% lost to follow-up rate implying 198 evaluated subjects. Based on these assumptions the study has 86% power to detect a 20% increase in the rate. For secondary outcomes the target sample size provides 80% power to detect a mean difference of 0.40 standard deviations.
2.3. Analysis
Analysis of the primary outcomes will use a one-sided test comparing the proportion of subjects with an identified causal variant in the WES arm to the proportion in the usual care arm using standard two-sample tests of proportions. Secondary analyses will compare quantitative patient outcomes that are detailed below using two-sample comparison of means, or alternatively, rank-based methods (Mann– Whitney–Wilcoxon) for measures with strong floor and/or ceiling effects or for highly skewed measures. These tests are two sided, and pre-specified subgroup analyses based on the adenomatous vs. hyperplastic polyp type.
3. Comparative effectiveness and patient-centered outcome measures
3.1. Patient reported outcomes (PROs)
Patient-centered outcomes research emphasizes the patient's perspective and as such, methods to incorporate patient-centered and patient reported outcomes (PROs) are necessary. By definition, a PRO is any report of the status of a patient's health condition that comes directly from the patient, without interpretation of the patient's response by a clinician or anyone else [7]. Patient reported outcomes have become a standard measure of patient-centered outcomes for RCTs [24]. General examples of PROs are disease-specific patient questionnaires such as the Functional Assessment of Cancer—Colorectal Cancer Module, domain-specific measures of pain and other symptoms, and the more generic measures of health-related quality of life and functional status.
We selected PRO instruments with the goal of capturing psychosocial impacts from both CRCP results and IFs. Focus groups and cognitive interviews also conducted by our group show that patient preferences for incidental genomic findings are likely influenced by a complex set of diverse attributes [25]. As anxiety about possible IF results from genomic tests has been noted in patients in our clinic, we hypothesized that participants in the arm receiving IFs will report more anxiety symptoms. However, because WES is a more comprehensive test with presumptively a higher efficacy to detect pathogenic variants than usual care, we also hypothesized those patients receiving WES will be overall more satisfied with the testing process than those who do not. Taking into consideration that participants in this study may have CRCP syndromes, we used previous literature and the taxonomy of Patrick and Erickson [26] to identify potential psychosocial impacts on patients receiving CRCP and IF results [26–30]. These include: 1) symptoms of anxiety or depressive mood, 2) perceptions such as the worry associated with genetic testing results, worry about indecision, satisfaction with the testing process, and 3) feelings of resilience such as having a positive affect toward the testing. Measures used previously to assess the impact of genetic testing on patient outcomes have been relatively lengthy or not specific to genetic testing [29,30].
The selected measures (Table 1) are as brief as possible for minimal respondent burden and are administered via surveys through the University of Washington's Research Electronic Data Capture (REDCap) system [31]. These surveys are provided during the initial visit, two weeks after the first return visit, and then at two weeks, four months and ten months after the second return visit.
Table 1.
Patient reported outcomes (PRO) measures. Patients are asked to complete a series of survey instruments that assess multiple dimensions of their quality of life and functional status at several time points throughout the trial. The surveys are administered during the initial visit, 2 weeks after the first return visit, and at 2 weeks, 4 months and 10 months after the second return visit. Abbreviations: Generalized Anxiety Disorder 7-item instrument (GAD-7); Patient Health 9-item Questionnaire (PHQ-9); Feelings About genomiC Testing Results (FACToR); Veterans RAND 12-item (VR12); Multidimensional Impact of Cancer Risk Assessment (MICRA); Big Five Inventory short 10-item questionnaire (BFI-10).
We are using use the Generalized Anxiety Disorder 7-item instrument (GAD-7) [32] to assess anxiety severity and related impairment [32]. Depressive symptoms are measured using the Patient Health Questionnaire (PHQ-9) [33], a validated measure based on DSM-IV diagnostic criteria [34,35].
To capture worry and positive psychological effects associated specifically with genetic testing, we developed the Feelings About genomiC Testing Results (FACToR), which was adapted from the Multidimensional Impact of Cancer Risk Assessment (MICRA) [36]. We adapted the MICRA by modifying the items to be relevant to genetic test results more broadly. Specifically, we used patient focus groups to identify novel concepts from the target population that were not originally included in the MICRA pool of items and cognitive interviews to assess the relevance and comprehension of items and refine phrasing and item response options.
To assess health-related quality of life we are applying the Veterans RAND 12-item health survey [37], a brief, generic, multi-use tool that summarizes health in two scores: a physical health summary measure and a mental health summary measure. We are also measuring personality using the Big Five Inventory short 10-item questionnaire (BFI-10) to assess the mediating impact of personality and an optimistic psychological orientation (resilience) on negative symptoms and other measures above. The BFI-10 [38] is a shorter version of a 44-item tool developed to assess the multiple dimensions of personality [38].
In summary, we have selected a diverse yet brief set of PRO instruments designed to capture the key psychosocial effects associated with genetic testing in this population.
3.2. Patient preferences (the value of knowing)
From an economic standpoint the value of a genetic test result can go beyond clinical outcomes and may extend to knowledge of cause of disease — this has been defined as ‘personal utility’ [39]. Quantitatively valuing patients' personal utility of testing for information surrounding the risk of CRCP and the return of IFs can be accomplished using a preference-based approach, particularly if the research aims to inform the cost–utility of different testing strategies. An increasingly popular method to utility assessment is the discrete choice experiment (DCE) approach [40–42].
To robustly quantify the personal utility associated with the different types of information received in the WES or usual care arms, two separate DCE questionnaires are employed to elicit patients' preferences and willingness to pay to receive information on CRCP or IFs. In each DCE, patients are asked to complete a series of 16 choice tasks, where each task queries the individual to choose between two testing alternatives that represent the possible results and costs of the testing approaches. The development of the IF DCE survey (IMPRINT — Instrument to Measure PReferences for Information from Next-generation Testing) instrument is fully described in the publication by Bennette et al. [25]. The DCE is administered at baseline and two weeks following the return of relevant results: CRCP-related results after the first visit, and IFs after the second visit. Because returning IFs within a clinical context are particularly novel, we are separating the return of CRCP-related results from those of IFs to isolate the effects of returning IFs in the WES group compared to usual care.
4. Economic outcomes
We describe below the strategies deployed to assess economic outcomes in this study: trial-based cost analysis, cost–utility analysis, and cost–benefit analysis.
4.1. Trial-based cost analysis
Healthcare-related resource utilization and patient behavior data are collected using a patient survey (Table 2, see survey in Appendix A) [43]. These surveys are administered during the initial visit, one month after the first return visit, and then at one, four, seven, and ten months after the second return visit. Patients are asked about the use of medical services such as physician visits, hospitalizations, prescription and non-prescription drug use, screening, ancillary care, and mental health services. Patients asked how many family members they have informed of their test results, and what actions their family members have taken to their knowledge — e.g., received genetic testing or CRCP screening. Patients are also asked actual or intended changes to their health and life insurance policies.
Table 2.
Measures of health, insurance behaviors and health resource utilization. Patients are asked to answer a brief series of questions that assess any changes in behavior, healthcare utilization, or insurance coverage made in response to receiving a genetic test result. The surveys are administered during the initial visit, 1 month after the first return visit, and at 1 month, 4 months, 7 months and 10 months after the second return visit. Abbreviations: Rapid Assessment of Physical Activity (RAPA); CDC-Behavioral Risk Factor Surveillance System, year 2011 (BRFSS 2011).
| Measure | Number of items | |
|---|---|---|
| Health behaviors | ||
| Physical activity | RAPA [57] | 1 |
| Diet, smoking, alcohol intake | BRFSS 2011 | 7 |
| Insurance behaviors | ||
| Health/life insurance, disability | Novel items | 2 |
| Healthcare resource utilization | ||
| Medical services, procedures | Novel items | 5 |
Costs will be assigned to these resources using established sources such as Medicare reimbursement and market prices when available [44]. The cost for usual care CRCP testing will be derived from reimbursed insurance amounts and patient out of pocket expenses. The cost for exome sequencing will be estimated from several sources. The average cost incurred in the research study will be evaluated, as well as the cost of clinical WES, which is currently performed at a higher price than research WES. We will use the market price of exome sequencing as a proxy for the opportunity cost.
We will follow guidelines for the conduct of economic evaluations alongside RCT [44]. Averages, standard errors, and 95% confidence intervals for total costs will be computed for each treatment arm with the use of methods to account for dependent patient censoring [45]. Generalized linear models will be used to estimate group differences in use and costs by medical care utilization categories and overall. The models will include patient clinical and demographic variables, group indicator, and other control variables.
4.2. Cost–utility and cost–benefit analyses
In cost-effectiveness analysis, the added costs and health outcomes associated with an intervention or program are used to calculate the incremental cost-effectiveness ratio relative to the next best comparator. A common measured outcome in health economics is the amount of life years gained, when adjusted for the quality of life, also referred as QALYs. The particular type of cost-effectiveness analysis that uses QALYs as a unit of measure is also called a cost–utility analysis. In cost–benefit analysis, the incremental consequences are expressed in dollar terms, so the overall analysis of a program's costs and effects can be conducted entirely in dollars. Cost–utility analysis and cost–benefit analysis are not mutually exclusive, and to a certain point they represent a continuum in the analysis process [46].
We will develop a disease-based decision model to evaluate the incremental cost–utility (cost per quality-adjusted life-year (QALY) gained) of identifying patients with a genetic cause of CRCP and another model to evaluate IFs. After these two cost– utility analyses are done, we will use the results from the Patient preferences (the value of knowing) section and plug the willingness to pay values to do cost–benefit analyses for CRCP and IFs independently. We will use approaches similar to our previous work in this area [47–49], and build upon the work of Mvundura and colleagues [20], who have shown that the primary clinical benefit of testing patients with CRC is mediated through an increase in CRC screening in family members and thus avoidance of CRC and other related cancers [20,50,51]. The analyses will be conducted from the societal perspective; a payer perspective will be assessed in a scenario analysis. Estimates of the behavior of family members will be derived from the trial, as well as healthcare costs of the patients and the increase in CRC findings with exome analysis.
Parameter estimates for long-term outcomes will be derived from systematic searches of the literature, as will data not directly derived from the trial. The value of obtaining information about the potential genomic causes of their CRCP syndrome to a patient will be incorporated as a willingness to pay derived from the DCE study described above. We will analyze the model using probabilistic sensitivity analysis to generate cost-effectiveness acceptability curves [52]. We hypothesize that the incremental cost per QALY gained will be less than $100,000 in 95% of simulations.
5. Discussion
To illustrate the incorporation of comparative effectiveness and patient-centered outcomes research concepts to the evaluation of next generation sequencing technologies in clinical care, we describe the design of a randomized controlled trial intended to measure the impact of WES on diagnosis of CRCP as well as the impact of incidental findings arising from the use of WES. This study is the first to include CER design elements and PCOR measures to assess the impact of next generation sequencing for disease diagnosis. We characterized different components of PCOR, specifically the integration of PRO in a prospective trial to evaluate the impact of a genomic diagnostic tool, and to that end we have used previously validated tools and created a new one to address the psychological impact of genetic testing. We will utilize two forms of cost-effectiveness analysis in this study (cost–utility and cost–benefit analyses), and have developed novel instruments to assess the direct and indirect costs of the interventions, as well as the benefits in terms of quality of life, costs avoided, and to quantify personal utility. The instruments have been created to evaluate the impact of genetic test results related to hereditary colorectal cancer and polyposis, and incidental findings separately.
Our study may shed light on the benefits and adverse effects of the use of genomic sequencing at a personal level, complementing traditional outcomes like the impact on morbidity, mortality, or drug treatment outcomes in the case of pharmacogenetic variants. Although this is not the first randomized trial evaluating genetic testing interventions, the emphasis on patient-centered outcomes and comparative effectiveness sets this study apart from others [29,53]. It is also distinguished from previous studies by evaluating the return of IFs, which represent the majority of information that will be obtained by WES.
This study has implications beyond the quantification of diagnostic effectiveness in genomics. Understanding the impact of the return of IFs is a priority in the field of genomic medicine [54], and will help determine the role that genomic technologies such as WES will play in clinical care. The evidence obtained from this study and other parallel efforts underway within the CSER (Clinical Sequencing Exploratory Research) consortium [8] will inform the engagement of stakeholders like public/private payers, the medical community and our patients, who are the ultimate recipients of this information. Obtaining this evidence will inform reimbursement procedures and, if found to be beneficial, will place medical geneticists and genetic counselors in more extensive roles for delivering healthcare.
There are challenges applying comparative effectiveness research to the study of next generation sequencing. First, although recruitment is an almost universal problem of RCT designs, it can be particularly problematic for genetic conditions, which have a low prevalence. Although CRCP is a common inherited condition, it is only 3–5% of colorectal cancer cases. Our patient base can also be affected by referral volume, which is limited by the referring physician's knowledge of the indication for referral and the clinical care setting where they practice.
Second, the return of IFs is a vibrant topic in ethical discussions [55], and sometimes it is not an easy task to discriminate what type of variant in which gene should be returned. These situations can be alleviated to a certain degree by standardized approaches to the return of IFs, like the actionable gene panel proposed by the American College of Medical Genetics [21].
Third, the challenges with the incorporation of PROs and patient preferences when assessing genomic sequencing studies are also tangible for the subjects. For example, we found that distinguishing between findings associated with CRCP and IFs not pertinent to the chief complaint was a difficult task for the patients, leading us to design two sets of DCE, one for CRCP findings and another for IFs [25]. Furthermore, the complexity of the DCE choice task and respondent burden might be a concern for user fatigue for our participants, especially if these tests are administered with other surveys at the same time. Lastly, assessing the clinical and economic impact of returning IFs to patients is challenging for several reasons. First, if only the clinically actionable IFs (the ones that will change medical care) are returned, these are relatively rare [22] and the clinical impact can vary drastically among mutations. Second, the clinical effect of these variants might not be evident until years after the presentation to the clinic. Finally, the evidence needed to interpret many of the identified variants in CRCP and IF genes remains challenging without even considering variants of uncertain significance that are not returned for IFs for NEXT Medicine study participants.
In summary, we designed a randomized controlled trial to evaluate whole exome sequencing in the evaluation of patients with inherited colorectal cancer and polyposis syndromes. We incorporated patient reported outcomes by modifying a validated questionnaire to address the psychological effects of genetic testing, and evaluated patient preferences by creating two new discrete choice experiment instruments. This paper may help guide other investigators in clinical genomics to identify useful outcome measures and strategies to address comparative effectiveness questions on the extensive information resulting from the identification of genomic incidental findings. Although the application of comparative effectiveness and patient-centered outcomes research to the clinical implementation of next generation sequencing is challenging, we believe that these approaches offer valuable evidence to inform patient, provider, and payer decisions.
Supplementary Material
Abbreviations
- CER
comparative effectiveness research
- PCOR
patient-centered outcomes research
- WES
whole exome sequencing
- IFs
incidental findings
- CRCP
colorectal cancer and polyposis syndromes
- MMR
mismatch repair
- RCT
randomized controlled trial
- PRO
patient reported outcomes
- DCE
discrete choice experiment
- QALY
quality-adjusted life-year
Footnotes
Funding: Grants U01 HG0006507 and U01HG007307 from the National Human Genome Research Institute (GPJ) and grant number K12 HS021686 Patient-Centered Outcomes Research Career Development Program from the Agency for Healthcare Research and Quality/University of Washington (CJG).
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.cct.2014.06.016.
Contributor Information
Carlos J. Gallego, Email: carlosjg@uw.edu.
Caroline S. Bennette, Email: cb11@u.washington.edu.
Patrick Heagerty, Email: heagerty@uw.edu.
Bryan Comstock, Email: bac4@uw.edu.
Martha Horike-Pyne, Email: mjpyne@u.washington.edu.
Fuki Hisama, Email: fmh2@uw.edu.
Laura M. Amendola, Email: lauraa7@uw.edu.
Robin L. Bennett, Email: robinb@uw.edu.
Michael O. Dorschner, Email: mod@uw.edu.
Peter Tarczy-Hornoch, Email: pth@u.washington.edu.
William M. Grady, Email: wgrady@fhcrc.org.
S. Malia Fullerton, Email: smfllrtn@u.washington.edu.
Susan B. Trinidad, Email: sbtrini@uw.edu.
Dean A. Regier, Email: dregier@bccrc.ca.
Deborah A. Nickerson, Email: debnick@u.washington.edu.
Wylie Burke, Email: wburke@u.washington.edu.
Donald L. Patrick, Email: donald@u.washington.edu.
Gail P. Jarvik, Email: pair@uw.edu.
David L. Veenstra, Email: veenstra@uw.edu.
References
- 1.Gonzaga-Jauregui C, Lupski JR, Gibbs RA. Human genome sequencing in health and disease. Annu Rev Med. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manolio TA, Chisholm RL, Ozenberger B, Roden DM, Williams MS, Wilson R, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med. 2013;15:258–267. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson L, Goldsmith L, O'Connor A, Skirton H. Incidental findings in genetic research and clinical diagnostic tests: a systematic review. Am J Med Genet A. 2012;158A:3159–3167. doi: 10.1002/ajmg.a.35615. [DOI] [PubMed] [Google Scholar]
- 4.Jarvik GP, Amendola LM, Berg JS, Brothers K, Clayton EW, Chung W, et al. Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am J Hum Genet. 2014;94:818–826. doi: 10.1016/j.ajhg.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Machini K, Douglas J, Braxton A, Tsipis J, Kramer K. Genetic counselors' views and experiences with the clinical integration of genome sequencing. J Genet Couns. 2014 doi: 10.1007/s10897-014-9709-4. [DOI] [PubMed] [Google Scholar]
- 6.Institute of Medicine. Initial national priorities for comparative effectiveness research. The National Academies Press. 2009 [Google Scholar]
- 7.Patient-Centered Outcomes Research Institute. Patient-centered outcomes research working definition. 2012 [Google Scholar]
- 8.National Human Genome Research Institute. Clinical Sequencing Exploratory Research (CSER) [Google Scholar]
- 9.Woolf CM. A genetic study of carcinoma of the large intestine. Am J Hum Genet. 1958;10:42–47. [PMC free article] [PubMed] [Google Scholar]
- 10.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 11.Hampel H, Frankel W, Panescu J, Lockman J, Sotamaa K, Fix D, et al. Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res. 2006;66:7810–7817. doi: 10.1158/0008-5472.CAN-06-1114. [DOI] [PubMed] [Google Scholar]
- 12.Wijnen JT, Vasen HF, Khan PM, Zwinderman AH, van der Klift H, Mulder A, et al. Clinical findings with implications for genetic testing in families with clustering of colorectal cancer. N Engl J Med. 1998;339:511–518. doi: 10.1056/NEJM199808203390804. [DOI] [PubMed] [Google Scholar]
- 13.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasen HF, Moslein G, Alonso A, Bernstein I, Bertario L, Blanco I, et al. Guidelines for the clinical management of Lynch syndrome (hereditary non-polyposis cancer) J Med Genet. 2007;44:353–362. doi: 10.1136/jmg.2007.048991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindor NM, Petersen GM, Hadley DW, Kinney AY, Miesfeldt S, Lu KH, et al. Recommendations for the care of individuals with an inherited predisposition to Lynch syndrome: a systematic review. JAMA. 2006;296:1507–1517. doi: 10.1001/jama.296.12.1507. [DOI] [PubMed] [Google Scholar]
- 16.Lynch HT, Boland CR, Gong G, Shaw TG, Lynch PM, Fodde R, et al. Phenotypic and genotypic heterogeneity in the Lynch syndrome: diagnostic, surveillance and management implications. Eur J Hum Genet. 2006;14:390–402. doi: 10.1038/sj.ejhg.5201584. [DOI] [PubMed] [Google Scholar]
- 17.Kovacs ME, Papp J, Szentirmay Z, Otto S, Olah E. Deletions removing the last exon of TACSTD1 constitute a distinct class of mutations predisposing to Lynch syndrome. Hum Mutat. 2009;30:197–203. doi: 10.1002/humu.20942. [DOI] [PubMed] [Google Scholar]
- 18.Johansen Taber KA, Dickinson BD, Wilson M. The promise and challenges of next-generation genome sequencing for clinical care. JAMA. 2013 doi: 10.1001/jamainternmed.2013.12048. [DOI] [PubMed] [Google Scholar]
- 19.Evaluation of Genomic Applications in Practice Prevention Working Group. Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med. 2009;11:35–41. doi: 10.1097/GIM.0b013e31818fa2ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mvundura M, Grosse SD, Hampel H, Palomaki GE. The cost-effectiveness of genetic testing strategies for Lynch syndrome among newly diagnosed patients with colorectal cancer. Genet Med. 2010;12:93–104. doi: 10.1097/GIM.0b013e3181cd666c. [DOI] [PubMed] [Google Scholar]
- 21.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorschner MO, Amendola LM, Turner EH, Robertson PD, Shirts BH, Gallego CJ, et al. Actionable, pathogenic incidental findings in 1,000 participants' exomes. Am J Hum Genet. 2013;93:631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritchard CC, Smith C, Salipante SJ, Lee MK, Thornton AM, Nord AS, et al. ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J Mol Diagn. 2012;14:357–366. doi: 10.1016/j.jmoldx.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Speight J, Barendse SM. FDA guidance on patient reported outcomes. BMJ. 2010;340:c2921. doi: 10.1136/bmj.c2921. [DOI] [PubMed] [Google Scholar]
- 25.Bennette CS, Trinidad SB, Fullerton SM, Patrick D, Amendola L, Burke W, et al. Return of incidental findings in genomic medicine: measuring what patients value-development of an instrument to measure preferences for information from next-generation testing (IMPRINT) Genet Med. 2013;15:873–881. doi: 10.1038/gim.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patrick D, Erickson P. Health status and health policy. New York, NY: Oxford University Press. 1993 [Google Scholar]
- 27.Claes E, Denayer L, Evers-Kiebooms G, Boogaerts A, Philippe K, Tejpar S, et al. Predictive testing for hereditary nonpolyposis colorectal cancer: subjective perception regarding colorectal and endometrial cancer, distress, and health-related behavior at one year post-test. Genet Test. 2005;9:54–65. doi: 10.1089/gte.2005.9.54. [DOI] [PubMed] [Google Scholar]
- 28.Collins VR, Meiser B, Ukoumunne OC, Gaff C, St John DJ, Halliday JL. The impact of predictive genetic testing for hereditary nonpolyposis colorectal cancer: three years after testing. Genet Med. 2007;9:290–297. doi: 10.1097/gim.0b013e31804b45db. [DOI] [PubMed] [Google Scholar]
- 29.Green RC, Roberts JS, Cupples LA, Relkin NR, Whitehouse PJ, Brown T, et al. Disclosure of APOE genotype for risk of Alzheimer's disease. N Engl J Med. 2009;361:245–254. doi: 10.1056/NEJMoa0809578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloss CS, Schork NJ, Topol EJ. Effect of direct-to-consumer genomewide profiling to assess disease risk. N Engl J Med. 2011;364:524–534. doi: 10.1056/NEJMoa1011893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap) — a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spitzer RL, Kroenke K, Williams JB, Lowe B. A brief measure for assessing generalized anxiety disorder: the GAD-7. Arch Intern Med. 2006;166:1092–1097. doi: 10.1001/archinte.166.10.1092. [DOI] [PubMed] [Google Scholar]
- 33.Kroenke K, Spitzer RL, Williams JB. The PHQ-9: validity of a brief depression severity measure. J Gen Intern Med. 2001;16:606–613. doi: 10.1046/j.1525-1497.2001.016009606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroenke K, Spitzer RL, Williams JB, Lowe B. The Patient Health Questionnaire Somatic, Anxiety, and Depressive Symptom Scales: a systematic review. Gen Hosp Psychiatry. 2010;32:345–359. doi: 10.1016/j.genhosppsych.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Martin A, Rief W, Klaiberg A, Braehler E. Validity of the Brief Patient Health Questionnaire Mood Scale (PHQ-9) in the general population. Gen Hosp Psychiatry. 2006;28:71–77. doi: 10.1016/j.genhosppsych.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Cella D, Hughes C, Peterman A, Chang CH, Peshkin BN, Schwartz MD, et al. A brief assessment of concerns associated with genetic testing for cancer: the Multidimensional Impact of Cancer Risk Assessment (MICRA) questionnaire. Health Psychol. 2002;21:564–572. [PubMed] [Google Scholar]
- 37.Selim AJ, Rogers W, Fleishman JA, Qian SX, Fincke BG, Rothendler JA, Updated US, et al. population standard for the Veterans RAND 12-item Health Survey (VR-12) Qual Life Res Int J Qual Life Asp Treat Care Rehab. 2009;18:43–52. doi: 10.1007/s11136-008-9418-2. [DOI] [PubMed] [Google Scholar]
- 38.John O, Donahue E, Kentle R. The Big Five Inventory — versions 4a and 54. Berkeley, CA: University of California, Berkeley, Institute of Personality and Social Research. 1991 [Google Scholar]
- 39.Von Neumann J, Morgenstern O. Theory of games and economic behavior. New York: John Wiley & Sons. 1964 [Google Scholar]
- 40.Herbild L, Bech M, Gyrd-Hansen D. Estimating the Danish populations' preferences for pharmacogenetic testing using a discrete choice experiment. The case of treating depression. Value Health. 2009;12:560–567. doi: 10.1111/j.1524-4733.2008.00465.x. [DOI] [PubMed] [Google Scholar]
- 41.Grosse SD, Wordsworth S, Payne K. Economic methods for valuing the outcomes of genetic testing: beyond cost-effectiveness analysis. Genet Med. 2008;10:648–654. doi: 10.1097/gim.0b013e3181837217. [DOI] [PubMed] [Google Scholar]
- 42.Payne K, Fargher EA, Roberts SA, Tricker K, Elliott RA, Ratcliffe J, et al. Valuing pharmacogenetic testing services: a comparison of patients' and health care professionals' preferences. Value Health. 2011;14:121–134. doi: 10.1016/j.jval.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 43.Patrick DL, Grembowski D, Durham M, Beresford SA, Diehr P, Ehreth J, et al. Cost and outcomes of Medicare reimbursement for HMO preventive services. Health Care Financ Rev. 1999;20:25–43. [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsey S, Willke R, Briggs A, Brown R, Buxton M, Chawla A, et al. Good research practices for cost-effectiveness analysis alongside clinical trials: the ISPOR RCT-CEA Task Force report. Value Health. 2005;8:521–533. doi: 10.1111/j.1524-4733.2005.00045.x. [DOI] [PubMed] [Google Scholar]
- 45.Doshi JA, Glick HA, Polsky D. Analyses of cost data in economic evaluations conducted alongside randomized controlled trials. Value Health. 2006;9:334–340. doi: 10.1111/j.1524-4733.2006.00122.x. [DOI] [PubMed] [Google Scholar]
- 46.Gold M, Siegel J, Russell L, Weinstein MC. Cost-effectiveness in health and medicine. 1996 [Google Scholar]
- 47.Carlson JJ, Garrison LP, Ramsey SD, Veenstra DL. The potential clinical and economic outcomes of pharmacogenomic approaches to EGFR-tyrosine kinase inhibitor therapy in non-small-cell lung cancer. Value Health. 2009;12:20–27. doi: 10.1111/j.1524-4733.2008.00415.x. [DOI] [PubMed] [Google Scholar]
- 48.Oestreicher N, Ramsey SD, Linden HM, McCune JS, van't Veer LJ, Burke W, et al. Gene expression profiling and breast cancer care: what are the potential benefits and policy implications? Genet Med. 2005;7:380–389. doi: 10.1097/01.gim.0000170776.31248.75. [DOI] [PubMed] [Google Scholar]
- 49.Regier DA, Friedman JM, Marra CA. Value for money? Array genomic hybridization for diagnostic testing for genetic causes of intellectual disability. Am J Hum Genet. 2010;86:765–772. doi: 10.1016/j.ajhg.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramsey SD, Burke W, Clarke L. An economic viewpoint on alternative strategies for identifying persons with hereditary nonpolyposis colorectal cancer. Genet Med. 2003;5:353–363. doi: 10.1097/01.GIM.0000086626.03082.B5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramsey SD, Clarke L, Etzioni R, Higashi M, Berry K, Urban N. Cost-effectiveness of microsatellite instability screening as a method for detecting hereditary nonpolyposis colorectal cancer. Ann Intern Med. 2001;135:577–588. doi: 10.7326/0003-4819-135-8_part_1-200110160-00008. [DOI] [PubMed] [Google Scholar]
- 52.Briggs A. Probabilistic analysis of cost-effectiveness models: statistical representation of parameter uncertainty. Value Health. 2005;8:1–2. doi: 10.1111/j.1524-4733.2005.08101.x. [DOI] [PubMed] [Google Scholar]
- 53.Knowles JW, Assimes TL, Kiernan M, Pavlovic A, Goldstein BA, Yank V, et al. Randomized trial of personal genomics for preventive cardiology: design and challenges. Circ Cardiovasc Genet. 2012;5:368–376. doi: 10.1161/CIRCGENETICS.112.962746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berg JS, Amendola LM, Eng C, Allen EV, Gray SW, Wagle N, et al. Processes and preliminary outputs for identification of actionable genes as incidental findings in genomic sequence data in the Clinical Sequencing Exploratory Research Consortium. Genet Med. 2013;15:860–867. doi: 10.1038/gim.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burke W, Matheny Antommaria AH, Bennett R, Botkin J, Clayton EW, Henderson GE, et al. Recommendations for returning genomic incidental findings? We need to talk! Genet Med. 2013;15:854–859. doi: 10.1038/gim.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rammstedt B, John O. Measuring personality in one minute or less: a 10-item short version of the Big Five Inventory in English and German. J Res Pers. 2007;41:203–212. [Google Scholar]
- 57.Topolski TD, LoGerfo J, Patrick DL, Williams B, Walwick J, Patrick MB. The rapid assessment of physical activity (RAPA) among older adults. Prev Chronic Dis. 2006;3:A118. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.