Background: TRPM1 is essential for the light response of retinal depolarizing bipolar cells.
Results: Recombinant purified TRPM1 is mostly dimeric, and a low resolution cryo-EM structure is presented.
Conclusion: Because most TRP channels function as tetramers, active TRPM1 channels likely require additional partner subunits.
Significance: The results suggest a novel paradigm for structure and regulation within the TRP channel family.
Keywords: Cryoelectron Microscopy, Ion Channel, Membrane Protein, Transient Receptor Potential Channels (TRP Channels), Vision, G Protein Signaling
Abstract
Transient receptor potential melastatin-1 (TRPM1) is essential for the light-induced depolarization of retinal ON bipolar cells. TRPM1 likely forms a multimeric channel complex, although almost nothing is known about the structure or subunit composition of channels formed by TRPM1 or any of its close relatives. Recombinant TRPM1 was robustly expressed in insect cells, but only a small fraction was localized to the plasma membrane. Similar intracellular localization was observed when TRPM1 was heterologously expressed in mammalian cells. TRPM1 was affinity-purified from Sf9 cells and complexed with amphipol, followed by detergent removal. In blue native gels and size exclusion chromatography, TRPM1 migrated with a mobility consistent with detergent- or amphipol-bound dimers. Cross-linking experiments were also consistent with a dimeric subunit stoichiometry, and cryoelectron microscopy and single particle analysis without symmetry imposition yielded a model with approximate 2-fold symmetrical features. Finally, electron microscopy of TRPM1-antibody complexes revealed a large particle that can accommodate TRPM1 and two antibody molecules. Taken together, these data indicate that purified TRPM1 is mostly dimeric. The three-dimensional structure of TRPM1 dimers is characterized by a small putative transmembrane domain and a larger domain with a hollow cavity. Blue native gels of solubilized mouse retina indicate that TRPM1 is present in two distinct complexes: one similar in size to the recombinant protein and one much larger. Because dimers are likely not functional ion channels, these results suggest that additional partner subunits participate in forming the transduction channel required for dim light vision and the ON pathway.
Introduction
ON bipolar cells of the vertebrate retina relay information from rod and cone photoreceptors to downstream neurons, predominantly A-II amacrine cells and OFF bipolar cells, where it is subject to further signal processing before being communicated to retinal ganglion cells and their target areas in the brain (1). Postsynaptic signaling in ON bipolar cells begins with the metabotropic glutamate receptor mGluR6, which, via the heterotrimeric G protein Go, ultimately controls the open state of a non-selective cation channel (2–8). In the dark, this pathway is chronically activated by the constitutive release of glutamate from photoreceptor terminals, maintaining the postsynaptic transduction channel in a closed state. Light-induced hyperpolarization of photoreceptors leads to a decrease or cessation of glutamate release and consequent deactivation of the mGluR6 pathway (7, 9–11). Goα is rapidly inactivated through GTPase-accelerating protein complexes including RGS7/RGS11 and Gβ5 (12–15), and the transduction channel is opened, resulting in depolarization.
TRPM1,2 a member of the transient receptor potential family of cation channels, is essential for expression of the light-regulated conductance in ON bipolar cells. TRPM1 is an ∼180-kDa protein localized to the somas and dendrites of both rod and cone ON bipolar cells and colocalized with mGluR6 at the dendritic tips (16–20). TRPM1 knock-out mice have a no-b-wave (nob) electroretinogram phenotype (17, 21) and defective ON bipolar cell signaling in whole-cell recordings (17, 22, 23). In addition, a nob mouse derived from a mutagenesis screen was found to have a point mutation in the predicted pore region of TRPM1 (24). Genetic lesions in the TRPM1 gene are also associated with complete congenital stationary night blindness in humans (25–28). Complete congenital stationary night blindness patients with mutations in TRPM1 have electroretinography results consistent with a defective ON pathway in both rods and cones (25, 26). The OFF pathway is spared, which is why mutation of TRPM1 (or other complete congenital stationary night blindness genes such as GRM6 or nyctalopin (NYX)) does not result in a more severe blinding phenotype.
Virtually nothing is known about the structure of any of the TRPM family proteins apart from the isolated coiled coil domain of TRPM7, which forms an antiparallel four-stranded coiled coil (29). Single particle reconstructions of the distantly related TRPV1 (30, 31), TRPV2 (32), TRPV4 (33), and TRPA1 (34) indicated 4-fold symmetric complexes with a small transmembrane domain and a larger basket-shaped cytoplasmic domain. Homology between TRPM1 and the TRPV family is strictly limited to the transmembrane domain (∼40% similarity), which accounts for only ∼20% of the TRPM1 sequence.
To study the biochemical and structural properties of TRPM1, we developed an expression and purification procedure capable of generating large amounts of folded and soluble protein. The purified protein was found to be mostly dimeric, and cryoelectron microscopy was used to determine the shape of TRPM1 dimers. In addition, highly specific TRPM1 monoclonal antibodies (mAbs) were generated and used for detection of native TRPM1-containing complexes in retina.
EXPERIMENTAL PROCEDURES
Cells
Spodoptera frugiperda clone 9 (Sf9) cells were maintained in InsectXpress medium (Lonza) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (HyClone), 50 units/ml penicillin, and 50 μg/ml streptomycin (HyClone). Human embryonic kidney (HEK) cells were maintained in Dulbecco's modified Eagle medium (Corning) supplemented with 10% FBS and penicillin/streptomycin as above. 1D4 hybridomas (35) were cultured in serum-free Cell mAb Medium (BD Biosciences) supplemented with penicillin/streptomycin as above. TRPM1 hybridomas were cultured in Iscove's Modification of DMEM (Corning) with either 15% FBS for expansion or 10% low IgG serum (Sigma) for antibody purification and penicillin/streptomycin.
Molecular Cloning and Baculovirus Construction
Full-length Trpm1 (GenBankTM accession number NM_001039104.2) was cloned from mouse retina cDNA (36) using primers 5′-ACTCTCTTACCTCAGCTGACCAG-3′ and 5′-ATGGTCTGGCTGTTGAGTGCTTG-3′ and 15 ng of template DNA. The open reading frame with a C-terminal 1D4 tag (TETSQVAPA) (37) was subsequently cloned into the SpeI/KpnI sites of the pFb1 vector (Invitrogen) for baculovirus construction and KpnI/NotI of pCDNA3.1 (Invitrogen) for expression in mammalian cells. Nyx (GenBank accession number NM_173415.4) was cloned from mouse retina cDNA using primers 5′-ACAGCTGAGTGGCTCTAAGCCACC-3′ and 5′-GTGGTTTGGGTCACCTCAGTGTGG-3′ and 100 ng of template DNA. A product with GST placed behind the Nyx signal sequence (Nyx(1–19)-GST-Nyx(20–476)) was constructed by overlap extension PCR and cloned into the SpeI/KpnI sites of pFb1. 5% DMSO was required for PCR amplification of Nyx. Human Gαo (splice variant A) cDNA was purchased from the Missouri University of Science and Technology cDNA resource center and cloned into XbaI/EcoRI sites of pFb1.
Recombinant bacmids were made using the Bac-to-Bac procedure (Invitrogen). Bacmid DNA was prepared after a second round of colony purification and screened by PCR with M13 primers. Sf9 cells in 25-cm2 flasks were transfected with 15 μl of bacmid DNA and 12 μl of Cellfectin or Cellfectin II (Invitrogen) in Grace's unsupplemented medium according to the manufacturer's instructions. The P1 virus stock was harvested after 4 days. Viruses were passaged blind: P2 stocks were made by infecting Sf9 cells in a 75-cm2 flask with 1 ml of P1 virus; P3 stocks were made by infecting 150-cm2 flasks with 0.5 ml of P2. All infections were initiated with a 1-h adsorption in minimal volume at room temperature followed by addition of medium and incubation at 27 °C for 3–4 days. P3 virus was typically used for protein expression.
Gel Electrophoresis and Western Blotting
SDS-PAGE was performed according to the method of Laemmli (38). TRPM1 samples were incubated in SDS loading buffer for 5–15 min at 42 °C and typically resolved with 8% acrylamide gels. Proteins were transferred to nitrocellulose in TG (25 mm Tris, 192 mm glycine), and membranes were blotted with primary antibody as follows: 1D4, 0.1 μg/ml; TRPM1 274G7, 1 μg/ml; TRPM1 545H5, 1 μg/ml; TRPM1 biotin-545H5, 1:500; actin (Developmental Studies Hybridoma Bank, University of Iowa, JLA20), 1:1,000; Gαo (Millipore MAB3073), 0.3 μg/ml. Anti-mouse IRDye680 (LI-COR Biosciences) at 1:10,000, anti-mouse horseradish peroxidase (HRP) (Jackson ImmunoResearch Laboratories) at 1:2,000, or streptavidin-HRP (Cell Signaling Technology) at 1:3,000 was used for detection with the Odyssey infrared imager (LI-COR Biosciences) or SuperSignal West Pico chemiluminescent substrate (Thermo) and exposure to film.
Cross-linking reactions were resolved using hybrid SDS gels with 3–3.5% acrylamide and 0.5% agarose in a Laemmli buffer system as described (39). Proteins were transferred in TG to polyvinylidene fluoride (PVDF) membrane and blotted as above. Markers were Coomassie-stained on the blot after transfer. Unstained HiMark (Invitrogen) was used for the 500-, 290-, and 240-kDa marker bands, and nebulin from mouse muscle extract was used as a 730-kDa marker (40). Soleus/gastrocnemius muscle was dissected from hind limbs of CD1 mice, minced in PBS with 2× Complete protease inhibitor mixture (PIC) (Roche Applied Science) and 2% SDS, homogenized, and centrifuged at 1,000 × g for 15 min. ∼20 μg of solubilized muscle extract was used for the marker lane, and the identity of the nebulin band was confirmed by Western blotting with nebulin antibody (Santa Cruz Biotechnology, sc-28286; 1 μg/ml).
Blue native (BN-) PAGE (41) was performed with Bis-Tris, 3–12% acrylamide gels (Invitrogen) according to the manufacturer's instructions with cathode buffer containing 0.02% Coomassie G-250. Before loading, samples were mixed with 2.5 μl of 4× NativePAGE loading dye (Invitrogen) and 1 μl of 5% Coomassie G-250 in a total volume of 10 μl. NativeMark (Invitrogen) soluble molecular weight markers were used without additive according to the manufacturer's instructions. Electrophoresis was performed with the tank submersed in ice water. For Western blotting, the gel was first soaked for 30 min in TG with 0.1% SDS and then transferred in TG to PVDF membrane at ∼65 mA overnight. The membrane was fixed for 15 min in 8% acetic acid, rinsed in water, allowed to air dry, then rewet, and destained in methanol. The marker lane was cut off and stained with Coomassie; the rest of the membrane was subjected to Western blotting as described above. For estimation of nominal molecular weight, a linear fit to log(molecular mass) as a function of relative mobility was constructed using only the apoferritin, phycoerythrin, and lactate dehydrogenase bands (720, 480, 242, and 146 kDa), which comprised the most linear portion of the curve (see Fig. 4C). Estimation using a polynomial fit to all seven bands in the standard yielded similar results (not shown). It should be noted that mobilities of membrane proteins measured by this procedure have been found to deviate systematically from those expected from calibration with soluble proteins (42, 43).
FIGURE 4.
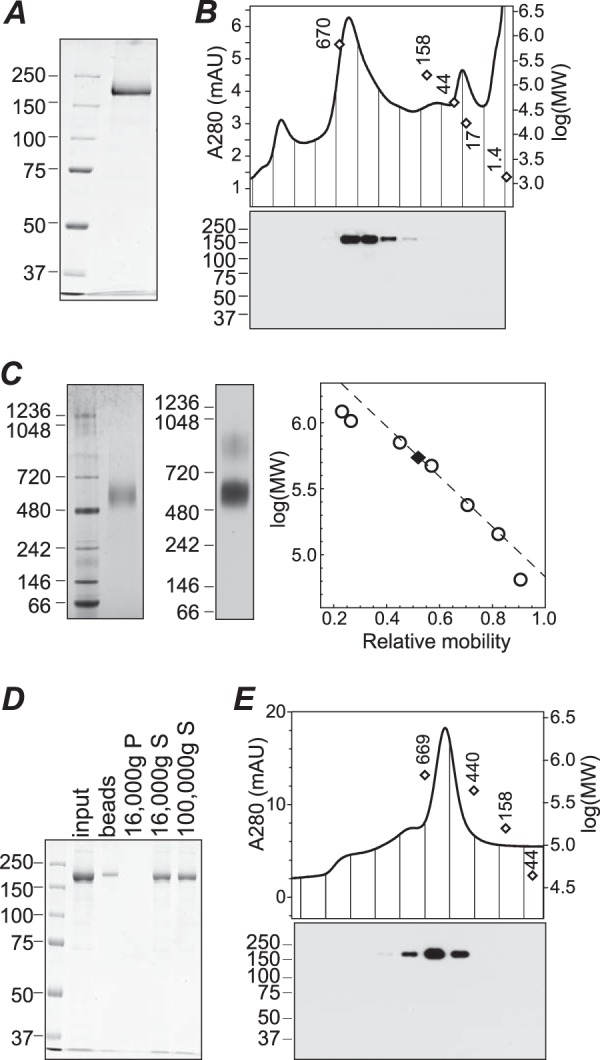
Purification of TRPM1 from Sf9 cell membranes. A, SDS-PAGE and Coomassie staining of purified protein. B, size exclusion chromatography of purified protein in buffer containing fos-choline-14. Fractions were subjected to SDS-PAGE and Western blotting (bottom). C, BN-PAGE of purified protein in fos-choline-14 followed by Coomassie staining (left) or 1D4 Western blotting (middle). The calibration curve used for determining the apparent molecular mass (right, dashed line) was constructed from the 720-, 480-, 242-, and 146-kDa markers. D, Coomassie-stained SDS-PAGE of input protein, protein stuck to BioBeads after amphipol exchange and detergent removal, and supernatant (S) and pellet (P) fractions after centrifugation at 16,000 × g. Approximately 50% of the input protein was recovered in the supernatant; further centrifugation at 100,000 × g did not result in additional loss of protein. E, size exclusion chromatography of TRPM1-amphipol complexes in detergent-free buffer. Fractions were subjected to 1D4 Western blotting (bottom) to confirm the identity of the peak. Peak positions of molecular weight markers in B and E are shown as open diamonds. mAU, milli-absorbance units.
1D4 Antibody Production and Conjugation
1D4 antibody (1D4Ab) (35) was purified from hybridoma culture supernatant supplemented with 50 mm Tris-Cl, pH 8 using protein G-Sepharose 4 Fast Flow (GE Healthcare) and/or HiTrap protein G HP (GE Healthcare). The column was washed in 20 mm sodium phosphate, pH 7 and eluted with 0.1 m glycine, pH 3. 1D4Ab-conjugated Sepharose was prepared with 4 g of CNBr-activated Sepharose 4B (GE Healthcare) and 50 mg of Ab according to the manufacturer's instructions. 1D4Ab-Sepharose was stored at 4 °C as a 30% slurry in 0.1 m NaHCO3, 0.5 m NaCl, 0.02% sodium azide, pH 8.5 for up to ∼1 year.
Protein Expression and Purification
Sf9 cells were infected in 150-cm2 flasks with 0.5 ml of P3 virus. Adsorption was carried out in a 2.5-ml total volume at room temperature for 1 h with frequent agitation. Alternatively, batches of 5 × 108 cells were infected in suspension with 5 ml of P3 virus. Adsorption was carried out at 2.5 × 107 cells/ml at room temperature for 1 h with frequent agitation, and then cells were diluted to 1 × 106 cells/ml in spinner bottles. Cells were grown at 27 °C in InsectXpress medium + FBS as described above. At ∼48 h postinfection (hpi), cells were pelleted and washed in cracking buffer (CB; 25 mm Tris, 300 mm sucrose, 15 mm EDTA, 2 mm MgCl2, pH 8.1). Cell pellets were frozen in liquid nitrogen and stored at −80 °C. Cells were lysed in CB with 1× Complete PIC using 25 passes through a microfluidizer (Microfluidics) at ∼100 p.s.i. Cell debris and nuclei were removed by centrifugation at 4,000 × g for 5 min and 10,000 × g for 25 min, and membranes were pelleted by centrifugation at 100,000 × g for 2.5 h. Membrane pellets were homogenized in CB + PIC with a Potter-Elvehjem Teflon homogenizer, frozen in liquid nitrogen, and stored at −80 °C. Membranes were solubilized in 25 mm Tris, 150 mm sucrose, 200 mm NaCl, 7.5 or 15 mm EDTA, 1 mm MgCl2, 1.5× PIC, 2 mm DTT, and 2.3 mg/ml fos-choline-14 (Anatrace), pH 8.1 with incubation for 90 min on a Nutator. Following centrifugation at 100,000 × g for 1 h, the supernatant was incubated with 1D4Ab-Sepharose for 4 h on a Nutator, and the resin was washed in a column with ∼30–50 bed volumes of wash buffer (25 mm Tris-Cl, 200 mm NaCl, 1× PIC, 2 mm DTT, 0.7 mg/ml fos-choline-14, pH 8.1). To elute TRPM1, the resin was suspended in wash buffer containing 5 mg/ml 1D4 peptide (GenScript) with pH adjusted to ∼8 by addition of 0.01 n NaOH and incubated for 1 h. Purified protein was quantified by densitometry of Coomassie-stained gels including a BSA standard curve.
Purified protein in elution buffer was mixed with amphipol A835 (Anatrace) at a 1:1 (w/w) ratio and incubated with end-over-end mixing for 3 h. BioBeads SM-2 (Bio-Rad) were prepared by stirring in methanol for 15 min, washing extensively with methanol followed by water, and degassing. Detergent was removed from the protein-amphipol complexes by addition of ∼0.2 g/ml BioBeads, mixing for 90 min, addition of another ∼0.2 g/ml BioBeads, and mixing for an additional 60–90 min. The sample was separated from the beads and centrifuged to remove aggregates. All steps were performed at 4 °C.
Detergent Screens
Sf9 cell membranes were prepared as described above for protein purification. HEK cells were transfected with polyethylenimine Max (PEI) (molecular weight, 40,000; Polysciences). 12 μg of pCDNA3.1-TRPM1–1D4 were mixed with 0.3 ml of 1 mg/ml PEI, pH 7 in 1.5 ml of serum-free DMEM supplemented with 20 mm HEPES, pH 7.4; vortexed; incubated for 10 min; and then added to 10-cm dishes containing cells ∼50–60% confluent in 15 ml of serum-free medium. The medium was replaced with serum-containing medium after 2–3 h. At ∼50 h post-transfection, cells were collected with a cell scraper and washed in CB. Cells from five dishes were suspended in CB + PIC, passed 30 times through a 23-gauge needle, and centrifuged at 4,000 × g for 5 min. Membranes were pelleted as for Sf9 cells, homogenized in 2 ml of CB + PIC, and frozen in liquid nitrogen.
Membranes were solubilized as for protein purification except using the following detergents: decyl β-maltoside, 4.3%, 50× CMC; dodecyl β-maltoside, 0.44%, 50× CMC; CYMAL-6, 1.4%, 50× CMC; Triton X-100, 0.75%, 50× CMC; octyl β-glucoside, 5.3%, 10× CMC; fos-choline-12, 2.4%, 50× CMC; fos-choline-14, 0.23%, 50× CMC; and CHAPS, 4.9%, 10× CMC. All detergents were from Anatrace except Triton X-100 (OmniPur) and CHAPS (Mallinckrodt Baker). Following centrifugation at 100,000 × g for 1 h, pellets were homogenized in SDS sample buffer, and pellet and supernatant fractions were subjected to SDS-PAGE and 1D4 Western blotting. The detergent screen of retina TRPM1 was as for Sf9 cells except that whole clarified lysate prepared as described below was used, and solubilization was performed in 25 mm Tris, 200 mm NaCl, 15 mm EDTA, 2 mm DTT, 2× PIC.
Size Exclusion Chromatography
Purified protein was subjected to gel filtration with Bio-Sil 400 (Bio-Rad) or Superose 6 PC 3.2/30 (GE Healthcare) columns using an HPLC instrument (Shimadzu). The column was equilibrated in 25 mm Tris, 200 mm NaCl, 15 mm EDTA, 2 mm DTT, 0.3 mg/ml fos-choline-14, pH 8.2 for protein-detergent complexes or in 25 mm Tris, 200 mm NaCl, 2 mm DTT, pH 8.1–8.2 for protein-amphipol complexes. Molecular mass markers were as follows: thyroglobulin, 670 kDa; γ-globulin, 158 kDa; ovalbumin, 44 kDa; myoglobin, 17 kDa; vitamin B12, 1.4 kDa (all from Bio-Rad); thyroglobulin, 669 kDa; ferritin, 440 kDa (both from GE Healthcare); and bovine IgM, 970 kDa (Sigma). For estimation of nominal molecular weight, a linear fit to log(molecular mass) as a function of retention time was constructed using only the thyroglobulin and ferritin peaks.
Cross-linking
Prior to cross-linking, protein was dialyzed extensively against 25 mm HEPES-NaOH, 200 mm NaCl, 2 mm DTT, pH 8.1 with a dash of solid PMSF. 10× stocks of disuccinimidyl tartrate (DST) (Thermo) were prepared in anhydrous DMSO immediately before use. Cross-linking reactions contained 0, 0.25, or 2.5 mm DST as indicated and 10% DMSO, and protein concentrations were ∼50–200 μg/ml unless indicated otherwise. Reactions were incubated for 30 min at room temperature; then stopped by addition of 0.1 m Tris-Cl, pH 8.2; and further incubated for 15 min at room temperature.
Immunofluorescence Microscopy
Sf9 cells plated on glass coverslips in 12-well plates were infected with 60 μl of P3 virus. At ∼42–46 hpi, cells were fixed in 2% paraformaldehyde in PBS for 10 min, washed in PBS, and permeabilized in PBSAT (PBS + 1% BSA + 0.1% Triton X-100) for 15–30 min. Cells were labeled with 1 μg/ml 1D4Ab in PBSAT for 45 min followed by donkey anti-mouse Alexa Fluor 488 (Invitrogen) at 1:1,000 in PBSAT along with DAPI for 30 min. Coverslips were mounted with Prolong Gold (Invitrogen). For labeling membranes, live cells were washed in HBSS, incubated in HBSS containing 10 μg/ml rhodamine-labeled wheat germ agglutinin (WGA; Vector Laboratories) for 15 min, washed in HBSS, then fixed, and labeled with 1D4Ab as described above. All steps were carried out at room temperature. HEK293 cells grown in DMEM were plated on poly-d-lysine-coated coverslips and transiently transfected with TRPM1–1D4 using Lipofectamine 2000 (Invitrogen). At ∼36 h post-transfection, coverslips were immersed in warm HBSS containing 5 μg/ml rhodamine-labeled WGA, incubated at 37 °C for 10 min, and then washed in warm HBSS. Post-WGA labeling, cells were fixed in 2% paraformaldehyde in PBS for 10 min, washed in PBS, and permeabilized in PBSAT for 15 min. Cells were labeled as described for Sf9 cells, and coverslips were mounted with Prolong Gold. Fixation and all subsequent steps were performed at room temperature. Cells were imaged with a TCS-SP5 confocal microscope (Leica). For Fig. 3B, transfected cells were trypsinized and replated on poly-d-lysine-coated coverslips at ∼20 h post-transfection, labeled as for Sf9 cells at ∼42 h, and imaged with a DeltaVision deconvolution microscope.
FIGURE 3.
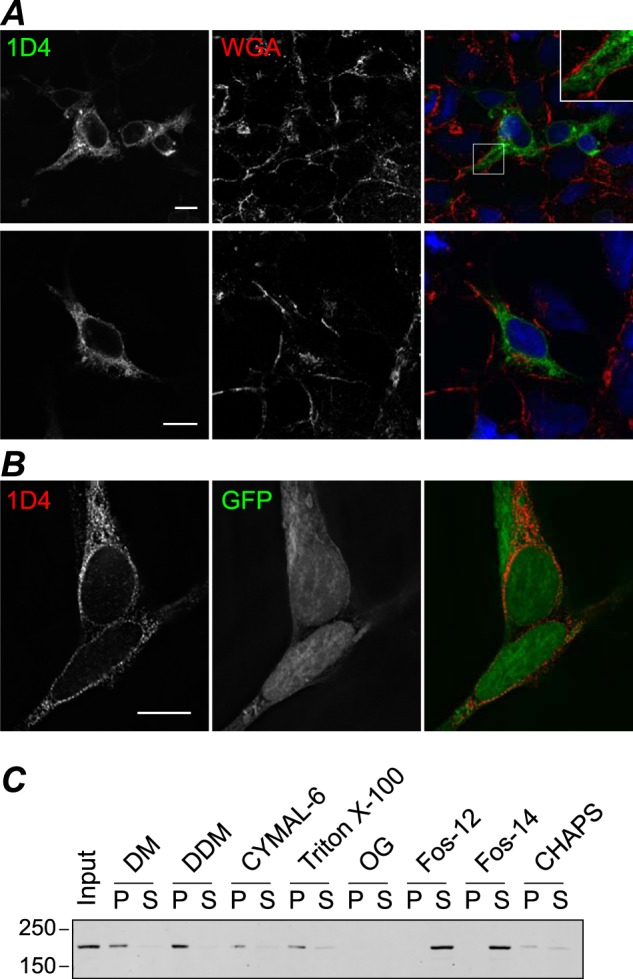
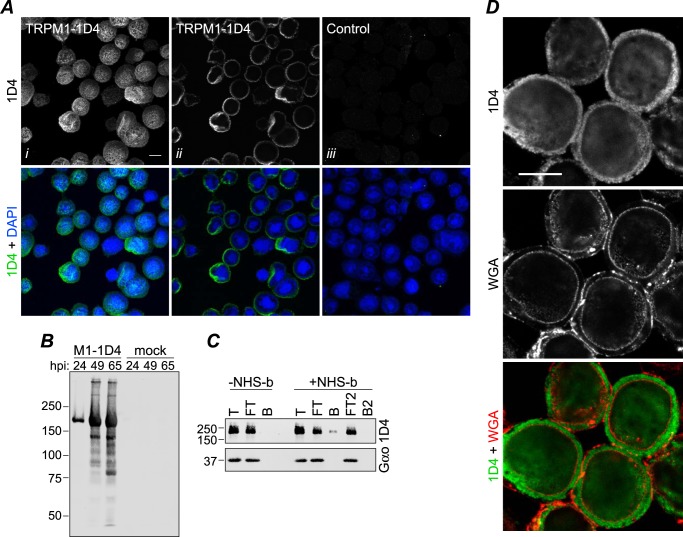
Expression of TRPM1 in mammalian cells. HEK cells transiently transfected with TRPM1–1D4 (A) or co-transfected with TRPM1–1D4 and GFP (B) were fixed and permeabilized at 36–42 h post-transfection and labeled with 1D4Ab (A and B) and WGA (A) as a membrane marker. Single optical sections (A) or single slices through the reconstructed volume after deconvolution (B) are shown. Scale bar, 10 μm. C, detergent screen with membranes from transfected HEK cells as described in Fig. 2. Fos-12, fos-choline-12; Fos-14, fos-choline-14; DM, decyl β-maltoside; DDM, dodecyl β-maltoside; OG, octyl β-glucoside; P, pellet; S, supernatant.
Surface Biotinylation
Sf9 cells plated in 6-well plates were infected with 100 μl of P3 virus. At ∼45–46 hpi, cells were dislodged and washed three times in PBS, pH 8.1. Cells were labeled in suspension with 2 mm NHS-PEG4-biotin (Thermo) in PBS, pH 8.1 for 30 min at room temperature with end-over-end mixing. The reaction was quenched by adding 6 volumes of blocking buffer (PBS with 0.1 m glycine, pH 8) and washing twice more in blocking buffer. Cells were lysed in cold lysis buffer (25 mm Tris, 200 mm NaCl, 15 mm EDTA, 2 mm DTT, 2× PIC, 0.23% fos-choline-14) for 30 min on ice and centrifuged at 16,000 × g for 10 min to remove insoluble material. The supernatant was incubated with streptavidin-agarose beads (Thermo) at 4 °C for 2.5 h to precipitate biotin-labeled proteins, and beads were washed five times in lysis buffer. Samples were denatured by sequential 10-min treatments at 42 and 100 °C in Laemmli sample buffer containing 2% SDS, 8 m urea, 3.6 mm biotin and subjected to SDS-PAGE.
Electron Microscopy and Image Processing
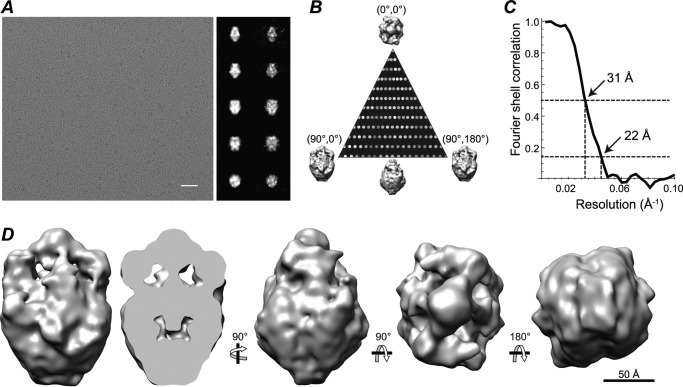
For cryoelectron microscopy, purified TRPM1 protein solution (2 μl) was applied onto Quantifoil holey grids with a thin continuous carbon film coating. The grids were blotted, rapidly frozen in liquid ethane using a Vitrobot IV (FEI Co.), and stored in liquid nitrogen for imaging. Frozen-hydrated TRPM1 specimens were imaged using a JOEL JEM2010F electron microscope at 200 kV equipped with a Gatan liquid nitrogen cryoholder and 4,000 × 4,000 charge-coupled device camera. Images were acquired using a dose of 15–18e/Å2 and magnification of 60,000× (1.81 Å/pixel). ∼8,500 ice-embedded TRPM1 particles were picked and corrected for the contrast transfer function using the Ctfit routine in EMAN (44). An initial three-dimensional model was generated based on the class averages sorted from aligned images using a reference-free procedure in EMAN. To avoid model bias, the structure was re-refined from a cylinder starting model with C1 symmetry. The refined models were essentially the same. The final C2 three-dimensional map was generated and refined from a set of ∼7,900 particles using standard iterative projection matching, class averaging, and Fourier reconstruction in EMAN. Models generated independently from even and odd halves of the data were used to generate a “gold standard” Fourier shell correlation curve, which indicated a resolution of 22 Å at the 0.143 threshold and 31 Å at the 0.5 threshold.
For negative stain imaging of TRPM1, specimens were applied to carbon-coated copper grids and stained with 2% uranyl acetate. Data were collected with a JEM2100 microscope at 200 kV at a dose of 15–18e/Å2 and a magnification of 60,000×. A low resolution map was reconstructed from 3,000 particles as described above for ice images and low-pass filtered to 30 Å.
For imaging of TRPM1-1D4Ab complexes, 1D4-amphipol was dialyzed extensively to remove the elution peptide and incubated with 1D4Ab in a 1:1 molar ratio of IgG to TRPM1 dimers for 30 min on ice. Grids with continuous carbon film coating were applied to a sample drop for ∼1 min, washed with three drops of water, and stained using Nanovan (Nanoprobes) for 2 × ∼30 s. ∼3,500 particles were picked from 66 frames, and three-dimensional maps were constructed by iterative refinement using the model of TRPM1 alone from negatively stained samples as the initial model. The final maps were low-pass filtered to 30 Å.
Animals
TRPM1 knock-out mice (Trpm1tm1Lex; Lexicon Pharmaceuticals) were obtained from the European Mutant Mouse Archive and back-crossed to C57BL/6 for 15 generations. Age-matched wild-type C57BL/6 mice (Baylor College of Medicine Center for Comparative Medicine) were used in experiments with the knock-outs. Detergent screens were performed with wild-type CD1 mice (Charles River). All procedures were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee.
Monoclonal Antibody Generation
Hybridoma clones were generated by the Baylor College of Medicine Baculovirus/Monoclonal Antibody core facility. Briefly, mice were immunized with full-length purified TRPM1 complexed with amphipol. Hybridoma culture supernatants were screened by ELISA with purified TRPM1, Western blotting of retina lysate, and immunohistochemistry of retina sections. Selected hybridomas were cloned by limiting dilution and rescreened as above. Two clones (545H5 and 274G7) were used in this study with indistinguishable results. TRPM1 mAbs were purified from culture supernatant as described above for 1D4. Purified antibody (1 mg/ml) was biotinylated by dialyzing in PBS, incubating with 20-fold molar excess NHS-PEG4-biotin at room temperature for 30 min, and then dialyzing again in PBS to remove free biotin.
Retina Lysate
Retinas were collected into cold PBS, washed several times in PBS, and homogenized in 25 mm Tris, 200 mm NaCl, 15 mm EDTA, 2 mm DTT, 2× PIC. Cells were lysed by 30–100 passes through a 23-gauge needle and brief sonication in an ice/ethanol bath, and debris and nuclei were removed by centrifugation at 4,700 × g for 10 min. For BN-PAGE, the clarified lysate was solubilized with 2.3 mg/ml fos-choline-14 for 30–60 min and centrifuged at 100,000 × g for 30 min. Total protein was determined by BCA assay (Thermo).
Sequence Analysis and Homology Modeling
TMHMM 2.0 (45) was used for predictions of transmembrane (TM) helices. Pairwise alignment of profile hidden Markov models and homology modeling were performed using HHpred (46) and Modeller (47). The rat TRPV1 atomic resolution structure (Protein Data Bank code 3J5P (30)) was used as the template.
RESULTS
Expression and Purification of TRPM1
When murine TRPM1 with a C-terminal 1D4 tag was overexpressed in Sf9 cells using recombinant baculovirus, Western blots of infected cells revealed robust expression of a 1D4-tagged protein consistent with the expected molecular mass of 184 kDa (Fig. 1B). By immunofluorescence confocal microscopy, TRPM1 appears to be uniformly localized in infected cells with no obvious aggregates (Fig. 1A). Analysis of subcellular localization is problematic in Sf9 cells because most of the cell volume is occupied by the nucleus. However, co-staining with WGA, a lectin marker for membranes, indicates that TRPM1 is predominantly intracellular with little or no co-localization with the plasma membrane (Fig. 1D). To determine whether any TRPM1 could be detected at the plasma membrane, surface-localized proteins were biotinylated in live cells followed by cell lysis and streptavidin-agarose precipitation (Fig. 1C). Only a small fraction of TRPM1 was present in the bead fraction: quantification of band intensities yielded an estimate of ∼5% for the amount of TRPM1 that is accessible to surface labeling. In contrast, baculovirus-expressed Gαo, which is expected to be entirely intracellular, was not detected in the bead fraction. Immunofluorescence microscopy of transfected HEK cells likewise indicates that the vast majority of expressed TRPM1 is intracellular (see Fig. 3, A and B).
FIGURE 1.
Expression of TRPM1 in Sf9 cells. A, Sf9 cells were infected with baculovirus expressing TRPM1–1D4 (panels i and ii) or a control virus (GST-nyctalopin; panel iii). At 46 hpi, cells were fixed, permeabilized, and labeled with 1D4Ab (green) and DAPI (blue). Cells were imaged by confocal fluorescence microscopy; projections (panel i) or single optical slices near the middle of the cells (panels ii and iii) are shown. Scale bar, 10 μm. B, 1D4 Western blot of cells infected with TRPM1–1D4-expressing baculovirus or mock-infected and harvested at the indicated time point. C, live cells infected with baculovirus expressing TRPM1–1D4 or Gαo were treated with biotinylation reagent (+NHS-b) or mock-treated (−NHS-b) at ∼46 hpi. The reaction was quenched, cells were lysed in fos-choline-14, and biotinylated proteins were precipitated with streptavidin-agarose. Total (T), flow-through (FT), and bead-bound (B) fractions are shown. The flow-through was subjected to a second round of binding with fresh beads. No TRPM1 was detected in the second bound fraction (B2), indicating that the capacity of the beads was not exceeded. Similar results were obtained from two independent experiments. D, Sf9 cells infected with baculovirus expressing TRPM1–1D4 were labeled with WGA (red) at 42 hpi, then fixed, permeabilized, and labeled with 1D4Ab (green). A single optical slice is shown. Scale bar, 10 μm.
Several detergents were screened for the ability to solubilize TRPM1 from isolated Sf9 cell membranes (Fig. 2A). None of the detergents tested without charged moieties were useful for solubilizing TRPM1; only two relatively harsh zwitterionic detergents, fos-choline-12 and fos-choline-14, yielded significant amounts of solubilized protein. Similar results were obtained with protein expressed in Saccharomyces cerevisiae yeast (not shown) and HEK cells (Fig. 3C) and with native TRPM1 in mouse retina tissue (see Fig. 9B). BN-PAGE and a 1D4 Western blot of the solubilized Sf9 membranes are shown in Fig. 2B. A distinct band with an apparent mobility of ∼500–600 kDa as well as a diffuse band at higher molecular mass was observed. The relative distribution of these bands was highly variable in different preparations of solubilized membranes. These results suggest that the solubilized membranes may contain multiple oligomeric species of TRPM1. Alternatively, the higher molecular mass band may represent aggregates, complexes with incompletely solubilized lipids, or binding with endogenous cellular component(s). TRPM1 was affinity-purified from fos-choline-14-solubilized Sf9 membranes by binding to 1D4Ab-conjugated Sepharose followed by elution with a 9-amino acid peptide corresponding to the 1D4 epitope (Fig. 4A).
FIGURE 2.
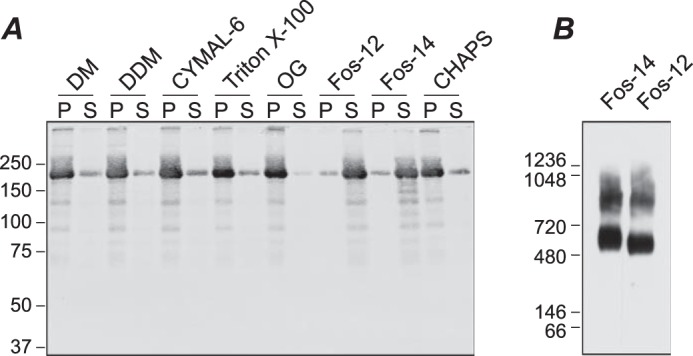
Detergent screen. Sf9 cell membranes were incubated with the indicated detergent for 90 min at 4 °C followed by centrifugation at 100,000 × g for 1 h. A, SDS-PAGE and 1D4 Western blot of pellet (P) and supernatant (S) fractions. B, BN-PAGE and 1D4 Western blot of supernatant fractions from membranes solubilized in fos-choline-12 (Fos-12) and fos-choline-14 (Fos-14). DM, decyl β-maltoside; DDM, dodecyl β-maltoside; OG, octyl β-glucoside.
FIGURE 9.
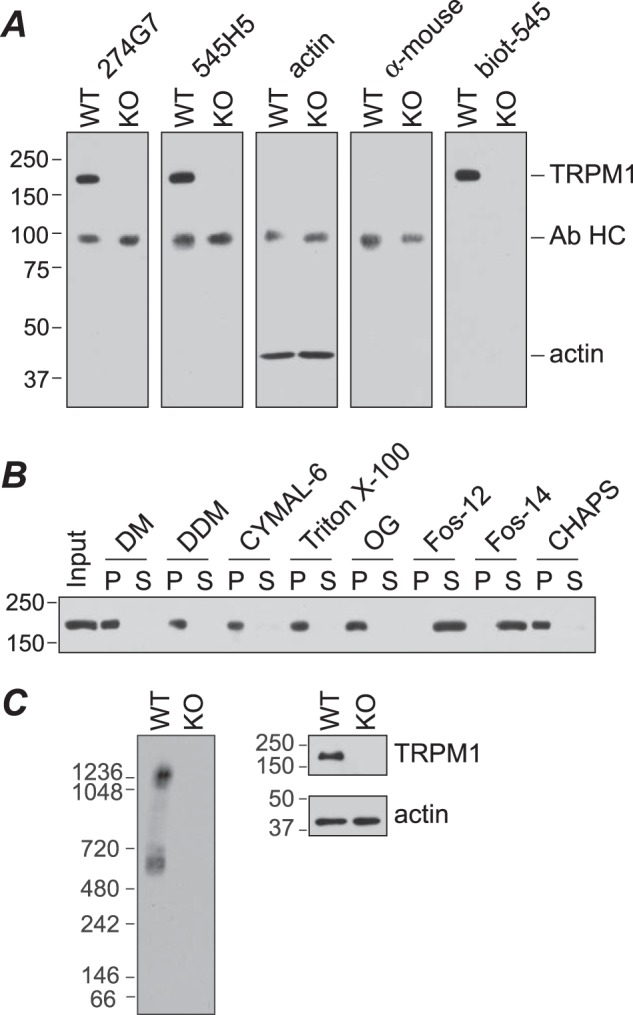
Analysis of TRPM1 in mouse retina. A, retina lysates (∼50 μg) from wild-type (WT) or Trpm1−/− (KO) mice were subjected to SDS-PAGE and Western blotting for TRPM1 with mAbs 274G7 and 545H5, or actin antibody, followed by anti-mouse HRP secondary antibody, secondary antibody only, or biotinylated 545H5 (biot-545) followed by streptavidin-HRP. Endogenous antibody heavy chain dimers (Ab HC) are detected by the secondary antibody. B, detergent screen. WT retina lysate was incubated with the indicated detergent for 90 min at 4 °C followed by centrifugation at 100,000 × g for 1 h. Pellet (P) and supernatant (S) fractions were analyzed by SDS-PAGE and Western blotting with 274G7. C, retina lysates from WT and KO mice were solubilized with fos-choline-14 (Fos-14) and analyzed by BN-PAGE and Western blot with biotinylated 545H5 (left) or SDS-PAGE and Western blot with 545H5 or actin antibody (right). Fos-12, fos-choline-12; DM, decyl β-maltoside; DDM, dodecyl β-maltoside; OG, octyl β-glucoside.
Purified TRPM1 Is a Dimer
BN-PAGE of purified protein yielded a single major band with an apparent molecular mass of ∼564 ± 20 kDa (Fig. 4C; see “Experimental Procedures”). In contrast to the BN-PAGE results with solubilized membranes, only a very small fraction of the purified protein was observed in higher molecular mass bands. This behavior was consistent among 12 preparations of purified protein and was highly stable, with indistinguishable results seen after 8 weeks of storage at 4 °C. Protein purified in fos-choline-12 also had a similar behavior in BN-PAGE (not shown). In size exclusion chromatography with buffer containing fos-choline-14, purified TRPM1 migrated in a peak with an apparent size of ∼500–600 kDa (Fig. 4B), consistent with the BN-PAGE results. Higher order oligomeric species were not evident by gel filtration.
For analysis by electron microscopy, purified protein was complexed with amphipol A835 (78) followed by detergent removal with polystyrene adsorbent beads. Approximately 50% of the input protein was recovered in the supernatant after centrifugation (Fig. 4D). In size exclusion chromatography with detergent-free buffer, the TRPM1-A835 complexes migrated as a single peak with mobility similar to that of the detergent-solubilized protein (∼530 ± 20 kDa) (Fig. 4E).
The molecular mass of the purified protein calculated from standard curves generated from soluble proteins in both BN-PAGE and gel filtration is very close to that of a TRPM1 trimer. However, both of these techniques are likely to overestimate the mass of membrane proteins due to the binding of detergent, amphipol, and/or Coomassie Brilliant Blue dye molecules (42, 43, 48). For example, using n-[14C]dodecyl β-d-maltoside, Heuberger et al. (42) determined that ∼200 detergent molecules bound to the monomeric form of the lactose transporter LacS, adding nearly 100 kDa to the mass, and when washed with the same concentration of Coomassie dye used for BN-PAGE, all of the detergent molecules were displaced by bound Coomassie dye. By performing BN-PAGE of several well characterized small membrane proteins, they determined that the molecular mass calibration of the membrane proteins was greater than that of soluble proteins by a factor of 1.8. AMPA receptor complexes were also found to migrate in BN-PAGE at an apparent molecular mass ∼1.5–2 times the calculated mass (49, 50). Similarly, binding of amphipol A835 to bacteriorhodopsin was determined to add 54 kDa to the apparent molecular mass (51). In addition to binding of detergent, dye, and amphipol molecules to membrane proteins, a hollow interior cavity or elongated shape will also affect the mobility. Therefore the migration of TRPM1 in BN-PAGE and gel filtration is more likely indicative of a dimer or possibly a monomer, rather than a trimer or tetramer, of TRPM1.
To further assess the oligomeric nature of the TRPM1 complex, purified protein samples in detergent or amphipol were cross-linked with the lysyl-directed reagent DST and resolved by SDS-polyacrylamide/agarose gel electrophoresis (PAAGE) (Fig. 5A). The major cross-linked species migrated at ∼300–400 kDa, almost certainly representing a dimer. In some experiments, appearance of the cross-linked band was accompanied by significant depletion of the monomer band (see for example Fig. 5, A and B), suggesting that the vast majority of molecules can be cross-linked. The efficiency of cross-linking was variable between experiments. The reason for the variability is unknown, but because BN-PAGE and gel filtration results were highly reproducible, it likely reflects experimental variability in the cross-linking procedure rather than variability in the oligomerization state of the protein. A very small amount of higher molecular mass cross-linked complexes was also sometimes detected. To rule out the possibility that the observed cross-linked products are the result of collisional cross-linking at high protein concentrations, reactions were performed with protein at a series of dilutions over a 100-fold range (Fig. 5B). The cross-linked dimer, accompanied by depletion of the monomer band, was evident at all concentrations.
FIGURE 5.
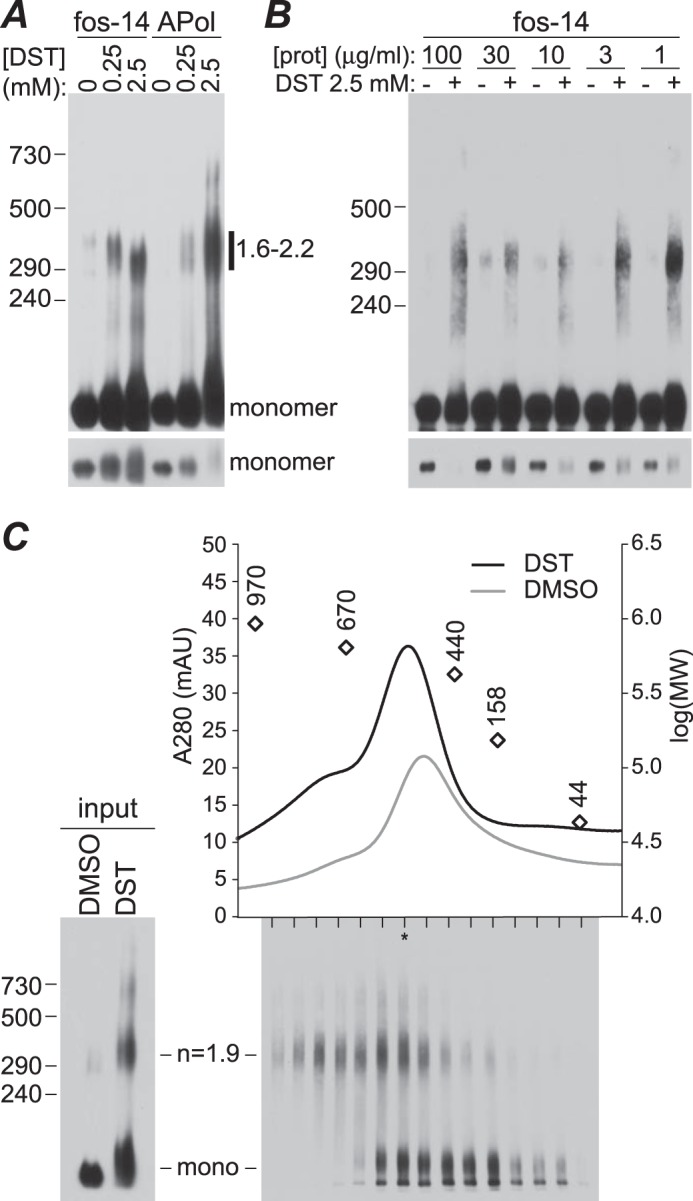
Cross-linking of purified TRPM1. A, purified protein in fos-choline-14 (fos-14) or complexed with amphipol (APol) was treated with 0.25 or 2.5 mm DST or mock-treated with DMSO and analyzed by SDS-PAAGE and 1D4 Western blotting. Calculated sizes of the cross-linked bands relative to the monomer molecular mass (184 kDa) are indicated. A shorter exposure of the same blot (bottom) is included to show the depletion of the monomer band in cross-linked samples. B, protein (prot) in fos-choline-14 was cross-linked at the indicated concentration and then diluted to equivalent concentrations for SDS-PAAGE and 1D4 Western blotting. C, size exclusion chromatography of cross-linked TRPM1. Protein-amphipol complexes cross-linked with 2.5 mm DST or mock-treated with DMSO were separated on a size exclusion chromatography column, and column fractions from the cross-linked sample were subjected to SDS-PAAGE and 1D4 Western blotting (bottom). The lane with the highest total protein, determined by quantification of total signal intensity in each lane, is indicated with an asterisk. Peak positions of molecular weight markers are shown as open diamonds. mAU, milli-absorbance units.
To confirm that the major species is not a monomer, cross-linking reactions were subjected to size exclusion chromatography, and fractions were analyzed by SDS-PAAGE and 1D4 Western blotting (Fig. 5C). Control and cross-linked samples migrated in similar peaks, with the apparent molecular mass of the peak being ∼10% larger in cross-linked samples. The peak shift may be due to mass added by cross-linkers and/or changes to the hydrodynamic radius imposed by cross-linking. Importantly, cross-linked dimers and total protein migrated in similar positions, suggesting that the majority of the protein is in at least a dimeric state. The migration of TRPM1 in BN-PAGE and size exclusion chromatography, which suggests that the protein is smaller than a trimer, together with the cross-linking data, which suggest that the subunit stoichiometry is at least 2, strongly indicates that the purified protein is mostly dimeric.
Electron cryomicroscopy images of ∼3,700 particles embedded in vitreous ice were used to generate preliminary three-dimensional reconstructions with either no symmetry (C1) or 2-fold rotational symmetry (C2) imposed (Fig. 6). Reconstruction without any assumption about symmetry can result in noise erroneously contributing to the alignments and producing symmetry-breaking artifacts (44). However, it should still yield a model in which symmetrical features, if they exist, are detectable. Imposing the correct symmetry during reconstruction should then improve the model without grossly altering its shape. Indeed, the strong similarity of the C1 and C2 models indicates that the particles do in fact exhibit C2 symmetry, providing further evidence that the protein is a dimer.
FIGURE 6.
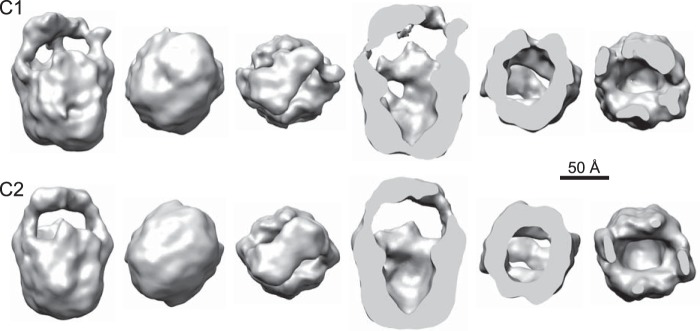
Comparison of TRPM1 models reconstructed from ∼3,700 ice images with C1 (top) or C2 (bottom) symmetry. Front, bottom, and top views and corresponding section views are shown.
Finally, three-dimensional reconstructions from images of negatively stained TRPM1 complexed with 1D4 IgG further confirm the dimeric nature of the purified protein (Fig. 7). Without symmetry imposition, extra density is evident on two sides (Fig. 7C). Reconstruction using C2 symmetry revealed a large complex that can accommodate TRPM1 and two IgG molecules (Fig. 7, C and E). The central portion of the complex resembles TRPM1 alone, indicating that binding of the 1D4Ab does not alter the oligomerization state. The assignment of Fab and Fc domains is uncertain as is which of the two IgG domains apparently connected to the TRPM1 body is bound to the 1D4 epitope. However, the bottom-most IgG domain (Fig. 7, D and E, indicated by arrows) has a significantly stronger density connecting it to TRPM1 (Fig. 7D), suggesting that this is the 1D4-binding Fab and therefore the likely location of the TRPM1 C terminus. Density is visible for the other Fab and the Fc region despite significant flexibility between the IgG domains perhaps because very weak interactions with other parts of the IgG are stabilized by 1D4 binding.
FIGURE 7.
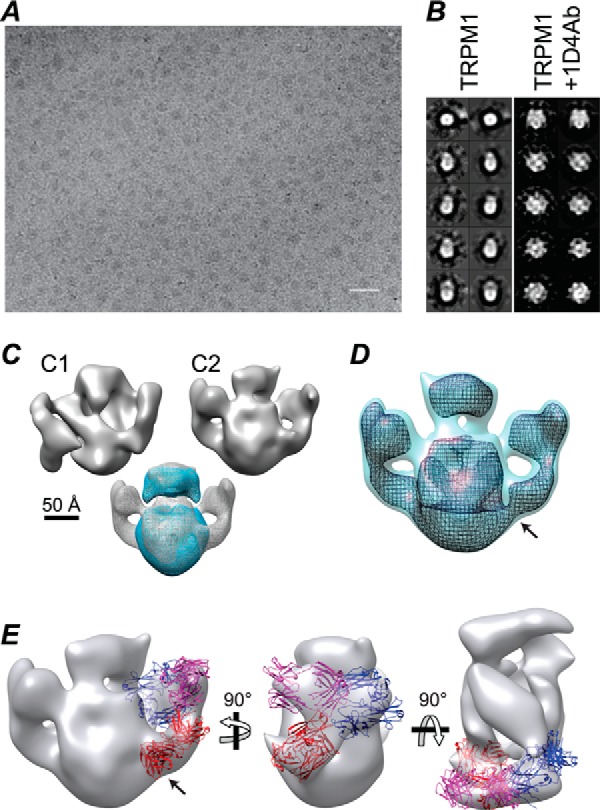
Electron microscopy of TRPM1–1D4Ab complexes. Scale bar, 500 Å. A, sample field of TRPM1–1D4Ab complexes stained with Nanovan. B, three-dimensional maps were reconstructed from ∼3,500 TRPM1–1D4Ab particles, and for comparison, a three-dimensional map of TRPM1 alone was reconstructed from ∼3,000 images of negatively stained TRPM1. Class averages (left) and corresponding projections (right) are shown. C, top, reconstructions with C1 or C2 symmetry imposed. Bottom, superposition of TRPM1 (blue) and TRPM1–1D4Ab (gray) maps. Maps were low-pass filtered to 30 Å and displayed at a threshold corresponding to ∼360 (TRPM1 dimer) and ∼700 kDa (TRPM1 dimer plus two IgG1 molecules), respectively. D, TRPM1–1D4Ab map rendered at two different thresholds. The arrow indicates the highest density connection between the IgG and the TRPM1 body. E, the IgG portion of the map was fit with two Fab (red and magenta) and one Fc (blue) domains (Protein Data Bank code 1IGY (77)). The proposed 1D4-binding Fab is indicated with an arrow.
Three-dimensional Structure of TRPM1
Biochemical experiments (Figs. 4 and 5), electron microscopy of TRPM1-1D4Ab complexes (Fig. 7), and preliminary three-dimensional reconstruction without symmetry imposed (Fig. 6) indicated that the purified TRPM1 is a dimer. Therefore, C2 symmetry was imposed for the final TRPM1 model.
An example electron micrograph of TRPM1 on carbon film and embedded in vitreous ice is shown in Fig. 8A. A three-dimensional reconstruction was obtained from ∼7,900 particles in ice images (Fig. 8D). TRPM1 contains a smaller “top” domain and a larger basket-shaped “bottom” domain with a hollow cavity. The top domain comprises ∼20% of the total volume of the model, which is in good agreement with the ∼16–25% of TRPM1 predicted to form the TM domain. The top-to-bottom thickness of the top domain (∼45 Å) is also consistent with the thickness of a lipid bilayer. These results suggest the assignment of this portion of the complex as the putative TM domain.
FIGURE 8.
Cryoelectron microscopy reconstruction of TRPM1. A, sample field of TRPM1-amphipol embedded in vitreous ice with continuous carbon film. Scale bar, 500 Å. The three-dimensional map was reconstructed from ∼7,900 particles with C2 symmetry imposed. Projections from the three-dimensional map (first column) and corresponding class averages (second column) are shown. B, distribution of particle orientations in the asymmetric unit (minimum, 12; maximum, 92). C, Fourier shell correlation curve, determined from independent maps made using even and odd halves of the data, indicates a resolution of ∼22 Å (at a Fourier shell correlation of 0.143). D, different views of the final three-dimensional reconstruction of TRPM1 presented at a threshold corresponding to a molecular mass of 360 kDa.
Analysis of Endogenous TRPM1 in Retina
To detect native TRPM1, mAbs were generated against full-length purified TRPM1-amphipol complexes. Western blotting of mouse retina lysate with two mAb clones (274G7 and 545H5) detected a band consistent with the ∼180-kDa molecular mass of TRPM1 (Fig. 9A). To avoid detection of endogenous IgG by the anti-mouse secondary antibody, 545H5 was biotinylated and detected with streptavidin-HRP. Comparison of samples from wild-type and Trpm1−/− mice demonstrated that these antibodies are highly specific.
Screening detergents for effective solubilization of native TRPM1 (Fig. 9B) yielded virtually identical results to those obtained with recombinant TRPM1 in Sf9 cell membranes (Fig. 2). Native TRPM1-containing complexes were analyzed by BN-PAGE of retina lysate solubilized with fos-choline-14 (Fig. 9C). Two bands were detected: one with an apparent molecular mass ∼20% larger (∼667 ± 38 kDa) than that of purified recombinant TRPM1 and one significantly larger (>1 MDa). Thus a substantial portion of TRPM1 in solubilized retina is in a complex that is too small to be a homotetramer. Given its similarity in size to the purified protein, this complex is likely to be a homodimer. The larger complex likely contains other subunits or components of the signal transduction pathway.
TRPM1 TM Domain Homology Model
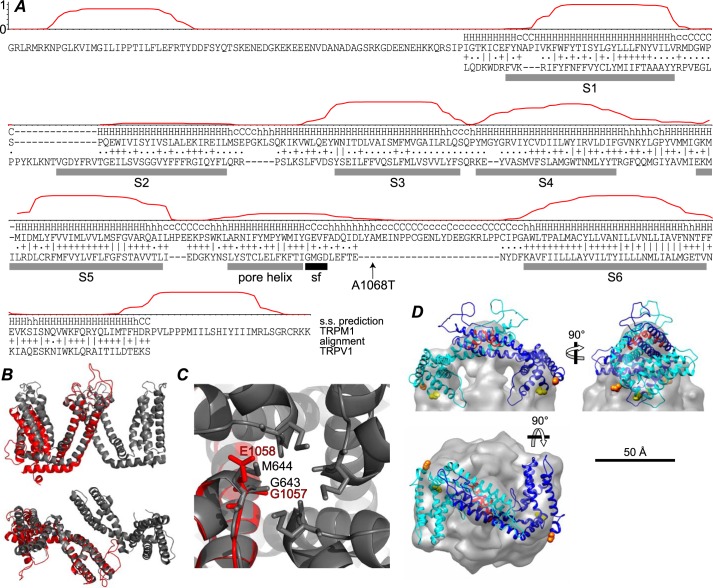
Although it has been assumed by analogy with other transient receptor potential channels that TRPM1 has both termini in the cytoplasm and six TM passes, its topology is not known with certainty. TM helix predictions (45) are shown for residues 784–1176 of TRPM1 in Fig. 10A, along with the pairwise hidden Markov model (46) alignment with TRPV1. Detectable homology is limited to the TRPV1 TM domain and residues 861–1149 of TRPM1. With the exception of the S2 helix, the TRPV1 TM helices (30) are in good agreement with the predicted TRPM1 TM helices. TRPM3, which has ∼75% identity to TRPM1 in this region, yields a reasonable TM prediction (∼0.5 probability) for S2 (not shown). Using the TRPV1 structure (30) as a template, a homology model of the TRPM1 TM domain was constructed using Modeller (47). The homology model and two C2-related subunits of TRPV1 are shown in Fig. 10B. A magnified view of the pore region of the TRPV1 tetramer shows the positions of the TRPV1 selectivity filter residues and the analogous residues in TRPM1 (Fig. 10C), which may be candidate selectivity filter residues. Two subunits of the TRPM1 TM model fit reasonably well in the putative transmembrane domain of the EM map (Fig. 10D), although fitting required a significant change from the orientation of the subunits in TRPV1.
FIGURE 10.
Alignment with TRPV1 and homology model. A, the posterior probability of location in a TM helix, calculated using TMHMM (45), is plotted for residues 784–1176 of TRPM1. The HHpred (46) secondary structure (s.s.) prediction and alignment with TRPV1 is shown below. The ∣, +, and . represent very good, good, and poor matches, respectively. The rat TRPV1 sequence from the structure lacking pore loop residues 604–626 (Protein Data Bank code 3J5P (30)) was used. TRPV1 TM helices S1–S6 and pore helix (gray bars) and selectivity filter (sf) (black bar) are indicated. The location of the TRPM1 A1068T nob mutation (24) is also indicated. B, side view (top) and extracellular view (bottom) of two C2-related (diagonally opposite) TRPV1 TM domains (gray) superposed with the TRPM1 homology model (red) aligned at the S5 helix. C, extracellular view of the TRPV1 tetramer pore region (gray) and TRPM1 homology model (red). TRPV1 selectivity filter residues Gly-643 and Met-644 are indicated as sticks along with candidate TRPM1 selectivity filter residues Gly-1057 and Glu-1058. D, two subunits of the TRPM1 TM domain homology model were fit into the EM map. The N and C termini of the model are shown as orange and yellow spheres, respectively, and the pore helix is red.
DISCUSSION
In this study, TRPM1 purified from Sf9 cells was found to be a nearly homogeneous and well-behaved population of dimers. Single particle reconstruction revealed that the overall shape of the TRPM1 dimer is somewhat similar to that of TRPV1 (30, 31), TRPV4 (33), and TRPA1 (34) tetramers. The recent high resolution cryo-EM structure of TRPV1 (30) revealed a tetrameric TM domain with remarkable structural similarity to voltage-gated potassium channels. Despite their low overall sequence similarity and apparent difference in architecture, TRPM1 and TRPV1 do share detectable homology of their TM domains (22% identity in the TM domain; the rest of the sequence contains no significant similarity). Prediction of TRPM1 TM helices is also in good agreement with the location of the S1–S6 helices in TRPV1 (see Fig. 10). Furthermore, the TRPM1-dependent bipolar cell transduction channel shares pharmacological features with TRPV1 including activation by capsaicin and anandamide and inhibition by capsazepine and SB366791 (21). These similarities suggest that the functional TRPM1 pore probably requires contributions from four subunits.
It is possible that TRPM1 forms higher order oligomers that were disrupted by our purification procedure. If this is the case, the fact that dimers were the major species after purification suggests that there is a stable dimer interface that is distinct from interactions forming higher order oligomers, and it is therefore unlikely that TRPM1 forms a tetramer with 4-fold symmetry such as that reported for TRPV1, TRPV4, and TRPA1. In cross-linking experiments, a very faint higher order cross-linked band was often detected (see Fig. 5); the molecular mass of this band was intermediate between that of a trimer and a tetramer. Larger complexes may have been disrupted by the rather harsh detergent conditions required for solubilization. In addition, the asymmetry in reduction potential in the native membrane environment is by necessity lost during solubilization. In this study, all purification steps from solubilization onward were carried out in the presence of reducing agent, which could disrupt extracellular disulfide bonds required for some subunit interactions. Purification in the absence of reducing agent resulted in profound reductions in the efficiency of solubilization and overall protein yield even in the presence of iodoacetamide, suggesting the formation of aggregates (not shown). The TRPM1 isoform used here contains 25 cysteines at least 20 of which are predicted to be intracellular and therefore reduced. The inability to maintain both reduction of intracellular cysteines and oxidation of extracellular cysteines, both of which may be important for structure and function, is a significant caveat of membrane protein purification in general. It is also possible that the 1D4 epitope tag affects the structure of the complex. However, the presence of a similarly sized complex in retina (see Fig. 9) suggests that this is not the case.
Another possibility is that in its native environment TRPM1 is part of a heteromeric channel complex along with one or more other proteins. This is thought to be the case for TRPP2, which forms complexes containing three TRPP2 molecules and one PKD1 molecule (52). PKD1 contains a small region homologous to the TRPP2 pore loop, which was speculated to participate as the fourth subunit in forming the pore (52). In addition, several transient receptor potential proteins have been shown to form functional heteromers with other members of the same family including proteins of the TRPM (53, 54), TRPC (55–57), and TRPV (58, 59) families. Several other integral membrane proteins are required for the depolarizing light response of ON bipolar cells, namely mGluR6 (4), GPR179 (60–62), and LRIT3 (63, 64). Nyctalopin, which is an integral membrane protein in mice but not humans (65, 66), is also required (67–70). Aside from mGluR6, the functions of these proteins are mostly unknown, although none of them are immediately obvious candidates for contribution to the TRPM1 pore. GPR179 does appear to play a role in expression of a functional transduction channel: responses to capsaicin are significantly reduced in the GPR179nob mouse even though TRPM1 protein is present at ∼75% of wild-type levels and appears to be correctly localized (61). It is conceivable that one of these partners could stabilize homotetramer formation by TRPM1. Alternatively, other as-yet-unidentified proteins or perhaps other splice variants of TRPM1 may be required for complete channel assembly.
BN-PAGE of solubilized mouse retina revealed the presence of two distinct TRPM1-containing complexes (see Fig. 9). The smaller one is likely a TRPM1 homodimer; the ∼20% size discrepancy may be due to different post-translational modification or association with a small protein. The larger complex is at least double the size of the smaller one and may represent a fully formed channel. This complex is significantly larger than the minor band sometimes observed in BN-PAGE of solubilized Sf9 membranes (see Fig. 2B). The presence of putative TRPM1 dimers in retina lysate suggests that assembly may occur through a dimeric intermediate.
Two subunits of the TRPM1 TM domain homology model (or TRPV1, the template on which it is based) can be fit reasonably well into the putative TM domain of TRPM1 (see Fig. 10), although the low resolution of the map precludes definitive placement. The orientation of the two subunits with respect to each other and with respect to the plane of the membrane appears to be significantly different from that of TRPV1 (30). Orienting the two subunits with the pore loops close together while respecting 2-fold symmetry places the pore loop in the center of the TM domain. This would suggest that the two subunits in the dimer correspond to diagonally opposed subunits in the fully assembled channel complex. Alternatively, the purified dimers may not have exact 2-fold symmetry despite the similarity of the C1 and C2 models (see Fig. 6), or binding of other subunits may induce a conformational change that rearranges the pore loops.
Virtually nothing is known about the determinants of TRPM1 oligomerization. A conserved C-terminal coiled coil domain has been implicated in assembly of other members of the TRPM family. This domain is required for TRPM2 assembly and function (71), and the TRPM7 domain was crystallized as a four-stranded antiparallel coiled coil (29), which if preserved in the full-length protein would be incompatible with 4-fold symmetry. However, whereas isolated coiled coil domains of TRPM2, TRPM3, TRPM6, TRPM7, and TRPM8 formed multimers in solution (four or more), that of TRPM1 appeared to be monomeric (72). This suggests that the coiled coil domain is not sufficient for oligomerization of TRPM1, although it does not preclude its involvement.
Only a small fraction of TRPM1 expressed in Sf9 cells was localized to the plasma membrane (see Fig. 1). Similarly, when heterologously expressed in mammalian cells, most or all TRPM1 is localized in internal structures (73, 74) (see Fig. 3). Consistent with these observations, many attempts in our laboratory to demonstrate a calcium flux activity mediated by TRPM1 heterologously expressed in Sf9 cells, HEK cells, and CHO cells have failed. Nevertheless, there have been reports of cation currents induced by TRPM1 expression in mammalian cells (22, 54, 74). These currents are likely derived from the small fraction of TRPM1 that is localized at the plasma membrane. However, the currents have been quite small, and it is not clear that these expressed channels have the same properties as the ON bipolar cell transduction channel, nor is it clear that they are formed without participation of endogenous proteins. Interestingly, TRPM1 has been reported to form relatively robust channels when it is co-expressed with TRPM3 or when selected regions have been substituted with the corresponding regions of TRPM3 (54). Most recently, it was reported (23) that in HEK293 cells transiently transfected with TRPM1 the calcium influx in response to addition of extracellular calcium was not increased over that of untransfected cells. However, expression of TRPM1 rendered the calcium flux activity sensitive to capsazepine, which is an inhibitor of the rod bipolar cell transduction current (21). These results are consistent with TRPM1 expression conferring capsazepine sensitivity on an endogenous Ca2+ current.
A link between subunit assembly and trafficking has been demonstrated for several ion channels. For example, the AMPA receptor subunit GluA2 (formerly GluR2) is largely endoplasmic reticulum-retained and unassembled until incorporated into tetramers with other GluA subunits, and it was speculated that tetramerization masks an endoplasmic reticulum retention signal in GluA2 (49, 75). Similarly, heteromeric assembly of the G protein-gated inwardly rectifying potassium channel subunit Kir3.1 is assured by the requirement of an endoplasmic reticulum export signal from Kir3.2 or -3.4 (76). Assembly of TRPM1-containing channel complexes and forward trafficking to the plasma membrane may be coordinately regulated in such a manner.
In conclusion, we have presented here a low resolution structure of TRPM1 dimers. The structure suggests that TRPM1 forms an unusual channel complex lacking 4-fold symmetry and perhaps including subunits from one or more other proteins. Future studies are needed to characterize the quaternary structure of the functional TRPM1-dependent channel in its native environment in the retina.
Acknowledgments
We thank Vera Moiseenkova-Bell, Jared C. Gilliam, Irina Serysheva, Timothy L. Dosey, and Kurt R. Christensen for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 EY07981 and R01 EY11900 (to T. G. W.); T32 EY007102 and F32 EY200672 (to M. A. A.); K12 GM084897 (to S. J. W.); T32 DK007696 (to J. M.); P30 EY002520, a Vision Research Core grant; P30 CA125123; and P41 GM103832, a Center grant. This work was also supported by the Welch Foundation and the Knights Templar Eye Foundation.
- TRPM
- transient receptor potential melastatin
- nob
- no-b-wave
- NYX
- nyctalopin
- TRPV
- transient receptor potential vanilloid
- Sf9
- S. frugiperda clone 9
- DMSO
- dimethyl sulfoxide
- PIC
- protease inhibitor cocktail
- BN
- blue native
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Ab
- antibody
- hpi
- h postinfection
- CB
- cracking buffer
- CMC
- critical micelle concentration
- DST
- disuccinimidyl tartrate
- PAAGE
- polyacrylamide/agarose gel electrophoresis
- HBSS
- Hanks' balanced salt solution
- WGA
- wheat germ agglutinin
- NHS
- N-hydroxysuccinimide
- TM
- transmembrane.
REFERENCES
- 1. Lukasiewicz P. D. (2005) Synaptic mechanisms that shape visual signaling at the inner retina. Prog. Brain Res. 147, 205–218 [DOI] [PubMed] [Google Scholar]
- 2. Dhingra A., Lyubarsky A., Jiang M., Pugh E. N., Jr., Birnbaumer L., Sterling P., Vardi N. (2000) The light response of ON bipolar neurons requires Gαo. J. Neurosci. 20, 9053–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Masu M., Iwakabe H., Tagawa Y., Miyoshi T., Yamashita M., Fukuda Y., Sasaki H., Hiroi K., Nakamura Y., Shigemoto R., Takada M., Nakamura K., Nakao K., Katsuki M., Nakanishi S. (1995) Specific deficit of the ON response in visual transmission by targeted disruption of the mGluR6 gene. Cell 80, 757–765 [DOI] [PubMed] [Google Scholar]
- 4. Nakajima Y., Iwakabe H., Akazawa C., Nawa H., Shigemoto R., Mizuno N., Nakanishi S. (1993) Molecular characterization of a novel retinal metabotropic glutamate receptor mGluR6 with a high agonist selectivity for l-2-amino-4-phosphonobutyrate. J. Biol. Chem. 268, 11868–11873 [PubMed] [Google Scholar]
- 5. Nawy S. (1999) The metabotropic receptor mGluR6 may signal through Go, but not phosphodiesterase, in retinal bipolar cells. J. Neurosci. 19, 2938–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nomura A., Shigemoto R. (1994) Developmentally regulated postsynaptic localization of a metabotropic glutamate receptor in rat rod bipolar cells. Cell 77, 361–369 [DOI] [PubMed] [Google Scholar]
- 7. Shiells R.A., Falk G., Naghshineh S., (1981) Action of glutamate and aspartate analogues on rod horizontal and bipolar cells. Nature 294, 592–594 [DOI] [PubMed] [Google Scholar]
- 8. Vardi N. (1998) Alpha subunit of Go localizes in the dendritic tips of ON bipolar cells. J. Comp. Neurol. 395, 43–52 [PubMed] [Google Scholar]
- 9. Nawy S., Jahr C. E. (1990) Suppression by glutamate of cGMP-activated conductance in retinal bipolar cells. Nature 346, 269–271 [DOI] [PubMed] [Google Scholar]
- 10. Slaughter M. M., Miller R. F. (1981) 2-Amino-4-phosphonobutyric acid: a new pharmacological tool for retina research. Science 211, 182–185 [DOI] [PubMed] [Google Scholar]
- 11. Snellman J., Kaur T., Shen Y., Nawy S. (2008) Regulation of ON bipolar cell activity. Prog. Retin. Eye Res. 27, 450–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cao Y., Masuho I., Okawa H., Xie K., Asami J., Kammermeier P. J., Maddox D. M., Furukawa T., Inoue T., Sampath A. P., Martemyanov K. A. (2009) Retina-specific GTPase accelerator RGS11/Gβ5S/R9AP is a constitutive heterotrimer selectively targeted to mGluR6 in ON-bipolar neurons. J. Neurosci. 29, 9301–9313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cao Y., Pahlberg J., Sarria I., Kamasawa N., Sampath A. P., Martemyanov K. A. (2012) Regulators of G protein signaling RGS7 and RGS11 determine the onset of the light response in ON bipolar neurons. Proc. Natl. Acad. Sci. U.S.A. 109, 7905–7910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mojumder D. K., Qian Y., Wensel T. G. (2009) Two R7 regulator of G-protein signaling proteins shape retinal bipolar cell signaling. J. Neurosci. 29, 7753–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang J., Jeffrey B. G., Morgans C. W., Burke N. S., Haley T. L., Duvoisin R. M., Brown R. L. (2010) RGS7 and -11 complexes accelerate the ON-bipolar cell light response. Invest. Ophthalmol. Vis. Sci. 51, 1121–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cao Y., Posokhova E., Martemyanov K. A. (2011) TRPM1 forms complexes with nyctalopin in vivo and accumulates in postsynaptic compartment of ON-bipolar neurons in mGluR6-dependent manner. J. Neurosci. 31, 11521–11526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Morgans C. W., Zhang J., Jeffrey B. G., Nelson S. M., Burke N. S., Duvoisin R. M., Brown R. L. (2009) TRPM1 is required for the depolarizing light response in retinal ON-bipolar cells. Proc. Natl. Acad. Sci. U.S.A. 106, 19174–19178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nakajima Y., Moriyama M., Hattori M., Minato N., Nakanishi S. (2009) Isolation of ON bipolar cell genes via hrGFP-coupled cell enrichment using the mGluR6 promoter. J. Biochem. 145, 811–818 [DOI] [PubMed] [Google Scholar]
- 19. Orlandi C., Cao Y., Martemyanov K. A. (2013) Orphan receptor GPR179 forms macromolecular complexes with components of metabotropic signaling cascade in retina ON-bipolar neurons. Invest. Ophthalmol. Vis. Sci. 54, 7153–7161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pearring J. N., Bojang P., Jr., Shen Y., Koike C., Furukawa T., Nawy S., Gregg R. G. (2011) A role for nyctalopin, a small leucine-rich repeat protein, in localizing the TRP melastatin 1 channel to retinal depolarizing bipolar cell dendrites. J. Neurosci. 31, 10060–10066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shen Y., Heimel J. A., Kamermans M., Peachey N. S., Gregg R. G., Nawy S. (2009) A transient receptor potential-like channel mediates synaptic transmission in rod bipolar cells. J. Neurosci. 29, 6088–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koike C., Obara T., Uriu Y., Numata T., Sanuki R., Miyata K., Koyasu T., Ueno S., Funabiki K., Tani A., Ueda H., Kondo M., Mori Y., Tachibana M., Furukawa T. (2010) TRPM1 is a component of the retinal ON bipolar cell transduction channel in the mGluR6 cascade. Proc. Natl. Acad. Sci. U.S.A. 107, 332–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shen Y., Rampino M. A., Carroll R. C., Nawy S. (2012) G-protein-mediated inhibition of the Trp channel TRPM1 requires the G βγ dimer. Proc. Natl. Acad. Sci. U.S.A. 109, 8752–8757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peachey N. S., Pearring J. N., Bojang P., Jr., Hirschtritt M. E., Sturgill-Short G., Ray T. A., Furukawa T., Koike C., Goldberg A. F., Shen Y., McCall M. A., Nawy S., Nishina P. M., Gregg R. G. (2012) Depolarizing bipolar cell dysfunction due to a Trpm1 point mutation. J. Neurophysiol. 108, 2442–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Audo I., Kohl S., Leroy B. P., Munier F. L., Guillonneau X., Mohand-Saïd S., Bujakowska K., Nandrot E. F., Lorenz B., Preising M., Kellner U., Renner A. B., Bernd A., Antonio A., Moskova-Doumanova V., Lancelot M. E., Poloschek C. M., Drumare I., Defoort-Dhellemmes S., Wissinger B., Léveillard T., Hamel C. P., Schorderet D. F., De Baere E., Berger W., Jacobson S. G., Zrenner E., Sahel J. A., Bhattacharya S. S., Zeitz C. (2009) TRPM1 is mutated in patients with autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 85, 720–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Z., Sergouniotis P. I., Michaelides M., Mackay D. S., Wright G. A., Devery S., Moore A. T., Holder G. E., Robson A. G., Webster A. R. (2009) Recessive mutations of the gene TRPM1 abrogate ON bipolar cell function and cause complete congenital stationary night blindness in humans. Am. J. Hum. Genet. 85, 711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakamura M., Sanuki R., Yasuma T. R., Onishi A., Nishiguchi K. M., Koike C., Kadowaki M., Kondo M., Miyake Y., Furukawa T. (2010) TRPM1 mutations are associated with the complete form of congenital stationary night blindness. Mol. Vis. 16, 425–437 [PMC free article] [PubMed] [Google Scholar]
- 28. van Genderen M. M., Bijveld M. M., Claassen Y. B., Florijn R. J., Pearring J. N., Meire F. M., McCall M. A., Riemslag F. C., Gregg R. G., Bergen A. A., Kamermans M. (2009) Mutations in TRPM1 are a common cause of complete congenital stationary night blindness. Am. J. Hum. Genet. 85, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fujiwara Y., Minor D. L. (2008) X-ray crystal structure of a TRPM assembly domain reveals an antiparallel four-stranded coiled-coil. J. Mol. Biol. 383, 854–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liao M., Cao E., Julius D., Cheng Y. (2013) Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moiseenkova-Bell V. Y., Stanciu L. A., Serysheva I. I., Tobe B. J., Wensel T. G. (2008) Structure of TRPV1 channel revealed by electron cryomicroscopy. Proc. Natl. Acad. Sci. U.S.A. 105, 7451–7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huynh K. W., Cohen M. R., Chakrapani S., Holdaway H. A., Stewart P. L., Moiseenkova-Bell V. Y. (2014) Structural insight into the assembly of TRPV channels. Structure 22, 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shigematsu H., Sokabe T., Danev R., Tominaga M., Nagayama K. (2010) A 3.5-nm structure of rat TRPV4 cation channel revealed by Zernike phase-contrast cryoelectron microscopy. J. Biol. Chem. 285, 11210–11218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cvetkov T. L., Huynh K. W., Cohen M. R., Moiseenkova-Bell V. Y. (2011) Molecular architecture and subunit organization of TRPA1 ion channel revealed by electron microscopy. J. Biol. Chem. 286, 38168–38176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Molday R. S., MacKenzie D. (1983) Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry 22, 653–660 [DOI] [PubMed] [Google Scholar]
- 36. Gilliam J. C., Wensel T. G. (2011) TRP channel gene expression in the mouse retina. Vision Res. 51, 2440–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. MacKenzie D., Arendt A., Hargrave P., McDowell J. H., Molday R. S. (1984) Localization of binding sites for carboxyl terminal specific anti-rhodopsin monoclonal antibodies using synthetic peptides. Biochemistry 23, 6544–6549 [DOI] [PubMed] [Google Scholar]
- 38. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 39. Tatsumi R., Hattori A. (1995) Detection of giant myofibrillar proteins connectin and nebulin by electrophoresis in 2% polyacrylamide slab gels strengthened with agarose. Anal. Biochem. 224, 28–31 [DOI] [PubMed] [Google Scholar]
- 40. Labeit S., Ottenheijm C. A., Granzier H. (2011) Nebulin, a major player in muscle health and disease. FASEB J. 25, 822–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schägger H., von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 42. Heuberger E. H., Veenhoff L. M., Duurkens R. H., Friesen R. H., Poolman B. (2002) Oligomeric state of membrane transport proteins analyzed with blue native electrophoresis and analytical ultracentrifugation. J. Mol. Biol. 317, 591–600 [DOI] [PubMed] [Google Scholar]
- 43. Wittig I., Beckhaus T., Wumaier Z., Karas M., Schägger H. (2010) Mass estimation of native proteins by blue native electrophoresis: principles and practical hints. Mol. Cell. Proteomics 9, 2149–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ludtke S. J., Baldwin P. R., Chiu W. (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 45. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 46. Söding J. (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics 21, 951–960 [DOI] [PubMed] [Google Scholar]
- 47. Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M., Pieper U., Sali A. (2006) Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics 15, 5.6.1–5.6.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Le Maire M., Aggerbeck L. P., Monteilhet C., Andersen J. P., Møller J. V. (1986) The use of high-performance liquid chromatography for the determination of size and molecular weight of proteins: a caution and a list of membrane proteins suitable as standards. Anal. Biochem. 154, 525–535 [DOI] [PubMed] [Google Scholar]
- 49. Greger I. H., Khatri L., Kong X., Ziff E. B. (2003) AMPA receptor tetramerization is mediated by Q/R editing. Neuron 40, 763–774 [DOI] [PubMed] [Google Scholar]
- 50. Kim K. S., Yan D., Tomita S. (2010) Assembly and stoichiometry of the AMPA receptor and transmembrane AMPA receptor regulatory protein complex. J. Neurosci. 30, 1064–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gohon Y., Dahmane T., Ruigrok R. W., Schuck P., Charvolin D., Rappaport F., Timmins P., Engelman D. M., Tribet C., Popot J.-L., Ebel C. (2008) Bacteriorhodopsin/amphipol complexes: structural and functional properties. Biophys. J. 94, 3523–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yu Y., Ulbrich M. H., Li M. H., Buraei Z., Chen X. Z., Ong A. C., Tong L., Isacoff E. Y., Yang J. (2009) Structural and molecular basis of the assembly of the TRPP2/PKD1 complex. Proc. Natl. Acad. Sci. U.S.A. 106, 11558–11563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chubanov V., Waldegger S., Mederos y Schnitzler M., Vitzthum H., Sassen M. C., Seyberth H. W., Konrad M., Gudermann T. (2004) Disruption of TRPM6/TRPM7 complex formation by a mutation in the TRPM6 gene causes hypomagnesemia with secondary hypocalcemia. Proc. Natl. Acad. Sci. U.S.A. 101, 2894–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lambert S., Drews A., Rizun O., Wagner T. F., Lis A., Mannebach S., Plant S., Portz M., Meissner M., Philipp S. E., Oberwinkler J. (2011) Transient receptor potential melastatin 1 (TRPM1) is an ion-conducting plasma membrane channel inhibited by zinc. J. Biol. Chem. 286, 12221–12233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hofmann T., Schaefer M., Schultz G., Gudermann T. (2002) Subunit composition of mammalian transient receptor potential channels in living cells. Proc. Natl. Acad. Sci. U.S.A. 99, 7461–7466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Strübing C., Krapivinsky G., Krapivinsky L., Clapham D. E. (2001) TRPC1 and TRPC5 form a novel cation channel in mammalian brain. Neuron 29, 645–655 [DOI] [PubMed] [Google Scholar]
- 57. Strübing C., Krapivinsky G., Krapivinsky L., Clapham D. E. (2003) Formation of novel TRPC channels by complex subunit interactions in embryonic brain. J. Biol. Chem. 278, 39014–39019 [DOI] [PubMed] [Google Scholar]
- 58. Cheng W., Yang F., Takanishi C. L., Zheng J. (2007) Thermosensitive TRPV channel subunits coassemble into heteromeric channels with intermediate conductance and gating properties. J. Gen. Physiol. 129, 191–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hoenderop J. G., Voets T., Hoefs S., Weidema F., Prenen J., Nilius B., Bindels R. J. (2003) Homo- and heterotetrameric architecture of the epithelial Ca2+ channels TRPV5 and TRPV6. EMBO J. 22, 776–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Peachey N. S., Ray T. A., Florijn R., Rowe L. B., Sjoerdsma T., Contreras-Alcantara S., Baba K., Tosini G., Pozdeyev N., Iuvone P. M., Bojang P., Jr., Pearring J. N., Simonsz H. J., van Genderen M., Birch D. G., Traboulsi E. I., Dorfman A., Lopez I., Ren H., Goldberg A. F., Nishina P. M., Lachapelle P., McCall M. A., Koenekoop R. K., Bergen A. A., Kamermans M., Gregg R. G. (2012) GPR179 is required for depolarizing bipolar cell function and is mutated in autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 90, 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ray T. A., Heath K. M., Hasan N., Noel J. M., Samuels I. S., Martemyanov K. A., Peachey N. S., McCall M. A., Gregg R. G. (2014) GPR179 is required for high sensitivity of the mGluR6 signaling cascade in depolarizing bipolar cells. J. Neurosci. 34, 6334–6343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Audo I., Bujakowska K., Orhan E., Poloschek C. M., Defoort-Dhellemmes S., Drumare I., Kohl S., Luu T. D., Lecompte O., Zrenner E., Lancelot M. E., Antonio A., Germain A., Michiels C., Audier C., Letexier M., Saraiva J. P., Leroy B. P., Munier F. L., Mohand-Saïd S., Lorenz B., Friedburg C., Preising M., Kellner U., Renner A. B., Moskova-Doumanova V., Berger W., Wissinger B., Hamel C. P., Schorderet D. F., De Baere E., Sharon D., Banin E., Jacobson S. G., Bonneau D., Zanlonghi X., Le Meur G., Casteels I., Koenekoop R., Long V. W., Meire F., Prescott K., de Ravel T., Simmons I., Nguyen H., Dollfus H., Poch O., Léveillard T., Nguyen-Ba-Charvet K., Sahel J. A., Bhattacharya S. S., Zeitz C. (2012) Whole-exome sequencing identifies mutations in GPR179 leading to autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 90, 321–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zeitz C., Jacobson S. G., Hamel C. P., Bujakowska K., Neuillé M., Orhan E., Zanlonghi X., Lancelot M. E., Michiels C., Schwartz S. B., Bocquet B., Congenital Stationary Night Blindness Consortium, Antonio A., Audier C., Letexier M., Saraiva J. P., Luu T. D., Sennlaub F., Nguyen H., Poch O., Dollfus H., Lecompte O., Kohl S., Sahel J. A., Bhattacharya S. S., Audo I. (2013) Whole-exome sequencing identifies LRIT3 mutations as a cause of autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 92, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Neuillé M., El Shamieh S., Orhan E., Michiels C., Antonio A., Lancelot M.-E., Condroyer C., Bujakowska K., Poch O., Sahel J.-A., Audo I., Zeitz C. (2014) Lrit3 deficient mouse (nob6): a novel model of complete congenital stationary night blindness (cCSNB). PLoS One 9, e90342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. O'Connor E., Eisenhaber B., Dalley J., Wang T., Missen C., Bulleid N., Bishop P. N., Trump D. (2005) Species specific membrane anchoring of nyctalopin, a small leucine-rich repeat protein. Hum. Mol. Genet. 14, 1877–1887 [DOI] [PubMed] [Google Scholar]
- 66. Bojang P., Jr., Gregg R. G. (2012) Topological analysis of small leucine-rich repeat proteoglycan nyctalopin. PLoS One 7, e33137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bech-Hansen N. T., Naylor M. J., Maybaum T. A., Sparkes R. L., Koop B., Birch D. G., Bergen A. A., Prinsen C. F., Polomeno R. C., Gal A., Drack A. V., Musarella M. A., Jacobson S. G., Young R. S., Weleber R. G. (2000) Mutations in NYX, encoding the leucine-rich proteoglycan nyctalopin, cause X-linked complete congenital stationary night blindness. Nat. Genet. 26, 319–323 [DOI] [PubMed] [Google Scholar]
- 68. Gregg R. G., Mukhopadhyay S., Candille S. I., Ball S. L., Pardue M. T., McCall M. A., Peachey N. S. (2003) Identification of the gene and the mutation responsible for the mouse nob phenotype. Invest. Ophthalmol. Vis. Sci. 44, 378–384 [DOI] [PubMed] [Google Scholar]
- 69. Gregg R. G., Kamermans M., Klooster J., Lukasiewicz P. D., Peachey N. S., Vessey K. A., McCall M. A. (2007) Nyctalopin expression in retinal bipolar cells restores visual function in a mouse model of complete X-linked congenital stationary night blindness. J. Neurophysiol. 98, 3023–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pusch C. M., Zeitz C., Brandau O., Pesch K., Achatz H., Feil S., Scharfe C., Maurer J., Jacobi F. K., Pinckers A., Andreasson S., Hardcastle A., Wissinger B., Berger W., Meindl A. (2000) The complete form of X-linked congenital stationary night blindness is caused by mutations in a gene encoding a leucine-rich repeat protein. Nat. Genet. 26, 324–327 [DOI] [PubMed] [Google Scholar]
- 71. Mei Z.-Z., Xia R., Beech D. J., Jiang L.-H. (2006) Intracellular coiled-coil domain engaged in subunit interaction and assembly of melastatin-related transient receptor potential channel 2. J. Biol. Chem. 281, 38748–38756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tsuruda P. R., Julius D., Minor D. L. (2006) Coiled coils direct assembly of a cold-activated TRP channel. Neuron 51, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dhingra A., Fina M. E., Neinstein A., Ramsey D. J., Xu Y., Fishman G. A., Alexander K. R., Qian H., Peachey N. S., Gregg R. G., Vardi N. (2011) Autoantibodies in melanoma-associated retinopathy target TRPM1 cation channels of retinal ON bipolar cells. J. Neurosci. 31, 3962–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Oancea E., Vriens J., Brauchi S., Jun J., Splawski I., Clapham D. E. (2009) TRPM1 forms ion channels associated with melanin content in melanocytes. Sci. Signal. 2, ra21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Greger I. H., Khatri L., Ziff E. B. (2002) RNA editing at Arg607 controls AMPA receptor exit from the endoplasmic reticulum. Neuron 34, 759–772 [DOI] [PubMed] [Google Scholar]
- 76. Ma D., Zerangue N., Raab-Graham K., Fried S.R., Jan Y. N., Jan L. Y. (2002) Diverse trafficking patterns due to multiple traffic motifs in G protein-activated inwardly rectifying potassium channels from brain and heart. Neuron 33, 715–729 [DOI] [PubMed] [Google Scholar]
- 77. Harris L. J., Skaletsky E., McPherson A. (1998) Crystallographic structure of an intact IgG1 monoclonal antibody. J. Mol. Biol. 275, 861–872 [DOI] [PubMed] [Google Scholar]
- 78. Popot J. L., Althoff T., Bagnard D., Banères J. L., Bazzacco P., Billon-Denis E., Catoire L. J., Champeil P., Charvolin D., Cocco M. J., Crémel G., Dahmane T., de la Maza L. M., Ebel C., Gabel F., Giusti F., Gohon Y., Goormaghtigh E., Guittet E., Kleinschmidt J. H, Kühlbrandt W., Le Bon C., Martinez K. L., Picard M., Pucci B., Sachs J. N., Tribet C., van Heijenoort C., Wien F., Zito F., Zoonens M. (2011) Amphipols from A to Z. Annu. Rev. Biophys. 40, 379–408 [DOI] [PubMed] [Google Scholar]