Abstract
Genome engineering with targetable nucleases depends on cellular pathways of DNA repair after target cleavage. Knowledge of how those pathways work, their requirements and their active factors, can guide experimental design and improve outcomes. While many aspects of both homologous recombination (HR) and nonhomologous end joining (NHEJ) are shared by a broad range of cells and organisms, some features are specific to individual situations. This article reviews the influence of repair mechanisms on the results of gene targeting experiments, with an emphasis on lessons learned from experiments with Drosophila.
Keywords: ZFNs, TALENs, DNA repair, gene targeting, NHEJ, homologous recombination
1. Introduction
The targetable nucleases have created a small revolution in genetics by providing the means to make very specific changes in genomic DNA with high efficiency. First zinc-finger nucleases (ZFNs) [1–3], then transcription activator-like effector nucleases (TALENs) [4, 5], and most recently CRISPR/Cas RNA-guided nucleases (CRISPRs) [6, 7] have become powerful tools for genome engineering [8–10]. In fact, the only thing these reagents do is make breaks in chromosomal DNA at designed locations. After that, everything is left up to the cells in which the breaks have been made. The reason this works well is that cells detect double-strand breaks (DSBs) as potentially lethal damage and activate mechanisms to repair them (Figure 1).
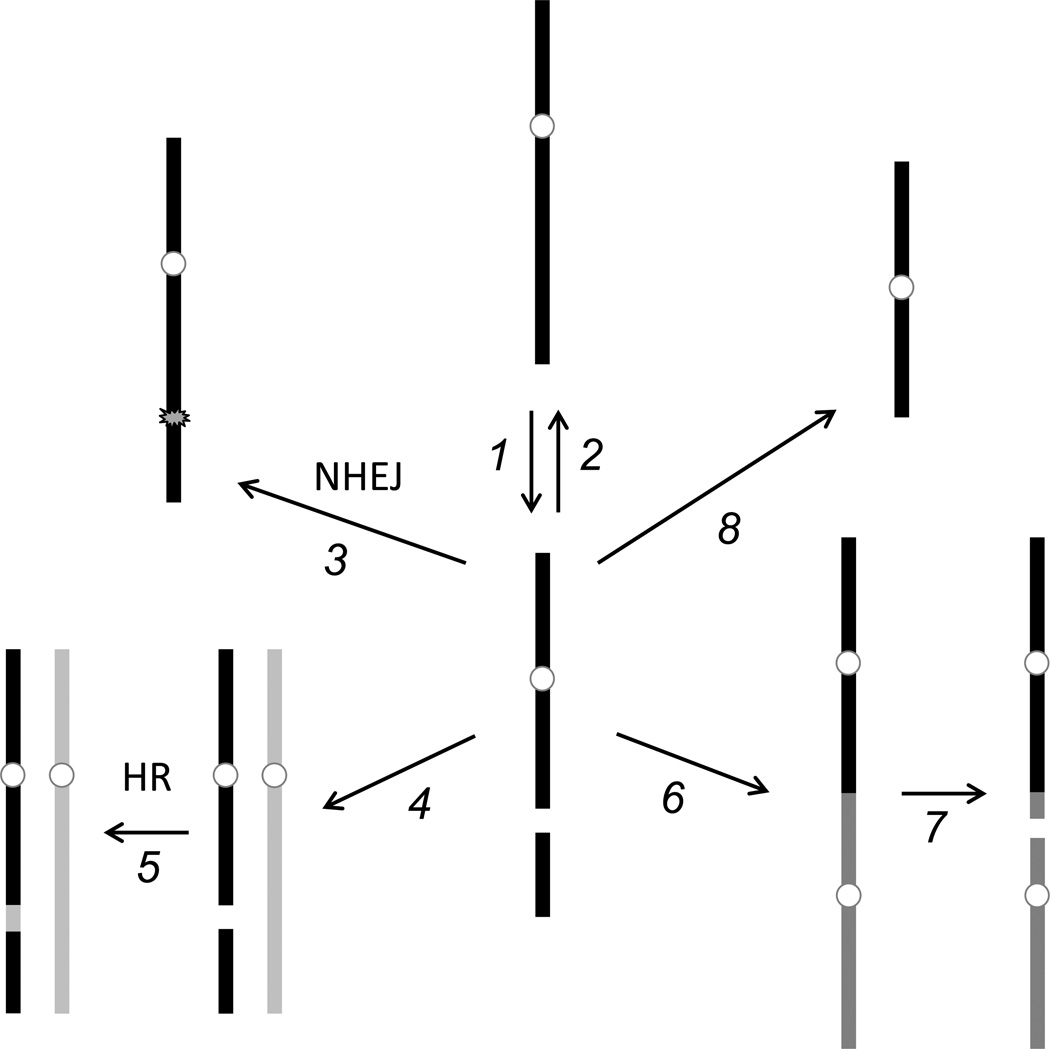
Figure 1.
Fates of double-strand breaks. An intact chromosome (top, middle) may suffer a double-strand break (1). This can be repaired by accurate religation to restore the original sequence (2), or rejoining may be inaccurate and introduce a sequence change (3, NHEJ). Pairing with a sister chromatid or homologous chromosome (4) allows accurate repair by homologous recombination (5, HR). If left unrepaired, the broken end may be inappropriately joined to another chromosome fragment (6). If the product has two centromeres, a new break may be introduced at mitosis due to centromeres being pulled in opposite directions (7). If the unrepaired chromosome enters mitosis, the fragment lacking a centromere may be lost (8).
Cellular DSB repair pathways fall into two broad categories – ones that depend on extensive sequence homology and ones that do not [11]. The latter are typically classified as nonhomologous end joining (NHEJ). A characteristic of homology-independent repair is that it is frequently inaccurate, since it has no template from which to gather instructions. Genome engineers rely on NHEJ to generate local mutations at sites of nuclease cleavage.
Homology-mediated repair, often simply called homologous recombination (HR), is a more orderly process designed to restore the interrupted sequence precisely. This mechanism is the principal one used during the S and G2 phases of the mitotic cell cycle, and the sister chromatid is the preferred template [11]. Fortunately, cells will also use DNA provided exogenously as a template for repair. Thus, experimenters can introduce desired sequence changes by delivering an engineered donor DNA with sufficient homology flanking a targeted DSB.
It is difficult to measure absolute frequencies of repair by HR and NHEJ in most situations. First, there are repair events – HR with the sister chromatid, accurate religation of the ends at the break – that restore the original sequence and are therefore invisible. These products can, of course, be recut by the targeted nuclease. In many cases, NHEJ products are resistant to recutting, but this is not always true, particularly with TALENs, which allow variable spacer lengths between paired binding sites. Donor DNAs can be engineered to resist recleavage following HR. Any products that are uncuttable constitute a sink that accumulates mutants, and this is what we ultimately measure.
Overall frequencies of mutation stimulated by targetable nucleases can be very high, depending on many parameters. Levels well over 50% have been reported with the best TALENs and CRISPR nucleases [12, 13]. When a donor is supplied and HR is desired, cells will still repair a proportion of DSBs by NHEJ. In many cell types, NHEJ is the main pathway of repair, and this can frustrate attempts to make very specific sequence changes at the target.
2. Cellular DSB repair mechanisms
2.1 Nonhomologous end joining (NHEJ)
We have an incomplete understanding of all the ways that DNA ends can be rejoined, but it is common practice to specify a canonical pathway and one or more secondary or alternative pathways [14]. Canonical NHEJ in many organisms requires a common set of factors, including a dedicated DNA ligase (ligase IV or Lig4) and its associated Xrcc4 protein, and the end-binding dimer, Ku70/Ku80. When any of these factors is disabled, NHEJ is reduced to a variable extent, depending on the organism or cell type [11]. Genetic requirements for alternative NHEJ are poorly defined, although DNA ligase III is implicated, at least in some situations [15].
Not only do the factors and efficiencies of canonical and alternative NHEJ differ, so do the junctions produced. While both processes yield short sequence insertions and/or deletions at the break site, events occurring in the absence of canonical factors appear to rely more heavily on short sequence matches at or near the break, called microhomologies [14]. The molecular details of microhomology-mediated end joining are not fully worked out, but several plausible models exist. One idea is that the short matches provide a transient primer-template complex that can be captured and extended by DNA polymerase (Figure 2) [16–18]. DNA ligase I could complete such a process by sealing nicks left after DNA synthesis, obviating a need for Lig4.
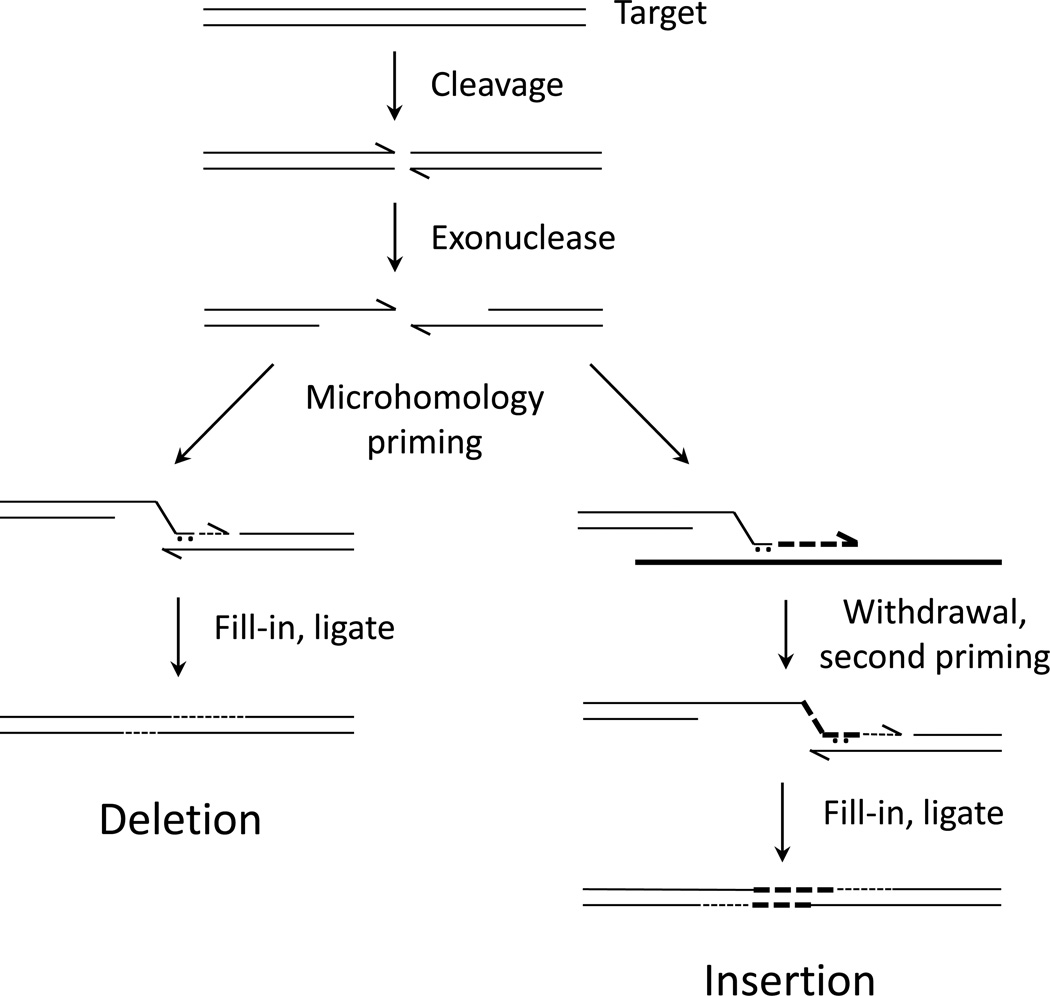
Figure 2.
Possible mechanisms of microhomology-mediated NHEJ. After cleavage, 5’ ends are resected, exposing single-stranded 3’ ends. One such end may pair with the other from the same break using a microhomology (left side). If that very short duplex is captured and extended by DNA polymerase, the junction is extended and stabilized. Filling in on the other side and ligation completes the process, resulting in a deletion. On the right side of the figure, the possibility of using some other template (thick line) is illustrated. The initial microhomology is extended on that template, the extended strand withdraws and pairs with the other end from the original break, perhaps again using a microhomology. The process is completed as on the left, resulting in an insertion of sequences from the first template.
2.2 Homologous recombination (HR)
Although some aspects may differ, the basic features of break-stimulated HR in mitotically dividing cells (meiosis is special) are shared by essentially all eukaryotic organisms [11]. At the DNA level, each end at the break is resected by 5’->3’ nuclease activity, producing a single strand with a free 3’ end (Figure 3). This end invades homologous sequence and is then extended by DNA polymerase. The extended strand withdraws and pairs with the exposed 3’-ending single strand from the other end of the original break. DNA polymerase and DNA ligase activities restore DNA duplex integrity. Additional protein requirements for the process, which is called synthesis-dependent strand annealing (SDSA), are also known [19]. The Rad51 protein mediates invasion; Rad54 is involved at several steps, including extension of the invading end; Rad52 is a critical HR factor in yeast, but plays a much more minor role in mammalian cells [20], and is not present in Drosophila.
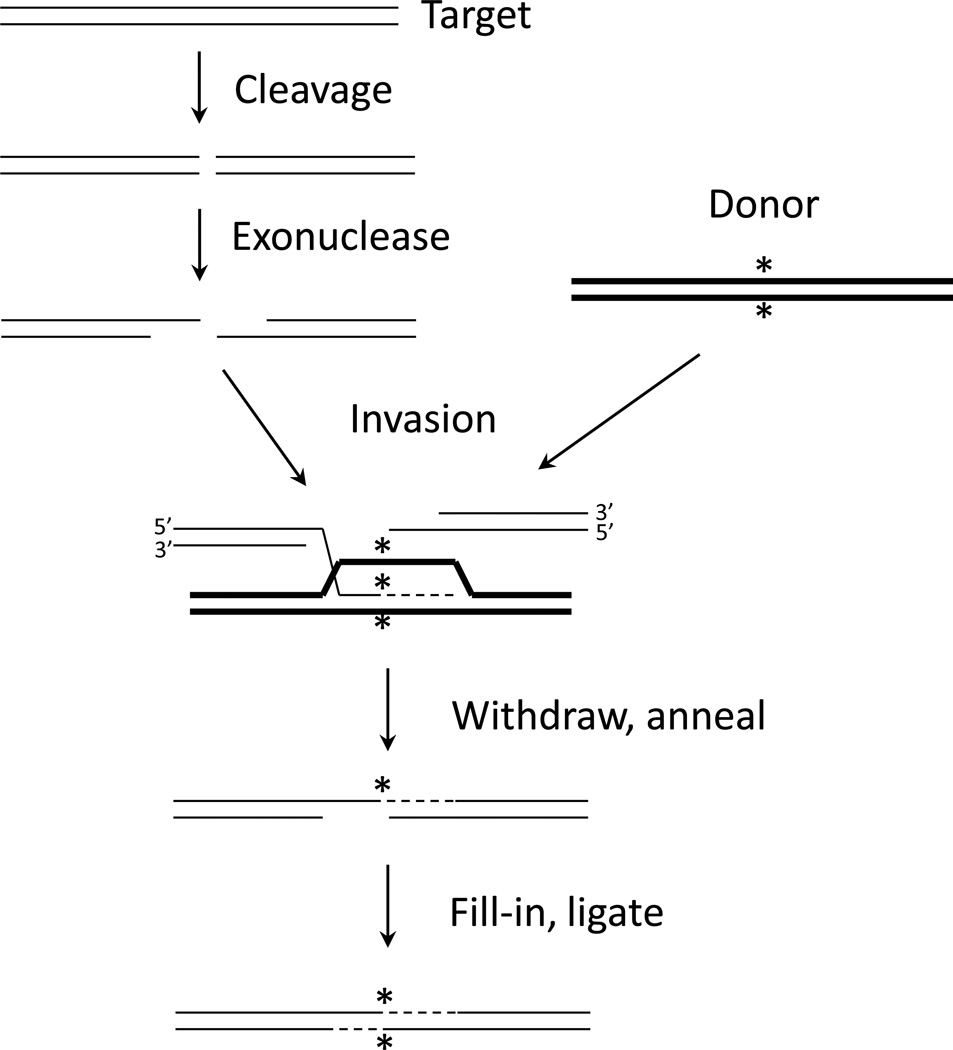
Figure 3.
Synthesis-dependent strand annealing (SDSA) mechanism of homologous recombination. As in Figure 2, 5’ ends are resected after the break. In this case, one 3’ single-stranded tail invades homologous sequence in the double-stranded donor (thick lines), which carries some distinguishing characteristic (*). The invading end is extended by DNA polymerase, using the donor as template. The extended strand withdraws and pairs with the other end from the original break. Polymerase and ligase activities complete the repair.
3. Experimental observations in Drosophila
Our group has studied how various genetic and molecular parameters influence the outcome of gene targeting experiments in Drosophila. Many of the experiments were done with ZFNs, although some aspects have been confirmed with TALENs. Because all the targetable nucleases make DSBs and the basic features of DNA repair pathways are shared among eukaryotic organisms, we expect that the lessons learned will be broadly applicable. We have recently published a description of procedures for using targetable nucleases in Drosophila [21], and the reader is referred to that article for experimental details.
3.1. Genetic manipulation of DNA repair
Working with Drosophila offered us the opportunity to explore the effects of disabling specific repair functions. Using well-characterized mutations, we showed that generation of most of the HR products depended on Rad51 and Rad54, as expected [22]. A minority of products appeared to have arisen from an alternative, single-strand annealing (SSA) pathway. Since SSA requires ends on both the donor and the target, this class was eliminated when the donor was presented as a circle.
Eliminating Lig4 sharply reduced the number of NHEJ products, and was accompanied by a rise in HR products [22]. Surprisingly, when both Rad51 and Lig4 were disabled, the frequency of NHEJ events returned to approximately wild type levels. In the absence of Lig4, the NHEJ sequence alterations were somewhat different from those recovered in its presence [22, 23]. Rather common products of blunt end joining were eliminated in the lig4− background. Deletions were somewhat more common and larger without Lig4, and there was a slight increase in use of microhomologies at the junctions. Elimination of Lig4 in other systems causes a larger shift to the use of microhomologies. In the nematode, Caenorhabditis elegans, we found that lig4 mutants produced very few NHEJ products at all [24]. This highlights the fact that even common components may behave differently in various systems.
The initial experiments in Drosophila were done with a rather complex method for delivering ZFNs and donor DNA [25]. We found that the effect of mutating lig4 was reproduced in a simpler, direct embryo injection protocol [23] and was effective for both ZFN- and TALEN-mediated targeting (Figure 4). Thus, when HR products are desired, a lig4 mutant host strain can be used to favor that outcome. Lig4 is dispensable in Drosophila. The flies are not completely healthy, but are viable and fertile. This is not the case in many other organisms. For example, a lig4 knockout is lethal in mouse embryos [26]. To take advantage of Lig4 depletion in such cases either a conditional mutation or a small molecule inhibitor will be needed.
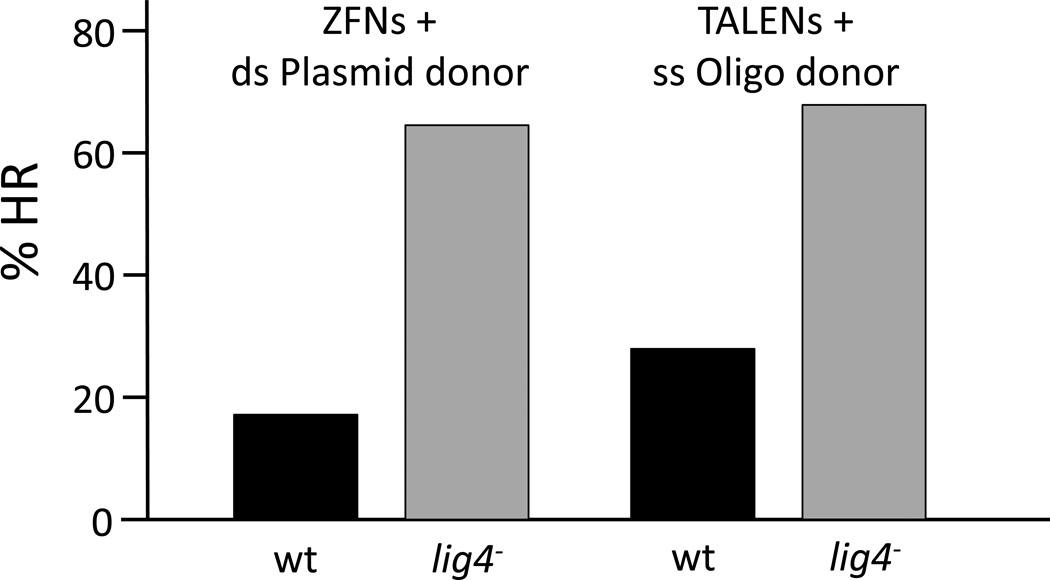
Figure 4.
Effect of lig4 mutation on ratio of NHEJ and HR repair. Nucleases for targets in the rosy (ry) gene were introduced into wild type (wt) or lig4 mutant Drosophila embryos by mRNA injection, along with appropriate donor DNAs. New germline ry mutants were recovered among offspring of the injected flies, and they were characterized by molecular analysis as resulting from HR or NHEJ. The data on the left are for ZFNs and double-stranded plasmid donors with several kb of homology to the target and are the average of 11 separate experiments. The data on the right are for a TALEN pair injected with a 111-nucleotide single-stranded oligonucleotide donor. These data are selected from more extensive experiments reported in references [29] and [32].
3.2. Homology requirements
Repair by HR requires good sequence matching between the donor DNA and the target. In the absence of target cleavage in mammalian cells, longer homologies, up to 10 kb and beyond, increase the frequency of recombination [27]. With target cleavage, however, shorter matches are quite effective. In mammalian cells, although no systematic study has been done, several hundred base pairs on either side of the break are sufficient to support HR with full efficiency – e.g., [28].
We explored homology requirements in Drosophila. We should note that we typically use donor DNAs that carry sequence changes that prevent nuclease recognition and recleavage, but this may not be necessary. We found that double-stranded DNAs injected into embryos as linear molecules were not effective as donors [23], but circular donors worked well. In this context, total homology of 2 kb was required for full efficiency, both in wild type and lig4 mutant flies [29]. Without lig4, homologies of 0.5 and 1.0 kb were nearly half as effective as longer ones, but donors with only 200 bp of homology were totally ineffective. As long as the donor carried homology close to at least one end at the nuclease-induced break, large insertions and deletions were readily introduced at the target [29]. This suggests that little degradation takes place at the 3’ ends at the break, and one end must lie very close to donor homology to initiate successful SDSA repair.
The issue of linear vs. circular donors is an interesting one. Upon injection into Drosophila embryos, we speculate that linear molecules are either degraded by exonucleases or converted by end joining activities into long, linear concatemers that are ineffective as donors [23]. In other systems, the fate of linear or circular donors will depend on the mode of delivery and the presence of activities that modify DNA configuration. In mammalian cells, for instance, one cannot be certain that the form introduced is the one maintained. End ligation of transfected linear molecules certainly occurs, and circular DNAs can be broken during transit to the nucleus [30].
An approach that has not been explored adequately is attempting to enhance HR by providing increased concentrations of required activities. At present we do not know what factors might be limiting – Rad51 or exonuclease, perhaps. Beyond increasing the levels of endogenous factors, one might consider providing components of simpler recombination systems. For example, recombineering in bacteria makes use of proteins from the bacteriophage λ Red recombination system [31], and these factors might help in some eukaryotes as well.
3.3. Oligonucleotide donors
In Drosophila, as in other systems, synthetic, single-stranded DNA oligonucleotides have proven to be effective donors for HR [29, 32]. The mechanism and requirements in this case are likely to be different, since invasion of duplex is not involved and homologies are shorter; but no one has investigated this directly. In mammalian cells, oligos as short as 40 nucleotides work well [33], but lengths in the range of 100 nt are more common in most applications. While donors this short can only introduce changes over their length, they can carry short functional sequences, like epitope coding sequences, integrase landing sites and/or restriction enzyme recognition sites to help with later analysis. In addition, oligos with one “foot” matching a sequence at the break and the other matching sequence some distance away will stimulate deletion of the region between those homologies [34]. Deletions as large as tens of kb have been produced by this approach [33].
A few mismatches in the homologies with the target do not significantly reduce HR frequency, and the donor polymorphisms are often found in the target [29]. As with long double-stranded donors, the absence of Lig4 encourages repair by HR with oligo donors [29, 32]. In our experience, the oligo can match either strand of the target equally effectively [29].
The products of oligo-mediated HR should be examined carefully in any new application. In Drosophila and in mammalian cells, clean homologous repair seems to be operating. In zebrafish embryos, however, the products often appear to be homologous on one end, but ragged, or nonhomologous on the other [35]. This may be a consequence of the dominance of NHEJ in this developmental stage, and similar outcomes might be encountered elsewhere.
Oligos are commonly provided in single-stranded form. As noted above, long linear double-stranded DNAs are not effective donors. We have recovered rare HR products from injections of double-stranded oligos [29], but they may have been produced by an excess of one single strand. In mammalian cells, double-stranded oligos are captured at nuclease cleavage sites by dual NHEJ [36], but this has not been reported in Drosophila.
3.4. Conversion tracts
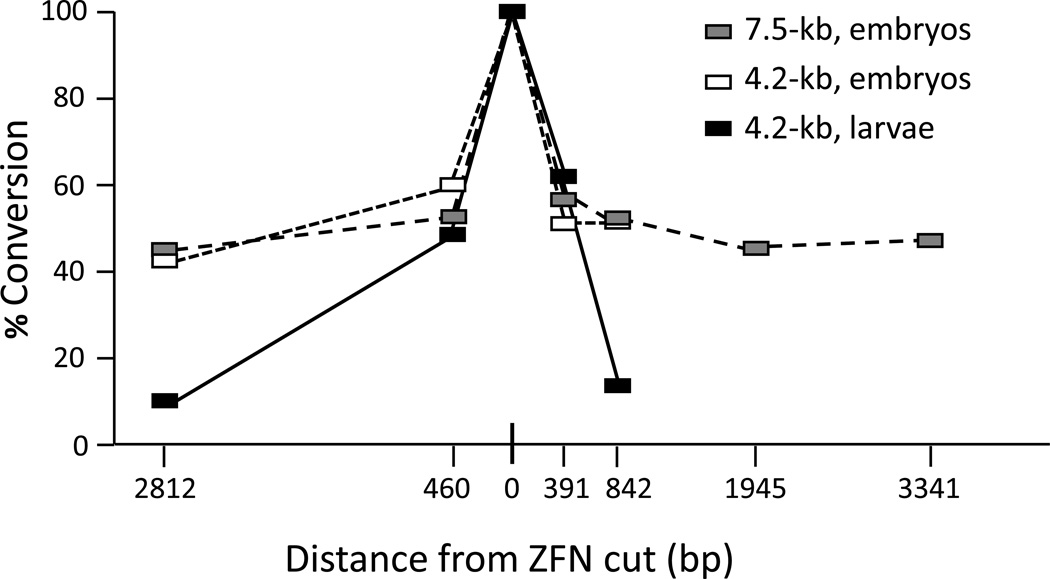
A key issue in HR repair is how much donor sequence is transferred to the target. Recombination initiates at one end at the break and travels some distance into the donor (see Figure 3). How far synthesis goes and how that new sequence replaces target sequence determines what are called conversion tracts. We tested this parameter in Drosophila in two different experimental protocols [29]. In the larval germ line after induction of ZFN expression and donor excision, transfer of donor sequence fell by half about 500 bp on either side of the break and continued to decrease over longer distances (Figure 5), similarly to earlier results with breaks induced by P element excision [37, 38]. In the embryo injection protocol, however, conversion frequency was remarkably still at 50% 3 kb or more from the break.
Figure 5.
Conversion tracts during homologous recombination in Drosophila. % Conversion indicates the proportion of HR products that had donor sequences in the genomic target at each of several locations at various distances from the targeted ZFN cut site. The value at the cut site is necessarily 100%. Three different experiments are illustrated: The ZFNs were induced in larvae along with excision of a 4.2-kb donor, or ZFN mRNAs were injected into embryos along with circular, double-stranded 4.2-kb or 7.5-kb donors. Modified from [29].
When conversion tracts are long, as in Drosophila embryos, a single nuclease can be used to introduce sequence changes throughout an extensive genomic target, simply by providing different donors. In cultured mammalian cells, unfortunately, conversion tracts are very short, and donor sequences more than 100–200 bp from the break are rarely recovered in the target [39]. This means that the cleavage reagent must be designed to cut quite close to the desired conversion site. For example, to produce a translational fusion at the N- or C-terminus of a natural protein, nuclease cleavage must be directed to a site very close to the corresponding genomic coding sequences. It would be very valuable to know what limits conversion tracts in mammalian cells or what makes them long in Drosophila embryos.
4. Donor design
Based on the above considerations, we can offer recommendations on constructing donor DNAs (Table 1). These principles apply directly to experiments with Drosophila, but they are generalizable to some extent to other organisms and situations.
Of course, if all you want is a targeted sequence change that is likely to disrupt the function of the gene is question, you need only design one of the nuclease types to a promising site in that gene. NHEJ repair after cleavage will produce the desired result. The specific cleavage target will be determined by the particular situation, but for protein coding sequences, a target near the 5’ end of the gene may be best.
For specific, very local sequence changes, a synthetic oligonucleotide donor will often suffice. This requires no cloning, and silent coding changes can be introduced to prevent recleavage after HR. Of course, the oligo must match the target on both sides of the cut.
When larger changes are required, a double-stranded circular donor with at least 1–2 kb of total homology to the target is preferred. Sequences very close to one end of the break at the target must be homologous to the donor for effective initiation of HR. Using this approach, gene-sized insertions and deletions are readily copied from the donor into the target.
Using a long donor, both subtle and substantial sequence changes can be introduced to the target even at distances several kb from the nuclease-induced break. Thus, a single donor can be used to introduce multiple changes simultaneously, or separate donors can be used to alter sequences throughout a locus one at a time. Conversion frequency decreases to some extent with distance from the break, so candidate products must be checked carefully.
Both oligo donors and plasmid donors can be used to produce large deletions, when they carry homologies to sequences at the ends of the desired deletion. Indications so far are that one end at the break must match oligo donor sequences [33], just as with the longer double-stranded donor.
Large deletions can also be produced by NHEJ, relying on expression of two nucleases targeted to the ends of the desired deletion [32]. Inversions of the intervening DNA have also been observed with this approach in mammalian cells [40].
Table 1.
Experimental design and outcomes of nuclease-mediated engineering
| Desired change | Donor | Repair |
|---|---|---|
| Simple knockout | None | NHEJ |
| Specific knock-in | ssOligo | HR |
| Long dsDNA | HR | |
| Long insertion | Long dsDNA | HR |
| Short insertion | Long dsDNA | HR |
| dsOligo | NHEJ | |
| Long deletion | Long dsDNA | HR |
| ssOligo | HR | |
| None – 2 nucleases | NHEJ | |
| Inversion | None – 2 nucleases | NHEJ |
In each case a targeted nuclease is assumed – a ZFN pair, a TALEN pair, or Cas9 protein plus a guide RNA. More detail is provided in the text, section 4.
Abbreviations: ss, single-stranded; ds, double-stranded; NHEJ, nonhomologous end joining; HR, homologous recombination.
5. Final comments
Knowledge of DNA repair functions can guide the design of nuclease-mediated genome engineering projects to enhance the desired outcome. Providing donor DNAs for HR repair in the proper format and with adequate homology to the target is one example. Repair capabilities and preferences will differ among experimental organisms, cell types and situations. In favorable circumstances, repair activities can be manipulated to promote specific processes. As targetable nucleases are applied to more and more demanding engineering tasks, these considerations will take on greater importance.
Acknowledgments
We are grateful to the people in our lab, who have worked on targetable nucleases over many years, and to our collaborators, who have enriched our experience with introductions to a variety of organisms and scientific questions. Our research has been supported by grants from the NIH, most recently R01 GM078571.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carroll D. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porteus MH, Carroll D. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 3.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 4.Bogdanove AJ, Voytas DF. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 5.Joung JK, Sander JD. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sampson TR, Weiss DS. Bioessays. 2014;36:34–38. doi: 10.1002/bies.201300135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terns RM, Terns MP. Trends Genet. 2014;30:111–118. doi: 10.1016/j.tig.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll D. Ann. Rev. Biochem. 2014;83 doi: 10.1146/annurev-biochem-060713-035418. in press. [DOI] [PubMed] [Google Scholar]
- 9.Gaj T, Gersbach CA, Barbas CF., 3rd Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segal DJ, Meckler JF. Annu Rev Genomics Hum Genet. 2013;14:135–158. doi: 10.1146/annurev-genom-091212-153435. [DOI] [PubMed] [Google Scholar]
- 11.Chapman JR, Taylor MR, Boulton SJ. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 12.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. Cell Reports. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boboila C, Alt FW, Schwer B. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 15.Decottignies A. Front Genet. 2013;4:48. doi: 10.3389/fgene.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehman CW, Trautman JK, Carroll D. Nucleic Acids Res. 1994;22:434–442. doi: 10.1093/nar/22.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merrihew RV, Marburger K, Pennington SL, Roth DB, Wilson JH. Mol Cell Biol. 1996;16:10–18. doi: 10.1128/mcb.16.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu AM, McVey M. Nucleic Acids Res. 2010;38:5706–5717. doi: 10.1093/nar/gkq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Symington LS. Microbiol. Mol. Biol. Revs. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jasin M, Rothstein R. Cold Spring Harb Perspect Biol. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beumer KJ, Carroll D. Methods. 2014 [Google Scholar]
- 22.Bozas A, Beumer KJ, Trautman JK, Carroll D. Genetics. 2009;182:641–651. doi: 10.1534/genetics.109.101329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beumer KJ, Trautman JK, Bozas A, Liu JL, Rutter J, Gall JG, Carroll D. Proc Natl Acad Sci U S A. 2008;105:19821–19826. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morton J, Davis MW, Jorgensen EM, Carroll D. Proc Natl Acad Sci U S A. 2006;103:16370–16375. doi: 10.1073/pnas.0605633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bibikova M, Beumer K, Trautman JK, Carroll D. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 26.Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 27.Deng C, Capecchi MR. Mol Cell Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Urnov FD, Miller JC, Lee Y-L, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 29.Beumer KJ, Trautman JK, Mukherjee K, Carroll D. G3 (Bethesda) 2013;3:657–664. doi: 10.1534/g3.112.005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wake CT, Vernaleone F, Wilson JH. Mol Cell Biol. 1985;5:2080–2089. doi: 10.1128/mcb.5.8.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Nat Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beumer KJ, Trautman JK, Christian M, Dahlem TJ, Lake CM, Hawley RS, Grunwald DJ, Voytas DF, Carroll D. G3. 2013;3:1717–1725. doi: 10.1534/g3.113.007260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, Collingwood TN, Frodin M, Davis GD. Nat Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O'Connor-Giles KM. Genetics. 2013;194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, Tan W, Penheiter SG, Ma AC, Leung AY, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orlando SJ, Santiago Y, DeKelver RC, Freyvert Y, Boydston EA, Moehle EA, Choi VM, Gopalan SM, Lou JF, Li J, Miller JC, Holmes MC, Gregory PD, Urnov FD, Cost GJ. Nucleic Acids Res. 2010;38:e152. doi: 10.1093/nar/gkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gloor GB, Nassif NA, Johnson-Schlitz DM, Preston CR, Engels WR. Science. 1991;253:1110–1117. doi: 10.1126/science.1653452. [DOI] [PubMed] [Google Scholar]
- 38.Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee HJ, Kweon J, Kim E, Kim S, Kim JS. Genome Res. 2012;22:539–548. doi: 10.1101/gr.129635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]