Abstract
RecQ-like helicases are a highly conserved family of proteins which are critical for preserving genome integrity. Genome instability is considered a hallmark of cancer and mutations within three of the five human RECQ genes cause hereditary syndromes that are associated with cancer predisposition. The human RecQ-like helicase BLM has a central role in DNA damage signaling, repair, replication, and telomere maintenance. BLM and its budding yeast orthologue Sgs1 unwind double-stranded DNA intermediates. Intriguingly, BLM functions in both a pro- and anti-recombinogenic manner upon replicative damage, acting on similar substrates. Thus, BLM activity must be intricately controlled to prevent illegitimate recombination events that could have detrimental effects on genome integrity. In recent years it has become evident that post-translational modifications (PTMs) of BLM allow a fine-tuning of its function. To date, BLM phosphorylation, ubiquitination, and SUMOylation have been identified, in turn regulating its subcellular localization, protein-protein interactions, and protein stability. In this review, we will discuss the cellular context of when and how these different modifications of BLM occur. We will reflect on the current model of how PTMs control BLM function during DNA damage repair and compare this to what is known about post-translational regulation of the budding yeast orthologue Sgs1. Finally, we will provide an outlook towards future research, in particular to dissect the cross-talk between the individual PTMs on BLM.
Keywords: RecQ, BLM, PTM, Replication, SUMO, Ubiquitin, Homologous Recombination (HR), DNA repair, Phosphorylation, Cancer, Sgs1, Genome Integrity
1. Introduction: The RecQ-like helicase BLM regulates multiple diverse cellular processes
Genome integrity is continuously challenged by different types of DNA damage, which are caused by endogenous sources as well as exogenous agents. Accurate and timely DNA repair is crucial to preserve genome integrity and therefore human health, as genome instability is closely associated with cancer predisposition and aging. One family of highly conserved proteins critical for error-free DNA damage repair is the homologs of the bacterial RecQ DNA helicase. While budding yeast encodes two RecQ-like helicases, Sgs1 and Hrq1, five members have been identified in humans: BLM, WRN, RECQL1, RECQL4, and RECQL5. Underlining the cellular importance of RecQ-like helicases in maintaining genome integrity is the fact that mutations within three of the five human RECQ genes (BLM, WRN, RECQL4) lead to distinct heritable diseases that have the common characteristic of cancer predisposition.
In humans, mutations within BLM cause the rare genetic disorder Bloom syndrome (BS). Patients with BS have a strong growth deficiency, develop facial skin lesions after sun-exposure, a moderate immunodeficiency, neurological defects, reduced fertility, amongst other symptoms. The major clinical complications that arise in BS are cancer and diabetes [1]. Most pathologic mutations within BLM result in premature translation termination and mRNA destabilization, resulting in a complete absence of detectable BLM protein levels. A second group of mutations target highly conserved residues within the helicase domain, leading to the expression of a mutant BLM protein which is unable to unwind DNA and thus considered a catalytic null [1]. On a molecular level the genome instability of BS cells is marked by an increased frequency of sister chromatid exchanges (SCE), quadriradial chromosome configurations (Qrs) [2, 3], elevated mutation rates, as well as defects in replication that include a slowed rate of DNA synthesis [4] and the accumulation of aberrant replication intermediates [5]. Understanding the molecular mechanisms of how loss of BLM function is linked to genome instability and in turn cancer predisposition, has been the focus of many studies over the past decades. Significant progress has been made, in part by making use of yeast as a model organism to study RecQ-like helicase function. Importantly, S. cerevisiae Sgs1 is considered to be most homologous to mammalian BLM. sgs1A mutants display several phenotypes very similar to BS cells, such as hyper-recombination and replication defects [6].
BLM and Sgs1 function during multiple cellular processes that require the unwinding of double-stranded DNA, such as double-strand break (DSB) repair by homologous recombination (HR), telomere maintenance, and replication [7, 8]. In wild-type cells DSBs are repaired by two major pathways: through re-ligation of the two broken DNA-ends by non-homologous end joining (NHEJ) or by making use of a homologous template during HR, which is considered the error-free pathway. Even though NHEJ is predominantly used to repair DSBs in human cells, mis-regulation and errors during HR have severe consequences that can lead to genome instability. When a DSB occurs during late S or G2 phases of the cell cycle, it can be repaired by HR, making use of the homologous sister chromatid. In this case, the broken DNA ends are resected to yield 3′ overhangs of ssDNA. The exposed ssDNA is rapidly coated by the ssDNA binding protein RPA, which is then displaced by RAD51 in a process that requires RAD52 and several other mediators. The RAD51 nucleo-filament mediates homology search and strand invasion at a homologous template forming a displacement (D)-loop. After second end capture, double Holliday junctions (dHJ) can form and are subsequently resolved to complete the repair process.
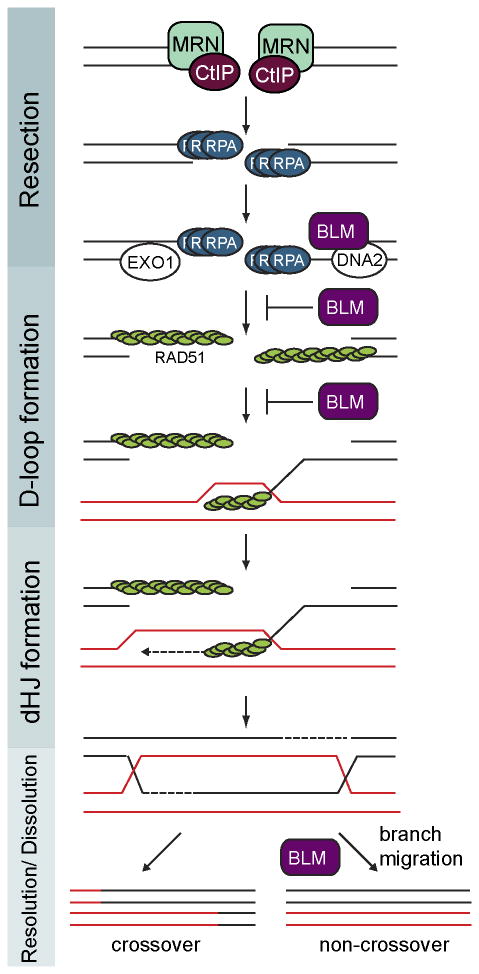
Intriguingly, BLM acts at several steps, early and late, to control HR (Figure 1). BLM bound to DNA2 promotes the extensive resection that occurs after the MRE11-mediated initial resection of the DSB DNA ends. While BLM's anti-recombinogenic activity negatively affects nucleo-filament formation and disrupts D-loops to suppress HR in vitro [9, 10], BLM also promotes the association of RAD51 with damaged replication forks in vivo [11]. In later steps, BLM is involved in branch migration of recombination intermediates [12]. A central function of BLM is the resolution of secondary DNA structures, including dHJs and replication intermediates, as well as G-quadruplexes and other hemi-catenene-like structures (reviewed in [7, 8]). The increase of SCEs in BS cells is thought to arise from the lack of BLM function during the dissolution of HJs, underling the importance of BLM during this process. During replicative stress BLM is additionally targeted by the checkpoint response [13-17] and acts to stabilize the DNA polymerase and prevent premature HR at stalled replication forks. Furthermore, BLM functions during telomere replication to prevent sister telomere loss and telomere defects ([18], discussed in [8]). Overall, BLM functions at many key steps in preserving genome integrity, highlighting the cellular importance of this central DNA helicase.
Figure 1. Schematic overview of BLM's roles in homologous recombination (HR).
A DSB is detected and bound by the MRN (MRE11-RAD50-NBS1) complex, which performs the initial resection in conjunction with CtIP. The produced 3′ single stranded DNA (ssDNA) is rapidly coated by the ssDNA binding protein RPA. BLM with DNA2 as well as EXO1 then perform extensive resection. The mediators RAD52 and BRCA2 along with several other mediators facilitate displacement of RPA and RAD51 nucleo-filament formation. The RAD51 filament mediates the homology search and strand invasion of the homologous template (red) forming a displacement (D)-loop. BLM's anti-recombinogenic activity displaces RAD51 and inhibits D-loop formation. Upon DNA-synthesis double Holliday junctions (dHJ) form. BLM acts to promote branch migration and dissolution of dHJ without crossovers. Resolution of dHJs yields crossover products. Adapted from [7, 9, 68].
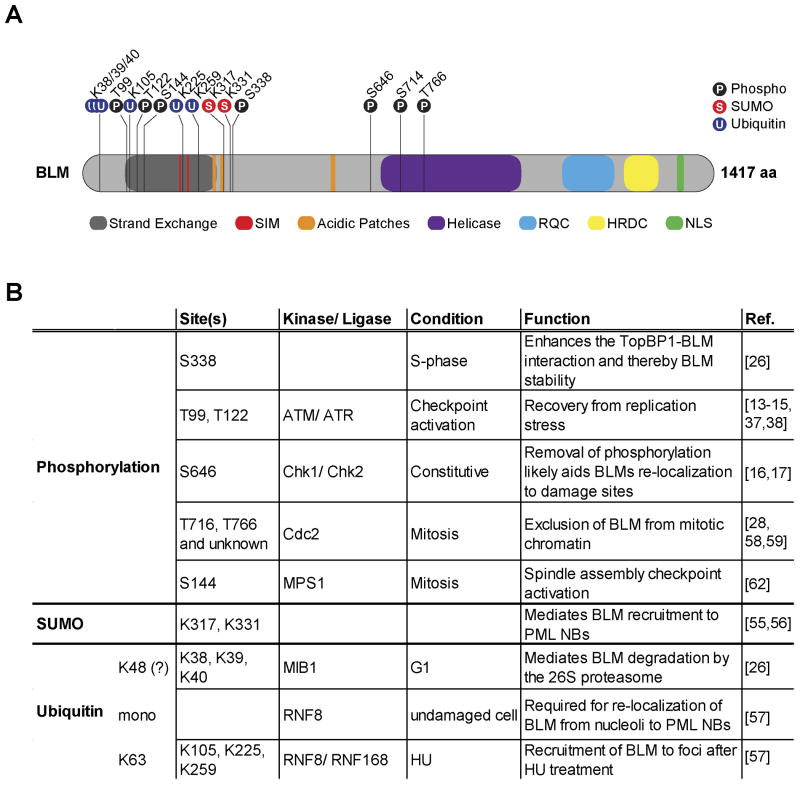
Human BLM is a 1417 amino acid protein that localizes to the nucleus and is comprised of several conserved domains (Figure 2A). Characteristic of the RecQ protein family is the central helicase domain, which contains ATP-binding and DEAH box motifs, and the adjacent RecQ helicase C-terminal domain (RQC), which is thought to mediate protein-protein interactions [7]. C-terminally flanking this highly conserved region are the helicase and RNase D C-terminal (HRDC) domain and a nuclear localization sequence (NLS). The N-terminus of BLM contains several short acid patches as well as a region required for strand exchange [19]. Many of BLM's interaction partners bind to the N-terminus, among them TOP3α [20, 21], RMI1/2 [22-24], and RAD51 [25]. Furthermore, the majority of the currently known post-translational modifications (PTM) also occur within the N-terminus of BLM, leading to the possibility that PTMs may regulate physical interactions targeting this region (see following sections and Figure 2).
Figure 2. Summary of BLM post-translational modifications and their cellular function.
A) Cartoon depicting the domain architecture and post-translational modification (PTM) sites of the human BLM protein. SIM: SUMO interacting motif. RQC: RecQ C-terminal. HRDC: Helicase and RNAse C-terminal. NLS: Nuclear localization sequence. (P): Phosphorylation, (S): SUMOylation, (U): Ubiquitination. B) Table summarizing the post-translational modifications of BLM. K48/K63: ubiquitin K48/K63-chain linkage. HU: Hydroxyurea. PML-NBs: PML nuclear bodies. (For references please see Ref. column)
Due to the crucial involvement of BLM and its homologues in multiple distinct cellular pathways, RecQ-like helicase activity must be intricately controlled to prevent illegitimate DNA damage processing, which can lead to genomic rearrangements and mutations. In this review, we will discuss the cellular context of BLM regulation by phosphorylation, ubiquitination, and SUMOylation. We will reflect on the current understanding of how differential PTMs of BLM affect its sub-cellular localization, protein stability, and ultimately function, in particular during replicative stress. In contrast to BLM, surprisingly little is known about the regulation of budding yeast Sgs1. We will compare the regulatory mechanisms of these two RecQ-like helicases and summarize the currently prevalent open questions, whose investigation will further advance the molecular understanding of RecQ-like helicase function in maintaining genome integrity and preventing cancer.
2. Cellular regulation of BLM function
As described in the introduction, BLM critically regulates numerous different steps of DNA damage signaling, repair, and replication. Most of these processes require BLM helicase function to act on secondary DNA structures. Intriguingly, BLM functions in opposing pathways to intricately regulate DNA damage repair and maintain genome integrity. For example, BLM is a critical factor for DNA damage repair by HR, which normally occurs at DSBs in late S and G2-phases of the cell cycle. HR at other times during the cell cycle or at illegitimate substrates, for example a stalled replication fork, can have detrimental outcomes such as chromosome translocations or deletions. BLM acts on similar substrates to initiate HR in some cases, while inhibiting HR in other contexts, and making it clear that BLM activity must be differentially regulated depending on the cellular context. Recent advances have uncovered multiple layers of BLM regulation, in particular through different forms of PTMs. These cellular mechanisms control BLM stability, as well as its localization, and interaction with binding partners.
2.1. Cell cycle regulation of BLM protein levels by TopBP1 and MIB1
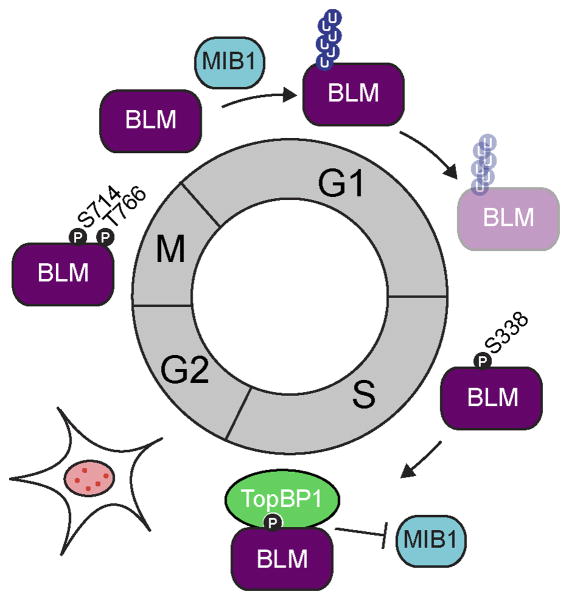
Like many HR proteins, BLM protein levels are cell cycle regulated. Consistent with a role in HR and DNA replication, BLM expression peaks during S/G2 cell cycle phases [26-28] and remains low during G1. A very recent study by Wang and colleagues uncovered a novel mechanism regulating BLM protein levels during the cell cycle, involving phosphorylation, MIB1-mediated ubiquitination, and binding of TopBP1 [26] (Figure 3).
Figure 3. Cell cycle regulation of BLM protein levels by TopBP1 and MIB1.
BLM is ubiquitinated by MIB1 at K38, K39, K40 during G1, which leads to degradation by the 26 S proteasome. In S-phase, BLM is phosphorylated at S338, thereby enhancing its interaction with TopBP1 and preventing BLM degradation. In S and G2 cell cycle phases BLM protein levels are high and BLM forms nuclear foci that co-localize with PML [26]. In mitosis, BLM is phosphorylated at S714 and T776 as well as other sites, preventing BLM's localization to chromatin [28, 58]. BLM is only found on chromatin in late anaphase within structures called ultra-fine anaphase bridges (UFBs) [60].
2.1.1. The E3 ubiquitin ligase MIB1
The E3 ubiquitin-protein ligase Mindbomb 1 (MIB1) was first identified for its function in Notch-Delta signaling in human cells, where it promotes ubiquitination and internalization of Delta [29]. Upon further analysis, the survival motor neuron (SMN) protein was identified as an additional MIB1 substrate [30]. MIB1 primarily localizes to the cytoplasm [29], but is also detectable in the nucleus of U2OS cells, consistent with its function in regulating BLM [26]. Interestingly, MIB1 was identified in a large-scale screen for proteins localizing to centrosomes [31], where TopBP1 is also found in mitotic cells [32].
2.1.2. TopBP1
The highly conserved DNA topoisomerase 2β-binding protein 1 (TopBP1) is essential for replication [33] and checkpoint signaling [34]. TopBP1 levels increase during S-phase, correlating with its role in replication, and TopBP1 nuclear foci form at stalled replication forks [33]. In addition to its role during DNA replication, TopBP1 is also recruited to sites of DNA damage in response to IR, where it frequently co-localizes with PML nuclear bodies [35] (discussed in section 2.3.1.).
2.1.3. Regulation of BLM by MIB1 and TopBP1
In their recent study, Wang et al. identify a multi-layered mechanism that controls BLM protein levels during the cell cycle. In G1, BLM is degraded by the 26 S proteasome upon ubiquitination at lysines 38, 39, and/or 40 by the ubiquitin E3 ligase MIB1. Conversely, BLM is stabilized through its interaction with TopBP1 throughout S-phase. The interaction of BLM with TopBP1 during S-phase is strongly enhanced upon phosphorylation of BLM at serine 338 [26] (Figure 3), thereby protecting BLM from ubiquitination and subsequent proteasomal degradation. Consistent with a requirement for stabilization of BLM protein during S-phase, depletion of TopBP1 or mutation of the TopBP1 BLM-interacting domain leads to an increase in SCEs [26], a characteristic of BS cells [2, 3]. In line with a model where MIB1 and TopBP1 compete for BLM binding, BLM protein levels, as well as the increase of SCEs in cells depleted of TopBP1, can be rescued by co-depletion of MIB1. Conversely, an increase in BLM protein levels in G1 sensitizes cells to DSBs and the authors suggest that this is due to increased end-resection mediated by BLM, which would interfere with DSB repair by NHEJ [26]. In summary, regulation of BLM protein levels during the cell cycle occurs in multiple steps, BLM is phosphorylated in S-phase enhancing its interaction with TopBP1, which shields BLM from ubiquitination by the E3 ligase MIB1 [26] (Figure 3). It will be interesting to uncover further mechanistic details of this regulatory process, for example, if the phosphorylation on BLM S338 is actively removed by a protein phosphatase during the transition to mitosis.
2.2. Phosphorylation of BLM during replication checkpoint signaling is important for recovery from replication stress
In addition to its role in HR, BLM also has important functions during DNA replication checkpoint signaling. The replication or intra S-phase checkpoint responds to replicative stress and halts cell cycle progression, inhibits late firing origins, induces the expression of damage response genes, and inhibits HR (reviewed in [36]). The two central kinases that control this checkpoint in mammalian cells are ATM and ATR. Interestingly, BLM is a target of both ATM upon irradiation [14] and ATR during replication stress [13], where it is phosphorylated at threonines 99 and 122 [15] (Figure 2). Cells expressing a mutant non-phosphorylate-able version of BLM do not properly recover from a replication arrest induced by hydroxyurea (HU) treatment. Furthermore, blocking phosphorylation of BLM at both T99 and T122 does not impair BLM's ability to suppress SCEs, nor does it regulate BLM's localization into DNA damage foci [13]. Conversely, a study investigating the phosphorylation only at T99 of BLM upon camptothecin treatment finds that this modification increases the co-localization of BLM with γ-H2AX [37]. Consistent with this result is the finding that BLM phosphorylation at T99 upon replication fork stalling with HU increases its interaction and co-localization with 53BP1 [38]. This indicates a potential function of BLM phosphorylation during the cellular response to replication stress.
To complicate matters, BLM is also phosphorylated by the checkpoint kinases Chk1 [16] and Chk2 [17] (Figure 2), which act downstream during the replication checkpoint response (reviewed in [39]). Chk1 and Chk2 are responsible for constitutive phosphorylation of BLM at serine 646 which, when reduced upon DNA damage [17], likely aids in BLM's re-localization to damage sites. Overall, the different kinases that phosphorylate BLM and the location of these sites act to regulate BLM's function during replication stress.
2.3. Regulation of BLM's sub-nuclear localization by SUMO and phosphorylation
Without damage, the majority of BLM foci co-localize with PML (Promyelocytic leukemia) in PML nuclear bodies (PML-NBs) [40]. BLM localization is abolished in mouse PML -/- cells and human promyelocytic NB4 cells lacking normal PML-NBs [40]. Mouse PML -/- cells notably display an elevated frequency of SCEs [40], at levels intermediate of normal and BS cells [2, 3]. These findings suggest a functional requirement for the localization of BLM to PML-NBs to maintain genome integrity.
2.3.1. PML nuclear bodies are large stress response centers associated with SUMO
The PML protein localizes to distinct nuclear structures called PML-NBs [41-43]. PML-NBs are thought to be general stress sensors that can form as a result of different insults to a cell, such as telomere shortening, DNA damage, proteasome inhibition, senescence, aggregation of mis-folded proteins, and alterations in ribosome biogenesis [44]. The involvement of PML-NBs in DNA damage repair, as well as telomere maintenance, makes them crucial factors in preserving genome integrity. Furthermore, PML-NBs contain several proteins important for the DNA damage response [45], including the MRN complex (MRE11, RAD50, NBS1) [46, 47], RAD51 [48], RAD52 [48], TOP3α [49], p53 [50-52], TopBP1 [35] and BLM itself [40]. A shared feature of PML and its associated proteins is that they can almost all be modified with the small ubiquitin-like modifier SUMO [44]. SUMOylation at an internal lysine of the substrate occurs through the sequential action of an E1 activating enzyme, the E2 SUMO-conjugating enzyme Ubc9, and an E3 SUMO ligase. In addition, many PML-binding proteins contain one or more SUMO-interacting motifs (SIMs), making it likely that their recruitment to PML-NBs is at least in part mediated through binding of their SIM to SUMOylated PML [44]. For telomeres and for DNA breaks, it has been proposed that PML may be recruited onto preassembled protein aggregates containing SUMO at the site of damage [53, 54].
2.3.2. SUMO mediates BLM localization to PML nuclear bodies
Consistent with its localization to PML-NBs, BLM is modified by SUMO in vitro and in vivo at lysines 317 and 331 [55, 56] (Figure 2). Blocking BLM SUMOylation at these sites leads to a redistribution of BLM from PML-NBs to repair foci, as marked by the presence of the DNA damage response markers γ-H2AX and BRCA1 [55]. Therefore, BLM modification by SUMO is likely involved in the induction of repair foci as well as altering protein subcellular localization to PML-NBs.
Furthermore, in line with the hypothesis that recruitment and retention of BLM in PML-NBs is mediated by SUMO- SIM interactions is the finding that BLM possesses two SIMs at amino acids 217-220 and 235-238, which are crucial for its localization to PML-NBs [56] (Figure 2). Simultaneous mutation of both SIMs nearly abolishes SUMO binding and SUMOylation of BLM in vitro. Consistently, a truncated BLM variant with intact SIMs localizes to the nucleoplasm and PML-NBs, while a construct lacking both SIMs does not co-localize with PML [56]. These findings suggest that binding of BLM to SUMO, or a SUMOylated protein, potentially Ubc9 [56], is required for its retention in PML-NBs.
2.3.3. Ubiquitination of BLM and the presence of 53BP1 are required for BLM localization from nucleoli to PML nuclear bodies
As well as being post-translationally modified by SUMO, BLM is furthermore ubiquitinated [57] (Figure 2). Mono-ubiquitination of BLM is required for its re-localization from nucleoli to PML-NBs [57]. Interestingly, the DNA repair protein 53BP1 is also critical for the re-localization of BLM from nucleoli to PML-NBs [16]. One possibility is that mono-ubiquitination of BLM is important for its protein-protein interaction with 53BP1, which may aid in recruiting or retaining BLM within PML-NBs. Alternatively, the mono-ubiquitination of BLM may make it a substrate for SUMOylation, which is also required for its localization to PML-NBs. Future studies will be needed to differentiate between these models for BLM recruitment and retention in PML-NBs.
2.3.4. Exclusion of BLM from mitotic chromosomes by phosphorylation
In addition to a fluctuation of overall protein levels, the localization of BLM is also regulated throughout the cell cycle. BLM is phosphorylated during mitosis in undamaged cells [28] (Figure 2, 3) and excluded from the chromatin fraction. However, this modification does not directly affect helicase activity or its association with TOP3α [58]. BLM is phosphorylated by the mitotic kinase Cdc2 on multiple sites including serine 714 and threonine 766 [58, 59]. In late anaphase however, BLM localizes to thin structures that link the DNA of the two nuclei in dividing cells, called ultra-fine bridges (UFBs) [60]. While the nature, regulation, and resolution of UFBs is a topic of current investigation, a role for BLM helicase is likely, as these structures presumably reflect persistent DNA catenanes formed by earlier replication or recombination events [61] or a partial lack of replication.
In addition, BLM is phosphorylated at serine 144 by MPS1, an event that is not required to prevent SCEs, but plays a role during the spindle assembly checkpoint (SAC), which ensures faithful chromosome segregation in mitosis [62]. Proper function of the SAC is also crucial to prevent chromosome loss and aneuploidy and therefore to maintain genome integrity. Hence, this role of BLM in mitosis may further contribute to the severe genetic instability phenotype observed in BS cells.
2.4. Regulation of the pro- and anti-recombinogenic role of BLM by post-translational modifications
While BS cells exhibit elevated recombination rates and SCEs, BLM also has important functions during replication and the repair of replicative damage (reviewed in [63]). Importantly, indicative of a central role for BLM in replication, patient derived BS cells exhibit a lower rate of DNA elongation and maturation of replication intermediates [2, 4, 5]. BLM is recruited to damaged replication forks [50, 64-66] and is thought to stabilize the polymerase at a stalled replication fork, likely to suppress illegitimate recombination events and to allow replication forks to re-start. Alternatively, if the stalled replication fork collapses, resulting in DSB formation, BLM functions to repair the DNA damage by promoting HR. Thus, BLM acts on similar double-stranded DNA substrates in two opposing fashions, to either repress or promote HR, depending upon the cellular context. BLM activity at these substrates is tightly regulated to prevent illegitimate recombination events. Recent work has uncovered post-translational modification of BLM by ubiquitin and/or SUMO to be crucial in this regulatory process [11, 55, 57, 67].
2.4.1. The pro-recombinogenic function of BLM promotes DNA-end resection and dissolves secondary DNA structures
To allow DSB repair by HR, broken DNA-ends are resected to produce 3′ ssDNA overhangs, which are then coated by the ssDNA binding protein RPA. Multiple factors including the central HR mediators RAD52 and BRCA2 facilitate RPA displacement and RAD51 filament formation on these ssDNA ends. The subsequent homology search is carried out by the RAD51 filament, followed by strand invasion, D-loop formation, and branch migration (reviewed in [68]; Figure 1). The BLM-TOP3-RMI1/2 complex resolves double Holliday junctions (dHJ), one structure that can be formed after branch migration.
In addition to this later function during HJ dissolution, BLM also acts early during HR. In parallel to promoting the function of the exonuclease EXO1 [69], BLM, together with the nuclease DNA2, promotes extensive end-resection at DSBs [69, 70] after initial resection by the MRN complex and CtIP. Extensive DNA-end resection is not only important for processing of HR directly, but also enhances the activation of the checkpoint response, which depends on ssDNA generation during S/G2 cell cycle phases. Consistently, co-depletion of BLM and EXO1 impairs ssDNA/RPA- activated phosphorylation by ATR and leads to hypersensitivity towards the DNA damaging agent camptothecin [70].
The mechanism of BLM regulation during resection has not been completely uncovered. However, several factors of resection are CDK [71] and DNA damage response kinase substrates [72, 73], and mammalian BLM is itself phosphorylated in a cell cycle dependent manner (see section 2.3.4.) and upon DNA damage (see section 2.2.). Since phosphorylation by Chk1 and Chk2 affects BLM's recruitment to sites of DNA damage, this modification may also alter its ability to facilitate resection, for example by affecting BLM's interaction with DNA2.
Intriguingly, recent work [74] demonstrates that BLM, in association with 53BP1 and RIF1, functions to prevent the CtIP/MRE11- initiated alternative end-joining (A-EJ) pathway, a highly error-prone mechanism that causes chromosomal translocations. Interestingly, the phosphorylation of BLM at T99 appears to be important for resection and 53BP1 focus formation upon IR treatment [74], while RIF1 promotes BLM focus formation and localization to DSB sites after irradiation [75].
Furthermore, work from the Ellis lab has demonstrated that BLM SUMOylation is required for HR-mediated repair of stalled replication forks. Expression of a non-SUMOylate-able BLM variant (SM-BLM) in BS cells causes defects in HR-mediated repair marked by an increase in γ-H2AX foci, more HU-induced DSBs and impaired recruitment of the HR mediators RAD52 and BRCA2 [55, 67]. While BLM SUMOylation is not required for its recruitment to a stalled replication fork or for its helicase activity, it enhances the interaction with the RAD51 recombinase [11]. Thus, enhancing the interaction of BLM with RAD51 by SUMOylation could function to recruit or retain RAD51 and its accessory factors at ssDNA to promote the homology search and strand invasion steps of HR.
2.4.2. The anti-recombinogenic function of BLM during replication stress is mediated by poly-ubiquitination and potentially SUMOylation
When a replication fork stalls, as is the case during HU treatment, BLM is recruited to the fork, where it functions to prevent illegitimate recombination [50, 76]. BLM's anti-recombinogenic function suppresses HR at early steps by displacing RAD51 from ssDNA [9] and disrupting D-loops [9, 10, 77]. This regulation could occur either by translocation of BLM along the ssDNA and/or through a physical interaction of BLM with RAD51 [25]. The finding that SUMOylation of BLM enhances its interaction with RAD51 to promote, not suppress, HR [67] is at first somewhat puzzling in this context. However, SUMOylation could alter the BLM- RAD51 interaction in a manner that promotes RAD51 nucleo-filament formation rather than inhibiting it. Intriguingly, RAD51 interacts with two regions of BLM [25], and the SUMOylation sites are adjacent to the N-terminal region which binds RAD51 [25]. It remains to be determined if two different modes of RAD51 binding to BLM exist depending on which region of BLM is targeted and how they are affected by post-translational modification of BLM.
The recruitment of BLM to stalled forks also depends on 53BP1 [16] as well as BLM ubiquitination [57]. K63-linked poly-ubiquitination of BLM at lysines 105, 225, and 259 (Figure 2) is induced by HU treatment and is necessary for its re-localization into HU-induced foci [57]. This poly-ubiquitination by RNF8/RNF168 is required to suppress excessive HR, likely by inducing RAP80- dependent BLM re- localization to damage sites. Intriguingly, RAP80 is also necessary for BLM stability [57], suggesting that RAP80 could also exhibit a similar regulatory mechanism as TopBP1- mediated stabilization of BLM (see section 2.1.). However, it remains to be determined if a physical interaction between BLM and RAP80, mediated by ubiquitination of BLM, is required for BLM stability, or if RAP80's function in DNA repair, which also depends on its ubiquitin interacting motifs [78], indirectly regulates BLM protein levels.
In summary, ubiquitination of BLM facilitates its recruitment to HU- induced foci and prevents excessive HR. The suppression of HR could potentially occur through enhancement of BLM's activity in disrupting RAD51 nucleo-filament formation as well as through the stabilization of the stalled polymerase. It appears equally plausible that SUMOylation could inhibit this function, as ubiquitination may enhance it.
2.5. BLM function at telomeres
Telomeres are protein- DNA complexes at chromosome ends that play important roles in maintaining genome integrity, as well as having effects on cell survival and proliferation (reviewed in [79]). Unless telomere ends are actively extended, their length shortens during normal somatic cell divisions and ultimately leads to cellular senescence or apoptosis. In the absence or inactivation of telomerase, cells can use a recombination and DNA repair-based mechanism to maintain telomere length called the ALT-pathway (alternative lengthening of telomeres) [80]. Importantly, as the other RecQ-like helicases WRN and RECQL4, BLM interacts with telomeric proteins [81-83] and functions in telomere maintenance (reviewed in [8]). BLM deficient cells exhibit more telomere defects, such as sister telomere loss and telomere free ends [18], and display an increase in telomere associations between homologous chromosome arms, which may arise from unresolved replication intermediates or entangled telomeres [81].
Typically, PML-NBs do not contain nucleic acid, however, a subpopulation of ALT-positive cells (∼5-20%) contain telomeric DNA that resides within PML-NBs forming ALT-associated PML bodies (APBs) ([48], and reviewed in [84]). APBs contain PML-NB components, DNA damage response factors including BLM, as well as the SMC5/6-complex [53]. Interestingly, the co-localization of BLM with the telomeric shelterin protein TRF2 increases in the G2/M cell cycle phases and a physical interaction of BLM with TRF2 is observed in ALT positive cells [81]. In ALT cells, PML-NBs contain the SMC5/6 complex, which is required for telomere recombination and APB formation. According to current models, it is likely that SUMOylation of the shelterin complex by the SMC5/6-complex E3 SUMO ligase MMS21 mediates the recruitment of telomeres to PML-NBs and thereby facilitates the formation of APBs [53]. Furthermore, similar to PML-NBs [85], a combination of SUMO- SIM interactions is likely to be critical for APB formation. The molecular function of APBs in the ALT pathway is still largely unknown and further studies are needed to clarify their exact role in telomere maintenance. For example, one possibility is that APBs cluster telomeres and facilitate telomeric HR ([48, 86], reviewed in [87]). Typically, BLM loss is associated with hyper-recombination phenotypes, exemplified by the increase of SCEs in BS cells [2, 3]. In contrast, depletion of BLM in an ALT-activating background prevents the formation of APBs and C-circles, which form as a byproduct of telomere recombination [88]. Thus, BLM is thought to promote HR at telomeres in ALT cells. Given BLM's function in recombination and SUMOylation-dependent localization to PML-NBs, it is tempting to speculate that BLM SUMOylation, possibly by MMS21, may mediate its pro-recombinogenic function at the telomere in ALT positive cells.
3. Regulation of the budding yeast RecQ-like helicase Sgs1
In contrast to BLM, surprisingly little is known about post-translational regulation of the budding yeast orthologue Sgs1. In addition to functioning in analogous pathways that require the unwinding of double-stranded DNA, Sgs1 is considered most homologous to the mammalian RecQ-like helicase BLM due to the very similar domain architecture and the presence of the evolutionarily conserved Sgs1-Top3-Rmi1 complex. Similar to BLM, Sgs1 can function in both a pro- and anti-recombinogenic manner to facilitate accurate DNA repair. Furthermore, sgs1Δ mutant cells exhibit phenotypes reminiscent of BS syndrome cells, including hyper-recombination and the accumulation of replication intermediates [6].
A fraction of Sgs1 is SUMOylated [89] at lysine 621 upon DSB-inducing DNA damage such as γ-irradiation or EMS treatment [90]. Yet in contrast to BLM, Sgs1 SUMOylation, at least at K621, does not appear to regulate Sgs1's function in HR. Conversely, SUMOylation of Sgs1 is important for telomere- telomere recombination and a yeast cell only expressing Sgs1-K621R exhibits defects in type II recombination events [90], which are thought to resemble the mammalian ALT pathway due to the amplification of telomeric repeats [80, 91]. Interestingly, SUMOylation of the fission yeast RecQ-like helicase Rqh1 also controls its role at dysfunctional telomeres [92]. One hypothesis is that Sgs1 SUMOylation may trigger its recruitment to telomeres to facilitate telomere recombination [90]. While Sgs1 forms nuclear foci [93, 94], it is currently unclear what these foci are, how focus formation is regulated, and if PTMs play a role in this process. Taking into account what is known about BLM and the formation of APBs, it is possible that Sgs1 nuclear foci may resemble similar structures, where telomeric DNA is clustered for recombination and that SUMOylation of Sgs1 is important for the maintenance of these structures. The function of BLM in APBs in mammalian cells is not fully understood and further investigation of Sgs1 PTMs and how they may regulate its focus formation could uncover additional conserved mechanistic details.
In addition to being SUMOylated, Sgs1 is also phosphorylated by Mec1 (ATR) in a similar manner to BLM upon activation of the intra S-phase checkpoint. Phosphorylation of Sgs1 likely promotes the response to HU- induced replicative damage by further enhancing the activation of the budding yeast Chk2 homolog Rad53 [95]. In contrast to BLM, ubiquitination of Sgs1 has not been reported to date. However, since Sgs1 is cell cycle regulated [93], mechanisms regulating Sgs1 cellular protein levels remain to be determined.
It is also interesting to note that most PTMs of BLM occur within a confined region of the N-terminus (Figure 2), while the known PTM sites in Sgs1 are closer to the central helicase domain. Intriguingly, in order for BLM to complement some sgs1 phenotypes, the N-terminus of Sgs1 must be present in a chimeric protein [96], suggesting that the mechanisms of regulation for these two conserved proteins may differ substantially. Nonetheless, further clarification of the regulatory mechanisms that control Sgs1 function will surely provide novel insight in the analysis of BLM function in higher eukaryotes.
4. Conclusions and future directions
As discussed, BLM is phosphorylated, ubiquitinated, and SUMOylated differentially in response to various forms of cellular stress involving DNA damage. While many aspects of BLM regulation by PTMs have been uncovered, the physiological function of each type of modification has primarily been studied individually. Future analysis will need to address the cross-talk and interdependence of different forms of modification and how they fine-tune BLMs function in different pathways with sometimes opposing functions, as becomes evident during replicative repair.
It is possible that the modification of BLM acts as a molecular switch to determine if a cell engages in HR or not. SUMOylation of BLM could promote HR, while ubiquitination may function to inhibit HR, for example at a stalled fork. In this scenario, the two types of post-translational modification could occur in a mutually exclusive manner. Ubiquitination of BLM takes place at K105, K225, and K259 [57], and is needed to recruit BLM to stalled forks and to repress HR [57], possibly by enhancing BLM's function in disrupting RAD51 nucleo-filaments. Interestingly, K225 is within the short stretch between the two proposed SIMs, which are important for BLM localization to PML-NBs [56]. It seems possible that ubiquitination could interfere with the SIM and thereby alter BLM localization.
It is furthermore intriguing that the sites of post-translational modification are almost all clustered within a confined region of the N- terminus of BLM (Figure 2), again suggesting that there may be significant cross- talk between adjacent sites of different modifications. At least for SUMOylation and mitotic phosphorylation, there currently is no evidence that post-translational modification affects BLM helicase activity directly [11, 58]. Therefore, it is more likely that SUMOylation alters BLM activity through other mechanisms and it is conceivable that protein- protein interactions and/or the strand-exchange activity of BLM are controlled by PTMs within the N-terminus.
Fully understanding the molecular mechanisms of BLM function is highly relevant for human health. While Bloom syndrome is a rare heritable autosomal recessive disease, it has been speculated that heterozygous mutations within BLM may also predispose individuals to breast and colorectal cancer [97-99]. Furthermore, uncovering molecular details of the cellular response to DNA damage will likely reveal novel drug targets and biomarkers that may further advance cancer treatment. In addition, telomere maintenance has become of increasing interest in the development of cancer therapies. Cancer cells activate mechanisms to maintain telomere length and enhance proliferation. While most cancers reactivate telomerase, a significant proportion initiate the ALT-pathway [100], and links of ALT activation to tumors with complex karyotypes have been reported [101-103]. A study by Reddel and colleagues [101] identifies a correlation between patient age and ALT activation, which is seen mostly in young individuals [101]. Furthermore, it has also been discussed that treatment of telomerase-positive tumors with telomerase inhibitors may select for cells which activate ALT [104, 105]. Thus, fully understanding the role of BLM and its modification on APB formation and telomere recombination could potentially provide novel therapeutic targets, particularly because significant progress has been made in developing small molecule inhibitors of BLM [106].
Acknowledgments
We thank Nathan Ellis, Roderick O'Sullivan, Patricia Opresko, and Meghan Sullivan for critical reading of the manuscript and helpful suggestions. This work was supported by the National Institutes of Health grant (GM088413) and the Ellison Medical Foundation (AG-NS-0935-12) to K.A.B.
Footnotes
Conflict of interest: The authors confirm there is no conflict of interest, financial or otherwise, in this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.German J, Sanz MM, Ciocci S, Ye TZ, Ellis NA. Syndrome-causing mutations of the BLM gene in persons in the Bloom's Syndrome Registry. Hum Mutat. 2007;28:743–753. doi: 10.1002/humu.20501. [DOI] [PubMed] [Google Scholar]
- 2.German J, Crippa LP, Bloom D. Bloom's syndrome. III. Analysis of the chromosome aberration characteristic of this disorder. Chromosoma. 1974;48:361–366. doi: 10.1007/BF00290993. [DOI] [PubMed] [Google Scholar]
- 3.Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc Natl Acad Sci U S A. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hand R, German J. A retarded rate of DNA chain growth in Bloom's syndrome. Proc Natl Acad Sci U S A. 1975;72:758–762. doi: 10.1073/pnas.72.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lonn U, Lonn S, Nylen U, Winblad G, German J. An abnormal profile of DNA replication intermediates in Bloom's syndrome. Cancer Res. 1990;50:3141–3145. [PubMed] [Google Scholar]
- 6.Bernstein KA, Shor E, Sunjevaric I, Fumasoni M, Burgess RC, Foiani M, Branzei D, Rothstein R. Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. EMBO J. 2009;28:915–925. doi: 10.1038/emboj.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu Rev Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ Helicases in DNA Repair, Recombination, and Replication. Annu Rev Biochem. 2014:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bugreev D, Yu X, Egelman E, Mazin A. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Brabant AJ, Ye T, Sanz M, German IJ, Ellis NA, Holloman WK. Binding and melting of D-loops by the bloom syndrome helicase. Biochemistry (Mosc) 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- 11.Ouyang KJ, Woo LL, Zhu J, Huo D, Matunis MJ, Ellis NA. SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol. 2009;7:e1000252. doi: 10.1371/journal.pbio.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karow J, Constantinou A, Li J, West S, Hickson I. The Bloom's syndrome gene product promotes branch migration of holliday junctions. Proc Natl Acad Sci U S A. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beamish H, Kedar P, Kaneko H, Chen P, Fukao T, Peng C, Beresten S, Gueven N, Purdie D, Lees-Miller S, Ellis N, Kondo N, Lavin MF. Functional link between BLM defective in Bloom's syndrome and the ataxia-telangiectasia-mutated protein, ATM. J Biol Chem. 2002;277:30515–30523. doi: 10.1074/jbc.M203801200. [DOI] [PubMed] [Google Scholar]
- 15.Ababou M, Dutertre S, Lecluse Y, Onclercq R, Chatton B, Amor-Gueret M. ATM-dependent phosphorylation and accumulation of endogenous BLM protein in response to ionizing radiation. Oncogene. 2000;19:5955–5963. doi: 10.1038/sj.onc.1204003. [DOI] [PubMed] [Google Scholar]
- 16.Sengupta S, Robles AI, Linke SP, Sinogeeva NI, Zhang R, Pedeux R, Ward IM, Celeste A, Nussenzweig A, Chen J, Halazonetis TD, Harris CC. Functional interaction between BLM helicase and 53BP1 in a Chk1-mediated pathway during S-phase arrest. J Cell Biol. 2004;166:801–813. doi: 10.1083/jcb.200405128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaur S, Modi P, Srivastava V, Mudgal R, Tikoo S, Arora P, Mohanty D, Sengupta S. Chk1-dependent constitutive phosphorylation of BLM helicase at serine 646 decreases after DNA damage. Mol Cancer Res. 2010;8:1234–1247. doi: 10.1158/1541-7786.MCR-10-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barefield C, Karlseder J. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res. 2012;40:7358–7367. doi: 10.1093/nar/gks407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CF, Brill SJ. An essential DNA strand-exchange activity is conserved in the divergent N-termini of BLM orthologs. EMBO J. 2010;29:1713–1725. doi: 10.1038/emboj.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID. The Bloom's syndrome gene product interacts with topoisomerase III. J Biol Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 21.Gangloff S, McDonald J, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, Delgado-Cruzata L, Rothstein R, Freyer G, Boone C, Brown G. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci U S A. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J Biol Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- 25.Wu L, Davies S, Levitt N, Hickson I. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Chen J, Gong Z. TopBP1 controls BLM protein level to maintain genome stability. Mol Cell. 2013;52:667–678. doi: 10.1016/j.molcel.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gharibyan V, Youssoufian H. Localization of the Bloom syndrome helicase to punctate nuclear structures and the nuclear matrix and regulation during the cell cycle: comparison with the Werner's syndrome helicase. Mol Carcinog. 1999;26:261–273. doi: 10.1002/(sici)1098-2744(199912)26:4<261::aid-mc5>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 28.Dutertre S, Ababou M, Onclercq R, Delic J, Chatton B, Jaulin C, Amor-Gueret M. Cell cycle regulation of the endogenous wild type Bloom's syndrome DNA helicase. Oncogene. 2000;19:2731–2738. doi: 10.1038/sj.onc.1203595. [DOI] [PubMed] [Google Scholar]
- 29.Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- 30.Kwon DY, Dimitriadi M, Terzic B, Cable C, Hart AC, Chitnis A, Fischbeck KH, Burnett BG. The E3 ubiquitin ligase mind bomb 1 ubiquitinates and promotes the degradation of survival of motor neuron protein. Mol Biol Cell. 2013;24:1863–1871. doi: 10.1091/mbc.E13-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, Falkenby LG, Bennetzen M, Westendorf J, Nigg EA, Uhlen M, Hyman AA, Andersen JS. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 2011;30:1520–1535. doi: 10.1038/emboj.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reini K, Uitto L, Perera D, Moens PB, Freire R, Syvaoja JE. TopBP1 localises to centrosomes in mitosis and to chromosome cores in meiosis. Chromosoma. 2004;112:323–330. doi: 10.1007/s00412-004-0277-5. [DOI] [PubMed] [Google Scholar]
- 33.Makiniemi M, Hillukkala T, Tuusa J, Reini K, Vaara M, Huang D, Pospiech H, Majuri I, Westerling T, Makela TP, Syvaoja JE. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J Biol Chem. 2001;276:30399–30406. doi: 10.1074/jbc.M102245200. [DOI] [PubMed] [Google Scholar]
- 34.Yamane K, Chen J, Kinsella TJ. Both DNA topoisomerase II-binding protein 1 and BRCA1 regulate the G2-M cell cycle checkpoint. Cancer Res. 2003;63:3049–3053. [PubMed] [Google Scholar]
- 35.Xu ZX, Timanova-Atanasova A, Zhao RX, Chang KS. PML colocalizes with and stabilizes the DNA damage response protein TopBP1. Mol Cell Biol. 2003;23:4247–4256. doi: 10.1128/MCB.23.12.4247-4256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 37.Rao VA, Fan AM, Meng L, Doe CF, North PS, Hickson ID, Pommier Y. Phosphorylation of BLM, dissociation from topoisomerase IIIalpha, and colocalization with gamma-H2AX after topoisomerase I-induced replication damage. Mol Cell Biol. 2005;25:8925–8937. doi: 10.1128/MCB.25.20.8925-8937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tripathi V, Kaur S, Sengupta S. Phosphorylation-dependent interactions of BLM and 53BP1 are required for their anti-recombinogenic roles during homologous recombination. Carcinogenesis. 2008;29:52–61. doi: 10.1093/carcin/bgm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stracker TH, Usui T, Petrini JH. Taking the time to make important decisions: the checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair (Amst) 2009;8:1047–1054. doi: 10.1016/j.dnarep.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong S, Hu P, Ye T, Stan R, Ellis N, Pandolfi P. A role for PML and the nuclear body in genomic stability. Oncogene. 1999;18:7941–7947. doi: 10.1038/sj.onc.1203367. [DOI] [PubMed] [Google Scholar]
- 41.Dyck JA, Warrell RP, Jr, Evans RM, Miller WH., Jr Rapid diagnosis of acute promyelocytic leukemia by immunohistochemical localization of PML/RAR-alpha protein. Blood. 1995;86:862–867. [PubMed] [Google Scholar]
- 42.Koken MH, Puvion-Dutilleul F, Guillemin MC, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, et al. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 44.de The H, Le Bras M, Lallemand-Breitenbach V. The cell biology of disease: Acute promyelocytic leukemia, arsenic, and PML bodies. J Cell Biol. 2012;198:11–21. doi: 10.1083/jcb.201112044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dellaire G, Bazett-Jones DP. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays. 2004;26:963–977. doi: 10.1002/bies.20089. [DOI] [PubMed] [Google Scholar]
- 46.Wu G, Lee WH, Chen PL. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J Biol Chem. 2000;275:30618–30622. doi: 10.1074/jbc.C000390200. [DOI] [PubMed] [Google Scholar]
- 47.Mirzoeva OK, Petrini JH. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol Cell Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- 49.Hu P, Beresten SF, van Brabant AJ, Ye TZ, Pandolfi PP, Johnson FB, Guarente L, Ellis NA. Evidence for BLM and Topoisomerase IIIalpha interaction in genomic stability. Hum Mol Genet. 2001;10:1287–1298. doi: 10.1093/hmg/10.12.1287. [DOI] [PubMed] [Google Scholar]
- 50.Sengupta S, Linke SP, Pedeux R, Yang Q, Farnsworth J, Garfield SH, Valerie K, Shay JW, Ellis NA, Wasylyk B, Harris CC. BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J. 2003;22:1210–1222. doi: 10.1093/emboj/cdg114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fogal V, Gostissa M, Sandy P, Zacchi P, Sternsdorf T, Jensen K, Pandolfi PP, Will H, Schneider C, Del Sal G. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 2000;19:6185–6195. doi: 10.1093/emboj/19.22.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo A, Salomoni P, Luo J, Shih A, Zhong S, Gu W, Pandolfi PP. The function of PML in p53-dependent apoptosis. Nat Cell Biol. 2000;2:730–736. doi: 10.1038/35036365. [DOI] [PubMed] [Google Scholar]
- 53.Potts PR, Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nature Struct & Mol Biol. 2007;14:581–590. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- 54.Chung I, Leonhardt H, Rippe K. De novo assembly of a PML nuclear subcompartment occurs through multiple pathways and induces telomere elongation. J Cell Sci. 2011;124:3603–3618. doi: 10.1242/jcs.084681. [DOI] [PubMed] [Google Scholar]
- 55.Eladad S, Ye TZ, Hu P, Leversha M, Beresten S, Matunis MJ, Ellis NA. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum Mol Genet. 2005;14:1351–1365. doi: 10.1093/hmg/ddi145. [DOI] [PubMed] [Google Scholar]
- 56.Zhu J, Zhu S, Guzzo CM, Ellis NA, Sung KS, Choi CY, Matunis MJ. Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J Biol Chem. 2008;283:29405–29415. doi: 10.1074/jbc.M803632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tikoo S, Madhavan V, Hussain M, Miller ES, Arora P, Zlatanou A, Modi P, Townsend K, Stewart GS, Sengupta S. Ubiquitin-dependent recruitment of the Bloom syndrome helicase upon replication stress is required to suppress homologous recombination. EMBO J. 2013;32:1778–1792. doi: 10.1038/emboj.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dutertre S, Sekhri R, Tintignac LA, Onclercq-Delic R, Chatton B, Jaulin C, Amor-Gueret M. Dephosphorylation and subcellular compartment change of the mitotic Bloom's syndrome DNA helicase in response to ionizing radiation. J Biol Chem. 2002;277:6280–6286. doi: 10.1074/jbc.M105735200. [DOI] [PubMed] [Google Scholar]
- 59.Bayart E, Dutertre S, Jaulin C, Guo RB, Xi XG, Amor-Gueret M. The Bloom syndrome helicase is a substrate of the mitotic Cdc2 kinase. Cell Cycle. 2006;5:1681–1686. doi: 10.4161/cc.5.15.3122. [DOI] [PubMed] [Google Scholar]
- 60.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan KL, Hickson ID. New insights into the formation and resolution of ultra-fine anaphase bridges, Semin. Cell Dev Biol. 2011;22:906–912. doi: 10.1016/j.semcdb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 62.Leng M, Chan DW, Luo H, Zhu C, Qin J, Wang Y. MPS1-dependent mitotic BLM phosphorylation is important for chromosome stability. Proc Natl Acad Sci U S A. 2006;103:11485–11490. doi: 10.1073/pnas.0601828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 64.Davalos AR, Kaminker P, Hansen RK, Campisi J. ATR and ATM-dependent movement of BLM helicase during replication stress ensures optimal ATM activation and 53BP1 focus formation. Cell Cycle. 2004;3:1579–1586. doi: 10.4161/cc.3.12.1286. [DOI] [PubMed] [Google Scholar]
- 65.Rassool FV, North PS, Mufti GJ, Hickson ID. Constitutive DNA damage is linked to DNA replication abnormalities in Bloom's syndrome cells. Oncogene. 2003;22:8749–8757. doi: 10.1038/sj.onc.1206970. [DOI] [PubMed] [Google Scholar]
- 66.Davalos AR, Campisi J. Bloom syndrome cells undergo p53-dependent apoptosis and delayed assembly of BRCA1 and NBS1 repair complexes at stalled replication forks. J Cell Biol. 2003;162:1197–1209. doi: 10.1083/jcb.200304016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ouyang KJ, Yagle MK, Matunis MJ, Ellis NA. BLM SUMOylation regulates ssDNA accumulation at stalled replication forks. Front Genet. 2013;4:167. doi: 10.3389/fgene.2013.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Renkawitz J, Lademann CA, Jentsch S. Mechanisms and principles of homology search during recombination. Nat Rev Mol Cell Biol. 2014;15:369–383. doi: 10.1038/nrm3805. [DOI] [PubMed] [Google Scholar]
- 69.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci U S A. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferretti LP, Lafranchi L, Sartori AA. Controlling DNA-end resection: a new task for CDKs. Front Genet. 2013;4:99. doi: 10.3389/fgene.2013.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–693. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Makharashvili N, Tubbs AT, Yang SH, Wang H, Barton O, Zhou Y, Deshpande RA, Lee JH, Lobrich M, Sleckman BP, Wu X, Paull TT. Catalytic and Noncatalytic Roles of the CtIP Endonuclease in Double-Strand Break End Resection. Mol Cell. 2014:1022–1033. doi: 10.1016/j.molcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grabarz A, Guirouilh-Barbat J, Barascu A, Pennarun G, Genet D, Rass E, Germann SM, Bertrand P, Hickson ID, Lopez BS. A role for BLM in double-strand break repair pathway choice: prevention of CtIP/Mre11-mediated alternative nonhomologous end-joining. Cell Rep. 2013;5:21–28. doi: 10.1016/j.celrep.2013.08.034. [DOI] [PubMed] [Google Scholar]
- 75.Feng L, Fong KW, Wang J, Wang W, Chen J. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J Biol Chem. 2013;288:11135–11143. doi: 10.1074/jbc.M113.457440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tripathi V, Nagarjuna T, Sengupta S. BLM helicase-dependent and -independent roles of 53BP1 during replication stress-mediated homologous recombination. J Cell Biol. 2007;178:9–14. doi: 10.1083/jcb.200610051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bachrati C, Borts R, Hickson I. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yan J, Yang XP, Kim YS, Jetten AM. RAP80 responds to DNA damage induced by both ionizing radiation and UV irradiation and is phosphorylated at Ser 205. Cancer Res. 2008;68:4269–4276. doi: 10.1158/0008-5472.CAN-07-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lillard-Wetherell K, Machwe A, Langland GT, Combs KA, Behbehani GK, Schonberg SA, German J, Turchi JJ, Orren DK, Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum Mol Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- 82.Stavropoulos DJ, Bradshaw PS, Li X, Pasic I, Truong K, Ikura M, Ungrin M, Meyn MS. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet. 2002;11:3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- 83.Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 84.Chung I, Osterwald S, Deeg KI, Rippe K. PML body meets telomere: the beginning of an ALTernate ending? Nucleus. 2012;3:263–275. doi: 10.4161/nucl.20326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seeler JS, Dejean A. SUMO: of branched proteins and nuclear bodies. Oncogene. 2001;20:7243–7249. doi: 10.1038/sj.onc.1204758. [DOI] [PubMed] [Google Scholar]
- 86.Draskovic I, Arnoult N, Steiner V, Bacchetti S, Lomonte P, Londono-Vallejo A. Probing PML body function in ALT cells reveals spatiotemporal requirements for telomere recombination. Proc Natl Acad Sci U S A. 2009;106:15726–15731. doi: 10.1073/pnas.0907689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 88.O'Sullivan RJ, Arnoult N, Lackner DH, Oganesian L, Haggblom C, Corpet A, Almouzni G, Karlseder J. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat Struct Mol Biol. 2014;21:167–174. doi: 10.1038/nsmb.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 90.Lu CY, Tsai CH, Brill SJ, Teng SC. Sumoylation of the BLM ortholog, Sgs1, promotes telomere-telomere recombination in budding yeast. Nucleic Acids Res. 2009:488–498. doi: 10.1093/nar/gkp1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rog O, Miller KM, Ferreira MG, Cooper JP. Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres. Mol Cell. 2009;33:559–569. doi: 10.1016/j.molcel.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 93.Frei C, Gasser SM. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 94.Tkach JM, Yimit A, Lee AY, Riffle M, Costanzo M, Jaschob D, Hendry JA, Ou J, Moffat J, Boone C, Davis TN, Nislow C, Brown GW. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol. 2012;14:966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hegnauer AM, Hustedt N, Shimada K, Pike BL, Vogel M, Amsler P, Rubin SM, van Leeuwen F, Guenole A, van Attikum H, Thoma NH, Gasser SM. An N-terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks. EMBO J. 2012;31:3768–3783. doi: 10.1038/emboj.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mirzaei H, Schmidt KH. Non-Bloom syndrome-associated partial and total loss-of-function variants of BLM helicase. Proc Natl Acad Sci U S A. 2012;109:19357–19362. doi: 10.1073/pnas.1210304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thompson ER, Doyle MA, Ryland GL, Rowley SM, Choong DY, Tothill RW, Thorne H, Barnes DR, Li J, Ellul J, Philip GK, Antill YC, James PA, Trainer AH, Mitchell G, Campbell IG. Exome sequencing identifies rare deleterious mutations in DNA repair genes FANCC and BLM as potential breast cancer susceptibility alleles. PLoS Genet. 2012;8:e1002894. doi: 10.1371/journal.pgen.1002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Goss KH, Risinger MA, Kordich JJ, Sanz MM, Straughen JE, Slovek LE, Capobianco AJ, German J, Boivin GP, Groden J. Enhanced tumor formation in mice heterozygous for Blm mutation. Science. 2002;297:2051–2053. doi: 10.1126/science.1074340. [DOI] [PubMed] [Google Scholar]
- 99.Gruber SB, Ellis NA, Scott KK, Almog R, Kolachana P, Bonner JD, Kirchhoff T, Tomsho LP, Nafa K, Pierce H, Low M, Satagopan J, Rennert H, Huang H, Greenson JK, Groden J, Rapaport B, Shia J, Johnson S, Gregersen PK, Harris CC, Boyd J, Rennert G, Offit K. BLM heterozygosity and the risk of colorectal cancer. Science. 2002;297:2013. doi: 10.1126/science.1074399. [DOI] [PubMed] [Google Scholar]
- 100.Siddiqa A, Cavazos D, Chavez J, Long L, Marciniak RA. Modulation of telomeres in alternative lengthening of telomeres type I like human cells by the expression of werner protein and telomerase. J Oncol. 2012;2012:806382. doi: 10.1155/2012/806382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Henson JD, Hannay JA, McCarthy SW, Royds JA, Yeager TR, Robinson RA, Wharton SB, Jellinek DA, Arbuckle SM, Yoo J, Robinson BG, Learoyd DL, Stalley PD, Bonar SF, Yu D, Pollock RE, Reddel RR. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin Cancer Res. 2005;11:217–225. [PubMed] [Google Scholar]
- 102.Montgomery E, Argani P, Hicks JL, DeMarzo AM, Meeker AK. Telomere lengths of translocation-associated and nontranslocation-associated sarcomas differ dramatically. Am J Pathol. 2004;164:1523–1529. doi: 10.1016/S0002-9440(10)63710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JH, Sung PA, Jasin M, Rosenbluh J, Zwang Y, Weir BA, Hatton C, Ivanova E, Macconaill L, Hanna M, Hahn WC, Lue NF, Reddel RR, Jiao Y, Kinzler K, Vogelstein B, Papadopoulos N, Meeker AK. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 105.Siddiqa A, Cavazos DA, Marciniak RA. Targeting telomerase. Rejuvenation Res. 2006;9:378–390. doi: 10.1089/rej.2006.9.378. [DOI] [PubMed] [Google Scholar]
- 106.Nguyen GH, Dexheimer TS, Rosenthal AS, Chu WK, Singh DK, Mosedale G, Bachrati CZ, Schultz L, Sakurai M, Savitsky P, Abu M, McHugh PJ, Bohr VA, Harris CC, Jadhav A, Gileadi O, Maloney DJ, Simeonov A, Hickson ID. A small molecule inhibitor of the BLM helicase modulates chromosome stability in human cells. Chem Biol. 2013;20:55–62. doi: 10.1016/j.chembiol.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]