Abstract
Background
The recent X-ray crystal structure of the murine μ opioid receptor (MUR) allowed us to reengineer a previously designed water-soluble variant of the transmembrane portion of the human MUR (wsMUR-TM).
Methods
The new variant of water soluble MUR (wsMUR-TM_v2) was engineered based upon the murine MUR crystal structure. This novel variant was expressed in E. coli and purified. The properties of the receptor were characterized and compared with those of wsMUR-TM.
Results
Seven residues originally included for mutation in the design of the wsMUR-TM, were reverted to their native identities. wsMUR-TM_v2 contains 16% mutations of the total sequence. It was overexpressed and purified with high yield. Although dimers and higher oligomers were observed to form over time, the wsMUR-TM_v2 stayed predominantly monomeric at concentrations as high as 7.5 mg/ml in buffer within a 2-month period. Its secondary structure was predominantly helical and comparable with those of both the original wsMUR-TM variant and the native MUR. The binding affinity of wsMUR-TM_v2 for naltrexone (Kd ~ 70 nM) was in close agreement with that for wsMUR-TM. The helical content of wsMUR-TM_v2 decreased cooperatively with increasing temperature, and the introduction of sucrose was able to stabilize the protein.
Conclusions
A novel functional wsMUR-TM_v2 with only 16% mutations was successfully engineered, expressed in E. coli and purified based on information from the crystal structure of murine MUR. This not only provides a novel alternative tool for MUR studies in solution conditions, but also offers valuable information for protein engineering and structure function relationships.
Introduction
The G-protein-coupled receptor (GPCR) superfamily contains many major pharmacological targets.1 The study of the molecular mechanisms of GPCRs is hampered by limited information on structure-function relationships, the difficulties of working with membrane proteins, various conformational states of the receptors, and the complexity in the mechanisms of selectivity and activation. Solving the crystal structures of GPCRs is a major scientific and technologic undertaking and often requires substantial molecular engineering.2 Revealing the crystal structures of the four major opioid receptors has been a major milestone in opioid related research and drug development.3–6
From the opioid family, the μ opioid receptor (MUR) is clinically important since it is target of opioid analgesics. MUR is a class-A GPCR widely expressed in the central nervous system, and its signaling pathways are relevant in mediating analgesia, sedation, and euphoria. Despite the recent advances, the detailed mechanisms of MUR activation and related protein dynamics are still missing due to the many technical limitations including the difficulty in obtaining large quantities of the functional receptor and the inherent low stability when these proteins are isolated from the membrane environment.
To overcome the difficulty of studying the MUR in a lipid environment, we successfully applied computational protein design to create the first version of a water-soluble variant of the human MUR (wsMUR-TM). The protein was expressed in E. coli and retained structural and functional features consistent with those of the native MUR.7 wsMUR-TM was engineered using a comparative (homology) model based on the crystal structures of human β2 adrenergic receptor and bovine rhodopsin.7 Although the homology model of the human MUR superimposed well with the crystal structure of murine MUR overall (root-mean-square deviation of the Cα atoms = 2.60 Å),8 there are several structural differences, which would impact the selection of sites for designing to confer water solubility. Such differences are expected since there is only 27% sequence similarity between 288-residue transmembrane portions of the human β2 adrenergic receptor and the human MUR7. The murine MUR crystal structure provides critical information to locate the mutated positions in wsMUR-TM since the sequence similarity between the human and the mouse MURs is 94% for the entire sequence and 99% for the segment solved in the crystal structure. Such critical information is used in the present study to reengineer the wsMUR-TM by eliminating mutations that may be unnecessary to confer water solubility and avoid toxicity to the bacterial expression system. This restoration of native amino acid identities allow us to explore possible deleterious expression, solubility, structure and function, and probe the sensitivity of structural and functional features of the designed variants to the choice of template structure. The resulting watersoluble variant (wsMUR-TM_v2) was characterized; structural and functional properties were compared to that of wsMUR-TM. Important findings regarding the stability of these water-soluble analogs were analyzed in detail as well as their relevance as suitable surrogates to study properties of the native MUR.
Materials and Methods
The SNAP-Lumi4-Tb-μ-opioid receptor (Cisbio Bioassays, Bedford, MA), which is composed of a SNAP-tag® enzyme labeled with terbium cryptate (Lumi4®-Tb, a long lifetime fluorescence resonance energy transfer donor) fused at the N-terminus of a native MUR, was expressed on HEK293 cells. Red-naltrexone, an analog of naltrexone, the MUR antagonist that contains the d2 dye, was also obtained from Cisbio Bioassays for receptor affinity determination. All other chemicals with high purity were obtained from Sigma-Aldrich (St. Louis, MO) except when specified otherwise.
Structural geometrical analysis and wsMUR re-engineering
The structure of the murine MUR (PDB accession code: 4DKL)6 was obtained from the protein data bank (PDB)a). The sequence of the previously designed water-soluble variant was mapped onto the crystal structure of the murine MUR and structural analysis was performed using Visual Molecular Dynamics program(Beckman Institute, Urbana, IL)9 and PyMOL (Version 1.3, Schrödinger, LLCb)). Residue positions selected to be mutated to the native human residue identities were identified based on their relative location in the context of the murine mu opioid crystal structure. Herein the Ballesteros and Weinstein indexing is adopted as a superscript in identifying particular residue positions in the receptors.10
Protein expression and purification
The synthetic complementary deoxyribonucleotides encoding of the wsMUR-TM (previous version reported7), wsMUR-TM_v2 (the modified new version) were constructed by DNA2.0 Inc. (Menlo Park, CA) and GenScript Inc. (Piscataway, NJ), respectively. The sequences were subcloned between the NdeI and XhoI restriction sites of the expression plasmid pET-28b(+) (EMD/Novagen, Billerica, MA). This cloning strategy resulted in placement of a His-tag at the amino terminus of the protein and was not removed in subsequent characterization. Protein expression using E. coli BL21(DE3) cells (EMD/Novagen) and purification were performed as described previously.7
Mass spectrometry
The purified proteins were run in a sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) gel. The protein band of the expected molecular weight was excised and digested with trypsin for verification purposes. NanoLC proteomics experiments were run at 200 nL/min for 60 min with gradient elution by injecting digested peptides into a nano-LC mass spectrometry (10 cm C18 capillary column) to be separated by Eksigent. Separated peptides were sprayed into LTQ by Nanospray (Thermo Fisher Scientific, MA). Xcalibur (Xcalibur, Inc., Arlington, VA) was used for raw data acquisition. Sequest (Thermo Fisher Scientificc)) was used to search the database Uniprotd). Scaffold 2.6 (Proteome Software, Inc., Portland, OR) was used to analyze the data quantitatively.
High-resolution atomic force microscopy
Tapping mode atomic force microscopy in air (MFP-3D, Asylum Research, CA) was used to visualize the protein particles under various solution conditions as we described previously.11 Similar technology has been used to observe Alzheimer β-amyloid oligomers and monomers in solution conditions.12 To prepare a sample, protein solution was applied deposited onto the surface of an oxidized silicon wafer and allowed to incubate in a humid atmosphere for 1 h. The sample was then washed in two separate baths of deionized water for 3 min each in order to remove weakly bound protein molecules. From this series of experiments, we tested whether the protein remained predominantly in a monomeric state in the buffer condition.
Circular dichroism and thermal stability
Circular dichroism was used to determine the secondary structure content and the thermostability of the protein as described previously.13 Chirascan circular dichroism Spectrometer (AppliedPhotophysics Limited, Leatherhead, United Kingdom) was used with 1 mm path length and a scan speed of 1 nm/s. The experiments were performed at least three times to ensure repeatability of the experiments. Data sets were recorded three times and averaged to increase the signal-to-noise ratio. The CDNN circular dichroism spectra deconvolution software14 was utilized to determine the content of the secondary structure. Circular dichroism spectra for wsMUR-TM_v2 were also determined from 10 °C to 90 °C with increments of 2 °C per min (6 µM of the receptor in 5 mM sodium phosphate, pH=7.0) in the absence and presence of 1 M sucrose. The effect of sucrose on the helical content of the wsMUR-TM_v2 was also tested in the presence and absence of 3 M urea. The cuvette was sealed with oil to avoid errors from evaporation. The helical content is plotted against temperature using GraphPad Prism (v 5.02, GraphPad Software, Inc., La Jolla, CA).
Interaction with naltrexone: binding assay and docking calculations
To determine the binding affinities of the modified new protein constructs, we utilized the same homogeneous time-resolved fluorescence methodology described in our previous report7. Briefly, Tag-lite ® MUR cells (Cisbio Bioassays, Bedford, MA) in culture medium were dispensed in white low-volume microplates (384-well, Greiner Bio-One North America, Monroe, NC) at 3700 cells/10 µL/well Tag-lite μ opioid labeled cells with 60 nM of Tag-lite opioid receptors red-naltrexone, 5 µL of the engineered MUR with 1:2 serial dilutions from the µM to the nM range was added into the sample with the final volume of 20 µl. After incubation at room temperature for 2 h, homogeneous time-resolved fluorescence signals were determined by FLUOstar plate reader (BMG, Cary, NC) with excitation at 337 nm at 620 and 665 nm emission. The experiments were repeated at least three times. Affinities for wsMURs were determined by fitting to concentration–responses curves using the GraphPad Prism program (v 5.02, GraphPad Software, Inc., La Jolla CA).
To interpret the experimental ligand-binding data after the seven mutations were introduced, docking calculations for the ligand naltrexone using the structure of the murine MUR (4DKL)6 were carried out using the DockingServere) 15 as previously described16,17. Affinity (grid) maps of 30×30×30 Å grid points with a spacing of 0.375 Å were used for the docking calculations.
Statistical analysis
The shift of the thermostability curve is determined by comparing the temperature difference where the normalized mean residue molar ellipticity is at 50%. Three repeats of data are obtained and analyzed using the t test. The affinities are presented as mean ± one standard deviation from three repeated measurements. The results were analyzed using GraphPad Prism (version 5.02 Windows version). p-values smaller than 0.05 are considered statistically significant.
Results
Structural analysis suggests candidate residues for back-mutation in water-soluble MUR
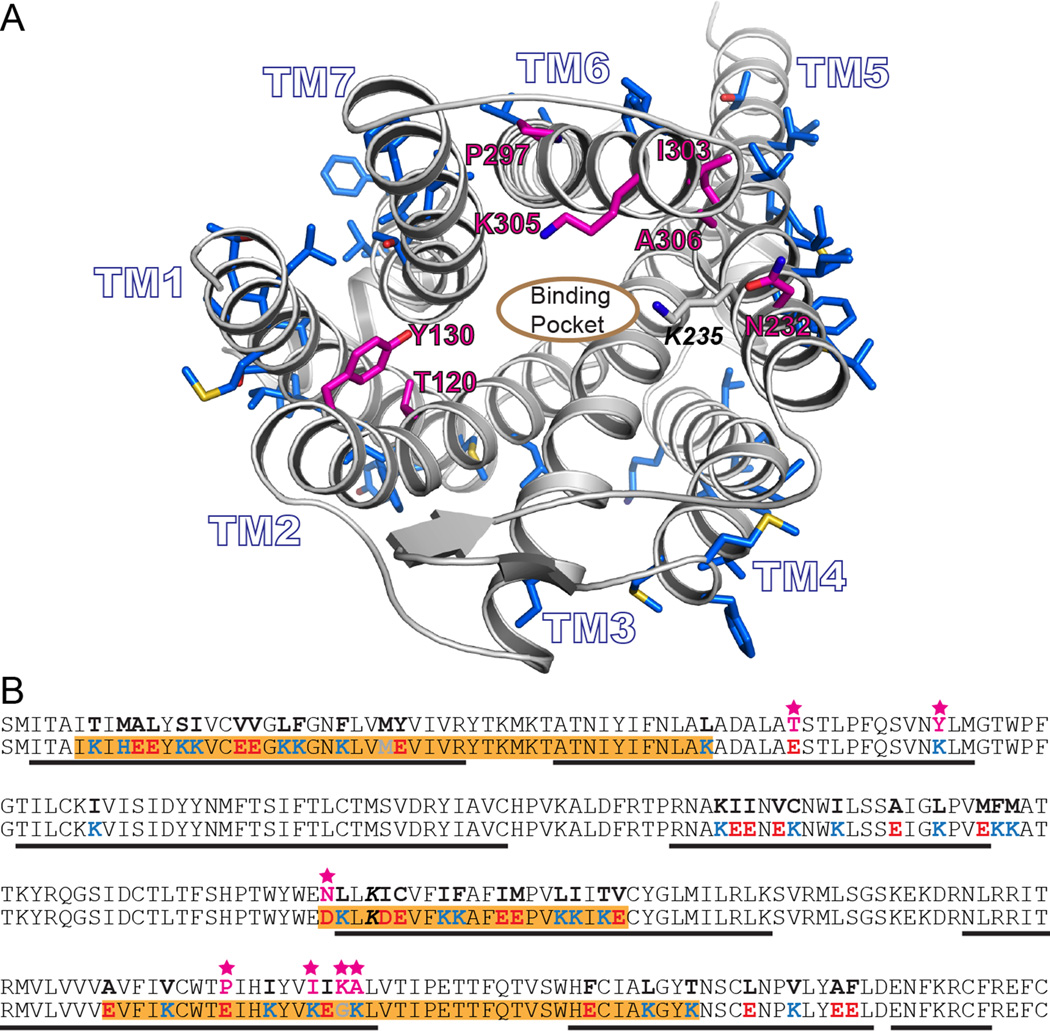
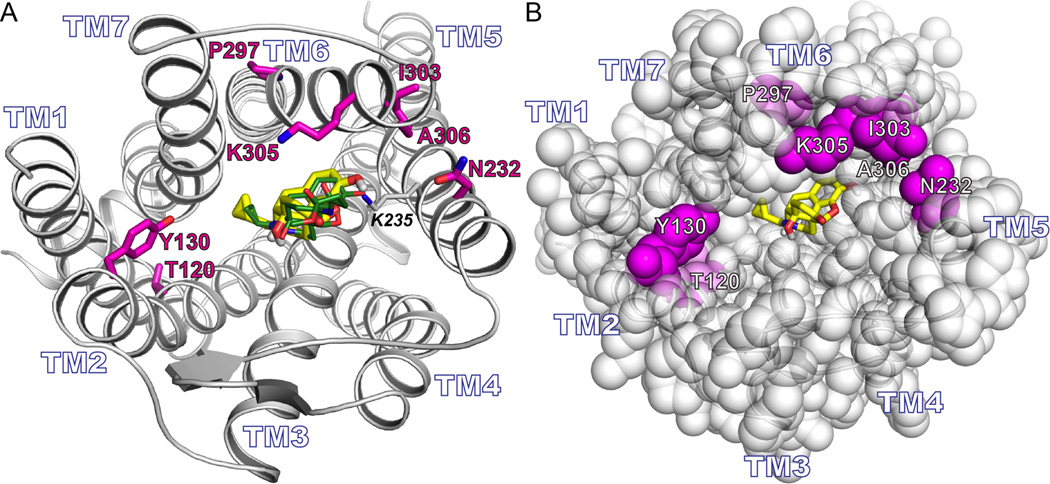
From a comparative structural analysis of wsMUR-TM and the murine crystal structure, five positions (E1202.54, K1302.64, D2325.36, G3056.58, and K3066.59) were identified whose variation could possibly disrupt either ligand binding or internal packing of amino acids (fig. 1A). Based on previous binding studies carried out in wsMUR-TM with a naltrexone analog, we found no significant disruption in the binding capabilities of wsMUR-TM compared to the native receptor. Nonetheless, native MUR also binds larger ligands, and these five mutations in the vicinity of the orthosteric binding site could affect the receptor’s binding to other ligands.7 In addition, two residues were mutated back to their native residue identities based on structural considerations (E2976.50 and K3036.56). Position E2976.50 was originally a proline residue in wsMUR-TM, and its presence could be important in retaining the right geometry of transmembrane helix 6. At position K3036.56, the native isoleucine appears to play an important role in the complementary packing of the transmembrane 5 and transmembrane 6 helices (fig. 1A). None of the seven positions were directly in contact with the beta-Funaltrexamine (β-FNA) ligand that was cocrystalized with murine MUR. These seven residues in wsMUR-TM, located in the transmembrane helical core and near the binding pocket, were mutated back to their native identities (E120T2.54, K130Y2.64, D232N5.36, E297P6.50, K303I6.56, G305K6.58, and K306A6.59) to generate a modified new analog denominated wsMUR-TM_v2 (fig. 1A and B).
Figure 1.
A) Rendering of a model based on the murine mu opioid receptor crystal structure. The side chains of residues computationally designed to generate wsMUR-TM are rendered blue. The root-mean-square deviation of the Cα atoms between this previous comparative model model and the murine structure involving transmembrane domain residues is 2.60Å.8 The designed positions selected to be mutated back to the human native identities are shown in magenta (E120T2.54, K130Y2.64, D232N5.36, E297P6.50, K303I6.56, G305K6.58, and K306A6.59, see Materials and Methods for description about Ballesteros and Weinstein indexing.10 B) Sequences of the transmembrane domain of wild-type human μ opioid receptor (top) and the designed water-soluble variant wsMUR-TM (bottom) are displayed. Highlighted in orange are the segments of the designed sequence (wsMUR-TM) identified to be important to preserve protein expression. Seven residues that have been restored to native identities are starred and colored in magenta. wsMUR-TM = first variant of the water-soluble human μ opioid receptor transmembrane portion; wsMUR-TM_v2 = modified (second) variant of the water-soluble human μ opioid receptor transmembrane portion; TM = transmembrane; M/Z = mass-to-charge ratio.
Overexpression, purification, and verification of wsMURs
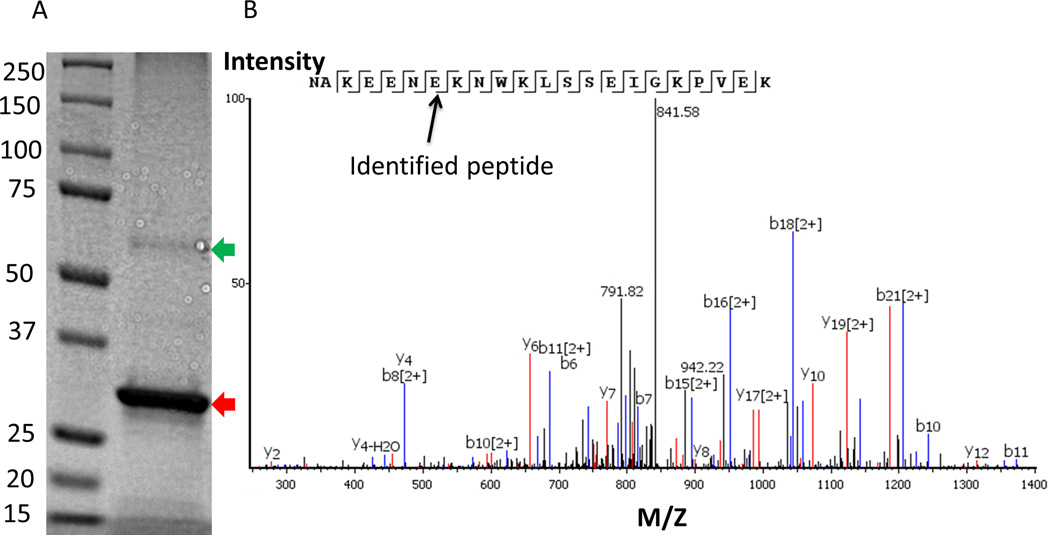
Both variants of the wsMURs expressed well and were isolated with high purity using nickel affinity chromatography. There was no appreciable difference in the bacteria growth (as monitored by optical density [OD]) and the yields of both variants (~20 mg/L of shake flask culture). ~0.1% SDS was initially required to solubilize and purify the receptors. After removing nonbound SDS via dialysis, both variants were soluble at ~7.5 mg/mL in a buffer containing 130 mM NaCl, 20 mM sodium phosphate with a pH of 7.0 without appreciable precipitation. A thick dark band in the SDS-PAGE presented in figure 2A belonged to wsMUR-TM_v2, as confirmed by mass spectroscopy. The coverage of the identified peptide segment was as high as 85%. One of the protein fragments of the wsMUR-TM_v2 variant, identified by mass spectrometry, is presented in figure 2B. It is important to note that there was a very faint band in the gel with a molecular weight corresponding to a dimer of the receptor (fig. 2A). This band was also studied by mass spectrometry and corroborated to be the sequence of wsMUR-TM_v2, suggesting the potential existence of covalent dimer of the protein.
Figure 2.
Overexpression and verification of wsMUR-TM_v2. A) A sodium dodecyl sulfate polyacrylamide gel electrophoresis for wsMUR-TM_v2 is shown, where lane 1 corresponds to the MW standard, lane 2 to purified wsMUR-TM_v2. The band corresponding to the molecular weight of wsMUR-TM_v2 is indicated by a red arrow. A faint band around twice the molecular weight of wsMUR-TM_v2 is also noted (green arrow). Both bands were excised for mass spectrometry and confirmation of protein identity. Each band was identified as wsMUR-TM_v2 with around 85% coverage of the sequence. The faint band represents the dimer of wsMUR-TM_v2. B) Representative mass spectrometry data for fingerprinting of an identified peptide fragment is displayed and data are consistent with the following subsequence (NAKEENEKNWKLSSEIGKPVEK). wsMUR-TM_v2 = modified (second) variant of the water-soluble human μ opioid receptor transmembrane portion; M/Z = mass-to-charge ratio.
Atomic force microscopy
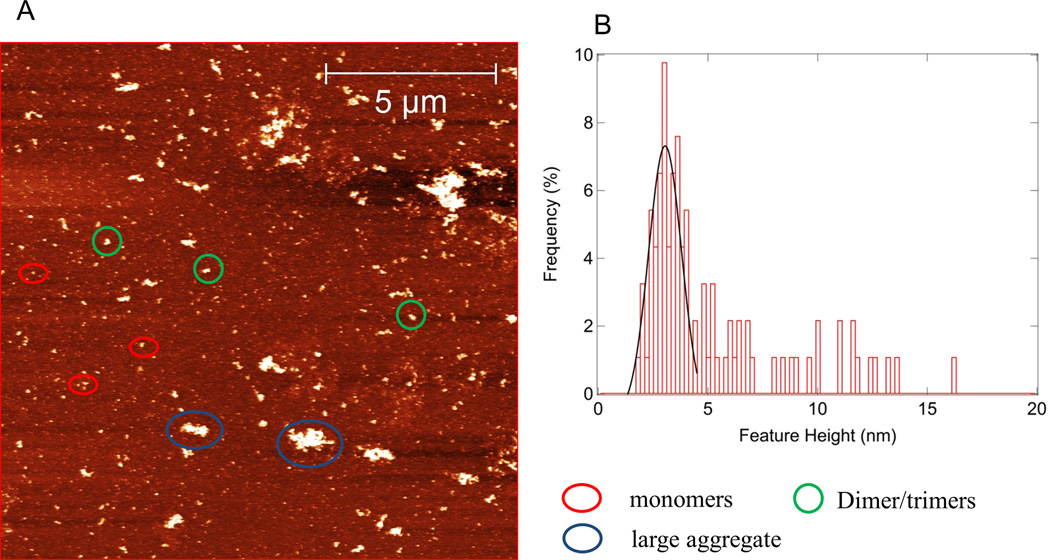
From a series of atomic force microscopy experiments, we found that both wsMURs are could be dispersed using buffer conditions and were predominantly monomeric (80~90%) when freshly prepared within a 1-week period. Consistent with the observation in SDS-PAGE, the existence of features whose heights are consistent with those of the dimer were observed in the atomic force microscopy experiments. The receptor was observed to aggregate as a function of time. 0.02% SDS and 5 mM 2-mercaptoethanol could reduce such aggregation significantly. As indicated in figure 3A and B, 70% of the surface features are ascribed to be monomeric for a 2-month old sample of wsMUR-TM_v2 in a buffer condition of 20 mM sodium phosphate, 130 mM sodium chloride, 0.02% SDS, and 5 mM 2-mercaptoethanol, pH=7.0.
Figure 3.
A) Atomic force microscopy image of a sample wsMUR-TM_v2 after stored at 4 °C for about 2 months. Protein concentration is 7.5 mg/mL. Buffer is 20mM sodium phosphate, 130mM NaCl, 0.02% sodium dodecyl sulfate, 5mM 2ME, pH=7.0. Monomer, dimer, and large aggregates are present. B) Histogram of the feature of the protein in the solution. The estimated number of features present on surface consistent with the dimensions of the monomer is ~70%. wsMUR-TM = first variant of the water-soluble human μ opioid receptor transmembrane portion; wsMUR-TM_v2 = modified (second) variant of the water-soluble human μ opioid receptor transmembrane portion.
Secondary structure content of wsMURs
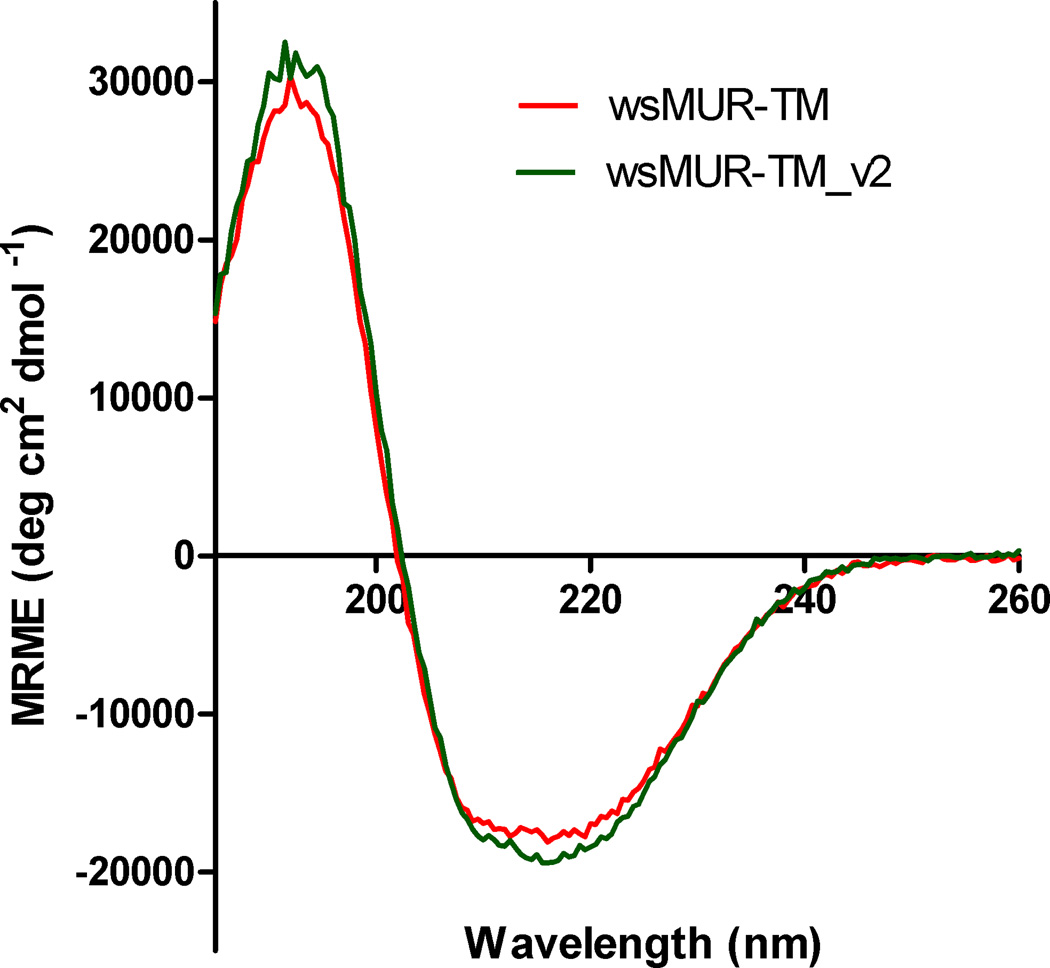
The circular dichroism spectra of wsMUR-TM_v2 indicated predominantly helical structure with a helical content of 57% estimated by the molar ellipticity over the range 205 to 260 nm. This value is consistent with similar measurements on wsMUR-TM (fig. 4). The helical content of wsMUR-TM_v2 and wsMUR-TM is presented in table 1. Upon addition of 1 M sucrose, no significant change in the helical content was observed. Also, the inclusion of 3 M urea reduced the helical content significantly but such reduction was restored by the presence of 1 M sucrose (data not shown).
Figure 4.
Circular dichroism spectra of the wsMUR-TM_v2 compared with the originally designed wsMUR-TM at buffer condition of 5 mM sodium phosphate, pH = 7.0 at temperature of 25 °C. The two proteins displayed similar spectra. Estimates of helical content obtained using the two spectra are presented in table 1. MRME, mean residue molar ellipticity; wsMUR-TM_v2 = modified (second) variant of the water-soluble human mu opioid receptor transmembrane portion.
Table 1.
Comparison of the Secondary Structure of the Two Variants of wsMURs
| 205–260 nm | wsMUR-TM | wsMUR-TM_v2 |
|---|---|---|
| Helix | 53.7% | 55.6% |
| Antiparallel | 4.6% | 4.4% |
| Parallel | 4.9% | 4.6% |
| Beta-Turn | 13.9% | 13.6% |
| Others | 22.9% | 21.8% |
wsMUR-TM = first variant of the water-soluble human μ opioid receptor transmembrane portion; wsMUR-TM_v2 = modified (second) variant of the water-soluble human μ opioid receptor transmembrane portion. Buffer condition: 0.01% Sodium dodecyl sulfate, 5 mM sodium phosphate, pH = 7.0 in room temperature.
Thermal stability of wsMURs
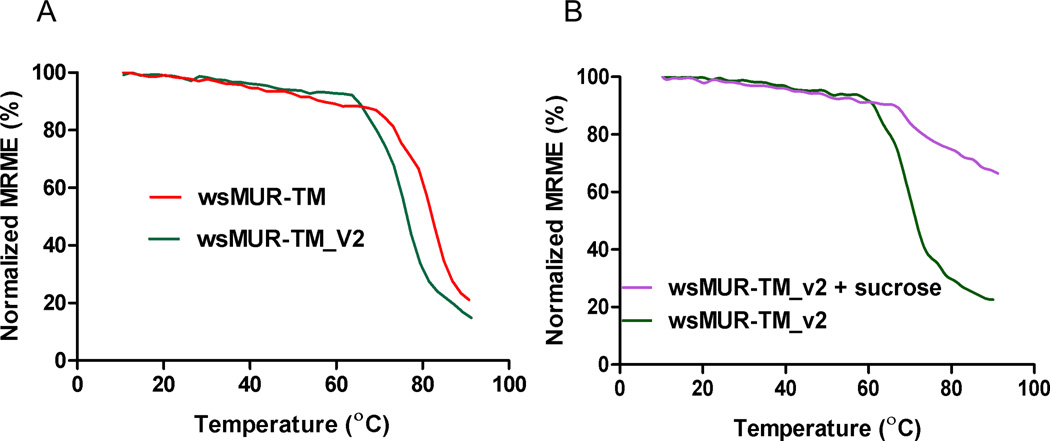
As monitored by circular dichroism, wsMURs started to lose ellipticity with increasing temperature and were nearly fully unfolded at 90 °C (fig. 5A). The thermal stability curve of wsMUR-TM_v2 shifted to the left (lower temperature) by 5.8 ± 1.5 °C relative to the wsMUR-TM (p = 0.007). It is important to note that the thermal stability curve shifted to higher temperature significantly in the presence of sucrose (fig. 5B).
Figure 5.
Molar circular dichroism derived percentage of the original helical content (determined at 222 nm) of wsMUR-TM and wsMUR-TM_v2 in buffer solution (5 mM sodium phosphate, pH = 7.0) as functions of the temperature. A) It demonstrated that wsMUR-TM is thermodynamically more stable than wsMUR-TM_v2. B) The addition of sucrose stabilized the wsMUR-TM_v2 as indicated by the significant rightward shift of the thermostability curve. MRME = mean residue molar ellipticity; wsMUR-TM = first variant of the water-soluble human μ opioid receptor transmembrane portion; wsMUR-TM_v2 = modified (second) variant of the water-soluble human μ opioid receptor transmembrane portion.
Interaction with opioid antagonist naltrexone
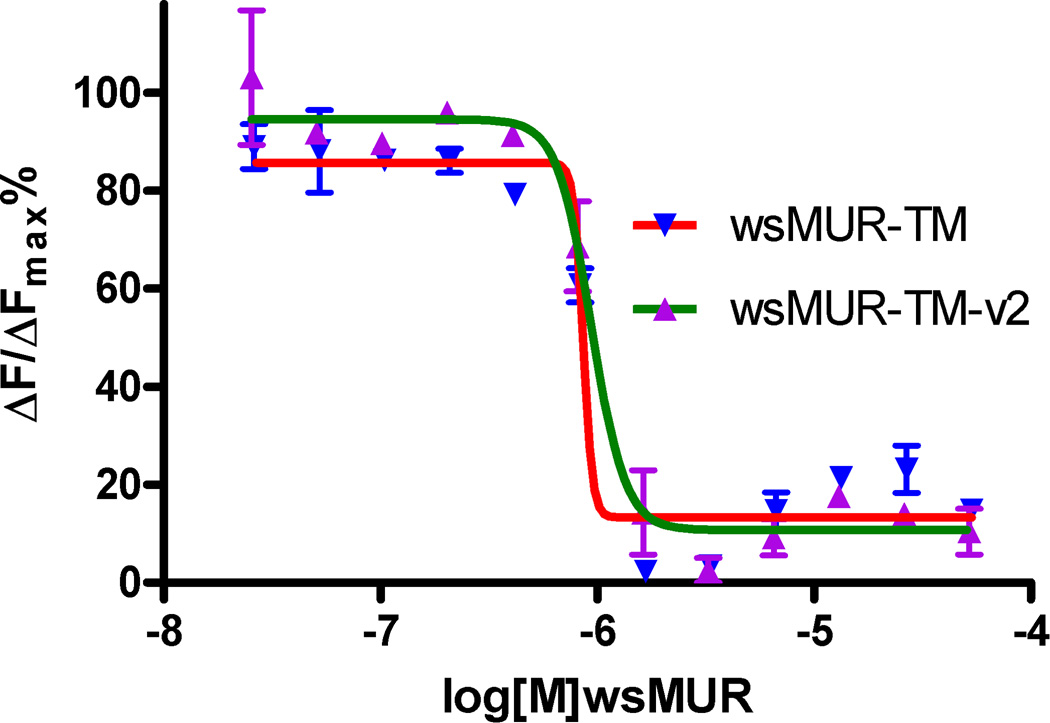
Naltrexone binding was monitored using a competitive time-resolved fluorescence resonance energy transfer based assay with fluorescently labeled wild type MUR and a naltrexone-derived antagonist. The ratio of fluorescence emission at 620 and 665 nm decreased in a concentration-dependent manner with increasing concentrations of both variants of wsMUR. There was no significant difference for the affinity with naltrexone for both wsMURs (fig. 6). The Kd value for wsMUR-TM was similar to the previously reported value (67 ± 4 nM), while the Kd value for wsMUR-TM_v2 was 70 ± 7 nM. Both values are in closed agreement with values for native MUR reported using other techniques.7,18
Figure 6.
Binding competition assay between the human μ opioid receptor expressed in HEK293 cells and the μ opioid water-soluble variants. Inhibition of the native μ opioid receptor constitutive signal from the interaction with naltrexone in the presence of increasing concentrations of wsMUR-TMs in sodium phosphate buffer is demonstrated which is essentially in a similar pattern, indicating a similar affinity of naltrexone for both of the variants. The Kd values are 67 ± 4 nM for wsMUR-TM and 70 ± 7 nM for wsMUR-TM_v2. The Kd value indicates the average value of three measurements. The error bars in the figure are obtained as the standard deviation of three independent measurements. ΔF is used for the comparison of different runs of the same assay which reflects the signal to background of the assay. F=[(Ratiosample−Ratiobackgroud)/Ratiobackgroud](%). As we described previously for the homogeneous time-resolved fluorescence binding assay,7 “Ratio” was the ratio of fluorescence emission at 665 nm and 620 nm, and Kd values were determined by fitting the dose–response curves using Prism (GraphPad Software, San Diego, CA).The Kd value indicates the average value of three measurements. wsMUR-TM = first variant of the water-soluble human μ opioid receptor transmembrane portion; wsMUR-TM_v2 = modified (second) variant of the water-soluble human μ opioid receptor transmembrane portion.
To interpret the findings from the binding assay at a molecular level, docking calculations were carried out using the mouse crystal structure and naltrexone. Docking calculation results indicated that the naltrexone binding site overlapped well with that of the β-FNA as demonstrated in figure 7A. The consistency in the affinity values for both wsMURs could be ascribed to the fact that there is no direct contact of any of the seven mutated residues with the ligand naltrexone (fig. 7B).
Figure 7. The relationship of the naltrexone with the mutated residues.
A) Rendering of the murine μ opioid receptor. The seven residues that were mutated back to their native residue identities are in magenta (E120T2.54, K130Y2.64, D232N5.36, E297P6.50, K303I6.56, G305K6.58, and K306A6.59). The docking pose of naltrexone (yellow) overlaps with the pose of β-Funaltrexamine (green). B) Similar view as (A) but with the protein rendered in space filling representation. The docked posed of naltrexone (yellow) does not directly contact any of the seven mutated positions (magenta).
Discussion
The crystal structure of the murine MUR complexed with the irreversible antagonist β-FNA6 provides valuable information to reengineer our previously designed water-soluble variant wsMUR-TM.7 Based on structural analysis, seven positions in wsMUR-TM were returned to their native residue identities. Residues were selected based upon their relative position to the surface of the protein and the orthosteric binding pocket in the structure of the murine MUR. The resulting protein, wsMUR-TM_v2, expressed well in E. coli and was purified in large amounts sufficient to perform biophysical experiments without significant change in the protein’s water solubility. Interestingly, while the affinity of the protein for naltrexone did not change significantly, the thermostability of the protein decreased slightly. Addition of the osmolyte sucrose increased the thermal stability of the protein significantly.
Computational design of wsMURs
In the quest to facilitate the characterization of different membrane proteins, computational design has been used to engineer water-soluble variants of membrane proteins. Previously, variants have been engineered of the potassium channel from Streptomyces lividans (KcsA)19–21 and the transmembrane domain of the α1 subunit of the nicotinic acetylcholine receptor (nAChR),22 with mutations ranging from 18% to 30% of the total number of residues. In our first version of the water-soluble analog of the human MUR, wsMUR-TM, 18% of the residues (53 out of 288) was mutated. An important question that remained was to what extent to which the number of mutations could be reduced without compromising desired properties such as water solubility and protein production in E. coli. As we discussed previously, the toxic effects of the human MUR to cell lines have limited the capability to express the native receptor in this heterologous bacterial expression system.7 The cytotoxicity of the native MUR is possibly a result of hydrophobic residues located within the receptor’s transmembrane region, as mutations of mainly hydrophobic residues in the designed water-soluble variants significantly reduced the cytotoxicity and facilitated over-expression in E. coli. While we cannot conclude that all of the mutated residues in the present study were required for high protein expression and formation of a variant with structural and functional properties comparable to those observed in the native receptor, we achieved the lowest number of mutations for any water-soluble variants we have designed so far (16%, 46 mutated residues out of 288), an important technical achievement.
The production of a soluble and functional water-soluble MUR for its biophysical characterization involves computationally redesigning the protein, and residues in the transmembrane region are the focus. Mutating residues, including mutating redesigned sites back to their native identities, could disrupt any of the steps associated with production of native-like protein, including: cytotoxicity of the heterologous system (failure of expression); protein purification; protein stability; water solubility; and receptor’s binding properties. However, even after mutating seven residues located in transmembrane helical core of the receptor, the protein can still be over-expressed in E. coli and retains intrinsic structural and binding features as a MUR. The protein variant created using a previously created homology model alone (wsMUR-TM) and that created by an analysis of the crystallographic murine receptor and restoration of wild-type identities at seven residues (wsMUR-TM_v2) had essentially identical helical content and naltrexone affinities. The information of the mutations introduced in wsMUR-TM_v2 offers additional structure-sequence details to identify the features involved in cytotoxicity, solubility, and sequence-structural- function relationship studies for MUR and other GPCRs in the same family. These observations further confirm our previous finding that, at least for the GPCR family of proteins, homology modeling can yield high quality structures suitable for the computational design of water-soluble variants. While the properties of the two designed proteins are similar, it is unclear whether there are significant tertiary structure differences. Since both are water soluble that can be over expressed in E. coli, thus, they can be used as parallel alternative tools to access sequence-structure-function relationships using nuclear magnetic resonance in solution conditions, which is an ongoing effort within our group.
wsMURs oligomeric states in solution condition
Atomic force microscopy provides a very useful tool to evaluate our water-soluble proteins. The data are consistent with previous work indicating that the engineered wsMURs are soluble in buffer conditions and stay predominantly monomeric.7 Though the protein exists largely in the monomeric form, the formation of oligomers, particularly dimers, is also confirmed. The presence of different oligomeric states is consistent with the SDS-PAGE electrophoresis data. It is known that the MUR can form SDS resistant dimers in living organisms, and dimerization may play critical roles in physiological and pharmacological functions of the receptor.23,24 This self-association appears not to be eliminated by the large numbers of mutations involving exposed hydrophobic residues described in our study. Such dimerization persists but is diminished in the presence of 5 mM of the reducing agent 2-mercaptoethanol. The presence of SDS and 2-mercaptoethanol appear to reduce aggregation to improve the overall shelf life of the protein sample. Using crystallography, we have previously demonstrated that low concentrations of SDS do not change the protein structure in the natural mammalian protein, apoferritin.25 Nonetheless, further studies are needed to determine the extent to which the tertiary structure of the wsMUR is altered in such low SDS concentrations. Further studies are also needed to eliminate SDS in the purification process.
The structure and thermostability
The secondary structure of the modified new version wsMUR-TM_v2 is consistent with that of the previously reported wsMUR-TM variant. As intended, the introduction of the mutations K130Y2.64, K303I6.56, and K306A6.59 seem to induce better protein packing as the helical content improved slightly in wsMUR-TM_v2 when compared with wsMUR-TM. The remaining mutations could also play similar roles in improving the overall protein packing of the new water-soluble variant. However, it is interesting to note that the thermal stability curve of the modified new design shifted to lower temperature by 5 °C as compared to that of the first version, suggesting slight decrease of the thermal stability. One possible explanation is the reintroduction of a proline residue at position 2976.50. Although this mutation was intended to restore and facilitate interactions that stabilize the irregularities in transmembrane helix 6, the helix propensity of proline is the lowest of all the amino acid residues and in the absence of the lipid bilayer, this could decrease the thermal stability of the protein.
With the exception of rhodopsin, GPCRs are generally unstable, one of the major obstacles for structural function relationship studies.26 We recently demonstrated that the hydrophobic molecule cholesterol improves the thermostability of the wsMUR-TM.7 It is critical to identify a water-soluble compound that could be used to stabilize the protein thermodynamically. Sucrose stabilizes some proteins and forces some unfolded protein to folded states.27 Consistent with these reports, sucrose was able to restore the helical content reduction due to presence of urea. Moreover, addition of 1 M sucrose increased the thermal stability of wsMUR-TM_v2 significantly. This finding is critical for further structural and functional studies of these opioid receptor analogs, since the addition of this osmolyte is able to stabilize the protein without precipitation. Sucrose also does not generally interference with protein nuclear magnetic resonance signals, which is desired for the study of the receptor’s structure and dynamics in solution conditions.
Ligand Binding Properties of the wsMURs
In this study, we adopted the same methodology reported recently to quantify the affinity of the wsMURs with naltrexone, a potent antagonist of MUR.7 The results indicated no significant change in the affinity of wsMUR-TM_v2 relative to wsMUR-TM for naltrexone despite the introduction of seven mutations in proximity to the binding pocket. These findings are consistent with the observations that none of these residues have direct contact with β-FNA in the murine crystal structure of the MUR. Additionally, our docking calculations suggest that naltrexone binds in essentially the same binding pocket as the morphian group of β-FNA. These findings corroborate our conclusions from our previous report that demonstrated a close agreement of the naltrexone affinity of the wsMUR-TM with that of the native MUR.
Conclusions and Future Directions
The study presented here demonstrated that available crystal structure allowed us to reliably reduce number of mutated residues in the water-soluble variant wsMUR-TM_v2 without affecting the protein production in E. coli, the affinity for naltrexone, and the solubility of the protein in the buffer solution relative to its predecessor wsMUR-TM. However, the thermostability study revealed that some of the mutated residues play important roles in the protein stability, which is an important consideration for future mutagenesis studies and for structure-function investigations. Sucrose could be an important alternative molecule to be used to stabilize these water-soluble variants for structural studies. This study not only provides a novel variant of water soluble MUR as a useful tool for MUR studies, but also offers valuable information to engineer water-soluble versions of GPCR, a clinically relevant membrane protein superfamily.
Acknowledgments
The authors appreciate the outstanding technical support from Qingchen Meng, Ph.D. and Jingyuan Ma, B.A. candidate, at the Department of Anesthesiology and Critical Care at the University of Pennsylvania (Philadelphia, Pennsylvania); Dr. Renyu Liu thanks the mentorship and valuable discussions from Roderic G. Eckenhoff, M.D. at the Department of Anesthesiology and Critical Care at the University of Pennsylvania and the support from the Department of Anesthesiology and Critical Care at the University of Pennsylvania
Funding: This work was supported by the Foundation for Anesthesia Education and Research, Rochester, Minnesota, National Institute of Health, Bethesda, Maryland: K08-GM-093115-01, the Mary Elizabeth Groff Foundation (Radnor, Pennsylvania), and the Department of Anesthesiology and Critical Care at the University of Pennsylvania (Philadelphia, Pennsylvania) to Dr. Liu. This work was also supported by the National Science Foundation, Arlington, Virginia: PFI-AIR 13-12202 to Johnson and Saven), MRSEC DMR-1120901 to Saven, and NSEC DMR-0425780 to Johnson.
Footnotes
Competing interests: The authors declare no competing interests.
http://www.rcsb.org/pdb/explore/explore.do?structureId=4DKL; last date accessed April 19, 2014.
http://www.pymol.org/; last date accessed April 19, 2014.
http://fields.scripps.edu/sequest; last date accessed April 19, 2014.
http://www.uniprot.org; last date accessed April 19, 2014.
http://www.dockingserver.com; last date accessed April 19, 2014.
References
- 1.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol pharmaco. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 2.Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 3.Granier S, Manglik A, Kruse AC, Kobilka TS, Thian FS, Weis WI, Kobilka BK. Structure of the delta-opioid receptor bound to naltrindole. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson AA, Liu W, Chun E, Katritch V, Wu H, Vardy E, Huang XP, Trapella C, Guerrini R, Calo G, Roth BL, Cherezov V, Stevens RC. Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature. 2012;485:395–399. doi: 10.1038/nature11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, Mascarella SW, Westkaemper RB, Mosier PD, Roth BL, Cherezov V, Stevens RC. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012;485:327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez-Aguilar JM, Xi J, Matsunaga F, Cui X, Selling B, Saven JG, Liu R. A computationally designed water-soluble variant of a g-protein-coupled receptor: The human mu opioid receptor. PLOS ONE. 2013;8:e66009. doi: 10.1371/journal.pone.0066009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez-Aguilar JM, Saven JG, Liu R. Human mu opioid receptor models with evaluation of the accuracy using the crystal structure of the murine mu opioid receptor. J Anesth Clin Res. 2012;3:218. doi: 10.4172/2155-6148.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 10.Ballesteros JA, Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 11.Goldsmith BR, Mitala JJ, Josue J, Castro A, Lerner MB, Bayburt TH, Khamis SM, Jones RA, Brand JG, Sligar SG, Luetje CW, Gelperin A, Rhodes PA, Discher BM, Johnson AT. Biomimetic chemical sensors using nanoelectronic readout of olfactory receptor proteins. ACS Nano. 2011;5:5408–5416. doi: 10.1021/nn200489j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shekhawat GS, Lambert MP, Sharma S, Velasco PT, Viola KL, Klein WL, Dravid VP. Soluble state high resolution atomic force microscopy study of Alzheimer's beta-amyloid oligomers. Appl Phys Lett. 2009;95:183701. doi: 10.1063/1.3251779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poschner BC, Reed J, Langosch D, Hofmann MW. An automated application for deconvolution of circular dichroism spectra of small peptides. Anal Biochem. 2007;363:306–308. doi: 10.1016/j.ab.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 15.Bikadi Z, Hazai E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J Cheminform. 2009;1:15. doi: 10.1186/1758-2946-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu R, Perez-Aguilar JM, Liang D, Saven JG. Binding site and affinity prediction of general anesthetics to protein targets using docking. Anesth Anal. 2012;114:947–955. doi: 10.1213/ANE.0b013e31824c4def. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui X, Yeliseev A, Liu R. Ligand interaction, binding site and G protein activation of the mu opioid receptor. Eur J Pharmaco. 2013;702:309–315. doi: 10.1016/j.ejphar.2013.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zwier JM, Roux T, Cottet M, Durroux T, Douzon S, Bdioui S, Gregor N, Bourrier E, Oueslati N, Nicolas L, Tinel N, Boisseau C, Yverneau P, Charrier-Savournin F, Fink M, Trinquet E. A fluorescent ligand-binding alternative using Tag-lite(R) technology. J Biomol Screen. 2010;15:1248–1259. doi: 10.1177/1087057110384611. [DOI] [PubMed] [Google Scholar]
- 19.Slovic AM, Kono H, Lear JD, Saven JG, DeGrado WF. Computational design of water-soluble analogues of the potassium channel KcsA. Proc Natl Acad Sci U S A. 2004;101:1828–1833. doi: 10.1073/pnas.0306417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bronson J, Lee OS, Saven JG. Molecular dynamics simulation of WSK-3, a computationally designed, water-soluble variant of the integral membrane protein KcsA. Biophys J. 2006;90:1156–1163. doi: 10.1529/biophysj.105.068965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Aguilar JM, Saven JG. Computational design of membrane proteins. Structure. 2012;20:5–14. doi: 10.1016/j.str.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui T, Mowrey D, Bondarenko V, Tillman T, Ma D, Landrum E, Perez-Aguilar JM, He J, Wang W, Saven JG, Eckenhoff RG, Tang P, Xu Y. NMR structure and dynamics of a designed water-soluble transmembrane domain of nicotinic acetylcholine receptor. Biochimica et Biophysica Acta - Biomembranes. 2011;1818:617–626. doi: 10.1016/j.bbamem.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li-Wei C, Can G, De-He Z, Qiang W, Xue-Jun X, Jie C, Zhi-Qiang C. Homodimerization of human mu-opioid receptor overexpressed in Sf9 insect cells. Protein Pept Lett. 2002;9:145–152. doi: 10.2174/0929866023408850. [DOI] [PubMed] [Google Scholar]
- 24.Decaillot FM, Rozenfeld R, Gupta A, Devi LA. Cell surface targeting of mu-delta opioid receptor heterodimers by RTP4. Proc Natl Acad Sci U S A. 2008;105:16045–16050. doi: 10.1073/pnas.0804106105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu R, Loll PJ, Eckenhoff RG. Structural basis for high-affinity volatile anesthetic binding in a natural 4-helix bundle protein. FASEB J. 2005;19:567–576. doi: 10.1096/fj.04-3171com. [DOI] [PubMed] [Google Scholar]
- 26.Cherezov V, Abola E, Stevens RC. Recent progress in the structure determination of GPCRs, a membrane protein family with high potential as pharmaceutical targets. Methods Mol Biol. 2010;654:141–168. doi: 10.1007/978-1-60761-762-4_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cioni P, Bramanti E, Strambini GB. Effects of sucrose on the internal dynamics of azurin. Biophys J. 2005;88:4213–4222. doi: 10.1529/biophysj.105.060517. [DOI] [PMC free article] [PubMed] [Google Scholar]