Background: DNA mimic proteins prevent DNA-binding proteins from binding to DNA.
Results: T4 phage DNA mimic protein Arn disrupts H-NS·DNA binding and neutralizes the gene-silencing effect of H-NS.
Conclusion: Arn participates in viral anti-host defense system by its DNA mimicking properties.
Significance: This anti-H-NS function of Arn represents a novel battle mechanism between phage and bacteria.
Keywords: Crystal Structure, DNA-binding Protein, Protein Structure, Protein-Protein Interaction, Protein-DNA Interaction, DNA Mimic Protein, Molecular Mimicry
Abstract
The T4 phage protein Arn (Anti restriction nuclease) was identified as an inhibitor of the restriction enzyme McrBC. However, until now its molecular mechanism remained unclear. In the present study we used structural approaches to investigate biological properties of Arn. A structural analysis of Arn revealed that its shape and negative charge distribution are similar to dsDNA, suggesting that this protein could act as a DNA mimic. In a subsequent proteomic analysis, we found that the bacterial histone-like protein H-NS interacts with Arn, implying a new function. An electrophoretic mobility shift assay showed that Arn prevents H-NS from binding to the Escherichia coli hns and T4 p8.1 promoters. In vitro gene expression and electron microscopy analyses also indicated that Arn counteracts the gene-silencing effect of H-NS on a reporter gene. Because McrBC and H-NS both participate in the host defense system, our findings suggest that T4 Arn might knock down these mechanisms using its DNA mimicking properties.
Introduction
Research in the last decade has revealed several examples of proteins that can mimic DNA (1–3). These DNA mimic proteins prevent DNA-binding proteins from binding to DNA using their DNA-like surfaces. To date only a few DNA mimic proteins have been reported and functionally characterized because the amino acid sequences and protein structures of reported DNA mimic proteins are divergent (1, 4). To extend our knowledge of this novel type of control factor, we searched for new DNA mimic proteins from selected species including bacteriophage, white spot syndrome virus, Neisseria, and Staphylococcus. In the past five years, we have successfully identified four DNA mimic proteins, ICP11, DMP19, DMP12, and SAUGI, using structural and proteomic approaches (4–7). In the course of discovering these new DNA mimic proteins, gene mining from the whole genome has been the first step to explore the potential targets. To our knowledge the genes encoding small and acidic proteins have a higher chance to be DNA mimics. Another important criterion is the functions of these potential targets. The annotation of these gene products usually specifies “DNA-related” functions such as regulating transcription, inhibiting DNA metabolic enzyme, or binding to DNA binder. To screen the undiscovered functions of these potential DNA mimics, a pulldown assay is one of the proteomic approaches to characterize them. Based on these preliminary studies we found that the bacteriophage T4 protein Arn (Anti-restriction nuclease) is likely to be a DNA mimic. After analyzing the three-dimensional structure and the function of Arn, we believe that Arn could be added to the list of DNA mimics.
In previous reports Arn was identified as an auxiliary protein that blocks McrBC restriction nuclease activity and prevents the T4 genome from host R/M (restriction modification) system digestion (8, 9). Because Arn has the patterns of negatively charged amino acids reminiscent of the DNA mimic proteins Ocr and DinI, Putnam and Tainer (2) proposed that this protein is a potential DNA mimic. In the present paper we tested this hypothesis by determining the crystal structure of Arn. The structural analysis showed that this dimeric protein has a crescent-like shape that is similar to a bent, double-stranded DNA molecule (dsDNA). Additionally, a dsDNA-like negative charge distribution could be found on the surface of Arn dimer. Arn may, therefore, use these DNA-like properties to control the restriction nuclease activity of McrBC. Interestingly, we found that the bacterial histone-like protein H-NS can interact with Arn in our proteomic studies. We also present evidence that Arn can disrupt H-NS·DNA binding and attenuate H-NS-induced gene silencing. Together, these observations suggest that Arn might serve as an antagonist of the host anti-phage defense systems (e.g. McrBC and H-NS) by its DNA mimic properties.
EXPERIMENTAL PROCEDURES
Preparation of Recombinant Arn-His and H-NS
The full-length T4 phage Arn gene (residues 1–92) was synthesized by Genomics (Taipei, Taiwan) and cloned into a pET21b expression vector (Novagen); the resulting plasmid was called pET21b-Arn. The recombinant Arn protein contained a C-terminal His6 tag (100 amino acids) referred to as Arn-His. This protein was expressed in Escherichia coli BL21 (DE3) cells at 37 °C for 4 h after induction with 1 mm isopropyl β-d-1-thiogalactopyranoside. Soluble Arn-His was purified by immobilized metal-ion chromatography with a nickel-nitrilotriacetic acid column followed by anion exchange with a Q column and gel filtration using Superdex 200pg (GE Healthcare).
The full-length hns gene with a stop codon was obtained by direct PCR from E. coli BL21 (DE3) cells and cloned into a pET21b expression vector (Novagen). The tag-free H-NS was overexpressed in BL21 (DE3) cells at 37 °C after induction with 1 mm isopropyl β-d-1-thiogalactopyranoside for 4 h. The total lysate of H-NS-overexpressing BL21 (DE3) cells was treated with Benzonase (Merck) before centrifugation. Soluble H-NS was purified by a cation exchange SP column and a heparin column (GE Healthcare). The DNA fragment encoding H-NS DNA binding domain (H-NS91–137) was cloned into a pET16b expression vector (Novagen). The recombinant H-NS DNA binding domain contained an N-terminal His10 tag referred to as His-H-NS91. His-H-NS91 was expressed in E. coli BL21 (DE3) cells at 30 °C for 4 h after induction with 0.5 mm isopropyl β-d-1-thiogalactopyranoside. Soluble His-H-NS91 was purified by immobilized metal-ion chromatography with a nickel-nitrilotriacetic acid column followed by anion exchange with a heparin column and gel filtration using Superdex 200pg (GE Healthcare).
Recombinant Arn-His Crystallization and Data Collection
Purified Arn-His was concentrated in a crystallization buffer (50 mm Tris at pH 8.0, 100 mm NaCl) to 20 mg/ml. For crystallization, 2 μl of the Arn-His solution was mixed with 2-μl of a reservoir (3.6 m sodium formate, 0.1 m Tris at pH 8.0) with 1 μl of additive (Bis-Tris2 at pH 5.2). The mixture was then equilibrated with the reservoir by the sitting drop method at 25 °C. The PtII-containing compound terpyridine-PtIICl2 was used in derivatizing the crystals to determine the phase. Both native and single wavelength anomalous diffraction x-ray diffraction data from the Arn-His crystals were collected using beamline BL13B1 at the National Synchrotron Radiation Research Center in Hsinchu, Taiwan and beamline BL12B2 at SPring-8 in Japan. The data were processed using HKL2000 (10). The space group of the Arn-His crystals is H32. Other details are shown in Table 1.
TABLE 1.
Data collection and refinement of Arn-His
| Arn-His (native) | Arn-His (Pt) | |
|---|---|---|
| Data collection | ||
| Space group | H32 | H32 |
| Unit cell a, b, c (Å) | 81.96, 81.96, 149.42 | 81.79, 81.79, 149.84 |
| Wavelength (Å) | 0.97622 | 1.0716 (peak) |
| Resolution range (Å) | 23.66–1.90 (1.97–1.90)a | 40.89–1.93 (2.00–1.93)a |
| Number of reflections | ||
| Observed | 118,227 | 148,891 |
| Unique | 15,419 | 14,531 |
| Completeness (%) | 99.4 (100.0) | 98.0 (82.6) |
| Rmerge (%) | 4.1 (47.8) | 4.4 (46.6) |
| I/σ (I) | 45.07 (6.15) | 50.34 (2.76) |
| Refinement | ||
| Rwork (%) | 20.8 | |
| Rfree (%) | 23.2 | |
| Geometry deviations | ||
| Bond length (Å) | 0.0183 | |
| Bond angles (°) | 1.7674 | |
| Ramachandran plot (%)b | ||
| Most favored | 93.3 | |
| Additionaly allowed | 6.7 | |
| Average B (Å2)/no. of atoms | ||
| Protein | 38.33/828 | |
| Water | 55.97/155 | |
a Numbers in parentheses are for the highest resolution shells.
b The stereochemistry of the models was validated with PROCHECK.
Structure Determination and Refinement
The Arn-His structure was solved using the single wavelength anomalous diffraction phasing method and the program SHELX CDE (11). The native and peak datasets in the range of 23–1.90 and 40–1.93 Å resolution were collected at wavelengths of 0.97622 and 1.0716 Å, respectively. Two PtII sites were found in an asymmetric unit. As a result of employing the program DM (12), a clear electron density map allowed automatic model building by the program BUCCANEER (13). The programs CNS (14), Refmac5 (15), and COOT (16) were used in the refinement, and the relevant statistics are shown in Table 1. The subsequent structural analysis, figure drawing, and model making were conducted with the CCP4 package (17) and PyMOL (18).
His Pulldown Assay and Protein Identification
To determine the Arn-interacting proteins from the host, we used the His pulldown method with E. coli BL21 lysate. E. coli BL21 (DE3) cells were cultured overnight, harvested (the pellet was ∼0.6 g from a 100-ml culture), and directly resuspended in 2.4 ml of B-PER bacterial protein extraction reagent (Thermo Scientific) with an EDTA-free protease inhibitor mixture (Roche Applied Science) and Benzonase (Merck). After centrifugation at 4000 × g for 30 min, the soluble protein fraction was mixed with Arn-His and nickel-nitrilotriacetic acid beads; the salt concentration was adjusted to 100 mm NaCl and 40 mm imidazole. After incubation at 4 °C for 16 h, the beads were washed with binding buffer (20 mm Tris at pH 7.5, 100 mm NaCl, and 40 mm imidazole; 10× bead volume) 3 times. The proteins remaining on the beads were eluted with buffer containing 250 mm imidazole and analyzed by SDS-PAGE. Other His pulldown assays were performed by incubating the two purified proteins, Arn-His and H-NS, at room temperature for 10 min under conditions of different pH or ionic strength.
The protein bands of interest were manually excised from the SDS-PAGE gel followed by in-gel trypsin digestion. Using LC-MS/MS to resolve protein fragments and combining data with the search program MASCOT (19), several fragments were matched as significant hits and were regarded as positive identifications.
Analytical Ultracentrifugation (AUC) Analysis
Sedimentation velocity analysis was performed at 60,000 rpm using a 4-hole An-60 Ti rotor at 20 °C in a Beckman Optima XL-I AUC equipped with absorbance optics. Each sample was adjusted to a suitable concentration (280-nm absorption between 0.4 and ∼0.6) in 20 mm Tris at pH 7.5 and 50 mm NaCl. Data were analyzed with the c(M) distribution of the Lamm equation solutions calculated by the program SEDFIT (created at the National Institutes of Health). The SEDFIT parameters were: buffer density, 1.0087 g/ml; buffer viscosity, 0.01013 poise (g/cm/s); protein partial specific volume, 0.73.
Electrophoresis Mobility Shift Assay (EMSA)
The oligonucleotides were synthesized by Mission Biotech (Taipei, Taiwan). The short dsDNA was prepared by mixing equal amounts of complementary oligonucleotides, heating to 95 °C for 15 min, and then reducing to 25 °C for annealing. For the competitive binding assay, the chronological order of H-NS·DNA complex formation and Arn protein emergence was taken into consideration for the purpose of imitating the in vivo dynamics. The purified H-NS and dsDNA were first mixed in the reaction buffer (20 mm Tris at pH 8.0, 100 mm NaCl) and incubated at 25 °C for 10 min, and then purified recombinant Arn-His was added at 25 °C for another 10 min. All reactions were analyzed on a 0.8% agarose (Yeastern) gel and stained with SYBR Green I (Sigma).
In Vitro Gene Regulation Assay
The assay was performed in a cell-free expression system using an EasyXpress protein synthesis kit (QIAgen). The plasmid containing inducible gfp as the reporter gene driven by the T7 promoter was previously constructed, and its use in the gene regulation assay was described previously (6). To execute the assay, the plasmid was mixed with (or without) purified H-NS protein at 25 °C for 10 min followed by incubation with different amounts of purified recombinant Arn-His for another 10 min. After the preincubation step, the mixture was combined with the E. coli extract containing the protein synthesis reaction buffer that was provided in the kit, and the reaction was initiated by the addition of 1 mm isopropyl β-d-1-thiogalactopyranoside. During the 1-h induction at 37 °C, the relative amount of GFP expressed was detected at 395-nm excitation and 508-nm emission wavelengths.
Transmission Electron Microscopy Imaging
DNA·H-NS complexes were prepared for EM using the aqueous drop spreading method (20). The DNA·H-NS (DNA 100 ng/μl, H-NS 2 μm) was mixed with 200 μg/ml cytochrome c in 2.5 m ammonium acetate and incubated for 3 min. A 50-μl drop was placed on a clean Parafilm surface, and DNA·H-NS was picked up on a Parlodion-covered grid, dehydrated, and air-dried. Dehydration was carried out in 70% ethanol followed by 90% ethanol. Finally, the grids were blotted on filter paper and imaged using a rotary shadow at an angle of 8° with platinum:palladium (80%:20%) in a high vacuum. The samples were examined in an FEI Tecnai F20 transmission electron microscopy using 200 kV accelerating voltage, and the images were recorded using a Gatan (Pleasanton, CA) slow scan 4k × 4k CCD camera. The preparation and imaging of Arn-His (20 μm) containing DNA·H-NS transmission electron microscopy samples were identical to the methods used for the DNA·H-NS samples.
RESULTS
The Crystal Structure of Arn Shows DNA Mimic Properties
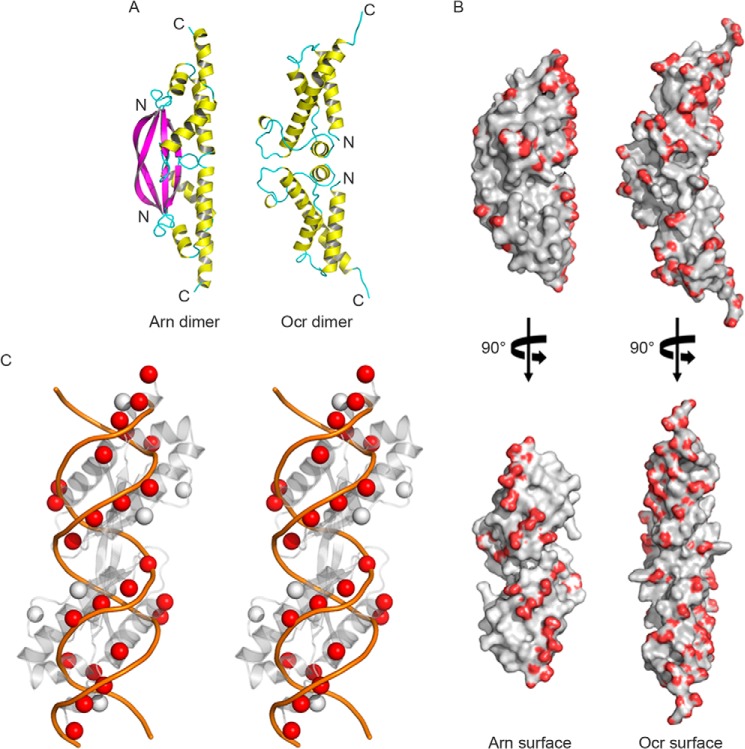
To further understand the biological properties of Arn, the protein was crystallized. Its structure was solved by single wavelength anomalous diffraction. Each asymmetric unit contains an Arn monomer, but the monomer forms a homodimer with a dyad-related partner. Despite a high solvent content of >60%, the structure refines quite well (Table 1, PDB code 3WX4). The Arn monomer folds into a three-stranded, anti-parallel β-sheet, and bundled on the other side are four α-helices (Fig. 1A). Part of the His-tag propagates as an extension of the C-terminal helix. At the N terminus, the β-strand stretches out and associates with its counterpart on the other monomer, forming a long β-ribbon of 14 residues and expanding the β-sheet across the dimer interface. Dimerization of Arn buries 1240 Å2 of the 7090 Å2 surface area of each monomer.
FIGURE 1.
The crystal structure of T4 Arn and the DNA mimic protein T7 Ocr. A, the Arn dimer and Ocr dimer are shown as schematics. B, the distributions of the negative surface charges on the Arn and Ocr dimers are compared. The carboxyl groups of the acidic amino acids, Glu and Asp, are colored red. C, the negatively charged groups (i.e. C-β carbons on Glu and Asp) of the Arn dimer (gray spheres) and double-stranded B-DNA (orange lines) are compared in the wall-eyed stereo view, and the dsDNA-matched C-β carbons are colored in red spheres.
The overall structure of Arn revealed a crescent-shaped dimer that resembles T7 Ocr (21), another dimeric DNA mimic protein (PDB code 1S7Z; Fig. 1A). Numerous acidic residues located in the helical bundles are found on the concave side of the dimer. The distribution of these negatively charged amino acids is more concentrated on this side in Arn when compared with Ocr (Fig. 1B). On the convex side, the formation of an extended β-sheet across the dimer interface in Arn suggests a much more stable dimer than Ocr, which despite its larger size has a dimer interface of only 500 Å2. As expected for a DNA mimic protein, the acidic residues of Arn are arranged in a symmetric and similar way as the phosphate groups in dsDNA (Fig. 1C).
E. coli H-NS Interacts with Arn
In a preliminary analysis, we performed a His pulldown assay that used His-tagged Arn as bait to search for potential interaction partners whose functions are DNA-related in an E. coli extract. Several distinct proteins were pulled down by the Arn-His-bound resin, as shown in the SDS-PAGE results (Fig. 2). We selected the five most obvious bands (bands 1–5) and analyzed them by LC-MS/MS (Fig. 2 and Table 2). All of the major protein species were identified. However, many of them are metabolic enzymes or bear the function of protein synthesis or trafficking. Interestingly, two potential interaction partners of Arn that bear DNA binding ability were found in band 1 and band 5. Although DNA topoisomerase 1 was found in band 5, this protein usually appears in our other His pulldown assays, and we regard this protein as a nonspecific binding to our His pulldown assay system. In band 1, we found the histone-like nucleoid structuring protein H-NS, which is a major protein species that bears DNA-related function; thus, H-NS is a potential new target of Arn. In addition to H-NS original function, it also serves as a gene silencer against exogenous DNA molecules such as the phage genome, and thus it is a potential target of bacteriophages (22–24).
FIGURE 2.
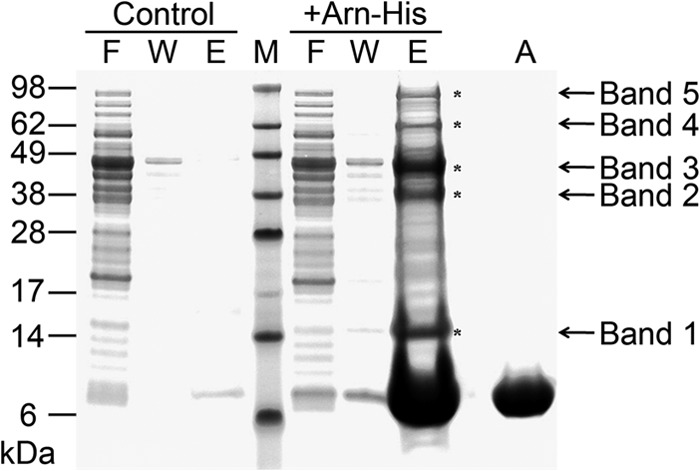
The His pulldown of Arn-His from E. coli lysate. Compared with the control group, there are five bands representing pulled down proteins, and each is marked with an asterisk. F, flow-through; W, washing fractions; E, eluates from nickel beads; M, marker; A, Arn-His protein only.
TABLE 2.
Protein identification by LC-MS/MS
PTS, carbohydrate phosphotransferase system.
| Protein name | Accession no. | MS/mpsa | Sequence coverage |
|---|---|---|---|
| % | |||
| Band 1 | |||
| UPF0325 protein YaeH | P62768 | 4,007/72 | 96 |
| DNA-binding protein H-NS | P0ACF8 | 2,766/55 | 89 |
| Band 2 | |||
| Galactitol-1-phosphate 5-dehydrogenase | P0A9S3 | 7,461/121 | 67 |
| PTS system mannose-specific EIIAB component | P69797 | 4,165/72 | 86 |
| Elongation factor Tu 1 | A7ZSL4 | 2,766/45 | 80 |
| Band 3 | |||
| Elongation factor Tu 1 | A7ZSL4 | 5,536/115 | 87 |
| Isocitrate dehydrogenase [NADP] | P08200 | 2,976/63 | 68 |
| Band 4 | |||
| Glutamine-fructose-6-phosphate aminotransferase | P17169 | 11,124/156 | 81 |
| Succinate dehydrogenase flavoprotein subunit | P0AC41 | 8,543/138 | 80 |
| d-Lactate dehydrogenase | P06149 | 3,687/64 | 72 |
| Band 5 | |||
| Aconitate hydratase 1 | P25516 | 10,868/179 | 68 |
| Alanine-tRNA ligase | B1XCM5 | 4,534/66 | 72 |
| DNA topoisomerase 1 | P06612 | 4,327/80 | 59 |
| NADH-quinone oxidoreductase subunit G | P33602 | 4,242/70 | 60 |
| Protein translocase subunit SecA | C4ZRJ3 | 3,471/61 | 56 |
| Aldehyde-alcohol dehydrogenase | P0A9Q7 | 2,969/47 | 60 |
a Mowse score/number of significant peptide matches.
To confirm the Arn·H-NS interaction, purified Arn-His and H-NS recombinant proteins were used in another His pulldown assay. Indeed, Arn-His and H-NS interacted with each other in this assay (Fig. 3A). AUC analysis further confirmed the Arn·H-NS interaction (Fig. 3B). The AUC result clearly indicated that the H-NS DNA binding domain, named His-H-NS91, binds to an Arn-His dimer in solution and forms a larger complex.
FIGURE 3.
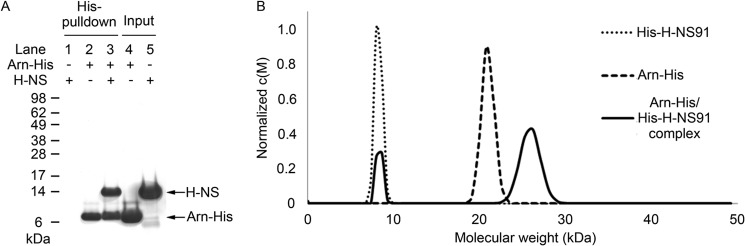
The His pulldown assay and analytical ultracentrifugation analysis both confirm the Arn·H-NS interaction. A, C-terminal His6-tagged Arn serves as bait, and tag-free H-NS serves as prey in this His pulldown assay. The His pulldown lanes, lane 1–3, are His pulldown eluates that represent the status of H-NS, Arn-His, and H-NS/Arn-His binding to nickel beads or protein complex pulled down, respectively. The input lanes, lane 4 and 5, are purified Arn-His and H-NS protein that are used in the assay respectively. B, under the buffer conditions of 20 mm Tris at pH 7.5 and 50 mm NaCl, the protein distribution of AUC analysis was as follows: the H-NS DNA binding domain (we used the protein His-H-NS91; the theoretical molecular mass is 8 kDa) only exhibits an 8-kDa peak as a monomer; the Arn protein (we used Arn-His; the theoretical molecular mass is 12 kDa) exhibits a single 21.6 kDa peak as a dimer; the His-H-NS91 and Arn-His mixture shows an ∼8-kDa peak that overlaps with the peak of the His-H-NS91 monomer and an ∼26.2 kDa peak that is significantly larger than the dimer peak of Arn-His.
Arn·H-NS Binding Is Affected by Salt Concentration and/or pH
To further understand the interaction between Arn and H-NS, we performed the His pulldown assay under different salt concentrations and pH values. We found that H-NS could be pulled down by Arn-His only in low salt conditions (Fig. 4A). When the NaCl concentration was increased to 200 mm, the interaction between these two proteins was disrupted. Ambient pH is another factor that modulates the formation of the Arn·H-NS complex. We performed a pH-dependent His pulldown assay using pH values ranging from 5.5 to 8.5, and the Arn-His protein level in each eluent was found to be similar (Fig. 4B). The Arn·H-NS complex was most stable at pH 7.5 without interference from the Tris and Hepes buffer systems. When the pH was lowered to 5.5, virtually no Arn·H-NS complexes were formed. Therefore, the stability of the Arn·H-NS complex is strongly influenced by increasing the salt concentration and suggests that ionic bonds play an important role in the interaction between the surfaces of these two proteins.
FIGURE 4.
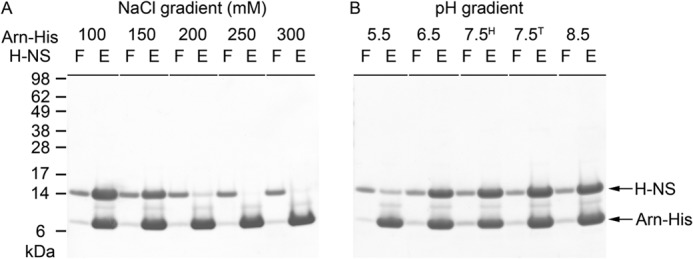
Characterizes the interaction between Arn-His and H-NS. A, the His pulldown assay under different salt concentrations, with NaCl ranging from 100 mm to 300 mm. F, flow-through; E, eluates from the nickel beads. B, the His-pulldown assay under different pH conditions, from pH 5.5 to 8.5. The buffers used in each experiment are as follows: citrate for pH 5.5, MES for pH 6.5, Hepes for pH 7.5H, Tris for pH 7.5T, and pH 8.5.
Arn Disrupts H-NS·DNA Binding
To verify that Arn serves as a DNA-like molecule and competes with the DNA binding site of H-NS, an EMSA was used. Previous studies had shown that several H-NS binding sequences occur in the promoter regions of many E. coli genes, such as the proV, virF, OriC, and fisP promoters and even the hns promoter (25, 26). Here we used a 32-mer DNA fragment, PHns-32, from the hns promoter region (5′-CAATTTTGAATTCCTTACATTCCTGGCTATTG-3′),which contains an H-NS DNA binding consensus sequence (underlined). The EMSA showed a significant band shift of PHns-32 dsDNA after the addition of 10 μm H-NS (Fig. 5A, left four lanes). In addition, H-NS·DNA complex formation could be disrupted by the addition of Arn-His, indicating that this competitive effect was dose-dependent (Fig. 5A, right five lanes). Furthermore, two other small and acidic proteins were used as control proteins: the histone binding DNA mimic protein Icp11 (9.2 kDa, pI 4.20) (7) and Neisseria NMO-0455 (accession number YP_003082676; 22.2 kDa, pI 4.85). Neither of these proteins affected the H-NS·DNA complex (Fig. 5B).
FIGURE 5.
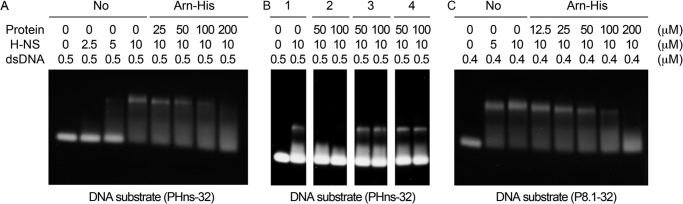
An EMSA of H-NS with the DNA mimic protein Arn-His as competition. A, the EMSA result shows that H-NS binds to its own promoter, the hns promoter (32-mer), and H-NS·DNA binding can be antagonized with the DNA mimic protein Arn-His. B, the EMSA result shows that the H-NS/DNA complex is broken only by the addition of Arn-His but not by another DNA mimic protein (i.e. Icp11) or another small and acidic protein (i.e. Neisseria NMO-0455). Group 1: without addition of DNA mimic or small and acidic protein in EMSA; group 2: with addition of Arn-His; group 3: with addition of Icp11; group 4: with addition of NMO-0455. C, H-NS also binds to the early gene promoter p8.1 of the T4 phage, and this DNA-binding property can be disrupted by Arn-His.
Subsequently we performed another EMSA using the DNA consensus sequence from T4 phage early promoters as the dsDNA substrate to validate the effect of H-NS on T4 phage DNA. The design was based on the strong T4 phage early gene promoter p8.1 (27). This 32-mer dsDNA (5′-ACGATAAAAAGTTGTTTACTTCCTCGGTTAGT-3′),called P8.1–32, contains the consensus sequence (underlined) and its flanking regions. The EMSA showed that the H-NS protein binds to the P8.1–32 dsDNA substrate (Fig. 5C, left three lanes), and the H-NS·DNA complex bands become progressively diminished with increasing concentrations of the Arn-His protein (Fig. 5C, right six lanes). These EMSA experiments demonstrated that the Arn protein competes with bacteria and phage DNA fragments for binding to H-NS, supporting the DNA mimic characteristic of the Arn protein.
A Proposed Binding Model Shows How Arn Interacts with the DNA Binding Domain of H-NS
To validate the Arn·H-NS complex, we docked the H-NS DNA binding domain (PDB codes 1HNR) to an Arn (PDB code 3WX4) dimer by ClusPro (28–31) and proposed a model of the Arn·H-NS complex to speculate on the DNA mimic surface of the Arn dimer. This model suggests that the Arn dimer utilizes its negative charge-dense side to interact with the H-NS DNA binding domain (Fig. 6A). The interface of the model shows that the three-dimensional shape and electrostatic surfaces of these two proteins are complementary to each other (Fig. 6B).
FIGURE 6.

A model of the Arn·H-NS complex by ClusPro docking. A, in this docking result of the Arn·H-NS complex, Arn is shown with its surface potential, and the H-NS DNA binding domain is shown in green. B, the interface of the Arn·H-NS complex is shown with the surface potential and presented in an open book view. On each interaction surface of the Arn monomer and the H-NS DNA binding domain, four regions are marked with circles that have either a specific complementary electrostatic attraction or a matching hydrophobic attraction.
The Arn Protein Neutralizes the Gene-silencing Effect of the H-NS Protein
H-NS serves as a bacterial defense against foreign DNA by its gene-silencing effect; thus, we performed an in vitro gene regulation assay (6). Using green fluorescent protein (gfp) as the reporter gene, we tested whether T4 phage Arn could neutralize the gene-repressing activity of H-NS. Prior to this assay we had confirmed that H-NS also targets the T7 promoter, and Arn can disrupt H-NS binding to the T7 promoter (Fig. 7). Thus, it is likely that H-NS directly affects GFP expression.
FIGURE 7.
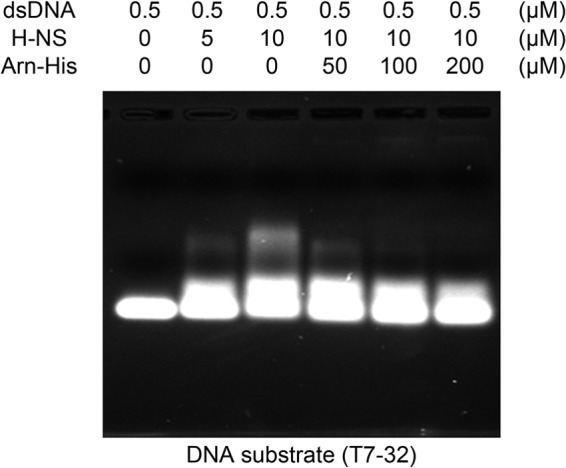
An EMSA of H-NS with the DNA mimic protein Arn-His shows competition even when the DNA template is the T7 promoter. The EMSA result shows that the T7 promoter region of the plasmid (T7–32) can also be bound by H-NS, and these band shifts can be disrupted by adding Arn protein. T7–32 is a 32-mer T7 promoter-containing DNA fragment (5′-CGCGAAATTAATACGACTCACTATAGGGGAAT-3′; the T7 promoter is underlined) and is the region on the plasmid used in in vitro translation assays.
The assay is described under “Experimental Procedures.” Upon the addition of 10 μm H-NS to the reaction mixture, the GFP expression level dropped to only 15.6% that of the original level (Fig. 8). As a control, the presence of 100 μm Arn-His alone had no significant effect on GFP expression (92.4% of the original level). However, in reactions containing 10 μm H-NS, the addition of 10 or 100 μm Arn-His reversed the gene-silencing effect of H-NS and partially restored the GFP expression levels to 37.73 and 51.45%, respectively (Fig. 8).
FIGURE 8.
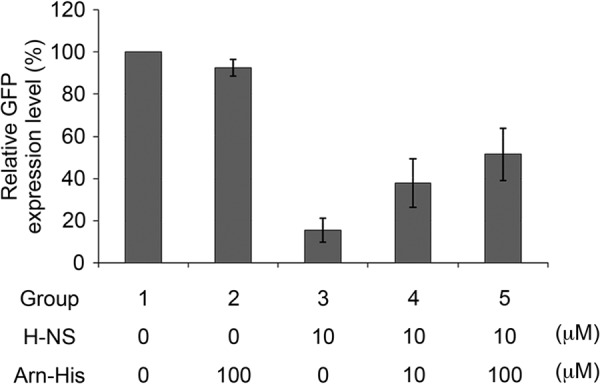
Analyzing the anti-gene silencing function of Arn by an in vitro gene expression assay. The relative levels of GFP expression were normalized to the control level of GFP expression (group 1), which was set as 100%. The experiments were repeated three times, and the corresponding S.D. are shown. An additional 100 μm Arn-His was added to group 2, and the relative GFP expression level was 92.40 ± 3.98%. Group 3 was analyzed with 10 μm H-NS, and the relative GFP expression level was 15.61 ± 5.69%. A total of 10 μm H-NS was added in groups 4 and 5 with an additional 10 and 100 μm Arn-His, respectively. The relative GFP expression levels of groups 4 and 5 were 37.73 ± 11.54% and 51.45 ± 12.36%, respectively.
Two proteins with low pI values, Icp11 (pI 4.2) and BSA (pI 4.9), were selected as additional controls. However, these proteins had no influence on gene regulation by H-NS (data not sown). These results demonstrate that Arn specifically antagonizes the gene-silencing effect of H-NS.
The Arn Protein Affects the Higher Order Structure of Plasmid DNA Induced by H-NS
Previous studies showed that H-NS binds DNA and forms a higher order structure, thereby silencing the expression of many genes. To investigate whether Arn disrupts the H-NS-induced higher order DNA·H-NS structure, we monitored the formation of plasmid DNA·H-NS complexes using electron microscopy (EM). When plasmid DNA was incubated with H-NS, H-NS induced a large amount of plasmid DNA to form supercoil-like structures (Table 3 and Fig. 9A), similar to previous observations (32). In contrast, the higher order plasmid DNA·H-NS structures disappeared with the addition of Arn at a level of 10 times the amount of H-NS, generating more relaxed DNA as the major DNA form (Table 3 and Fig. 9B). Taken together, these results support the H-NS-dependent, anti-host gene-silencing role of Arn.
TABLE 3.
Statistics of DNA plasmid forms
| EM sample | DNA forms |
Number of molecules | ||
|---|---|---|---|---|
| Supercoiled | Intermediatea | Relaxed | ||
| % | % | % | ||
| DNA + H-NS | 74.8 | 6.9 | 18.3 | 377 |
| DNA + H-NS + Arn-His | 31.5 | 21.0 | 47.5 | 238 |
a The intermediate form indicates that the structure of plasmid DNA contains partially ordered structures in some regions that are not similar to highly compact supercoiled or fully relaxed form plasmid DNA.
FIGURE 9.
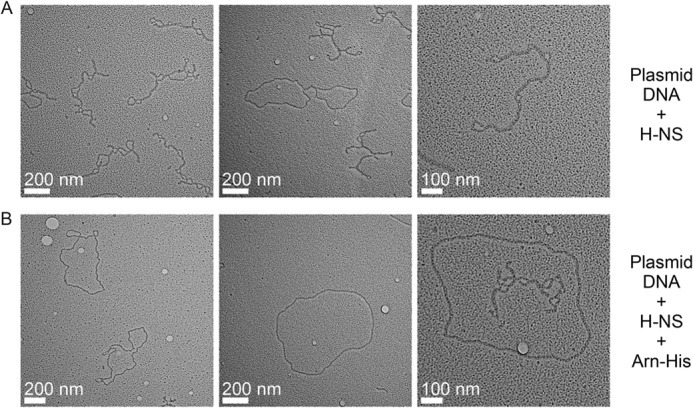
H-NS binding to plasmid DNA with or without Arn as viewed under an electron microscope. A, the plasmid DNA was mixed with H-NS. B, Arn was added to the plasmid DNA·H-NS mixture.
DISCUSSION
The functions of DNA mimic proteins are diverse and range from inhibiting DNA metabolic enzymes to regulating transcriptional factors (2). DNA mimic proteins (or domains) share some common features, namely their small size and acidity, but they have various shapes and negative charge distributions on their surfaces. Their functions depend on the DNA conformation and which part of DNA they mimic. For example, Ugi strongly inhibits uracil-DNA glycosylase (UDG); it mimics the DNA-phosphate backbone and has a uracil nucleotide recognition pocket at the transition state of UDG flipping uracil nucleotides in DNA (33). Ocr (21) and ArdA (34) are inhibitors of restriction enzymes and are roughly rod-shaped. Their negative charges are uniformly spread on the protein surface, and they mimic longer DNA substrates.
Interestingly, our newly identified T4 Arn dimer has a crescent-like shape that is similar to T7 Ocr. The negative charges of the Arn dimer cluster on one side of the surface, whereas the arrangement on Ocr dimer surfaces is uniform and reflects a DNA double-helix pattern (Fig. 1, A and B). These distinct negative charge distributions imply different binding modes between Arn and Ocr with their respective binding partners. In models of the DNA·M.EcoKI and Ocr·M.EcoKI complex (35), the clamp-like operation of M.EcoKI holds the DNA substrate as well as the double-helix negative charge arrangement of the Ocr dimer. Conversely, the binding model between the H-NS DNA binding domain and the Arn dimer suggests that the Arn dimer utilizes the side with a dense negative charge distribution (Fig. 6) to mimic the DNA-phosphate backbone interacting with the H-NS DNA binding domain, which presents a different binding fashion from that of the Ocr·M.EcoKI complex.
In addition to the H-NS/Arn binding model, we also proposed a McrB DNA binding domain·Arn complex by aligning the β-carbons of Asp and Glu on Arn and the DNA-phosphate groups on the McrB·DNA complex structure (Fig. 10) (36). Interestingly, Arn could replace the DNA substrate in the McrB·DNA complex structure, and the resulting model of McrB/Arn shows a good three-dimensional space matching between McrB and Arn (Fig. 10C). This model implies that Arn inhibits McrBC possibly by mimicking DNA with another structural attribute. Thus, our models provide a probable answer to how the DNA mimic protein Arn can be recognized by and inhibit two different categories of DNA-binding proteins: H-NS, a gene silencer, in translation events, and McrBC, a restriction enzyme, in DNA metabolic process.
FIGURE 10.
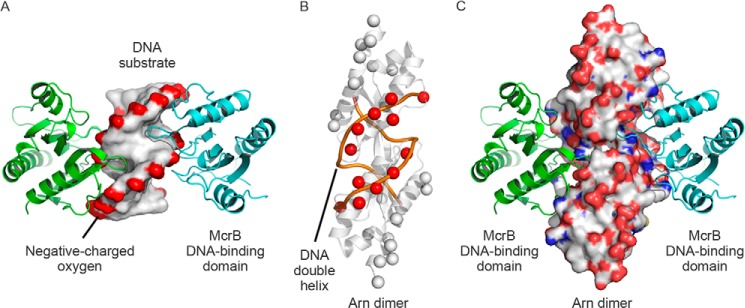
A proposed model of the Arn dimer·McrB DNA binding domain complex. A, the McrB DNA binding domain·DNA complex structure (PDB code 3SSC) contains two McrB DNA binding domains (shown in green and cyan) and a DNA substrate (shown as the surface). The complex structure revealed that the DNA-associating loops of two McrB DNA binding domains protrude into the DNA minor groove on two lateral parts of DNA. B, the β-carbons of Asp and Glu on Arn (showed as spheres) were manually aligned with the DNA-phosphate backbone (orange lines), and the DNA-matched β-carbons are highlighted in red. C, the Arn dimer in the Arn dimer·McrB DNA binding domain complex structure is shown in the surface mode and color-coded according to the atom type (white, carbon; red, oxygen; blue, nitrogen) to emphasize the three-dimensional space arrangement.
A newly identified Arn target, the bacterial histone-like protein H-NS, is known to be a global gene silencer as well as a bacterial defense protein that acts by inducing higher order H-NS·DNA structure. Similar to the histone proteins in eukaryotic cells, H-NS is involved in chromosome organization in bacteria (37–39) as it silences gene expression of certain loci without obvious specific recognition sites (40, 41). Previous studies also indicated that H-NS acts as a global repressor and regulates >200 genes that mostly respond to environmental change or bacterial virulence (42–45). Although H-NS binds DNA sequences promiscuously, further studies revealed that H-NS preferentially binds AT-rich sequences and represses the expression of the corresponding genes. Thus, H-NS is a xenogeneic DNA silencer and is involved in the bacterial defense system (22–24).
As a member of the anti-phage system, H-NS is a reasonable target for the phage anti-host system. For example, the product of the T7 phage 5.5 gene (gp5.5T7), the 5.5 protein, inhibits H-NS function by disrupting the higher order H-NS·DNA complex and thus blocks H-NS-mediated gene repression (46). H-NS may also target the T4 phage genome due to the low GC content (35%) of T4 genomic DNA compared with that of the E. coli host genome (∼50%) (47). The interaction between Arn and H-NS may, therefore, benefit T4 phage replication.
In summary, Arn is an anti-host defense system protein of the T4 phage with DNA mimic properties. Arn was first identified as an anti-restriction protein that acts by binding to and inhibiting restriction enzymes. Here we show that Arn also binds to H-NS to inhibit H-NS-induced gene silencing. Our findings show that T4 phage DNA encodes Arn as an anti-H-NS agent that participates in the phage-bacteria battle using a novel DNA mimic mechanism.
Acknowledgments
The heavy metal complex for phase determination, terpyridine-PtIICl2, was synthesized by Dr. Yan-Chung Lo. We thank Dr. Meng-Ru Ho in the Biophysics Core Facility at the Scientific Instrument Center at Academia Sinica for performing the AUC experiments. LC-MS/MS data were acquired at the Academia Sinica Common Mass Spectrometry Facilities located at the Institute of Biological Chemistry. X-ray diffraction data were collected on beamline BL13B1 at the National Synchrotron Radiation Research Center in Hsinchu, Taiwan and beamline BL12B2 at the Spring-8 Synchrotron Radiation Research Center in Hyogo, Japan. We are also grateful for the assistance of Dr. Li-Tzu Li and Hui-Ju Huang during preparation of the electron microscopy samples and for the use of the Tecnai F20 in the Cryo-EM Core Facility at the Scientific Instrument Center at Academia Sinica.
This work was supported by the Core Facilities for Protein Structural Analysis funded by National Core Facilities Program for Biotechnology (NSC 100-2325-B-001-029 and NSC 101-2319-B-001-003).
The atomic coordinates and structure factors (code 3WX4) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- AUC
- analytical ultracentrifugation.
REFERENCES
- 1. Dryden D. T., Tock M. R. (2006) DNA mimicry by proteins. Biochem. Soc. Trans. 34, 317–319 [DOI] [PubMed] [Google Scholar]
- 2. Putnam C. D., Tainer J. A. (2005) Protein mimicry of DNA and pathway regulation. DNA Repair 4, 1410–1420 [DOI] [PubMed] [Google Scholar]
- 3. Wang H. C., Ho C. H., Hsu K. C., Yang J. M., Wang A. H. (2014) DNA mimic proteins: functions, structures, and bioinformatic analysis. Biochemistry 53, 2865–2874 [DOI] [PubMed] [Google Scholar]
- 4. Wang H. C., Hsu K. C., Yang J. M., Wu M. L., Ko T. P., Lin S. R., Wang A. H. (2014) Staphylococcus aureus protein SAUGI acts as a uracil-DNA glycosylase inhibitor. Nucleic Acids Res. 42, 1354–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang H. C., Wu M. L., Ko T. P., Wang A. H. (2013) Neisseria conserved hypothetical protein DMP12 is a DNA mimic that binds to histone-like HU protein. Nucleic Acids Res. 41, 5127–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang H. C., Ko T. P., Wu M. L., Ku S. C., Wu H. J., Wang A. H. (2012) Neisseria conserved protein DMP19 is a DNA mimic protein that prevents DNA binding to a hypothetical nitrogen-response transcription factor. Nucleic Acids Res. 40, 5718–5730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang H. C., Wang H. C., Ko T. P., Lee Y. M., Leu J. H., Ho C. H., Huang W. P., Lo C. F., Wang A. H. (2008) White spot syndrome virus protein ICP11: a histone-binding DNA mimic that disrupts nucleosome assembly. Proc. Natl. Acad. Sci. U.S.A. 105, 20758–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dharmalingam K., Revel H. R., Goldberg E. B. (1982) Physical mapping and cloning of bacteriophage T4 anti-restriction endonuclease gene. J. Bacteriol. 149, 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim B. C., Kim K., Park E. H., Lim C. J. (1997) Nucleotide sequence and revised map location of the arn gene from bacteriophage T4. Mol. Cells 7, 694–696 [PubMed] [Google Scholar]
- 10. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 11. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 12. Cowtan K. D., Main P. (1996) Phase combination and cross validation in iterated density-modification calculations. Acta Crystallogr. D Biol. Crystallogr. 52, 43–48 [DOI] [PubMed] [Google Scholar]
- 13. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 14. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 15. Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. (2004) REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 16. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 17. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 18. DeLano W. L. (2008) The PyMOL Molecular Graphics System, Version 1.1 DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 19. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 20. Thresher R., Griffith J. (1992) Electron microscopic visualization of DNA and DNA-protein complexes as adjunct to biochemical studies. Methods Enzymol. 211, 481–490 [DOI] [PubMed] [Google Scholar]
- 21. Walkinshaw M. D., Taylor P., Sturrock S. S., Atanasiu C., Berge T., Henderson R. M., Edwardson J. M., Dryden D. T. (2002) Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell 9, 187–194 [DOI] [PubMed] [Google Scholar]
- 22. Ali S. S., Xia B., Liu J., Navarre W. W. (2012) Silencing of foreign DNA in bacteria. Curr. Opin. Microbiol. 15, 175–181 [DOI] [PubMed] [Google Scholar]
- 23. Navarre W. W., Porwollik S., Wang Y., McClelland M., Rosen H., Libby S. J., Fang F. C. (2006) Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238 [DOI] [PubMed] [Google Scholar]
- 24. Navarre W. W., McClelland M., Libby S. J., Fang F. C. (2007) Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 21, 1456–1471 [DOI] [PubMed] [Google Scholar]
- 25. Bouffartigues E., Buckle M., Badaut C., Travers A., Rimsky S. (2007) H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 14, 441–448 [DOI] [PubMed] [Google Scholar]
- 26. Lang B., Blot N., Bouffartigues E., Buckle M., Geertz M., Gualerzi C. O., Mavathur R., Muskhelishvili G., Pon C. L., Rimsky S., Stella S., Babu M. M., Travers A. (2007) High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 35, 6330–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sommer N., Salniene V., Gineikiene E., Nivinskas R., Rüger W. (2000) T4 early promoter strength probed in vivo with unribosylated and ADP-ribosylated Escherichia coli RNA polymerase: a mutation analysis. Microbiology 146, 2643–2653 [DOI] [PubMed] [Google Scholar]
- 28. Comeau S. R., Gatchell D. W., Vajda S., Camacho C. J. (2004) ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 32, W96–W99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Comeau S. R., Gatchell D. W., Vajda S., Camacho C. J. (2004) ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 20, 45–50 [DOI] [PubMed] [Google Scholar]
- 30. Kozakov D., Brenke R., Comeau S. R., Vajda S. (2006) PIPER: an FFT-based protein docking program with pairwise potentials. Proteins 65, 392–406 [DOI] [PubMed] [Google Scholar]
- 31. Kozakov D., Hall D. R., Beglov D., Brenke R., Comeau S. R., Shen Y., Li K., Zheng J., Vakili P., Paschalidis I. Ch., Vajda S. (2010) Achieving reliability and high accuracy in automated protein docking: ClusPro, PIPER, SDU, and stability analysis in CAPRI rounds 13–19. Proteins 78, 3124–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dame R. T., Wyman C., Goosen N. (2000) H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 28, 3504–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Putnam C. D., Shroyer M. J., Lundquist A. J., Mol C. D., Arvai A. S., Mosbaugh D. W., Tainer J. A. (1999) Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J. Mol. Biol. 287, 331–346 [DOI] [PubMed] [Google Scholar]
- 34. McMahon S. A., Roberts G. A., Johnson K. A., Cooper L. P., Liu H., White J. H., Carter L. G., Sanghvi B., Oke M., Walkinshaw M. D., Blakely G. W., Naismith J. H., Dryden D. T. (2009) Extensive DNA mimicry by the ArdA anti-restriction protein and its role in the spread of antibiotic resistance. Nucleic Acids Res. 37, 4887–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kennaway C. K., Obarska-Kosinska A., White J. H., Tuszynska I., Cooper L. P., Bujnicki J. M., Trinick J., Dryden D. T. (2009) The structure of M. EcoKI Type I DNA methyltransferase with a DNA mimic antirestriction protein. Nucleic Acids Res. 37, 762–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sukackaite R., Grazulis S., Tamulaitis G., Siksnys V. (2012) The recognition domain of the methyl-specific endonuclease McrBC flips out 5-methylcytosine. Nucleic Acids Res. 40, 7552–7562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Falconi M., Gualtieri M. T., La Teana A., Losso M. A., Pon C. L. (1988) Proteins from the prokaryotic nucleoid: primary and quaternary structure of the 15-kDa Escherichia coli DNA binding protein H-NS. Mol. Microbiol. 2, 323–329 [DOI] [PubMed] [Google Scholar]
- 38. Browning D. F., Grainger D. C., Busby S. J. (2010) Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 13, 773–780 [DOI] [PubMed] [Google Scholar]
- 39. Arold S. T., Leonard P. G., Parkinson G. N., Ladbury J. E. (2010) H-NS forms a superhelical protein scaffold for DNA condensation. Proc. Natl. Acad. Sci. U.S.A. 107, 15728–15732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dame R. T., Wyman C., Goosen N. (2001) Structural basis for preferential binding of H-NS to curved DNA. Biochimie 83, 231–234 [DOI] [PubMed] [Google Scholar]
- 41. Kahramanoglou C., Seshasayee A. S., Prieto A. I., Ibberson D., Schmidt S., Zimmermann J., Benes V., Fraser G. M., Luscombe N. M. (2011) Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 39, 2073–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ono S., Goldberg M. D., Olsson T., Esposito D., Hinton J. C., Ladbury J. E. (2005) H-NS is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem. J. 391, 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hansen A. M., Qiu Y., Yeh N., Blattner F. R., Durfee T., Jin D. J. (2005) SspA is required for acid resistance in stationary phase by downregulation of H-NS in Escherichia coli. Mol. Microbiol. 56, 719–734 [DOI] [PubMed] [Google Scholar]
- 44. Laaberki M. H., Janabi N., Oswald E., Repoila F. (2006) Concert of regulators to switch on LEE expression in enterohemorrhagic Escherichia coli O157:H7: interplay between Ler, GrlA, HNS, and RpoS. Int. J. Med. Microbiol. 296, 197–210 [DOI] [PubMed] [Google Scholar]
- 45. Stella S., Falconi M., Lammi M., Gualerzi C. O., Pon C. L. (2006) Environmental control of the in vivo oligomerization of nucleoid protein H-NS. J. Mol. Biol. 355, 169–174 [DOI] [PubMed] [Google Scholar]
- 46. Ali S. S., Beckett E., Bae S. J., Navarre W. W. (2011) The 5.5 protein of phage T7 inhibits H-NS through interactions with the central oligomerization domain. J. Bacteriol. 193, 4881–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zuber S., Ngom-Bru C., Barretto C., Bruttin A., Brüssow H., Denou E. (2007) Genome analysis of phage JS98 defines a fourth major subgroup of T4-like phages in Escherichia coli. J. Bacteriol. 189, 8206–8214 [DOI] [PMC free article] [PubMed] [Google Scholar]