Background: Chronic ethanol consumption induces pancreatic β-cell dysfunction and metabolic syndrome.
Results: Ethanol-induced Atf3 inhibits glucokinase transcriptional activity through direct binding or Atf3/Pdx-1/Hdac1 axis on glucokinase promoter.
Conclusion: ATf3 fosters β-cell dysfunction via Gck down-regulation and triggers T2D, which is ameliorated by in vivo Atf3 silencing.
Significance: The presented data uncover a new role for Atf3 as a potential therapeutic target in treating type 2 diabetes.
Keywords: Gene Expression, Gene Therapy, Glucokinase, Metabolic Disease, Pancreatic Islet, Type 2 Diabetes
Abstract
Chronic ethanol consumption induces pancreatic β-cell dysfunction through glucokinase (Gck) nitration and down-regulation, leading to impaired glucose tolerance and insulin resistance, but the underlying mechanism remains largely unknown. Here, we demonstrate that Gck gene expression and promoter activity in pancreatic β-cells were suppressed by chronic ethanol exposure in vivo and in vitro, whereas expression of activating transcription factor 3 (Atf3) and its binding to the putative Atf/Creb site (from −287 to −158 bp) on the Gck promoter were up-regulated. Furthermore, in vitro ethanol-induced Atf3 inhibited the positive effect of Pdx-1 on Gck transcriptional regulation, enhanced recruitment of Hdac1/2 and histone H3 deacetylation, and subsequently augmented the interaction of Hdac1/Pdx-1 on the Gck promoter, which were diminished by Atf3 siRNA. In vivo Atf3-silencing reversed ethanol-mediated Gck down-regulation and β-cell dysfunction, followed by the amelioration of impaired glucose tolerance and insulin resistance. Together, we identified that ethanol-induced Atf3 fosters β-cell dysfunction via Gck down-regulation and that its loss ameliorates metabolic syndrome and could be a potential therapeutic target in treating type 2 diabetes. The Atf3 gene is associated with the induction of type 2 diabetes and alcohol consumption-induced metabolic impairment and thus may be the major negative regulator for glucose homeostasis.
Introduction
Chronic ethanol consumption is well established as an independent risk factor for type 2 diabetes (T2D)2 and has been suggested to cause increased adiposity, impaired glucose homeostasis, and insulin resistance (1, 2). Several studies have demonstrated that light-to-moderate ethanol consumption is associated with a lower risk of T2D through increased glucose-stimulated insulin secretion and insulin sensitivity in peripheral tissues (3, 4). However, it is generally believed that the administration of moderate amounts of ethanol to rodents with metabolic disorders may increase the risk of complications in metabolic diseases or diabetes (5, 6). Based on several previous studies, pancreatic β-cells may be sensitive to ethanol-induced oxidative stress associated with the production of reactive oxygen species, which is one of the earliest events in glucose intolerance and is associated with mitochondrial dysfunction (7, 8). However, the mechanism by which chronic ethanol consumption induces pancreatic β-cell dysfunction and how such induced β-cell dysfunction contributes to the progression of T2D remain largely unknown.
Glucokinase plays a critical role in blood glucose homeostasis, serving as a glucose sensor for glucose-stimulated insulin secretion in pancreatic β-cells and as the major regulator of glucose uptake in hepatocytes (9, 10). Recently, we suggested that ethanol consumption may induce pancreatic β-cell dysfunction and apoptosis through Gck nitration and down-regulation, correlated with impaired glucose responsiveness and insulin resistance (11). Although we demonstrated that ethanol-induced Atf3 in pancreatic β-cells may function as an upstream regulator of Gck down-regulation, little is known about the exact role and regulatory mechanism of Atf3 in ethanol-induced Gck down-regulation.
Atf3, a member of the Atf/Creb family of transcription factors, regulates gene expression by binding to the consensus Atf/Creb cis-regulatory element via a basic leucine zipper domain (12). Given its frequent induction by various cellular stressors, ectopic expression of Atf3 in heart, liver, and pancreatic β-cells causes cardiac enlargement, liver or pancreatic β-cell dysfunction and apoptosis, impaired glucose metabolism, and diabetes (13). Although pancreatic duodenal homeobox-1 (Pdx-1) and sterol regulatory element-binding protein-1c directly bind to specific elements of the pancreatic and liver Gck promoter, respectively, and are positive regulators for Gck gene expression (14, 15), the relevant upstream activator or repressor regulators involved in Gck transcriptional regulation are little known. We previously suggested that lipotoxicity-induced Atf3 may be associated with the inhibition of Pdx-1-mediated transcriptional activity (16), but the precise action mechanisms of Atf3 are still not clear. Generally, transcription is regulated by various complex processes that require cooperation between transcription factors and co-activators or co-repressors that modulate histone structure (17). Histone modification via acetylation, phosphorylation, and methylation has been implicated in increased or decreased accessibility to transcription machinery, thereby leading to the repression or activation of gene expression (18). The β-cell-specific transcription factor Pdx-1 has been shown to interact with the histone acetyltransferase p300/Cbp, and this interaction has been demonstrated to be important for Gck-mediated insulin and Glut2 expression (19). We also demonstrated that endoplasmic reticulum stress-induced Atf3 inhibits Pdx-1-mediated PBE/Luc activity or Gck/Luc activity by inhibiting the interaction between Pdx-1 and p300, which leads to β-cell dysfunction (20). These results prompted us to investigate whether ethanol-induced Atf3 may also act as a transcriptional repressor of Gck gene expression via histone modification, leading to pancreatic β-cell dysfunction and apoptosis. This study provides molecular insight into the mechanism by which chronic ethanol-induced Atf3 inhibits the transcriptional activity of Gck through direct binding to the consensus Atf/Creb-binding site and the formation of an Atf3/Pdx-1/Hdac1/2 axis at the Gck promoter with the deacetylation of histone H3. Clear evidence for the amelioration of these events by Atf3 silencing using in vivo-jetPEI siRNA delivery system may offer a therapeutic strategy to combat the impaired glucose metabolism, insulin insensitivity, and hyperglycemia associated with chronic ethanol consumption.
EXPERIMENTAL PROCEDURES
Cell Lines and Isolated Islet Cells
Rat INS-1 pancreatic β-cells were cultured in RPMI 1640 medium containing 11.1 mm glucose supplemented with 10% fetal bovine serum, 2 mm glutamine, 1 mm sodium pyruvate, 55 μm β-mercaptoethanol, 10 mm HEPES, 100 IU/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). All experiments were performed between passage P4 and P20. Islet cells were isolated from overnight-fasted C57BL/6 mice using a previously described collagenase digestion technique (11). All antibodies were obtained from Cell Signaling Technology (Beverly, MA) or Santa Cruz Biotechnology Inc. (Santa Cruz, CA), and chemicals were purchased from Sigma.
In Vitro Treatment of Ethanol
To determine the concentrations and time points of ethanol showing positive effects on cell physiology (insulin content and ATP production) and the changes of phenotypes (apoptosis), we have treated the INS-1 cells or isolated islet cells with ethanol at the different concentrations (0, 25, 50, 100, and 200 mm) and time points (0, 6, 12, 24, and 48 h). Thereafter, the effective concentration (100 mm) and time (24 h) of ethanol, which show the most effective effects on the decrease of insulin content and ATP production and the increase of apoptosis, were selected. The treatment of ethanol with less than 50 mm or more than 100 mm did not have effective effects. Although there are still conflicting claims in the relative calculation between millimolar and in vivo percentage (%) of blood alcohol levels (22), several previous studies have shown that the selected 100 mm ethanol actually corresponds to about 0.46% (23), which can yield signs of intoxication in in vivo organs. The selected concentration (100 mm) and time (24 h) of ethanol is currently accepted and considered as an acute ethanol consumption in an in vitro model (24, 25). When cells were treated with 100 mm ethanol, the final media contained the volume of treated ethanol. However, when cells were treated with ethanol, alcohol exposure of cells may be hampered by evaporation of the alcohol. The fluctuation of alcohol concentration and ethanol effects on the cells was due to evaporation. To avoid this, investigators used settings where ethanol was added into the culture media and the cell culture plates were maintained for the entire duration of stimulation in a microclimate chamber at 37 °C with a gas mixture and an alcohol atmosphere (26).
Animals
C57BL/6J male mice (6 weeks old) originally purchased from The Jackson Laboratory (Bar Harbor, ME) were used in all experiments. Individually caged mice were placed on a Lieber-Decarli regular liquid diet (Dyets; control diet number 710027 or ethanol diet number 710260). Mice were pair-fed with the control versus 5% (v/v) ethanol diet for 8 weeks as reported previously (11). All animal experiments were conducted in accordance with guidelines from the Korean National Institute of Health Animal Facility.
Plasmids
Human wild-type ATF3 and ATF3(ΔC,101–181) with a C-terminal deletion and cDNA expression vectors were generous gifts from Dr. T. Hai (Ohio State University). ATF3(ΔN, 1–100) cDNA was amplified by PCR using the primer forward 5′-GGAATTCGCAAAGTGCCGAAACAAG-3′ and reverse 5′-ACGCGTCGACTTAGCTCTGCAATGTTCC-3′ and cloned into pcDNA3 or pEGFP-C2 (Invitrogen). Pdx-1 expression vector (pcDNA3/Pdx-1) was provided by Dr. T. Stein (Ohio State University). Rat Gck promoter reporter pRGP-1003/Luc, pRGP-404/Luc, pRGP-287/Luc, and pRGP-84/Luc were kindly provided by Dr. Y. Ahn (Yonsei University College of Medicine). pRGP-158/Luc vector, deletion mutant, or site-mutated plasmids were constructed in the pGL3-Gck/Luc plasmid by the two-step PCR method or QuikChange site-directed mutagenesis (Stratagene) according to the manufacturer's instructions. The used primer sets for each mutated construct are follows: pRGP-158/Luc, forward 5′-TTTCTCTATCGATAGGTACCGGTGACAGGAGTAGAGAG-3′ and reverse 5′-CCAAGCTGATCTCGAGCCCG-3′; mCREB1, forward 5′-CTGCTCCTTAGTAAGTGATACAGGCACTAAGGCAC-3′ and reverse 5′-GTGCCTTAGTGCCTGTATCACTTACTAAGGAGCAG-3′; mCREB2, forward 5′-GTAACAGGCACTAAGGCACAAACCTGGGAACTGAGCAG-3′ and reverse 5′-CTGCTCAGTTCCCAGGTTTGTGCCTTAGTGCCTGTTAC-3′; mCREB3, forward 5′-CACTGACCTGGGAACTGGCCAGGTGGTAATGTCTAC-3′ and reverse 5′-GTAGACATTACCACCTGGCCAGTTCCCAGGTCAGTG-3′; mCREB4, forward 5′-CTGGCAGTCACTGCAGATACAGGGTGACAGAGTG-3′ and reverse 5′-CACTCTGTCACCCTGTATCTGCAGTGACTGCCAG-3′; mCREB5, forward 5′-CACTGCAGTGACAGGGATACAGAGTGGTCCCATG-3′ and reverse 5′-CATGGTGACCACTCTGTATCCCTGTCACTGCAGTG-3′; mCREB6, forward 5′-CACTGCAGATACAGGGATACAGAGTGGTCACCAT-3′ and reverse 5′-ATGGTGACCACTCTGTATCCCTGTATCTGCAGTG-3′; mAP1, forward 5′-GTCTACCAGGCTGGCATACACTGCAGTGACAGGG-3′ and reverse 5′-CCCTGTCACTGCAGTGTATGCCAGCCTGGTAGAC-3′; mAP2, forward 5′-GACAGGGTGACAG AGTGTACACCATGGTGACAGGAG-3′ and reverse 5′-CTCCTGTCACCATGGTGTACACTCTGTCACCCTGTC-3′; mPdx-1, forward 5′-CTGCATGGTGGCTCTAGATATAGAATGGTCACCATAG-3′ and reverse 5′-CTATGGTGACCATTCTATATCTAGAGCCACCATGCAG-3′; ΔPdx-1, forward 5′-GTTTTCTGCATGGTGGAATGGTCACCATAGAAAC-3′ and reverse 5′-GTTTCTATGGTGACCATTCCACCATGCAGAAAAC-3′; and ΔATF, forward 5′-GGCTGGCAGTCACTGCAGTGGTCACCATGGTG-3′ and reverse 5′-CACCATGGTGACCACTGCAGTGACTGCCAGCC-3′.
Luciferase Activity Analysis
After transfection or treatment, the cells were lysed, and luciferase activity was measured using the Luciferase Assay System (Promega, Madison, WI).
RNA Interference and Transient Transfection
The human (sc-29757) or rat Atf3- (sc-72029) and mouse (sc-38761) or rat Pdx-1 (sc-10840) siRNAs were purchased from Santa Cruz Biotechnology. The cells were plated at 60–70% confluence and transfected with siRNA complexes or only with transfection reagents using Lipofectamine 2000 (Invitrogen) following the protocol recommended by the manufacturer.
RT-PCR and Quantitative RT-PCR
Total RNA was extracted from INS-1 cells and islet cells using TRIzol reagent (Invitrogen) and was subjected to reverse transcription using reverse transcriptase (Promega) at 42 °C for 1 h, and the resulting cDNA was amplified by PCR using gene-specific primers. The primers were designed using the Primer 3 program and cross-checked by a BLAST search of the NCBI database. The primer sets are as follows: mouse Gck (NM_010292), forward 5′-GTGCTGCTCAAGCTGGTAGA-3′ and reverse 5′-GCGATTTATGACCCCCGCTA-3′; rat Gck (NM_001270849), forward 5′-GACAGTC CTCACCTGCAACA-3′ and reverse 5′-GCATTTGTTGGTGCCCAGAG-3′; mouse Glut2 (NM_031197), forward 5′-ACCGGGATGATTGGCATGTT-3′ and reverse 5′-GAACACGTAAGGCCCAAGGA-3′; rat Glut2 (NM_012879), forward 5′-ACTGGCACATCCTACTTGGC-3′ and reverse 5′-CAGTCGACGCCTCTTCCTTT-3′; mouse Atf3 (NM_007498), forward 5′-GGAAAACTGGCTTCCTGTGC-3′ and reverse 5′-TGGCCATTGGACAACCTCAA-3′; rat Atf3 (NM_012912), forward 5′-CTCTAGCCGCTCTCTGGACC-3′ and reverse 5′-CGGCATTCACACTCTCCAGT-3′; mouse Gapdh (NM_008084), forward 5′-GGTTGTCTCCTGCGACTTCA-3′ and reverse 5′-TAGGGCCTCTCTTGCTCAGT-3′; and rat Gapdh (NM_017008), forward 5′-GGACCAGGTTGT CTCCTGTG-3′ and reverse 5′-ATTCGAGAGAAGGGAGGGCT-3′. The quantitative reverse transcriptase-PCR analysis was performed by using SYBR Green PCR Supermix (Bio-Rad) to detect the real time quantitative PCR products of reverse-transcribed cDNA according to the manufacturer's instructions (24). The primers are as follows: rat Gck, forward 5′-TCTACTTCCCCAACGACCCC-3′ and reverse 5′-GTTCATGTGCCCGTTGTGAG-3′; and rat Glut2, forward 5′-GTCCCAGTTTTCTGCAT GGT-3′ and reverse 5′-GAGTAGATGCCTCCCGTCAG-3′.
Electrophoretic Mobility Shift Assay (EMSA)
DNA mobility shift assays were performed as described previously (27). In brief, EMSA was performed in 20-μl volumes with 20 mm Tris-HCl, pH 7.9, 1.5% glycerol, 50 μg/ml BSA, 1 mm DTT, 0.5 mm phenylmethylsulfonyl fluoride, 2 μg of poly(dI-dC), 1 ng of 32P-labeled probe (5′-TGCAGTGACAGGGTGACAGA-3′), and 10 μg of nuclear extract. Reactions were incubated at 25 °C for 20 min and subsequently analyzed by electrophoresis through nondenaturing stock polyacrylamide gels (6 or 10%) in 0.5× TBE buffer containing 44.5 mm Tris-HCl, pH 8.2, 44.5 mm boric acid, and 1 mm EDTA. After the gel was pre-run at 100 V for 2 h, electrophoresis was performed at 270 V for 2 h at 4 °C. The gels were exposed to x-ray films using two intensifying screens at −70 °C.
Biochemical Analysis
Blood was collected by cardiac puncture from mice that had fasted overnight. Sera and relevant peripheral tissues were stored at −20 °C and used subsequently to assess the levels of triglycerides. Triglycerides were measured by colorimetric assays. For glucose tolerance test and insulin tolerance test, the mice were fasted at 8:00 a.m. for 6 h and then injected intraperitoneally with 1g/kg glucose or 1.5 units/kg regular human insulin, respectively. Blood samples were collected from the tail vein at time 0 (before injection), 30, 60, 90, and 120 min after injection, and the blood glucose level was measured using a portable glucose meter (Glucocard II Arkray, Kyoto, Japan) (11). The levels of insulin, ATP, and nitrite were also determined as described previously (11). Intracellular ROS generation was measured by using dichlorodihydrofluorescein diacetate (Molecular Probes, Eugene, OR) (11).
Immunoblots and Coimmunoprecipitation
Cells were lysed in RIPA buffer at 4 °C, vortexed, and then centrifuged at 16,000 rpm for 10 min at 4 °C. The supernatant was mixed in Laemmli loading buffer, boiled for 4 min, and then subjected to SDS-PAGE. For endogenous complexes, mitochondrial lysates (1 mg) fractionated from the cells were immunoprecipitated with 2 μg of antibody and immunoblotted.
Terminal Deoxynucleotidyltransferase-mediated dUTP Nick-end Labeling (TUNEL)
Apoptotic cells were detected using Apop Tag, an in situ apoptosis detection kit (Oncor, Gaithersburg, MD) as described previously (11).
Histopathology
For immunohistochemistry analysis, after pancreatic tissues were fixed, deparaffinized, and washed, sections were treated with diluted blocking serum for 20 min. Slides were incubated overnight at 4 °C in a humidified chamber with specific antibodies diluted in blocking serum. For immunocytochemistry analysis, the INS-1 cells were grown on glass coverslips for 24 h in a 6-well plate. After treatment, the cells were washed with cold PBS and fixed in paraformaldehyde (4% in PBS) for 15 min. All procedures were performed as described previously (11).
Chromatin Immunoprecipitation (ChIP)
The ChIP assays were performed according to the manufacturer's protocol (Upstate Biotechnology, Lake Placid, NY). In brief, after treatment, cells were cross-linked with 1% formaldehyde in medium for 15 min at 37 °C and then washed with ice-cold PBS and resuspended in 200 μl of SDS sample buffer containing protease inhibitor mixture. The suspension was sonicated 10 times for 30 s with a 1-min cooling period on ice between times and pre-cleared with 20 μl of protein A-agarose beads blocked with sonicated salmon sperm DNA for 30 min at 4 °C. The beads were removed, and the chromatin solution was immunoprecipitated overnight with anti-Atf3, Pdx-1, Stat5, Ac-K18 histone H3 (1766-1, Epitomics), Ac-K8 histone H4 (1796–1, Epitomics), and polymerase II (2035-1, Epitomics) antibodies at 4 °C, followed by incubation with protein A-agarose beads for an additional hour at 4 °C. The immunocomplexes were eluted with 100 μl of elution buffer (1% SDS and 0.1 mol/liter NaHCO3), and formaldehyde cross-links were reversed by heating at 65 °C for 6 h. Proteinase K was added to the reaction mixtures and incubated at 45 °C for 1 h. DNAs of the immunoprecipitates and control input DNAs were purified using a QIAquick PCR purification kit (Qiagen Inc.) and then analyzed by regular PCR using rat Gck promoter-specific primers. Alternatively, quantitative PCR was performed using the same primers in the presence of SYBR Green PCR Supermix (Bio-Rad) to detect the real time quantitative PCR products of chromatin immunoprecipitation samples according to the manufacturer's instructions (20, 28). The primer sequences for ChIP and qChIP are as follows: rat Gck promoter (−283 to −135), forward 5′-CGTAGAGGGCTCTGCTCCTT-3′ and reverse 5′-CAAAGGCCTCTCTACTCCTGTC-3′; rat Gck promoter (−127 to −28), forward 5′-GTCCCAGTTTTCTGCATGGT-3′ and reverse 5′-GAGTAGATGCCTCCCGTCAG-3′; and qChIP analysis of rat Gck promoter (−125 to +15), forward 5′-CCCAGTTTTCTGCATGGTGG-3′ and reverse 5′-CGAGCTCAGTCACAGTCGAT-3′.
In Vivo Atf3 Gene Silencing
Chemically synthetic siRNA against Atf3 was synthesized by Dharmacon RNA Technology. A commercially available cationic polymer transfection reagent (in vivo-jetPEITM, PolyPlus-Transfection, Illkirch, France) was used to deliver siRNA via intravenous injection (29–31). Briefly, 150 μg of siRNA diluted in 200 μl of 5% glucose solution was mixed with in vivo-jetPEI solution including transfection reagent jetPEI. The mixture was incubated for 15 min at room temperature to allow the complexes to form. This mixture was then injected into the ethanol-fed mice for 5 weeks once to six times with a 3-day interval between injections and continuously exposed to ethanol by 8 weeks. The scrambled siRNA solution mixed with in vivo-jetPEI solution served as a control siRNA. Three-day interval injection of siRNA was found to be optimum to sustain the effects of siRNA via preliminary experiments. The in vivo delivery efficiency of siRNA was assessed by performing RT-PCR and Western blotting in the removed tissues, lung, liver or pancreas, and isolated islet cells. The four siRNA candidates for mouse-specific Atf3 are as follows: Atf3 mouse-736, forward 5′-GCGGCGAGAAAGAAAUAATT-3′ and reverse 5′-UAAAGAGGUUCCUCUCGUCTT-3′; Atf3 mouse-587, forward 5′-GCAGAAAGAGUCAGAGAAATT-3′ and reverse 5′-UUUCUCUGACUCUUUCUGCTT-3′; Atf3 mouse-516, forward 5′-GCGGCGAGAAAGA AAUAAATT-3′ and reverse 5′-UUUAUUUCUUUCUCGCCGCTT-3′; and Atf3 mouse-356, forward 5′-CACCCUUUGUCAAGGAAGATT-3′ and reverse 5′-UCUUCCUUGACAAAGGGUGTT-3′.
Statistical Analysis
For comparing values obtained in three or more groups, one-factor analysis of variance was used, followed by Turkey's post hoc test, and p < 0.05 was taken to imply statistical significance.
RESULTS
Chronic Ethanol Consumption Represses Gck Transcriptional Activity in Pancreatic β-Cells
In agreement with previous reports (11), Gck expression was reduced in the liver and pancreas of ethanol-fed mice and in ethanol-exposed pancreatic β-cells, and this reduction was correlated with increased hepatic steatosis and reduced islet cell mass (Fig. 1, A–C). Similarly, mRNA levels (Fig. 1, D–F) of Gck and Glut2, a glucose transporter, and the Luc activity (Fig. 1, G and H) of rat pancreatic Gck promoter pRGP-1003/Luc were significantly reduced in islet cells or INS-1 cells exposed to 100 mm ethanol, which was ameliorated by 4-methylpyrazole, an inhibitor of cytochrome P4502E1 (32), or peroxynitrite scavengers such as l-NMMA, uric acid, and deferoxamine (11). These results suggest that ethanol metabolism and peroxynitrites play a critical role in ethanol-mediated reduction of Gck transcriptional activity.
FIGURE 1.
Down-regulation of Gck expression in pancreatic β-cells of ethanol-fed mice. 6-Week-old male C57BL/6J mice (n = 8, each group) were fed an ethanol diet for 8 weeks. A, liver and pancreas tissues of ethanol-fed mice were subjected to Western blotting for anti-Gck antibody. B, H&E staining (left, scale bars, 50 μm (liver) and 100 μm (pancreas)) and immunohistochemistry analysis for Gck (right, scale bars, 100 μm). C, INS-1 (upper panel) and isolated islet cells (lower panel) were treated with ethanol for the indicated time periods and subjected to Western blotting. D, RT-PCR in islet cells isolated from ethanol-fed mice. E and F, representative real time PCR analysis. INS-1 cells were treated with 100 mm ethanol in the presence or absence of 100 μm 4-methylpyrazole (E) or l-NMMA (NMMA) (100 μm), uric acid (UA) (100 μm), or deferoxamine (DFO) (50 μm) (F) and subjected to real time-PCR analysis (*, p < 0.01; **, p < 0.05). Sal, saline. G and H, Gck promoter activity. Twenty four hours after transfection, INS-1 cells were treated with 50 or 100 mm ethanol for 24 h in the presence or absence of 4-methypyrazole (G) and l-NMMA, uric acid, or deferoxamine (H), and then luciferase activity was measured (*, p < 0.01; **, p < 0.05). The data were normalized to β-galactosidase activity. All data are representative of three independent experiments. CTL, control.
To determine the region of Gck promoter crucial for ethanol-mediated Gck down-regulation, we overexpressed rat Gck promoter constructs with 5′-serial deletions and analyzed their responsiveness to ethanol treatment in INS-1 cells (Fig. 2A). Ethanol treatment significantly decreased the Luc activity of the Gck promoter between −1003 and −287 bp, although we did not detect significant reductions between −158 and −84 bp, suggesting that the consensus binding site for a repressive transcription factor induced by ethanol was located between −287 and −158 bp. These results were confirmed using the pRGP-287/Luc and pRGP-158/Luc constructs (Fig. 2B). Also, we know that de novo protein synthesis as a repressive transcriptional factor is required for ethanol-repressed Gck transcriptional activity using cycloheximide, a de novo protein synthesis inhibitor (Fig. 2, C and D).
FIGURE 2.
De novo protein synthesis is required for ethanol-reduced Gck expression. A, INS-1 cells were transfected with constructs containing various lengths of serial deletions of the 5′-flanking region on the rat glucokinase promoter and then treated with 100 mm ethanol. After treatment for 24 h, Gck promoter activities were measured (*, p < 0.01; **, p < 0.05). B, Gck-Luc activity. Cells transfected with pRGP-287/Luc or pRGP-158/Luc construct were treated with ethanol and/or l-NMMA (NMMA) (*, p < 0.01; **, p < 0.05). CTL, control. C, stability of Gck mRNA. INS-1 cells were pretreated with cycloheximide (CHX) (10 μm) and then treated with ethanol. The samples were subjected to RT-PCR. D, cycloheximide inhibits ethanol-reduced Gck promoter activity (*, p < 0.01; **, p < 0.05). All data are representative of three independent experiments.
Ethanol-enhanced Atf3 Plays a Critical Role in the Down-regulation of Gck Transcriptional Activity
To understand the molecular mechanisms by which ethanol inhibits Gck promoter activity, we investigated how Gck promoter activity is regulated by searching a 129-bp consensus sequence between −287 and −158 bp of the 5′-flanking region of the Gck promoter using the TRANSFAC transcription factor binding database. Illustrated in Fig. 3A, five potential binding sites for Atf/Creb that contained the TAAC, TGAA, TGAG, or TGAC element, in addition to two binding sites for Ap-1 (GTCA), were found in the Gck promoter region (Fig. 3A). Because stress-inducible Atf3 is still considered as a dual-face transcription factor that activates or represses a gene expression (13), we first determined the role of Atf3 in repressing Gck transcriptional activity. Gck expression and the pRGP-1003/Luc or pRGP-287/Luc promoter activities were decreased by Atf3 overexpression but not by Atf3 siRNA or C-terminal domain-deleted ATF3(ΔC) (Fig. 3, B–F). So, we wondered whether ethanol exposure affects Atf3 gene expression in vivo and in vitro (Fig. 3, G and H). Atf3 was strongly increased in pancreatic tissue and islet cells of ethanol-fed mice or ethanol-treated INS-1 cells, indicating that de novo protein synthesis of Atf3 protein by ethanol may act as a negative transcription regulator to reduce Gck gene expression.
FIGURE 3.
Ethanol-mediated down-regulation of Gck gene expression depends on Atf3 induction. A, schematic representation of rat Gck promoter (−287/−158). The putative binding sites for Atf/Creb and AP-1 are underlined. Their putative binding sites were searched using the TRANSFAC transcription factor binding database. B, RT-PCR analysis after transfection with ATF3 cDNA into the INS-1 cells. CTL, empty vector-transfected cells. C, Atf3 inhibits Gck promoter activity. INS-1 cells were cotransfected with pRGP-1003/Luc or pRGL-287/Luc vectors and Atf3 or Atf3 siRNA (*, p < 0.01; **, p < 0.05). D, RT-PCR analysis in the full-length (FL) ATF3, N-terminal domain-deleted (ΔN) or C-terminal domain-deleted ATF3 (ΔC)-transfected cells (*, p < 0.01; **, p < 0.05). E, RT-PCR analysis. INS-1 cells were transfected with ATF3 cDNA or Atf3 siRNA and then exposed to 100 mm ethanol (E) (*, p < 0.01; **, p < 0.05). F, Gck-luciferase activity. Atf3 potentiates ethanol-reduced pRGP-1003/Luc activity (*, p < 0.01; **, p < 0.05; &, p < 0.005). G and H, Atf3 expression in pancreas tissues and islet cells isolated from ethanol-fed mice (G) or ethanol-treated INS-1 cells (H). The results represent the average ± S.E. from three independent experiments. CTL, control.
Atf3 Directly Binds to the Putative Atf/Creb Site on the Gck Promoter
Fig. 4A shows that ATF3 overexpression repressed Gck transcriptional activity through a direct interaction with the consensus binding sites of Atf/Creb located between −287 and −158 bp of the Gck promoter. The specific role of Atf3 within the proximal −287-bp region of the Gck promoter was confirmed by cotransfection with ATF3 or inactive ATF3(ΔC) and pRGP-287/Luc or pRGP-158/Luc (Fig. 4B). The pRGP-287/Luc activity was significantly decreased by Atf3, but there was a small reduction by Atf3 in pRGP-158/Luc-transfected cells. A TRANSFAC search revealed that the pRGP-158/Luc promoter also has two potential binding sites for Atf/Creb (TGAC) and one for Ap-1 (GTCA), but mutations of their consensus elements were not responsive to Atf3 (data not shown). Therefore, to confirm the functional activity of putative binding sites for Atf3 in pRGP-287/Luc, plasmids containing the putative binding sites were reconstructed by point mutagenesis (Fig. 4C). As shown in Fig. 4D, only pRGP-287(mCreb6)/Luc, which contains point mutations at the neighboring sites for mCreb4 and mCreb5, did not respond to ATF3 overexpression or ethanol treatment, whereas other constructs containing potential Atf3-binding site mutations still responded to them, suggesting that Creb4 and Creb5, located between −185 and −174 bp of the Gck promoter, are essential for the inhibitory responsiveness of ethanol-induced Atf3. These are also confirmed by electrophoretic mobility shift assays (EMSAs) using a labeled oligonucleotide covering the −190 to −171-bp region (Creb4/5(W)) in cells overexpressing ATF3, ATF3(ΔC), or ATF3(ΔN) (Fig. 4E). Binding complexes were observed in nuclear extracts of ATF3- or ATF3(ΔN)-transfected cells but not in those of ATF3(ΔC)-transfected cells. Addition of an excess amount of the unlabeled competitor Creb4/5(W) (50- or 100-fold) completely blocked the binding of Atf3 to the Gck promoter. To increase the specificity of the −190 to −171-bp region, three 20-bp wild-type oligonucleotides (Ap-1(W), Creb1(W), and Creb2/3(W)) that cover the putative binding site containing the sequence of response elements for Ap-1 or Atf/Crebs, respectively, and the mutated oligonucleotide Creb4/5(M), originating from wild-type Creb4/5(W), were designed (Fig. 4G). A 100-fold molar excess of unlabeled oligonucleotide for wild-type Creb4/5(W) (2nd lane) completely abolished the formation of the binding complex in nuclear extracts of ethanol-treated cells, whereas any oligonucleotides for mutated Creb4/5(M), including wild-type Ap-1(W), Creb1(W), and Creb2/3(W), did not compete (Fig. 4F). The specificity of Atf3 in the binding complexes was also confirmed in the results of a supershift assay using an anti-Atf3 antibody. The Atf3-DNA binding was much stronger in ethanol-treated INS-1 cells (no competitor lane) than those in nontreated cells (control). The recruitment of Atf3 to the Gck promoter was confirmed in vivo by chromatin immunoprecipitation. A 149-bp PCR product covering the putative Atf/Creb (from −283 to −135) of Gck promoter was detected in the nuclear extracts of ethanol-treated cells, but not in untreated cells, immunoprecipitated with the anti-Atf3 antibody (Fig. 4, H and I). There was no binding in chromatin immunoprecipitated with the anti-Stat5 antibody. As well, chromatin binding in anti-Atf3 immunoprecipitates was significantly increased in Atf3-transfected or ethanol-treated cells, which was strongly decreased by Atf3 siRNA (Fig. 4J), indicating that Atf3 is specifically recruited to Atf/Creb binding region of the Gck promoter to result in a decrease of its transcriptional activity in ethanol-treated cells.
FIGURE 4.
Atf3 directly binds to the putative Atf/Creb site in the Gck promoter. A, serially deleted constructs of Gck promoter were cotransfected with ATF3 and measured luciferase activity (*, p < 0.01; **, p < 0.05). B, Gck-Luc activities were measured in cells cotransfected with pRGP-287 or pRGP-158 and ATF3 or ATF3(ΔC) (*, p < 0.01; **, p < 0.05). C, point-mutated sequences of putative Atf3-binding sites in the Gck promoter. D, effects of Atf3 or ethanol on the activities of point-mutated pRGP-287/Luc promoter constructs (*, p < 0.01; **, p < 0.05). E and F, after transfection or ethanol treatment, EMSA was performed using Creb4/5(W) oligonucleotide. NE, nuclear extracts; Ab, antibody. G, oligonucleotides for EMSA and competition experiments. H, diagram showing the region between −283 and −135 of Gck promoter in the ChIP assay. I and J, ChIP assays. After transfection or treatment, immunoprecipitated (IP) DNA was amplified by PCR using the specific primers covering the Atf/Creb-binding sites (from −283 to −135). The results represent the average ± S.E. from three independent experiments. CTL, control.
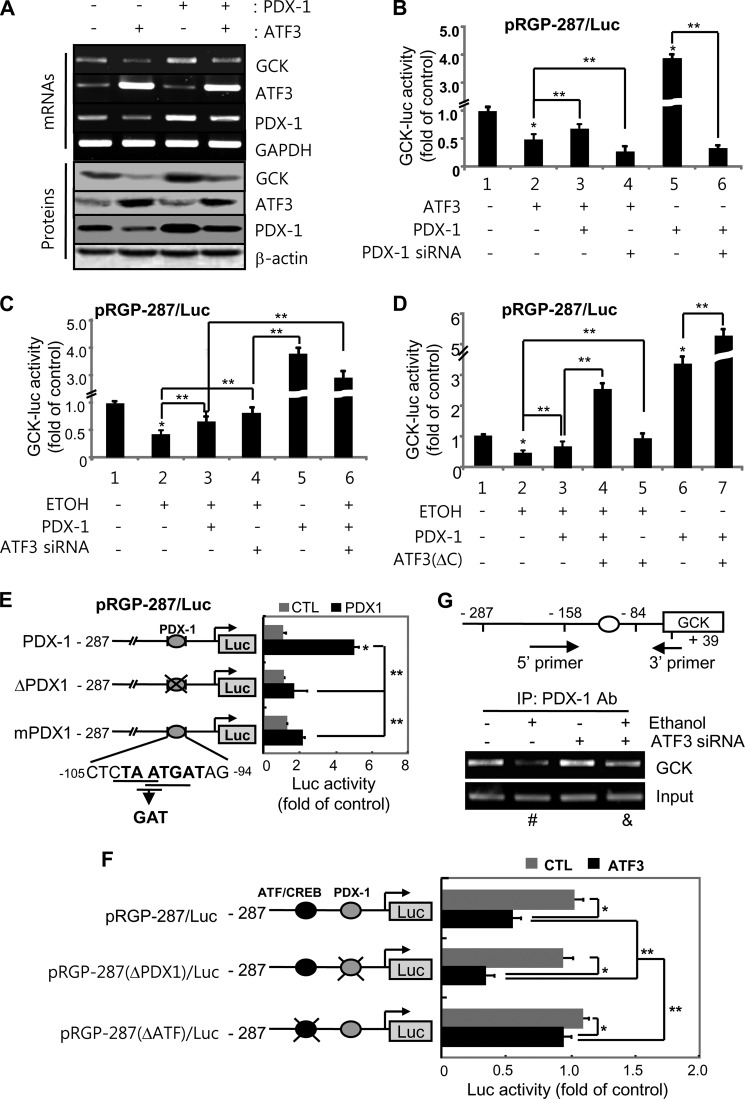
Ethanol and Atf3 Counteract the Positive Effect of Pdx-1 on Gck Transcriptional Regulation
Although the effects of Pdx-1 on Gck expression are controversial (33), our data show the functional significance of the inter-regulation between Atf3 and Pdx-1 in Gck expression (Fig. 5A). The reduction of the Gck promoter activity by Atf3 or ethanol was potently increased by Pdx-1 depletion (Fig. 5B), whereas the recovering effects of Pdx-1 were potentiated by Atf3 silencing or ATF3(ΔC) (Fig. 5, C and D), suggesting that Pdx-1 may counteract the repressive effects of Atf3 or ethanol on Gck transcriptional activity. Two putative Pdx-1-binding sites, TAAT and TGAT from −105 to −94 bp, were 72 bp from the core Atf/Creb binding sequence in the pRGP-287/Luc reporter (Fig. 5E). To determine whether these sites include the binding region for Pdx-1, we generated constructs via the deletion (−105/−94 bp, ΔPdx-1) or point mutation (ATG→GAT, mPdx-1) of putative binding sites for Pdx-1 in the pRGP-287/Luc-promoter. Pdx-1-enhanced pRGP-287/Luc activity was significantly decreased in pRGP-287(ΔPdx-1)/Luc or pRGP-287(mPdx-1)/Luc-transfected cells, indicating that the conserved binding site of Pdx-1 is located between −105 and −94 bp of the Gck promoter. The counteracting roles of Atf3 and Pdx-1 on Gck transcriptional regulation were further confirmed by using the construct in which Atf3-binding sites in Gck promoter are deleted (−190/−171, ΔAtf; Fig. 5F). The reduction of its activity by Atf3 was potentiated by the deletion of the Pdx-1-binding site (ΔPdx-1), which was rescued by the deletion of the Atf3-binding site, indicating that Atf3 and Pdx-1 may have opposite effects on Gck transcriptional activity through a direct interaction. Next, we hypothesized that the negative effect of ethanol on Pdx-1-mediated Gck transcriptional activation may be caused via the up-regulation of Atf3, which prevents Pdx-1 recruitment to its putative binding site in the Gck promoter. To this, cells exposed to ethanol in the presence of Atf3 siRNA were assayed by ChIP analysis using primer sequences covering the Pdx-1-binding site (from −127 to −28). The binding of Pdx-1 to the putative binding region of the Gck promoter was reduced in ethanol-treated cells but not in Atf3 siRNA (Fig. 5G) or ATF3(ΔC)-transfected cells (data not shown), suggesting that induced Atf3 may indirectly suppress Gck transcriptional activity by preventing Pdx-1 recruitment to its putative binding site.
FIGURE 5.
Atf3 counteracts Pdx-1-mediated Gck transcriptional activity. A, INS-1 cells were transfected with Pdx-1 and/or ATF3 and subjected to the semiquantitative RT-PCR (upper panel) and Western blotting (bottom panel). B–D, Pdx-1 restored Atf3 or ethanol-reduced pRGP-287/Luc activity. Twenty four hours after transfection with Pdx-1 siRNA (B), Atf3 siRNA (C), or ATF3(ΔC) (D), the luciferase activity was measured (*, p < 0.01; **, p < 0.05). E, identification of Pdx-1-binding sites on pRGP-287/Luc promoter. Deletion- or point-mutated constructs of putative Pdx-1-binding sites within −105 and −94 region of pRGP-287/Luc vector were generated and transfected into the cells (*, p < 0.01; **, p < 0.05). F, ATF3 cDNA was cotransfected into the cells together with pRGP-287(ΔPdx-1)/Luc or pRGP-287(ΔAtf)/Luc vector and measured the luciferase activity (*, p < 0.01; **, p < 0.05). G, ChIP assay for Pdx-1 binding to the Gck promoter. The immunoprecipitated (IP) DNAs were used as templates for PCR with primers covering Pdx-1-binding sites from −127 to −28 bp. The results represent the average ± S.E. from three independent experiments. Ab, antibody; CTL, control.
Ethanol-induced Atf3 Interacts with Pdx-1 and Subsequently Promotes the Recruitment of Hdac1/2 and Histone H3 Deacetylation on Gck Promoter
Next, to examine whether Atf3 may inhibit Pdx-1-mediated Gck transcription by decreasing histone acetylation on the Gck promoter, we have quantified the amount of Gck gene promoter associated with acetylated histone H3 or H4. To this, ChIP analysis was performed by using the primers covering Pdx-1-binding sites from −127 to −28 bp. As expected, histone H3 acetylation on the Gck promoter was lower in ethanol-treated (Fig. 6A) or ATF3-overexpressed cells (Fig. 6B), correlated with reduction in p300 binding, whereas the recruitment of histone deacetylase, Hdac1 and -2, to the Pdx-1 binding region of Gck promoter was increased in ethanol-treated cells, which were markedly attenuated and reversely rescued by Atf3 depletion or trichostatin A (TSA), an inhibitor of histone deacetylase (27). Concomitantly, Gck mRNA was significantly decreased by ethanol or ATF3 overexpression, which was restored by TSA or Atf3 siRNA (Fig. 6C). Also, quantitative PCR using ChIP samples revealed that ethanol-induced Atf3 directly binds to the Gck promoter despite no Atf3-binding sites, whereas recruitment of Pdx-1, Ac-H3, and RNA polymerase II to the promoter was significantly reduced in ethanol-treated cells, which were reversely attenuated by Atf3 depletion (Fig. 6D), demonstrating the specificity of their recruitment by Atf3. Additionally, the inhibition of histone deacetylase activity by TSA may partially restore the recruitment of Pdx-1, Ac-H3, and polymerase II to the Gck promoter after ethanol treatment, although there was no impact on Atf3 recruitment (Fig. 6E), indicating that Atf3 may be an upstream regulator of Hdac1/2 activation.
FIGURE 6.
Atf3 recruitment inhibits histone H3 acetylation and induces sequential recruitment of Hdac1/2. A and B, ChIP assays were performed in ethanol-treated cells (A) and ATF3-transfected cells (B) exposed to Atf3 siRNA or TSA (10 nm), respectively. The immunoprecipitated DNA was amplified by PCR using the specific primers covering the Pdx-1-binding sites. C, quantitative RT-PCR analysis for Gck expression (*, p < 0.01; **, p < 0.05). D and E, ChIP analyses for Atf3, Pdx-1, Ac-H3, Ac-H4, or polymerase II (Pol II) in cells treated with ethanol in the presence or absence of Atf3 siRNA (D) or TSA (E). The immunoprecipitated chromatin was analyzed by quantitative PCR (*, p < 0.01; **, p < 0.05). The results represent the average ± S.E. from three independent experiments. CTL, control.
Atf3 Attenuates Pdx-1-dependent Gck Transcriptional Activity by Enhancing the Physical Interaction with Hdac1
We next investigated whether ethanol-induced Atf3 could inhibit Pdx-1-dependent Gck transcriptional activity by enhancing the activity of histone deacetylase (Fig. 7A). The increase of pRGP-158/Luc activity by Pdx-1 was significantly decreased by ethanol treatment or ATF3 overexpression, which was restored by TSA or Atf3 siRNA, indicating that the activated histone deacetylase may play an essential role in the inhibitory effects of Atf3 on Pdx-1-mediated Gck transcriptional activity. Also, ethanol-induced Atf3 inhibits Pdx-1-mediated Gck transcriptional activity by triggering a physical in vitro and in vivo interaction of Pdx-1 with Hdac1 rather than p300 (Fig. 7, B and C). Immunoprecipitated Pdx-1 directly interacted with endogenous Atf3 induced by ethanol or overexpressed GFP-ATF3 but not with GFP-ATF3(ΔC). Conversely, the interaction of p300 with Flag-Pdx-1 was significantly decreased by ethanol treatment or ATF3 overexpression. Additionally, Gck down-regulation in ethanol-treated β-cells was correlated with the induction of Atf3 and the reduction of nuclear localization of Pdx-1, which were reversely rescued by Atf3 siRNA (Fig. 7D).
FIGURE 7.
Atf3 inhibits Gck transcriptional activity via physical interaction with HDAC1/Pdx-1. A, Gck-Luc activity. Atf3 inhibits Pdx-1-mediated pRGP-158/Luc promoter activity, which is rescued by TSA or Atf3 siRNA (*, p < 0.01; **, p < 0.05). B, interaction assay for Flag-Pdx-1. After ethanol treatment into the cells cotransfected with Flag-Pdx-1, GFP-ATF3, or GFP-ATF3(ΔC), the lysates were immunoprecipitated (IP) with FLAG antibody (Ab) and subjected to Western blotting. C, in vivo binding assay for endogenous Pdx-1. INS-1 cells were treated with 100 mm ethanol in the presence or absence of Atf3 siRNA, and then we performed the interaction assay. D, immunocytochemistry analyses for Gck, Atf3, Pdx-1, and insulin were performed in the cells treated with ethanol and/or Atf3 siRNA (scale bars, 100 μm). The results represent the average ± S.E. from three independent experiments.
In Vivo Atf3 Silencing Ameliorates Metabolic Syndrome and β-Cell Damage in Ethanol-fed Mice
To confirm the specific upstream role of Atf3 on ethanol consumption-induced Gck down-regulation and metabolic syndrome, a loss-of-function study was performed through in vivo Atf3 siRNA injection. Among four siRNA candidates, we selected 356-Atf3 siRNA, which had the highest efficiency for Atf3 silencing in various tissues (Fig. 8, A and B). As expected, the impaired glucose tolerance (GTT and OGTT; Fig. 8, C–E) and insulin tolerance (ITT; Fig. 8F) induced by ethanol consumption were markedly inhibited by the administration of Atf3 siRNA. Triglyceride accumulation and reduced plasma insulin levels in ethanol-fed mice were also attenuated by Atf3 knockdown (Fig. 8, G and H), correlated with restoring the reduced ATP (Fig. 8I). Similarly, the reduction in glucose-stimulated insulin secretion (GSIS; Fig. 8J) and the production of nitrite or ROS (Fig. 8K) in islet cells of ethanol-fed mice also depended on the presence of Atf3. Together with the amelioration of pancreatic β-cell dysfunction, in vivo Atf3 silencing inhibited ethanol-induced β-cell apoptosis, which was associated with the restoration of Gck, insulin, and Pdx-1 expression (Fig. 8, L and M). These results suggest that the Atf3 gene is associated with the induction of T2D and alcohol consumption-induced metabolic impairment and thus may play a major negative regulator for glucose homeostasis.
FIGURE 8.
In vivo Atf3 knock-out ameliorates the impairment of glucose metabolism and β-cell damage in ethanol-fed mice. Male 6-week-old C57BL/6J mice (n = 8, each group) were fed an ethanol diet, and after 2 weeks, the mice were intravenously injected with Atf3 siRNA six times with a 3-day interval between injections, then maintained for 14 weeks, and sacrificed. A, expression of Atf3 mRNA and proteins by RT-PCR (left panel) and Western blotting (right panel) in the tissues of lung and liver of the sacrificed mice. B, in vivo silencing of Atf3 induced the increase of Gck expression down-regulated in ethanol-fed mice. RT-PCR (upper panel) and Western blotting (lower panel) were performed in pancreas tissues (left panel) and isolated islet cells (right panel) of Atf3 siRNA-injected mice. C and D, Atf3 silencing improves glucose tolerance (C). Areas under the curve (D) are shown. *, p < 0.01; **, p < 0.05. E, oral glucose tolerance test (OGTT, 1g/kg; *, p < 0.01; **, p < 0.05). F, insulin tolerance (ITT; *, p < 0.01; **, p < 0.05). G, plasma triglyceride levels (*, p < 0.01; **, p < 0.05). H, plasma insulin levels (*, p < 0.01; **, p < 0.05). I, cellular ATP production in islet cells isolated from each group mice (*, p < 0.01; **, p < 0.05). J, glucose-stimulated insulin release was measured in islets exposed to 5.5 and 16 mm/liter glucose. GSIS, glucose-stimulated insulin secretion. *, p < 0.01; **, p < 0.05. K, NO and ROS production (*, p < 0.01; **, p < 0.05). L, apoptosis-related protein expression. M, TUNEL assay in isolated islet cells (top panel, ×100). Casp., caspase. TUNEL-positive apoptotic cells were quantified (bottom panel, *, p < 0.01; **, p < 0.05). The results represent the average ± S.E. from three independent experiments.
DISCUSSION
In this study, we provide evidence that ethanol-induced Atf3 suppresses Gck gene expression through direct interaction with the Atf/Creb-binding site (−185/−174) of the Gck promoter. Additionally, Atf3 directly interacts with Pdx-1 and specifically recruits Hdac1/2 to the Gck promoter in ethanol-treated β-cells, thereby repressing functional Pdx-1 on Gck transcriptional regulation. In vivo Atf3 silencing ameliorates ethanol-mediated metabolic syndrome and pancreatic β-cell dysfunction. All of these findings are summarized in Fig. 9.
FIGURE 9.
Proposed model by which Atf3 induces Gck down-regulation and metabolic syndrome in chronic ethanol-fed mice. A, in normal conditions Pdx-1 interacts with p300 and binds to the putative binding site of the Gck promoter, followed by the acetylation of histone H3 and H4, and then activates Gck transcription. B, ethanol induces Atf3 via the production of peroxynitrite and ROS, which depends on ethanol metabolism. C, ethanol-induced Atf3 directly binds to Atf/Creb-binding site of Gck promoter and then inhibits Gck transcription. D, as well as the direct binding of Atf3 to the Atf/Creb-binding site of the Gck promoter, ethanol-induced Atf3 also facilitates ethanol-reduced Gck transcriptional activity via the direct interaction with Pdx-1 and the recruitment of Hdac1/2 rather than p300. E, in vivo Atf3 silencing ameliorates Gck down-regulation and metabolic syndrome induced by ethanol-induced Atf3 (red).
This study demonstrates a functional role of Atf3 as a major upstream regulator that negatively regulates Gck transcriptional activity in ethanol-fed mice, based on the following observations. (i) The reduction of Gck transcriptional activity by ethanol was potentiated by Atf3, which was inhibited by Atf3 depletion. (ii) Two putative Atf3-binding sites that are essential for the inhibitory responsiveness by Atf3 were identified between −185 and −174 bp of the Gck promoter. (iii) Atf3 also binds to the Pdx-1-binding sites between −102 and −92 of the Gck promoter, resulting in chromatin remodeling via histone H3 deacetylation.
Several studies have suggested that Pdx-1 is a β-cell master gene that regulates the expression of β-cell-specific genes such as insulin, Glut2, and Gck (34–36). Although we previously suggested that activated Atf3 directly interacted with Pdx-1 (20), the correlation of Pdx-1 function with Gck expression in the transcriptional hierarchy is unclear, and the upstream regulator that serves as the activator or repressor for Pdx-1-mediated gene expression is also unknown. This study suggested the opposing effects of Atf3 on Pdx-1-mediated Gck transcriptional regulation, which were caused by their direct interaction in vitro and in vivo (Figs. 5–7). Pdx-1 overexpression reversely attenuated or rescued the inhibitory effects of Atf3 on Gck expression, but how Pdx-1 counteracts Atf3-repressed Gck transcriptional activity in response to ethanol is still unclear. Our data show that two putative Pdx-1-binding sites may be essential for counteracting the inhibitory effects of Atf3 on Gck transcriptional activity by using a pRGP-287/Luc with a deleted or point-mutated Pdx-1-binding site (Fig. 5D). Despite the lack of an Atf3-binding site, Atf3 still induced a small reduction in pRGP-287(ΔAtf)/Luc activity, which was similar to the small reduction in the pRGP-158/Luc promoter (Fig. 4), suggesting that Atf3 may also inhibit Pdx-1 functional activity within the −158-bp region. The association of Pdx-1 with insulin promoter is enhanced by its interaction with p300, and the initial acetylation of histone H4 may stabilize Pdx-1 binding (37). However, when glucose levels are low, Pdx-1 interacts with Hdac1/2 rather than p300, and the complexes are recruited to the insulin promoter, thus leading to histone H4 deacetylation and insulin gene down-regulation (38). Similarly, ethanol-induced Atf3 may first interact with Pdx-1 and subsequently prevent the formation of Pdx-1/p300, followed by chromatin remodeling via alternative recruitment of histone deacetylase to the consensus Pdx-1-binding sites of the Gck promoter. Furthermore, because the primers used in ChIP assay cover the putative Pdx-1-binding sites (−28/−127) on the pRGP-158/Luc promoter, the product does not contain Atf3-binding sites and may present only low responsiveness for Atf3. Therefore, it is important to determine how Atf3 acts as a negative upstream regulator for Pdx-1 functional activity at the −158-bp proximal region of the Gck promoter. Quantitative ChIP assay revealed that ethanol-induced Atf3 binds to the proximal −158-bp region, whereas recruitments of Pdx-1, acetylated H3, and RNA polymerase II were significantly decreased in ethanol-treated cells, which were ameliorated by Atf3 depletion (Fig. 6D), demonstrating that Atf3 association with Gck promoter is a critical step for histone H3 deacetylation. Despite the decreased recruitment to the Gck promoter and expression of Pdx-1 in ethanol-treated cells, how the formation of Atf3/Pdx-1/Hdac1/2 complexes is increased in response to ethanol is unknown. Previously, it was demonstrated that Pdx-1 phosphorylation may determine the interaction of Pdx-1 with p300/Cbp or Hdac1/2 in the regulation of the insulin gene, which may depend on the concentration of glucose (35–37). Similarly, it is possible that ethanol-induced Atf3 directly interacts with Pdx-1, subsequently decreasing Pdx-1 phosphorylation and recruiting Hdac1/2 rather than p300 (Fig. 7); this possibility should be explored further.
We also confirmed the specific upstream role of Atf3 in chronic ethanol-induced Gck down-regulation and pancreatic β-cell dysfunction by performing a loss-of-function study using the in vivo Atf3 siRNA delivery system (Fig. 8). In vivo Atf3 silencing ameliorated the impaired glucose metabolism and pancreatic β-cell dysfunction in ethanol-treated cells, and therefore prevented the development of T2D by reducing RNS production. However, in our model, the injected Atf3 siRNA may act on several organs such as liver and lung and is not specific to pancreatic β-cells (Fig. 8, A and B). Therefore, to define the precise role of Atf3 in ethanol-mediated Gck down-regulation and β-cell dysfunction, additional studies using pancreas-specific knock-out models are required. Nevertheless, we propose that in vivo Atf3 silencing may be sufficient to improve metabolic syndrome and pancreatic β-cell dysfunction by ameliorating ethanol-induced Gck down-regulation. In contrast with the ameliorating effects of Atf3-silencing mice in vivo, it will be possible that Atf3 knock-out mice may conversely potentiate ethanol-mediated Gck down-regulation and β-cell dysfunction. Because stress-inducible Atf3 is a dual-face transcription factor that activates or represses gene expression (13), it may initially develop a compensatory or adaptive response for the acute oxidative stress or endoplasmic reticulum stress. Although the data are not shown here, we have determined genetic susceptibility of Atf3 to T2D risk by performing conditional analyses using our previous GWAS data (KARE study) based on the Korean population (39, 40). Furthermore, we have identified a noncoding variant in the Atf3 gene as a novel predisposing factor to T2D after controlling for alcohol consumption in T2D groups (data not shown). From these results, we propose that Atf3 may play an important role in ethanol-mediated metabolic alteration, and it may be strongly associated with T2D.
In this study, the direct interaction of Atf3 with the Atf/Creb-binding site of the Gck promoter may have been a major molecular mechanism by which ethanol reduced Gck expression and pancreatic β-cell dysfunction and apoptosis. Also, Atf3-dependent interaction of Pdx-1 with Hdac1/2 and subsequent inactivation of Pdx-1 and chromatin remodeling may represent an additional molecular mechanism by which ethanol-induced Atf3 decreases Gck down-regulation (Fig. 9). Taken together, these results show for the first time that Atf3 may act as an upstream negative regulator for Gck transcriptional activation via direct binding to the putative binding site and by indirect binding with Pdx-1/Hdac1/2 on the Gck promoter. Additionally, our findings suggest that strategies based on the inhibition of Atf3 may be of benefit in the treatment of ethanol-mediated glucose impairment and metabolic syndrome.
Acknowledgments
cDNA expression vectors, human wild-type Atf3 and Atf3(ΔC,101–181) with a C-terminal deletion, were the generous gift from Dr. T. Hai (Ohio State University). Pdx-1 expression vector (pcDNA3/Pdx-1) was provided by Dr. T. Stein (Ohio State University). Rat Gck promoter reporter pRGP-1003/Luc, pRGP-404/Luc, pRGP-287/Luc, and pRGP-84/Luc were kindly provided by Dr. Y. Ahn (Yonsei University College of Medicine).
This work was authored, in whole or in part, by National Institutes of Health staff. This work was supported by Research Grant 4845-302-210-13 from the Korean National Institutes of Health.
- T2D
- type 2 diabetes
- Gck
- glucokinase (hexokinase IV)
- ROS
- reactive oxygen species
- Hdac
- histone deacetylase
- Creb
- cAMP-response element-binding protein
- Luc
- luciferase
- TSA
- trichostatin A.
REFERENCES
- 1. Teli M. R., Day C. P., Burt A. D., Bennett M. K., James O. F. (1995) Determinants of progression to cirrhosis or fibrosis in pure alcoholic fatty liver. Lancet 346, 987–990 [DOI] [PubMed] [Google Scholar]
- 2. Dam-Larsen S., Franzmann M., Andersen I. B., Christoffersen P., Jensen L. B., Sørensen T. I., Becker U., Bendtsen F. (2004) Long term prognosis of fatty liver: risk of chronic liver disease and death. Gut 53, 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fueki Y., Miida T., Wardaningsih E., Ito M., Nakamura A., Takahashi A., Hanyu O., Tsuda A., Saito H., Hama H., Okada M. (2007) Regular alcohol consumption improves insulin resistance in healthy Japanese men independent of obesity. Clin. Chim. Acta 382, 71–76 [DOI] [PubMed] [Google Scholar]
- 4. Conigrave K. M., Hu B. F., Camargo C. A., Jr., Stampfer M. J., Willett W. C., Rimm E. B. (2001) A prospective study of drinking patterns in relation to risk of type 2 diabetes among men. Diabetes 50, 2390–2395 [DOI] [PubMed] [Google Scholar]
- 5. Roh W. G., Shin H. C., Choi J. H., Lee Y. J., Kim K. (2009) Alcohol consumption and higher incidence of impaired fasting glucose or type 2 diabetes in obese Korean men. Alcohol 43, 643–648 [DOI] [PubMed] [Google Scholar]
- 6. Adaramoye O. A., Oloyede G. K. (2012) Effect of moderate ethanol administration on biochemical indices in streptozotocin-diabetic Wistar rats. West Indian Med. J. 61, 3–9 [DOI] [PubMed] [Google Scholar]
- 7. Onishi Y., Honda M., Ogihara T., Sakoda H., Anai M., Fujishiro M., Ono H., Shojima N., Fukushima Y., Inukai K., Katagiri H., Kikuchi M., Oka Y., Asano T. (2003) Ethanol feeding induces insulin resistance with enhanced PI 3-kinase activation. Biochem. Biophys. Res. Commun. 303, 788–794 [DOI] [PubMed] [Google Scholar]
- 8. Zhao L. N., Hao L. P., Yang X. F., Ying C. J., Yu D., Sun X. F. (2009) The diabetogenic effects of excessive ethanol: reducing beta-cell mass, decreasing phosphatidylinositol 3-kinase activity and GLUT-4 expression in rats. Br. J. Nutr. 101, 1467–1473 [DOI] [PubMed] [Google Scholar]
- 9. Matschinsky F. M., Magnuson M. A., Zelent D., Jetton T. L., Doliba N., Han Y., Taub R., Grimsby J. (2006) The network of glucokinase-expressing cells in glucose homeostasis and the potential of glucokinase activators for diabetes therapy. Diabetes 55, 1–12 [PubMed] [Google Scholar]
- 10. Iynedjian P. B. (2009) Molecular physiology of mammalian glucokinase. Cell. Mol. Life Sci. 66, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim J. Y., Song E. H., Lee H. J., Oh Y. K., Park Y. S., Park J. W., Kim B. J., Kim D. J., Lee I., Song J., Kim W. H. (2010) Chronic ethanol consumption-induced pancreatic β-cell dysfunction and apoptosis through glucokinase nitration and its down-regulation. J. Biol. Chem. 285, 37251–37262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hai T., Hartman M. G. (2001) The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 273, 1–11 [DOI] [PubMed] [Google Scholar]
- 13. Hai T., Wolford C. C., Chang Y. S. (2010) Atf3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: is modulation of inflammation a unifying component? Gene Exp. 15, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chakrabarti S. K., Mirmira R. G. (2003) Transcription factors direct the development and function of pancreatic beta cells. Trends Endocrinol. Metab. 14, 78–84 [DOI] [PubMed] [Google Scholar]
- 15. Kim S. Y., Kim H. I., Kim T. H., Im S. S., Park S. K., Lee I. K., Kim K. S., Ahn Y. H. (2004) SREBP-1c mediates the insulin-dependent hepatic glucokinase expression. J. Biol. Chem. 279, 30823–30829 [DOI] [PubMed] [Google Scholar]
- 16. Lee J. W., Kim W. H., Lim J. H., Song E. H., Song J., Choi K. Y., Jung M. H. (2009) Mitochondrial dysfunction: glucokinase downregulation lowers interaction of glucokinase with mitochondria, resulting in apoptosis of pancreatic beta-cells. Cell. Signal. 21, 69–78 [DOI] [PubMed] [Google Scholar]
- 17. Struhl K. (2001) Gene regulation. A paradigm for precision. Science 293, 1054–1055 [DOI] [PubMed] [Google Scholar]
- 18. Berger S. L. (2002) Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12, 142–148 [DOI] [PubMed] [Google Scholar]
- 19. Mosley A. L., Corbett J. A., Ozcan S. (2004) Glucose regulation of insulin gene expression requires the recruitment of p300 by the beta-cell-specific transcription factor Pdx-1. Mol. Endocrinol. 18, 2279–2290 [DOI] [PubMed] [Google Scholar]
- 20. Kim W. H., Jang M. K., Kim C. H., Shin H. K., Jung M. H. (2011) Atf3 inhibits Pdx-1-stimulated transactivation. Biochem. Biophys. Res. Commun. 414, 681–687 [DOI] [PubMed] [Google Scholar]
- 21. Deleted in proof.
- 22. Dolganiuc A., Szabo G. (2009) In vitro and in vivo models of acute alcohol exposure. World J. Gastroenterol. 15, 1168–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pochareddy S., Edenberg H. (2012) Chronic alcohol exposure alters gene expression in HepG2 cells. Alcohol. Clin. Exp. Res. 36, 1021–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tang Y., Banan A., Forsyth C. B., Fields J. Z., Lau C. K., Zhang L. J., Keshavarzian A. (2008) Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol. Clin. Exp. Res. 32, 355–364 [DOI] [PubMed] [Google Scholar]
- 25. Dolganiuc A., Bakis G., Kodys K., Mandrekar P., Szabo G. (2006) Acute ethanol treatment modulates Toll-like receptor-4 association with lipid rafts. Alcohol. Clin. Exp. Res. 30, 76–85 [DOI] [PubMed] [Google Scholar]
- 26. Szabo G., Mandrekar P. (2008) Human monocytes, macrophages, and dendritic cells: alcohol treatment methods. Methods Mol. Biol. 447, 113–124 [DOI] [PubMed] [Google Scholar]
- 27. Kim J. W., Jang S. M., Kim C. H., An J. H., Kang E. J., Choi K. H. (2012) New molecular bridge between ReIA/p65 and NF-κB target genes via histone acetyltransferase TIP60 cofactor. J. Biol. Chem. 287, 7780–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim J. Y., Song E. H., Lee H. J., Oh Y. K., Choi K. H., Yu D. Y., Park S. I., Seong J. K., Kim W. H. (2010) HBx-induced hepatic steatosis and apoptosis are regulated by TNFR1- and NF-κB-dependent pathways. J. Mol. Biol. 397, 917–931 [DOI] [PubMed] [Google Scholar]
- 29. Kim J. Y., Park K. J., Kim G. H., Jeong E. A., Lee D. Y., Lee S. S., Kim D. J., Roh G. S., Song J., Ki S. H., Kim W. H. (2013) In vivo Atf3 silencing ameliorates the AMPK compensatory effects for ER stress-mediated β-cell dysfunction during the progression of type 2 diabetes. Cell. Signal. 25, 2348–2361 [DOI] [PubMed] [Google Scholar]
- 30. Bolcato-Bellemin A. L., Bonnet M. E., Creusat G., Erbacher P., Behr J. P. (2007) Sticky overhangs enhances siRNA-mediated gene silencing. Proc. Natl. Acad. Sci. U.S.A. 104, 16050–16055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sioud M. (2005) siRNA delivery in vivo. Methods Mol. Biol. 309, 237–249 [DOI] [PubMed] [Google Scholar]
- 32. Kim W. H., Hong F., Jaruga B., Hu Z., Fan S., Liang T. J., Gao B. (2001) Additive activation of hepatic NF-κB by ethanol and hepatitis B protein X (HBx) or HCV core protein: involvement of TNF-α receptor I-independent and -dependent mechanisms and implication in cooperative effects of alcohol and viral hepatitis in liver disease. FASEB J. 15, 2551–2553 [DOI] [PubMed] [Google Scholar]
- 33. Brissova M., Shiota M., Nicholson W. E., Gannon M., Knobel S. M., Piston D. W., Wright C. V., Powers A. C. (2002) Reduction in pancreatic transcription factor Pdx-1 impairs glucose-stimulated insulin secretion. J. Biol. Chem. 277, 11225–11232 [DOI] [PubMed] [Google Scholar]
- 34. Kaneto H., Miyatsuka T., Kawamori D., Yamamoto K., Kato K., Shiraiwa T., Katakami N., Yamasaki Y., Matsuhisa M., Matsuoka T. A. (2008) Pdx-1 and MafA play a crucial role in pancreatic β-cell differentiation and maintenance of mature β-cell function. Endocr. J. 55, 235–252 [DOI] [PubMed] [Google Scholar]
- 35. Watada H., Kajimoto Y., Umayahara Y., Matsuoka T., Kaneto H., Fujitani Y., Kamada T., Kawamori R., Yamasaki Y. (1996) The human glucokinase gene beta-cell-type promoter: an essential role of insulin promoter factor 1/Pdx-1 in its activation in HIT-T15 cells. Diabetes 45, 1478–1488 [DOI] [PubMed] [Google Scholar]
- 36. Mosley A. L., Ozcan S. (2004) The pancreatic duodenal homeobox-1 protein (Pdx-1) interacts with histone deacetylases Hdac-1 and Hdac-2 on low levels of glucose. J. Biol. Chem. 279, 54241–54247 [DOI] [PubMed] [Google Scholar]
- 37. Mosley A. L., Ozcan S. (2003) Glucose regulates insulin gene transcription by hyperacetylation of histone H4. J. Biol. Chem. 278, 19660–19666 [DOI] [PubMed] [Google Scholar]
- 38. Jin Q., Yu L. R., Wang L., Zhang Z., Kasper L. H., Lee J. E., Wang C., Brindle P. K., Dent S. Y., Ge K. (2011) Distinct role of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cho Y. S., Go M. J., Kim Y. J., Heo J. Y., Oh J. H., Ban H. J., Yoon D., Lee M. H., Kim D. J., Park M., Cha S. H., Kim J. W., Han B. G., Min H., Ahn Y., Park M. S., Han H. R., Jang H. Y., Cho E. Y., Lee J. E., Cho N. H., Shin C., Park T., Park J. W., Lee J. K., Cardon L., Clarke G., McCarthy M. I., Lee J. Y., Lee J. K., Oh B., Kim H. L. (2009) A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet. 41, 527–534 [DOI] [PubMed] [Google Scholar]
- 40. Cho Y. S., Chen C. H., Hu C., Long J., Ong R. T., Sim X., Takeuchi F., Wu Y., Go M. J., Yamauchi T., Adari L. S., Aung T., Chang Y. C., Kwak S. H., Ma R. C., Yamamoto K., Adair L. S., Aung T., Cai Q., Chang L. C., Chen Y. T., Gao Y., Hu F. B., Kim H. L., Kim S., Kim Y. J., Lee J. J., Lee N. R., Li Y., Liu J. J., Lu W., Nakamura J., Nakashima E., Ng D. P., Tay W. T., Tsai F. J., Wong T. Y., Yokota M., Zheng W., Zhang R., Wang C., So W. Y., Ohnaka K., Ikegami H., Hara K., Cho Y. M., Cho N. H., Chang T. J., Bao Y., Hedman Å. K., Morris A. P., McCarthy M. I., DIAGRAM Consortium, MuTHER Consortium, Takayanagi R., Park K. S., Jia W., Chuang L. M., Chan J. C., Maeda S., Kadowaki T., Lee J. Y., Wu J. Y., Teo Y. Y., Tai E. S., Shu X. O., Mohlke K. L., Kato N., Han B. G., Seielstad M. (2012) Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat. Genet. 44, 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]