Background: Soluble oligomers of α-synuclein, rather than the amyloid fibrils, are presumed to be more neurotoxic in Parkinson disease.
Results: A site-specific fluorescence approach is used to unravel the internal architecture of α-synuclein oligomers.
Conclusion: α-Synuclein oligomers are organized aggregates with a defined network of intermolecular contacts.
Significance: This contact map can be used for developing molecular models, essential for mechanistic studies and drug design.
Keywords: Alpha-synuclein (a-Synuclein), Amyloid, Fluorescence, Parkinson Disease, Protein Aggregation
Abstract
The presence of intraneuronal deposits mainly formed by amyloid fibrils of the presynaptic protein α-synuclein (AS) is a hallmark of Parkinson disease. Currently, neurotoxicity is attributed to prefibrillar oligomeric species rather than the insoluble aggregates, although their mechanisms of toxicity remain elusive. Structural details of the supramolecular organization of AS oligomers are critically needed to decipher the structure-toxicity relationship underlying their pathogenicity. In this study, we employed site-specific fluorescence to get a deeper insight into the internal architecture of AS oligomeric intermediates. We demonstrate that AS oligomers are ordered assemblies possessing a well defined pattern of intermolecular contacts. Some of these contacts involve regions that form the β-sheet core in the fibrillar state, although their spatial arrangement may differ in the two aggregated forms. However, even though the two termini are excluded from the fibrillar core, they are engaged in a number of intermolecular interactions within the oligomer. Therefore, substantial structural remodeling of early oligomeric interactions is essential for fibril growth. The intermolecular contacts identified in AS oligomers can serve as targets for the rational design of anti-amyloid compounds directed at preventing oligomeric interactions/reorganizations.
Introduction
Parkinson disease (PD)3 is a devastating neurodegenerative movement disorder affecting >2% of the population over 65. One signature of PD is the presence of eosinophilic, intraneuronal deposits of fibrillar and misfolded proteins in the affected brain areas. These inclusions are mainly formed by amyloid fibrils of the 140-amino acid presynaptic protein α-synuclein (AS) (1). Currently, prefibrillar soluble amyloid oligomers, rather than the insoluble aggregates, are pointed as the most neurotoxic species (1). Similar to other neurodegenerative diseases, the different pathways of oligomer pathobiology are thought to involve the establishment of aberrant protein interactions (2), the impairment of biomembranes (3), and the interference with the protein quality and clearance mechanisms (4). It appears that toxicity mediated by oligomeric intermediates is a general phenomenon related to their structure (5). In addition, certain AS aggregates may participate in the prion-like spreading of the pathology (6). Despite the relevance of prefibrillar intermediates in neurodegeneration and disease, detailed information about the internal architecture of AS oligomers (oAS) is still scarce. In addition to its importance for understanding the structural bases of oligomer-induced toxicity, obtaining molecular details of oAS is urgently needed for developing therapeutic agents/strategies aimed at ameliorating synucleinopathies.
Structurally, AS is a natively disordered monomer, whose fold as a helical tetramer in vivo is still matter of debate (7, 8). A complex network of transient tertiary contacts has been mapped in the monomeric disordered protein (9). In the fibrillar state, the monomers adopt a parallel, in-register five-layered (β1 to β5) β-sandwich arrangement spanning residues Leu38–Val95 (10) with the two termini protruding from the amyloid core (11), although polymorphs differing in the number and distribution of secondary structure elements have been reported (11, 12). In regard to oligomeric intermediates, it is recognized that oAS contain a substantial amount of β-sheet structure (13), lacking the canonical cross-β-fibrillar fold (14). Indeed, we demonstrated that oAS adopt a distinctive antiparallel β-sheet structure as opposed to the parallel organization found in the fibrillar state (15). Tryptophan (Trp) fluorescence and deuterium exchange studies showed that the core of oAS spans residues ∼Phe4–Ala90, where distinctive segments from the N-terminal and central part of the protein exhibit stable hydrogen-bonded or solvent-shielded structures (16, 17). In this study, we employed a number of engineered single cysteine AS variants selectively labeled with thiol-reactive fluorophores with the aim of identifying cooperatively folded units and intermolecular proximities within oAS with residue-specific resolution.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Several of the plasmids encoding for single cysteine variants of AS were a kind gift of Dr. C. Bertoncini (Institute for Research in Biomedicine, Barcelona, Spain) and Dr. T. Jovin (Max Planck Institute for Biophysical Chemistry, Göttingen, Germany). Other mutants were obtained from GeneScript Corp. (Piscataway, NJ). In all cases, the codon 136 was changed from TAC to TAT to prevent cysteine misincorporation upon bacterial expression (18). The single cysteine variants were expressed and purified as the wild-type protein (15), with the addition of 5 mm DTT in the elution buffers to avoid disulfide bridge formation. Proteins were dialyzed against 5 mm DTT in Milli-Q water and stored at −20 °C until further use. Protein concentrations were determined by absorbance (ϵ275 = 5600 m−1 cm−1).
Protein Labeling
The single-cysteine AS variants were labeled with acrylodan or N-(1-pyrene)-maleimide (Molecular Probes, Invitrogen) after removal of DTT by passage through a Sephadex G-25, PD-10 desalting column (General Electric) using 25 mm NaH2PO4/Na2HPO4, pH 6.5, as elution buffer. The working pH was maintained at 6.5 because N-(1-pyrene)-maleimide shows a tendency to undergo aminolysis at pH ≥ 7.5, especially in the presence of free amines (19). Protein fractions were identified by tyrosine fluorescence. A 4-fold excess of the corresponding dye in dimethyl sulfoxide was added to 300 μm protein solutions (the final dimethyl sulfoxide concentration was <1%). The samples were incubated overnight at 4 °C in the dark, and the reactions were quenched by the addition of 10 mm DTT. The labeled proteins were purified with PD-10 columns, and protein fractions were identified by fluorescence emission of the attached probes. The samples were centrifuged to remove any precipitate, concentrated in Amicon Ultra-4 10 KDa cut-off filters (Millipore), and buffer exchanged into Milli-Q water. Because the dye absorption bands strongly overlap with the protein absorption at 275 nm, protein concentrations were determined by the Lowry assay using wild-type AS (quantified by absorbance) as standard for the calibration curve. The labeling efficiencies, typically 70–85%, were calculated using ϵ372 = 16,400 m−1 cm−1 for acrylodan (20) and ϵ343 = 36000 m−1 cm−1 for pyrene (19). The labeled proteins were stored at −20 °C until further use. For control experiments, glutathione (GSH) was chosen as a mimetic of a polypeptide chain. Labeling of GSH was achieved by incubation of 100:1 GSH/dye solutions overnight at 4 °C in phosphate buffer. Solutions were centrifuged at 14,000 × g to remove any particle and lyophilized.
Oligomer Formation
Monomeric AS solutions (300 μm) were prepared by mixing the adequate amounts of wild-type and labeled proteins to give an initial label density of 5% acrylodan-tagged or 25% pyrene-tagged AS variant, unless stated otherwise. Typical initial volumes were 1 ml and 400 μl per independent experiment of the acrylodan- and pyrene-containing mixtures, respectively. The samples were freeze-dried and redissolved in phosphate buffer. Due to the stochastic nature of aggregation, labeled monomers are expected to be randomly incorporated in the oligomer (21, 22). Samples were centrifuged (20,000 × g, 30 min, 4 °C) to remove big particles. oAS were separated from the monomer using Amicon Ultra-0.5 100-kDa cut-off filters (Millipore). Diafiltration was carried out at 4 °C to diminish the tendency of fibril formation, which can be enhanced by the presence of the hydrophobic probe moieties (23) due to π-π stacking interactions between aromatic rings. The procedure was repeated until the fluorescence intensity of the probe detected in the flow-through was negligible as compared with the intensity measured in the retentate. oAS were in a final volume of 50–100 μl in phosphate buffer. (Typical final concentrations were ∼100–150 μm as measured by the Lowry assay.) The removal of the monomeric protein was confirmed by native gradient PAGE. The absence of amyloid fibrils was verified by Thioflavin T (ThioT) fluorescence. If needed, additional ultracentrifugation steps (50,000 × g, 30 min, 4 °C) were performed to settle out ThioT positive aggregates of the oAS solution. oAS duplicate samples were prepared independently of each other.
For the pyrene-labeled oAS, an additional guanidinium chloride (GdmCl) cold denaturation step was implemented to disrupt possible preformed protein aggregates that could be present in the highly concentrated labeled protein stock solutions. If present, these aggregates would result in biased excimer signals. Then, 300 μm stock AS solutions were kept overnight at −20 °C in the presence of 0.1 m GdmCl, a condition that suffices to disaggregate AS oligomers as appraised by red-shifted emission spectra and native gradient PAGE of acrylodan-labeled oAS (data not shown). GdmCl was exchanged into Milli-Q water before lyophilization, and oligomeric species were purified as described above.
Native Gradient PAGE
Native polyacrylamide gels were cast with a linear gradient from 4 to 15%. HMW native marker (GE Healthcare) was loaded as molecular mass marker. Electrophoresis was performed under non-denaturing conditions at a constant 60 V. Gels were silver-stained.
Transmission Electron Microscopy
Aliquots of oAS (5 μl) were adsorbed onto Formvar-coated carbon grids (200 mesh), washed with Milli-Q water, and stained with 1% (w/v) uranyl acetate. The samples were imaged in a JEM-1200 Ex (Jeol) transmission electron microscope equipped with a GATAN camera, model 785.
Attenuated Total Reflectance FTIR
IR spectra were acquired on an Equinox 55 IR spectrophotometer (Bruker Optics) equipped with a single reflection diamond reflectance accessory (Golden Gate, Specac) and purged with dry air. Aliquots of oAS (∼2 μl) were spread on a diamond crystal, flushed with nitrogen. A total of 256 accumulations were recorder at 21 °C using a resolution of 2 cm−1. The water vapor was subtracted, and the spectra were baseline-corrected between 1600 and 1700 cm−1.
Fluorescence Measurements
Emission spectra were acquired with a Cary Eclipse spectrofluorometer (Agilent Technologies) equipped with a thermally controlled multi-cuvette holder. Ten spectra were acquired at 25 °C using a 0.3-mm path cuvette and averaged. Acrylodan was excited at 380 nm, with excitation/emission slit widths of 5/5 nm. Pyrene was excited at 343 nm, with excitation/emission slit widths of 5/2.5 nm. ThioT was excited using λexc = 446 nm, spectral bandwidths of 10 nm and a 1-cm path cuvette. For denaturation experiments, each acrylodan-labeled sample was equilibrated for 30 min at room temperature by increasing GdmCl concentrations prior to spectral acquisition. Same spectral features were obtained in samples incubated up to 17 h, indicating that incubation for 30 min was enough to reach equilibrium (data not shown). Matched amounts of monomeric labeled proteins at 4 m GdmCl exhibited the same fluorescence yields regardless the position of the probe (data not shown). Therefore, after subtracting the corresponding blank curves, the acrylodan fluorescence spectra were scaled relative to the absolute intensity obtained at 4 m GdmCl for an easier comparison among the different variants. Denaturation profiles were expressed as fractional changes at 480 nm. For proximity analysis, pyrene spectra were normalized to the intensity of the peak at 375 nm (monomer signal). The ratio between the fluorescence intensities measured at 460 nm (excimer signal) and 375 nm were computed for each labeled oAS.
Bioinformatic Tools
The profiles describing the β-sheet and amyloid aggregation propensities were generated using the sequence-based algorithms TANGO (24), WALTZ (high sensitivity filter) (25), and Zyggregator (26). Averaged hydropathy indexes were calculated from hydropathy plots predicted according to the Kyte-Doolittle scale using a seven-amino acid window (27).
RESULTS
Sub-stoichiometrically Labeled oAS Behave Similar to Wild-type oAS
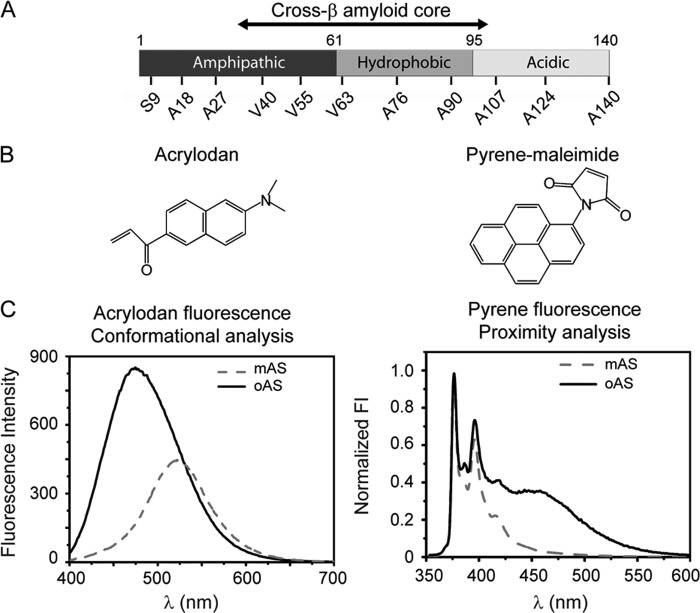
We capitalized on the absence of cysteine residues in wild-type AS for selectively labeling a number of engineered individual cysteine variants with thiol-reactive fluorophores. The mutations cover the three functional domains of AS (Fig. 1A), i.e. the amphipatic N-terminal domain (residues 1–60), which holds the familial mutations linked to early onset of PD, the hydrophobic aggregation-prone central region (residues 61–95) and the acidic C terminus (residues 96–140) that modulates fibrillogenesis. We selected the environment-sensitive fluorescent dye acrylodan to report on the conformational status of different segments of AS after protein assembly (Fig. 1C). In addition, we exploited the ability of pyrene to form distance-dependent excimers (excited state dimers) to map intermolecular proximities in neighboring monomers within oAS. Typical spectral responses of the dyes employed in this work (Fig. 1B) are represented in Fig. 1C.
FIGURE 1.
Shedding light on oAS structural features. A, functional domains of AS highlighting the amyloid β-sheet core (residues ∼Leu38–Val95) and the amino acids changed to cysteine for the specific labeling with thiol-reactive fluorophores. B, chemical structure of the fluorescent dyes used in this work. C, representative spectra of acrylodan-labeled (left) and pyrene-labeled (right) AS monomers (mAS) and oligomers (oAS). Information on the conformational status of different segments of AS was garnered from the solvatochromic response of acrylodan, whereas pyrene excimer emission reported on intermolecular proximities within oAS.
A number of methods for producing oAS have been reported in the literature (13, 14, 28–30). We have previously used a simple method based on freeze-drying of concentrated protein samples (15) to generate stable β-sheet -rich wild-type oAS exhibiting vesicle disruption properties (13, 29). As described under “Experimental Procedures,” we optimized here the protocol to avoid undesirable dye-dye stacking interactions before lyophilization and fibril formation during the purification steps of labeled oAS. We then characterized the oligomers by complementary techniques and representative results obtained for oAS labeled with acrylodan at position Ala76 are presented in Fig. 2. The results obtained for the wild-type protein are also shown for comparison.
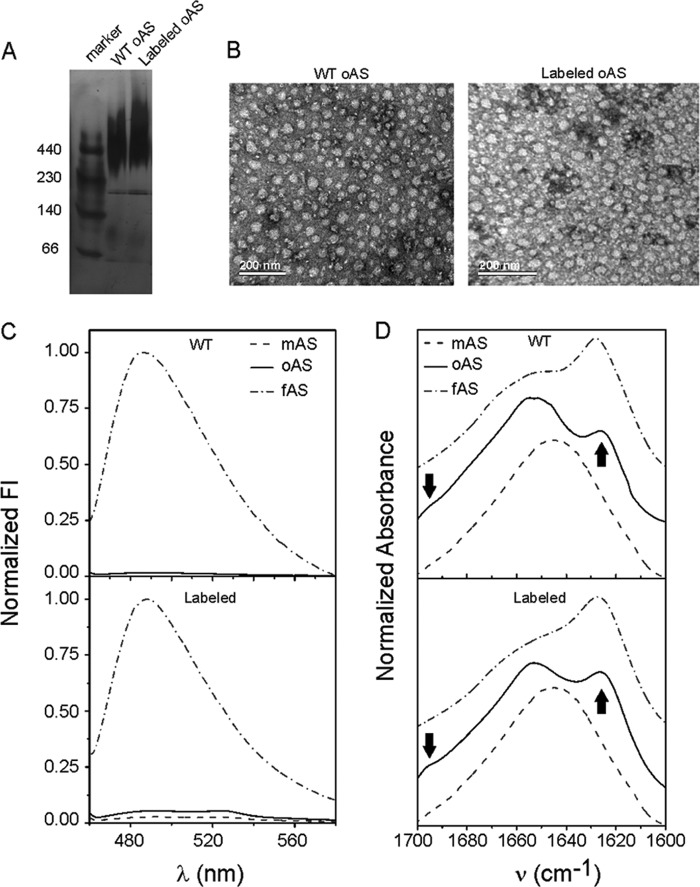
FIGURE 2.
Validating general properties of labeled oAS. A, native gradient PAGE with a polyacrylamide gradient from 4 to 15% stained with silver nitrate. B, uranyl acetate-stained transmission electron microscopy images of wild-type and labeled oAS. C, fluorescence emission spectra of ThioT in the presence of wild-type (upper panel) and labeled (lower panel) oAS along with the corresponding signals for AS monomers (mAS) and fibrils (fAS). D, normalized FTIR spectra in the amide I region of wild-type (upper panel) and labeled (lower panel) mAS, oAS, and fAS. Arrows point the characteristic IR bands at 1625 cm−1 and 1695 cm−1 assigned to the antiparallel β-sheet structure. Shown is the acrylodan-labeled Ala76 variant as a representative case.
Labeled oAS showed the typical smeared band in a native gradient PAGE gel migrating concomitantly with the molecular mass marker ferritin of ∼440 kDa, as the wild-type oligomers (Fig. 2A). The incorporation of the labeled protein does not alter the morphology of the oligomers, as both labeled and unlabeled species appeared as polydisperse spheroidal aggregates (Fig. 2B). In addition, neither labeled nor unlabeled oAS enhanced the fluorescence of ThioT, which became highly fluorescent upon intercalation into the amyloid core of fibrils (Fig. 2C) indicating that both oAS lack the canonical cross-β-amyloid structure. Finally, the distinctive IR signatures of the different aggregation states were present in both the labeled and wild-type proteins (15), i.e. the spectra of the monomeric proteins were centered at ∼1645 cm−1, as expected for predominantly unordered conformations; the amide I band of the oligomeric species presented a band at 1625 cm−1 along with a shoulder at 1695 cm−1, the hallmark of antiparallel β-sheet structure; lastly, the FTIR spectra of fibrils displayed prominent maxima at ∼1627 cm−1, typical of parallel β-sheets (Fig. 2D). Therefore, we concluded that labeled oAS recapitulate the morphology, polydispersity, tinctorial properties, and secondary structure signatures of amyloid oligomers formed by the wild-type protein (15).
Site-specific Environmental Polarity and Conformational Stability
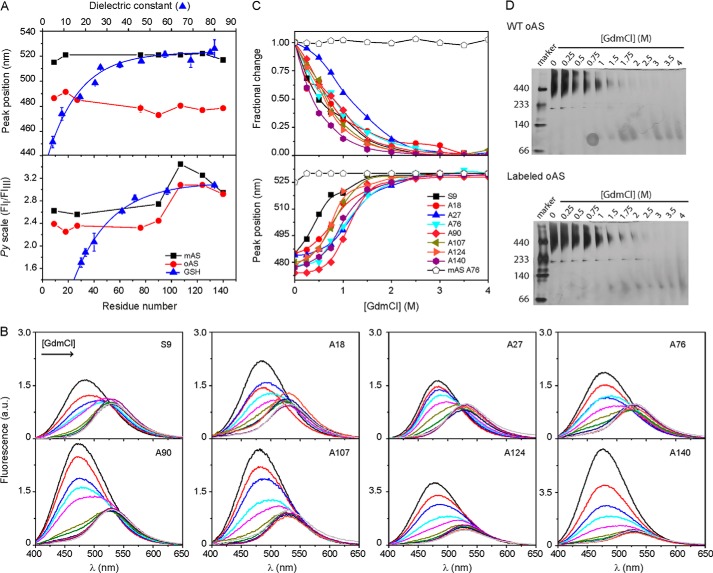
We mapped the environmental properties of labeled oAS with site-specific resolution. All of the tested positions of acrylodan-labeled oAS showed hyperchromic blue-shifted emission spectra (λmax = 476–491 nm) as compared with the monomeric proteins (λmax = 515–528 nm) indicating sequestration from the aqueous solvent (Figs. 1C and 3A, upper panel). In the framework of a continuous dielectric model, the solvatochromic response of acrylodan spans a polarity range defined by a dielectric constant (ϵ) of ∼5–40, as judged by the reference curve obtained from the fluorescence spectra of acrylodan-labeled GSH in neat solvents (upper panel of Fig. 3A). Based on the spectral features of this dye, all of the labeled positions in oAS experience low-polarity microenvironments with apparent ϵ ∼ 10–20, substantially lower than those (ϵ ≥ 50) found for the same positions in the monomeric form. The ϵ values appraised for oAS are in the same range as those estimated from the spectral analysis of Trp residues selectively positioned at the N-terminal and central domains of the protein in single-Trp oAS mutants (ϵ ∼ 10–30) (16). For the sake of comparison, we note that an apparent ϵ value of ∼11 was estimated for wild-type AS fibrils when bound to the extrinsic dual-emission probe 4′-(diethylamino)-3-hydroxyflavone (31).
FIGURE 3.
Mapping environmental properties. A, spectral signatures of acrylodan- and pyrene-labeled AS monomers (■) and oligomers (●). The peak wavelength of acrylodan emission (upper panel) and the Py scale of pyrene (lower panel) defined as the ratio of the intensities of the first (375 nm) and third (385 nm) emission bands, are plotted as a function of the labeled position. Initial label density for oAS preparation was 5% in both cases. The spectral variation of the dyes conjugated to GSH in solvents of increasing dielectric constants (▴) are shown for comparison. The polarity reference curves were obtained from the spectra of labeled GSH redissolved in the following: Cl3CH, Cl3CH:ethanol (12.5%), n-butanol, ethanol, mixtures of H2O:dioxane (50, 45, 35, 25, 15, and 5%) and H2O for acrylodan, and the protic solvents n-butanol, n-propanol, ethanol, ethylene glycol, glycerine, formic acid, and H2O for pyrene. Dye concentration was ∼ 1 μm. B, acrylodan fluorescence spectra of oAS labeled at different positions along the protein sequence at increasing GdmCl concentrations. C, site-specific GdmCl denaturation profiles of acrylodan-labeled oAS. Fractional changes in fluorescence intensity at 480 nm (upper panel) and peak wavelength (lower panel) of acrylodan emission are plotted as a function of GdmCl concentrations. The spectral changes of the monomer labeled at position Ala76 as a function of denaturant concentration is also shown for comparison (open symbols). D, native gradient (4–15%) PAGE of wild-type (upper panel) and acrylodan-labeled (lower panel) oAS at increasing GdmCl concentrations. The gels were overstained to reveal the presence, if any, of low-populated partially dissociated intermediates.
In addition, we employed the relative changes in intensity of characteristic vibronic bands of pyrene monomer fluorescence (region between ∼370 and 410 nm, Fig. 1C) as a complementary tool to evaluate the polarity of the probe surroundings. The Py scale of polarity is defined by the intensity ratio between the first (375 nm) and third (385 nm) emission bands. The higher the Py value, the higher the polarity of the environment (32). A drop of the Py value observed upon oligomerization, attested to an increase in the local hydrophobicity around the probe for all the labeled positions except for Ala140 (Fig. 3A, lower panel). Comparison with the calibration curve measured for pyrene-labeled GSH dissolved in protic solvents yielded apparent ϵ values of ∼ 40 and 30 for the labeled positions within the N-terminal and central parts of the protein in its monomeric and oligomeric form, respectively. The three most C-terminal labeled residues in both AS monomers and oligomers exhibited Py values that lie within the polarity range where the reference curve approximates a plateau. At these positions, the pyrene probe senses much more polar microenvironments with apparent ϵ values ≥60. These raised Py values correlates with the high net charge (−16) and the strong predicted hydrophilicity of the C-terminal domain (averaged hydropathy indexes are −0.14, +0.80, and −1.48 for the N-terminal, central, and C-terminal domains, respectively). It is worth noting that the shape of the polarity profiles of pyrene-labeled AS species resembled those of single Trp-AS variants (16). In this sense, it appears that the complex photophysical cycles behind Trp and pyrene fluorescence are influenced by the specific chemical nature of this region of the protein because the same trend was observed for AS monomers and oligomers. In the case of the reported Trp variants, apparent ϵ values between 50 and 65 can be estimated for oAS labeled at two sites within the C-terminal domain (16).
The discrepancies in the absolute apparent ϵ values estimated from the spectral response of different solvatochromic dyes (acrylodan and pyrene in this work and Trp in Ref. 16) may arise from additional parameters such as hydration, viscosity, and specific interactions, which influence the microenvironment relaxation of the excited state of the probes (32). These parameters, tough to predict in the highly anisotropic protein environment, make an unequivocal correlation between the steady state response of the probes and polarity in terms of macroscopic ϵ values, difficult to achieve. Due to these experimental limitations, we consider the relative changes between two different scenarios, i.e. the monomeric and oligomeric forms of the protein, more informative than the absolute values per se. Overall, our data indicate that all the tested positions are in low-polarity surroundings upon oligomerization, independently of the probe employed in the assay.
We then evaluated the chemical stability of oAS by performing site-specific equilibrium denaturation studies. The steady-state acrylodan fluorescence spectra of each labeled oAS at increasing GdmCl concentrations are displayed in Fig. 3B. The emission spectra of acrylodan-labeled variants decreased in intensity and shifted to the red (from 477–485 to 530 nm) at increasing denaturant concentrations, consistent with the removal of the probe from a low-polarity interior into the solvent environment (Fig. 3B). Because fluorescence intensity is directly related to the mole fraction of states (33), we used the changes in intensity at 480 nm as an observable parameter for probing the conformational status of different regions of the protein in the aggregates (Fig. 3C, upper panel). We choose 480 nm due to the larger sensitivity of this spectral region, but the same trend was obtained by analyzing the intensity changes at 510 nm. The red-shifts in λmax are also shown for comparison (Fig. 3C, lower panel).
At 0 m GdmCl, the positions toward the C-terminal domain, i.e. Ala124 and Ala140, exhibited higher yields than the other positions, which might reflect the absence of quenching interactions at this region (Fig. 3B). The lack of clearly defined isosbestic points in the fluorescence emission spectra indicates that the chemical equilibrium unfolding transitions of oAS is more complex than a two-state mechanism. The transitions started at the lowest (0.25 m) GdmCl concentration and finished at 1.75 m with apparent midpoints at 0.65 m (Fig. 3C, upper panel). Positions Ala140 and Ala27 exhibited somehow lower and higher stabilities with apparent midpoints at 0.35 and 1.1 m GdmCl, respectively (Fig. 3C, upper panel). The shifts in λmax did not correlate with the changes in intensity, probably due to the non-two state nature of the unfolding transitions (Fig. 3C, lower panel). However, one should keep in mind that peak position is not, a priori, proportional to the population of the states and could lead to a biased assignment toward the state with the higher quantum yield (33) such as the aggregated native state in our case. The global unfolding transitions were estimated at ∼1 m GdmCl for both labeled and unlabeled oAS from native gradient PAGE (Fig. 3D), close to that determined for the Ala27 variant (Fig. 3C, upper panel). Therefore, the complex chemical-induced unfolding behavior observed by fluorescence might be due to local changes around the attached probes prior to global unfolding.
oAS Display a Well Defined Network of Intermolecular Contacts
Then, we exploited the ability of pyrene to form distance-dependent excimers to map intermolecular proximities in neighboring monomers within oAS. When two pyrene moieties are positioned 4–10 Å apart from each other, the long fluorescent lifetime of pyrene allows the interaction of one molecule in the excited state to interact with an unexcited molecule before energy emission, giving rise to a red-shifted band in the 430–470 nm region. Thus, the excimer ratio (ratio of the excimer/monomer fluorescence intensities, FI460/FI375) is a relative indicator of the extent of excimer formation and, therefore, the spatial proximity between two pyrene moieties.
Oligomers carrying pyrene at single sites (25% labeled, 75% wild-type protein) displayed excimer fluorescence (λmax ∼ 460 nm) depending on the labeled position (Fig. 4A). The observation of weak excimer signals at some positions indicates that pyrene moieties do not artificially induce intermolecular interactions, but rather report on the spatial arrangement of AS monomers within the oligomers. As a further confirmation, we performed additional experiments at varying initial label densities of the Ala76 variant, mutant that exhibited the highest excimer ratio (see below). The decrease in excimer signals with the increase in the proportion of wild-type protein (inset in Fig. 4A) demonstrated that pyrene moieties do not show preferential association/proximity with each other upon aggregation and that low excimer ratios are obtained when the probability of two pyrene molecules of being spatially close is reduced.
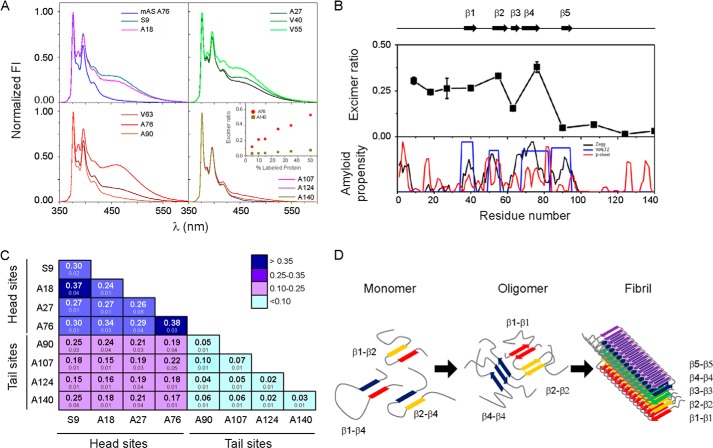
FIGURE 4.
Identifying intermolecular contacts in oAS. A, fluorescence emission spectra of oAS labeled with pyrene at different positions along the protein sequence. The monomer labeled at position Ala76 is shown for comparison. The spectra were normalized to the intensity of the monomeric peak (375 nm). The inset shows the excimer ratio of oAS labeled at position Ala76 (●) and Ala140 (■) at increasing initial label density. B, proximity analysis assessed by excimer fluorescence in oAS labeled with pyrene at single sites (upper panel) along with AS aggregation propensity predicted by Zyaggregator (black line) and WALTZ (blue line), and β-sheet propensity (red line) computed by TANGO (lower panel). β-Strands defined within the cross-β-fibrillar amyloid core are represented as black arrows and numbered β1 to β5. C, excimer fluorescence of oAS assembled from 1:1 mixtures of two pyrene-labeled AS variants. Mean values of independent duplicates ± S.D. are indicated. Head and tail sites are marked with lines. D, schematic representation of structural reorganization of segments participating in the cross-β-amyloid core during AS fibril formation. Transient tertiary contacts in the monomeric protein (9) can foster aggregation. Rearrengements of the network of antiparallel β-sheet interactions (Ref. 15 and this work) lead to the fibrillar state, where five-stranded β-sheet monomers are packed in a parallel, in-register fashion (10).
The profile of excimer ratios as a function of residue number (Fig. 4B) revealed high values for residues at the N-terminal and central parts of the protein (N-terminal to-Ala76, head sites), with position Ala76 bearing the highest signal, denoting close proximity (<10 Å) between the same positions on nearby molecules in oAS. On the contrary, the weak excimer bands at the C-terminal portion (Ala90-to-C-terminal, tail sites) point that labeled residues are not proximal (distances >10 Å) between each other. These results stress the importance of the N-terminal region in amyloid self-assembly.
Finally, we mixed a number of selected pyrene-labeled AS monomers in all equimolar pair-wise combinations to identify potential intermolecular interactions involving residues located in different parts of the protein (Fig. 4C). In this case, the amount of each of the labeled protein was reduced to half (12.5% each labeled protein, 75% wild-type protein) to diminish excimer signals expected from self-self-interactions of proteins labeled within the head and tail regions. For those variants exhibiting high excimer fluorescence (head sites in Fig. 4B), a decrease by half in the initial label density resulted in an ∼50% reduction of the excimer signal (data not shown), similar to the trend previously shown for the Ala76 variant (inset in Fig. 4A). As for the oligomers tagged in single positions, all oAS labeled at the tail region produced very low excimer signals (Fig. 4C), suggesting that these positions do not contribute to intermolecular contacts per se. The strongest signal arose from oAS labeled at head sites, whereas intermediates values were measured for combined tail and head labeled oAS. Thus, excimer signals revealed no new contributions from the Head region, except likely for the Ser9–Ala18 combination, and no interactions between sites assayed at the head and tail loci (Fig. 4C).
DISCUSSION
Elucidating the supramolecular arrangement of oligomeric prefibrillar intermediates of AS remains elusive. This task has been hampered by the transient nature and inherent polydispersity of such species and the low amount they can be produced in vitro. Several methods have been developed to generate stable oligomers within a short time frame at the required yields to allow their characterization (13, 14, 28, 30). These methods could lead to structurally and functionally distinct oligomeric species. For instance, it has been shown that oligomers differing in their ability to induce membrane cell permeability and intracellular seeding (28) or aggregation number (21, 22) can be obtained depending on the aggregation conditions. However, with the knowledge that the driving force of aggregation is the same for all species, that the toxicity associated to amyloid oligomers appears to be related to their structure (34), and that a variety of prefibrillar intermediates share structural epitopes (35), one can expect common structural features among the different oligomer populations. Despite the experimental difficulties detailed above, some progress has been made on our current knowledge of the structure of oAS. Indeed, it has been demonstrated that oAS contain a substantial amount of non-fibrillar β-sheets (13, 14) and that they are arranged in an antiparallel orientation (15, 30). Within the oligomeric core spanning residues ∼Phe4–Ala90 (16), discrete segments Phe4–Ala17, Tyr39–Thr54, and Val70–Ala89 exhibit stable hydrogen-bonded or solvent-shielded structures (17). In this work, we relied on the rational site-specific positioning of fluorescent tags along the protein sequence to obtain a more detailed representation of the internal architecture of oAS. Site-specific fluorescence has proven valuable to garner structural features on amyloid assemblies (36).
The solvatochromic sensitivities of acrylodan and pyrene reported on the microenvironmental changes upon AS oligomerization. All of the tested positions in oAS were shielded from the solvent as compared with the monomeric proteins (Fig. 3A). This fact points a significant difference on the structural arrangement of the monomeric units in oligomers and amyloid fibrils because the two termini are excluded from the fibrillar amyloid core (11). The global unfolding transition of oAS, reflecting the conformational stability of the aggregated species, was estimated at ∼1 m GdmCl (Fig. 3D). As judged from the apparent midpoint denaturation values, the site-specific unfolding transition of variant Ala27 was found to be similar to the global stability of the oligomer assembly, whereas the stabilities of the remaining variants were somehow lower (Fig. 3C). Thus, it appears that local instabilities give rise to the buildup of intermediates preceding the global unfolding/dissociation of the aggregates.
Unlike the most commonly used FRET pairs, which allow the estimation of distances between ∼20 and 80 Å, pyrene excimer fluorescence reports on a much shorter distance range, typically 4–10 Å. This ability of pyrene has been successfully employed to gather structural details of amyloid fibrils formed by a yeast prion (37) and toxic amyloid oligomers assembled from a non-disease related protein (38) as well as for monitoring α-synuclein amyloid formation (23). Inspired by these works, we relied on excimer signals arising from pyrene-labeled oAS to infer about intermolecular proximities among the monomers.
Among the amino acid segments that form the β-strands within the AS fibrillar fold (10), positions Val40, Val55, and Ala76 exhibited the highest excimer ratios in oAS carrying pyrene at single sites (Fig. 4B), indicating close proximity between the labeled residues and the same residues on one or more nearby molecules. Notably, these residues belong to β1 (Val37–Lys43), β2 (Val52–Thr59) and β4 (Gly68–Val77) strands (10), regions that contain a critical anchoring point (Tyr39) of amyloid inhibitors (39), a mutation site (A53T) linked to early-onset PD and faster oligomerization and fibrillation rates in vitro (40, 41), and the hydrophobic stretch (Val71–Val82) critical for fibril formation (42), respectively. Therefore, we postulate that fibril-like β1-β1, β2-β2, and β4-β4 contacts may be engaged in oligomeric antiparallel β-sheet interactions (15). Although we cannot exclude that excimer signals were due to layered β-strands, our interpretation is in line with the high propensity of these segments to form β-sheets and aggregate into amyloids (Fig. 4B). However, based on the extremely low excimer intensity of the Ala90 variant, we suggest that β5 strand, if formed, would not be involved in β5-β5-sheet contacts in such a way that favor excimer formation (Fig. 4B).
High excimer signals were also determined for oAS assembled from proteins labeled at head sites that are not involved in fibrillar β-sheet contacts (Fig. 4B) (10, 11). These results strongly suggest a somehow tight packing at least for the region encompassing residues Ser9–Ala76 and likely reflect the adoption of a well defined structure. These findings correlate with previous studies delimiting the core of oAS between residues ∼Phe4–Ala90 (16), where specific discrete stretches of amino acids present hampered backbone dynamics (17). Moreover, our data highlight the key role of the N-terminal part of the protein on early oligomeric interactions. In this regard, we observe that a number of small-molecule anti-amyloid compounds preferentially target the regions enclosing residues ∼Val3–Lys23 and/or Leu38–Gly51 (39, 43).
As discussed previously, our excimer profile suggests that β5 region is not proximal in oAS (Fig. 4B). Taking into account that position Ala90 experiences low polarity surroundings upon oligomerization (Fig. 3A) and bears high amyloid propensity (Fig. 4B), we expect the locus enclosing position Ala90 be structured and establishing interchain interactions. Although we did not find indications of spatial proximity between Ala90 and the other sites at the head and tail loci (Fig. 4C), it still could be interacting with other residues in the protein. This hypothesis is in line with the fact that β5 region establishes long range tertiary contacts in the monomeric protein (9) and that position 90 is shielded from the solvent and protected from a soluble quencher in Trp-engineered oligomers (16).
A similar behavior was observed for other residues located in the tail region, i.e. they appeared to be spatially apart from each other (Fig. 4B) and from other residues from the head region (Fig. 4C), but they experienced a more hydrophobic environment as compared with the monomeric protein (Fig. 3A). Based on these results and taking in consideration the avidity of the C terminus for establishing intra- (9, 44–46) and interchain (47) tertiary N-/C-terminal interactions and the sensitivity of a variety of fluorophores positioned at the two termini of the protein to early stages of amyloid formation (23, 48–50), we expect tail sites to be packed against other regions of the protein and, likely, participating in regular interactions/structures.
As discussed above, AS amyloid formation demands significant structural reorganization during self-assembly. Such reorganization, particularly for those regions involved in the cross-β fibrillar core, is schematically depicted in Fig. 4D. A number of transient tertiary contacts are present in the soluble monomeric protein, which could direct the formation of early aggregation-prone species (9). Then, antiparallel β-sheet-rich oligomeric intermediates are buildup (15), probably involving β1-β1, β2-β2, and β4-β4 intermolecular interactions (this work). A remodeling of the network of these early β-sheet interactions leads to the formation of the parallel in-register amyloid fibril where each monomer is folded as a 5-layered β-sandwich (10).
Our fluorescence-based approach in conjunction with determinations of structural features (14, 15), dynamics patterns (17), and aggregation numbers (21, 22) indicate that oAS are ordered assemblies with distinct sizes. Our current data provide structural constraints for the development of molecular models of oAS, which is crucial for improving the understanding of the structural bases of oligomer-induced toxicity and for the rational design of anti-amyloidogenic drugs directed at preventing oligomeric interactions/reorganizations.
Acknowledgments
We thank C. Bertoncini (Institute for Research in Biomedicine, Barcelona) and T. Jovin (Max Planck Institute for Biophysical Chemistry, Göttingen) for several of the cysteine-AS plasmids. We also thank our colleagues at Centro de Investigaciones en Química Biológica de Córdoba for many helpful discussions.
This work was supported in part by grants from Agencia FONCYT PICT 1913, CONICET PIP-2011-2013, SECyT-UNC, and ISN-CAEN-2012.
- PD
- Parkinson disease
- AS
- α-synuclein
- oAS
- AS oligomer(s)
- GSH
- glutathione
- ThioT
- tHioflavin T.
REFERENCES
- 1. Lashuel H. A., Overk C. R., Oueslati A., Masliah E. (2013) The many faces of α-synuclein: from structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 14, 38–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Olzscha H., Schermann S. M., Woerner A. C., Pinkert S., Hecht M. H., Tartaglia G. G., Vendruscolo M., Hayer-Hartl M., Hartl F. U., Vabulas R. M. (2011) Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144, 67–78 [DOI] [PubMed] [Google Scholar]
- 3. Butterfield S. M., Lashuel H. A. (2010) Amyloidogenic protein-membrane interactions: mechanistic insight from model systems. Angew. Chem. Int. Ed. Engl. 49, 5628–5654 [DOI] [PubMed] [Google Scholar]
- 4. Morimoto R. I. (2008) Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 22, 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bucciantini M., Giannoni E., Chiti F., Baroni F., Formigli L., Zurdo J., Taddei N., Ramponi G., Dobson C. M., Stefani M. (2002) Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416, 507–511 [DOI] [PubMed] [Google Scholar]
- 6. Frost B., Diamond M. I. (2010) Prion-like mechanisms in neurodegenerative diseases. Nat. Rev. Neurosci. 11, 155–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bartels T., Choi J. G., Selkoe D. J. (2011) α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 477, 107–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fauvet B., Mbefo M. K., Fares M. B., Desobry C., Michael S., Ardah M. T., Tsika E., Coune P., Prudent M., Lion N., Eliezer D., Moore D. J., Schneider B., Aebischer P., El-Agnaf O. M., Masliah E., Lashuel H. A. (2012) α-Synuclein in central nervous system and from erythrocytes, mammalian cells, and Escherichia coli exists predominantly as disordered monomer. J. Biol. Chem. 287, 15345–15364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Esteban-Martín S., Silvestre-Ryan J., Bertoncini C. W., Salvatella X. (2013) Identification of fibril-like tertiary contacts in soluble monomeric α-synuclein. Biophys. J. 105, 1192–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vilar M., Chou H. T., Lührs T., Maji S. K., Riek-Loher D., Verel R., Manning G., Stahlberg H., Riek R. (2008) The fold of α-synuclein fibrils. Proc. Natl. Acad. Sci. U.S.A. 105, 8637–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heise H., Hoyer W., Becker S., Andronesi O. C., Riedel D., Baldus M. (2005) Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid-state NMR. Proc. Natl. Acad. Sci. U.S.A. 102, 15871–15876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bousset L., Pieri L., Ruiz-Arlandis G., Gath J., Jensen P. H., Habenstein B., Madiona K., Olieric V., Böckmann A., Meier B. H., Melki R. (2013) Structural and functional characterization of two α-synuclein strains. Nat. Commun. 4, 2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Volles M. J., Lee S. J., Rochet J. C., Shtilerman M. D., Ding T. T., Kessler J. C., Lansbury P. T., Jr. (2001) Vesicle permeabilization by protofibrillar α-synuclein: implications for the pathogenesis and treatment of Parkinson's disease. Biochemistry 40, 7812–7819 [DOI] [PubMed] [Google Scholar]
- 14. Kim H. Y., Cho M. K., Kumar A., Maier E., Siebenhaar C., Becker S., Fernandez C. O., Lashuel H. A., Benz R., Lange A., Zweckstetter M. (2009) Structural properties of pore-forming oligomers of α-synuclein. J. Am. Chem. Soc. 131, 17482–17489 [DOI] [PubMed] [Google Scholar]
- 15. Celej M. S., Sarroukh R., Goormaghtigh E., Fidelio G. D., Ruysschaert J. M., Raussens V. (2012) Toxic prefibrillar α-synuclein amyloid oligomers adopt a distinctive antiparallel β-sheet structure. Biochem. J. 443, 719–726 [DOI] [PubMed] [Google Scholar]
- 16. van Rooijen B. D., van Leijenhorst-Groener K. A., Claessens M. M., Subramaniam V. (2009) Tryptophan fluorescence reveals structural features of α-synuclein oligomers. J. Mol. Biol. 394, 826–833 [DOI] [PubMed] [Google Scholar]
- 17. Mysling S., Betzer C., Jensen P. H., Jorgensen T. J. (2013) Characterizing the dynamics of α-synuclein oligomers using hydrogen/deuterium exchange monitored by mass spectrometry. Biochemistry 52, 9097–9103 [DOI] [PubMed] [Google Scholar]
- 18. Masuda M., Dohmae N., Nonaka T., Oikawa T., Hisanaga S., Goedert M., Hasegawa M. (2006) Cysteine misincorporation in bacterially expressed human α-synuclein. FEBS Lett. 580, 1775–1779 [DOI] [PubMed] [Google Scholar]
- 19. Wu C. W., Yarbrough L. R. (1976) N-(1-pyrene)maleimide: a fluorescent cross-linking reagent. Biochemistry 15, 2863–2868 [DOI] [PubMed] [Google Scholar]
- 20. Prendergast F. G., Meyer M., Carlson G. L., Iida S., Potter J. D. (1983) Synthesis, spectral properties, and use of 6-acryloyl-2-dimethylaminonaphthalene (Acrylodan). A thiol-selective, polarity-sensitive fluorescent probe. J. Biol. Chem. 258, 7541–7544 [PubMed] [Google Scholar]
- 21. Zijlstra N., Claessens M. M., Blum C., Subramaniam V. (2014) Elucidating the aggregation number of dopamine-induced α-synuclein oligomeric assemblies. Biophys. J. 106, 440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zijlstra N., Blum C., Segers-Nolten I. M., Claessens M. M., Subramaniam V. (2012) Molecular composition of sub-stoichiometrically labeled α-synuclein oligomers determined by single-molecule photobleaching. Angew Chem. Int. Ed. Engl. 51, 8821–8824 [DOI] [PubMed] [Google Scholar]
- 23. Thirunavukkuarasu S., Jares-Erijman E. A., Jovin T. M. (2008) Multiparametric fluorescence detection of early stages in the amyloid protein aggregation of pyrene-labeled α-synuclein. J. Mol. Biol. 378, 1064–1073 [DOI] [PubMed] [Google Scholar]
- 24. Fernandez-Escamilla A. M., Rousseau F., Schymkowitz J., Serrano L. (2004) Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 22, 1302–1306 [DOI] [PubMed] [Google Scholar]
- 25. Maurer-Stroh S., Debulpaep M., Kuemmerer N., Lopez de la Paz M., Martins I. C., Reumers J., Morris K. L., Copland A., Serpell L., Serrano L., Schymkowitz J. W., Rousseau F. (2010) Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat. Methods 7, 237–242 [DOI] [PubMed] [Google Scholar]
- 26. Tartaglia G. G., Vendruscolo M. (2008) The Zyggregator method for predicting protein aggregation propensities. Chem. Soc. Rev. 37, 1395–1401 [DOI] [PubMed] [Google Scholar]
- 27. Kyte J., Doolittle R. F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 28. Danzer K. M., Haasen D., Karow A. R., Moussaud S., Habeck M., Giese A., Kretzschmar H., Hengerer B., Kostka M. (2007) Different species of α-synuclein oligomers induce calcium influx and seeding. J. Neurosci. 27, 9220–9232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Rooijen B. D., Claessens M. M., Subramaniam V. (2009) Lipid bilayer disruption by oligomeric α-synuclein depends on bilayer charge and accessibility of the hydrophobic core. Biochim Biophys. Acta 1788, 1271–1278 [DOI] [PubMed] [Google Scholar]
- 30. Lorenzen N., Nielsen S. B., Buell A. K., Kaspersen J. D., Arosio P., Vad B. S., Paslawski W., Christiansen G., Valnickova-Hansen Z., Andreasen M., Enghild J. J., Pedersen J. S., Dobson C. M., Knowles T. P., Otzen D. E. (2014) The role of stable α-synuclein oligomers in the molecular events underlying amyloid formation. J. Am. Chem. Soc. 136, 3859–3868 [DOI] [PubMed] [Google Scholar]
- 31. Celej M. S., Caarls W., Demchenko A. P., Jovin T. M. (2009) A triple emission fluorescent probe reveals distinctive amyloid fibrillar polymorphism of wild-type α-synuclein and its familial Parkinson's disease mutants. Biochemistry 48, 7465–7472 [DOI] [PubMed] [Google Scholar]
- 32. Valeur B. (2001) Molecular Fluorescence: Principles and Applications, 1st Ed., pp. 200–225, Wiley-VCH, Weinheim, Germany [Google Scholar]
- 33. Eftink M. R. (1994) The use of fluorescence methods to monitor unfolding transitions in proteins. Biophys. J. 66, 482–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bucciantini M., Calloni G., Chiti F., Formigli L., Nosi D., Dobson C. M., Stefani M. (2004) Prefibrillar amyloid protein aggregates share common features of cytotoxicity. J. Biol. Chem. 279, 31374–31382 [DOI] [PubMed] [Google Scholar]
- 35. Glabe C. G. (2008) Structural classification of toxic amyloid oligomers. J. Biol. Chem. 283, 29639–29643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bertoncini C. W., Celej M. S. (2011) Small molecule fluorescent probes for the detection of amyloid self-assembly in vitro and in vivo. Curr. Protein Pept Sci. 12, 205–220 [DOI] [PubMed] [Google Scholar]
- 37. Krishnan R., Lindquist S. L. (2005) Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435, 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Campioni S., Mannini B., Zampagni M., Pensalfini A., Parrini C., Evangelisti E., Relini A., Stefani M., Dobson C. M., Cecchi C., Chiti F. (2010) A causative link between the structure of aberrant protein oligomers and their toxicity. Nat. Chem. Biol. 6, 140–147 [DOI] [PubMed] [Google Scholar]
- 39. Lamberto G. R., Binolfi A., Orcellet M. L., Bertoncini C. W., Zweckstetter M., Griesinger C., Fernández C. O. (2009) Structural and mechanistic basis behind the inhibitory interaction of PcTS on α-synuclein amyloid fibril formation. Proc. Natl. Acad. Sci. U.S.A. 106, 21057–21062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Conway K. A., Harper J. D., Lansbury P. T. (1998) Accelerated in vitro fibril formation by a mutant α-synuclein linked to early-onset Parkinson disease. Nat. Med. 4, 1318–1320 [DOI] [PubMed] [Google Scholar]
- 41. Conway K. A., Lee S. J., Rochet J. C., Ding T. T., Williamson R. E., Lansbury P. T., Jr. (2000) Acceleration of oligomerization, not fibrillization, is a shared property of both α-synuclein mutations linked to early-onset Parkinson's disease: implications for pathogenesis and therapy. Proc. Natl. Acad. Sci. U.S.A. 97, 571–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Giasson B. I., Murray I. V., Trojanowski J. Q., Lee V. M. (2001) A hydrophobic stretch of 12 amino acid residues in the middle of α-synuclein is essential for filament assembly. J. Biol. Chem. 276, 2380–2386 [DOI] [PubMed] [Google Scholar]
- 43. Rao J. N., Dua V., Ulmer T. S. (2008) Characterization of α-synuclein interactions with selected aggregation-inhibiting small molecules. Biochemistry 47, 4651–4656 [DOI] [PubMed] [Google Scholar]
- 44. Bertoncini C. W., Jung Y. S., Fernandez C. O., Hoyer W., Griesinger C., Jovin T. M., Zweckstetter M. (2005) Release of long-range tertiary interactions potentiates aggregation of natively unstructured α-synuclein. Proc. Natl. Acad. Sci. U.S.A. 102, 1430–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dedmon M. M., Lindorff-Larsen K., Christodoulou J., Vendruscolo M., Dobson C. M. (2005) Mapping long-range interactions in α-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J. Am. Chem. Soc. 127, 476–477 [DOI] [PubMed] [Google Scholar]
- 46. Lee J. C., Gray H. B., Winkler J. R. (2005) Tertiary contact formation in α-synuclein probed by electron transfer. J. Am. Chem. Soc. 127, 16388–16389 [DOI] [PubMed] [Google Scholar]
- 47. Wu K. P., Baum J. (2010) Detection of transient interchain interactions in the intrinsically disordered protein α-synuclein by NMR paramagnetic relaxation enhancement. J. Am. Chem. Soc. 132, 5546–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kaylor J., Bodner N., Edridge S., Yamin G., Hong D. P., Fink A. L. (2005) Characterization of oligomeric intermediates in α-synuclein fibrillation: FRET studies of Y125W/Y133F/Y136F α-synuclein. J. Mol. Biol. 353, 357–372 [DOI] [PubMed] [Google Scholar]
- 49. Yushchenko D. A., Fauerbach J. A., Thirunavukkuarasu S., Jares-Erijman E. A., Jovin T. M. (2010) Fluorescent ratiometric MFC probe sensitive to early stages of α-synuclein aggregation. J. Am. Chem. Soc. 132, 7860–7861 [DOI] [PubMed] [Google Scholar]
- 50. Yap T. L., Pfefferkorn C. M., Lee J. C. (2011) Residue-specific fluorescent probes of α-synuclein: detection of early events at the N- and C-termini during fibril assembly. Biochemistry 50, 1963–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]