Supplemental Digital Content is Available in the Text.
Key Words: broadly neutralizing antibodies, HIV, vaccine, coreceptor tropism
Abstract
Background:
Recently discovered broadly neutralizing antibodies have revitalized hopes of developing a universal vaccine against HIV-1. Mainly responsible for new infections are variants only using CCR5 for cell entry, whereas CXCR4-using variants can become dominant in later infection stages.
Methods:
We performed a statistical analysis on two different previously published data sets. The first data set was a panel of 199 diverse HIV-1 isolates for which IC50 neutralization titers were determined for the broadly neutralizing antibodies VRC01, VRC-PG04, PG9, and PG16. The second data set contained env sequences of viral variants extracted from HIV-1–infected humanized mice treated with the antibody PGT128 and from untreated control mice.
Results:
For the panel of 199 diverse HIV-1 isolates, we found a statistically significant association between viral resistance to PG9 and PG16 and CXCR4 coreceptor usage (P = 0.0011 and P = 0.0010, respectively). Our analysis of viral variants from HIV-1–infected humanized mice under treatment with the broadly neutralizing antibody PGT128 indicated that certain antibodies might drive a viral population toward developing CXCR4 coreceptor usage capability (P = 0.0011 for the comparison between PGT128 and control measurement).
Conclusions:
These analyses highlight the importance of accounting for a possible coreceptor usage bias pertaining to the effectiveness of an HIV vaccine and to passive antibody transfer as therapeutic approach.
INTRODUCTION
With more than 2.5 million new HIV-1 infections each year, there is an urgent need for additional protective measures against this virus. A natural approach to preventing HIV infections would be to vaccinate people with a universal HIV vaccine. One alternative is to induce neutralizing antibodies that interfere with the process of viral cell entry like for hepatitis B vaccine. Unfortunately, because of the high genomic diversity of HIV, a universal HIV vaccine that elicits broad neutralization responses against most of the existing HIV strains has not yet been discovered.1 Most recent approaches to isolate broadly neutralizing antibodies from patient sera2–4 have demonstrated promising results. One of these new antibodies, PG9, has been shown to interact with the glycan shield and specific sites of the variable loops 1 and 2 as well as the variable loop 3 (V3)2,5 of the Env protein of the virus. The structure of these loops largely determines which coreceptor the virus can use for entering and infecting new cells. The two coreceptors that are mainly relevant in vivo are the chemokine receptors CCR5 and CXCR4. Viruses that can only bind to the CCR5 coreceptor are called R5 viruses, and viruses that can only use the CXCR4 coreceptor are called X4 viruses. Generally, viruses capable of binding to both coreceptors are called dual-tropic viruses, however, the discrimination of dual-tropic and X4 viruses is difficult to make both by genotypic and (commercially available) phenotypic assays. Here, we refer to X4-capable viruses for both, X4 viruses and dual-tropic viruses. These have very low capacity of newly infecting humans—at least by sexual transmission—as evidenced by the highly significant underrepresentation of individuals with a homozygous Δ32 variant of the CCR5 structural gene among seropositive white individuals.6 These people lack a functional CCR5 coreceptor and therefore cannot be infected by R5 viruses. The fact that only very few individuals with this mutation were HIV seropositive as compared with the mutation frequency in the population indicates that R5 viruses might be more relevant regarding new infections than X4-capable viruses. Thus, a vaccine needs to elicit responses against R5 viruses, but responses against X4-capable viruses are less important for preventing HIV infection.
Beyond using knowledge about broadly neutralizing antibodies for vaccine design, it might be possible to effectively treat HIV-1–infected patients with a combination of these antibodies. Klein et al7 showed that HIV-1–infected humanized mice could be effectively treated by a combination of broadly neutralizing antibodies and proposed to re-examine this approach as a treatment modality in HIV-1–infected patients. Recent studies in SHIV-infected Rhesus macaques also yielded promising results.8,9
In this work, we show that there is a significant association between CXCR4 coreceptor usage and resistance to PG9 and PG16.2 We provide evidence that this may have important implications both for vaccination approaches and for therapeutic approaches: The first implication is while configuring a panel assessing neutralization capacities of antibodies against HIV-1, one should take coreceptor usage of the strains into account for an unbiased evaluation of an antibody's capacity to neutralize strains that can establish an infection. The second implication is that certain antibodies such as PGT128, which was one of the antibodies Klein et al7 used in their study, might drive the viral population toward developing CXCR4 coreceptor usage capability. This is supported by our analysis on data from their study showing a significant difference between coreceptor usage of the variants emerging under treatment with PGT128 as compared with the control variants at similar time points. Treatment with a combination of such antibodies might therefore be problematic in patients with weak immune systems harboring a substantially higher risk of carrying X4-capable viruses.10 Therefore, it seems advantageous to place similar restrictions on treatment with neutralizing antibodies causing the selection of X4-capable viruses as those already in use for CCR5 antagonists. We suggest that this issue be investigated further before testing these potent monoclonal antibodies in this group of people.
MATERIALS AND METHODS
To investigate the association between coreceptor usage and neutralization capabilities of specific broadly neutralizing antibodies, we evaluated coreceptor usage on a large HIV-1 isolate panel introduced recently.11 In that study, the neutralization capabilities of the broadly neutralizing antibodies VRC01, VRC-PG04, PG9, and PG16 were evaluated against a panel of 208 different HIV isolates from different subtypes, also including recombinant forms. Env sequences that covered the V3 region were available for only 199 of the 208 strains (see additional file 2, http://links.lww.com/QAI/A550). The coreceptor usage of these 199 strains was determined with the widely used prediction tool geno2pheno[coreceptor].12 The prediction model is based on the V3 sequence only and has been shown to have good performance13 predicting both, phenotypic tropism and clinical response to treatment with R5 antagonists. geno2pheno[coreceptor] uses a linear support vector machine to predict whether a sequence is from an X4-capable or an R5 virus. The distance of the sequence to the separating hyperplane is transformed into a false-positive rate (FPR) to facilitate interpretability in terms of the confidence of the prediction. geno2pheno[coreceptor] reports the minimal FPR at which the sequence would be classified as X4-capable. In this calculation, X4-capable viruses are considered positive samples and R5 viruses are considered negative samples. It is general clinical practice to determine coreceptor usage based on this notion of FPR and on expert knowledge. In Europe, interpretation of tropism by geno2pheno[coreceptor] has been included in the guidelines for determining HIV-1 coreceptor tropism.14 We adhered to the 10% FPR cutoff to distinguish X4-capable viruses from R5 viruses as recommended by the European Consensus Group on clinical management of HIV-1 tropism testing.14 With the cutoffs of 5% and 15% as recommended by the German treatment guidelines, we achieved similar results. These guidelines suggest to classify all viruses with an FPR of less than 5% as X4-capable and all viruses with an FPR of 15% or more as R5 disregarding the viruses with FPRs between 5% and 15%. The numbers of sensitive and resistant strains, respectively, for the different antibodies using the 5%/15% cutoffs are shown in the Supplemental Digital Content (see Table S1, Supplemental Digital Content, http://links.lww.com/QAI/A550).
For 15 of the 199 isolates from the test panel, tropism information from biological experiments was available in the Los Alamos HIV database (see Table S2, Supplemental Digital Content, http://links.lww.com/QAI/A550). Using the 10% FPR threshold for geno2pheno[coreceptor], 13 of 15 HIV variants are assigned the same tropism label. The two discordances are for borderline cases (between 5% and 15% FPR), but they do not change the numbers calculated from data presented in Table 1 because both variants are resistant to PG9/PG16 and tropisms are R5 and X4-capable, respectively. Note that the analysis in the Supplemental Digital Content does not include variants in the borderline region and still finds a similarly strong association between coreceptor tropism and PG9/PG16 resistance (see Table S1, Supplemental Digital Content, http://links.lww.com/QAI/A550).
TABLE 1.
Coreceptor Usage of HIV Variants Resistant and Susceptible to 4 Different Broadly Neutralizing Antibodies
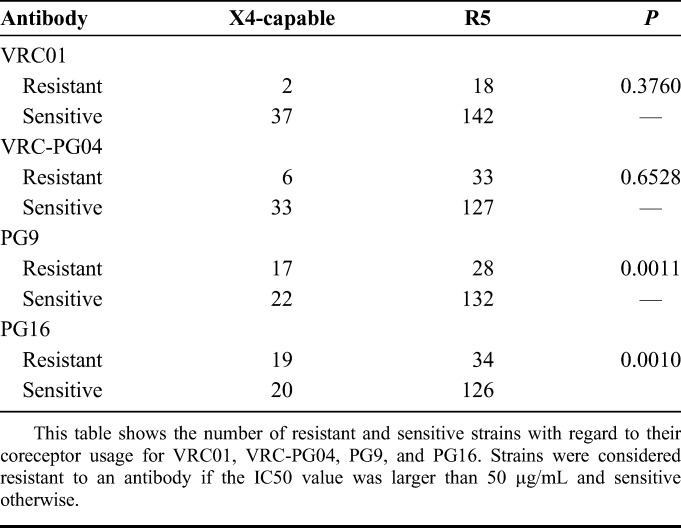
To test whether the sensitivity/resistance to an antibody or serum is significantly different with regard to viral tropism, we computed P values using the two-sided Fisher exact test for the two-by-two contingency tables with resistant/sensitive as the row label and X4-capable/R5 as the column label using significance level α = 0.05. The null hypothesis is that there is no difference.
To investigate which amino acids at which position of the Env protein are associated with PG9 sensitivity, we performed the Fisher exact test for all observed amino-acid polymorphisms at all sequence locations. The q values were calculated according to Storey and Tibshirani15 by conservatively assuming π0 to be equal to 1. We performed the same analysis also for finding associations between observed amino-acid polymorphisms and predicted coreceptor usage (with the 10% FPR threshold introduced above).
To calculate the potential N-glycosylation sites, we used the tool N-glycosite.16 The numbering of the positions is according to the Env amino-acid sequence of the HIV strain HXB2.
For comparing the effect of treatment by passive antibody transfer with broadly neutralizing antibodies, we computed the coreceptor usage FPR values for each variant of each treatment using geno2pheno[coreceptor].12 For each comparison between treatments (e.g., PGT128 versus 45-46GW), we tested whether there is a significant difference between the 2 sets of FPRs according to a Wilcoxon rank-sum test at a significance level of α = 0.05.
RESULTS AND DISCUSSION
Neutralization Capabilities of PG9 and PG16 Are Biased Toward R5 Viruses
The glycan shield of HIV-1 differs considerably between R5 viruses and X4-capable viruses with fewer glycans being attached to X4-capable viruses. Additionally, there are specific sites in V3 that are highly predictive of coreceptor usage. Therefore, we evaluated whether there is a significant difference between the viruses that can be neutralized by PG9 (sensitive strains) and viruses that cannot be neutralized by PG9 (resistant strains) with regard to their coreceptor usage (viral tropism) based on data from a recent neutralization study.11 We found a statistically significant difference (P = 0.0011) between the sensitive and the resistant strains according to the Fisher exact test, with a higher fraction of X4-capable viruses in the group of resistant strains. This means that a significantly higher percentage of HIV strains resistant to PG9 are X4-capable than expected by chance. An analogous analysis for the broadly neutralizing antibody PG16, which is known to bind to variable loops 1 and 2 as well as to V3, led to a similarly strong P value (0.0010). We call this property of an antibody to neutralize R5 viruses more effectively than X4-capable viruses R5-bias from now on. For comparison, the same evaluation for the broadly neutralizing antibodies VRC01 and VRC-PG04 that bind to the CD4 binding site did not show a significant association (P = 0.3760 and P = 0.6528, respectively). The exact numbers are given in Table 1. Only 56% and 51%, respectively, of X4-capable viruses could be neutralized by PG9 and PG16, compared with 95% and 85%, respectively, of X4-capable viruses that could be neutralized by VRC01 and VRC-PG04. This implies that the higher the fraction of X4-capable viruses in the panel, the worse will antibodies with an R5-bias perform, unless coreceptor usage of the viral strains is taken into account. Analyzing only R5-tropic strains, the percentage of strains sensitive to PG9 increased from 77% to 83% and from 73% to 79% for PG16, whereas it remained at similar levels for VRC01 (89% instead of 90%) and VRC-PG04 (80% for both evaluations). Therefore, when evaluating the breadth of antibodies with regard to preventing new HIV infections, the coreceptor usage of the viruses should be taken into account. Because R5 viruses are driving primary HIV infections almost exclusively, the unknown use of X4-capable viruses in a panel may lead to underestimation of the breadth of neutralizing capacities of R5-biased antibodies.
Positions in the V3 Loop Are Predictive of PG9 Resistance
Additionally, we investigated whether certain amino acids at specific positions of the Env protein are associated with sensitivity/resistance to PG9 as described in the Methods section. The most significant association was observed between an asparagine at position 160 of the Gp120 protein (P = 1.90e−10) and PG9 sensitivity. This association is supported by a recent structural analysis by McLellan et al,5 which found that the N-linked glycan at position 160 is critical for PG9 recognition. They also hypothesized that N-linked glycans at positions 156 and 173 are important for PG9 recognition. The frequencies of potential N-linked glycosylation sites (PNG) among sensitive and resistant viruses, respectively, for the region between positions 152 and 173 are shown in Figure 1. All sensitive viruses had a PNG at position 160, whereas only about 62% of the PG9-resistant viruses possessed a PNG at that position. There was no significant difference between PNG frequencies at positions 156 and 173 for sensitive and resistant viruses. The association analysis on the complete Env protein sequence resulted in 4 associations that were significant after correction for multiple testing (q < 0.05). Additional to the asparagine at position 160, we found a lysine at position 432 (P = 8.27e−06), a valine at position 372 (P = 1.38e−05), and a glutamine at position 315 (P = 2.07e−05) to be significantly associated with PG9 sensitivity/resistance. This supports the hypothesis that more positions than just the ones responsible for the glycan interaction are important for determining sensitivity to PG9. Furthermore, there were 27 PG9-resistant viruses in the panel, for which the PNGs at position 160 and either at position 156 or at position 173 could be observed. This underlines that viruses can be resistant to PG9 although they possess the N-glycosylation sites important for PG9 binding discovered by McLellan et al.5 Another association, a threonine at position 319 with P value of 0.0001 came out just slightly above the q value threshold (q value = 0.055). Positions 315 and 319 are located in the V3 region of HIV and are highly predictive of coreceptor usage (arginine at position 315: P = 2.80e−05; and alanine at position 319: P = 1.74e−08). This supports the hypothesis that there is a relation between PG9 sensitivity and coreceptor usage. A similar but slightly weaker association could be observed for PG16 shown in the Supplemental Digital Content (see Appendix S1, Supplemental Digital Content, http://links.lww.com/QAI/A550).
FIGURE 1.
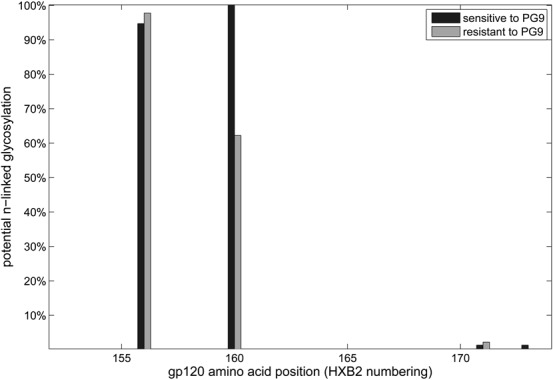
Differences in glycosylation between HIV strains that are sensitive and resistant to PG9, respectively. All viruses sensitive to PG9 had a PNG at position 160 (black), whereas this was not the case for viruses resistant to PG9 (gray).
PGT Antibodies Might Have an Even Stronger R5-Bias and Could Drive HIV Variants Toward Developing CXCR4-Usage Capability
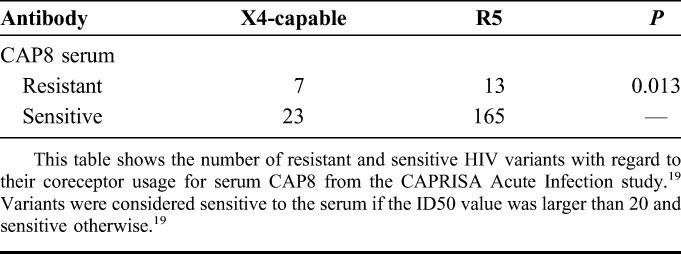
It has been shown that the highly potent PGT-neutralizing antibodies interact with V3 sites 323, 324, 325, and 327 as well as N-linked glycans at position 301 and/or 332.17,18 Thus, these antibodies might also exhibit stronger neutralization capabilities against R5 viruses than against X4-capable viruses. In previous studies, many of these antibodies (e.g., PGT128 and PGT121) were almost 10-fold more potent than PG9, PG16, and VRC01, but their neutralization capabilities were less broad. Unfortunately, Walker et al17 do not provide the env sequences from the test panel, which is why we could not predict coreceptor usage for the test panel they used. However, we could test for an R5-bias on neutralization data from sera of patients of the CAPRISA Acute Infection study with PGT-like neutralization potential.19 For one of the sera, the authors showed computationally and through a mutation study that position 316 was highly relevant for neutralization. This was the only serum for which they showed that there was an important position in the V3 loop. We tested whether there is a significant difference between the viruses that can be neutralized by CAP8 serum (sensitive strains) and viruses that cannot be neutralized by CAP8 serum (resistant strains) with regard to their coreceptor usage (viral tropism) as described in the Methods section and found a significant difference (P = 0.013). The counts can be found in Table 2. The P value is slightly larger than the P values for the tests between PG9/PG16 resistance and viral tropism described above, but this might be due to the use of whole sera to test the neutralization capability instead of monoclonal antibodies. Likely, some of the sera used contain antibodies with an R5-bias and additional antibodies that are able to neutralize X4-capable viruses reducing in the overall R5-bias of the serum. One could also test for the effect of PGT121 using the test panel of Mouquet et al20; but because the number of viruses in this panel is 119 compared with 208 and 225 in the other two panels, it could be that the sample size is too small to reach significance.
TABLE 2.
Coreceptor Usage of HIV Variants Resistant and Sensitive to CAP8 Serum According to the 10% FPR Cutoff
Additionally, we could test whether PGT128 exhibits R5-bias on data from Klein et al.7 In their study, the researchers monitored HIV-infected humanized mice that were under treatment with broadly neutralizing antibodies for several weeks. This monitoring included the sequencing of the viral populations at various time points (between 6 and 80 days after beginning of treatment). Because all mice were infected with the same HIV variant, we could test whether there was a significant difference in the coreceptor usage of the variants emerging after beginning of treatment as described in the Methods section. When comparing PGT128 with 45-46GW, which is an antibody that binds to the CD4 binding site, we found a significant difference (P = 0.0008) with a trend toward CXCR4-usage for the variants extracted from the mice treated with PGT128. A similar result could be found for the comparison between PGT128 and the control measurement (P = 0.0011). The basis for this analysis were the V3 loop sequences of 32, 84, and 66 HIV variants extracted from the PGT128-treated mice, the 45-46GW-treated mice, and the control mice, respectively. For the mice treated with the penta-mix that also contained PGT128, we could not find a similar trend, but this might be due to the lower viral load of these mice throughout the course of treatment indicating a higher antiviral efficacy of the combined antibody treatment. Note that PGT128 was not contained in the tri-mix that Klein et al. tested.
We could not find a significant difference between treatment with PG16 and treatment with 45-46GW (also not compared with the control measurements). This may be due to the lower antiviral efficacy of PG16 in comparison with PGT12817 resulting in a lower selective pressure, such that the time interval of treatment was too short for escape variants to emerge. Alternatively, the R5-bias itself might be stronger for PGT128 than for PG16. This hypothesis deserves further investigation.
CONCLUSION
We have shown that PG9 and PG16 neutralize a substantially higher percentage of strains when excluding X4-capable viruses, whereas there is no such effect for the CD4-binding site antibodies VRC01 and VRC-PG04. Additionally, we could show that certain amino acids at specific positions of the Env protein are significantly associated with PG9 sensitivity/resistance. We observed the already known relevance of the amino acid asparagine at position 160 in this context, but we also discovered previously unknown associations. Some of these amino-acid variants are also significantly associated with coreceptor usage, supporting the hypothesis that resistance to the broadly neutralizing antibodies PG9 and PG16 can be associated with coreceptor usage. Because neutralization of X4-capable viruses is less important for preventing new infections, further HIV vaccine studies should account for coreceptor usage of the viral populations in the target isolates, to avoid underestimating the power of R5-biased broadly neutralizing antibodies in this setting. It would also be worth evaluating whether there are antibodies with a bias against HIV variants harboring other properties related to be more or less relevant in HIV primary infection, such as low-level monocyte-derived macrophages replication, because enhanced monocyte-derived macrophage replication is related to mortality among infected patients.21
It has to be noted that our coreceptor assignments were based on predictions of the tool geno2pheno[coreceptor]12 instead of phenotypic assays such as Trofile or ESTA.22 Genotypic coreceptor usage tests are general practice in several regions of the world for the determination of viral tropism. For Europe, Sanger sequencing of the V3 loop in combination with geno2pheno[coreceptor] was included into the guidelines for HIV-1 coreceptor tropism determination,14 underlining the confidence in this method. In recent reanalyses of the early maraviroc clinical trials, genotypic methods were at least as clinically predictive as Trofile.23 Finally, for sequences of the panel for which phenotypic information is available from biological experiments, there was a high agreement between the phenotypic assay and the bioinformatic prediction method (see Table S2, Supplemental Digital Content, http://links.lww.com/QAI/A550).
It has been shown recently that viral escape from glycan-dependent broadly neutralizing antibodies can lead to changes in glycosylation patterns of HIV.24 We have shown in our analysis that PGT128 might drive the HIV population of infected individuals faster toward developing CXCR4 coreceptor usage capability. It can be expected that similar antibodies to PGT128 have a comparable effect. Of note, there is no clear evidence to believe that X4-capable viruses induced by an R5 virus-blocking treatment are causing faster disease progression. A careful strategy would be either to exclude antibodies with an R5-bias from or to include an antibody with good neutralization capabilities against X4 viruses into antibody treatment combinations to reduce the chance of a coreceptor switch. This might not be necessary in pre-exposure prophylaxis settings, where it can be expected that enhancing the immune system's capability of neutralizing R5 viruses is much more important than supporting the neutralization of X4-capable viruses.25
Furthermore, the finding that PGT128 can drive coreceptor switch adds to the discussion about why certain HIV-infected patients never develop X4-capable viruses. It could be that a certain antibody response against parts of the V3 loop is necessary for a coreceptor switch or at least makes it more likely.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Nicole Doria-Rose and John Mascola for providing Genbank accession numbers for env sequences and Ari Halper-Stromberg and Florian Klein for providing the env sequences of the HIV-1 variants from HIV-1–infected mice.
Footnotes
Supported by the BMBF project HIV Cell Entry, grant no. 0315480A. N.P. was an employee of Microsoft until 8/2011. The Max Planck Institute for Informatics collaborates with a company in the area of medical diagnostics on the topic of viral tropism of HIV. In its context, a version-controlled implementation of geno2pheno[coreceptor] with secure access is maintained by Max Planck Institute for Informatics under a paid service agreement.
N.P. designed the study, performed the experiments and interpreted the results. T.L. contributed ideas to the design and interpretation of the study. H.W. contributed ideas to the interpretation of the study. N.P., H.W., and T.L. wrote and approved the manuscript.
The authors have no further conflicts of interest to disclose.
Part of the data was presented at the AIDS Vaccine Conference 2013, October 7-10, 2013, Barcelona, Spain.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.jaids.com).
REFERENCES
- 1.Virgin HW, Walker BD. Immunology and the elusive AIDS vaccine. Nature. 2010;464:224–231 [DOI] [PubMed] [Google Scholar]
- 2.Walker LM, Phogat SK, Chan-Hui PY, et al. Broad and potent neutralizing antibodies from an African Donor Reveal a new HIV-1 vaccine target. Science. 2009;326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu X, Yang ZY, Li Y, et al. Rational design of envelope Identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton DR, Poignard P, Stanfield RL, et al. Broadly neutralizing antibodies Present new prospects to counter highly Antigenically diverse viruses. Science. 2012;337:183–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLellan JS, Pancera M, Carrico C, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean M, Carrington M, Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a Deletion Allele of the CKR5 structural gene. Science. 1996;273:1856–1862 [DOI] [PubMed] [Google Scholar]
- 7.Klein F, Halper-Stromberg A, Horwitz JA, et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012;492:118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barouch DH, Whitney JB, Moldt B, et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shingai M, Nishimura Y, Klein F, et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013;503:277–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brumme ZL, Goodrich J, Mayer HB, et al. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J Infect Dis. 2005;192:466–474 [DOI] [PubMed] [Google Scholar]
- 11.Doria-Rose NA, Louder MK, Yang Z, et al. HIV-1 neutralization coverage is improved by combining monoclonal antibodies that target independent epitopes. J Virol. 2012;86:3393–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lengauer T, Sander O, Sierra S, et al. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol. 2007;25:1407–1410 [DOI] [PubMed] [Google Scholar]
- 13.Prosperi MC, Bracciale L, Fabbiani M, et al. Comparative determination of HIV-1 co-receptor tropism by Enhanced Sensitivity Trofile, gp120 V3-loop RNA and DNA genotyping. Retrovirology. 2010;7:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandekerckhove LP, Wensing AMJ, Kaiser R, et al. European guidelines on the clinical management of HIV-1 tropism testing. Lancet Infect Dis. 2011;11:394–407 [DOI] [PubMed] [Google Scholar]
- 15.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Gaschen B, Blay W, et al. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14:1229–1246 [DOI] [PubMed] [Google Scholar]
- 17.Walker LM, Huber M, Doores KJ, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pejchal R, Doores KJ, Walker LM, et al. A potent and broad neutralizing antibody Recognizes and Penetrates the HIV glycan shield. Science. 2011;334:1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lacerda M, Moore PL, Ngandu NK, et al. Identification of broadly neutralizing antibody epitopes in the HIV-1 envelope glycoprotein using evolutionary models. Virol J. 2013;10:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mouquet H, Scharf L, Euler Z, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 2012;109:E3268–E3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuttle DL, Anders CB, Aquino-De Jesus MJ, et al. Increased replication of non-syncytium-inducing HIV type 1 isolates in monocyte-derived macrophages is linked to advanced disease in infected children. AIDS Res Hum Retroviruses. 2002;18:353–362 [DOI] [PubMed] [Google Scholar]
- 22.Reeves J, Han D, Wilkin T, et al. An Enhanced Version of the Trofile HIV Co-receptor Tropsim Assay Predicts Emergence of CXCR4 Use in ACTG5211 Vicriviroc Trial Samples. 15th CROI [Abstract 869]. 2008. Available at: http://www.natap.org/2008/CROI/croi_73.htm. Accessed August 18, 2014 [Google Scholar]
- 23.Swenson LC, Dong WW, Mo T, et al. Use of cellular HIV DNA to predict virologic response to maraviroc: performance of population-based and deep sequencing. Clin Infect Dis. 2013;56:1659–1666 [DOI] [PubMed] [Google Scholar]
- 24.Moore PL, Gray ES, Wibmer CK, et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med. 2012;18:1688–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen MS, Chen YQ, McCauley M, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365:493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


