Background: There is no report on Ndh subunits that destabilize the NDH-1 complex and repress activity.
Results: Deletion of ndhO in Synechocystis 6803 increased the activity of cyclic electron transport around photosystem I, whereas overexpression repressed the activity and destabilized NDH-1M complex.
Conclusion: NdhO destabilizes NDH-1M and represses the activity.
Significance: NdhO is a new type subunit that controls NDH-1M negatively.
Keywords: Cyanobacteria, Electron Transfer Complex, Electron Transport, Membrane Protein, Photosynthesis, NDH-1 Complexes, NDH-CET, NdhO, Synechocystis 6803, Stability
Abstract
Two mutants that grew faster than the wild-type (WT) strain under high light conditions were isolated from Synechocystis sp. strain PCC 6803 transformed with a transposon-bearing library. Both mutants had a tag in ssl1690 encoding NdhO. Deletion of ndhO increased the activity of NADPH dehydrogenase (NDH-1)-dependent cyclic electron transport around photosystem I (NDH-CET), while overexpression decreased the activity. Although deletion and overexpression of ndhO did not have significant effects on the amount of other subunits such as NdhH, NdhI, NdhK, and NdhM in the cells, the amount of these subunits in the medium size NDH-1 (NDH-1M) complex was higher in the ndhO-deletion mutant and much lower in the overexpression strain than in the WT. NdhO strongly interacts with NdhI and NdhK but not with other subunits. NdhI interacts with NdhK and the interaction was blocked by NdhO. The blocking may destabilize the NDH-1M complex and repress the NDH-CET activity. When cells were transferred from growth light to high light, the amounts of NdhI and NdhK increased without significant change in the amount of NdhO, thus decreasing the relative amount of NdhO. This might have decreased the blocking, thereby stabilizing the NDH-1M complex and increasing the NDH-CET activity under high light conditions.
Introduction
The NDH-13 complex is an energy-converting NAD(P)H:quinone oxidoreductase, also called complex I. The complex is identified in a majority of species spanning from bacteria to mammals (1–4). The cyanobacterial NDH-1 complex is involved in a variety of bioenergetic reactions, including respiration, cyclic electron transport around photosystem I, and CO2 uptake (5–7). Structurally, the cyanobacterial NDH-1 complexes closely resemble energy-converting complex I in eubacteria and the mitochondrial respiratory chain regardless of the absence of homologs of three subunits in cyanobacterial genomes that constitute the catalytically active core of complex I (1, 3, 8). Over the past few years, significant achievements have been made in resolving the subunit compositions and functions of the multiple NDH-1 complexes in several cyanobacterial strains (9–12). Four types of NDH-1 have been identified in the cyanobacterium Synechocystis sp. strain PCC 6803 (hereafter Synechocystis 6803), and all four types are involved in NDH-1-dependent cyclic electron transport around photosystem I (NDH-CET) (13). The NDH-CET plays a particularly important role in coping with various environmental stresses by increasing its activity and supplying additional ATP. For example, this function can greatly alleviate high light-sensitive growth phenotypes (14–16). Therefore, high light strategy can help in identifying the proteins that affect NDH-CET activity.
Cyanobacterial and chloroplastic NDH-1 complexes constitute a subclass of the complex I family (12). A common feature of this group, apart from the absence of three active subunits homologous to NuoE, NuoF, and NuoG of the Escherichia coli enzyme, is the presence of several subunits specific for complexes originating from cells performing oxygenic photosynthesis. Electron microscopy studies revealed that in Synechocystis 6803 these subunits, NdhL, NdhM, NdhN, and NdhO, are located together, comprising the oxygenic photosynthesis-specific domain of unknown function (12, 17). Recently, NdhP, NdhQ, and NdhS were proposed to be new members of the oxygenic photosynthesis-specific domain (14, 18, 19). In addition to these oxygenic photosynthesis-specific subunits, this group also contains 11 common subunits (NdhA to NdhK) that exist in complex I family as well (3). However, subunits that constitute the cyanobacterial NDH-1S complex are absent in higher plants, while subunits present in the chloroplastic subcomplex B and lumen subcomplex have no counterparts in cyanobacteria (12, 20, 21). The severe alteration of subunit components of the NDH-1 complexes suggests significant changes in the function of certain subunits of NDH-1 complexes during evolution of cyanobacteria to higher plants.
The absence of any Ndh subunits identified to date has been considered to inactivate and destabilize the NDH-1 complexes in cyanobacteria (9–12) and higher plants (20–22). We found that cyanobacterial NdhO is a new type of subunit, which destabilizes the NDH-1M complex and represses the NDH-CET activity. This study reports the effect of deletion and overexpression of ndhO on the stability of NDH-1 complexes and NDH-CET activity. The possible mechanism of the NdhO-induced destabilization of NDH-1 enzyme is discussed based on the effect of NdhO on the interaction of Ndh subunits.
EXPERIMENTAL PROCEDURES
Culture Conditions
Synechocystis 6803 glucose-tolerant strain (wild type) and its mutants, ΔndhO and OX-ndhO, were cultured at 30 °C in BG-11 medium (23) buffered with Tris-HCl (5 mm, pH 8.0) and bubbled with 2% (v/v) CO2 in air. Solid medium was the same BG-11 supplemented with 1.5% agar. Continuous illumination was provided by fluorescence lamps at 40 μmol photons m−2 s−1.
Isolation of High NDH-CET Mutants
A cosmid library of Synechocystis 6803 genome contained 105 clones with inserts of 35–38.5 kb. The library was subjected to in vitro transposon mutagenesis using EZ-Tn5TM 〈KAN-2〉 insertion kit (Epicenter Biotechnologies, Madison, WI) and then transformed into wild-type (WT) Synechocystis 6803 cells. Following transformation, cells were spread on 1.5% BG-11 agar plates containing 5 μg of kanamycin ml−1, and KamR mutants that grew better than the WT under high light but not under growth light were isolated. Genomic DNA isolated from each mutant was digested with HhaI and after self-ligation was used as a template for inverse PCR with primers (supplemental Table 1) complementary to the N- and C-terminal regions of the KamR cassette. The exact position of the cassette in the mutant genome was determined by sequencing the PCR product.
Deletion and Overexpression of ndhO
A fragment of 207 bp between BamHI and KpnI sites in the ssl1690 (ndhO) gene was replaced with a kanamycin resistance (KamR) cassette amplified by PCR using appropriate primers, ndhO-C and ndhO-D (Fig. 2A and supplemental Table 1). The vector thus constructed was used to transform the WT cells of Synechocystis 6803 to generate the ndhO deletion mutant (ΔndhO). The transformants were spread on agar plates containing BG-11 medium and kanamycin (10 μg ml−1) buffered at pH 8.0, which were incubated in 2% (v/v) CO2 in air under continuous illumination by fluorescent lamps at 40 μmol photons m−2 s−1. The mutated ndhO in the transformants was segregated to homogeneity (by successive streak purification) as determined by PCR amplification (data not shown) and immunoblotting (Fig. 2C).
FIGURE 2.
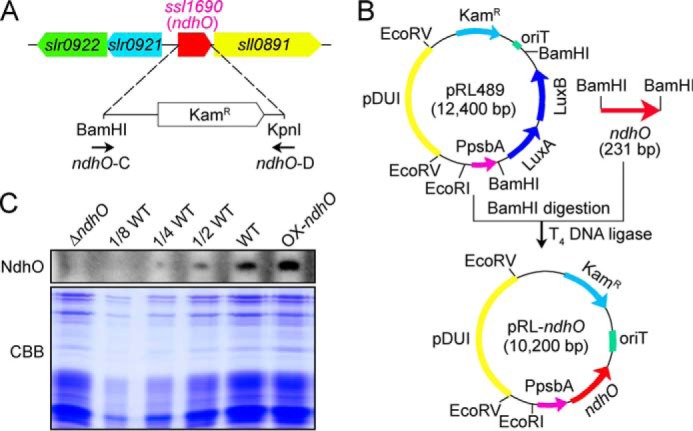
Construction of ΔndhO and OX-ndhO strains and their NdhO levels. A, construct used to generate the ΔndhO mutant. B, construction of a shuttle expression vector, pRL-ndhO, to generate the OX-ndhO mutant. C, Coomassie Brilliant Blue (CBB) staining profiles of total proteins from the ΔndhO, WT (including indicated serial dilutions), and OX-ndhO strains and their immunoblotting using the antibody against NdhO. Total protein corresponding to 3 μg of chlorophyll a was loaded onto each lane.
The pRL-489 shuttle expression vector (24) was used to generate ndhO-overexpression strain (OX-ndhO). A fragment containing the ndhO gene was amplified by PCR and then inserted into BamHI sites of pRL-489 to form the pRL-ndhO shuttle expression vector construct (Fig. 2B; primers are shown in supplemental Table 1), which was used to transform the WT cells of Synechocystis 6803 using triparental conjugative transfer method (24–29). Cells harboring pRL-ndhO plasmid were grown in BG-11 liquid medium supplemented with 100 μg ml−1 kanamycin. The overexpression level of NdhO in the transformants was estimated by protein blot (Fig. 2C).
Chlorophyll Fluorescence and P700 Analysis
The transient increase in chlorophyll fluorescence after actinic light had been turned off was monitored as described (30). The redox kinetics of P700 was measured according to methods described previously (14, 15). The re-reduction of P700+ in darkness was measured with a Dual-PAM-100 (Walz, Effeltrich, Germany) with an emitter-detector unit ED-101US/MD by monitoring absorbance changes at 830 nm and using 875 nm as a reference. Cells were kept in the dark for 2 min, and 10 μm 3-(3,4-dichlorophenyl)-1,1-dimethylurea was added to the cultures prior to the measurement. The P700 was oxidized by far-red light with a maximum at 720 nm from LED lamp for 30 s, and the subsequent re-reduction of P700+ in the dark was monitored.
Isolation of Crude Thylakoid Membranes
The cell cultures (800 ml) were harvested at the logarithmic phase of growth (A730 = 0.6–0.8) and washed twice by suspending in 50 ml of fresh BG-11 medium. Thylakoid membranes were isolated according to Gombos et al. (31) with some modifications as follows. Cells suspended in 5 ml of disruption buffer (10 mm HEPES-NaOH, 5 mm sodium phosphate, pH 7.5, 10 mm MgCl2, 10 mm NaCl, and 25% glycerol (v/v)) were supplemented by zirconia/silica beads and broken by vortexing 15 times at the highest speed for 20 s at 4 °C with 5 min cooling on ice between the runs. The crude extract was centrifuged at 5,000 × g for 5 min to remove the glass beads and unbroken cells. By further centrifugation at 20,000 × g for 30 min, we obtained crude thylakoid membranes from the precipitation.
Electrophoresis and Immunoblotting
Blue native (BN)-PAGE of Synechocystis 6803 membranes was performed as described previously (32) with slight modifications (14, 15). Thylakoid membranes were washed with 330 mm sorbitol, 50 mm BisTris, pH 7.0, and 0.5 mm PMSF (Sigma) and resuspended in 20% glycerol (w/v), 25 mm BisTris, pH 7.0, 10 mm MgCl2, 0.1 unit of RNase-free DNase RQ1 (Promega, Madison, WI), and 0.5 mm PMSF at a chlorophyll a concentration of 0.3 mg ml−1. The samples were incubated on ice for 10 min, and then an equal volume of 3% n-dodecyl β-d-maltoside was added. Solubilization was performed for 40 min on ice. Insoluble components were removed by centrifugation at 18,000 × g for 15 min. The collected supernatant was mixed with 0.1 volume of sample buffer, 5% Serva Blue G, 100 mm BisTris, pH 7.0, 30% sucrose (w/v), 500 mm ϵ-amino-n-caproic acid, and 10 mm EDTA. Solubilized membranes were then applied to a 0.75-mm-thick, 5–12.5% acrylamide gradient gel (Hoefer Mighty Small mini-vertical unit; San Francisco, CA). Samples were loaded on an equal chlorophyll a basis per lane. Electrophoresis was performed at 4 °C by increasing the voltage gradually from 50 to 200 V during the 5.5-h run. Several lanes of the BN gel were cut out and incubated in Laemmli SDS sample buffer containing 5% β-mercaptoethanol and 6 m urea for 1 h at 25 °C. SDS-PAGE of Synechocystis 6803 crude thylakoid membranes was carried out on 12% polyacrylamide gel with 6 m urea as described earlier (33).
For immunoblotting, the proteins were electrotransferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Millipore, Bedford, MA) and detected by protein-specific antibodies using an ECL assay kit (Amersham Biosciences) according to the manufacturer's protocol. Antibodies against NdhM, NdhO, and ATPβ proteins of Synechocystis 6803 were raised in our laboratory. Primer sequences used to amplify the ndhM, ndhO, and ATPβ genes are listed in supplemental Table 1. The PCR products were ligated into a vector, pET32a, and the constructs were amplified in E. coli DH-5α. The plasmids were used to transform E. coli strain BL21 (DE3) pLysS for protein expression. The expression products from E. coli were purified and used as antigens to immunize rabbits to produce corresponding polyclonal antibodies. The NDH-1 complexes were detected using the antibodies against NdhH, NdhI, and NdhK, respectively, which were previously raised in our laboratory (30).
Yeast Two-hybridization
Yeast two-hybridization was performed using the LexA system (Clontech). PCR-amplified ssl1690 (ndhO) or ndhI fragment was cloned in-frame into EcoRI and XhoI or XhoI sites of pLexA, respectively, to form the bait construct (primers are shown in supplemental Table 1). The fragments containing 14 ndh genes encoding NDH-1M subunits except NdhO and a GST gene were amplified by PCR and inserted into the EcoRI and XhoI or XhoI sites of pJG4-5 to form the prey construct (primers are shown in supplemental Table 1). The bait and prey constructs together with a reporter vector pSH18-34 were co-transformed into yeast strain EGY48 according to the manufacturer's instructions for the Matchmaker LexA two-hybrid system (Clontech). Transformed yeast was dropped into X-gal medium and then was grown at 30 °C in darkness as described previously (15).
Expression and Purification of Fusion Proteins
The target proteins NdhI, NdhK, and NdhO were fused to various tags as follows: (i) the fragments containing ndhI, ndhK, and ndhO genes were amplified by PCR and inserted between BamHI and SalI, EcoRI and XhoI, and BamHI and EcoRI sites of pET32a, respectively, to form the His-tagged fusion protein constructs (primers are shown in supplemental Table 1); (ii) PCR-amplified ndhO and ndhK genes were cloned in-frame into EcoRI and XhoI sites of pGEX-5X-1 to form the GST-tagged fusion protein constructs (primers are shown in supplemental Table 1); (iii) a PCR-amplified ndhI gene fragment was cloned in-frame into BamHI and XbaI of pMAL-p2x to form the MBP-tagged fusion protein construct (primers are shown in supplemental Table 1). Subsequently, these constructs were transformed into E. coli strain BL21 (DE3) pLysS and induced by 1 mm isopropyl β-d-thiogalactoside for 16 h at 16 °C to express His-, GST-, and MBP-tagged fusion proteins. These fusion proteins were purified at 4 °C using a nickel column (GE Healthcare), glutathione-Sepharose 4B (GE Healthcare), and amylose column with maltose (New England Biolabs), respectively, according to the manufacturers' instructions.
GST Pulldown Assay
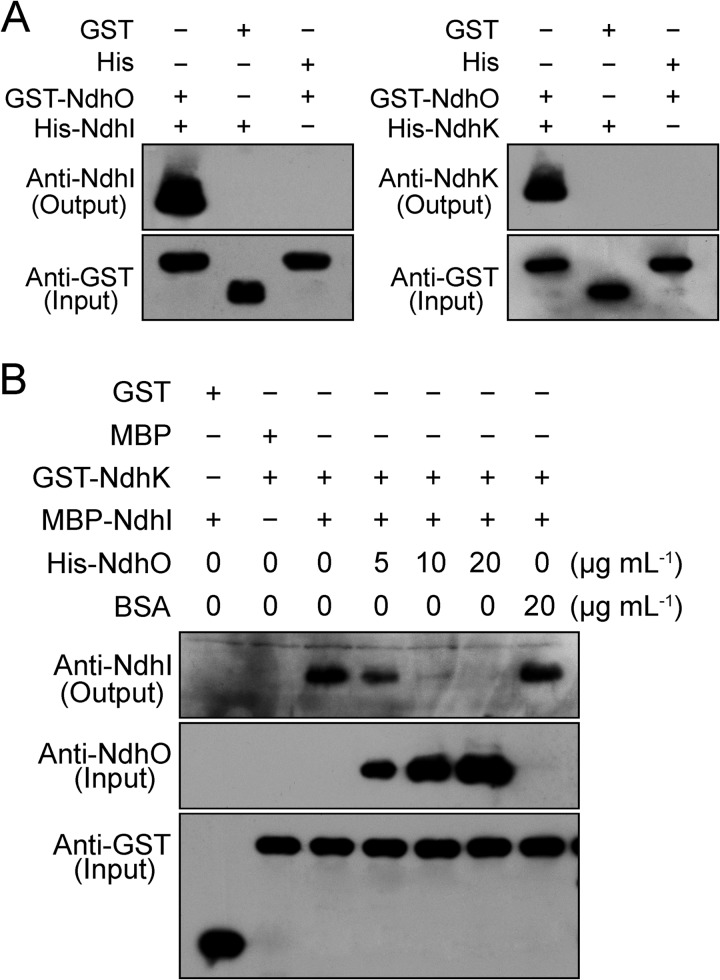
The GST pulldown assay was performed as described previously (34) with some modifications. (i) To confirm the interactions of NdhO with NdhI and NdhK, an equal amount of GST and GST-NdhO proteins was separately incubated with 20 μl of glutathione-Sepharose 4B beads (GE Healthcare) in 300 μl of binding buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 5 mm EDTA, 0.5% (v/v) Triton X-100, 1 mm phenylmethanesulfonyl fluoride and 1 mm DTT) at 4 °C for 1 h. The beads were washed three times with 1 ml of binding buffer and then an equal amount of His, His-NdhI, and His-NdhK was separately added and incubated in a 300-μl reaction system on a rotating shaker at 4 °C overnight. (ii) To confirm the interaction between NdhI and NdhK and, show the effect of NdhO on the interaction, the beads bound with an equal amount of GST and GST-NdhK proteins were, respectively, incubated with the MBP and the mixture of MBP-NdhI and His-NdhO or BSA (protein concentrations indicated in Fig. 6B) in a 300-μl reaction system on a rotating shaker at 4 °C overnight. After the overnight incubation, the beads were pelleted by centrifugation and washed four times with the ice-cold binding buffer described above. The washed pellets were resuspended in 30 μl of 1× SDS (2%) loading buffer and then boiled for 5 min. After centrifugation, the supernatant was collected and subjected to immunoblotting analysis.
FIGURE 6.
NdhO blocks the interaction between NdhI and NdhK. GST pulldown assays show the interaction of NdhO with NdhI and NdhK (A) and NdhI with NdhK (B). The effect of NdhO on the interaction between NdhI and NdhK is shown (B). The expressed proteins were mixed and incubated with GST beads on a rotating shaker at 4 °C overnight. A, after washing, interaction of GST-NdhO with His-NdhI and His-NdhK was detected using the antibodies against NdhI and NdhK. The interactions of GST (without NdhO) with His-NdhI and His-NdhK and of pET32a vector backbone with GST-NdhO were used as negative controls. B, after washing, interaction between MBP-NdhI and GST-NdhK and the effect of His-NdhO on the interaction were detected using the antibody against NdhI. Protein blot against NdhO was used to indicate the additional amount of His-NdhO to the reaction system, while interactions of GST (without NdhK) with MBP-NdhI and MBP (without NdhI) with GST-NdhK were used as negative controls. Immunoblotting against a GST antibody was used to show the loading and binding control.
RESULTS
Isolation of High NDH-CET Mutants
The NDH-CET has a protective role against high light stress in cyanobacteria (14, 15) and higher plants (16). Therefore, upon exposure of cells to high light, high NDH-CET mutants were expected to grow faster than wild type (WT) despite similar growth under moderate light irradiation. To screen the high NDH-CET mutants, we transformed WT cells with a transposon-bearing library, thus tagging and inactivating many genes randomly, and then we cultured the mutant cells under high light conditions. We isolated two mutants, which grew faster on plates under high light but similarly to the WT under growth light (Fig. 1A).
FIGURE 1.
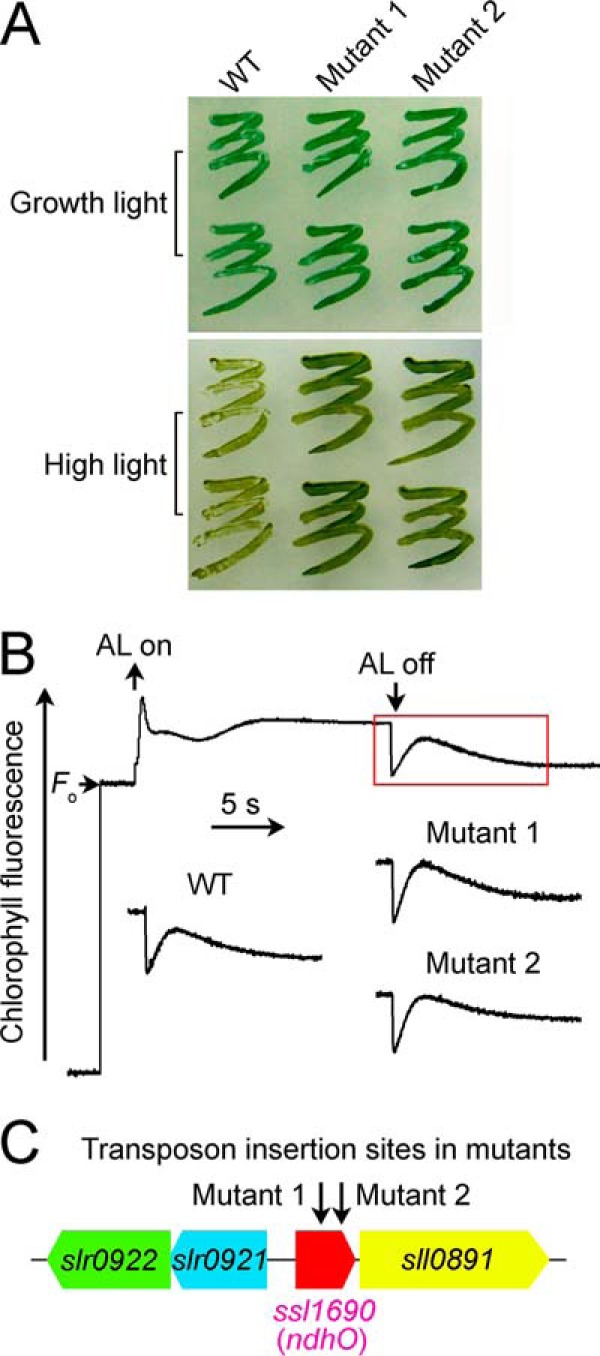
Characteristics of high light-tolerant mutants. A, growth of WT and mutants under normal light (40 μmol photons m−2s−1) and high light (200 μmol photons m−2s−1). B, monitoring of NDH-CET activity using chlorophyll fluorescence analysis. The top curve shows a typical trace of chlorophyll fluorescence in the WT Synechocystis 6803. See text for experimental details. C, transposon insertion sites in mutants 1 and 2 as probed by PCR analysis using the primers listed in supplemental Table 1.
To test whether the high light-dependent growth phenotype of the two mutants resulted from increased NDH-CET, we monitored the post-illumination rise in chlorophyll a fluorescence, which has been extensively used to evaluate the NDH-CET activity in cyanobacteria (14, 15, 30, 35, 36) and higher plants (37–46). As shown in Fig. 1B, the activity of NDH-CET was higher in both mutants than in the WT as judged by the height and relative rate of post-illumination increase in chlorophyll fluorescence. Evidently, the NDH-CET activity was increased in mutants 1 and 2.
To identify the genes inactivated by the transposon tagging, we analyzed the sites of transposon insertion in the mutants. Sequencing of the PCR products revealed that both mutants were tagged in the same known gene, ssl1690 (ndhO) (Fig. 1C). The transposon insertion occurred at positions 2,796,551 and 2,796,608 of the Synechocystis 6803 genome, respectively (NCBI gi: 16331673) (47). It seems likely that Ssl1690 (NdhO) represses the activity of NDH-CET in Synechocystis 6803.
NdhO Represses the NDH-CET Activity
To confirm that NdhO represses the NDH-CET activity, we constructed the ndhO deletion mutant (ΔndhO) (Fig. 2A) and overexpression strain (OX-ndhO) (Fig. 2B). As expected, NdhO was completely deleted in ΔndhO, but its amount was increased nearly to 2-fold of the WT value in the OX-ndhO strain (Fig. 2C). The activity of NDH-CET, as measured by the post-illumination rise in chlorophyll a fluorescence, was higher in ΔndhO but lower in OX-ndhO than in the WT (Fig. 3A). A similar result was obtained by measuring the oxidation of P700 by FR after AL illumination. When AL was turned off after a 30-s illumination by AL (800 μmol photons m−2 s−1) supplemented with FR, P700+ was transiently reduced by electrons from the plastoquinone (PQ) pool and subsequently reoxidized by the background FR. The operation of the NDH-1 complexes, which transfer electrons from the reduced cytoplasmic pool to PQ, hinders the re-oxidation of P700 (14, 15, 37). The re-oxidation of P700 was markedly slower in ΔndhO and faster in OX-ndhO as compared with that in the WT (Fig. 3B). We also measured the NDH-CET by monitoring the reduction rate of P700+ in darkness after illumination of cells with FR light. The re-reduction of P700+ was evidently faster in ΔndhO and slower in OX-ndhO compared with that in the WT (Fig. 3C). Under high light intensities, ΔndhO grew faster but OX-ndhO grew slower than the WT (Fig. 3E) despite similar growth under a growth light (Fig. 3D). Based on these results, we conclude that NdhO represses the NDH-CET activity in Synechocystis 6803.
FIGURE 3.
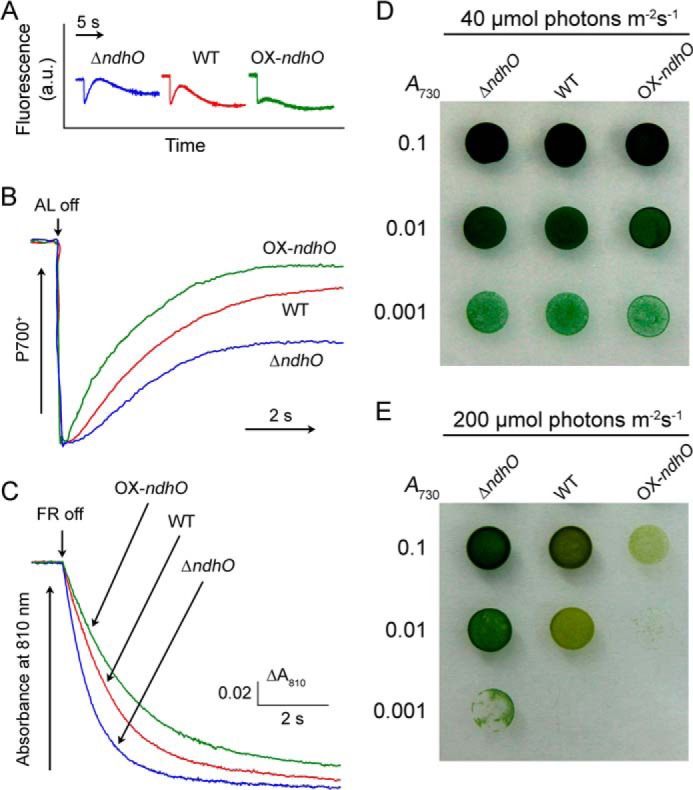
NDH-CET activities of ΔndhO, WT, and OX-ndhO strains and their growth. A, monitoring of NDH-CET activity by chlorophyll fluorescence. a.u. is arbitrary units. B, redox kinetics of P700. Cells were illuminated by AL supplemented with FR. After termination of AL illumination, P700+ was transiently reduced by electrons from the PQ pool and then reoxidized by background FR. C, kinetics of the P700+ re-reduction in darkness after turning off FR in the presence of 10 μm 3-(3,4-dichlorophenyl)-1,1-dimethylurea. The chlorophyll a concentration was adjusted to 20 μg ml−1, and curves are normalized to the maximal signal. D and E, 3 μl of cell suspensions with densities corresponding to A730 nm values of 0.1 (upper rows), 0.01 (middle rows), and 0.001 (lower rows) were spotted on agar plates. The plates were incubated under 2% CO2 in air (v/v) for 6 days at 40 μmol photons m−2 s−1 (D) and 3 days at 200 μmol photons m−2 s−1 (E), respectively.
NdhO Destabilizes the NDH-1M Complex
To reveal how NdhO represses the NDH-CET activity, we investigated the accumulation and assembly of NDH-1L and NDH-1M complexes in the thylakoid membranes of the ΔndhO, WT, and OX-ndhO strains. As deduced from the protein abundance of NdhH, NdhI, NdhK, and NdhM, deletion and overexpression of ndhO did not have a significant effect on the amount of these subunits in the thylakoid membranes (Fig. 4A). However, all of these subunits in the NDH-1M complex were significantly increased in the ΔndhO mutant but were scarcely visible in the OX-ndhO strain (Fig. 4C). The amount of these subunits in the NDH-1L complex did not change by deletion or overexpression of NdhO. This clearly indicates that NdhO destabilizes the NDH-1M complex, thereby repressing the NDH-CET activity.
FIGURE 4.
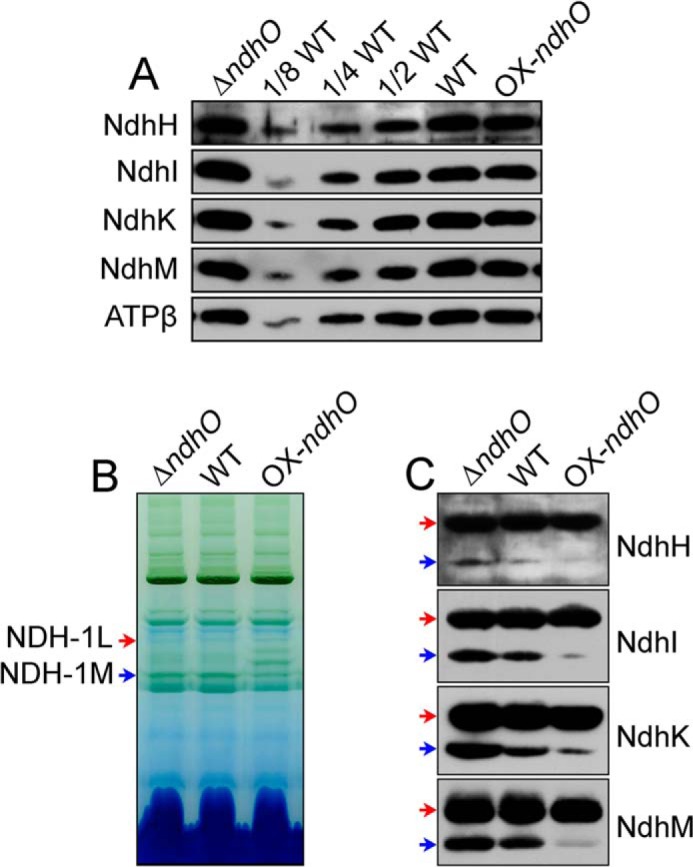
Amounts of Ndh subunits in the thylakoid membranes and NDH-1L and NDH-1M of ΔndhO, WT, and OX-ndhO strains. A, immunodetection of Ndh subunits in thylakoid membranes from the ΔndhO, WT (including indicated serial dilutions), and OX-ndhO strains. Protein blot was performed with antibodies against NdhH, NdhI, NdhK, and NdhM. Lanes were loaded with thylakoid membrane proteins corresponding to 1 μg of chlorophyll a. ATPβ was used as a loading control. B, profiles of BN-PAGE of thylakoid membranes from the WT and mutant strains. Each lane was loaded with thylakoid membrane extract corresponding to 9 μg of chlorophyll a. Red and blue arrows indicate the positions of NDH-1L and NDH-1M complexes, respectively. C, Western analyses of NDH-1L and NDH-1M using the antibodies against NdhH, NdhI, NdhK, and NdhM.
NdhO Interacts Specifically with NdhI and NdhK
To elucidate how NdhO destabilizes the NDH-1M complex, we analyzed the interaction of NdhO with 14 Ndh subunits that had been identified in the complex, using yeast two-hybrid system. The results indicated that NdhO strongly interacts with NdhI and NdhK but not with other 12 subunits of NDH-1M complex (Fig. 5A). As shown by Sazanov and Hinchliffe (48) in Thermus thermophilus on Nqo9 and Nqo6 (corresponding to cyanobacterial NdhI and NdhK, respectively), there was interaction between these subunits in Synechocystis 6803 (Fig. 5B). The interaction of NdhO with NdhI and NdhK was reinforced by the results of GST-pulldown assays (Fig. 6A). We therefore conclude that NdhO interacts specifically with NdhI and NdhK. These results suggest that NdhO may impede the interaction of NdhI with NdhK, thereby destabilizing the NDH-1M complex.
FIGURE 5.
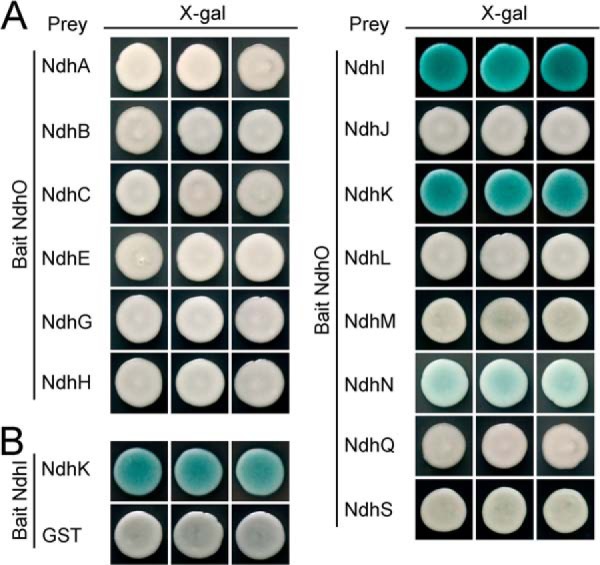
Interaction of NdhO with NdhI and NdhK and between NdhI and NdhK. Yeast two-hybrid experiments to test the interaction of NdhO with various Ndh subunits (A) and between NdhI and NdhK (B). The ndhO and ndhI genes were constructed into bait vector, while each of the ndh genes encoding NDH-1M subunits except ndhO and a GST gene were constructed into a prey vector. Subsequently, they were transformed into the yeast strain EGY48. Transformed yeast was dropped onto X-gal medium. Blue precipitate represents accumulated β-galactosidase activity resulting from the activation of the lacZ reporter gene by protein-protein interaction. The induction plate was grown at 30 °C for 23 h and then photographed. The interaction of NdhI-GST was assayed as a negative control. At least six independent experiments were performed, and the result of one representative is shown.
NdhO Blocks the Interaction of NdhI with NdhK
To test the above hypothesis, we analyzed the effect of various amounts of NdhO on the interaction between NdhI and NdhK using GST-pulldown assay strategy. Our results indicated that the interaction of NdhI with NdhK is weakened by addition of NdhO but not bovine serum albumin (BSA) to the reaction system (Fig. 6B). We therefore conclude that NdhO blocks the interaction of NdhI with NdhK, and the blocking may destabilize the NDH-1M complex and repress the NDH-CET activity.
Ratio of NdhO to NdhI and NdhK Drops under High Light Conditions
The NDH-CET has a particularly important role in coping with high light stress by increasing its activity and supplying additional ATP. To explore whether the increase in NDH-CET caused by high light stress was the result of decreased blocking, we analyzed the expression levels of NdhI, NdhK, and NdhO when WT cells were transferred from growth light to high light. As expected, the amounts of NdhI and NdhK were gradually increased along with the transfer process (Fig. 7). However, the level of NdhO did not change in this transfer process (Fig. 7). This was reinforced by the results of transcript levels of ndhI, ndhK, and ndhO during the transfer from growth light to high light (data not shown). Consequently, the ratio of NdhO to NdhK and NdhI dropped under high light conditions, which might have decreased the blocking and increased the NDH-CET activity.
FIGURE 7.
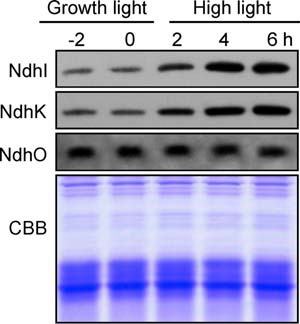
Amounts of NdhI, NdhK, and NdhO in WT cells grown under different irradiances. The protein levels of NdhI, NdhK, and NdhO were analyzed by Western blot on WT cells grown under growth light (40 μmol photons m−2 s−1) before (2 and 0 h) and after transfer to high light (200 μmol photons m−2 s−1) for 2, 4, and 6 h. Lanes were loaded with thylakoid membrane proteins corresponding to 1 μg of chlorophyll a. In the lower panel, a replicate gel stained with Coomassie Brilliant Blue (CBB) was used as a loading control.
DISCUSSION
The NdhO subunit exists exclusively in oxygenic photosynthetic organisms (49, 50). In higher plants, knock-out of ndhO resulted in a complete impairment of NDH-CET and entire collapse of subcomplex A of chloroplast NDH-1 complex (43, 50). NdhO was identified in Synechocystis 6803 by proteomics studies (49) and has been assumed to have a similar function as its homologs in higher plants. However, in Synechocystis 6803, deletion of ndhO greatly improved the assembly efficiency of the NDH-1M complex (Fig. 4) and raised the NDH-CET activity (Figs. 1 and 3). This was confirmed by the result that NDH-1M was almost completely decomposed when ndhO was overexpressed (Fig. 4C). Thus, cyanobacterial NdhO is a new type of Ndh subunit, which destabilizes the NDH-1M complex.
To our knowledge, this is the first case that a subunit of the NDH-1 complex destabilizes the enzyme and represses its activity. Except for this property, cyanobacterial NdhO possesses common properties with other Ndh subunits as follows: (i) the NdhO homologs are absent in Chlamydomonas reinhardtii, which lacks ndh genes (51); (ii) NdhO is associated with NDH-1L and NDH-1M complexes (49); (iii) NdhO is absent in M55 mutant (data not shown), which lacks NDH-1L and NDH-1M complexes (14, 15, 52); and (iv) cells grown under high CO2 and growth light contain roughly equimolar NdhO, NdhI, and NdhK (data not shown). These common properties of NdhO with other Ndh subunits consolidate the conclusion that cyanobacterial NdhO is a genuine subunit of NDH-1L and NDH-1M complexes, regardless of the severe alteration of the function of NdhO during evolution from cyanobacteria to higher plants.
Although NdhO is present both in NDH-1L and NDH-1M complexes, there was no significant effect of deletion or overexpression of ndhO on NDH-1L (Fig. 4C). NDH-1M is considered to be present as NDH-1MS, which is easily decomposed to NDH-1M and NDH-1S during solubilization with n-dodecyl β-d-maltoside (53). Recently, we found that the absence of NdhI and NdhK and their maturation factors such as Slr1097 predominantly destabilize the NDH-1M complex but scarcely influence the NDH-1L complex (data not shown). Furthermore, overexpression of ndhO might affect directly the stability of NdhI and NdhK both in NDH-1L and NDH-1M complexes because of a strong interaction of NdhO with NdhI and NdhK (Figs. 5A and 6A). Thus, the destabilization of NdhO on NDH-1M was most likely the result of influencing the stability of NdhI and NdhK in the complex. NDH-1L was evolved directly from complex I of E. coli and might have kept a relatively stable structure in the long term evolution process even in the absence of NdhI and NdhK, while NDH-1M, which was evolved after branching from the NDH-1L evolution tree, might have a structure more unstable than NDH-1L at least after destabilization of NdhI and NdhK in the NDH-1L and NDH-1M complexes. This may interpret the different effects of NdhO on the stability of NDH-1L and NDH-1M complexes.
Analysis of crystal structure of the hydrophilic domain of NDH-1 from T. thermophilus showed that Nqo9 and Nqo6, which correspond to NdhI and NdhK, respectively, in cyanobacterial NDH-1 complexes, bind three [4Fe-4S] clusters, N6a, N6b, and N2 (48, 54, 55), and form the terminal part of the electron transfer pathway to quinone (see red arrows in Fig. 8). We assumed that NdhO is localized between NdhI and NdhK based on the following reasons: (i) NdhO strongly interacts with NdhI and NdhK (Figs. 5A and 6A); (ii) NdhI interacts with NdhK (Figs. 5B and 6B); and (iii) NdhO blocks the interaction of NdhI with NdhK (Fig. 6B), as schematically represented in Fig. 8 (see pink letters). NdhO may block the electron transfer from NdhI to NdhK and destabilize the NDH-1M complex, thereby repressing the NDH-CET activity.
FIGURE 8.
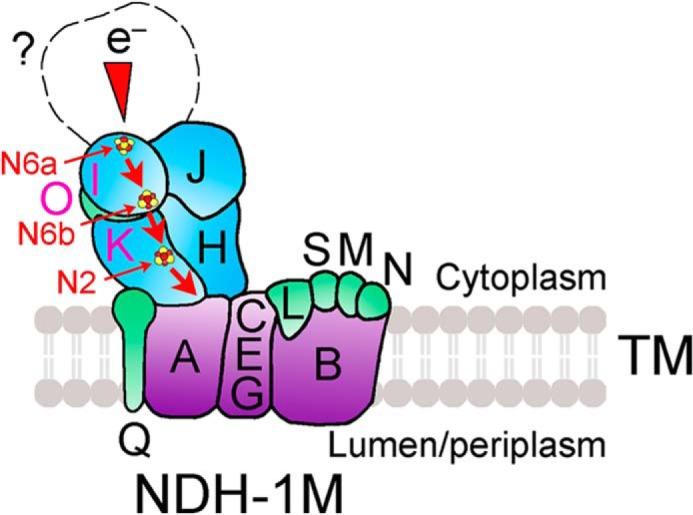
Schematic model of the localization of NdhO in NDH-1M complex. The electron transport route is shown in red. Electrons originating from unknown donor (question mark) are transferred via the chain of Fe-S clusters N6a, N6b and N2 (dark red-yellow spheres) in NdhI and NdhK. NdhO may be located between NdhI and NdhK and restrict the efficiency of electron transfer from N6b in NdhI to N2 in NdhK and destabilize the NDH-1M complex.
During the transfer of WT cells from growth light to high light, the expression levels of the majority of ndh genes were rapidly increased (56), thereby raising the NDH-CET activity (57). However, the level of ndhO did not change in this transfer process, and consequently the ratio of NdhO to NdhK and NdhI dropped (Fig. 7). It appears that NdhO was not directly involved in increasing the NDH-CET activity under high light, although the decrease of the relative amount of NdhO under such conditions might have assisted to increase the NDH-CET activity. Under high light, the increase in NDH-CET was more evident in ΔndhO but was less remarkable in OX-ndhO than in WT (data not shown). This may explain the different growth phenotypes of ΔndhO, WT, and OX-ndhO cells under high light (Fig. 3E).
Acknowledgments
We thank Professors C. Peter Wolk (Michigan State University) and Hongquan Yang (Shanghai Jiaotong University, China) for supplying pRL489 plasmid and yeast two-hybrid system, respectively.
This work was supported in part by National Natural Science Foundation of China Grant 31370270, National Basic Research Program of China Grant 2009CB118500, and Project of Shanghai Education Committee Grant 12ZZ132.

This article contains supplemental Table S1.
- NDH-1
- type I NADPH dehydrogenase
- NDH-1L
- large size NDH-1 complex
- NDH-1M
- medium size NDH-1 complex
- NDH-1S
- small size NDH-1 complex
- NDH-CET
- NDH-1-mediated CET
- OX-ndhO
- ndhO overexpression strain
- FR
- far-red light
- AL
- actinic light
- PQ
- plastoquinone
- BN
- Blue-Native
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MBP
- maltose-binding protein.
REFERENCES
- 1. Friedrich T., Steinmüller K., Weiss H. (1995) The proton-pumping respiratory complex I of bacteria and mitochondria and its homologue in chloroplasts. FEBS Lett. 367, 107–111 [DOI] [PubMed] [Google Scholar]
- 2. Yagi T., Yano T., Di Bernardo S., Matsuno-Yagi A. (1998) Procaryotic complex I (NDH-1), an overview. Biochim. Biophys. Acta 1364, 125–133 [DOI] [PubMed] [Google Scholar]
- 3. Friedrich T., Scheide D. (2000) The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 479, 1–5 [DOI] [PubMed] [Google Scholar]
- 4. Brandt U., Kerscher S., Dröse S., Zwicker K., Zickermann V. (2003) Proton pumping by NADH:ubiquinone oxidoreductase. A redox driven conformational change mechanism? FEBS Lett. 545, 9–17 [DOI] [PubMed] [Google Scholar]
- 5. Ogawa T. (1991) A gene homologous to the subunit-2 gene of NADH dehydrogenase is essential to inorganic carbon transport of Synechocystis PCC 6803. Proc. Natl. Acad. Sci. U.S.A. 88, 4275–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mi H., Endo T., Schreiber U., Ogawa T., Asada K. (1992) Electron donation from cyclic and respiratory flows to the photosynthetic intersystem chain is mediated by pyridine nucleotide dehydrogenase in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol. 33, 1233–1237 [Google Scholar]
- 7. Ohkawa H., Pakrasi H. B., Ogawa T. (2000) Two types of functionally distinct NAD(P)H dehydrogenases in Synechocystis sp. strain PCC6803. J. Biol. Chem. 275, 31630–31634 [DOI] [PubMed] [Google Scholar]
- 8. Arteni A. A., Zhang P., Battchikova N., Ogawa T., Aro E. M., Boekema E. J. (2006) Structural characterization of NDH-1 complexes of Thermosynechococcus elongatus by single particle electron microscopy. Biochim. Biophys. Acta 1757, 1469–1475 [DOI] [PubMed] [Google Scholar]
- 9. Battchikova N., Aro E. M. (2007) Cyanobacterial NDH-1 complexes: multiplicity in function and subunit composition. Physiol. Plant. 131, 22–32 [DOI] [PubMed] [Google Scholar]
- 10. Ogawa T., Mi H. (2007) Cyanobacterial NADPH dehydrogenase complexes. Photosynth. Res. 93, 69–77 [DOI] [PubMed] [Google Scholar]
- 11. Ma W. (2009) Identification, regulation and physiological functions of multiple NADPH dehydrogenase complexes in cyanobacteria. Front. Biol. China 4, 137–142 [Google Scholar]
- 12. Battchikova N., Eisenhut M., Aro E. M. (2011) Cyanobacterial NDH-1 complexes: Novel insights and remaining puzzles. Biochim. Biophys. Acta 1807, 935–944 [DOI] [PubMed] [Google Scholar]
- 13. Bernát G., Appel J., Ogawa T., Rögner M. (2011) Distinct roles of multiple NDH-1 complexes in the cyanobacterial electron transport network as revealed by kinetic analysis of P700+ reduction in various ndh-deficient mutants of Synechocystis sp. strain PCC6803. J. Bacteriol. 193, 292–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Battchikova N., Wei L., Du L., Bersanini L., Aro E. M., Ma W. (2011) Identification of a novel Ssl0352 protein (NdhS), essential for efficient operation of cyclic electron transport around photosystem I, in NADPH:plastoquinone oxidoreductase (NDH-1) complexes of Synechocystis sp. PCC 6803. J. Biol. Chem. 286, 36992–37001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dai H., Zhang L., Zhang J., Mi H., Ogawa T., Ma W. (2013) Identification of a cyanobacterial CRR6 protein, Slr1097, required for efficient assembly of NDH-1 complexes in Synechocystis sp. PCC 6803. Plant J. 75, 858–866 [DOI] [PubMed] [Google Scholar]
- 16. Endo T., Shikanai T., Takabayashi A., Asada K., Sato F. (1999) The role of chloroplastic NAD(P)H dehydrogenase in photoprotection. FEBS Lett. 457, 5–8 [DOI] [PubMed] [Google Scholar]
- 17. Birungi M., Folea M., Battchikova N., Xu M., Mi H., Ogawa T., Aro E. M., Boekema E. J. (2010) Possibilities of subunit localization with fluorescent protein tags and electron microscopy exemplified by a cyanobacterial NDH-1 study. Biochim. Biophys. Acta 1797, 1681–1686 [DOI] [PubMed] [Google Scholar]
- 18. Nowaczyk M. M., Wulfhorst H., Ryan C. M., Souda P., Zhang H., Cramer W. A., Whitelegge J. P. (2011) NdhP and NdhQ: two novel small subunits of the cyanobacterial NDH-1 complex. Biochemistry 50, 1121–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schwarz D., Schubert H., Georg J., Hess W. R., Hagemann M. (2013) The gene sml0013 of Synechocystis species strain PCC 6803 encodes for a novel subunit of the NAD(P)H oxidoreductase or complex I that is ubiquitous distributed among cyanobacteria. Plant Physiol. 163, 1191–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ifuku K., Endo T., Shikanai T., Aro E. M. (2011) Structure of the chloroplast NADH dehydrogenase-like complex: nomenclature for nuclear-encoded subunits. Plant Cell Physiol. 52, 1560–1568 [DOI] [PubMed] [Google Scholar]
- 21. Peng L., Yamamoto H., Shikanai T. (2011) Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim. Biophys. Acta 1807, 945–953 [DOI] [PubMed] [Google Scholar]
- 22. Suorsa M., Sirpiö S., Aro E. M. (2009) Towards characterization of the chloroplast NAD(P)H dehydrogenase complex. Mol. Plant 2, 1127–1140 [DOI] [PubMed] [Google Scholar]
- 23. Allen M. M. (1968) Simple conditions for growth of unicellular blue-green algae on plates. J. Phycol. 4, 1–4 [DOI] [PubMed] [Google Scholar]
- 24. Ma W., Wei L., Wang Q., Shi D., Chen H. (2007) Increased activity of the non-regulated enzymes fructose-1,6-bisphosphate aldolase and triose-phosphate isomerase in Anabaena sp. strain PCC 7120 increases photosynthetic yield. J. Appl. Phycol. 19, 207–213 [Google Scholar]
- 25. Elhai J., Wolk C. P. (1988) Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167, 747–754 [DOI] [PubMed] [Google Scholar]
- 26. Ma W., Shi D., Wang Q., Wei L., Chen H. (2005) Exogenous expression of the wheat chloroplastic fructose-1,6-bisphosphatase gene enhances photosynthesis in the transgenic cyanobacterium, Anabaena PCC7120. J. Appl. Phycol. 17, 273–280 [Google Scholar]
- 27. Ma W., Wei L., Wang Q., Shi D., Chen H. (2008) Increased activity of the tandem fructose-1,6-bisphosphate aldolase, triosephosphate isomerase and fructose-1,6-bisphosphatase enzymes in Anabaena sp. strain PCC 7120 stimulates photosynthetic yield. J. Appl. Phycol. 20, 437–443 [Google Scholar]
- 28. Ma W., Wei L., Long Z., Chen L., Wang Q. (2008) Increased activity of only an individual non-regulated enzyme fructose-1,6-bisphosphate aldolase in Anabaena sp. strain PCC 7120 stimulates photosynthetic yield. Acta Physiol. Plant. 30, 897–904 [Google Scholar]
- 29. Wei L., Ma W., Shi D., Wang Q. (2006) Modification of the N-terminal nucleotide sequence of mature hGM-CSF results in high expression in the foreign host, Anabaena sp. strain PCC 7120. J. Appl. Phycol. 18, 153–159 [Google Scholar]
- 30. Ma W., Mi H. (2005) Expression and activity of type-1 NAD(P)H dehydrogenase at different growth phases of cyanobacterium, Synechocystis PCC6803. Physiol. Plant. 125, 135–140 [Google Scholar]
- 31. Gombos Z., Wada H., Murata N. (1994) The recovery of photosynthesis from low-temperature photoinhibition is accelerated by the unsaturation of membrane lipids: a mechanism of chilling tolerance. Proc. Natl. Acad. Sci. U.S.A. 91, 8787–8791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kügler M., Jänsch L., Kruft V., Schmitz U. K., Braun H. P. (1997) Analysis of the chloroplast protein complexes by blue-native polyacrylamide gel electrophoresis (BN-PAGE). Photosynth. Res. 53, 35–44 [Google Scholar]
- 33. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 34. Sang Y., Li Q. H., Rubio V., Zhang Y. C., Mao J., Deng X. W., Yang H. Q. (2005) N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17, 1569–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mi H., Endo T., Ogawa T., Asada K. (1995) Thylakoid membrane-bound, NADPH-specific pyridine nucleotide dehydrogenase complex mediates cyclic electron transport in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 36, 661–668 [Google Scholar]
- 36. Deng Y., Ye J., Mi H. (2003) Effects of low CO2 on NAD(P)H dehydrogenase, a mediator of cyclic electron transport around photosystem I in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol. 44, 534–540 [DOI] [PubMed] [Google Scholar]
- 37. Shikanai T., Endo T., Hashimoto T., Yamada Y., Asada K., Yokota A. (1998) Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. U.S.A. 95, 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Burrows P. A., Sazanov L. A., Svab Z., Maliga P., Nixon P. J. (1998) Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 17, 868–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hashimoto M., Endo T., Peltier G., Tasaka M., Shikanai T. (2003) A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 36, 541–549 [DOI] [PubMed] [Google Scholar]
- 40. Wang P., Duan W., Takabayashi A., Endo T., Shikanai T., Ye J. Y., Mi H. (2006) Chloroplastic NAD(P)H dehydrogenase in tobacco leave functions in alleviation of oxidative damage caused by temperature stress. Plant Physiol. 141, 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peng L., Fukao Y., Fujiwara M., Takami T., Shikanai T. (2009) Efficient operation of NAD(P)H dehydrogenase requires the supercomplex formation with photosystem I via minor LHCI in Arabidopsis. Plant Cell 21, 3623–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peng L., Fukao Y., Myouga F., Motohashi R., Shinozaki K., Shikanai T. (2011) A chaperonin subunit with unique structures is essential for folding of a specific substrate. PLoS Biol. 9, e1001040. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43. Peng L., Fukao Y., Fujiwara M., Shikanai T. (2012) Multistep assembly of chloroplast NADH dehydrogenase-like subcomplex A requires several nucleus-encoded proteins, including CRR41 and CRR42, in Arabidopsis. Plant Cell 24, 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sirpiö S., Allahverdiyeva Y., Holmström M., Khrouchtchova A., Haldrup A., Battchikova N., Aro E. M. (2009) Novel nuclear-encoded subunits of the chloroplast NAD(P)H dehydrogenase complex. J. Biol. Chem. 284, 905–912 [DOI] [PubMed] [Google Scholar]
- 45. Yamamoto H., Peng L., Fukao Y., Shikanai T. (2011) An Src homology 3 domain-like fold protein forms a ferredoxin-binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 23, 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Armbruster U., Rühle T., Kreller R., Strotbek C., Zühlke J., Tadini L., Blunder T., Hertle A. P., Qi Y., Rengstl B., Nickelsen J., Frank W., Leister D. (2013) The photosynthesis affected mutant68–like protein evolved from a PSII assembly factor to mediate assembly of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell 25, 3926–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kaneko T., Sato S., Kotani H., Tanaka A., Asamizu E., Nakamura Y., Miyajima N., Hirosawa M., Sugiura M., Sasamoto S., Kimura T., Hosouchi T., Matsuno A., Muraki A., Nakazaki N., Naruo K., Okumura S., Shimpo S., Takeuchi C., Wada T., Watanabe A., Yamada M., Yasuda M., Tabata S. (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3, 109–136 [DOI] [PubMed] [Google Scholar]
- 48. Sazanov L. A., Hinchliffe P. (2006) Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311, 1430–1436 [DOI] [PubMed] [Google Scholar]
- 49. Battchikova N., Zhang P., Rudd S., Ogawa T., Aro E. M. (2005) Identification of NdhL and Ssl1690 (NdhO) in NDH-1L and NDH-1M complexes of Synechocystis sp. PCC 6803. J. Biol. Chem. 280, 2587–2595 [DOI] [PubMed] [Google Scholar]
- 50. Rumeau D., Bécuwe-Linka N., Beyly A., Louwagie M., Garin J., Peltier G. (2005) New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid Ndh complex functioning in higher plants. Plant Cell 17, 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Maul J. E., Lilly J. W., Cui L., dePamphilis C. W., Miller W., Harris E. H., Stern D. B. (2002) The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14, 2659–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang J., Gao F., Zhao J., Ogawa T., Wang Q., Ma W. (2014) NdhP is an exclusive subunit of large complex of NADPH dehydrogenase essential to stabilize the complex in Synechocystis sp. strain PCC 6803. J. Biol. Chem. 289, 18770–18781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang P., Battchikova N., Paakkarinen V., Katoh H., Iwai M., Ikeuchi M., Pakrasi H. B., Ogawa T., Aro E. M. (2005) Isolation, subunit composition and interaction of the NDH-1 complexes from Thermosynechococcus elongatus BP-1. Biochem. J. 390, 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Efremov R. G., Baradaran R., Sazanov L. A. (2010) The architecture of respiratory complex I. Nature 465, 441–445 [DOI] [PubMed] [Google Scholar]
- 55. Baradaran R., Berrisford J. M., Minhas G. S., Sazanov L. A. (2013) Crystal structure of the entire respiratory complex I. Nature 494, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hihara Y., Kamei A., Kanehisa M., Kaplan A., Ikeuchi M. (2001) DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13, 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mi H., Deng Y., Tanaka Y., Hibino T., Takabe T. (2001) Photo-induction of an NADPH dehydrogenase which functions as a mediator of electron transport to the intersystem chain in the cyanobacterium Synechocystis PCC6803. Photosynth. Res. 70, 167–173 [DOI] [PubMed] [Google Scholar]