Background: LIMK1 regulates F-actin dynamics through phosphorylation/inactivation of cofilin.
Results: LRAP25 forms a complex with MRCK and LIMK1 to promote LIMK1 phosphorylation/activation by MRCK.
Conclusion: Both LRAP25 and MRCK are required for LIMK1 activity in lamellipodial F-actin regulation.
Significance: LRAP adaptors are important determinants of MRCK cellular localization and downstream specificities.
Keywords: Actin, Cell Motility, Cofilin, Protein Phosphorylation, Small GTPase, F-actin Dynamics, LIM Kinase, MRCK, Lamellipodium
Abstract
Myotonic dystrophy kinase-related Cdc42-binding kinase (MRCK) has been shown to localize to the lamella of mammalian cells through its interaction with an adaptor protein, leucine repeat adaptor protein 35a (LRAP35a), which links it with myosin 18A (MYO18A) for activation of the lamellar actomyosin network essential for cell migration. Here, we report the identification of another adaptor protein LRAP25 that mediates MRCK association with LIM kinase 1 (LIMK1). The lamellipodium-localized LRAP25-MRCK complex is essential for the regulation of local LIMK1 and its downstream F-actin regulatory factor cofilin. Functionally, inhibition of either MRCK or LRAP25 resulted in a marked suppression of LIMK1 activity and down-regulation of cofilin phosphorylation in response to aluminum fluoride induction in B16-F1 cells, which eventually resulted in deregulation of lamellipodial F-actin and reorganization of cytoskeletal structures causing defects in cell polarization and motility. These biochemical and functional characterizations thus underline the functional relevance of the LRAP25-MRCK complex in LIMK1-cofilin signaling and the importance of LRAP adaptors as key determinants of MRCK cellular localization and downstream specificities.
Introduction
Cell migration is a fundamental event important for many physiological processes ranging from embryonic morphogenesis to wound healing. It entails a cycle of steps including polarization in response to extracellular signals, membrane protrusion coupled with adhesion to extracellular matrix, contractile activity for the translocation of cell body, and finally retraction of cell rear (1). Precise spatiotemporal regulation of these steps is critical for efficient movement (2). At the leading edge of the cell, membrane advancement is accomplished by protrusive cellular structures in lamellipodium and lamella. These are physically linked but biochemically distinct compartments characterized by F-actin meshwork and actomyosin bundles, respectively (3). Forces needed for membrane protrusion are generated by actin polymerization at the front region of lamellipodial actin meshwork (4). This process is regulated by the highly conserved actin filament nucleator Arp2/3 complex that functions to nucleate actin polymerization from existing filaments (5–7). Activation of Arp2/3 complex is achieved by the binding of nucleation-promoting factors such as Wiskott-Aldrich syndrome protein (WASP) and WASP-family verprolin-homologous (WAVE) proteins that are themselves downstream effectors of the small GTPases Rac and Cdc42 (4, 8).
This actin polymerization of the fast growing barbed end filaments at the tip of lamellipodia is kept in balance by the depolymerization of pointed end actin filaments located at the base of the network (4, 9). This process is critical in generating a sufficient supply of actin monomers to support the polymerization process at the tip. The operation of these two tightly coupled processes results in the rapid actin retrograde flow characteristic of the lamellipodial actin network (3, 10). The actin-depolymerizing factor/cofilin family proteins that are localized to the base of lamellipodia apparently play a major role in modulating F-actin dynamics at the leading edge (11). By virtue of their F-actin depolymerizing and filament severing activities, they have been shown to generate monomeric actin supply and free barbed ends capable of polymerization (12, 13). The activity of cofilin is tightly regulated by phosphorylation. LIM kinases (LIMK1/2) and testicular protein kinases (TESK1/2) phosphorylate cofilin at Ser-3 to inhibit its ability to bind and depolymerize F-actin (14–17), whereas dephosphorylation of this residue by the phosphatases slingshot and chronophin restores its biological activities (17–19). With respect to LIMK regulation, they are activated by the phosphorylation of Thr-508/Thr-505 (LIMK1/2) in the activation loop, under the regulation of Rac/Cdc42-PAK1 (20) and Rho-ROK/ROCK (21) signaling pathways, which eventually result in phosphorylation/inactivation of cofilin (13).
The effector kinase of Rac/Cdc42 GTPases, MRCK,3 has previously been shown to activate LIMKs (22), but the activating mechanism and the cellular relevance of this regulation are still obscure. We have previously shown that MRCK regulates myosin activity by phosphorylating myosin regulatory light chain (RMLC) (23) and myosin phosphatase binding subunit (24). Importantly, it interacts with adaptor protein LRAP35a, which allows it to form a tripartite complex with a myosin II-like myosin heavy chain MYO18A (25). This myosin-associated MRCK is targeted to the lamella where it regulates the activation of myosin IIA-enriched lamellar actomyosin network in a Cdc42-dependent manner (25). In this study, we identified LRAP25 as yet another adaptor protein of MRCK, and we found it to be important for MRCK localization to the lamellipodium. In addition, we also show that MRCK and LRAP25 form a complex with LIMK1, and they both are required for LIMK1 activation and cofilin activities in aluminum fluoride-induced cell migration. This study delineates a signaling cascade that involves LRAP25-MRCK and LIMK1-cofilin in the regulation of lamellipodial F-actin dynamics important for cell protrusion and migration.
EXPERIMENTAL PROCEDURES
cDNA Constructs, siRNAs, and Antibodies
Mammalian pXJ40 vectors with different N-terminal fusion tags, including FLAG, hemagglutinin (HA), glutathione S-transferase (GST), green fluorescent protein (GFP), red fluorescent protein, or mCherry, were used for the expression of all cDNA constructs used in this study. The cDNAs encoding MRCKα WT, deletion mutants of MRCKα, and LRAP35a were obtained as described (23–25). The cDNA encoding LRAP25 was cloned from rat brain cDNA by polymerase chain reaction (PCR) using primer pair 5′-CAG GA T CCA TGA ACG GGC TGC CCT CAA C-3′ (forward) and 5′-AGA AGC TTT TCA AAG GCT GAT TTG GAA G-3′ (reverse) and digested with BamHI and HindIII for ligating into BamHI/HindIII-digested pXJ40 and pcDNA vectors. Mammalian pcDNA vector containing an N-terminal HAmCherry tag was also used for the expression of LRAP25. The cDNAs encoding for LRAP25ΔLR and LRAP25 + 10 bp were obtained from human EST clones by PCR using the primer pair 5′-CAG GAT CCA TGA ACG GGC TGC CCT C-3′ (forward) and 5′-AGA AGC TTC AGA GGC TGA TGT GGA AG-3′ (reverse) and digested with BamHI and HindIII for ligating into BamHI/HindIII-digested pXJ40 vectors. All deletion constructs of both LRAP25 and LIMK1 were obtained by PCR. Two pairs of siRNA against mouse LRAP25 are as follows: siLRAP25–1 sense strand 5′-GCAUCUGUGCCAAGACCUGtt-3′ and siLRAP25–2 sense strand 5′ CCUCGACUCUGCACUAGCCtt-3′ (Invitrogen). Control siRNA is a dicer substrate RNAi duplex (Integrated DNA Technologies) as follows: sense strand, 5′-GGACUACAAACACCUGUGCCAAGAC-3′ and antisense strand 5′-GUCUUGGCACAGGUGUUUGUAGUCCUG-3′ (Integrated DNA Technologies). Sources of primary antibodies used in immunoblotting are as follows: polyclonal rabbit anti-FLAG (Sigma F7425) and monoclonal anti-FLAG M2-peroxidase (HRP) (Sigma A8592); monoclonal anti-HA F7-HRP (Santa Cruz Biotechnology sc-7392; 0.05–0.1 μg/ml); monoclonal anti-GST (Sigma G1160); rabbit anti-MRCKα (23); rabbit anti-MRCKβ (25); mouse monoclonal anti-MRCKα (25); anti-MYO18A (25); anti-LIMK1 and anti-pLIMK1(Thr(P)-508) (Cell Signaling Technology); anti-cofilin (Cytoskeleton Inc.); anti-Ser(P)-3-cofilin (Cell Signaling Technology 3311) and rabbit anti-phosphomyosin light chain 2 (Ser(P)-19) (Cell Signaling Technology 3675); mouse monoclonal anti-paxillin (ECM Biosciences PM1071); anti-β-actin (C4) (Santa Cruz Biotechnology sc-47778); rabbit polyclonal anti-ARPC2 (Upstate 07-227); and anti-Ser(P)-3-cofilin (4321) for immunostaining was a gift from J. Bamburg from Colorado State University. The rabbit polyclonal antibody against LRAP25 used in immunoblotting and immunofluorescence was obtained from rabbits immunized with a recombinant polypeptide corresponding to amino acids 1–189 of rat LRAP25. The antibody was affinity-purified by the corresponding polypeptide prior to use. Its applications in immunoblot analysis and immunoprecipitation are shown in Fig. 1F.
FIGURE 1.
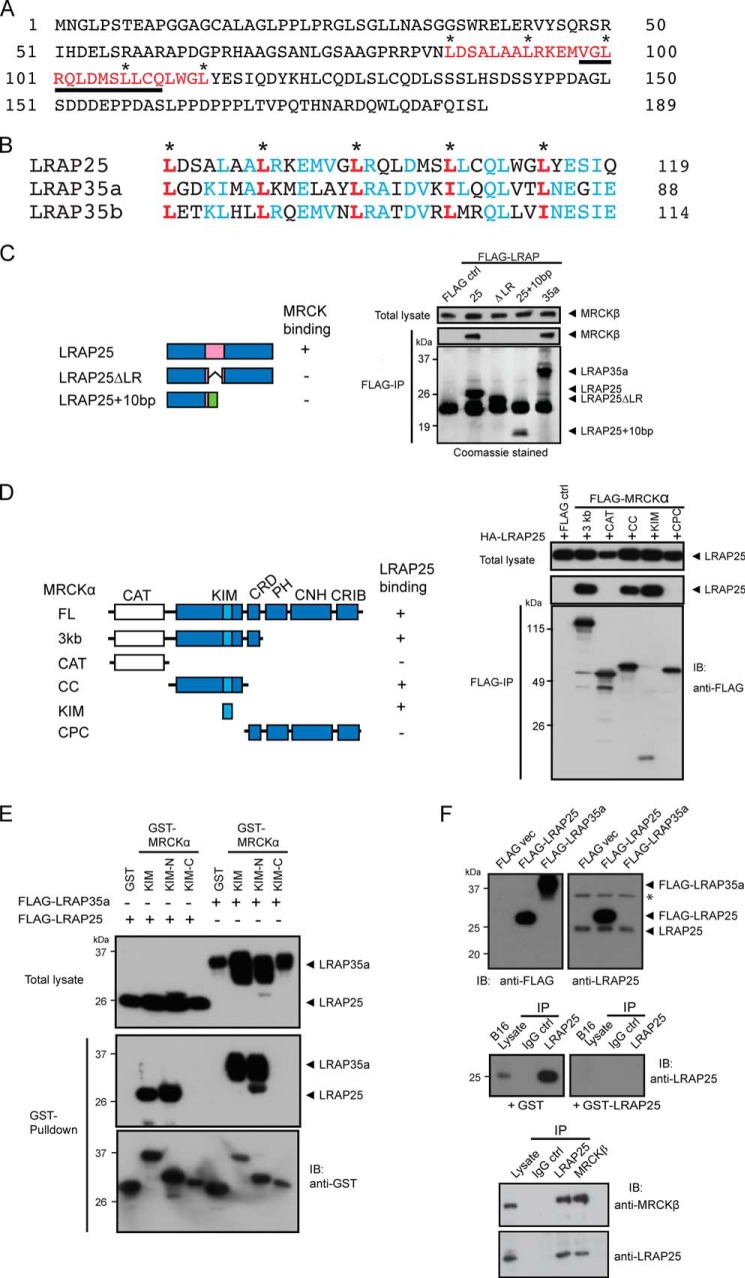
Identification of LRAP25 as an MRCK-interacting protein. A, amino acid sequence of LRAP25 (GenBankTM accession number NM_001015013.1). LR motif is in red. Conserved Leu residues in LR are marked by asterisks. Amino acids absent in alternative splice variant LRAP25ΔLR are underlined. B, comparison of leucine repeat sequences of LRAP25, LRAP35a, and LRAP35b. Leu/Ile residues in the leucine repeat are in red and marked by asterisks. Other conserved residues are in blue. C, schematic diagram of the three LRAP25 variants used in the mapping of the MRCK-binding site. Green box represents nonconserved amino acid sequence specific to LRAP25 + 10-bp variant. Binding activity of MRCK is shown on the right (left panel). COS7 cells were transfected with constructs of FLAG vector, FLAG-LRAP25, FLAG-LRAP25ΔLR, FLAG-LRAP25 + 10 bp, or FLAG-LRAP35a constructs. Immunoprecipitations (IP) were carried out using anti-FLAG antibody-coated agarose beads. The protein complexes precipitated were subjected to Coomassie Brilliant Blue staining or immunoblotting with anti-MRCKβ antibody (right panel). D, schematic representation of MRCKα and the various deletion mutants used in the mapping of the LRAP25-binding site. FL, full-length; 3kb, CAT-KIM-CRD domains; CAT, catalytic domain; CC, coiled-coil; KIM, kinase inhibitory motif; CPC, CRD-PH-CNH domains; CRD, cysteine-rich domain; PH, pleckstrin-homology domain; CNH, citron homology; CRIB, Cdc42/Rac1-interactive binding site. Binding activity of LRAP25 is shown on the right (left panel). HA-LRAP25 construct was co-expressed with various MRCKα fragments. Immunoprecipitations were carried out using anti-FLAG antibody-coated agarose beads. The protein complexes precipitated were subjected to immunoblotting (IB) with anti-FLAG or anti-HA antibodies (right panel). E, FLAG-LRAP25 and FLAG-LRAP35a constructs were separately co-expressed with either GST, GST-KIM, GST-KIM-N, or GST-KIM-C constructs in B16-F1 cells. Respective cell lysates were subjected to GST-pulldown using glutathione-Sepharose beads. The protein complexes precipitated were subjected to immunoblotting with anti-FLAG or anti-GST antibodies. F, lysates of B16 cells transfected with FLAG vector, FLAG-LRAP25, or FLAG-LRAP35a constructs were resolved on SDS-PAGE and immunoblotted with anti-FLAG or anti-LRAP25 antibodies. Asterisk marks a nonspecific band (top panel). Lysates of B16-F1 cells were subjected to immunoprecipitation with control IgG or anti-LRAP25 antibodies. Immunoprecipitates were immunoblotted for LRAP25 in the presence of 10 μg of GST or GST-LRAP25. Note that the recognition of LRAP25 by the antibody was specifically blocked by GST-LRAP25 (middle panel). B16-F1 cell lysates were subjected to immunoprecipitation using control IgG, anti-LRAP25, or anti-MRCKβ antibodies. Immunoprecipitates were immunoblotted for LRAP25 and MRCKβ (bottom panel).
Cell Culture and Transient and Stable Transfection
COS7 cells were grown in DMEM with 4500 mg/liter glucose supplemented with 10% fetal bovine serum (Hyclone). B16-F1 cells, a generous gift from Klemens Rottner (Braunschweig, Germany), were maintained in DMEM with 4500 mg/liter glucose supplemented with 10% fetal bovine serum (FBS) and 2 mm l-glutamine (Invitrogen). All cells were grown at 37 °C in the presence of 5% CO2. Transfection of COS7 cells and B16-F1 cells with plasmid DNA or siRNA was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. In knockdown experiments, cells were transfected with 20-40 nm of individual siRNA for 48 h before being further processed. A B16-F1 cell line stably expressing a low level of LIMK1-GFP was established by co-transfection of pXJ40-LIMK1-GFP and a puromycin-containing plasmid pBabe-puro and followed by puromycin selection at 2 μg/ml for 14 days.
Immunostaining and Image Analysis
B16-F1 cells intended for immunofluorescence analysis were plated fresh on laminin (Sigma; 20 μg/ml)-coated 35-mm coverslips. Cells were subjected to aluminum fluoride (AlF4−) treatment by adding B16F1 culture medium containing a final concentration of 50 μm AlCl3 and 30 mm NaF for 15–20 min. Treated B16F1 cells were fixed in either 4% paraformaldehyde for 20 min or methanol for 10 min. Antibodies including rabbit anti-LRAP25, mouse monoclonal anti-MRCKα, rabbit anti-MYO18A, anti-β-actin, anti-ARPC2, anti-Ser(P)-3-cofilin (number 4321) and anti-cofilin are compatible with methanol fixing. For immunostaining of endogenous proteins, cells were incubated with primary antibody at 4 °C overnight. Anti-mouse and anti-rabbit secondary antibodies conjugated with either Alexafluor-488, -546 (Invitrogen), or Cy3 (Jackson ImmunoResearch) were used to label primary antibodies for 1–2 h at room temperature. Alexafluor-647-conjugated phalloidin was used to visualize F-actin. All images were acquired using a CoolSNAP HQ camera (Roper Scientific) adapted to Zeiss Axioplan wide field epi-fluorescent microscope.
In Vivo Actin Incorporation Assay
AlF4−-treated B16F1 cells under study were permeabilized and labeled with 0.6 μm AlexaFluor-568-conjugated actin (Molecular Probes) in permeabilization buffer (20 mm HEPES-NaOH, 138 mm KCl, 4 mm MgCl2, 3 mm EGTA, 0.2 mg/ml saponin, 1% BSA, 1 mm ATP, 3 μm AlexaFluor-647-phalloidin) for 45 s at room temperature. After the labeling, cells were fixed with 4% paraformaldehyde/PBS for 20 min, rinsed with PBS 2–3 times, permeabilized with 0.2% Triton X-100 for 10 min, and finally counterstained with AlexaFluor-647-phalloidin for 1 h at room temperature. Analysis of AlexaFluor-568 actin incorporation was performed as mentioned for the fluorescence distribution profile of incorporated actin and total F-actin. The ratio of the intensity of incorporated actin to phalloidin was plotted as a function of distance from the leading edge.
Time-lapse Microscopy and Analysis
Time-lapse imaging was performed using Olympus Fluoview1000 confocal microscope equipped with a 30 milliwatt 488-nm multiline argon and 20-milliwatt 561-nm solid-state lasers, temperature control, and CO2 chamber. To plot the immunofluorescence intensity, a line perpendicular to the cell edge was drawn on merge images acquired by Olympus Fluoview1000 confocal microscope. Images were then analyzed using “Linescan” function in MetaMorph software. For determination of cell edge advancement speed, movies were acquired at 5-s intervals for 10 min and processed in MetaMorph software to generate kymographs with lines placed perpendicular to the protrusions. Cell edge advancement speeds were derived from kymographs.
Immunoprecipitations and Kinase Assays
Immunoprecipitations were performed as described previously (25). In brief, cells under study were lysed in cell lysis buffer supplemented with protease inhibitor mixture (Roche Applied Science). Crude cell lysates were precleared by centrifugation (10,000 × g, 4 °C, 10 min) and incubated with either mouse M2 anti-FLAG beads (Sigma), GFP-affinity beads (Chromotek), anti-LRAP-25 (amino acids 1–84), or anti-MRCKβ antibody in the presence of hydrated protein A-conjugated agarose beads for 3 h at 4 °C. After extensive washing with lysis buffer devoid of protease inhibitors, immunoprecipitates were boiled in sample buffer subjected to immunoblot analysis. B16-F1 cells subjected to immunoprecipitation/kinase assay experiments were first plated on a laminin-coated 90-mm dish for 1.5 h and treated with AlF for 10 min. These cells were lysed as described above in lysis buffer supplemented with 100 nm okadaic acid (LC Laboratories) and 100 nm calyculin A (Calbiochem). Kinase assays of the immunoprecipitated proteins were carried out as described previously (25). In experiments involving inhibitor treatment, B16-F1 cells were plated and treated with AlF4− for 10 min, followed by addition of specific inhibitors for another 30 min. To determine cellular level of Ser(P)-3-cofilin by immunoblotting, B16-F1 cells were treated with AlF4− as above. Cells were lysed immediately in 1.5× sample buffer containing 37.5 mm Tris-HCl, pH 6.8, 1.5% β-mercaptoethanol, 0.075% bromphenol blue, and 37.5% glycerol, heated at 100 °C for 5 min, and followed by a single 5-s pulse sonication at 15% power. Lysates were then precleared by centrifugation, and relative protein concentrations were estimated before SDS-PAGE.
Transwell Migration Assay
The lower sides of cell culture inserts (Corning Transwell; pore size of 8 μm) were precoated with 20 μg/ml laminin. The inserts were later placed in lower chambers containing 600 μl of culture medium. 1 × 105 cells in 300 μl of culture medium were added to the upper chambers. After 20 h of incubation, inserts were fixed in 4% paraformaldehyde/PBS for 20 min and rinsed with PBS. Cells on the upper side of the inserts were scraped off with cotton buds, and the fixed cells on the lower side were permeabilized with 0.2% Triton X-100, followed by mounting with Vectashield mounting medium supplemented with DAPI. Slides were viewed using ×20 objective, and cell count was based on stained nuclei.
RESULTS
Identification of LRAP25 as a Novel Interactor of MRCK
Our previous work detected two proteins of 35 and 25 kDa in the MRCK immunoprecipitate (25). The former was characterized as an adaptor protein LRAP35a that is involved in regulating the kinase-dependent nonmuscle myosin II activity (25). To identify the 25-kDa protein, we searched protein databases with the conserved leucine repeat 1 (LR1) sequence of LRAP35a that contains the MRCK-binding site. This revealed a protein known as C184M that contains a putative LR1-like motif (Fig. 1, A and B) (26, 27). Interestingly, it also shows an expected isoelectric point of 4.8 and a molecular mass that matches the 25-kDa MRCK-binding protein we had previously detected (Fig. 1, A and B). We thus termed this protein leucine repeat adaptor protein 25 (LRAP25) in virtue of its conserved LR motif and molecular mass.
Further analysis of database sequences shows that the gene encoding LRAP25 can give rise to three transcript variants, a full-length LRAP25 and two shorter variants that are devoid of a large part of the conserved LR motif (designated as ΔLR and +10 bp) due to alternative splicing events (Fig. 1, B and C). To determine whether LRAP25 interacts with MRCK as predicted, FLAG-tagged full-length LRAP25 and its two naturally occurring variants were overexpressed and subjected to immunoprecipitation. As shown in Fig. 1C, endogenous MRCK was found to only interact with the full-length protein but not with those lacking a complete LR, indicating the importance of LR for the interaction. From the reverse mapping experiment (Fig. 1D), the kinase inhibitory motif of MRCK was found to be responsible for this interaction, and further analysis showed that the N-terminal half of kinase inhibitory motif was sufficient, reminiscent of the LRAP35a-MRCK interaction (Fig. 1E) (25). Importantly, endogenous LRAP25 was readily co-immunoprecipitated with MRCK from B16-F1 melanoma cells (Fig. 1F) and mouse brain lysate (data not shown), with the use of affinity-purified antibodies (Fig. 1F). Taken together, our results show that LRAP25 interacts with MRCK through the conserved LR motif.
LRAP25 Interacts with MRCK and LIMK1 to Form a Tripartite Complex and Is Required for LIMK1 Activation
LIMKs are known to be activated by Rac/Cdc42-PAK1, Rho/ROCK, and MRCK signaling pathways through the phosphorylation of Thr-508 in the activation loop (20–22). As shown in Fig. 2A, the phosphorylation levels of LIMK1-Thr-508 are significantly elevated by co-expression of Rac1G12V, Cdc42G12V, RhoAG14V and the catalytic domain of MRCKα. Although PAK1 has been shown to interact with LIMK1 (20), the mechanism of how MRCK mediates LIMK1 activation is not entirely clear. Our previous study showed that LRAP35a plays the important role of bridging MRCK to MYO18A (25). This led us to investigate whether LRAP25 could be responsible for linking MRCK to its substrate LIMK1. To establish this, HA-tagged LRAP25 was co-expressed with wild type or various deletion mutants of LIMK1 fused to either a FLAG or a GFP tag and followed by immunoprecipitation with anti-FLAG antibodies or GFP affinity beads accordingly (Fig. 2, B and C). Our results show that LRAP25 specifically interacted with LIMK1 through the N-terminal LIM1 motif. In the reverse experiment, the LR motif of LRAP25 was shown to be the LIM1-binding site (Fig. 2D). To find out whether the three proteins are able to form a complex, we probed for the presence of both endogenous LRAP25 and LIMK1 in MRCK immunoprecipitates purified from the B16-F1 cells with or without prior aluminum fluoride (AlF4−) treatment, a well characterized Rac-inducing agent that is known to induce lamellipodial formation in B16 cells (28, 29), and could serve as a suitable tool for studying the regulation of LRAP25, MRCK, and LIMK1. The immunoprecipitation results show that both LRAP25 and LIMK1 could be readily detected in the MRCK immunocomplexes under both conditions with negligible differences (Fig. 2E), suggesting that the interactions could be constitutive and reminiscent of the interactions of MRCK, LRAP35a, and MYO18A (25). Our results thus indicate that MRCK, LRAP25, and LIMK1 have the capacity to form a tripartite complex in vivo.
FIGURE 2.
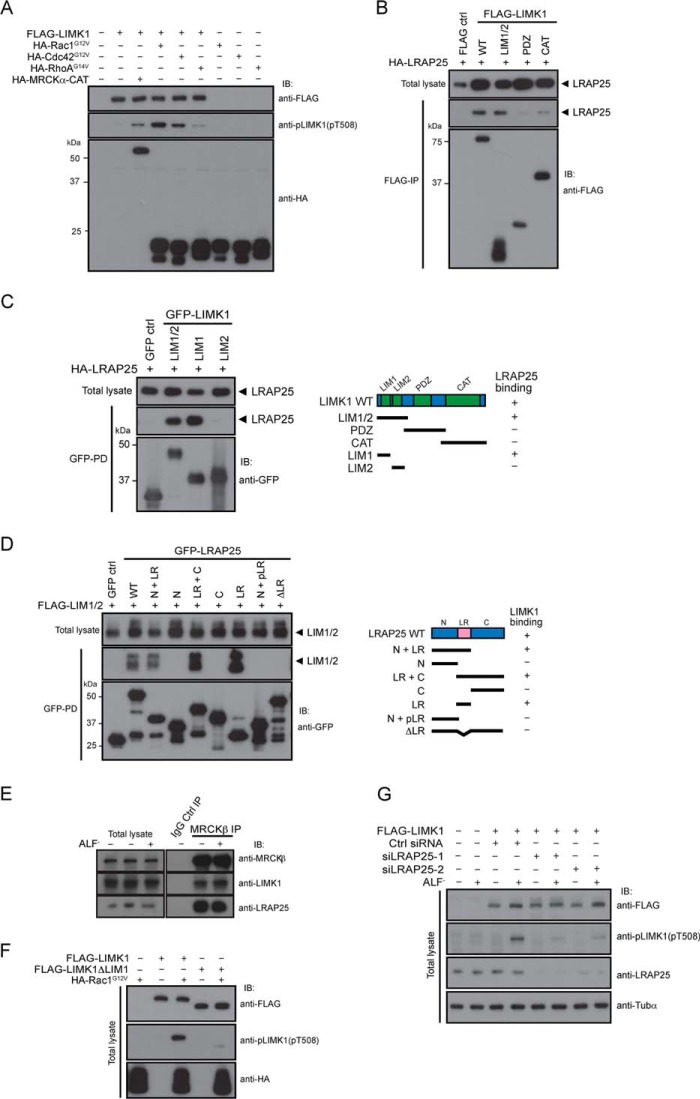
LRAP25 interacts with LIMK1 and MRCK to form a tripartite complex and is required for LIMK1 activation. A, FLAG empty vector and FLAG-LIMK1 constructs were separately co-expressed with either HA-Rac1G12V, HA-Cdc42G12V, HA-RhoAG14V, or HA-MRCKα-CAT constructs in COS7 cells as indicated. Respective lysates were subjected to immunoblotting (IB) with anti-FLAG, anti-HA, or anti-pLIMK1(Thr(P)-508) antibodies. B, HA-LRAP25 construct was co-expressed with either FLAG empty vector, FLAG-LIMK1 wild-type, LIM1/2, PDZ, or CAT constructs and followed by immunoprecipitation (IP) using anti-FLAG antibody-coated beads. The immunoprecipitates were subjected to immunoblotting with anti-FLAG or anti-HA antibodies. C, HA-LRAP25 construct was co-expressed with GFP empty vector and GFP-tagged LIM1/2, LIM1, or LIM2 regions of LIMK1 and followed by GST-pulldown using glutathione-Sepharose beads. The protein complexes recovered were subjected to immunoblotting with anti-HA or anti-GST antibodies (left). Schematic representation of LIMK1 and the various deletion mutants used in the mapping of the LRAP25-binding site. LIM1/2, LIM1 and LIM2 domains; PDZ, PSD95-Dlg-ZO1 domain; CAT, catalytic domain. The results of LRAP25 binding activity are shown (right). D, FLAG-LIM1/2 construct was co-expressed with either GFP empty vector, wild type, or various parts of LRAP25 as indicated. GFP-pulldown experiments were performed using GFP affinity beads. Recovered protein complexes were subjected to immunoblotting with anti-GFP or anti-HA antibodies. Schematic representation of LRAP25 and the various deletion mutants used in the mapping of LIMK1-binding site are shown. N, N terminus; LR, leucine repeat; pLR, partial leucine repeat; C, C terminus (left). The results of LIMK1 binding activity are shown (right). E, lysates of B16-F1 cells with or without prior AlF4− treatment were incubated with control IgG or anti-MRCKβ antibody. The immunoprecipitates were subjected to immunoblotting with anti-MRCK, anti-LIMK1, or anti-LRAP25 antibodies. F, FLAG-LIMK1 and FLAG-LIMK1-ΔLIM1 were expressed alone or co-expressed with HA-Rac1G12V as indicated. Respective lysates were immunoblotted with anti-FLAG, anti-pLIMK1(Thr(P)-508), or anti-HA antibodies. G, FLAG-LIMK1 construct was expressed in B16 cells prior to transfection with control, siLRAP25-1, or siLRAP25-2 siRNAs. Selected cells were treated with AlF4− as indicated. Respective lysates were immunoblotted with anti-FLAG, anti-pLIMK1(Thr(P)-508), anti-LRAP25, or anti-tubulin α (anti-Tubα) (loading control) antibodies.
Next, we tested the importance of LRAP25 in LIMK1 activation by comparing the levels of Rac1-induced phospho-Thr-508 signal between wild-type LIMK1 and the LIM1-deleted LIMK1 mutant that is incapable of interacting with LRAP25 (Fig. 2F). The results show that the phospho-Thr-508 level of LIMK1 without the LIM1 motif was markedly lower than that of the wild type, indicating an inefficient phosphorylation event likely due to the lack of LRAP25 interaction. These results give support on the involvement of the LRAP25-MRCK complex in LIMK1 activation. To further substantiate LRAP25 requirement in LIMK1 regulation, we analyzed the phospho-Thr-508 levels of wild-type LIMK1 in LRAP25-depleted B16 cells treated with AlF4−. As shown in Fig. 2G, the knockdown of LRAP25 by two separate pairs of LRAP25 siRNA resulted in significant reductions in the phospho-Thr-508 levels of LIMK1 induced by AlF4− treatment as compared with control. Taken together, these results underline the importance of LRAP25 in LIMK1 activation and support the view that LRAP25 acts as an adaptor molecule that tethers MRCK with LIMK1 for its activation.
LRAP25 Co-localizes with MRCK and LIMK1 at the Lamellipodia and Is Required for MRCK Lamellipodial Targeting
Live cell imaging has previously revealed that MRCKα accumulating in the lamella was mostly derived from the lamellipodium at the leading edge (25). To substantiate this, we re-examined the subcellular distribution of endogenous MRCKα in B16-F1 cells. As shown in Fig. 3A, besides its co-localization with phosphorylated RMLC in the lamella as described previously (25), MRCK was indeed found to concentrate in the F-actin-rich lamellipodium and co-distributed closely with the ARPC2 subunit of the heptameric Arp2/3 complex. Consistent with this, intense fluorescence signals of exogenously expressed GFP-MRCKα were detected within a 10-μm-wide region along cell edges that correspond to lamellipodium and lamella (Fig. 3B), as demarcated by the co-expressed β-actin-mCherry and RFP-RMLC that are known to mark lamellipodium and lamella, respectively (Fig. 3B) (3, 30). These results show that MRCK is localized to both lamellipodium and lamella of migrating B16-F1 cells.
FIGURE 3.
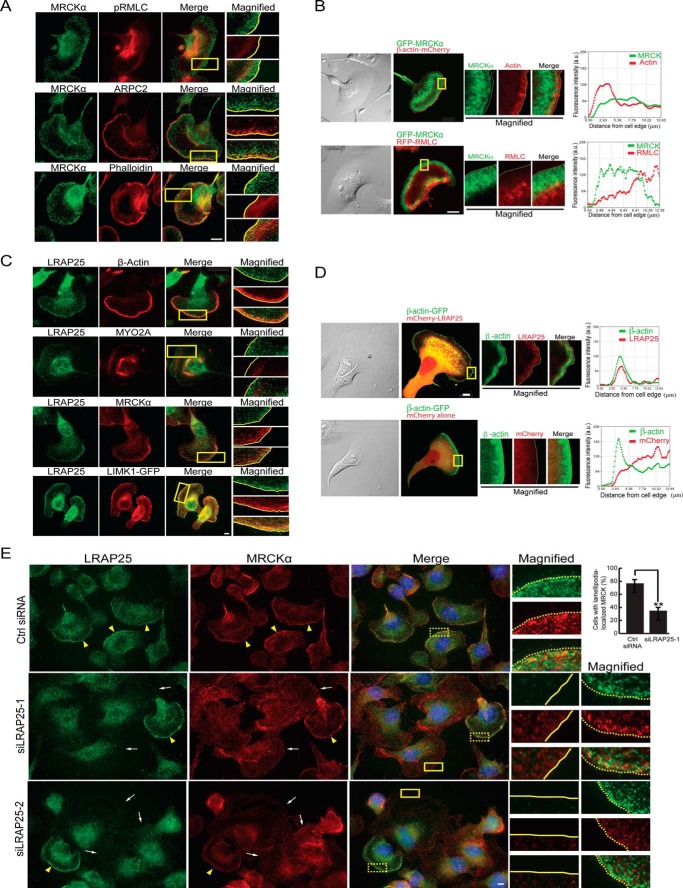
LRAP25 co-localizes with MRCK and LIMK1 at the lamellipodia and is required for MRCK lamellipodial targeting. A, B16-F1 cells were first treated with AlF4− and later fixed and stained for endogenous MRCKα, myosin light chain phosphorylated on Ser-19 (pRMLC), ARPC2, and F-actin (phalloidin) as indicated. Scale bar, 10 μm. Boxed regions are magnified and shown on the right. Yellow lines demarcate cell edges. B, distributions of GFP-MRCKα and the co-expressed β-actin-mCherry or RFP-myosin regulatory light chain (RMLC) in B16-F1 cells treated with AlF4−. Scale bar, 10 μm. Boxed regions are magnified (middle column). Yellow lines demarcate cell edges. Fluorescence intensity profiles of co-expressed proteins measured from cell edges (right column) are shown. Note the intense GFP-MRCKα signals across a 10-μm region. C, AlF4−-treated B16-F1 cells were fixed and stained for endogenous LRAP25, β-actin, nonmuscle myosin IIA (MYO2A), MRCKα, and LIMK1-GFP (transiently transfected) as indicated. Scale bar, 10 μm. Boxed regions are magnified and shown on the right. Yellow lines demarcate cell edges. D, co-localization of mCherry-LRAP25 and β-actin-GFP at the lamellipodia of B16-F1 cells. mCherry serves as a control. E, B16-F1 cells transfected with control, siLRAP25-1, or siLRAP25-2 siRNAs were treated with AlF4− and later immunostained for endogenous LRAP25, MRCKα, and nucleus (Hoechst 33342, blue). White arrows and solid boxes indicate LRAP25-depleted cells showing mislocalized MRCK; yellow arrowheads and dotted boxes indicate cells showing typical LRAP25 localization to the lamellipodium. Boxed regions are magnified. Scale bar, 10 μm. Percentage of B16-F1 cells with lamellipodium-localized MRCK following transfection with control or siLRAP25–1 siRNAs (graph). Mean values derived from three independent experiments are presented (±S.E.; n ≥250). Unpaired Student's t test indicates a significant difference between samples (** denotes p < 0.01).
We then proceeded to determine the intracellular distribution of LRAP25 and the extent of its co-localization with MRCK and LIMK1. At the leading edge, endogenous LRAP25 showed significant co-localization with β-actin, a marker for the lamellipodium, suggesting that LRAP25 was primarily targeted to the lamellipodium with some diffuse cytosolic distribution (Fig. 3C). Consistently, fluorescence intensity profiles of co-expressed mCherry-LRAP25 and β-actin-GFP also show their co-enrichment in the lamellipodium of B16-F1 cells (Fig. 3D). In contrast, a lack of overlapping signals with the lamella-concentrated nonmuscle myosin IIA staining indicates that LRAP25 was largely excluded from the lamella (Fig. 3C). The observed extensive co-localization of LRAP25 with MRCK and LIMK1 within the confines of lamellipodia is in agreement with their capacity to form a complex in vivo (Fig. 3C). These results thus support the conclusion that LRAP25 acts as the adaptor that mediates the specific recognition of LIMK1 by MRCK in the lamellipodia.
To determine whether LRAP25 is required for MRCK recruitment to the lamellipodium, we examined MRCK's distribution in LRAP25-depleted B16-F1 cells under AlF4− treatment. As shown in Fig. 3E, LRAP25 knockdown using two separate pairs of siRNAs consistently reduced the immunostaining of LRAP25 detected at cell edges and cytosol. Importantly, these siRNAs also resulted in a significant loss of MRCK staining along the leading edge and cell periphery in a large proportion of transfected cells (Fig. 3E). The lack of cell edge-localized MRCK observed in these cells suggests that the recruitment of MRCK to the leading edge is dependent on LRAP25.
MRCK and LRAP25 Regulate LIMK1 Activity and Lamellipodial Localization in Vivo
As MRCK is known to activate LIMK1 (22), we examined whether it is required for LIMK1 activity that is implicated in lamellipodial formation (15, 31). First, the activities of endogenous MRCK and LIMK1 in response to AlF4− treatment were analyzed. The two kinases were immunoprecipitated and subjected to in vitro kinase assays for a direct measurement of their activity. As shown in Fig. 4, A and B, AlF4− treatment consistently resulted in the activation of both endogenous MRCK and LIMK1. These observations are consistent with the reported involvement of LIMK1 activation for lamellipodial formation and cell migration induced by insulin (15).
FIGURE 4.
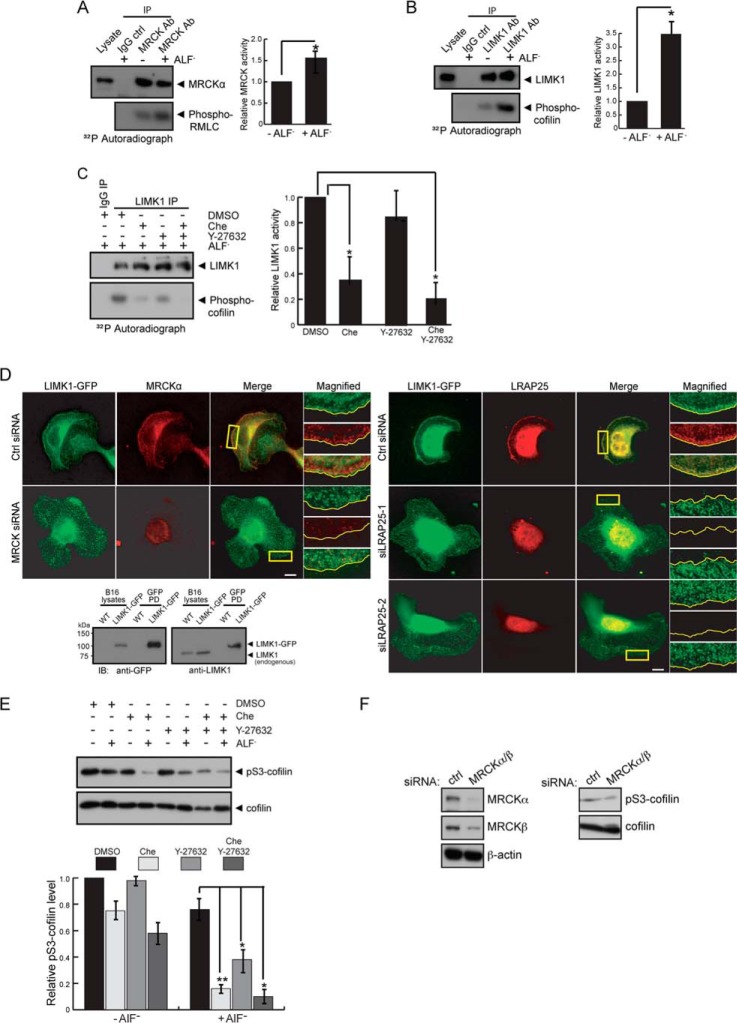
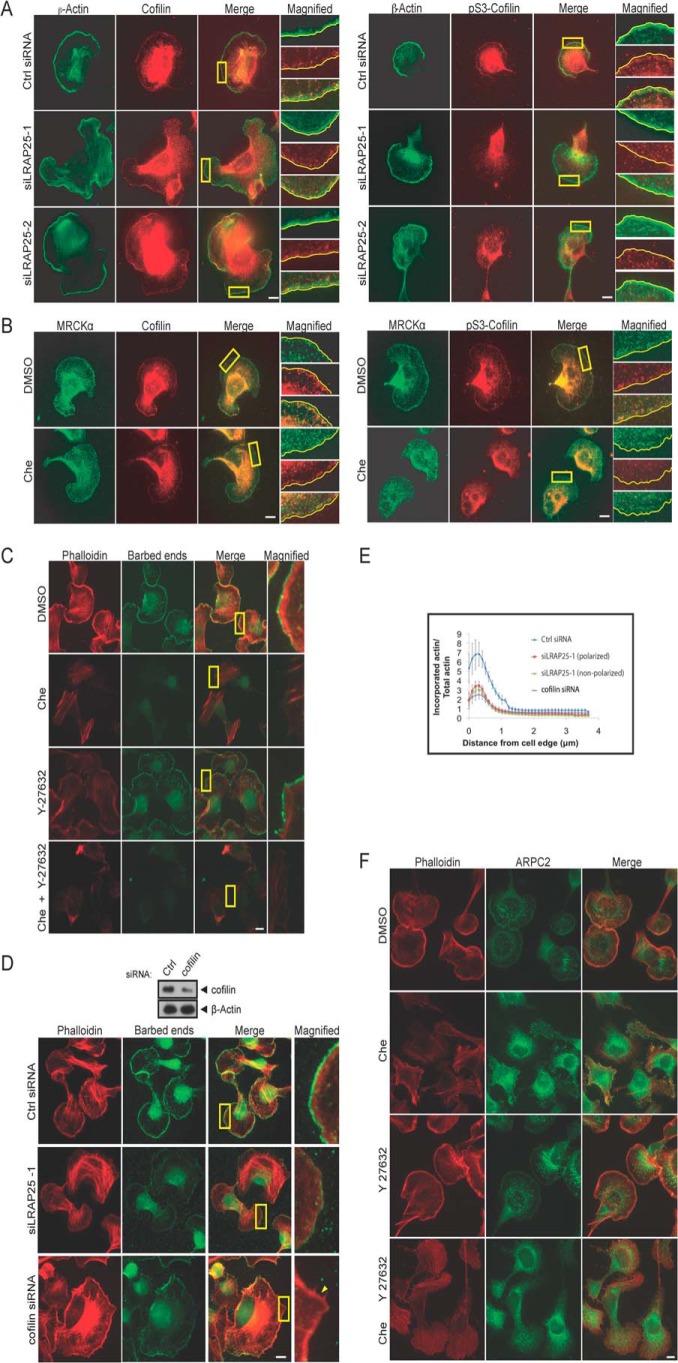
MRCK and LRAP25 are required for AlF4−-mediated LIMK1 activation and lamellipodial localization. A, B16-F1 cells with or without a prior 20-min AlF4− treatment were lysed and subjected to immunoprecipitations (IP) using anti-IgG or anti-MRCK antibody as indicated. Immunoprecipitated complexes were assayed for MRCK kinase activity using GST-RMLC as substrate. The 32P autoradiograph is shown. The immunoprecipitates were probed for MRCK (left top panel). Mean values derived from three independent experiments are presented (±S.E.) (right panel). Unpaired Student's t test indicates a significant difference between samples (* denotes p < 0.01). B, B16-F1 cells with or without prior 20 min AlF4− treatment were lysed and subjected to immunoprecipitations using anti-IgG or anti-LIMK1 antibody as indicated. Immunoprecipitated complexes were assayed for LIMK1 kinase activity using cofilin as substrate. The 32P autoradiograph is shown. The immunoprecipitates were probed for LIMK1 (left top panel). Mean values from three independent experiments are presented (±S.E.; n = 3) (right panel). Unpaired Student's t test indicates a significant difference between samples (* denotes p < 0.01). C, B16-F1 cells were treated with AlF4− for 10 min, followed by addition of either DMSO, 5 μm chelerythrine chloride (Che), 5 μm Y-27632 or a combination of Che and Y-27632 for 30 min. Treated cells were lysed and subjected to immunoprecipitations using anti-LIMK1 antibody and subsequently assayed for LIMK1 kinase activity using cofilin as substrate. The 32P autoradiograph is shown. The immunoprecipitates were probed for LIMK1 (left top panel). Mean values from three independent experiments are presented (±S.E.; n = 3) (right panel). Dunnett's multiple comparison test was used after one-way analysis of variance to generate p values (* denotes p < 0.05). D, B16-F1 cells transfected with control (Ctrl), MRCK, siLRAP25-1, or siLRAP25-2 siRNAs were treated with AlF4− and later immunostained for LIMK1-GFP, endogenous MRCKα, and LRAP25 as indicated. Boxed regions are magnified. Scale bar, 10 μm. Characterization of B16-F1 cells stably expressing LIK1-GFP is shown. Parental wild-type and the stable LIMK1-GFP cells were subjected to pull down (PD) with GFP affinity beads. Pulldown products were probed with either anti-GFP or anti-LIMK1 antibodies as indicated. Note that the GFP antibody recognized LIMK1-GFP present in both the total lysate and the pull down, although the LIMK1 antibody only detected LIMK-GFP present at a higher concentration in the pulldown fraction due to its low level in the lysate (bottom panel). E, B16-F1 cells with or without 10 min AlF4− treatment were added with either DMSO, 5 μm Che, 5 μm Y-27632 or a combination of Che and Y-27632 for 30 min. Lysates of treated cells were probed for endogenous Ser(P)-3-cofilin and cofilin. Quantification of relative Ser(P)-3-cofilin levels is shown (±S.E.; n = 3) (bottom panel). Newman-Keuls multiple comparison test was used after one-way analysis of variance to generate p values. * denotes p < 0.05; **, denotes p < 0.01. F, B16-F1 cells transfected with control or combined MRCKα and β siRNAs were treated with AlF4− before lysis. Lysates were probed with anti-MRCKα, anti-MRCKβ, anti-β-actin, anti-Ser(P)-3-cofilin, or anti-cofilin antibodies.
To further determine whether MRCK activity is essential for AlF4−-mediated LIMK1 activation, we measured the activity of endogenous LIMK1 from cells treated with either ROK inhibitor or the recently identified MRCK inhibitor, chelerythrine (32). In contrast to ROK inhibition by Y-27632, which resulted in a modest decrease (15%), inhibition of MRCK produced a marked reduction in AlF4−-mediated activation of LIMK1 (63%) (Fig. 4C), indicating that LIMK1 activation by AlF4− is largely MRCK-dependent. In addition, inhibition of both ROK and MRCK simultaneously appears to have an additive effect on LIMK1 suppression, suggesting that these two related kinases activate LIMK1 through separate mechanisms. As prolonged chelerythrine treatment at high concentrations has been shown to cause apoptosis, we ruled out the involvement of cell death in our experiments as cleaved caspase-3 was undetectable under our experimental conditions (data not shown) (32). Taken together, our findings suggest that AlF4− treatment activates MRCK, which in turn phosphorylates and activates LIMK1. These data thus substantiate the reported role of MRCK as the upstream activating kinase of LIMK1 that is important for lamellipodial formation.
Next, we determined whether LRAP25 and MRCK are important for the translocation of LIMK1 to the lamellipodium. As shown in Fig. 4D, the depletion of MRCK or LRAP25 both affected the lamellipodial localization of stably expressed LIMK1-GFP induced by AlF4−. Our findings thus indicate the importance of LRAP25 and MRCK and, accordingly, the formation of the tripartite complex in LIMK1 activation and its targeting to the lamellipodium.
We subsequently examined whether MRCK plays a role in the regulation of cofilin by measuring the level of cofilin phosphorylation at Ser-3 (Ser(P)-3-cofilin) in B16-F1 cells treated with chelerythrine or Y-27632, before and after AlF4− treatment. As shown in Fig. 4E, addition of AlF4− resulted in a drop (22%) in Ser(P)-3-cofilin levels in control cells, possibly due to Rac-mediated activation of cofilin phosphatases as reported previously (33, 34). However, the addition of chelerythrine and Y-27632, either separately or in combination, resulted in further reductions (Fig. 4E). Correspondingly, there is a more significant drop in Ser(P)-3-cofilin levels with MRCK inhibition (57.5%) than that of ROK (36.5%), and inhibition of both kinases simultaneously produced a similar additive effect observed with LIMK1 suppression. Consistently, siRNA-mediated depletion of MRCK also resulted in a drop in Ser(P)-3-cofilin levels (Fig. 4F). The lesser effect of MRCK depletion in comparison with chelerythrine treatment is likely attributable to our previous observation that prolonged down-regulation of MRCK activity by siRNAs resulted in up-regulation of stress fiber formation through the activation of Rho/ROK pathway, a process in which LIMKs and cofilin are also involved. Taken together, these results provide strong evidence that MRCK is an important upstream regulator of LIMK1-cofilin signaling.
Both MRCK Inhibition and LRAP25 Depletion Affect Cofilin Activity in Lamellipodia
Our findings thus far suggest a role for LRAP25 in targeting MRCK to the lamellipodia for LIMK1 regulation. It is thus important to find out whether LRAP25 is also involved in the regulation of cofilin in vivo. To examine this, we determined the localization of cofilin and the levels of Ser(P)-3-cofilin in the lamellipodia of B16-F1 cells depleted of LRAP25 or inhibited of MRCK activity, in response to AlF4− stimulation. From the measurements within a 3.5-μm-wide region along the leading edge, cofilin localization to cell edges was largely unaffected by LRAP25 depletion or MRCK inhibition; this was observed even in cells that were poorly polarized (Fig. 5, A and B). However, the levels of Ser(P)-3-cofilin detectable along the cell edges markedly diminished in response to LRAP25 and MRCK down-regulation (Fig. 5, A and B). These results reflect the importance of LRAP25 in MRCK-mediated LIMK1 activation and the subsequent regulation of cofilin phosphorylation.
FIGURE 5.
Inhibition of MRCK and LRAP25 affects cofilin activity. A, B16-F1 cells transfected with control, siLRAP25-1, or siLRAP25-2 siRNAs were treated with AlF4−. Cells were later fixed and stained for endogenous β-actin, cofilin, and Ser(P)-3-cofilin. Scale bar, 10 μm. Boxed regions are magnified and shown on the right. B, B16-F1 cells were first treated with AlF4− for 10 min followed by addition of either DMSO or 5 μm Che for 30 min. Cells were later fixed and stained for endogenous MRCK, cofilin, and Ser(P)-3-cofilin. Scale bar, 10 μm. Boxed regions are magnified and shown on the right. C, B16-F1 cells were induced with AlF4− for 10 min, followed by addition of either DMSO, 5 μm Che, and 5 μm Y-27632 or a combination of Che and Y-27632 for 30 min. Cells were then subjected to incorporation of fluorescent-labeled actin (green) for 45 s and counterstained with phalloidin (red). Scale bar, 10 μm. Boxed regions are magnified and shown on the right. D, lysates of B16-F1 cells transfected with control or cofilin siRNAs were probed for endogenous cofilin and β-actin (top panel). B16-F1 cells transfected with control siRNA, siLRAP25-1, or cofilin siRNAs were subjected to incorporation of fluorescently labeled actin (green) for 45 s following AlF4− treatment. Cells were counterstained with phalloidin (red). Scale bar, 10 μm. Boxed regions are magnified and shown on the right. E, fluorescence intensity ratio of incorporated actin relative to total F-actin (phalloidin-stained) of B16-F1 cells transfected with control siRNA, siLRAP25-1, or cofilin siRNAs. Measurements were made across a 3.4-μm-wide region beginning from the cell edge. LRAP25-depleted cells displaying both polarized and nonpolarized irregular morphology were analyzed. Data show results of a representative experiment (±S.E.; n ≥8). Similar results were obtained from three independent experiments. F, B16-F1 cells were first induced with AlF4− for 10 min, followed by addition of either DMSO, 5 μm Che, 5 μm Y-27632, or a combination of Che and Y-27632 for 30 min. Cells were later subjected to immunostaining with anti-ARPC2 antibody and phalloidin.
As cofilin contributes to the formation of free barbed ends at the leading edge through its F-actin severing activity (35, 36), we went on to examine the involvement of MRCK and LRAP25 in this process to further understand their functional link with cofilin. This was conducted by comparing the extent of labeled actin incorporation into control B16-F1 cells and those deprived of MRCK activity or LRAP25 expression, under AlF4− stimulation. As shown in Fig. 5, C–E, both MRCK inhibition and LRAP25 depletion consistently resulted in reduced incorporation of actin monomers into actin filaments at the leading edge, indicating a reduction in the density of free-barbed ends in the cell edge regions resulted from perturbed local F-actin dynamics. These results reflect the importance of dynamic phosphorylation of cofilin in lamellipodial F-actin regulation as increased levels of active dephosphorylated cofilin in response to MRCK and LRAP25 down-regulation were expected to induce barbed ends formation. Similar effects have been obtained for cofilin depletion (37) as well as the inhibition of its upstream regulators such as Rac, LIMK1, and PAK1 (31, 34, 37). In addition, ROK inhibition by Y-27632 did not result in a significant reduction in barbed end formation (Fig. 5C), a result that is consistent with the small effects of Y-27632 on LIMK1 activation observed earlier (Fig. 4C) and in a previous report (37). In further support, mislocalization of the subunit of the Arp2/3 complex ARPC2 from the cell edge was observed in cells treated with chelerythrine but not Y-27632 (Fig. 5F). We thus conclude that LRAP25 and MRCK are involved in the regulation of cofilin phosphorylation and F-actin dynamics at the leading edge, results that further substantiate their roles in LIMK1 activation.
LRAP25 Depletion Affects Cell Polarization and Motility
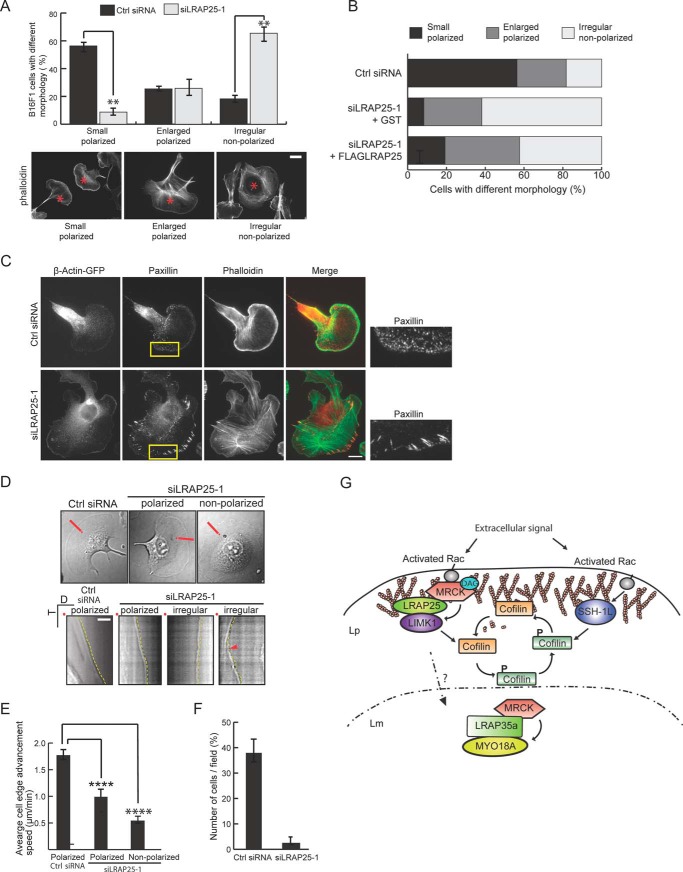
It was also noticeable that B16-F1 cells grown under standard conditions display mixed cellular morphology when treated with AlF4− (Fig. 6A). Although the majority of cells are small and polarized with broad lamellipodia (56%), the remaining populations are either nonpolarized cells with irregular morphology (18%) or polarized but untypically enlarged (25%). Interestingly, depletion of LRAP25 led to a significant drop in the percentage of cells with small and polarized morphology with corresponding increases in the population of nonpolarized cells (Fig. 6A). The cell polarization defect resulting from LRAP25 depletion could be partially reverted by expressing an siRNA-resistant LRAP25 at the expense of the nonpolarized population (Fig. 6B). These results indicate that LRAP25 is required for the process of cell polarization.
FIGURE 6.
LRAP25 depletion affects cell polarization and migration. A, B16-F1 cells were transfected with control siRNA or siLRAP25-1. Cells were treated with AlF4− and subjected to cell morphology analysis. Graph shows the percentage of B16-F1 cells exhibiting morphology corresponding to each of the following categories: small and polarized, enlarged and polarized, or irregular and nonpolarized. β-Actin-GFP expression from co-transfected plasmid marks transfectants for cell count (data not shown). Mean values derived from three independent experiments are shown (±S.E.; n ≥250 for each category). Unpaired Student's t test indicates a significant difference between samples (** denotes p < 0.01) (top panel). Representative cells in each category are marked by red asterisks (phalloidin-stained). B, B16-F1 cells were transfected with control siRNA, siLRAP25-1, plus a GST expression vector or siLRAP25-1 plus a FLAG-tagged siRNA-resistant LRAP25 expression vector. Graph shows the percentage of cells exhibiting morphology categorized as in A (n ≥150 for each category). C, B16-F1 cells were transfected with control siRNA or siLRAP25-1 and treated with AlF4−. Cells were later fixed and stained for F-actin (phalloidin) and paxillin. Cells were transfected with β-Actin-GFP to mark transfectants and lamellipodia. Scale bar, 10 μm. Boxed regions are magnified and shown on the right. D, kymograph analyses showing cell edge advancement dynamics taken from B16-F1 cells transfected with control (Ctrl) siRNA or siLRAP25-1. Dotted yellow lines mark cell edges. Arrowhead indicates the transition point between retraction and protrusion captured during image recording of LRAP25-depleted cells undergoing intermittent retractions (bottom panel). LRAP25-depleted cells displaying both polarized and nonpolarized irregular morphology were analyzed. Red lines indicate locations used to generate kymographs. Red dots indicate line origin (top panel). D and T denote distance and time, respectively. Scale bar, 10 μm. E, average cell edge advancement speed derived from B16-F1 cells transfected with siRNAs as in D. Mean values derived from two independent experiments are shown (±S.E.; n ≥17). Newman-Keuls multiple comparison test was used after one-way analysis of variance to generate p values (**** denotes p < 0.0001). F, migration of B16-F1 cells transfected with control siRNA or siLRAP25-1 was measured in a transwell assay using filters with pore diameters of 5 μm. Graph shows the percentage of cells migrated to the lower chamber derived from three independent experiments (±S.E.). G, model of MRCK complex regulation of LIMK1 and cofilin. Rac activation in response to extracellular signal targets MRCK complex to the cell membrane. Activation of MRCK by lipid hydrolysis (diacylglycerol) results in phosphorylation/activation of LIMK1, which in turn causes phosphorylation/inactivation of the F-actin depolymerizing/severing factor cofilin. Activation of the cofilin phosphatase SSH-1L in response to Rac activation results in dephosphorylation of cofilin. Coordination of the activities of the kinases and phosphatases are required to maintain the dynamic phosphorylation regulation of cofilin that is important for lamellipodial F-actin regulation. In the process of cell protrusion, MRCK translocates from the lamellipodium to the lamella with a corresponding change in complexity. The translocation is believed to be important for coordinating cytoskeletal regulations taking place in the two connected regions. Lp, lamellipodium; Lm, lamella; DAG, diacylglycerol.
As morphological changes usually correspond to changes in the underlying cytoskeleton, we further compared F-actin and focal adhesion structures in LRAP25-depleted and control B16-F1 cells. As shown in Fig. 6C, the population of LRAP25-knockdown cells, which displayed nonpolarized irregular morphology, also showed increased formation of stress fibers and enlarged focal adhesions (Fig. 6C), an indication of enhanced Rho activity. In addition, these poorly polarized cells lacked a well defined Rac-dependent lamellipodial F-actin meshwork, and the smaller focal complexes normally detectable in the leading edge of the control cells (Fig. 6C). These morphological changes are consistent with the previously observed lack of barbed end formation in the lamellipodia of these cells (Fig. 5D). Together, these results suggest that the loss of LRAP25 perturbed the balance of Rac-Rho activities seen in the control cells with an apparent down-regulation of Rac but a corresponding activation of Rho (38).
To determine whether the changes in cytoskeletal regulation affect leading-edge dynamics, the membrane displacement speed of migrating B16-F1 cells was measured. As shown by kymograph analyses derived from time-lapse imaging (Fig. 6, D and E), LRAP25 knockdown led to a significant reduction in cell edge advancement speed in B16-F1 cells. The leading edge of control cells was observed to advance steadily at an average of 1.76 μm/min, although LRAP25-depleted cells that are nonpolarized extended at a much slower rate at 0.53 μm/min. About 30% of these cells exhibited intermittent retraction during image recording, a sign of low persistence in membrane extension (Fig. 6E). Importantly, the small population of cells that remained morphologically polarized after LRAP25 depletion also showed a significantly compromised rate of 0.98 μm/min, suggesting a deficiency in cell migration. Indeed, the depletion of LRAP25 strongly suppressed cell migration in a transwell migration assay (Fig. 6F). Taken together, the defects in cell polarization, membrane protrusion, and migration as a result of LRAP25 depletion implicate LRAP25 playing a role in targeting MRCK for LIMK1 activation and the subsequent regulation of lamellipodial F-actin dynamics.
DISCUSSION
Here, we report the characterization of LRAP25 as a novel interactor of MRCK in both mammalian tissues and cultured cells. The interaction is through the conserved LR1 motif previously reported for LRAP35a (25). Together with LRAP35a and -b, these LRAP proteins form a unique group of MRCK adaptors that account for the majority of MRCK complex in cells and tissues. At the cellular level, the targeting of MRCK to the protrusive areas of migrating cells could be determined by its interaction with LRAP35a and LRAP25 as exemplified by the scaffolding role of adaptor molecules, such as protein kinase A-anchoring proteins and targeting subunits of protein phosphatase 1 (PP1c), in tethering cAMP-dependent protein kinase (PKA) and PP1c to specific downstream targets and subcellular sites (39, 40). Whereas LRAP35a interaction with MRCK directs it to the lamella through the formation of a tripartite complex with MYO18A for actomyosin activation, LRAP25 apparently is required for targeting MRCK to the lamellipodium possibly through mediating the MRCK-LIMK1 interaction. Hence, besides its reported functions in TGFβ signaling and as a cell surface receptor for the entry of mammary tumor virus (27, 41), LRAP25 apparently also has a role in cytoskeletal regulation.
The observation of a close association with ARPC2 and β-actin distribution in the lamellipodium provides further evidence that the LRAP25-MRCK complex may serve a role that is fundamentally distinct from that of LRAP35a-MRCK. Indeed, we show here that LRAP25-mediated MRCK targeting to LIMK1 and the lamellipodium are important for LIMK1 regulation in the region. MRCK has previously been shown to regulate LIMK1, but the cellular relevance of this event is obscure (22). Our observation that both MRCK and LIMK1 were up-regulated by AlF4− treatment is consistent with their involvement in the leading edge regulation of migrating cells. Although LIMK1 activation has been linked to Rac-mediated regulation (14, 15), up-regulation of MRCK may be directly induced by an increase in the level of diacylglycerol resulting from G protein coupled-phospholipase C activation downstream of AlF4− (42), in a manner similar to the effect of phorbol 12-myristate 13-acetate (an analog of diacylglycerol) binding to the C1 domain of MRCK (43). It is also important to note that MRCK can respond directly to Rac activation besides Cdc42, likely through the weaker Rac-binding CRIB domain or the unusual tandem CRIB motif in an MRCKα variant that could interact more strongly with Rac (44). The importance of MRCK activation playing a role in the AlF4− response was emphasized by its inhibition that could significantly suppress LIMK1 activation and the subsequent phosphorylation of cofilin. Because cofilin is the major LIMK1 substrate associated with lamellipodia (45), our data therefore strongly suggest that MRCK could be a key upstream regulator of LIMK1-cofilin signaling in the lamellipodia. This claim is further supported by the direct comparison of MRCK and ROK inhibition that showed MRCK as the major activator of LIMK1. The specificity of MRCK on LIMK1 activation observed here complies with previous results that discerned the differences in the regulation and functions of LIMK isoforms. Previous studies have shown that LIMK1 expression preferentially induced lamellipodial F-actin formation under Rac regulation (14, 15), whereas LIMK2 was shown to act downstream of Rho-ROCK signaling in the formation of stress fibers and focal adhesions (46, 47). The weak effect on the formation of barbed ends shown by ROK inhibition is also in keeping with the lesser role it plays in lamellipodial F-actin regulation (37, 48). The more significant reduction of the overall Ser(P)-3-cofilin level observed upon Y-27632 treatment is thus believed to be the effect of LIMK2 inhibition. Thus, these findings provide supportive evidence that the two LIMK isoforms are under the regulation of distinct signaling cascades for the spatial control of actin depolymerizing factor/cofilin activity at different cellular regions.
In addition to the biochemical observations, the suppression of MRCK and LRAP25 also led to severe defects in incorporation of actin into lamellipodial barbed ends, indicating a reduction in the density of polymerizing competent free barbed end filaments that is reminiscent of the reported effects of LIMK1 or cofilin suppression (34, 35, 37). Such defects were associated with the corresponding accumulations of stress fibers, enlarged focal adhesions, and more importantly failure of effective cell protrusion and migration that were also observed in perturbed LIMK1-cofilin signaling (49–51). As depicted in the model (Fig. 6G), our analyses thus reveal the biochemical and functional links between LRAP25-MRCK and LIMK1-cofilin signaling pathways in vivo and underline their importance in the regulation of lamellipodial F-actin dynamics and cell polarization.
The spatiotemporal coordination of cellular cytoskeletal activities and extracellular adhesion molecules is important for cell protrusion and movement (2, 52). PAK1 has recently been shown to couple leading edge actin dynamics to focal adhesions, possibly by coordinating its regulation on LIMK1 and focal adhesion turnover (53). MRCK's specific localization to the lamellipodium and lamella and its roles in both LIMK1 signaling and myosin activation also suggest that it could be involved in coordinating events in these two cellular compartments. It is thus possible that MRCK and PAK1 complement each other in this three-way process by coupling lamellipodial F-actin dynamics to myosin activity and focal adhesion turnover, respectively. The observation of a direct phosphorylation-dependent role of cofilin in actomyosin assembly further emphasizes the importance of functional coordination between F-actin dynamics and actomyosin assembly (51). Thus, with the lamellipodium-targeting MRCK and PAK1 that co-regulate LIMK1 at the cell front, and ROK being implicated for stress fibers that constitute the posterior of migrating cells (54, 55), it is conceivable that these two sets of kinases could be responsible for the regulation of separate F-actin dynamics at various cellular sites, and their coordination provides a concerted front-back regulatory mechanism for efficient cell migration.
Acknowledgment
We thank James Bamburg for the anti-Ser(P)-3-cofilin antibody.
This work was supported by funding from Agency of Science and Technology Research (A-STAR), Singapore.
- MRCK
- myotonic dystrophy kinase-related Cdc42-binding kinase
- RMLC
- myosin regulatory light chain
- Che
- chelerythrine chloride.
REFERENCES
- 1. Lauffenburger D. A., Horwitz A. F. (1996) Cell migration: a physical integrated molecular process. Cell 84, 359–369 [DOI] [PubMed] [Google Scholar]
- 2. Gupton S. L., Waterman-Storer C. M. (2006) Spatiotemporal feedback between actomyosin and focal adhesion systems optimizes rapid cell migration. Cell 125, 1361–1374 [DOI] [PubMed] [Google Scholar]
- 3. Ponti A., Machacek M., Gupton S. L., Waterman-Storer C. M., Danuser G. (2004) Two distinct actin networks drive the protrusion of migrating cells. Science 305, 1782–1786 [DOI] [PubMed] [Google Scholar]
- 4. Pollard T. D., Borisy G. G. (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 5. Mullins R. D., Heuser J. A., Pollard T. D. (1998) The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. U.S.A. 95, 6181–6186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Svitkina T. M., Borisy G. G. (1999) Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145, 1009–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goley E. D., Welch M. D. (2006) The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7, 713–726 [DOI] [PubMed] [Google Scholar]
- 8. Welch M. D., Mullins R. D. (2002) Cellular control of actin nucleation. Annu. Rev. Cell Dev. Biol. 18, 247–288 [DOI] [PubMed] [Google Scholar]
- 9. Wear M. A., Schafer D. A., Cooper J. A. (2000) Actin dynamics: Assembly and disassembly of actin networks. Curr. Biol. 10, R891–R895 [DOI] [PubMed] [Google Scholar]
- 10. Lai F. P., Szczodrak M., Block J., Faix J., Breitsprecher D., Mannherz H. G., Stradal T. E., Dunn G. A., Small J. V., Rottner K. (2008) Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 27, 982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bamburg J. R. (1999) Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15, 185–230 [DOI] [PubMed] [Google Scholar]
- 12. Oser M., Condeelis J. (2009) The cofilin activity cycle in lamellipodia and invadopodia. J. Cell Biochem. 108, 1252–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bernstein B. W., Bamburg J. R. (2010) ADF/Cofilin: a functional node in cell biology. Trends Cell Biol. 20, 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arber S., Barbayannis F. A., Hanser H., Schneider C., Stanyon C. A., Bernard O., Caroni P. (1998) Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393, 805–809 [DOI] [PubMed] [Google Scholar]
- 15. Yang N., Higuchi O., Ohashi K., Nagata K., Wada A., Kangawa K., Nishida E., Mizuno K. (1998) Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 393, 809–812 [DOI] [PubMed] [Google Scholar]
- 16. Toshima J., Toshima J. Y., Takeuchi K., Mori R., Mizuno K. (2001) Cofilin phosphorylation and actin reorganization activities of testicular protein kinase 2 and its predominant expression in testicular Sertoli cells. J. Biol. Chem. 276, 31449–31458 [DOI] [PubMed] [Google Scholar]
- 17. Van Troys M., Huyck L., Leyman S., Dhaese S., Vandekerkhove J., Ampe C. (2008) Ins and outs of ADF/cofilin activity and regulation. Eur. J. Cell Biol. 87, 649–667 [DOI] [PubMed] [Google Scholar]
- 18. Niwa R., Nagata-Ohashi K., Takeichi M., Mizuno K., Uemura T. (2002) Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell 108, 233–246 [DOI] [PubMed] [Google Scholar]
- 19. Huang T. Y., DerMardirossian C., Bokoch G. M. (2006) Cofilin phosphatases and regulation of actin dynamics. Curr. Opin. Cell Biol. 18, 26–31 [DOI] [PubMed] [Google Scholar]
- 20. Edwards D. C., Sanders L. C., Bokoch G. M., Gill G. N. (1999) Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat. Cell Biol. 1, 253–259 [DOI] [PubMed] [Google Scholar]
- 21. Maekawa M., Ishizaki T., Boku S., Watanabe N., Fujita A., Iwamatsu A., Obinata T., Ohashi K., Mizuno K., Narumiya S. (1999) Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science 285, 895–898 [DOI] [PubMed] [Google Scholar]
- 22. Sumi T., Matsumoto K., Shibuya A., Nakamura T. (2001) Activation of LIM kinases by myotonic dystrophy kinase-related Cdc42-binding kinase α. J. Biol. Chem. 276, 23092–23096 [DOI] [PubMed] [Google Scholar]
- 23. Leung T., Chen X. Q., Tan I., Manser E., Lim L. (1998) Myotonic dystrophy kinase-related Cdc42-binding kinase acts as a Cdc42 effector in promoting cytoskeletal reorganization. Mol. Cell. Biol. 18, 130–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tan I., Ng C. H., Lim L., Leung T. (2001) Phosphorylation of a novel binding subunit of protein phosphatase 1 reveals a conserved mechanism in the regulation of actin cytoskeleton. J. Biol. Chem. 276, 21209–21216 [DOI] [PubMed] [Google Scholar]
- 25. Tan I., Yong J., Dong J. M., Lim L., Leung T. (2008) A tripartite complex containing MRCK modulates lamellar actomyosin retrograde flow. Cell 135, 123–136 [DOI] [PubMed] [Google Scholar]
- 26. Sakuma-Takagi M., Tohyama Y., Kasama-Yoshida H., Sakagami H., Kondo H., Kurihara T. (1999) Novel related cDNAs (C184L, C184M, and C184S) from developing mouse brain encoding two apparently unrelated proteins. Biochem. Biophys. Res. Commun. 263, 737–742 [DOI] [PubMed] [Google Scholar]
- 27. Kokura K., Kim H., Shinagawa T., Khan M. M., Nomura T., Ishii S. (2003) The Ski-binding protein C184M negatively regulates tumor growth factor-β signaling by sequestering the Smad proteins in the cytoplasm. J. Biol. Chem. 278, 20133–20139 [DOI] [PubMed] [Google Scholar]
- 28. Hahne P., Sechi A., Benesch S., Small J. V. (2001) Scar/WAVE is localised at the tips of protruding lamellipodia in living cells. FEBS Lett. 492, 215–220 [DOI] [PubMed] [Google Scholar]
- 29. Steffen A., Rottner K., Ehinger J., Innocenti M., Scita G., Wehland J., Stradal T. E. (2004) Sra-1 and Nap1 link Rac to actin assembly driving lamellipodial formation. EMBO J. 23, 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Condeelis J., Singer R. H. (2005) How and why does b-actin mRNA target? Biol. Cell 97, 97–110 [DOI] [PubMed] [Google Scholar]
- 31. Nishita M., Tomizawa C., Yamamoto M., Horita Y., Ohashi K., Mizuno K. (2005) Spatial and teporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional migration. J. Cell Biol. 171, 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tan I., Lai J., Yong J., Li S. F., Leung T. (2011) Chelerythrine perturbs lamellar actomyosin filaments by selective inhibition of myotonic dystrophy kinase-related Cdc42-binding kinase. FEBS Lett. 585, 1260–1268 [DOI] [PubMed] [Google Scholar]
- 33. Nagata-Ohashi K., Ohta Y., Goto K., Chiba S., Mori R., Nishita M., Ohashi K., Kousaka K., Iwamatsu A., Niwa R., Uemura T., Mizuno K. (2004) A pathway of neuregulin-induced activation of cofilin-phosphatase Slingshot and cofilin in lamellipodia. J. Cell Biol. 165, 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun C. X., Magalhães M. A., Glogauer M. (2007) Rac1 and Rac2 differentially regulate actin free barbed end formation downstream of the fMLP receptor. J. Cell Biol. 179, 239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ghosh M., Song X., Mouneimne G., Sidani M., Lawrence D. S., Condeelis J. S. (2004) Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304, 743–746 [DOI] [PubMed] [Google Scholar]
- 36. Mouneimne G., Soon L., DesMarais V., Sidani M., Song X., Yip S. C., Ghosh M., Eddy R., Backer J. M., Condeelis J. (2004) Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J. Cell Biol. 166, 697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Delorme V., Machacek M., DerMardirossian C., Anderson K. L., Wittmann T., Hanein D., Waterman-Storer C., Danuser G., Bokoch G. M. (2007) Cofilin activity downstream of Pak1 regulates cell protrusion efficiency by organizing lamellipodium and lamella actin networks. Dev. Cell 13, 646–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rottner K., Hall A., Small J. V. (1999) Interplay between Rac and Rho in the control of substrate contact dynamics. Curr. Biol. 9, 640–648 [DOI] [PubMed] [Google Scholar]
- 39. Cohen P. T. (2002) Protein phosphatase 1-targeted in many directions. J. Cell Sci. 115, 241–256 [DOI] [PubMed] [Google Scholar]
- 40. Beene D. L., Scott J. D. (2007) A-kinase anchoring proteins take shape. Curr. Opin. Cell Biol. 19, 192–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Golovkina T. V., Dzuris J., van den Hoogen B., Jaffe A. B., Wright P. C., Cofer S. M., Ross S. R. (1998) A novel membrane protein is a mouse mammary tumour virus receptor. J. Virol. 72, 3066–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paris S., Pouysségur J. (1987) Further evidence for a phospholipase C-coupled G protein in hamster fibroblasts. Induction of inositol phosphate formation by fluoroaluminate and vanadate and inhibition by petussis toxin. J. Biol. Chem. 262, 1970–1976 [PubMed] [Google Scholar]
- 43. Tan I., Seow K. T., Lim L., Leung T. (2001) Intermolecular and intramolecular interactions regulate catalytic activity of myotonic dystrophy kinase-related Cdc42-binding kinase α. Mol. Cell. Biol. 21, 2767–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tan I., Cheong A., Lim L., Leung T. (2003) Genomic organization of human myotonic dystrophy kinase-related Cdc42-binding kinase a reveals multiple alternative splicing and functional diversity. Gene 304, 107–115 [DOI] [PubMed] [Google Scholar]
- 45. DesMarais V., Ghosh M., Eddy R., Condeelis J. (2005) Cofilin takes the lead. J. Cell Sci. 118, 19–26 [DOI] [PubMed] [Google Scholar]
- 46. Amano T., Tanabe K., Eto T., Narumiya S., Mizuno K. (2001) LIM-kinase 2 induces formation of stress fibres, focal adhesions and membrane blebs, dependent on its activation by Rho-associated kinase-catalysed phosphorylation at threonine-505. Biochem. J. 354, 149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sumi T., Matsumoto K., Nakamura T. (2001) Specific activation of LIM kinase 2 via phosphorylation of threonine 505 by ROCK, a Rho-dependent protein kinase. J. Biol. Chem. 276, 670–676 [DOI] [PubMed] [Google Scholar]
- 48. Song X., Chen X., Yamaguchi H., Mouneimne G., Condeelis J. S., Eddy R. J. (2006) Initiation of cofilin activity in response to EGF is uncoupled from cofilin phosphorylation and dephosphorylation in carcinoma cells. J. Cell Sci. 119, 2871–2881 [DOI] [PubMed] [Google Scholar]
- 49. Hotulainen P., Lappalainen P. (2006) Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 173, 383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sidani M., Wessels D., Mouneimne G., Ghosh M., Goswami S., Sarmiento C., Wang W., Kuhl S., El-Sibai M., Backer J. M., Eddy R., Soll D., Condeelis J. (2007) Cofilin determines the migration behaviour and turning frequency of metastatic cancer cells. J. Cell Biol. 179, 777–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wiggan O., Shaw A. E., DeLuca J. G., Bamburg J. R. (2012) ADF/cofilin regulates actomyosin assembly through competitive inhibition of myosin II binding to F-actin. Dev. Cell 22, 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Burnette D. T., Manley S., Sengupta P., Sougrat R., Davidson M. W., Kachar B., Lippincott-Schwartz J. (2011) A role for actin arcs in the leading-edge advance of migrating cells. Nat. Cell Biol. 13, 371–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Delorme-Walker V. D., Peterson J. R., Chernoff J., Waterman C. M., Danuser G., DerMardirossian C., Bokoch G. (2011) Pak1 regulates focal adhesion strength, myosin IIA distribution, and actin dynamics to optimize cell migration. J. Cell Biol. 193, 1289–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tan I., Leung T. (2009) Myosin light chain kinases: division of work in cell migration. Cell Adh. Migr. 3, 256–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vicente-Manzanares M., Ma X., Adelstein R. S., Horwitz A. R. (2009) Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10, 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]