Background: The transcription factor hASH1, encoded by the ASCL1 gene, has a crucial function in neurogenesis and tumor formation.
Results: Gene expression of ASCL1 is controlled by the RNA-binding protein hnRNP-A2/B1 in neuroblastoma cells grown at low oxygen tension.
Conclusion: ASCL1 mRNA is a new target of hnRNP-A2/B1.
Significance: These findings provide novel insights in oxygen-dependent gene regulatory mechanisms in neuroblastoma cells.
Keywords: Gene Expression, Hypoxia, Post-transcriptional Regulation, RNA Turnover, RNA-binding Protein, ASCL1, hASH1 (Mash1), hnRNP-A2/B1, hnRNP-R
Abstract
The basic helix-loop-helix transcription factor hASH1, encoded by the ASCL1 gene, plays an important role in neurogenesis and tumor development. Recent findings indicate that local oxygen tension is a critical determinant for the progression of neuroblastomas. Here we investigated the molecular mechanisms underlying the oxygen-dependent expression of hASH1 in neuroblastoma cells. Exposure of human neuroblastoma-derived Kelly cells to 1% O2 significantly decreased ASCL1 mRNA and hASH1 protein levels. Using reporter gene assays, we show that the response of hASH1 to hypoxia is mediated mainly by post-transcriptional inhibition via the ASCL1 mRNA 5′- and 3′-UTRs, whereas additional inhibition of the ASCL1 promoter was observed under prolonged hypoxia. By RNA pulldown experiments followed by MALDI/TOF-MS analysis, we identified heterogeneous nuclear ribonucleoprotein (hnRNP)-A2/B1 and hnRNP-R as interactors binding directly to the ASCL1 mRNA 5′- and 3′-UTRs and influencing its expression. We further demonstrate that hnRNP-A2/B1 is a key positive regulator of ASCL1, findings that were also confirmed by analysis of a large compilation of gene expression data. Our data suggest that a prominent down-regulation of hnRNP-A2/B1 during hypoxia is associated with the post-transcriptional suppression of hASH1 synthesis. This novel post-transcriptional mechanism for regulating hASH1 levels will have important implications in neural cell fate development and disease.
Introduction
The formation of neurons in the vertebrate central nervous system is a multistep procedure, which is initiated by the commitment of progenitor cells to their neuronal fate during the last mitotic division (1). Soon after exiting from the cell cycle, destined neural stem cells migrate out of the progenitor zone, begin to differentiate, and finally acquire a specific neuronal subtype identity (2). The complex neurogenic differentiation program is coordinated by basic helix-loop-helix (bHLH)2 transcription factors that are expressed in proliferating progenitor cells (3, 4). Among them, the repressor-type bHLH genes Hes1, Hes3, and Hes5 are considered to maintain neuronal stem cells in a proliferative state by antagonizing pro-differentiating bHLH molecules (5).
The human achaete-scute homolog-1 (hASH1) belongs to a group of proneural bHLH transcription factors, which directly control the neurogenic program (6). Human ASH1 (known as Mash1 in rodents) is encoded by the ASCL1 gene and activates transcription by binding to the E-box consensus motif (7). Candidate target genes of hASH1 in the developing ventral telencephalon mostly encode transcription factors, signal transduction components, and structural proteins (8). Functional annotation by gene ontology suggested a role of hASH1 during various phases of neurogenesis including cell proliferation, neuronal cell fate decision, and neurite outgrowth (9).
Targeted gene inactivation in mice demonstrated that Mash1 is necessary for the differentiation of autonomic neurons and for neural progenitor cell development, particularly in the ventral telencephalon and in the olfactory epithelium (10–13). Consistent with the phenotype abnormalities of the null-mutant mice, Mash1 is temporally and spatially expressed in the peripheral and central nervous system of wild-type embryos (14–16). Although hASH1 was barely detectable in adult tissues, high expression levels were found in neuroendocrine tumors including small lung cell cancer (17) and neuroblastoma (18).
Neuroblastoma is a malignant childhood tumor, which originates from the sympathetic cell lineage of the neural crest (19). Notably, patients with high expression of hASH1 in these tumors had significantly lower survival rates than those with low expression (20). The mechanistic reason for the association of high levels of hASH1 in neuroblastomas with poor clinical outcome is unknown. Mash1/hASH1 has recently been reported to up-regulate genes that promote cell cycle progression in neural progenitors (8, 21). It is therefore tempting to speculate that hASH1 enhances tumor growth by activating proliferation genes in neuroblastoma cells.
Interestingly, exposure of neuroblastoma-derived cell lines to a hypoxic atmosphere significantly reduced Mash1/hASH1 and other marker proteins of the sympathetic cell lineage (22, 23). Thus, ASCL1 gene expression in neuroblastoma cells appears to be regulated by the local tissue oxygenation, which may provide a critical link to tumor formation. However, little is known about the molecular mechanisms underlying oxygen-dependent control of ASCL1 gene expression.
We and others have previously shown that the ASCL1 gene is tightly regulated at both the transcriptional and post-transcriptional levels. For example, bHLH factor HES-1 strongly repressed the ASCL1 promoter in response to activation of the Notch signaling pathway in neuroblastoma cells (6, 24). Additionally, Notch signaling may regulate cellular hASH1 homeostasis also by inducing rapid protein degradation (25). On the other hand, the orphan nuclear receptor TLX, which has a role in self-renewal of neural stem cells (26) stimulated the promoter of the ASCL1 gene in hippocampus-derived progenitor cells (27). Post-transcriptional mechanisms of ASCL1 gene regulation include a decrease in mRNA half-life, which became evident after phorbol ester treatment of neuroblastoma cells and an increase in protein translation, which was mediated by the fragile X mental retardation protein (FMRP) (28, 29). How these complex mechanisms interact in control of the cellular hASH1 protein levels in prolonged hypoxia is not understood.
In view of this background, our study was aimed at elucidating the molecular signaling pathways responsible for oxygen-dependent expression of the ASCL1 gene in neuroblastoma cells. We report here that hypoxia strongly reduces the hASH1 protein in both primary neurons and neuroblastoma cells. Suppression of hASH1 protein synthesis in response to oxygen restriction is mediated by post-transcriptional control very early on and in addition by inhibition of the ASCL1 promoter activity in prolonged hypoxia. Post-transcriptional effects are mediated by a complex interplay of RNA-binding proteins hnRNP-A2/B1 and hnRNP-R that act at the 5′- and 3′-UTRs of the ASCL1 mRNA. In fact, hypoxic down-regulation of hnRNP-A2/B1 seems to be the main influence causing suppression of hASH1 synthesis during hypoxia.
EXPERIMENTAL PROCEDURES
Cell Culture and RNA/Protein Isolation
Human neuroblastoma-derived Kelly cells were grown at 37 °C, 5% CO2 in RPMI medium supplemented with 10% FBS. For hypoxic measurements, cells were maintained in a hypoxic chamber (1% O2, 37 °C, 5% CO2) for the indicated times. For cytoplasmic extract preparation, cells were lysed in 20 mm Tris (pH 7.4) buffer containing 140 mm NaCl, 1 mm EDTA, 0.1% SDS, 0.5% Nonidet P-40, 25% glucose, 100 units/ml of RNaseOUT (Invitrogen), 1 mm DTT, complete protease-inhibitor mixture (Roche Diagnostics), and phosphatase inhibitor mixtures 2 and 3 (Sigma). After a 10-min incubation on ice, cells were centrifuged at 10,000 × g at 4 °C for 10 min to obtain the S10 supernatant. Total cellular extracts were prepared by direct lysis of cells in 50 mm Tris (pH 6.8) buffer containing 4 m urea and 1% SDS. Total RNA was prepared using RNA-Bee (Biozol Diagnostica Vertrieb GmbH) according to the manufacturer's protocol. For RNA measurements after plasmid transfections, the RNA was first treated with DNase I and the RNA was isolated using the RNeasy mini kit (Qiagen).
Cell Cultures of Primary Cortex Neurons
Cell cultures of primary cortex neurons were prepared using E19 Wistar rats as described (30, 31) and according to the permit given by the Office for Health Protection and Technical Safety of the regional government of Berlin (LaGeSo, 0122/07). The initial cell density of cortex cells from E19 Wistar rats was 68,000/cm2. Cultured cells were maintained for 1 or 2 days in vitro under normoxic conditions (ambient O2) in B27- and 1% FCS-supplemented Neurobasal medium (32). After 1 or 2 days in vitro, cells were kept for 6 h under hypoxic (0.5%) conditions before they were fixed using paraformaldehyde and processed for immunochemistry as described (28, 29). The Mash1 antibody was used at a dilution of 1:500, and the MAP2 antibody at a dilution of 1:300. To visualize cell nuclei, stained preparations were mounted in 4,6-diamidino-2-phenylindole (DAPI)-containing Vectashield medium (Vector Laboratories, Burlingame, CA). Labeled neurons were visualized with a standard epifluorescence microscope (Olympus BX51, Olympus Deutschland GmbH, Hamburg, Germany) under U Plan FL N ×10.0 objective (Olympus). Appropriate filters (U-MSP100v2 MFISH DAPI and U-MSP102v1 MFISH Cy3; Olympus GmbH, Germany) allowed the detection and separation of fluorescent signals. Images were acquired and processed using a 14-bit cooled CCD camera (Spot PURSUIT, Visitron Systems GmbH, Puchheim, Germany) and using software Metamorph (Universal Imaging Corp., Downingtown, PA). Statistical significance was assessed using one-way analysis of variance followed by a post hoc Bonferroni test.
RNA Pulldown Assays
Biotin-labeled RNA was prepared from a PCR amplicon using the M13 forward sense primer and an ASCL1 mRNA-specific reverse primer. The amplicon contained the T7 promoter sequence and either the 5′-UTR or the 3′-UTR of ASCL1 mRNA. The PCR products were transcribed in vitro using recombinant T7 polymerase. Biotin-labeled RNAs were produced according to the suggestions of the manufacturer (Promega) with the modification that biotin-16-UTP (Roche Diagnostics) was included in a 3:1 ratio with UTP. Labeled RNAs were precipitated with LiCl and ethanol, and resuspended in diethyl pyrocarbonate water. The concentrations of the biotin-labeled RNAs were determined by absorbance at 260 nm.
For UV cross-linking experiments, 1 μg of RNA/mg of extract was used. The in vitro transcribed RNA was first incubated with extract for 20 min at room temperature with gentle rotation to allow binding to occur. Cross-linking was performed in 50-μl aliquots in a single well of a 96-well plate using the auto cross-link setting (120,000 μJ) (Stratagene, UV-Stratalinker 1800). Samples were then pooled and RNA pulldown was carried out using Streptavidin-agarose affinity beads (Sigma). Beads were washed extensively in wash buffer (20 mm Tris, pH 7.4, 3 mm MgCl2, 250 mm KCl, 0.5 mm DTT) with a final wash step in wash buffer containing 150 mm KCl. After the final wash step, the bound proteins were eluted by treatment with 10 μg/ml of RNase A and 10 units/ml of RNase T1 in wash buffer containing 150 mm KCl and 1 mm EDTA. The proteins were precipitated using 100% TCA and resuspended in SDS-PAGE loading buffer for Western blot analysis or submitted directly for MALDI-TOF mass spectrometry analysis as previously described (33).
Antibodies
The antibodies used were mouse monoclonal anti-Mash1 (BD Pharmingen), mouse monoclonal anti-hnRNP-A2/B1 (Acris Antibodies), mouse monoclonal anti-nucleolin (Santa Cruz Biotechnology), rabbit polyclonal anti-hnRNP-R (Abcam), rabbit polyclonal anti-PABP2 (Abcam), rabbit polyclonal anti-Tubb2B (Proteintech), rabbit polyclonal anti-hnRNP-K (Novus Biologicals), and guinea pig polyclonal anti-MAP2 (Synaptic Systems). Secondary antibodies used were donkey anti-rabbit IgG-HRP and goat anti-mouse IgG-HRP (Santa Cruz Biotechnology). Secondary antibodies coupled to carboxymethyl indocyanine or fluorescein isothiocyanate were used for immunofluorescence (Jackson ImmunoResearch Laboratories).
RNA Quantification by qPCR
Random hexamers were used to generate cDNA from 1 μg of total RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Quantitative real-time PCR (qPCR) experiments were carried out on an Applied Biosystems GeneAmp 5700 system using SYBR Green master mixture according to the manufacturer's instructions. Each sample was measured in triplicate. mRNA expression levels were normalized to 18 S rRNA using the ΔΔCt method. The following primer sequences were used: 18 S forward, 5′-GATCAAAACCAACCCGGTCA, 18 S reverse, 5′-CCGTTTCTCAGGCTCCCTCT; hASH1 forward, 5′-CGACTTCACCAACTGGTTCT, hASH1 reverse, 5′-CCGTGAATGATTGGAGTGC; hnRNP R forward, 5′-GCCCCTTTTTGAGAAGGCCG, hnRNP R reverse, 5′-ACGTCCACCAAACCCTCTGT; hnRNP A2/B1 forward, 5′-GTTATGGAGGAGGAAGAGGA, hnRNP A2/B1 reverse, 5′-CGTAGTTAGAAGGTTGCTGG; and Firefly luciferase forward, 5′-TCAAAGAGGCGAACTGTGTG, Firefly luciferase reverse, 5′-GGTGTTGGAGCAAGATGGAT.
RNA Interference
Transfection of control siRNA and siRNA targeted against hnRNP-R, hnRNP-A2/B1, and PABP2 into Kelly cells was performed using SilenceMag (OZ Biosciences) or DharmaFECT 2 (Thermo Scientific) as specified by the manufacturers. All siRNAs were purchased as SMARTpool siRNAs from Thermo Scientific Dharmacon. Cells were harvested 24 and 48 h post-transfection. For hypoxia treatment, 48 h post-transfection, one set of transfected cells was incubated at normoxia (21% O2) and one set was subjected to 24 h hypoxia (1% O2) prior to analysis.
Cell Transfections and Reporter Gene Assays
Reporter constructs bearing the ASCL1 promoter or ASCL1 mRNA UTRs (including the following mutants: U-rich (nucleotide 223–232), GA-rich element (nucleotide 304–330), and AU-rich element (nucleotide 397–416)) have been previously described (28). Each plasmid along with the Renilla luciferase phRL-TK vector (Promega) were co-transfected into 70% confluent Kelly cells grown in 96-well plates using Roti®-Fect (Carl-Roth GmbH & Co.). After an overnight incubation, the medium was changed. Cells were grown for an additional time at normoxia (21% O2; control) or hypoxia (1% O2) as indicated. For overexpression of hnRNP-B1 a commercially available expression vector was used (pReceiver-M02 vector, GeneCopoeiaTM, Inc., Ex F0171-M02) as described earlier (34). Luciferase activity was measured using the Dual-GloTM luciferase assay system and the data were collected using a luminometer (Labsystems Luminoskan RS).
Prediction of RNA-Protein Interaction
The online tool catRAPID fragments module (35, 36) was used to predict a putative hnRNP-A2/B1 binding site within the ASCL1 mRNA 5′-UTR. Accession numbers for hnRNP-A2/B1 and ASCL1 are ENST00000354667 and ENST00000266744, respectively.
mRNA Level Correlation Study
Normalized human mRNA expression levels for HNRNPA2B1, HNRNPR, and ASCL1 mRNAs were obtained from the Stanford Microarray database (37) as described by Stuart et al. (38). This set contains 12,435 gene entries that match to ENSEMBL genes and 1202 microarray experiments in total. The difference of the sum of all microarrays in the database and the number of data points used for the correlation analysis in this study results from the fact that some entries for correlated candidates were missing in the microarray data sets. Regression and correlation analysis was performed using the math module of the open source/GPL program xmgrace based on Grace-5.1.22 in the linux environment.
Statistics
If not indicated otherwise, all values are presented as mean ± S.D. Student's paired t test was applied to reveal statistical significances. p < 0.05 was considered significant.
RESULTS
Oxygen Deprivation Suppresses Mash1/hASH1 Expression
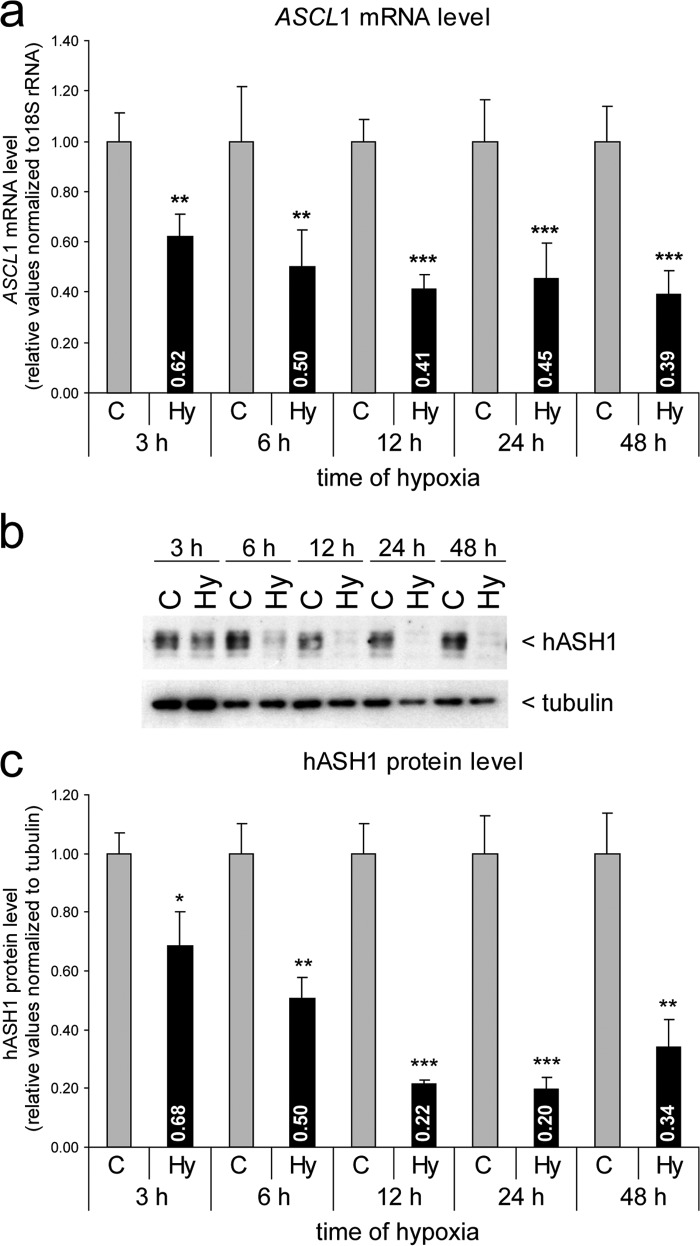
To explore whether expression of the rodent Ascl1 homolog Mash1 is sensitive to the local oxygen concentration, cortical neurons were isolated from rat embryos (E19) and kept either at 21 or 0.5% O2. Cellular Mash1 protein was visualized by immunofluorescence, and the integrated signal intensity was determined within nucleus-centered circular regions of interest, as described earlier (28, 39). Hypoxia diminished Mash1 immunoreactivity in primary cortical neurons in vitro (Fig. 1). Consistently, expression of the human ASCL1 gene in the Kelly neuroblastoma cell line was also significantly reduced in hypoxia (Fig. 2). In a time course up to 48 h of hypoxia, we observed a fast and persistent drop in ASCL1 mRNA (Fig. 2a) and protein levels (Fig. 2, b and c) that was detected as early as 3 h of hypoxia. Although the reduced ASCL1 mRNA level remained relatively stable at ∼40% compared with control conditions, the hASH1 protein level showed a much stronger decrease following 12 h of hypoxia (∼20% compared control). This stronger influence at the protein level could be attributed to the global suppression of mRNA translation during hypoxia (40). In line with this, protein stability measurements indicated no change in the half-life of hASH1 in hypoxia (data not shown).
FIGURE 1.
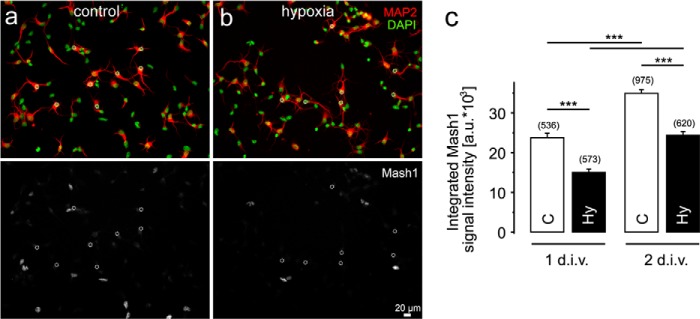
Hypoxia reduces Mash1 protein levels in primary cortical neurons. a and b, representative images in the top panels show signals corresponding to DAPI (green) and the neuronal marker MAP2 (red) in primary cortical neurons kept under control (21% O2) (a) and hypoxic (0.5% O2) (b) conditions. Corresponding minimum/maximum thresholded Mash1 signals are shown in the bottom panels. c, quantitation of the integrated Mash1 signal intensities (a.u., arbitrary units) measured in the circular regions of interest (dotted circles in the bottom panels a and b) projected on the nuclei of MAP2-positive neurons. Values shown are mean ± S.E. The numbers in parentheses indicate the total number of investigated neurons under each condition from three different cell cultures. 1 or 2 = 1 or 2 day(s) in vitro (d.i.v.); C, control; Hy, hypoxia. Significant differences are indicated with asterisks (***, p < 0.001).
FIGURE 2.
Hypoxia causes a time-dependent decrease in ASCL1 mRNA and hASH1 protein levels in Kelly neuroblastoma cells. a, Kelly cells were grown under control (21% O2, C) or hypoxic (1% O2, Hy) conditions for up to 48 h as indicated. The ASCL1 mRNA level was determined by qPCR and shows a constant decline during hypoxia (black bars) relative to control (gray bars). b, Western blot analysis was performed to assess relative hASH1 protein levels. Tubulin served as a loading control. c, quantitation and statistical analysis of the Western blot results (n = 6; *, p < 0.05; **, p < 0.01; ***, p < 0.001).
Regulation of ASCL1/hASH1 Expression during Hypoxia
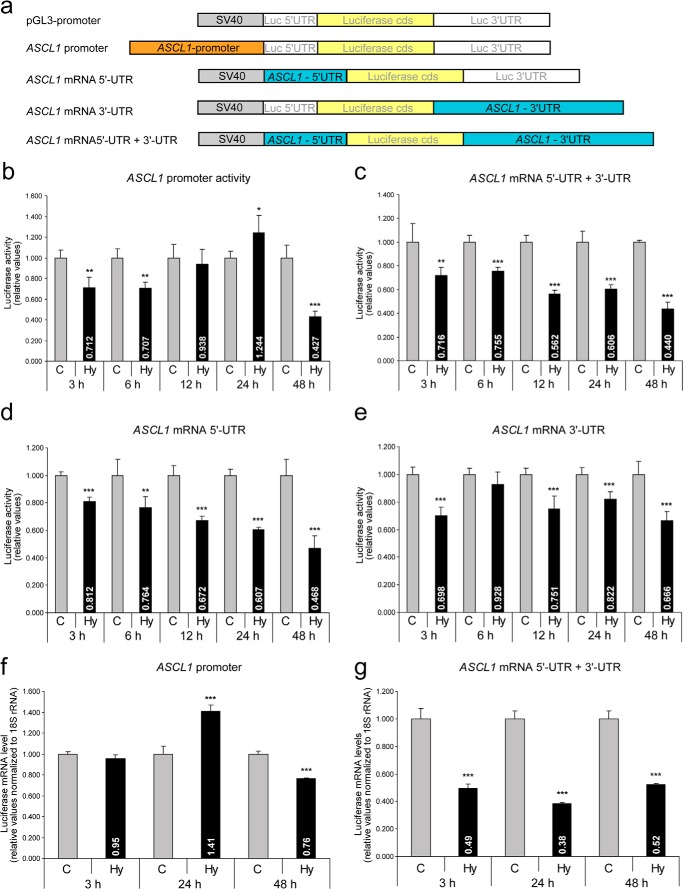
Gene expression regulated at the transcriptional level is primarily mediated by changes in promoter activity, whereas at the post-transcriptional level, for e.g. by alterations in mRNA stability, localization and/or translational efficiency, is mainly mediated by the mRNA 5′- and 3′-UTRs. To identify the mechanisms, whether transcriptional or post-transcriptional, regulating ASCL1/hASH1 expression during hypoxia, we performed reporter gene assays using either the ASCL1 promoter construct or constructs containing either or both the ASCL1 mRNA 5′- and 3′-UTRs. A schematic diagram of the constructs is shown in Fig. 3a. The constructs were transfected into Kelly cells and exposed to hypoxia for the indicated times.
FIGURE 3.
Transcriptional and post-transcriptional regulation of ASCL1 mRNA in hypoxia. Kelly cells were transfected with firefly-reporter constructs containing either the ASCL1 promoter or the ASCL1 mRNA 5′- and/or 3′-UTR and incubated under control (C, 21% O2) or hypoxic (Hy, 1% O2) conditions for up to 48 h. Values were normalized to Renilla activity and presented relative to the pGL3-promoter vector (SV40 promoter). a, schematic of the constructs used. b, influence of the ASCL1 promoter on luciferase activity in hypoxia. c-e, luciferase activity in the presence of both the ASCL1 mRNA 5′- and 3′-UTRs (c) and the individual influences of the ASCL1 mRNA 5′-UTR (d) and 3′-UTR (e) in hypoxia (n = 6; *, p < 0.05; **, p < 0.01; ***, p < 0.001). f and g, qPCR analysis to determine luciferase mRNA levels controlled either by the ASCL1 promoter (f) or ASCL1 mRNA 5′- and 3′-UTRs (g) under control and hypoxic conditions (n = 3; ***, p < 0.001).
At early time points (3 and 6 h of hypoxia), we observed a weak but significant decrease in ASCL1 promoter activity (∼70% compared with control) and a similar inhibition via the ASCL1 mRNA UTRs (∼75% compared with control) (Fig. 3, b and c). Strikingly, at 12 and 24 h of hypoxia, repression of ASCL1/hASH1 expression could be attributed only to the UTRs that showed an almost 50% decrease of UTR-dependent luciferase reporter activity (Fig. 3, b and c). At 48 h of hypoxia, both the ASCL1 promoter and mRNA UTRs resulted in decreased luciferase activity. We further determined the individual contributions of the ASCL1 mRNA 5′- and 3′-UTRs (Fig. 3, d and e). Although we observed that hypoxic inhibition is mediated by both the ASCL1 mRNA 5′- and 3′-UTRs, our data suggest a stronger influence of the 5′-UTR.
To ensure that these differences in luciferase activity directly reflect luciferase mRNA levels, we performed qPCR analysis to determine luciferase mRNA levels. In contrast to the promoter luciferase activity data (Fig. 3a), no significant alterations at the mRNA level were observed at early time points (Fig. 3f). However, we observed a weak but significant increase in the luciferase mRNA level after 24 h of hypoxia followed by a decrease at 48 h that was mediated by the ASCL1 promoter (Fig. 3f). These findings indicate that under prolonged hypoxic conditions transcriptional control also contributes to the inhibition of hASH1 synthesis. It is evident that for the ASCL1 mRNA UTRs the decrease in luciferase activity is in agreement with its luciferase mRNA level (Fig. 3g). Taken together, our findings suggest a repression of the ASCL1 promoter activity under prolonged hypoxic conditions, with a continuous level of repression provided by post-transcriptional mechanisms.
Trans-acting Factors Interacting with ASCL1 mRNA UTRs in Kelly Neuroblastoma Cells
We chose 24 h of hypoxia as the time for all future experiments to focus primarily on post-transcriptional mechanisms of ASCL1/hASH1 regulation.
Post-transcriptional control is mediated by trans-acting factors, such as RNA-binding proteins (RBPs) or micro-RNAs (miRNAs) that recognize cis-elements in the respective transcript. To identify RBPs that bind to the ASCL1 mRNA UTRs in neuroblastoma cells, we performed biotinylated RNA-streptavidin-based pulldown assays after UV cross-linking, and isolated RBPs bound directly to either the 5′- or 3′-UTRs from cytosolic extracts of cells cultivated either at 21 or 1% O2. Here it is important to note that the 3′-UTR construct lacked a poly(A) tail. A representative silver-stained gel of the isolated proteins is shown in Fig. 4a. We observed qualitative differences between factors binding at the 5′- and 3′-UTRs with subtle differences during hypoxia. Protein eluates were collected, subjected to MALDI/TOF-MS analysis, and compared with samples from the control pulldown. Identified factors bound either to the 5′- and/or the 3′-UTR and not to a control pulldown are shown in Table 1. We selected nucleolin, hnRNP-R, hnRNP-A2/B1, and PABP2 (PABPN1) for further verification by Western blot analysis, as they were identified by a good hit quality and are well known RBPs (Fig. 4b).
FIGURE 4.
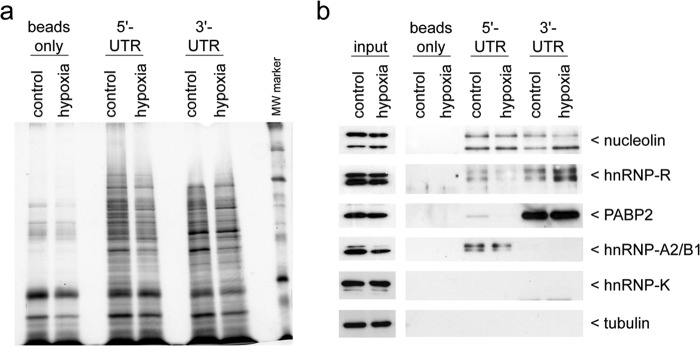
Affinity chromatography identification of RBPs interacting with the ASCL1 mRNA UTRs. Kelly neuroblastoma cells were subjected to control or hypoxic conditions for 24 h. Cellular extracts were prepared and used for affinity chromatography with in vitro transcribed biotinylated transcripts representing the ASCL1 mRNA 5′- or 3′-UTR as bait. a, representative silver-stained gel of the captured proteins binding to either the ASCL1 mRNA 5′- or 3′-UTR under control or hypoxic conditions. A no-transcript sample served as a negative control (beads only). b, Western blot confirmation of the trans-acting factors identified by MALDI/TOF-MS analysis (see Table 1). Cellular extracts that were used for affinity chromatography are shown in the input lanes. hnRNP-K and tubulin served as negative controls.
TABLE 1.
List of ASCL1 5′- and 3′-UTR interacting proteins identified by mass spectrometry
Proteins were identified by MALDI-TOF/MS analysis. Gene symbols corresponding to the identified candidates are given on the right side. Candidates identified with high confidence and being known RNA-binding proteins are marked in bold.
| Gene symbol | |
|---|---|
| 5′-UTR | |
| Histone H3.2 | HIST2H3A |
| Nucleolin | NCL |
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, ζ | YWHAZ |
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, β | YWHAB |
| Histone-lysine N-methyltransferase H3 lysine-36 and H4 lysine-20 specific | NSD1 |
| Y box binding proteins-1, -2, -3 | YBX1, -2, -3 |
| Heterogeneous nuclear ribonucleoproteins-A2/B1 | HNRNPA2B1 |
| Heterogeneous nuclear ribonucleoprotein-R | HNRNPR |
| RNA binding motif protein, X-linked-like 1 | RBMXL1 |
| Apolipoprotein A-I | APOA1 |
| Complement component 1, q subcomponent binding protein | C1QBP |
| U2 small nuclear RNA auxiliary factor 2 | U2AF2 |
| Serine/arginine-rich splicing factor 3 | SRSF3 |
| Histone cluster 1, H2ab | HIST1H2AB |
| Microtubule-associated protein 2 | MAP2 |
| 3′-UTR | |
| Heterogeneous nuclear ribonucleoprotein R | HNRNPR |
| Poly(A)-binding protein, nuclear 1 (PABP2) | PABPN1 |
| Microtubule-associated protein 2 | MAP2 |
| Nucleolin | NCL |
Nucleolin was found to interact with both the 5′- and the 3′-UTRs. However, nucleolin showed neither an alteration at the protein level in hypoxia nor a consistent change in its binding behavior, and thus was not considered for detailed functional analysis (Fig. 4b). The RBP hnRNP-R indicated a weaker binding at the ASCL1 mRNA 5′-UTR following hypoxia, in favor of an increased binding at the 3′-UTR (Fig. 4b). The cytoplasmic hnRNP-R protein level was not significantly changed during hypoxia (Fig. 4b). PABP2 preferentially interacted with the ASCL1 mRNA 3′-UTR. We observed no changes of the cytoplasmic protein level in hypoxia or of its binding ability (Fig. 4b). In contrast, the level of cytoplasmic hnRNP-A2/B1 protein was reduced in hypoxia, whereas the binding to the 5′-UTR was maintained (Fig. 4b). It is of interest that although the A2 isoform is more prominently expressed, it is the B1 variant that exhibited a higher RNA binding capacity. We confirmed that hnRNP-A2/B1 preferentially interacted with the ASCL1 mRNA 5′-UTR, which was identified to mediate a stronger influence in post-transcriptional inhibition of hASH1 synthesis in hypoxia (Fig. 3).
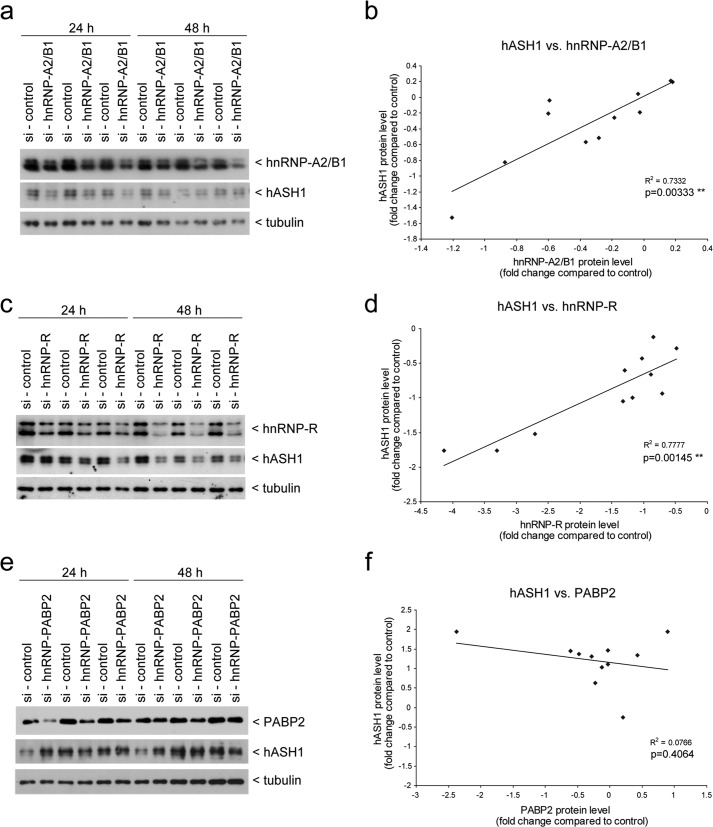
To test the functional relevance of the selected RBPs on hASH1 synthesis, we silenced each RBP expression by siRNA-mediated transient knockdown in Kelly cells. We first performed the knockdown for 24 and 48 h under control conditions and correlated the level of RBP knockdown with alteration in the hASH1 protein level for each sample (Fig. 5). We found a highly significant positive correlation between hASH1 and hnRNP-A2/B1 (Fig. 5, a and b) as well as between hASH1 and hnRNP-R expression levels (Fig. 5, c and d). In contrast, hASH1 protein levels did not correlate with PABP2 (Fig. 5, e and f). Although this suggests that PABP2 does not play a role in regulating hASH1 levels under these conditions, the lack of effect might also be explained by the fact that PABP4 and PABP5 can compensate for the function of PABP2 (41).
FIGURE 5.
hASH1 protein levels correlate with hnRNP-A2/B1 and hnRNP-R in Kelly cells. siRNA-mediated knockdown of hnRNP-A2/B1, hnRNP-R, and PABP2 was carried out in Kelly cells under control conditions. Cells were harvested following 24 and 48 h of siRNA transfection and subjected to Western blot analysis. a, c, and e, representative Western blot results and correlation analysis after hnRNP-A2/B1 (a and b), hnRNP-R (c and d), and PABP2 (e and f) knockdown. The blots were probed for hASH1 and the respective antibody. Tubulin served as a loading control. For the correlation analysis all values were normalized to tubulin. The regression lines, correlation coefficients, and p values are shown.
Role of hnRNP-A2/B1 and hnRNP-R in Inhibition of ASCL1/hASH1 Expression during Hypoxia
As we identified hnRNP-A2/B1 and hnRNP-R to correlate with hASH1 protein levels in neuroblastoma cells at 21% O2, we asked whether one or both of these RBPs mediate post-transcriptional control in hypoxia. We, thus, first silenced hnRNP-A2/B1 and hnRNP-R levels in Kelly cells for 48 h and then exposed the cells to 21 (control) or 1% (hypoxia) O2 for an additional 24 h. We then quantitated the mRNA levels of ASCL1 and both RBPs by qPCR (Fig. 6). Transient knockdown of hnRNP-A2/B1 caused a striking drop in steady-state ASCL1 mRNA levels to 53% under control conditions (Fig. 6a). hnRNP-R had a lesser but also significant effect on ASCL1 mRNA levels under control conditions (Fig. 6a). Hypoxia consistently caused a decrease in ASCL1 mRNA levels during control knockdown conditions. This level of decrease was maintained by the RBP knockdowns in hypoxia despite the already lowered levels of ASCL1 mRNA in these cells (Fig. 6a). Knockdown efficiency was confirmed by measurement of HNRNPA2B1 (Fig. 6b) and HNRNPR (Fig. 6c) mRNA levels. Interestingly, mRNA levels of both RBPs were significantly reduced during hypoxia in the presence of non-targeting siRNA. Of note is that the hnRNP-R knockdown caused a significant decrease in HNRNPA2B1 mRNA levels (Fig. 6b), and the hnRNP-A2/B1 knockdown resulted in a corresponding reduction of HNRNPR mRNA levels (Fig. 6c). These observations indicate that both hnRNP-A2/B1 and hnRNP-R not only influence hASH1 expression, but also mutually control the expression of each other.
FIGURE 6.
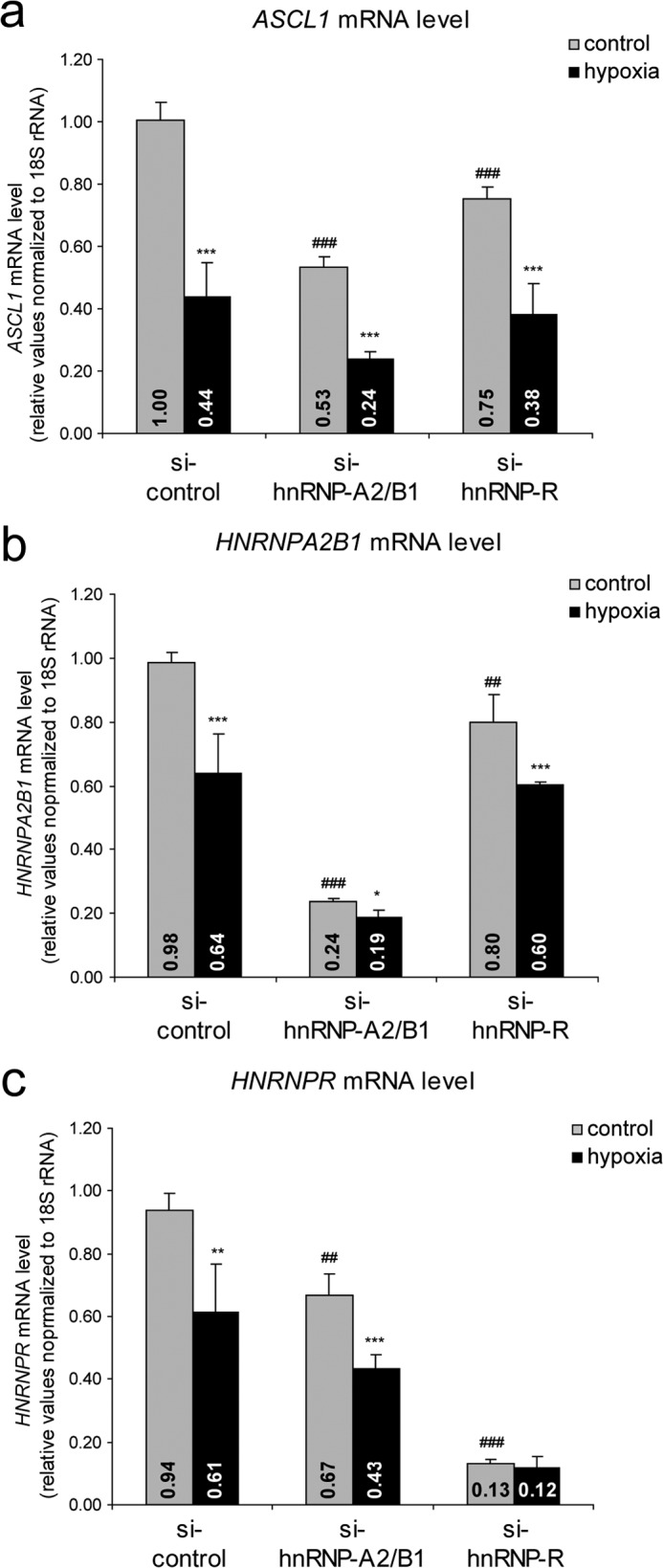
hnRNP-A2/B1 affects steady state levels of ASCL1 mRNA. Kelly neuroblastoma cells were transfected with either control siRNA (si-control) or siRNAs silencing the HNRNPA2B1 (si-hnRNP-A2/B1) or HNRNPR (si-hnRNP-R) genes and grown for 48 h. Cells were further cultivated for an additional 24 h under control (21% O2, gray bars) or hypoxic (1% O2, black bars) conditions. ASCL1 (a), HNRNPA2B1 (b), and HNRNPR (c) mRNA levels after control and specific knockdown conditions. Asterisks indicate significant differences between control and hypoxia for specific siRNA treatment. Number signs indicate a significant alteration by RBP knockdown compared with control si-RNA (n = 6; */#, p < 0.05; **/##, p < 0.01; ***/###, p < 0.001).
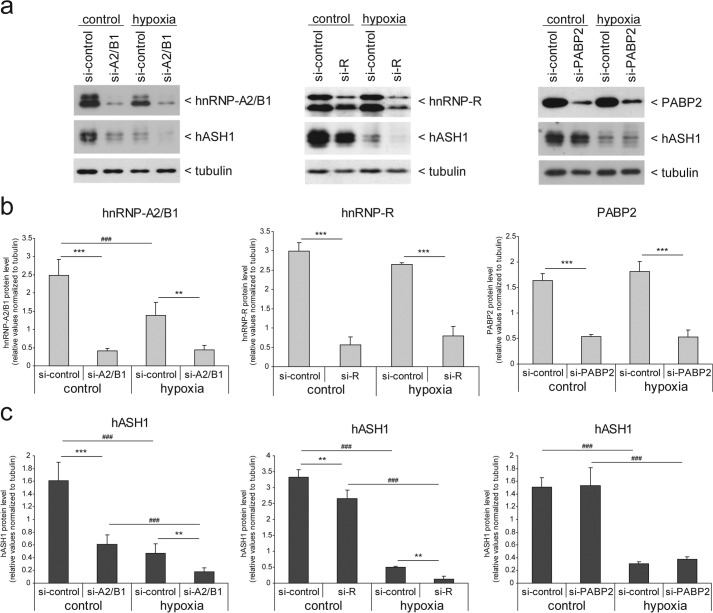
We further verified our observations at the protein level by Western blot analysis (Fig. 7). Consistent with the effect of the ASCL1 mRNA level, hnRNP-A2/B1 knockdown caused an ∼50% reduction in hASH1 protein levels (Fig. 7, a and c, left panels). Furthermore, in addition to hASH1, hnRNP-A2/B1 protein levels were also reduced by hypoxia, as observed in Kelly cells that were transfected with non-targeting siRNA (Fig. 7, a and b, left panels). hnRNP-R showed a weaker but significant effect on hASH1 protein levels, consistent with its effect on ASCL1 mRNA levels (Fig. 7, a and c, middle panels). We would like to point out that, in contrast to its mRNA level, hnRNP-R protein showed no significant decrease in hypoxia (Fig. 7, a and b, middle panels). We speculate that the hnRNP-R protein half-life is very high and thus would show a delayed response that is not observed under these conditions. Nevertheless, hASH1 levels were nearly non-detectable in hypoxia following hnRNP-A2/B1 or hnRNP-R knockdown. In the case of hnRNP-R knockdown, our data indicate a much stronger decrease of hASH1 at the protein level in hypoxia, suggesting that hnRNP-R delays the suppressive hypoxic influence on hASH1 synthesis.
FIGURE 7.
hnRNP-A2/B1 affects steady state levels of hASH1 protein. Kelly neuroblastoma cells were transfected with either control siRNA (si-control) or siRNAs silencing the HNRNPA2B1 (si-A2/B1), HNRNPR (si-R), or PABP2 (si-PABP2) genes and grown for 48 h. Cells were further cultivated for an additional 24 h under control (21% O2) or hypoxic conditions (1% O2). a, representative Western blot results for hASH1 and hnRNP-A2/B1 (left panel), hnRNP-R (middle panel), and PABP2 (right panel) following specific knockdown, respectively. Tubulin served as a loading control. b and c, statistical analysis of the Western blot results following knockdown of hnRNP-A2/B1, left; hnRNP-R, middle; and PABP2, right panel, are shown in b and the corresponding hASH1 protein levels in c. Number signs indicate a significant alteration between control and hypoxia. Asterisks indicate significant changes by RBP knockdown compared with control siRNA (n = 6; */#, p < 0.05; **/##, p < 0.01; ***/###, p < 0.001).
PABP2 knockdown had no effect on the ASCL1 mRNA level (data not shown) or at the protein level during control or during hypoxic conditions (Fig. 7, a and c, right panels). Taken together, these data show that hnRNP-A2/B1 knockdown causes a strong decrease in ASCL1/hASH1 levels that is further enhanced in hypoxia. It appears that the main effect on hASH1 synthesis under hypoxic conditions is due to the reduced expression of hnRNP-A2/B1 that directly affects ASCL1 mRNA and hASH1 protein levels. These data are in line with the observed time course of ASCL1/hASH1 suppression in hypoxia that indicated a strong post-transcriptional influence following 12 h or longer of hypoxia, highlighting the need first for the down-regulation of hnRNP-A2/B1.
Forced Expression of hnRNP-B1 Prevents hASH1 Suppression in Hypoxia
The data described above pointed to a key role of hnRNP-A2/B1 in regulating hASH1. The RNA pulldown experiments indicated a preferential binding of hnRNP-B1 to the ASCL1 mRNA 5′-UTR (Fig. 4b). We, therefore, separately quantified the decrease of hnRNP-A2 and -B1 by Western blot analysis following 24 h of hypoxia (Fig. 8a). The data revealed a stronger decrease of the B1 form, with hnRNP-B1 levels dropping to 38% and hnRNP-A2 to 75% compared with control conditions. We then focused on the mechanism by which a decrease in hnRNP-A2/B1 could influence hASH1 synthesis. Because steady-state measurements showed a decrease in ASCL1 mRNA levels under hnRNP-A2/B1 knockdown conditions (Fig. 6a), we wanted to examine whether ASCL1 mRNA stability is altered. To do this, cells were treated with actinomycin D for up to 4 h after silencing of hnRNP-A2/B1. Quantitation of ASCL1 mRNA levels showed an ∼30% decrease in ASCL1 mRNA stability (Fig. 8b). This finding supports the view that hnRNP-A2/B1 is a modulator of ASCL1 mRNA stability.
FIGURE 8.
Influence of hnRNP-B1 on ASCL1/hASH1 expression. a, Kelly cells were grown under control (21% O2, C) or hypoxic (1% O2, Hy) conditions for 24 h. Western blot analysis was performed to assess relative hnRNP-A2 and hnRNP-B1 protein levels. Tubulin served as a loading control. Shown is a representative Western blot and statistical quantification for hnRNP-A2 and -B1 protein levels following 24 h of hypoxia. b, ASCL1 mRNA half-life measurements following control (si-control) or hnRNP-A2/B1 (si-hnRNP-A2B1) knockdown conditions. c, schematic of the cis-element deletions within the ASCL1 mRNA 5′-UTR. The putative hnRNP-A2/B1 binding site as predicted by the catRAPID fragments module is indicated. d, Western blot analysis for hnRNP-A2/B1 after RNA pulldown using the ASCL1 mRNA 5′-UTR and the mutated variants as indicated in c. hnRNP-A2/B1 binding was severely impaired following deletion of the predicted binding site. A Coomassie Blue-stainable band served as a loading control. e, reporter gene assays following forced hnRNP-B1 expression. (For a schematic of constructs used see also Fig. 3a.) Values were normalized to Renilla activity. f, representative Western blot analysis following forced hnRNP-B1 expression under control and hypoxic conditions. g, statistical analysis for f. Asterisks indicate significant changes between mock and hnRNP-B1 transfection. Number signs indicate a significant alteration between control and hypoxia (n = 4; ***/###, p < 0.001; ##, p < 0.01).
We next aimed to identify the hnRNP-A2/B1 binding site within the ASCL1 mRNA 5′-UTR. By using the online tool catRAPID fragments module (35, 36), a GA-rich element (nucleotide 276–335) was predicted as a putative hnRNP-A2/B1 interaction site. To focus only on the GA-rich region, we deleted 25 nucleotides within the predicted binding site (del GA-rich). We also created deletion constructs in one upstream (del U-rich) and one downstream (del_AU-rich) element (Fig. 8c). Using these constructs in RNA pulldown experiments, we found that mutation of the GA-rich element prevented hnRNP-A2/B1 binding to the ASCL1 mRNA 5′-UTR (Fig. 8d). Functional verification using these constructs by reporter gene assays was performed following forced hnRNP-B1 expression as the hnRNP-B1 variant showed a stronger ASCL1 mRNA 5′-UTR binding capacity (Fig. 4b) and its decrease was more pronounced in hypoxia (Fig. 8a). We observed that overexpression of hnRNP-B1 had no effect on the ASCL1 promoter (Fig. 8e). Consistent with the finding that hnRNP-A2/B1 binds to the ASCL1 mRNA 5′-UTR (Fig. 4b), we found the strongest increase in luciferase activity for the ASCL1 mRNA 5′-UTR containing constructs. Deletion of the GA-rich element and thus loss of hnRNP-B1 binding abolished the increase in luciferase activity. In contrast, deletion of the other two cis-elements had no effect on hnRNP-A2/B1-dependent activation of ASCL1 mRNA 5′-UTR dependent luciferase activity (Fig. 8e). These data confirm that hnRNP-A2/B1 directly binds to the ASCL1 mRNA 5′-UTR and activates hASH1 expression.
Finally, we tested whether the decrease of hASH1 synthesis in hypoxia could be prevented or at least attenuated by forced expression of hnRNP-B1. Notably, up-regulation of hnRNP-B1 resulted in elevated hASH1 levels under control conditions (Fig. 8, f and g). Overexpression of hnRNP-B1 strongly attenuated the inhibition of hASH1 synthesis during hypoxia. In particular, following mock transfection, the hASH1 level declined to ∼17% in hypoxia, compared with 70% following forced hnRNP-B1 expression (Fig. 8g). These observations indicate that hnRNP-A2/B1, especially the B1 form, is a major determinant of hASH1 expression in hypoxia.
Global Positive Correlation of ASCL1 with HNRNPA2B1
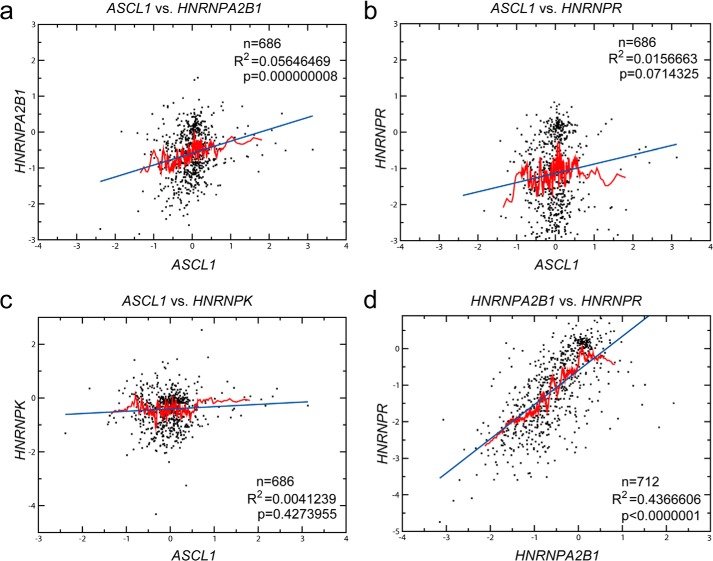
Finally, we aimed at confirming our findings of hnRNP-A2/B1 and hnRNP-R being trans-acting factors in hASH1 synthesis at the post-transcriptional level in a broader biological context. For this purpose, we correlated mRNA levels of ASCL1 as well as HNRNPA2B1 and HNRNPR using independent experimental data derived from >1200 microarray experiments available at the Stanford Microarray database (Fig. 9). Data were used as described by Stuart et al. (38). The data contain expression profiles of different in vitro experiments including studies of diverse biological processes such as cell cycle, stress, signaling, and apoptosis. Furthermore, this data collection contains expression profiles from tumor samples and different organ-specific tissue samples. A significant correlation can be expected if both candidates (i) are regulated by the same factor(s) or (ii) if one candidate is regulating the expression level of the other. In addition to the significance level (p value) that is derived from the correlation coefficient, a good correlation is also indicated by the moving average (red line in Fig. 9, a–d), which follows the regression line (blue line in Fig. 9, a–d).
FIGURE 9.
HNRNPA2B1 mRNA correlates globally with ASCL1 mRNA. Large-scale microarray expression data were obtained from the Stanford microarray database (37). The set contains genome-wide data from 1202 hybridization experiments from human tissues and cell lines. Correlation of HNRNPA2B1 (a), HNRNPR (b), and HNRNPK (c) mRNA levels with ASCL1 mRNA levels. d, correlation between HNRNPA2B1 and HNRNPR mRNAs. The numbers of matched values (n), coefficient of determination (R2), as well as the significance levels (p) are indicated. The regression line is shown in blue. The moving average is indicated as a red line and was performed on the x axis sorted values.
Analysis of these large-scale gene expression data confirmed a positive correlation between HNRNPA2B1 and ASCL1 mRNA levels that is seen by both, a highly significant p value as well as a good fit of the moving average to the regression line (Fig. 9a). In line with our experimental findings, these data indicate a general role of hnRNP-A2/B1 in regulating hASH1 expression. However, the correlation of ASCL1 mRNA and HNRNPR mRNA showed no significance (Fig. 9b). It is possible that the effect of hnRNP-R on hASH1 synthesis might be influenced by other factors and is more cell type or context specific. The effect we observed is also more pronounced at the protein level and would not be reflected here. As a negative control we used a correlation between HNRNPK mRNA and ASCL1 mRNA as PABP2 participates in a general regulation of all mRNAs via its interaction with the poly(A) tail and would likely result in a positive correlation. hnRNP-K was found not to interact with ASCL1 mRNA UTRs (Fig. 4b), and as expected showed no correlation (Fig. 9c). Because our data indicated that hnRNP-A2/B1 and hnRNP-R influence the expression level of each other, we asked whether this also holds true on a global scale. The strikingly strong positive correlation using large-scale gene expression data strongly hints to the mutual effect of hnRNP-A2/B1 and hnRNP-R on each other (Fig. 9d).
DISCUSSION
Oxygen depletion has been reported to cause a decrease in the expression of several neuronal and neuroendocrine marker genes including ASCL1/hASH1. Importantly, ASCL1/hASH1 can determine different cell fates by promoting either proliferation or differentiation of neural progenitors (8, 42). Thus, the local oxygen concentration may adjust ASCL1/hASH1 expression and associated pro-neural signaling pathways to the optimum level for the generation of different cell types in the developing central nervous system. In this study, we show that ASCL1/hASH1 expression during hypoxia is regulated by both, transcriptional and post-transcriptional mechanisms. Although transcriptional regulation via the ASCL1 promoter was observed under prolonged hypoxic conditions only, post-transcriptional control persists throughout and was already detectable during the early phase of hypoxia. We identified hnRNP-A2/B1 as one of the key post-transcriptional modulators of hASH1 synthesis during hypoxia.
The balance between pro-neural and repressor-type bHLH transcription factor signaling pathways under hypoxic conditions is crucial for neuronal fate. ASCL1/hASH1 expression during hypoxia has been attributed to such repressor-type signaling, for example, by Notch1-activated Hes1/5, leading to down-regulation of the ASCL1 promoter activity (25, 43). Moreover, increased Hes1/5 activity reduced the number of primary dendrites and promoted dendrite elongation instead of dendrite branching (44). These changes will inevitably impair synaptogenesis as well, because dendrite arborization precedes synapse formation at the dendritic tree (45, 46). There is a critical time window for dendrite arborization and synaptogenesis between the third trimester of pregnancy and the first few years of postnatal life where changes in the developmental program will have long-term consequences for neural network integrity that extends from puberty to adolescence and beyond (45, 47, 48). During the early critical period of neuronal differentiation changes in hASH1 levels, as occurring in hypoxia, will severely impact neural network function. Indeed, when neuroblastoma cells were induced to differentiate, hASH1 levels were rapidly down-regulated as an integral step to attaining a more differentiated phenotype, although whether constitutive expression of hASH1 would inhibit differentiation was not clearly resolved (49).
Among the major transcriptional effectors of ASCL1, Notch1 signaling was also shown to cause rapid degradation of hASH1 in overexpression studies (25). However, in our study we did not observe a reduced half-life of hASH1 protein during hypoxia. Consistent with the effect of HES-1 on ASCL1 regulation, we found a transcriptional inhibitory effect in prolonged hypoxia via the ASCL1 promoter and a more sustained influence by the ASCL1 mRNA UTRs during continuous hypoxia. Activation of Notch1/HES-1 has also been shown in cultured microglial cells exposed to hypoxia, although the response was slightly altered in primary cultured microglia (50).
Our data strongly support the view that hASH1 expression in hypoxia is determined by post-transcriptional control. Our findings reveal hnRNP-A2/B1 and hnRNP-R as important molecular determinants of hASH1 expression in hypoxia, through their direct interaction with the ASCL1 mRNA 5′- and 3′-UTRs. This may be relevant for both neuronal cell fate decision and neuroendocrine tumor formation.
The protein hnRNP-A2/B1 belongs to an RNA-binding protein family that plays important roles in the control of gene expression and mRNA processing. It is overexpressed in a variety of tumors, including breast, lung, and pancreas carcinomas (51–53). Altered expression of hnRNP-A2/B1 has also been reported during lung carcinogenesis where hASH1 seems to be involved (54, 55). Histochemical mapping of hnRNP-A2/B1 suggested a function in post-transcriptional regulation of neurons in the cerebral cortex and hippocampus in response to ischemia-reperfusion injury (56). Prolonged hypoxia has been reported to have a pronounced effect on cytoplasmic hnRNP-A2/B1 levels that is partially dependent on a higher mRNA turnover under hypoxic conditions (57). Our RNAi experiments under normoxic conditions revealed a significant co-regulation of hnRNP-A2/B1, hnRNP-R, and hASH1, suggesting that a complex network of molecular interactions governs mRNA regulation in response to hypoxia. Consistently, we found that down-regulation of hnRNP-A2/B1 by RNAi in hypoxia further decreased hASH1 expression revealing a synergistic effect that may serve to exacerbate the hypoxic influence on ASCL1/hASH1 levels. In keeping with this finding, forced expression of hnRNP-B1 prevented the decrease in hASH1 levels in hypoxia. Such complex patterns of regulation call for the existence of common structural elements in target mRNAs. Interestingly in this regard, a recent study utilized genome-wide mRNA stability data and identified structural elements that could play major roles in global mRNA regulation. Using one such structural motif found within the 3′-UTRs of transcripts, they identified hnRNP-A2/B1 as a functional regulator that binds this motif and regulates mRNA stability (58). Our UV cross-linking experiments indicate that hnRNP-A2/B1 directly interacts with a GA-rich element in the 5′-UTR of ASCL1 mRNA and promotes mRNA stability. It is likely that binding of hnRNP-A2/B1 at the 5′-UTR influences ASCL1 mRNA stability via cross-talk with the 3′-UTR. One possible scenario how hnRNP-A2/B1 might act via the ASCL1 mRNA 3′-UTR is through its influence on hnRNP-R expression.
The RBP hnRNP-R represents another ASCL1 mRNA UTR trans-acting factor identified in this study. hnRNP-R has ∼80% homology with hnRNP-Q proteins and both have been identified as components of mRNA granules transported in neuronal dendrites (59). hnRNP-R has been shown to enhance transcription from the c-fos promoter and regulate c-fos mRNA by interacting with an AU-rich element (60, 61).
We identified hnRNP-R to bind to both the 5′- and 3′-UTR of ASCL1 mRNA, with stronger binding to the 3′-UTR in hypoxia. Importantly, both factors, hnRNP-A2/B1 and hnRNP-R, are regulators of hASH1 synthesis. Therefore, we cannot exclude the possibility that the presence of hnRNP-A2/B1 influences the binding of hnRNP-R and vice versa. It is of interest that the shift in binding to the 3′-UTR is seen in hypoxia when hnRNP-A2/B1 levels decline. Moreover, a knockdown of hnRNP-A2/B1 was associated with a drop in hnRNP-R mRNA levels and vice versa, suggesting that the net effect on ASCL1/hASH1 expression is a result of a combined influence by hnRNP-A2/B1 and hnRNP-R. Collectively, these data indicate that both hnRNP-A2/B1 and hnRNP-R are involved in the post-transcriptional regulation of ASCL1/hASH1 expression in hypoxia. However, the profound effect the knockdown of hnRNP-A2/B1 had on ASCL1/hASH1 levels indicate that it is the dominant factor in its regulation. We cannot exclude the possibility that other trans-acting factors may exist that act in a cell-type/context-dependent manner and together exert a concerted influence on hASH1 synthesis.
The regulatory cues leading to ASCL1/hASH1 down-regulation during development and differentiation are not known. Because minor changes in its expression can potentially have a strong impact on its function, any regulatory mechanism affecting ASCL1/hASH1 levels will be of importance. Using hypoxia as an experimental means to modulate hASH1 levels, we identified novel post-transcriptional mechanisms involved in the regulation of hASH1. Whether such mechanisms are functional in tumor development and differentiation remain to be determined. In fact, the specific effect of hnRNP-A2/B1 on ASCL1/hASH1 can be exploited to define the function of hASH1 during differentiation.
Acknowledgments
We thank Jeannette Schmidt and Ursula Kastner for excellent technical assistance and Oliver Klein for mass spectrometry analysis.
This work was supported by Deutsche Forschungsgemeinschaft Grant FA845/2-2 (to M. F.) and the Bundesministerium für Bildung und Forschung BMBF ERA-Net NEURON II project CIPRESS (to J. C. M.).
- bHLH
- basic helix-loop-helix
- hASH1
- human achaete-scute homolog-1
- qPCR
- quantitative PCR
- RBP
- RNA-binding protein
- hnRNP
- heterogeneous nuclear ribonucleoprotein.
REFERENCES
- 1. Edlund T., Jessell T. M. (1999) Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell 96, 211–224 [DOI] [PubMed] [Google Scholar]
- 2. Marín O., Valiente M., Ge X., Tsai L. H. (2010) Guiding neuronal cell migrations. Cold Spring Harbor Perspect. Biol. 2, a001834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bertrand N., Castro D. S., Guillemot F. (2002) Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517–530 [DOI] [PubMed] [Google Scholar]
- 4. Powell L. M., Jarman A. P. (2008) Context dependence of proneural bHLH proteins. Curr. Opin. Genet. Dev. 18, 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kageyama R., Ohtsuka T., Hatakeyama J., Ohsawa R. (2005) Roles of bHLH genes in neural stem cell differentiation. Exp. Cell Res. 306, 343–348 [DOI] [PubMed] [Google Scholar]
- 6. Chen H., Thiagalingam A., Chopra H., Borges M. W., Feder J. N., Nelkin B. D., Baylin S. B., Ball D. W. (1997) Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc. Natl. Acad. Sci. U.S.A. 94, 5355–5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson J. E., Birren S. J., Saito T., Anderson D. J. (1992) DNA binding and transcriptional regulatory activity of mammalian achaete-scute homologous (MASH) proteins revealed by interaction with a muscle-specific enhancer. Proc. Natl. Acad. Sci. U.S.A. 89, 3596–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castro D. S., Martynoga B., Parras C., Ramesh V., Pacary E., Johnston C., Drechsel D., Lebel-Potter M., Garcia L. G., Hunt C., Dolle D., Bithell A., Ettwiller L., Buckley N., Guillemot F. (2011) A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 25, 930–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Castro D. S., Guillemot F. (2011) Old and new functions of proneural factors revealed by the genome-wide characterization of their transcriptional targets. Cell Cycle 10, 4026–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guillemot F., Lo L. C., Johnson J. E., Auerbach A., Anderson D. J., Joyner A. L. (1993) Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell 75, 463–476 [DOI] [PubMed] [Google Scholar]
- 11. Lo L., Tiveron M. C., Anderson D. J. (1998) MASH1 activates expression of the paired homeodomain transcription factor Phox2a, and couples pan-neuronal and subtype-specific components of autonomic neuronal identity. Development 125, 609–620 [DOI] [PubMed] [Google Scholar]
- 12. Horton S., Meredith A., Richardson J. A., Johnson J. E. (1999) Correct coordination of neuronal differentiation events in ventral forebrain requires the bHLH factor MASH1. Mol. Cell Neurosci. 14, 355–369 [DOI] [PubMed] [Google Scholar]
- 13. Cau E., Casarosa S., Guillemot F. (2002) Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development 129, 1871–1880 [DOI] [PubMed] [Google Scholar]
- 14. Lo L. C., Johnson J. E., Wuenschell C. W., Saito T., Anderson D. J. (1991) Mammalian achaete-scute homolog 1 is transiently expressed by spatially restricted subsets of early neuroepithelial and neural crest cells. Genes Dev. 5, 1524–1537 [DOI] [PubMed] [Google Scholar]
- 15. Guillemot F., Joyner A. L. (1993) Dynamic expression of the murine Achaete-Scute homologue Mash-1 in the developing nervous system. Mech. Dev. 42, 171–185 [DOI] [PubMed] [Google Scholar]
- 16. Torii Ma., Matsuzaki F., Osumi N., Kaibuchi K., Nakamura S., Casarosa S., Guillemot F., Nakafuku M. (1999) Transcription factors Mash-1 and Prox-1 delineate early steps in differentiation of neural stem cells in the developing central nervous system. Development 126, 443–456 [DOI] [PubMed] [Google Scholar]
- 17. Ball D. W., Azzoli C. G., Baylin S. B., Chi D., Dou S., Donis-Keller H., Cumaraswamy A., Borges M., Nelkin B. D. (1993) Identification of a human achaete-scute homolog highly expressed in neuroendocrine tumors. Proc. Natl. Acad. Sci. U.S.A. 90, 5648–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gestblom C., Grynfeld A., Ora I., Ortoft E., Larsson C., Axelson H., Sandstedt B., Cserjesi P., Olson E. N., Påhlman S. (1999) The basic helix-loop-helix transcription factor dHAND, a marker gene for the developing human sympathetic nervous system, is expressed in both high- and low-stage neuroblastomas. Lab. Invest. 79, 67–79 [PubMed] [Google Scholar]
- 19. Nakagawara A., Ohira M. (2004) Comprehensive genomics linking between neural development and cancer: neuroblastoma as a model. Cancer Lett. 204, 213–224 [DOI] [PubMed] [Google Scholar]
- 20. Isogai E., Ohira M., Ozaki T., Oba S., Nakamura Y., Nakagawara A. (2011) Oncogenic LMO3 collaborates with HEN2 to enhance neuroblastoma cell growth through transactivation of Mash1. PLoS One 6, e19297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. DeGregori J., Johnson D. G. (2006) Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr. Mol. Med. 6, 739–748 [DOI] [PubMed] [Google Scholar]
- 22. Jögi A., Øra I., Nilsson H., Lindeheim A., Makino Y., Poellinger L., Axelson H., Påhlman S. (2002) Hypoxia alters gene expression in human neuroblastoma cells toward an immature and neural crest-like phenotype. Proc. Natl. Acad. Sci. U.S.A. 99, 7021–7026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holmquist L., Löfstedt T., Påhlman S. (2006) Effect of hypoxia on the tumor phenotype: the neuroblastoma and breast cancer models. Adv. Exp. Med. Biol. 587, 179–193 [DOI] [PubMed] [Google Scholar]
- 24. Axelson H. (2004) The Notch signaling cascade in neuroblastoma: role of the basic helix-loop-helix proteins HASH-1 and HES-1. Cancer Lett. 204, 171–178 [DOI] [PubMed] [Google Scholar]
- 25. Sriuranpong V., Borges M. W., Strock C. L., Nakakura E. K., Watkins D. N., Blaumueller C. M., Nelkin B. D., Ball D. W. (2002) Notch signaling induces rapid degradation of achaete-scute homolog 1. Mol. Cell. Biol. 22, 3129–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shi Y., Chichung Lie D., Taupin P., Nakashima K., Ray J., Yu R. T., Gage F. H., Evans R. M. (2004) Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature 427, 78–83 [DOI] [PubMed] [Google Scholar]
- 27. Elmi M., Matsumoto Y., Zeng Z. J., Lakshminarasimhan P., Yang W., Uemura A., Nishikawa S., Moshiri A., Tajima N., Agren H., Funa K. (2010) TLX activates MASH1 for induction of neuronal lineage commitment of adult hippocampal neuroprogenitors. Mol. Cell. Neurosci. 45, 121–131 [DOI] [PubMed] [Google Scholar]
- 28. Benko E., Winkelmann A., Meier J. C., Persson P. B., Scholz H., Fähling M. (2011) Phorbol-ester mediated suppression of hASH1 synthesis: multiple ways to keep the level down. Front. Mol. Neurosci. 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fähling M., Mrowka R., Steege A., Kirschner K. M., Benko E., Förstera B., Persson P. B., Thiele B. J., Meier J. C., Scholz H. (2009) Translational regulation of the human achaete-scute homologue-1 by fragile X mental retardation protein. J. Biol. Chem. 284, 4255–4266 [DOI] [PubMed] [Google Scholar]
- 30. Eichler S. A., Förstera B., Smolinsky B., Jüttner R., Lehmann T. N., Fähling M., Schwarz G., Legendre P., Meier J. C. (2009) Splice-specific roles of glycine receptor α3 in the hippocampus. Eur. J. Neurosci. 30, 1077–1091 [DOI] [PubMed] [Google Scholar]
- 31. Förstera B., Belaidi A. A., Jüttner R., Bernert C., Tsokos M., Lehmann T. N., Horn P., Dehnicke C., Schwarz G., Meier J. C. (2010) Irregular RNA splicing curtails postsynaptic gephyrin in the cornu ammonis of patients with epilepsy. Brain 133, 3778–3794 [DOI] [PubMed] [Google Scholar]
- 32. Brewer G. J., Cotman C. W. (1989) Survival and growth of hippocampal neurons in defined medium at low density: advantages of a sandwich culture technique or low oxygen. Brain Res. 494, 65–74 [DOI] [PubMed] [Google Scholar]
- 33. Fähling M., Mrowka R., Steege A., Nebrich G., Perlewitz A., Persson P. B., Thiele B. J. (2006) Translational control of collagen prolyl 4-hydroxylase-α(I) gene expression under hypoxia. J. Biol. Chem. 281, 26089–26101 [DOI] [PubMed] [Google Scholar]
- 34. Fähling M., Mrowka R., Steege A., Martinka P., Persson P. B., Thiele B. J. (2006) Heterogeneous nuclear ribonucleoprotein-A2/B1 modulate collagen prolyl 4-hydroxylase, α(I) mRNA stability. J. Biol. Chem. 281, 9279–9286 [DOI] [PubMed] [Google Scholar]
- 35. Bellucci M., Agostini F., Masin M., Tartaglia G. G. (2011) Predicting protein associations with long noncoding RNAs. Nat. Methods 8, 444–445 [DOI] [PubMed] [Google Scholar]
- 36. Agostini F., Cirillo D., Bolognesi B., Tartaglia G. G. (2013) X-inactivation: quantitative predictions of protein interactions in the Xist network. Nucleic Acids Res. 41, e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Demeter J., Beauheim C., Gollub J., Hernandez-Boussard T., Jin H., Maier D., Matese J. C., Nitzberg M., Wymore F., Zachariah Z. K., Brown P. O., Sherlock G., Ball C. A. (2007) The Stanford Microarray Database: implementation of new analysis tools and open source release of software. Nucleic Acids Res. 35, D766–D770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stuart J. M., Segal E., Koller D., Kim S. K. (2003) A gene-coexpression network for global discovery of conserved genetic modules. Science 302, 249–255 [DOI] [PubMed] [Google Scholar]
- 39. Winkelmann A., Maggio N., Eller J., Caliskan G., Semtner M., Häussler U., Jüttner R., Dugladze T., Smolinsky B., Kowalczyk S., Chronowska E., Schwarz G., Rathjen F. G., Rechavi G., Haas C. A., Kulik A., Gloveli T., Heinemann U., Meier J. C. (2014) Changes in neural network homeostasis trigger neuropsychiatric symptoms. J. Clin. Invest. 124, 696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fähling M. (2009) Surviving hypoxia by modulation of mRNA translation rate. J. Cell Mol. Med. 13, 2770–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bhattacharjee R. B., Bag J. (2012) Depletion of nuclear poly(A) binding protein PABPN1 produces a compensatory response by cytoplasmic PABP4 and PABP5 in cultured human cells. PLoS One 7, e53036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wilkinson G., Dennis D., Schuurmans C. (2013) Proneural genes in neocortical development. Neuroscience 253, 256–273 [DOI] [PubMed] [Google Scholar]
- 43. Liu J., Lu W. G., Ye F., Cheng X. D., Hong D., Hu Y., Chen H. Z., Xie X. (2010) Hes1/Hes5 gene inhibits differentiation via down-regulating Hash1 and promotes proliferation in cervical carcinoma cells. Int. J. Gynecol. Cancer 20, 1109–1116 [DOI] [PubMed] [Google Scholar]
- 44. Salama-Cohen P., Arévalo M. A., Meier J., Grantyn R., Rodríguez-Tébar A. (2005) NGF controls dendrite development in hippocampal neurons by binding to p75NTR and modulating the cellular targets of Notch. Mol. Biol. Cell 16, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Juraska J. M. (1982) The development of pyramidal neurons after eye opening in the visual cortex of hooded rats: a quantitative study. J. Comp. Neurol. 212, 208–213 [DOI] [PubMed] [Google Scholar]
- 46. Khazipov R., Esclapez M., Caillard O., Bernard C., Khalilov I., Tyzio R., Hirsch J., Dzhala V., Berger B., Ben-Ari Y. (2001) Early development of neuronal activity in the primate hippocampus in utero. J. Neurosci. 21, 9770–9781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rakic P., Bourgeois J. P., Goldman-Rakic P. S. (1994) Synaptic development of the cerebral cortex: implications for learning, memory, and mental illness. Prog. Brain Res. 102, 227–243 [DOI] [PubMed] [Google Scholar]
- 48. Petanjek Z., Judas̆ M., S̆imic G., Rasin M. R., Uylings H. B., Rakic P., Kostovic I. (2011) Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc. Natl. Acad. Sci. U.S.A. 108, 13281–13286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Söderholm H., Ortoft E., Johansson I., Ljungberg J., Larsson C., Axelson H., Påhlman S. (1999) Human achaete-scute homologue 1 (HASH-1) is down-regulated in differentiating neuroblastoma cells. Biochem. Biophys. Res. Commun. 256, 557–563 [DOI] [PubMed] [Google Scholar]
- 50. Yao L., Kan E. M., Kaur C., Dheen S. T., Hao A., Lu J., Ling E. A. (2013) Notch-1 signaling regulates microglia activation via NF-κB pathway after hypoxic exposure in vivo and in vitro. PLoS One 8, e78439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou J., Allred D. C., Avis I., Martínez A., Vos M. D., Smith L., Treston A. M., Mulshine J. L. (2001) Differential expression of the early lung cancer detection marker, heterogeneous nuclear ribonucleoprotein-A2/B1 (hnRNP-A2/B1) in normal breast and neoplastic breast cancer. Breast Cancer Res. Treat. 66, 217–224 [DOI] [PubMed] [Google Scholar]
- 52. Tauler J., Zudaire E., Liu H., Shih J., Mulshine J. L. (2010) hnRNP A2/B1 modulates epithelial-mesenchymal transition in lung cancer cell lines. Cancer Res. 70, 7137–7147 [DOI] [PubMed] [Google Scholar]
- 53. Wang L., Liu H. L., Li Y., Yuan P. (2011) Proteomic analysis of pancreatic intraepithelial neoplasia and pancreatic carcinoma in rat models. World J. Gastroenterol. 17, 1434–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Teicher B. A. (2014) Targets in small cell lung cancer. Biochem. Pharmacol. 87, 211–219 [DOI] [PubMed] [Google Scholar]
- 55. Kosari F., Ida C. M., Aubry M. C., Yang L., Kovtun I. V., Klein J. L., Li Y., Erdogan S., Tomaszek S. C., Murphy S. J., Bolette L. C., Kolbert C. P., Yang P., Wigle D. A., Vasmatzis G. (2013) ASCL1 and RET expression defines a clinically relevant subgroup of lung adenocarcinoma characterized by neuroendocrine differentiation. Oncogene 33, 3776–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu Y., Gao Y., Wu Y., Wu Y., Wang H., Zhang C. (2010) Histochemical mapping of hnRNP A2/B1 in rat brain after ischemia-reperfusion insults. J. Histochem. Cytochem. 58, 695–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Garayoa M., Man Y. G., Martínez A., Cuttitta F., Mulshine J. L. (2003) Down-regulation of hnRNP A2/B1 expression in tumor cells under prolonged hypoxia. Am. J. Respir. Cell Mol. Biol. 28, 80–85 [DOI] [PubMed] [Google Scholar]
- 58. Goodarzi H., Najafabadi H. S., Oikonomou P., Greco T. M., Fish L., Salavati R., Cristea I. M., Tavazoie S. (2012) Systematic discovery of structural elements governing stability of mammalian messenger RNAs. Nature 485, 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bannai H., Fukatsu K., Mizutani A., Natsume T., Iemura S., Ikegami T., Inoue T., Mikoshiba K. (2004) An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1/NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites. J. Biol. Chem. 279, 53427–53434 [DOI] [PubMed] [Google Scholar]
- 60. Fukuda A., Nakadai T., Shimada M., Hisatake K. (2009) Heterogeneous nuclear ribonucleoprotein R enhances transcription from the naturally configured c-fos promoter in vitro. J. Biol. Chem. 284, 23472–23480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Huang J., Li S. J., Chen X. H., Han Y., Xu P. (2008) hnRNP-R regulates the PMA-induced c-fos expression in retinal cells. Cell Mol. Biol. Lett. 13, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]