Background: FOXP3 is a key transcription factor for the development and function of Tregs.
Results: PIM1-mediated phosphorylation of FOXP3 at serine 422 decreased its DNA binding activity.
Conclusion: PIM1 negatively regulates FOXP3-mediated transcriptional regulation and the suppressive activity of Tregs.
Significance: PIM1 is a newly identified negative regulator of the immunosuppressive activity of Tregs.
Keywords: Autoimmune Disease, DNA Binding Protein, Gene Regulation, Immunosuppression, Inflammation, Phosphorylation, FOXP3, PIM1, Tregs
Abstract
Previous reports have suggested that human CD4+ CD25hiFOXP3+ T regulatory cells (Tregs) have functional plasticity and may differentiate into effector T cells under inflammation. The molecular mechanisms underlying these findings remain unclear. Here we identified the residue serine 422 of human FOXP3 as a phosphorylation site that regulates its function, which is not present in murine Foxp3. PIM1 kinase, which is highly expressed in human Tregs, was found to be able to interact with and to phosphorylate human FOXP3 at serine 422. T cell receptor (TCR) signaling inhibits PIM1 induction, whereas IL-6 promotes PIM1 expression in in vitro expanded human Tregs. PIM1 negatively regulates FOXP3 chromatin binding activity by specifically phosphorylating FOXP3 at Ser422. Our data also suggest that phosphorylation of FOXP3 at the Ser418 site could prevent FOXP3 phosphorylation at Ser422 mediated by PIM1. Knockdown of PIM1 in in vitro expanded human Tregs promoted FOXP3-induced target gene expression, including CD25, CTLA4, and glucocorticoid-induced tumor necrosis factor receptor (GITR), or weakened FOXP3-suppressed IL-2 gene expression and enhanced the immunosuppressive activity of Tregs. Furthermore, PIM1-specific inhibitor boosted FOXP3 DNA binding activity in in vitro expanded primary Tregs and also enhanced their suppressive activity toward the proliferation of T effector cells. Taken together, our findings suggest that PIM1 could be a new potential therapeutic target in the prevention and treatment of human-specific autoimmune diseases because of its ability to modulate the immunosuppressive activity of human Tregs.
Introduction
Regulatory T cells (Tregs)4 that express the transcription factor forkhead box P3 (FOXP3) are essential for limiting inflammation and autoimmune responses in the immune system (1). Tregs are generally categorized into natural Tregs that originate in the thymus and induced Tregs that arise in the periphery (2). Continuous high-level expression of FOXP3 protein is indispensable for the maintenance of the suppressive function for both natural Tregs and induced Tregs (3). Adoptive transfer of in vitro expanded Tregs could be a potential approach to prevent overactivation of immune responses or to treat autoimmune diseases.
FOXP3 promotes the expression of a series of Treg marker genes, including cytotoxic T lymphocyte antigen-4 (CTLA-4), GITR, and CD25, and represses the expression of IL-2. Although the exact mechanisms by which Tregs maintain immune self-tolerance and homeostasis are still not fully understood, CTLA-4 expression and IL-2 repression have been shown to play a critical role (4). Therefore, understanding the mechanisms behind the regulation of FOXP3 activity in human Tregs should be of primary importance for identifying potential therapeutic targets and for the treatment of human autoimmune diseases.
The PIM kinase family is comprised of three proto-oncogene-encoded proteins acting as constitutively active serine/threonine protein kinases (PIM1, PIM2, and PIM3) (5). Although PIM kinases are functionally involved in cell proliferation, survival, apoptosis, and drug resistance (6), PIM kinase triple-deficient mice (pim1−/−pim2−/−pim3−/−) have no significant phenotypes compared with wild-type mice except for reduced body size (7). PIM1 localizes both in the cytoplasm and the nucleus, and its expression in lymphocytes can be induced by the STAT family of transcription factors in lymphocytes (8). In the nucleus, different transcription factors, including c-Myc (9) and NFATc1 (10), have been shown to recruit PIM1 to chromatin and coordinate to activate the transcription of their target genes.
Previous findings have shown that phosphorylation affects FOXP3 protein stability (11) and DNA binding activity (12). In this study, we unraveled a new mechanism by which human FOXP3 activity is regulated by PIM1. PIM1 expression could be down-regulated by TCR signaling and induced by the inflammatory cytokine IL-6 in in vitro expanded Tregs. We found that PIM1 phosphorylated human FOXP3 at Ser422 and prevented FOXP3 binding to its target genes, consequently decreasing its transcriptional activity. Upon treatment with the PIM1 specific inhibitor 3-cyano-4-phenyl-6-(3-bromo-6-hydroxy)phenyl-2(1H)-pyridone (PIM1i), in vitro expanded Treg cells showed enhanced suppressive activity in vitro. These findings reveal a hitherto unrecognized function of PIM1 in negatively regulating the transcriptional activity of human FOXP3, consequently dampening the suppressive function of in vitro expanded Tregs. Therefore, PIM1 could represent a promising therapeutic target for the treatment of diseases where the induction of Treg function is required.
EXPERIMENTAL PROCEDURES
Cell Culture, Transduction, and Transfection
Human HEK 293T and Jurkat T cells were cultured as described previously (13). Indicated plasmids were transfected into HEK 293T cells using Lipofectamine 2000 reagent (Invitrogen), whereas FuGENE 6 (Roche) was used in transfection experiments of Jurkat T cells or primary T cells according to the instructions of the manufacturer. For PIM knockdown experiments, shPIM1 or shPIM2 or its control shCK lentiviral vector-containing lentivirus were prepared as described previously (13). Cells were cultured at 37 °C in a 5% CO2-humidified incubator.
Isolation and in Vitro Expansion of Human Tregs
Human PBMCs were isolated from the buffy coat of healthy donors (Shanghai Blood Center). CD4+CD25hiCD127low Tregs were purified using a FACSAria II cell sorter (BD Biosciences). The purity of the sorted cells was 95–99%. In vitro expansion of isolated Tregs was performed in X-VIVO (Lonza) medium supplemented with 10% human AB serum, 1% GlutaMax (Invitrogen), 1% NaPyr (Invitrogen), and 100 μm rapamycin (Sigma) for the first 2 days and then including 500 units/ml rIL-2 (R&D Systems) and anti-human CD3/CD28-conjugated Dynabeads (Invitrogen) at a bead-to-cell ratio of 4:1 for further expansion. Freshly isolated Tregs were usually expanded for 7–10 days before experimentation. The phenotypes of the expanded cells were characterized by intracellular detection of FOXP3 expression by flow cytometry on a BD Fortessa flow cytometer (BD Biosciences).
Quantitative PCR (Q-PCR)
Total RNA was isolated from whole cells using TRIzol reagent (Invitrogen). Q-PCR was carried out on an ABI 7900HT sequence detection system. Relative target gene mRNA expression was normalized to β-actin. The following primers were used for detection: FOXP3, 5′-TCCCAGAGTTCCTCCACAAC- 3′ and 5′-ATTGAGTGTCCGCTGCTTCT-3′; PIM1, 5′-CCTGCTGTATGATATGGTGTG-3′ and 5′-GAGGTGGATCTCAGCAGTTTC-3′; PIM2, 5′-GTCACTGGGCATCCTCCTC-3′ and 5′-CCTCGGCTGGTGTTTGCATC-3′; PIM3, 5′-AAGCTCATCGACTTCGGTTC-3′ and 5′-AGGATCTCCTCGTCCTGCTC-3′; CTLA4, 5′-CTTCTCTTCATCCCTGTCTTC-3′ and 5′-AAGGTCAACTCATTCCCCATC-3′; CD25, 5′-GAGACGTCCATATTTACAACAG-3′ and 5′-CCTTTGATTTCACTTGGGCTTC-3′; GITR, 5′-AGTGGGACTGCATGTGTGTC-3′ and 5′-GCAGTCTGTCCAAGGTTTGC-3′; IL-2, 5′-GCAACTCCTGTCTTGCATTG-3′ and 5′-CAGTTCTGTGGCCTTCTTGG-3′; and β-actin, 5′-GGACTTCGAGCAAGAGATGG-3′ and 5′-AGCACTGTGTTGGCGTACAG-3′.
Immunoblot Analysis and Immunoprecipitation
Cells were lysed in radioimmune precipitation assay buffer containing 50 mm Tris/HCl (pH 7.4), 1% Nonidet P-40, 0.5% sodium deoxycholate, 150 mm NaCl, 1 mm EDTA, 1 mm PMSF, 1 mm Na3VO4, 1 mm NaF, and protease inhibitor (Sigma). Lysates were cleared by centrifugation, and the supernatants were immunoprecipitated with the indicated antibodies. Samples were then analyzed by Western blotting, and the protein expression level was quantified with National Institutes of Health ImageJ software.
Plasmids and Antibodies
FLAG-tagged FOXP3 or HA-tagged FOXP3 were constructed as described previously (14), and we cloned PIM1 with the same method. The shRNA-expressing vector pLKO.1 was purchased from Addgene. The shRNA sequence for PIM1 was 5′-GATACTCTCTTCTTCTCATAGC-3′. The antibodies used for Western blotting were as follows: anti-FLAG (catalog no. M2, Sigma), anti-HA (catalog no. F-7, Santa Cruz Biotechnology), anti-FOXP3 (hFOXY, eBioscience), anti-α-tubulin (catalog no. DM1A, Sigma), anti-β-actin (catalog no. 6G3, Sungene Biotech). Phospho-Ser422-FOXP3 site-specific polyclonal antibodies were generated by Abmart with the phosphorylated peptide 418SQRP(pS)RCSN426, and the following antibodies were used for flow cytometry: anti-CD25-PE (catalog no. BC96, Biolegend), anti-CD4-FITC (catalog no. RPA-T4, Biolegend), anti-CD127-PE-Cy7 (catalog no. eBioRDR5, eBioscience), anti-FOXP3-PE (catalog no. 236A/E7, eBioscience), and anti-CD8-APC (catalog no. RPA-T8, BD Biosciences).
In Vitro Kinase Assay
An in vitro kinase assay was performed as described previously (15) with some modifications. MBP-FOXP3 and His-PIM1 kinase or its K67M mutant were expressed in bacteria and affinity-purified with amylose resin for FOXP3 or nickel-nitrilotriacetic acid-agarose for PIM1 separately. Mixed purified MBP-FOXP3 (100 ng) and His-PIM1 or its K67M mutant (100 ng/each) were incubated in 50 μl of kinase reaction buffer, kept at room temperature for 30 min, and then stopped with 4× Laemmli buffer. Western blotting was performed and FOXP3 phosphorylation status was tested with phospho-Ser422-FOXP3-specific polyclonal antibodies.
Cytoplasmic, Nuclear, and Chromatin Fractionation
HA-FOXP3 or its mutants were coexpressed with or without FLAG-PIM1 in 10 million 293T cells for 48 h, and then cells were harvested and lysed with 400 μl of cytoplasm buffer (10 mm Tris-HCl, pH 7.5, 10 mm KCl, 0.1 mm EDTA, 1 mm DTT, and 0.6% Nonidet P-40) in the presence of protease inhibitor 1 mm PMSF on ice for 30 min. The pellet was spun down at 13,000 rpm for 30 s and the supernatant, constituting the cytoplasmic fraction, was recovered. A volume of 150 μl of nuclear extract buffer (20 mm Tris-HCl (pH 8.0), 400 mm NaCl, 1 mm EDTA, 1 mm DTT, 1 mm PMSF, 1 mm Na3VO4, 1 mm NaF, and protease inhibitor mixture (Sigma)) was added to the pellet and incubated under rotation in a cold room for 30 min. The supernatant, constituting the nuclear extract, was collected after centrifugation at 13,000 rpm for 10 min. The resulting pellet was added to 100 μl of 0.2 m HCl, rotated in a cold room for 30 min, and then spun down at 13,000 rpm for 10 min. The supernatant was collected and neutralized with 35 μl of 1 m Tris-HCl (pH 8.0). All samples collected from the above procedures were subjected to Western blot analysis.
Micro ChIP (μChIP) Analysis
μChIP analysis was performed as described previously (16). Briefly, 50,000–100,000 cells for each sample were cross-linked with 1% formaldehyde. Cells were then treated with lysis buffer, and chromatin was sonicated to produce fragments of 500–800 bp. The sheared chromatin was diluted with radioimmune precipitation assay ChIP buffer and then immunoprecipitated with 2 μg of FLAG M2 monoclonal antibody (Sigma) or anti-FOXP3 (hFOXY, eBioscience) separately. Mouse IgG was used as a negative control. The human IL-2 promoter primers used for Q-PCR detection were as follows: 5′-CTCTAGCTGACATGTAAGAAG-3′ and 5′-GCATATTGTGGTGGACAAGAG-3′.
In Vitro Suppression Assay
To determine the suppressive activity of human Tregs, the proliferation of CD8+ T cells in carboxyfluorescein succinimidyl ester (CFSE, Invitrogen)-labeled PBMCs was analyzed. Briefly, fresh human PBMCs were labeled with 5 μm CFSE for 10 min at 37 °C and then cocultured in 96-well plates with different ratios of PIM1i-pretreated Tregs in the presence of anti-CD3/CD28 beads. After culture for 3 days, cells were harvested and stained with viability dye (fixable viability dye eFluor® 780, eBioscience) and anti-CD8-APC antibody and then analyzed on a Fortessa flow cytometer (BD Biosciences).
Statistical Analysis
Unpaired Student's t test was used for statistical analyses on GraphPad Prism software.
RESULTS
PIM1 Interacts with FOXP3 and Phosphorylates FOXP3 at Ser422
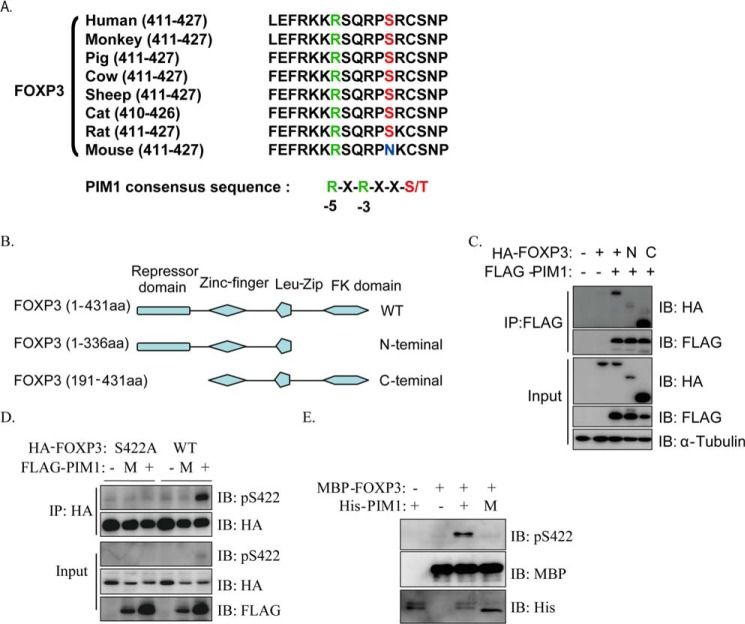
On the basis of the predicted result on the Kinexus Bioinformatics Corp. website, we identified Ser422 in human FOXP3 as a potential phosphorylation site for the kinase PIM1. Sequence alignment of FOXP3 from different species showed that Ser422 is highly conserved among mammals, except in mice, where all conserved sequences fulfilled the high selectivity of PIM1 for arginine at the 5 position relative to the potential phosphorylated serine/threonine within its target motif R-X-R-X-X-S/T (17) (Fig. 1A). Through ectopic expression of PIM1 together with FOXP3, N-terminal FOXP3 or C-terminal FOXP3 in HEK293T cells, followed by coimmunoprecipitation, we found that FOXP3 could indeed interact with PIM1 and that the interaction mainly happened on the C terminus of FOXP3 (Fig. 1, B and C). Next, we generated phospho-Ser422-FOXP3 (phospho-Ser422)-specific polyclonal antibodies by immunizing rabbits with the synthetic phosphopeptide 418SQRP(pS)RCSN426.
FIGURE 1.
PIM1 interacts with FOXP3 and phosphorylates FOXP3 at Ser422. A, sequence alignment of the potential PIM1 phosphorylation sites within FOXP3 orthologues from different species. B and C, different truncated FOXP3 constructs were generated as shown and were cotransfected with or without FLAG-PIM1 into 293T cells. Cell lysates were immunoprecipitated (IP) using anti-FLAG antibody, and FOXP3 levels were detected by Western blotting (IB). aa, amino acids. D, HA-FOXP3 or its S422A mutant was cotransfected with PIM1 or its kinase-deficient mutant (M, K67M) into 293T cells. Coimmunoprecipitation was performed with anti-HA antibody. E, His-PIM1 or its mutant and MBP-FOXP3 were purified from bacteria and incubated in MOPS buffer at 30 °C for 30 min. FOXP3 phosphorylation was detected with anti-phospho-Ser422-FOXP3-specific polyclonal antibody.
To determine whether FOXP3 could be phosphorylated by PIM1, we coexpressed HA-tagged FOXP3 or its S422A mutant with FLAG-tagged PIM1 or its kinase-deficient K67M mutant in HEK 293T cells. FOXP3 was immunoprecipitated with anti-HA antibody, subjected to Western blotting, and then the level of phospho-Ser422 was detected. The results indicated that PIM1 could phosphorylate FOXP3 at Ser422, whereas its kinase activity-deficient mutant K67M could not (Fig. 1D). MBP-FOXP3, His-PIM1, and its K67M mutant were expressed in bacteria to carry out a cell-free phosphorylation assay. The purified recombinant proteins were incubated with ATP, and Western blotting was performed. Wild-type PIM1, but not its kinase activity-deficient mutant, phosphorylated FOXP3 at Ser422 (Fig. 1E), indicating that PIM1 can phosphorylate FOXP3 directly. Together, our data indicate that PIM1 can interact and phosphorylate FOXP3 at the Ser422 site.
PIM1 Is Highly Expressed in Freshly Isolated Human Tregs
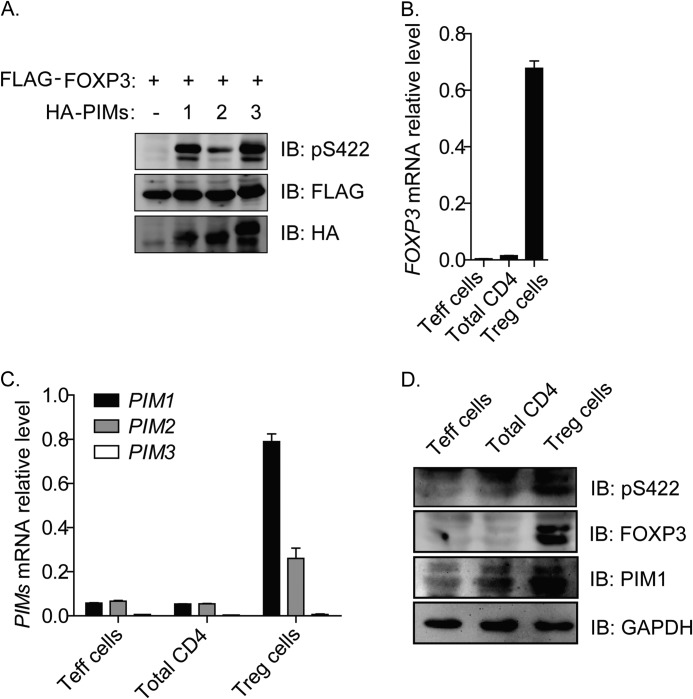
The three PIM kinase family members PIM1, PIM2, and PIM3 share similar substrate specificity (5). To test whether the three PIM kinases differently affect FOXP3 phosphorylation at the amino acid residue Ser422, we coexpressed FOXP3 with each PIM kinase family member and found that, although all the three members could phosphorylate FOXP3 at Ser422, the efficiency of PIM2 was lower than PIM1 and PIM3 (Fig. 2A).
FIGURE 2.
Comparison of PIM kinase activity on Ser422-FOXP3 phosphorylation and analysis of their expression profiles in freshly isolated human Tregs. A, FLAG-tagged FOXP3 constructs were cotransfected with the HA-tagged PIM kinase family members PIM1, PIM2, and PIM3 or control empty vector into 293T cells. FOXP3 phosphorylation was detected with anti-phospho-Ser422-FOXP3-specific polyclonal antibody by Western blotting (IB). B and C, FOXP3, PIM1, PIM2, and PIM3 mRNA expression profiles in freshly isolated total CD4+ T cells (Total CD4), CD4+CD25− T cells (Teff cells) and CD4+CD25hiCD127low Tregs (Treg cells). D, FOXP3 and PIM1 protein expression profiles and phosphorylation status of residue Ser422 in freshly isolated T cell subsets.
Next, we further tested the expression profile of PIM family members in human Tregs. Previous studies have found that PIM1 is a constitutively active kinase (15). We sorted fresh total CD4+ T cells, CD4+CD25− T cells, and CD4+CD25hiCD127low Tregs from human peripheral blood of healthy donors and performed Q-PCR and Western blot analysis of PIM1 expression. FOXP3 was shown to be highly expressed both at the mRNA level and protein level in freshly isolated Tregs (Fig. 2, B and D). Surprisingly, we found that PIM1 mRNA expression was higher than PIM2 in freshly isolated human CD4+CD25hiCD127low Tregs (Fig. 2C), although it has been reported previously that PIM2 is highly expressed (18). We also noted that PIM1, PIM2, and PIM3 were expressed at very low levels in freshly isolated human CD4+CD25− and total CD4+ T cells (Fig. 2C). Additionally, PIM1 was highly expressed at the protein level, and FOXP3 was highly phosphorylated at the Ser422 residue in freshly isolated Tregs without in vitro expansion, as shown by Western blot analysis (Fig. 2D).
TCR Signaling and IL-6 Signaling Regulate PIM1 Expression Reversely
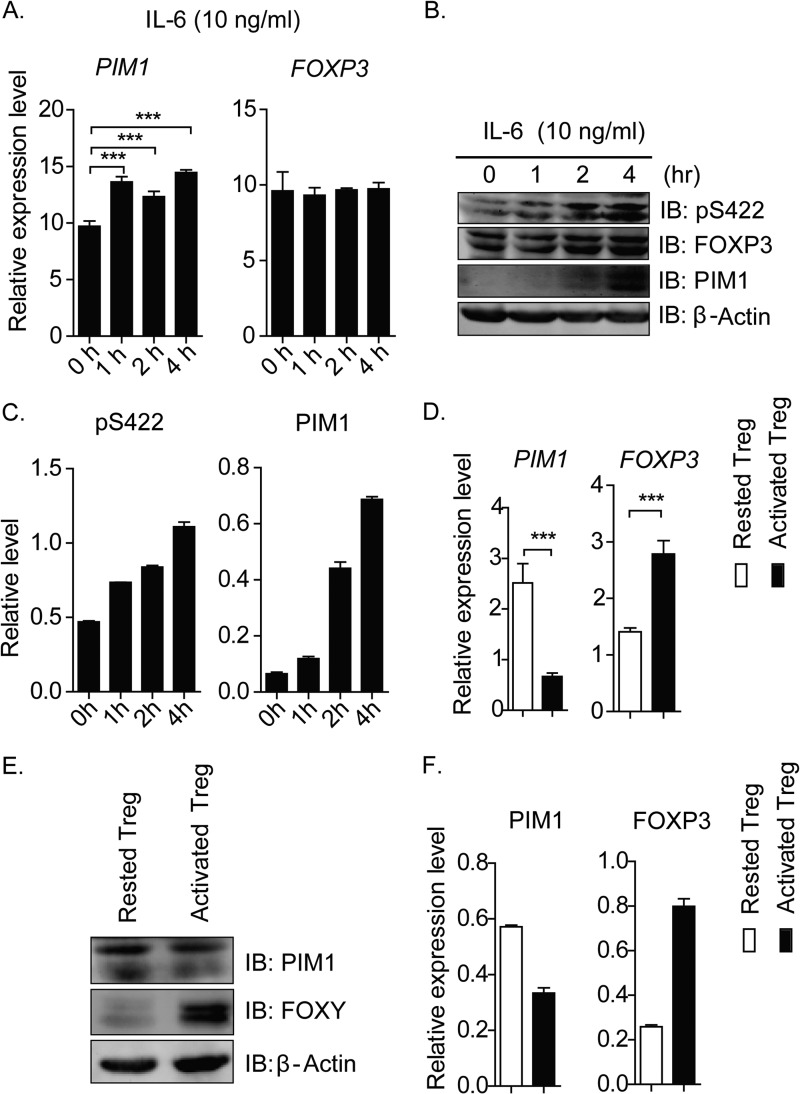
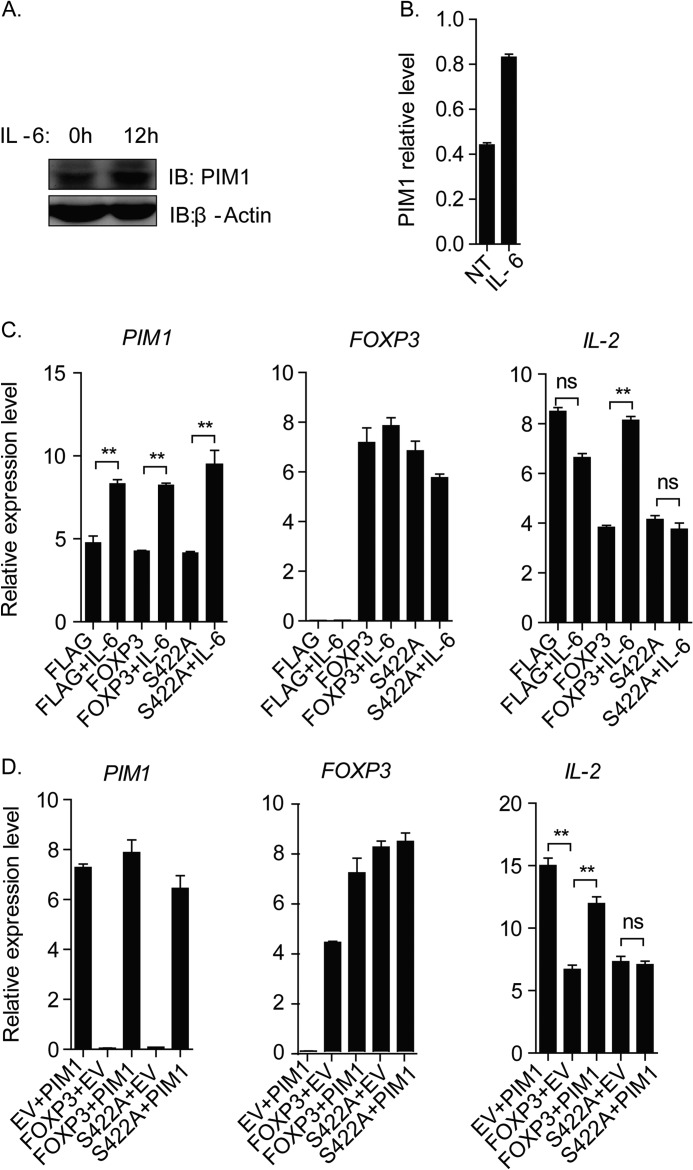
PIM1 is a major downstream target of the STAT proteins, and various inflammatory cytokines can induce its expression (19). Therefore, we took the classical inflammatory cytokine IL-6 as an example to test our hypothesis and found that the inflammatory cytokine IL-6 could induce PIM1 expression and FOXP3 phosphorylation at Ser422 in in vitro expanding human primary CD4+CD25hiCD127low Tregs, suggesting that the inflammatory cytokine IL-6 could potentially regulate human Treg activity through inducing PIM1 kinase expression (Fig. 3, A–C).
FIGURE 3.
TCR signaling and IL-6 signaling regulate PIM1 expression. A, in vitro expanded human Tregs were treated with IL-6 (10 ng/ml) for the indicated time points, and then samples were subjected to real-time PCR to detect PIM1 and FOXP3 mRNA expression level. (***, p < 0.001). B, the protein expression levels of PIM1 and FOXP3 and the phosphorylation status of FOXP3 were detected by incubating the membrane with the specified antibodies. IB, immunoblot. Phospho-Ser422 and PIM1 expression levels were normalized with the total FOXP3 level or β-actin level, respectively, and the statistical value was calculated and is shown in C. D, in vitro expanded CD4+CD25hiCD127low Tregs were rested by removing anti-CD3/CD28-conjugated Dynabeads for 2 days and subsequently reactivated or not reactivated with fresh beads for 12 h. PIM1 and FOXP3 mRNA level were detected by real-time PCR. (***, p < 0.001). E, PIM1 and FOXP3 protein expression levels were detected by Western blotting. PIM1 and FOXP3 protein levels were normalized with the corresponding level of β-actin, and the relative level of PIM1 and FOXP3 were quantified and are shown in F.
It has been documented that TCR signaling is essential for Treg development and activation (20). We also noticed that the PIM1 expression level is lower in in vitro expanding Tregs, which are activated by anti-CD3/CD28 Dynabeads, compared with resting Tregs in our experiment. For this reason, we decided to test how TCR signaling affected PIM1 expression in our system. In vitro expanded CD4+CD25hiCD127low Tregs were rested by removing anti-CD3/CD28-conjugated Dynabeads for 2 days and subsequently reactivating them with fresh beads for 12 h. Surprisingly, our data indicated that TCR signaling could significantly suppress PIM1 mRNA and its encoded protein expression level in activated Tregs, whereas FOXP3 mRNA and its encoded protein expression level were enhanced (Fig. 3, D, E, and F).
PIM1-mediated Phospho-Ser422 of FOXP3 Negatively Regulates Its DNA Binding Activity
A genome-wide binding assay has shown that human FOXP3 regulates the expression of hundreds of genes by directly binding to their promoters or 5′ regulatory regions (21). In Tregs, FOXP3 could directly bind to the promoter regions of genes encoding for cell surface markers, including CD25, CTLA4, and GITR, resulting in their transcriptional activation, whereas binding of FOXP3 to the promoters of IL-2 and IFN-γ facilitates their repression, finally conferring Treg suppressive function.
The crystal structure of the FOXP3 forkhead domain (FKH)-NFAT1-DNA ternary complex was described recently, showing some critical DNA binding residues in the C-terminal forkhead domain of human FOXP3 (22). Nie et al. (12) demonstrated that dephosphorylation of human FOXP3 at Ser418 negatively regulates its DNA binding activity and Treg function, which indicates that the activity of FOXP3 is sensitive to its posttranslational modification at its forkhead DNA-binding (FKD) domain. Given that the phosphorylated Ser422 site of FOXP3 is also located at the C-terminal region after its forkhead DNA-binding domain and near the residue of Ser418, we proposed that PIM1 could affect the FOXP3 DNA binding capacity by phosphorylating Ser422 and that the process may be affected by the phosphorylation status of the Ser418 residue.
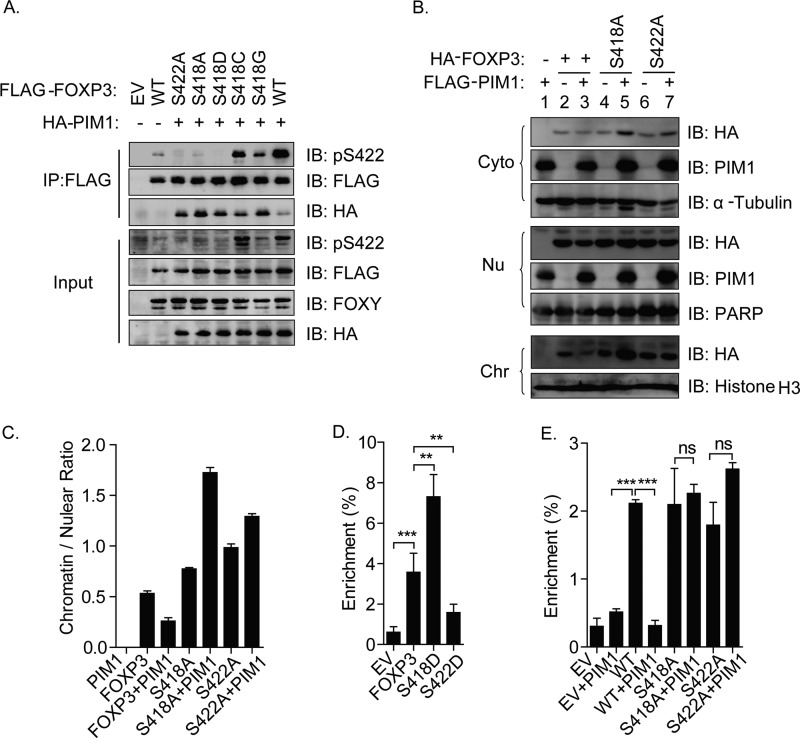
To validate our hypothesis, we first coexpressed PIM1 with FOXP3 or its mutants, including S422A, S418A, and S418D, and then detected FOXP3 phosphorylation at the Ser422 site. Surprisingly, we found that both the FOXP3 mutants S418A and S418D abolished PIM1-mediated phosphorylation at Ser422, suggesting a high sensitivity of PIM1-mediated phosphorylation of Ser422-FOXP3 to the phosphorylation of Ser418-FOXP3 (Fig. 4A). To exclude the possibility of non-specificity of the phospho-Ser422 antibody, we also mutated the residue of Ser418 to cysteine, which is more hydrophobic than serine, or glycine, which has the shortest side chain compared with other amino acids, and then coexpressed them with PIM1. Our results indicated that both of the mutations could rescue the phosphorylation of Ser422 mediated by PIM1, especially the S418G mutation (Fig. 4A), suggesting that both the hydrophobic character and the side chain effect of Ser422 are important for its phosphorylation mediated by PIM1 kinase.
FIGURE 4.
PIM1-mediated phosphorylation of FOXP3 at Ser422 negatively regulates its chromatin binding. A, FLAG-FOXP3 or its mutants, including S422A, S418A, S418D, S418C, and S418G, were cotransfected with or without PIM1 into 293T cells. Cell lysates were immunoprecipitated using anti-FLAG antibody, and then protein expression statuses were detected with the indicated antibodies in the input or immunoprecipitated fraction. EV, empty vector; IB, immunoblot. B, HA-FOXP3 or its mutants, including S418A and S422A, were cotransfected with or without PIM1 into 293T cells, and then cytoplasmic (Cyto), nuclear (Nu), and chromatin (Chr) fractionation was performed. Fractionated samples were detected by Western blot analysis with the indicated antibodies. The amount of FOXP3 protein localized in the nuclear fraction was normalized with the corresponding level of poly(ADP-ribose) polymerase 1 (PARP), whereas FOXP3 on chromatin was normalized by the protein level of histone H3. Then the ratio of FOXP3 on chromatin versus the nuclear fraction was quantified and is shown in C. D, FLAG-FOXP3 or its phosphorylation mimics, FOXP3 mutants S418D and S422D, were cotransfected into Jurkat T cells, and samples were harvested for μChIP after 24 h of incubation. μChIP analysis was performed with anti-FLAG antibody. E, FLAG-FOXP3 or its S418A and S422A mutants were cotransfected with HA-PIM1 or its control empty vector into Jurkat cells, followed by μChIP analysis using anti-FLAG antibody. Normal mouse IgG was used as a negative control. Data are mean ± S.E. of three separate experiments. **, p < 0.01; ***, p < 0.001; ns, not significant.
We noticed that we detected two Ser422 phosphorylation bands in the input of our N-terminal FLAG-tagged FOXP3 and HA-tagged PIM1 coexpression samples. We also found we can only detected one FOXP3 band with FLAG antibody. However, we can detected two bands with FOXY antibody in input samples. We also only detected one Ser422 phosphorylation band in samples immunoprecipitated with FLAG antibody, whereas there were two bands in the corresponding input samples. All these data confirmed previous finding that FOXP3 protein could be cleaved on its N terminus (23), and our phospho-Ser422-specific antibody could recognize both of bands (Fig. 4A).
To confirm whether PIM1 affects FOXP3 DNA binding mainly by promoting the phosphorylation of FOXP3 at Ser422, we transfected HEK 293T cells with vectors encoding FOXP3 or its Ser-to-Ala mutant(s) together with a PIM1 overexpression vector and performed cytoplasmic, nuclear, and chromatin fractionation to test the FOXP3 enrichment level on chromatin. The result shown in Fig. 4B indicates that FOXP3 mainly localized in the nuclear and chromatin fractions and that PIM1 expression decreased wild-type FOXP3 enrichment on chromatin but had no effect on the FOXP3 mutants S418A and S422A, which could not be modified at Ser422 by PIM1 (Fig. 4, B and C).
To further confirm the above results, we performed a μChIP assay. We transfected Jurkat T cells with plasmids encoding FOXP3 or its phosphorylation mimic mutants, including S418D and S422D, followed by μChIP to detect the percentage of enrichment on the IL-2 promoter. As predicted, the S418D FOXP3 mutant had stronger IL-2 promoter binding activity than wild-type FOXP3, whereas the binding activity of the S422D FOXP3 mutant on the IL-2 promoter was significantly less compared with wild-type FOXP3 (Fig. 4D). We also transfected Jurkat T cells with vectors encoding FOXP3 or its Ser-to-Ala mutants together with a PIM1 overexpression vector and performed a μChIP assay. Similar to Fig. 4, B and C, ectopic expression of PIM1 reduced the accumulation of wild-type FOXP3, but not the S422A and S418A mutants, on the IL-2 promoter (Fig. 4E).
PIM1 Negatively Regulates FOXP3-mediated Transcriptional Regulation and Treg Immunosuppressive Activity in in Vitro Expanded Tregs
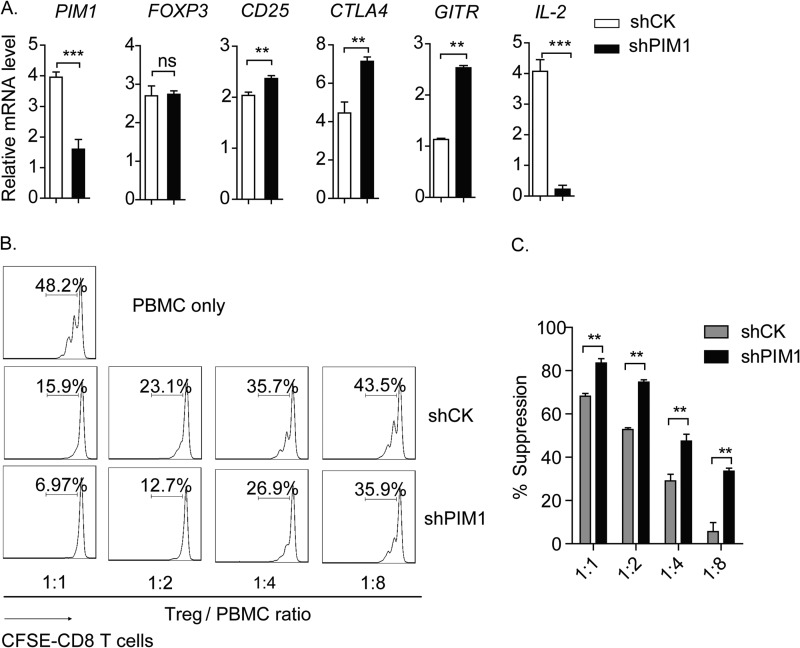
As mentioned above, a series of genes of Treg signature could be regulated by the transcription factor FOXP3, including CD25, CTLA4, GITR, and IL-2. We supposed that the expression level of the abovementioned genes could be regulated at the transcriptional level by PIM1, which regulates FOXP3 DNA binding activity. To verify this hypothesis, knockdown of PIM1 with shRNA was carried out in in vitro expanded CD4+CD25hiCD127low Tregs. As expected, we found that knockdown of PIM1 in Tregs strongly enhanced FOXP3-mediated transcriptional activation or repression of Treg-associated genes compared with control shRNA (shCK)-transduced Tregs, including CD25, CTLA4, GITR, and IL-2, whereas the FOXP3 expression level remained unchanged (Fig. 5A).
FIGURE 5.
PIM1 negatively regulates FOXP3-mediated transcriptional regulation. A, in vitro expanded primary Tregs were transduced with lentivirus delivering control shRNA (shCK) or shRNA targeting PIM1 (shPIM1) and then positively selected in the presence of puromycin for 3 days. Samples were harvested for real-time PCR to detect mRNA levels of PIM1, FOXP3, CD25, CTLA4, GITR, and IL-2. B, shCK or shPIM1 transduced human Tregs were coincubated with CFSE-labeled PBMCs at the indicated ratios for 3 days before detection of the proliferation status in the viable CD8+ T cell population by flow cytometry. C, percentages of suppression in each sample were analyzed. Data are mean ± S.E. of three separate experiments. **, p < 0.01; ns, not significant.
Given that the capacity to suppress the proliferation of effector cells is the most important feature of Tregs, we further tested the function of PIM1 on immunosuppression. We found that knockdown of PIM1 in Tregs enhanced their suppressive activity on the proliferation of CD8+ T cells in PBMCs (Fig. 5, B and C). Taken together, we confirmed that PIM1 negatively regulates FOXP3 activity on its target gene expression and the immunosuppressive activity of Tregs by phosphorylating FOXP3 at the Ser422 site.
FOXP3-Ser422 Is a Key Residue for Treg Response to IL-6 and PIM1-mediated FOXP3 Activity Down-regulation
To determine whether Ser422 is required for the Treg response to IL-6, we first tested PIM1 expression in CD4+T cells under IL-6 stimulation and confirmed that PIM1 expression could be induced by IL-6 treatment in CD4+T cells (Fig. 6, A and B). Then, we transduced wild-type FOXP3 or its S422A mutant into CD4+T cells and treated them with or without IL-6 for 12 h, and IL-2 mRNA expression levels were detected. Our data indicated that both the wild-type and the S422A mutant could suppress IL-2 transcription. However, IL-6 abolished the wild-type FOXP3 suppressive function on IL-2 transcription, whereas S422A mutant FOXP3 was not affected, suggesting that Ser422 is a key residue responsible for the Treg response to IL-6 (Fig. 6C). Furthermore, when we coexpressed FOXP3 or its S422A mutant with PIM1 in CD4+T cells, we found that PIM1 could attenuate the wild-type FOXP3-mediated repressive function on IL-2 expression, whereas PIM1 had no significant impact on the function of the S422A mutant (Fig. 6D).
FIGURE 6.
FOXP3-S422 is a key residue for the Treg response to IL-6. A, in vitro expanded primary CD4+T cells were treated with or without IL-6 for 12 h, and then cells were harvested for Western blot assay (IB) to detect the PIM1 expression level. The PIM1 expression level was quantified and normalized with the corresponding level of β-actin. The relative level of PIM1 is shown in B. C, FLAG-FOXP3, its S422A mutant, or an empty vector (EV) was transfected into 100,000–200,000 CD4+T cells and cultured for 24 h. Samples were treated with IL-6 for 12 h and harvested for mRNA extraction. mRNA levels of PIM1, FOXP3, and IL-2 were analyzed by Q-PCR. D, FLAG-FOXP3 or its S422A mutant were cotransfected with HA-PIM1 or its control empty vector into 100,000–200,000 CD4+T cells, followed by Q-PCR analysis to test PIM1, FOXP3, and IL-2 mRNA expression levels. Data are mean ± S.E. of three separate experiments. **, p < 0.01; ns, not significant.
The PIM1-specific Inhibitor Enhances Treg Immunosuppressive Function
From the perspective of translational medicine, we further tested the function of PIM1 on immunosuppression with its specific inhibitor (PIM1i). 3-cyano-4-phenyl-6-(3-bromo-6-hydroxy) phenyl-2(1H)-pyridone is an ATP-competitive PIM1 kinase inhibitor that displays specific selectivity over PIM2 (the IC50 values are 50 and >20,000 for PIM1 and PIM2, respectively). We first treated in vitro expanded Treg cells with the PIM1i for 12 h and then performed a μChIP assay to detect its effect on FOXP3 DNA binding activity on the IL-2 promoter. The result indicated that the inhibitor could dramatically enhance FOXP3 binding activity on the IL-2 promoter (Fig. 7A).
FIGURE 7.
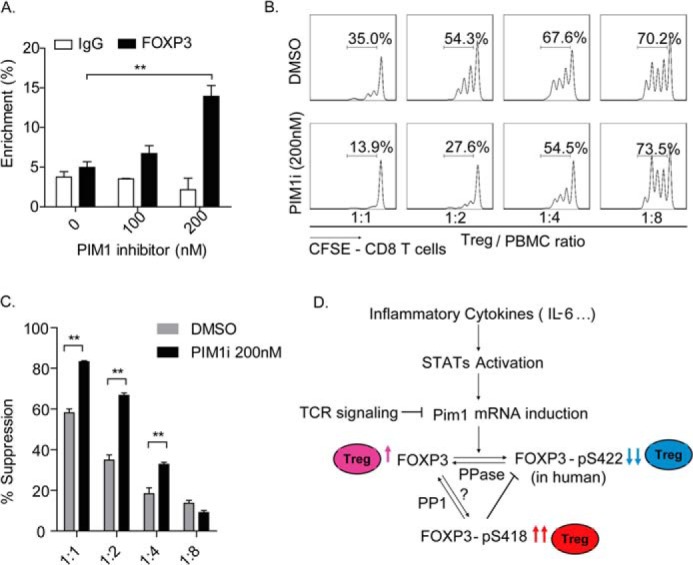
The PIM1-specific inhibitor enhances Treg suppressive function. A, in vitro expanded Tregs were treated with dimethyl sulfoxide (DMSO) or the PIM1-specific inhibitor at the indicated concentrations for 12 h. Samples were harvested for μChIP analysis, and the percent of enrichment of FOXP3 on the IL-2 promoter was detected with a specific anti-FOXP3 monoclonal antibody. **, p < 0.01. B, in vitro expanded human Tregs were pretreated with the PIM1-specific inhibitor (200 nm) or dimethyl sulfoxide for 2 days and then coincubated with CFSE-labeled PBMCs at the indicated ratios for an additional 3 days in the presence of the PIM1-specific inhibitor. CFSE dilution was determined for viable CD8+ T cells by flow cytometry. Data are from one representative experiment of three independent experiments with similar results. C, values indicate the percentages of suppression with mean ± S.E. of three independent experiments. **, p < 0.01. D, proposed working model for the mechanism of PIM1 regulation of human FOXP3 activity. Dephosphorylated human FOXP3 has normal standard DNA binding activity and confers immunosuppressive function to Tregs (pink). PIM1 could be induced by cytokines, including IL-6, through the STAT signaling pathway. Reversely, TCR signaling inhibits PIM1 mRNA transcription. The induced PIM1 phosphorylates human FOXP3 at its Ser422 site and negatively regulates its DNA binding activity, finally down-regulating the immunosuppressive function of Tregs (blue). On the other hand, phosphorylation at the Ser418 site of human FOXP3 would block phosphorylation of human FOXP3 at Ser422 by PIM1, therefore sustaining a high activity of human FOXP3 and the immunosuppressive function of Tregs (red).
Because TCR signaling has been shown to down-regulate PIM1 expression, we first rested the in vitro expanded Tregs and then treated the cells with PIM1 inhibitor for 2 days before using them in an in vitro suppressive assay, which was carried out in the presence of PIM1i. Notably, we found that PIM1 inhibitor treatment significantly enhanced the capacity of the Tregs to suppress the proliferation of CD8+ T cells (Fig. 7, B and C).
Together, our findings indicate that the Ser422 site is a key regulatory residue for FOXP3 and is fundamental for PIM1-mediated inhibition of FOXP3 transcriptional activity and immunosuppressive function of Tregs. PIM1 expression could be regulated at the transcriptional level by TCR signaling, which inhibits PIM1 mRNA induction, and by IL-6 signaling pathway, which facilitates PIM1 mRNA transcription. Additionally, we also confirmed the existence of cross-talk between Ser418 and Ser422 at the posttranslational level. Indeed, the phosphorylation status at Ser418 could prevent FOXP3 phosphorylation at Ser422 mediated by PIM1 (Fig. 7D). Our results are further supported by the previous finding that phosphorylation at Ser418 positively correlates with FOXP3 activity (12). Consistent with this observation, we found that the phosphorylation of FOXP3 at Ser418 may work through preventing the phosphorylation of FOXP3 at Ser422 mediated by PIM1.
DISCUSSION
FOXP3+ Tregs play a central role in the maintenance and balance of tolerance and immune activation, and the transcription factor FOXP3 is considered a master regulator for the development and function of Tregs. During graft transplantation and in autoimmune diseases, the enhancement of Treg activity would serve as a therapy. Conversely, for patients infected by pathogens or harboring tumors, Treg cell activity should be down-regulated to enhance immune function against the pathogen or the tumor. In this regard, it is critical to understand how Treg function is regulated to differentially treat these diseases.
Although the exact molecular mechanism of how Tregs maintain their immunosuppressive activity is complicated and still under considerable debate, many reported findings indicate that high expression of CTLA-4 and CD25 and repression of IL-2 in Tregs are critical to their function (24–26). Mice with Treg-specific CTLA-4 deficiency are impaired in in vivo and in vitro suppressive function of Tregs and harbor severe systemic lymphoproliferative disease despite having a minimal effect on Treg development (27). In our study, we found that the kinase PIM1 could regulate CTLA-4, CD25, and IL-2 expression through altering FOXP3 transcriptional activity and significantly affecting Treg immunosuppressive function. This finding provides a new therapeutic target for inflammatory diseases that need to be treated through modulation of Treg suppressive activity.
Human FOXP3+CD4+CD25hi Tregs may not be stable, encountering loss of FOXP3 expression or conversion into effector T cell-like cells under inflammatory conditions (28–30). The cytokine IL-2, in combination with IL-6, IL-21, or IL-23 significantly enhances Th17 conversion from CCR6+ CD4+CD25hi Tregs (29). The inflammatory cytokines IL-6, IL-21 (31), and IL-23 (32) could promote the downstream signaling pathway of STAT3. Our data suggest that STAT3 activation could be involved in PIM1 induction in Tregs. Further studies will be necessary to elucidate whether PIM1 induction is involved in Treg stability and conversion of Tregs into effector T cells during inflammation with disease models. Compared with human Tregs, murine CD4+CD25hiFoxp3+ Tregs have been reported to be rather stable and maintain their suppressive activity under inflammation (33, 34). Komatusu et al. (35) also showed that CD4+CD25loFoxp3+ T cells lose Foxp3 expression under arthritic conditions in mice and differentiate into Th17 cells, whereas CD4+CD25hiFoxp3+ T cells are a stable population in the pathological environment. Considering that there is no Ser422 residue in murine Foxp3, which is recognized by PIM1 kinase, we propose that this reveals a potential mechanism that may be accountable for the distinct responses between human and murine Tregs under inflammation.
Our data show that PIM1 phosphorylates human FOXP3 at Ser422 and that PIM1 may work as a negative regulator of human Tregs under inflammatory conditions. Expression levels of PIM1 in Tregs are strongly related to the intensity of TCR stimulation and the presence of the inflammatory cytokine IL-6. An increase of PIM1 expression occurred concomitantly with the enhancement of Ser422 phosphorylation upon IL-6 treatment. This could explain why IL-6- or IL-21-treated human Tregs lose their suppressive activity (36). However, in mice, which lack the conserved Foxp3 serine residue recognized by PIM1, excessive production of IL-6 has no apparent impact on the development and function of Tregs (37). Therefore, we propose that IL-6-mediated loss of Treg suppressive function occurs, at least partially, through PIM1 induction and subsequent phosphorylation of human FOXP3 at its Ser422 residue. Our data suggest that PIM1 kinase activity could represent a potential target for preventing and treating autoimmune diseases such as ankylosing spondylitis, systemic lupus erythematosus, and rheumatoid arthritis and for lowering the risk of transplant rejection.
Acknowledgments
We thank Dr. Mark I. Greene (University of Pennsylvania) for helpful discussions. We also thank the members of the flow cytometry facility at the Institut Pasteur of Shanghai for assistance.
This work was supported by National Basic Research Program of China (973 Program) Grants 2014CB541803, 2014CB541903, NSFC 81330072, 31370863, 31170825, 81271835, 31200646, 81302532, 31350110505, and SMCST 11ZR1404900; by Chinese Academy of Sciences Fellowship for Young International Scientists 2013Y1SB0005; and by Knowledge Innovation Program of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences Grant 2012KIP204.
- Treg
- regulatory T cell
- PBMC
- peripheral blood mononuclear cell
- TCR
- T cell receptor
- PIM1i
- 3-cyano-4-phenyl-6-(3-bromo-6-hydroxy)phenyl-2(1H)-pyridone
- Q-PCR
- quantitative PCR
- μChIP
- micro ChIP
- GITR
- glucocorticoid-induced tumor necrosis factor receptor
- CFSE
- carboxyfluorescein succinimidyl ester.
REFERENCES
- 1. Sakaguchi S., Miyara M., Costantino C. M., Hafler D. A. (2010) FOXP3+ regulatory T cells in the human immune system. Nat. Rev. Immunol. 10, 490–500 [DOI] [PubMed] [Google Scholar]
- 2. Curotto de Lafaille M. A., Lafaille J. J. (2009) Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity 30, 626–635 [DOI] [PubMed] [Google Scholar]
- 3. Williams L. M., Rudensky A. Y. (2007) Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat. Immunol. 8, 277–284 [DOI] [PubMed] [Google Scholar]
- 4. Wing J. B., Sakaguchi S. (2012) Multiple Treg suppressive modules and their adaptability. Front. Immunol. 3, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qian K. C., Wang L., Hickey E. R., Studts J., Barringer K., Peng C., Kronkaitis A., Li J., White A., Mische S., Farmer B. (2005) Structural basis of constitutive activity and a unique nucleotide binding mode of human Pim-1 kinase. J. Biol. Chem. 280, 6130–6137 [DOI] [PubMed] [Google Scholar]
- 6. Alvarado Y., Giles F. J., Swords R. T. (2012) The PIM kinases in hematological cancers. Expert Rev. Hematol. 5, 81–96 [DOI] [PubMed] [Google Scholar]
- 7. Mikkers H., Nawijn M., Allen J., Brouwers C., Verhoeven E., Jonkers J., Berns A. (2004) Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol. Cell. Biol. 24, 6104–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Z., Bhattacharya N., Weaver M., Petersen K., Meyer M., Gapter L., Magnuson N. S. (2001) Pim-1: a serine/threonine kinase with a role in cell survival, proliferation, differentiation and tumorigenesis. J. Vet. Sci. 2, 167–179 [PubMed] [Google Scholar]
- 9. Zippo A., De Robertis A., Serafini R., Oliviero S. (2007) PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat. Cell Biol. 9, 932–944 [DOI] [PubMed] [Google Scholar]
- 10. Rainio E. M., Sandholm J., Koskinen P. J. (2002) Cutting edge: transcriptional activity of NFATc1 is enhanced by the Pim-1 kinase. J. Immunol. 168, 1524–1527 [DOI] [PubMed] [Google Scholar]
- 11. Morawski P. A., Mehra P., Chen C., Bhatti T., Wells A. D. (2013) Foxp3 protein stability is regulated by cyclin-dependent kinase 2. J. Biol. Chem. 288, 24494–24502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nie H., Zheng Y., Li R., Guo T. B., He D., Fang L., Liu X., Xiao L., Chen X., Wan B., Chin Y. E., Zhang J. Z. (2013) Phosphorylation of FOXP3 controls regulatory T cell function and is inhibited by TNF-α in rheumatoid arthritis. Nat. Med. 19, 322–328 [DOI] [PubMed] [Google Scholar]
- 13. Chen Z., Barbi J., Bu S., Yang H. Y., Li Z., Gao Y., Jinasena D., Fu J., Lin F., Chen C., Zhang J., Yu N., Li X., Shan Z., Nie J., Gao Z., Tian H., Li Y., Yao Z., Zheng Y., Park B. V., Pan Z., Zhang J., Dang E., Li Z., Wang H., Luo W., Li L., Semenza G. L., Zheng S. G., Loser K., Tsun A., Greene M. I., Pardoll D. M., Pan F., Li B. (2013) The ubiquitin ligase stub1 negatively modulates regulatory T cell suppressive activity by promoting degradation of the transcription factor foxp3. Immunity 39, 272–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li B., Samanta A., Song X., Iacono K. T., Bembas K., Tao R., Basu S., Riley J. L., Hancock W. W., Shen Y., Saouaf S. J., Greene M. I. (2007) FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc. Natl. Acad. Sci. U.S.A. 104, 4571–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morishita D., Katayama R., Sekimizu K., Tsuruo T., Fujita N. (2008) Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 68, 5076–5085 [DOI] [PubMed] [Google Scholar]
- 16. Dahl J. A., Collas P. (2008) A rapid micro chromatin immunoprecipitation assay (microChIP). Nat. Protoc. 3, 1032–1045 [DOI] [PubMed] [Google Scholar]
- 17. Bullock A. N., Debreczeni J., Amos A. L., Knapp S., Turk B. E. (2005) Structure and substrate specificity of the Pim-1 kinase. J. Biol. Chem. 280, 41675–41682 [DOI] [PubMed] [Google Scholar]
- 18. Basu S., Golovina T., Mikheeva T., June C. H., Riley J. L. (2008) Cutting edge: Foxp3-mediated induction of pim 2 allows human T regulatory cells to preferentially expand in rapamycin. J. Immunol. 180, 5794–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bachmann M., Möröy T. (2005) The serine/threonine kinase Pim-1. Int. J. Biochem. Cell Biol. 37, 726–730 [DOI] [PubMed] [Google Scholar]
- 20. Sakaguchi S., Yamaguchi T., Nomura T., Ono M. (2008) Regulatory T cells and immune tolerance. Cell 133, 775–787 [DOI] [PubMed] [Google Scholar]
- 21. Sadlon T. J., Wilkinson B. G., Pederson S., Brown C. Y., Bresatz S., Gargett T., Melville E. L., Peng K., D'Andrea R. J., Glonek G. G., Goodall G. J., Zola H., Shannon M. F., Barry S. C. (2010) Genome-wide identification of human FOXP3 target genes in natural regulatory T cells. J. Immunol. 185, 1071–1081 [DOI] [PubMed] [Google Scholar]
- 22. Bandukwala H. S., Wu Y., Feuerer M., Chen Y., Barboza B., Ghosh S., Stroud J. C., Benoist C., Mathis D., Rao A., Chen L. (2011) Structure of a domain-swapped FOXP3 dimer on DNA and its function in regulatory T cells. Immunity 34, 479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Zoeten E. F., Lee I., Wang L., Chen C., Ge G., Wells A. D., Hancock W. W., Ozkaynak E. (2009) Foxp3 processing by proprotein convertases and control of regulatory T cell function. J. Biol. Chem. 284, 5709–5716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rudensky A. Y., Campbell D. J. (2006) In vivo sites and cellular mechanisms of T reg cell-mediated suppression. J. Exp. Med. 203, 489–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sakaguchi S., Wing K., Onishi Y., Prieto-Martin P., Yamaguchi T. (2009) Regulatory T cells: how do they suppress immune responses? Int. Immunol. 21, 1105–1111 [DOI] [PubMed] [Google Scholar]
- 26. Shevach E. M. (2009) Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity 30, 636–645 [DOI] [PubMed] [Google Scholar]
- 27. Wing K., Onishi Y., Prieto-Martin P., Yamaguchi T., Miyara M., Fehervari Z., Nomura T., Sakaguchi S. (2008) CTLA-4 control over Foxp3+ regulatory T cell function. Science 322, 271–275 [DOI] [PubMed] [Google Scholar]
- 28. Koenen H. J., Smeets R. L., Vink P. M., van Rijssen E., Boots A. M., Joosten I. (2008) Human CD25highFoxp3pos regulatory T cells differentiate into IL-17-producing cells. Blood 112, 2340–2352 [DOI] [PubMed] [Google Scholar]
- 29. Voo K. S., Wang Y. H., Santori F. R., Boggiano C., Wang Y. H., Arima K., Bover L., Hanabuchi S., Khalili J., Marinova E., Zheng B., Littman D. R., Liu Y. J. (2009) Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proc. Natl. Acad. Sci. U.S.A. 106, 4793–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Valmori D., Raffin C., Raimbaud I., Ayyoub M. (2010) Human RORγt+ TH17 cells preferentially differentiate from naive FOXP3+Treg in the presence of lineage-specific polarizing factors. Proc. Natl. Acad. Sci. U.S.A. 107, 19402–19407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zeng R., Spolski R., Casas E., Zhu W., Levy D. E., Leonard W. J. (2007) The molecular basis of IL-21-mediated proliferation. Blood 109, 4135–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Floss D. M., Mrotzek S., Klöcker T., Schröder J., Grötzinger J., Rose-John S., Scheller J. (2013) Identification of canonical tyrosine-dependent and non-canonical tyrosine-independent STAT3 activation sites in the intracellular domain of the interleukin 23 receptor. J. Biol. Chem. 288, 19386–19400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rubtsov Y. P., Niec R. E., Josefowicz S., Li L., Darce J., Mathis D., Benoist C., Rudensky A. Y. (2010) Stability of the regulatory T cell lineage in vivo. Science 329, 1667–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miyao T., Floess S., Setoguchi R., Luche H., Fehling H. J., Waldmann H., Huehn J., Hori S. (2012) Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity 36, 262–275 [DOI] [PubMed] [Google Scholar]
- 35. Komatsu N., Okamoto K., Sawa S., Nakashima T., Oh-hora M., Kodama T., Tanaka S., Bluestone J. A., Takayanagi H. (2014) Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat. Med. 20, 62–68 [DOI] [PubMed] [Google Scholar]
- 36. Goodman W. A., Young A. B., McCormick T. S., Cooper K. D., Levine A. D. (2011) Stat3 phosphorylation mediates resistance of primary human T cells to regulatory T cell suppression. J. Immunol. 186, 3336–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fujimoto M., Nakano M., Terabe F., Kawahata H., Ohkawara T., Han Y., Ripley B., Serada S., Nishikawa T., Kimura A., Nomura S., Kishimoto T., Naka T. (2011) The influence of excessive IL-6 production in vivo on the development and function of Foxp3+ regulatory T cells. J. Immunol. 186, 32–40 [DOI] [PubMed] [Google Scholar]