Background: Mitochondrial dynamics is involved in the regulation of apoptosis.
Results: p53 regulates mitochondrial dynamics by interacting with Prohibitin 1 and subsequent releasing Opa1 for Oma1-mediated processing, thereby promoting apoptosis and chemoresponsiveness.
Conclusion: CDDP resistance is in part due to dysregulation of p53-induced, Oma1-mediated L-Opa1 processing and mitochondrial fragmentation.
Significance: Understanding the regulation of mitochondrial dynamics may offer new strategies for overcoming chemoresistance.
Keywords: Apoptosis, Chemoresistance, Ovarian Cancer, p53, Protein-Protein Interaction, CDDP, Mitochondrial Fragmentation, Oma1, Opa1
Abstract
Mitochondria are highly dynamic organelles, and mitochondrial fission is a crucial step of apoptosis. Although Oma1 is believed to be responsible for long form Opa1 (L-Opa1) processing during mitochondrial fragmentation, whether and how Oma1 is involved in L-Opa1 processing and participates in the regulation of chemoresistance is unknown. Chemosensitive and chemoresistant ovarian (OVCA) and cervical (CECA) cancer cells were treated with cisplatin (CDDP). Mitochondrial dynamics and protein contents were assessed by immunofluorescence and Western blot, respectively. The requirements of Oma1 and p53 for CDDP-induced L-Opa1 processing, mitochondrial fragmentation, and apoptosis were examined by siRNA or cDNA. CDDP induces L-Opa1 processing and mitochondrial fragmentation in chemosensitive but not in chemoresistant cells. CDDP induced Oma1 40-kDa form increases in OV2008 cells, not in C13* cells. Oma1 knockdown inhibited L-Opa1 processing, mitochondrial fragmentation, and apoptosis. Silencing p53 expression attenuated the effects of CDDP in Oma1 (40 kDa) increase, L-Opa1 processing, mitochondrial fragmentation, and apoptosis in chemosensitive OVCA cells, whereas reconstitution of p53 in p53 mutant or null chemoresistant OVCA cells induced Oma1 (40 kDa) increase, L-Opa1 processing, mitochondrial fragmentation, and apoptosis irrespective of the presence of CDDP. Prohibitin 1 (Phb1) dissociates from Opa1-Phb1 complex and binds phosphorylated p53 (serine 15) in response to CDDP in chemosensitive but not chemoresistant CECA cells. These findings demonstrate that (a) p53 and Oma1 mediate L-Opa1 processing, (b) mitochondrial fragmentation is involved in CDDP-induced apoptosis in OVCA and CECA cells, and (c) dysregulated mitochondrial dynamics may in part be involved in the pathophysiology of CDDP resistance.
Introduction
Cervical (CECA) and ovarian (OVCA) cancer rank first and third in the number of new cases and are the leading causes of death in gynecologic cancer worldwide. Although radiotherapy is indicated in CECA, surgical debulking and cisplatin-based chemotherapy are important treatment methods for both diseases. Although the incidence of recurrence and resistance is lower in CECA than OVCA, chemoresistance severely limits treatment success in both cancer types. The underlying mechanism of chemoresistance is multifactorial and partly due to defects in drug-induced apoptosis (1). TP53 is frequently mutated in cancer cells and often associated with decreased chemoresponsiveness, suggesting that p53 is required for chemosensitivity (2). CDDP-induced, p53-mediated mitochondrial cell death is a determinant of chemosensitivity in gynecologic cancer cells (3). However, the mechanism by which p53 regulates mitochondrial-mediated cell death remains unclear.
Mitochondria are highly dynamic organelles that are constantly dividing and elongating to form a network (4). The dynamic nature of mitochondrial networks is due to two opposing processes, mitochondrial fission and fusion. The dynamic change of mitochondria allows the adjustment of mitochondrial morphologies to specific cellular processes and is also essential for mitochondrial quality control (5, 6). Upon extensive damage, mitochondria fragment from filamentous tubules into numerous small punctate particles, followed by cytochrome c release, which in turn activates different apoptotic pathway and causes cell death (7).
Opa1 is a key mitochondrial fusion protein, which exists in long and short forms generated by processing at specific sites (8). The presence of both long and short forms is necessary for generation of fusion competent mitochondria. In response to a proapoptotic stimulus, long form Opa1 (L-Opa1)5 is processed into short forms, thereby disrupting the balance of these forms, abolishing mitochondrial fusion, and inducing mitochondrial fragmentation and apoptosis (9).
Oma1 is a novel mitochondrial metallopeptidase responsible for L-Opa1 processing in mammalian cells. Significant L-Opa1 stabilization has been reported when Oma1 is down-regulated. Although Oma1 appears to be involved in the L-Opa1 processing during mitochondrial fragmentation and apoptosis (10–12), the mechanism of Oma1-mediated L-Opa1 processing is not clear, nor its significance in chemoresistance. Whether Oma1, Opa1, and mitochondrial dynamics play a role in regulation of chemosensitivity in OVCA and CECA cells is not known.
Prohibitins are multifunctional mitochondrial proteins. L-Opa1 processing and mitochondrial dynamics have been reported to be regulated by prohibitins (13, 14), although the mechanism involved is not clear. Recent publications have shown that p53 interacts with Prohibitin 1 (Phb1) upon apoptotic signaling, and the function of p53 is attenuated in the absence of Phb1 (15), suggesting that Phb1 may play an important role in p53 signaling pathways. Whether the interaction of p53 and Phb 1 is involved in the regulation of L-Opa1 processing is not known.
MATERIALS AND METHODS
Reagents
CDDP, DMSO, Hoechst 33258, PMSF, sodium orthovanadate (Na3VO4), and aprotinin were purchased from Sigma-Aldrich. Mouse monoclonal GAPDH, mouse monoclonal p-p53 (serine 20), mouse monoclonal HA tag antibody, mouse monoclonal Mfn1, mouse monoclonal Mfn2, and rabbit polyclonal Oma1 antibody were from Abcam (Cambridge, MA). Mouse monoclonal Tom20 antibody, mouse monoclonal p53 antibody, rabbit polyclonal p-p53 (serine 15) antibody, and rabbit polyclonal Phb1 antibody were obtained from Santa Cruz Biotechnology (Dallas, TX). Mouse monoclonal Opa1 and mouse monoclonal Drp1 were from BD Biosciences. Rabbit polyclonal Fis1 antibody was purchased from Life Span Biosciences (Seattle, WA). Oma1 plasmid, p53 plasmid, Oma1 siRNA, scramble siRNA, and mouse monoclonal Myc tag antibody were from Origene (Rockville, MD). p53 siRNA was from Qiagen (Valencia, CA). RNeasy Mini kits were purchased from Qiagen (Mississauga, Canada). Random decamer primers were from Ambion (Austin, TX). M-MLV Reverse Transcriptase was from Promega. Ribonuclease inhibitor and dNTP were from Fermentas (Burlington, Canada). Quantitative PCR primers were from Invitrogen.
Cell Lines and Cell Culture
The CDDP-sensitive cancer cell OV2008 (WT-p53) is of cervical origin. CDDP-resistant C13* (WT-p53) cell line is the isogenic resistant counterpart to OV2008, selected by chronic exposure of increasing concentrations of CDDP in vitro. CDDP-sensitive A2780s (WT-p53), its resistant variant A2780cp (p53 mutant), and CDDP-resistant HEY (WT-p53) human ovarian cancer cell lines were derived from serous cystadenocarcinomas of the ovary. SKOV3 cells (p53 null) are of clear cell carcinoma origin (16). These cell lines were gifts from Drs. Rakesh Goel and Barbara Vanderhyden (Ottawa Regional Cancer Centre, Ottawa, Ontario, Canada) and were cultured as reported previously (17–19).
Protein Extraction and Western Blot Analysis
Protein extraction and Western blot analysis were performed as described previously (20). Unless indicated otherwise, membranes were incubated overnight at 4 °C, with anti-Opa1 (1:1000), Mfn1 (1:1000), Mfn2 (1:1000), Drp1 (1:1000), Fis1 (1:1000), Oma1(1:1000), Phb1 (1:2000), p-p53 (serine 15) (1:2000), p-p53 (serine 20) (1:2000), p53 (1:5000), and HA tag (1:5000) and 1 h at room temperature for anti-GAPDH (1:10,000), vinculin (1:10,000) and horseradish peroxidase-conjugated rabbit or mouse secondary antibodies (1:5000–1:10,000), and band densities were analyzed (Scion Image software; Scion Corp., Frederick, MD).
Fluorescence Microscopy and Determination of Mitochondrial Phenotype
The procedures were performed as described previously (19). Cells were plated on poly-d-lysine-coated (0.05% w/v; Sigma) 8-well glass culture slides (BD Biosciences) and cultured in growth medium (48 h) prior to CDDP treatment. For immunostaining, cells were fixed in paraformaldehyde (4%, 1 h, room temperature), washed in PBS, and blocked with 1% BSA. Mitochondria were visualized by immunofluorescence microscopy, using a mouse monoclonal antibody anti-human Tom20 (1:100; Santa Cruz Biotechnology) and Alexa Fluor 488 goat anti-mouse secondary antibody (1:500; Invitrogen). Confocal images were obtained (×100 objective) on an Olympus IX81 inverted microscope with appropriate argon lasers (488 nm). Mitochondrial phenotype of each cell was categorized as tubular, intermediate, or fragmented as described previously (21–23). At least 100 cells were analyzed per treatment group.
RNA Interference
For gene knockdown studies, cells were transfected with p53 siRNA (0–100 nm; 48 h), Oma1 siRNA (0–40 nm; 48 h), or control siRNA (scrambled sequence) and were treated with CDDP (0–10 μm; 24 h) as described previously (19) and harvested for further analysis.
Transient cDNA Transfection
OV2008 were transfected with C terminus Oma1-Myc cDNA (0–1 μg, 24 h; Origene) as described previously (19). Empty vector served as a control. Following transfection, cells were treated with CDDP (10 μm) or vehicle (DMSO) and harvested for analyses or fixed for immunofluorescence studies. A2780cp and SKOV3 cells were transfected with p53 cDNA (0–2 μg, 24 h). Empty vector served as a control. Following transfection, cells were treated with CDDP (10 μm, 0–24 h) or vehicle (DMSO, 0–24 h) and fixed for immunofluorescence studies or Western blot.
HA-tagged wild type p53 in pcDNA3 was used as a template for site-directed mutagenesis, using the QuikChange site-directed kit from Stratagene (La Jolla, CA) as described previously (24). The presence of mutations was confirmed by direct sequencing at OHRI sequencing facility.
Reverse Transcriptase Polymerase Chain Reaction
Total RNAs were extracted according to the manufacturer's instructions, using the Qiagen RNeasy Mini kit. One μg total RNAs were reverse-transcribed into cDNA, and the mRNA abundances of target genes were analyzed by real-time quantitative PCR using LightCycler® 480 SYBR Green I Master (Roche Diagnostics). The variants of Opa1 were amplified using specific primer pairs based on the literature (25). Data were analyzed by 2−ΔΔCT method (26). The data are presented as the fold change in mRNA abundance normalized against the β-actin gene and expressed relative to the respective control.
Immunoprecipitation
One mg of protein sample was incubated (room temperature, 1 h) with 50 μl of protein G Dynabeads (Invitrogen) coated with rabbit polyclonal Phb1 antibody (2 μg, Santa Cruz Biotechnology) and immunoprecipitated. The beads were pelleted and resuspended in sample buffer, boiled, and loaded onto 9% SDS-PAGE. After protein was transferred to nitrocellulose, Phb1, phosphorylated p53, and Opa1 were examined by Western blotting using Clean Blot IP Detection Reagent (Thermo Scientific).
Determination of Apoptosis
Apoptosis was assessed morphologically with Hoechst 33258 nuclear dye (6.25 ng/ml). At least 400 cells/treatment groups were counted. Selected fields and blinded slides were determined randomly to avoid experimental bias (19).
Statistical Analysis
Results are expressed as the mean ± S.E. of at least three independent experiments. Statistical analysis was carried out by two-way analysis of variance, using PRISM software (version 5.0; GraphPad, San Diego, CA). Differences between multiple experimental groups were determined by the Bonferroni post hoc test. Statistical significance was inferred at p < 0.05.
RESULTS
CDDP Decreases Mitochondrial Fusion Protein Opa1 (Long Form) and Increases Mitochondria Fragmentation in Chemosensitive but Not Resistant Gynecologic Cancer Cells
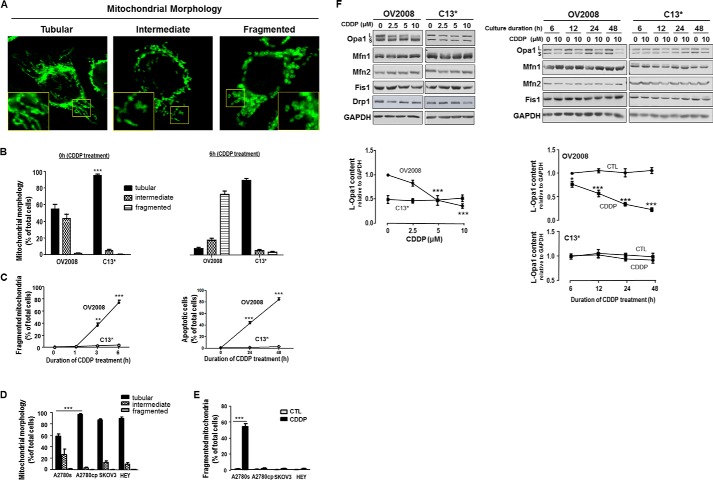
CDDP induces apoptosis in chemosensitive but not chemoresistant gynecologic cancer cells (3, 19, 20). To determine whether changes in mitochondrial dynamics play a role in the regulation of chemosensitivity in CECA cells, OV2008 and C13* cells were cultured in the absence and presence of CDDP (10 μm, 0–6 h), and mitochondrial phenotype was examined by immunofluorescence confocal microscopy. Three major mitochondrial phenotypes were found irrespective of the presence of CDDP: tubular, intermediate, and fragmented (Fig. 1A). Although only a few fragmented mitochondria were observed in these chemosensitive and chemoresistant cancer cells in the absence of CDDP, the percentage of cells bearing tubular mitochondria was much higher in chemoresistant C13* cells than in chemosensitive OV2008 cells (p < 0.001, Fig. 1B). CDDP induced mitochondrial fragmentation in the chemosensitive cells in a time-dependent manner, as evident by increased as early as 3 h of CDDP exposure the percentage of cells with fragmented mitochondria (p < 0.01 at 3 h, Fig. 1C). In contrast, in chemoresistant cells, CDDP failed to significantly influence these mitochondrial structural changes, implying the presence of stabilized and fused mitochondria in chemoresistant cells (Fig. 1, B and C). CDDP also induced apoptosis in chemosensitive OV2008 cells (p < 0.001 at 24 and 48 h) but not in chemoresistant C13* cells (Fig. 1C).
FIGURE 1.
Mitochondrial dynamics in chemosensitive and chemoresistant gynecologic cancer cells. A, mitochondrial phenotypes (tubular, intermediate, and fragmented) in CECA cells (OV2008 and C13*). Mitochondria were immunostained with Tom20 antibody. Mitochondrial phenotypes were examined by immunofluorescence confocal microscopy. The inset is an enlarged image of the boxed area. B, comparison of mitochondrial phenotypes in chemosensitive and chemoresistant CECA cells treated with CDDP. OV2008 and C13* cells were cultured with and without CDDP (10 μm, 0–6 h). Percentage of cells with tubular mitochondria in chemoresistant cells (C13*) is higher than in chemosensitive cells (OV2008) at 0 h. More than 100 cells were assessed in each experimental group (n = 3, ***, p < 0.001). C, comparison of mitochondrial fragmentation phenotype and apoptosis in chemosensitive and chemoresistant CECA cells treated with CDDP. OV2008 and C13* cells were cultured with CDDP (10 μm, 0–6 h). Mitochondrial phenotypes were examined by immunofluorescence confocal microscopy. OV2008 cells exhibited much higher level of fragmented mitochondria than C13* cells with CDDP treatment (n = 3, **, p < 0.01 at 3 h and ***, p < 0.001 at 6 h). At least 100 cells were assessed for each time point in each replicate. Apoptosis was examined by Hoechst assay. OV2008 cells exhibited higher apoptosis than C13* cells when treated with CDDP (n = 3, ***, p < 0.001 at 24 and 48 h). More than 300 cells were assessed in each experimental group. D, comparison of mitochondrial phenotypes in different OVCA cell lines. Chemoresistant A2780cp cells had more tubular mitochondria than chemosensitive A2780s cells (n = 3, ***, p < 0.001). Most SKOV3 and HEY cells exhibited tubular mitochondria. Over 100 cells were assessed in each experimental group. E, CDDP-induced mitochondria fragmentation in chemosensitive but not chemoresistant OVCA cells. A2780s, A2780cp, SKOV3, and HEY cells were cultured in the presence of CDDP (10 μm, 6 h) or control (CTL; DMSO) only. Mitochondrial phenotypes were examined by immunofluorescence confocal microscopy. CDDP induced significant mitochondria fragmentation in A2780s (***, p < 0.001, n = 3) but not in A2780cp, SKOV3, and HEY cells. At least 100 cells were assessed at each time point in each replicate. F, influence of CDDP on contents of mitochondrial fission proteins (Drp1 and Fis1) and mitochondrial fusion proteins (Mfn1, Mfn2, and Opa1) in CECA cells. OV2008 and C13* cells were cultured with CDDP (left panel, 0–10 μm, 24 h; right panel, 0–10 μm, 0–48 h), Opa1, Mfn1, Mfn2, Drp1, and Fis1 contents were examined by Western blot. Opa1 content (long form) significantly decreased in a concentration- and time-dependent manner in chemosensitive (OV2008) but not in chemoresistant (C13*) cells when treated with CDDP. L and S represent long and short form of Opa1, respectively. Contents of Mfn1, Mfn2, Drp1, and Fis1 were not altered by CDDP treatment. Representative Western blots of four independent experiments are shown.
To determine whether mitochondrial dynamics is dysregulated in chemoresistant OVCA cancer cells, and whether p53 status affects mitochondrial dynamics, we examined mitochondrial dynamics in chemosensitive A2780s cells (WT-p53) and its chemoresistant counterpart A2780cp cells (p53 mutant), SKOV3 cells (p53 null) and HEY cells (WT-p53). The percentage of cells bearing tubular mitochondria was much higher in chemoresistant A2780cp cells than in chemosensitive A2780s cells (p < 0.001, Fig. 1D). Most SKOV3 cells and HEY cells also exhibit tubular mitochondria (Fig. 1D). CDDP induced mitochondrial fragmentation in A2780s cells (p < 0.001, Fig. 1E), but not in A2780cp, SKOV3, and Hey cells.
To further examine the regulation of mitochondrial dynamics in CECA cells and its possible involvement in the control of chemosensitivity, we examined by Western blotting several key regulators of mitochondrial fission and fusion, including the contents of mitochondrial fission proteins Fis1 and Drp1 and mitochondrial fusion proteins Mfn1, Mfn2 and Opa1 in OV2008 and C13* cells following exposure to CDDP (0–10 μm, 0–48 h). CDDP decreased mitochondrial fusion protein L-Opa1 in OV2008 cells in a time- and concentration-dependent manner (p < 0.001, Fig. 1F). This response was, however, not apparent in C13* cells. The contents of other intracellular regulators of mitochondrial fission and fusion were not affected by CDDP under the same experimental conditions. Interestingly, the expression of Opa1 is lower in C13* cells than in OV2008 cells.
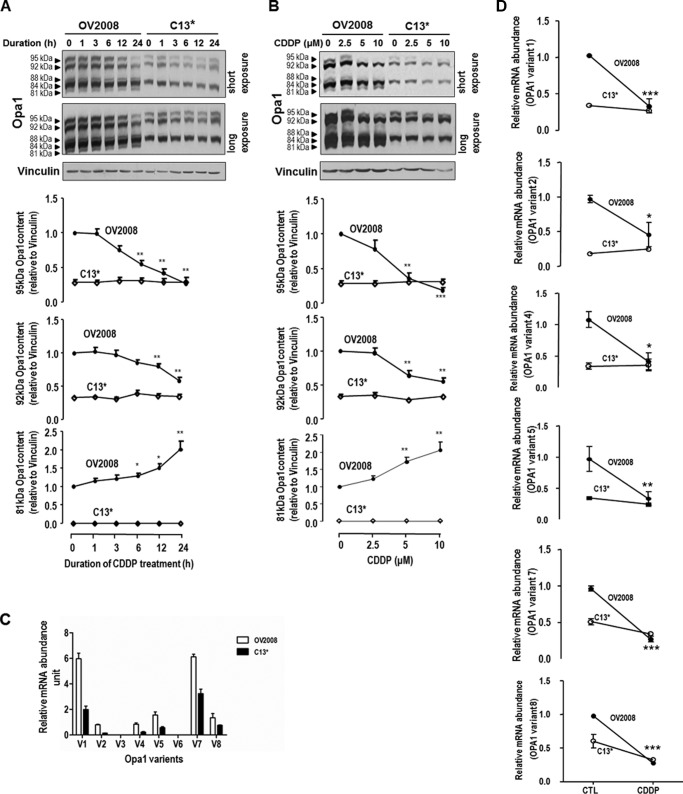
Chemosensitive CECA Cells Exhibit Five Opa1 Forms, Whereas only Three Are Present in the Chemoresistant Counterpart
At least five forms of Opa1 have been reported in mammalian cells (9). We thus examined the differences in the contents of different Opa1 forms when chemosensitive and chemoresistant CECA cells were challenged with CDDP (0–10 μm, 0–24 h). Interestingly, five forms of Opa1 (95, 92, 88, 84, and 81 kDa) could be recognized in OV2008 cells, whereas only three in C13* cells (95, 92, and 84 kDa) (Fig. 2). CDDP decreased the contents of Opa1 (95 and 92 kDa) forms (p < 0.001 and p < 0.01, respectively) but increased that of Opa1 (81 kDa) form (p < 0.01) in OV2008 cells in a time- and concentration-dependent manner (Fig. 2, A and B, respectively). However, the content of the Opa1 forms in C13* cells was not affected by the CDDP treatment.
FIGURE 2.
Different forms of Opa1 in CECA cells. Chemosensitive (OV2008) and chemoresistant (C13*) CECA cells were cultured with CDDP for different durations (A; 10 μm, 0–24 h) or at different concentrations (B; 0–10 μm, 24 h). Contents of different forms of Opa1 and vinculin (loading control) were examined by Western blot. Five forms of Opa1 (95, 92, 88, 84, and 81 kDa) were recognized in OV2008 cells, whereas only three forms (95, 92, and 84 kDa) were recognized in C13* cells. CDDP significantly decreased contents of L-Opa1 forms (95 and 92 kDa), whereas it significantly increased S-Opa1 content (81 kDa) in a time-dependent (A; *, p < 0.05; **, p < 0.01, n = 4) and concentration-dependent (B; *, p < 0.05; **, p < 0.01; ***, p < 0.001, n = 3) manner in OV2008 cells but not in C13* cells. C, OV2008 and C13* cells were cultured with CDDP (10 μm, 6 h; DMSO as control (CTL)), and the abundance of each Opa1 splice form was examined by quantitative PCR. Forms 1, 2, 4, 5, 7, and 8 were expressed in OV2008 and C13* cells, and the abundance of the above forms are higher in OV2008 cells than C13* cells (p < 0.05, n = 3). V with a number represents specific form of Opa1. D, CDDP decreased all Opa1 splice forms detected in OV2008 cells, not in C13* cells (p < 0.05, n = 3).
Opa1 has eight mRNA splice forms. The abundance of each mRNA splice form and their response to CDDP treatment in chemosensitive and chemoresistant CECA cells were examined by quantitative PCR. Variants 1, 2, 4, 5, 7, and 8 were present in OV2008 and C13* cells, and variants 1 and 7 are the most predominant forms. The abundance of the above variants are higher in the sensitive cells (OV2008) than in its resistant counterpart (C13*) (p < 0.05, Fig. 2C). CDDP significantly decreased the abundance of variants 1, 2, 4, 5, 7, and 8 in OV2008 cells, but not in C13* cells (p < 0.05, Fig. 2D).
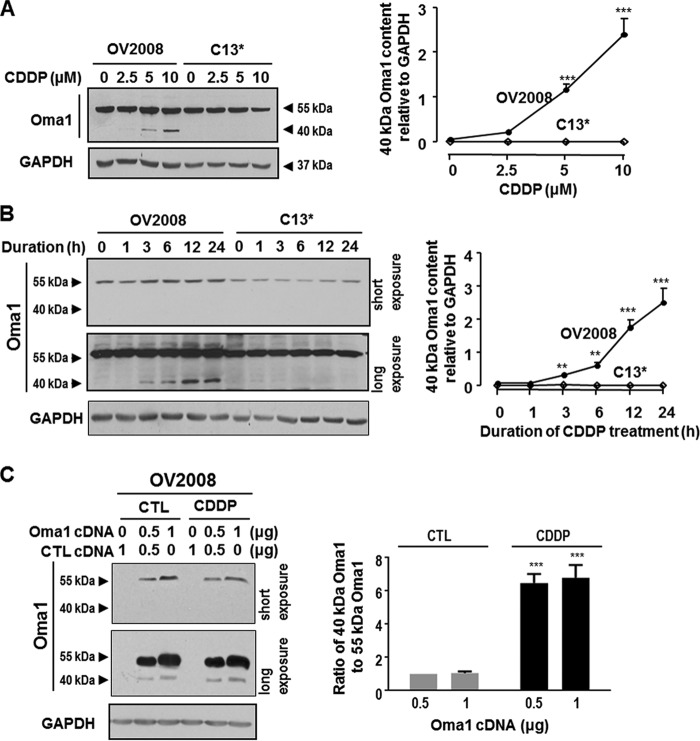
CDDP Increased Oma1 Content (40 kDa) in a Time- and Concentration-dependent Manner in Chemosensitive but Not Chemoresistant CECA Cells
Oma1 is a novel protease believed to be involved in L-Opa1 processing in mammalian cells (10–12). To examine expression of Oma1 and its possible role in regulating chemosensitivity, OV2008 cells and C13* cells were cultured with CDDP (0–10 μm, 0–24 h), and Oma1 content was examined by Western blot. Two Oma1 forms (55 and 40 kDa) were found in OV2008 cells. Oma1 content (40 kDa) significantly increased in a concentration-dependent (Fig. 3A, p < 0.001) and time-dependent (Fig. 3B, p < 0.001) manner in OV2008 cells in respond to CDDP. Interestingly, Oma1 (40 kDa) was not present in C13* cells irrespective of the presence of CDDP. In addition, CDDP failed to influence Oma1 content (55 kDa) in both OV2008 and C13* cells. To examine the relationship between the two Oma1 forms in OVCA cells, Oma1-Myc construct was transfected in OV2008 cells, which were subsequently challenged with CDDP. The expression of Myc was investigated by Western blot. Whereas two bands (55 and 40 kDa) were recognized using an anti-Myc antibody, CDDP increased the ratio of the 40-kDa band to 55-kDa band (p < 0.001, Fig. 3C).
FIGURE 3.
Different forms of Oma1 in chemosensitive and chemoresistant CECA cells. A and B, OV2008 and C13* cells were cultured with CDDP at different concentrations (A; 0–10 μm, 24 h) or for different duration (B; 0–24 h, 10 μm). Contents of Oma1 and GAPDH (loading control) were examined by Western blotting (n = 3). Two forms of Oma1 (55 and 40 kDa) were present in OV2008 cells but only one (55 kDa) in C13* cells when cultured in the presence of CDDP. Oma1 contents (40 kDa) in OV2008 cells but not in C13* cells significantly increased in the presence of CDDP in a concentration- and time-dependent manner. C, OV2008 cells were transfected with Myc-tagged Oma1 cDNA or control cDNA (CTL; vector) and subsequently cultured with CDDP (10 μm, 24 h) or control (DMSO). Two Myc bands (55 and 40 kDa) were detected by Western blot with anti-Myc antibody, with the ratio of 40-kDa band to 55-kDa band significantly increased in the presence of CDDP (***, p < 0.001, n = 3).
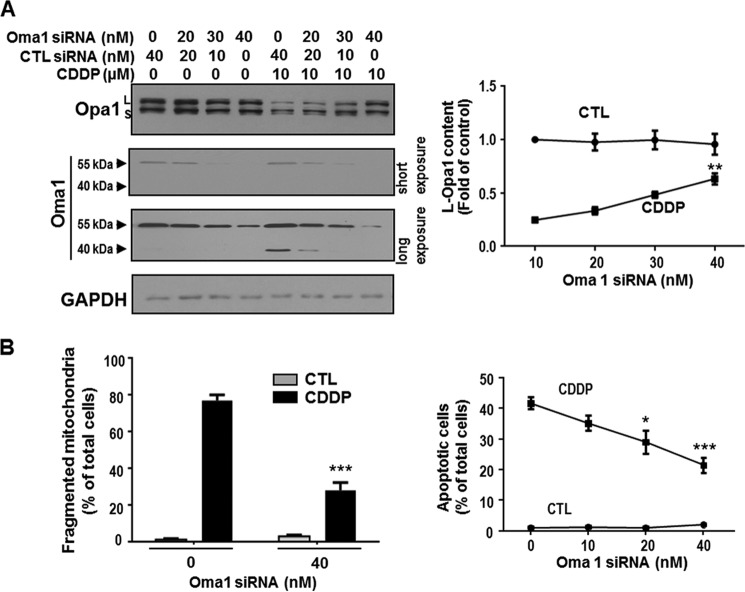
Oma1 Is Responsible for CDDP-induced L-Opa1 Processing, Mitochondrial Fragmentation, and Apoptosis in Chemosensitive CECA Cells
To determine whether Oma1 is required for CDDP-induced L-Opa1 processing in chemosensitive CECA cells, Oma1 expression in OV2008 cells was silenced by RNA interference (scramble siRNA as control) and then treated with CDDP in vitro (0–10 μm, 24 h). Oma1 and L-Opa1 contents were examined by Western blot. Treatment of OV2008 cells with Oma1 siRNA significantly decreased CDDP-induced L-Opa1 loss (p < 0.01, Fig. 4A) and inhibited CDDP-induced mitochondrial fragmentation by 45% (a reduction of apoptotic rate from 42 to 25%) at 6 h (p < 0.001, Fig. 4B) and apoptosis by 40% at 24 h (p < 0.001, Fig. 4B).
FIGURE 4.
Oma1 knockdown attenuated CDDP-induced loss in L-Opa1 content, mitochondrial fragmentation, and apoptosis in OV2008 cells. A, OV2008 cells were treated with Oma1 siRNA or control siRNA (CTL; scramble siRNA) (0–40 nm, 24 h) and cultured with CDDP (10 μm, 24 h) or control (DMSO). L-Opa1 and Oma1 contents were examined by Western blot. Oma1 siRNA (40 nm) significantly attenuated the loss of L-Opa1 content induced by CDDP. B, left: OV2008 cells were treated with Oma1 siRNA or control siRNA (scramble siRNA) (0–40 nm, 24 h) and subsequently with CDDP (10 μm, 6 h) or control (DMSO). Mitochondria were immunostained with anti-Tom20 antibody and examined by immunofluorescence confocal microscopy. Oma1 siRNA significantly decreased mitochondrial fragmentation induced by CDDP (***, p < 0.001. n = 3). More than 100 cells were assessed in each experimental group. Right: OV2008 cells were treated with Oma1 siRNA or control siRNA (scramble siRNA) (40 nm, 24 h) and subsequently with CDDP (10 μm, 24 h) or control (DMSO). Apoptosis was examined by Hoechst assay. Oma1 siRNA significantly decreased apoptosis induced by CDDP (***, p < 0.001; n = 3). More than 300 cells were assessed in each experimental group.
p53 Is Required for CDDP-induced, Oma1-mediated L-Opa1 Processing, Mitochondrial Fragmentation, and Apoptosis
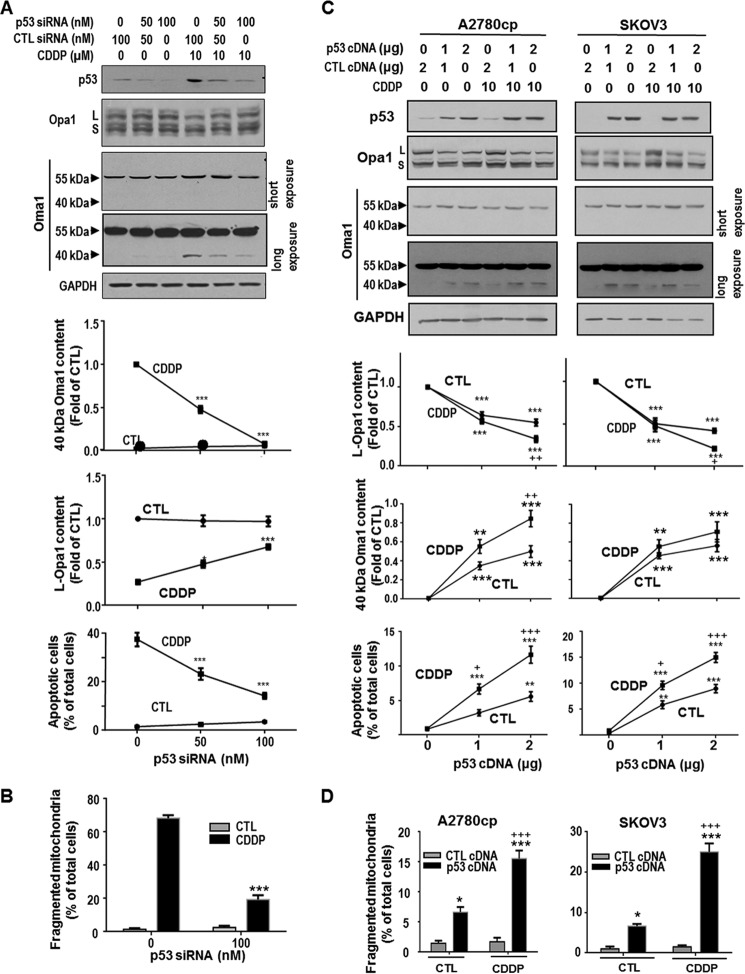
Because p53 targets mitochondria rapidly during CDDP-induced apoptosis in gynecologic cancer cells (3, 19), we hypothesized that p53 is required for Oma1 (40 kDa) increase, L-Opa1 processing, and mitochondria fragmentation in OV2008 cells induced by CDDP. To test this hypothesis, OV2008 cells were transfected with p53 siRNA (scramble siRNA as control), followed by CDDP treatment (0–10 μm, 24 h), and changes in p53, Oma1, and L-Opa1 contents were examined. Knockdown of p53 by siRNA inhibited CDDP-induced increase of Oma1 content (40 kDa; p < 0.001) and L-Opa1 loss (p < 0.001, Fig. 5A). Treatment of p53 siRNA also inhibited CDDP-induced mitochondrial fragmentation at 6 h (p < 0.001, Fig. 5B) and apoptosis at 24 h (p < 0.001, Fig. 5A).
FIGURE 5.
p53-dependent action of CDDP on Oma1 (40 kDa) and L-Opa1 contents, mitochondrial fragmentation, and apoptosis in gynecologic cancer cells. A, chemosensitive CECA cells (OV2008) were transfected with p53 siRNA or control siRNA (CTL; scramble siRNA) (0–100 nm, 24 h) and subsequently cultured with CDDP (10 μm, 24 h) or control (DMSO). The contents of p53, Oma1, Opa1, and GAPDH (loading control) were examined by Western blotting. Apoptosis was examined by Hoechst assay. Knockdown of p53 inhibited CDDP-induced increase in Oma1 content (40 kDa; ***, p < 0.001) and loss in L-Opa1 content (*, p < 0.05; ***, p < 0.001; n = 3). p53 siRNA significantly decreased apoptosis induced by CDDP (***, p < 0.001, n = 3). More than 300 cells were assessed in each experimental group. B, OV2008 cells were treated with p53 siRNA or control siRNA as described in A and subsequently cultured with CDDP (10 μm, 6 h) or control (DMSO). Mitochondria were immunostained with Tom20 antibody. Mitochondrial phenotypes were examined by immunofluorescence confocal microscopy. Knockdown of p53 significantly decreased CDDP-induced mitochondrial fission (***, p < 0.001, n = 3). More than 100 cells were assessed in each experimental group. C, chemoresistant A2780cp (p53 mutant) and SKOV3 (p53 null) cells were transfected with WT-p53 cDNA or control cDNA (empty vector) (0–2 μg, 24 h) and subsequently treated with CDDP (10 μm, 24 h) or control (DMSO). Contents of p53, L-Opa1, and Oma1 were examined by Western blot. Apoptosis was examined by Hoechst assay. Reconstitution of WT-p53 not only induced Oma1 cleavage, L-Opa1 processing, and apoptosis in both CDDP-resistant OVCA cell lines (***, p < 0.001; *, p < 0.05, versus control plasmid, n = 3), but markedly sensitized them to CDDP-induced Oma1 cleavage, L-Opa1 processing, and apoptosis (+, p < 0.05; ++, p < 0.01, +++, p < 0.001, versus control). More than 300 cells were assessed in each experimental group). D, A2780cp (p53 mutant) and SKOV3 (p53 null) cells were transfected with WT-p53 cDNA or control cDNA (empty vector) (0.44 μg/well on 8-well chamber slide, 24 h) and subsequently treated with CDDP (10 μm, 6 h) or control (DMSO). Mitochondria were immunostained with Tom20 antibody and examined by immunofluorescence confocal microscopy. Reconstitution of WT-p53 not only increased percentage of cells with fragmented mitochondria in both CDDP-resistant OVCA cell lines (***, p < 0.001; *, p < 0.05, versus control plasmid, n = 3), but markedly sensitized them to CDDP-induced mitochondrial fragmentation (+++, p < 0.001, versus DMSO). More than 100 cells were assessed in each experimental group.
To further explore the requirement of p53 in CDDP-induced mitochondria fragmentation in OVCA cells, WT-p53 was overexpressed in chemoresistant A2780cp (p53 mutant) and SKOV3 (p53 null) cells with transfection of p53 cDNA. p53 overexpression induced Oma1 increase (40 kDa), L-Opa1 processing, mitochondrial fragmentation, and apoptosis in these two cells lines (Fig. 5, C and D). Whereas CDDP alone had no effects on mitochondrial morphology in these p53-deficient CDDP resistant cells, reconstitution of wild type-p53 sensitize them to CDDP treatment (apoptosis, p < 0.001, Fig. 5C).
Phb1 Dissociates from Opa1 and Binds Phosphorylated p53 in Response to CDDP in Chemosensitive but Not Chemoresistant CECA Cells
Our previous publication has showed that CDDP-induced p53 phosphorylation at Ser-15 and Ser-20 is essential for apoptosis in chemosensitive CECA cells (20, 24). Recent publication suggested a link between Phb1 and p53 apoptotic pathway (15).
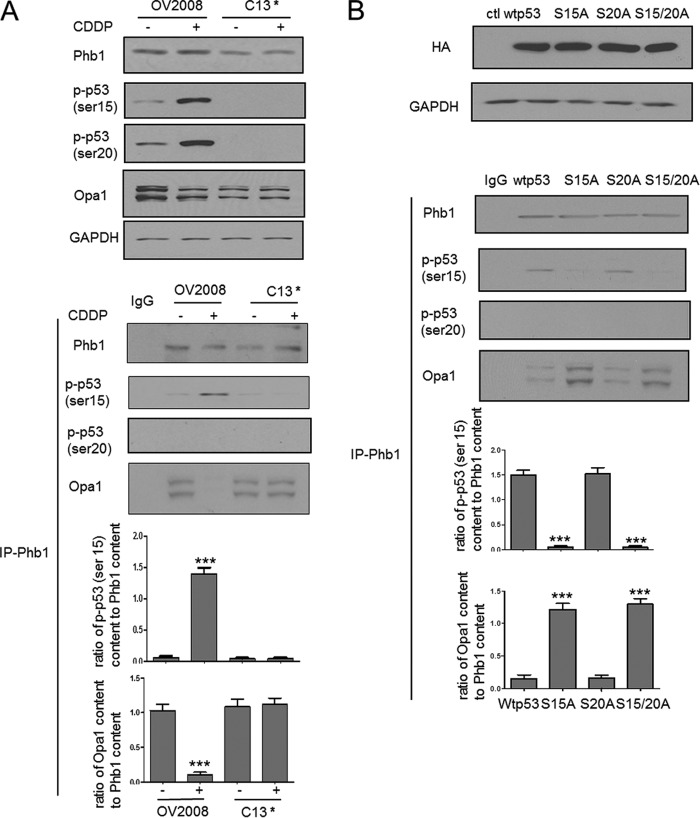
We therefore investigated the interaction between Phb1 and phosphorylated p53 (phospho-p53). OV2008 and C13* cells were treated with CDDP (0–10 μm, 6 h). Phb1, p-p53 (Ser-15), p-p53 (Ser-20), Opa1, and GAPDH contents were examined in the whole cell lysates. CDDP increased the content of both phospho-p53 (serine 15 and 20) in the chemosensitive cells but not in their resistant counterpart. Phb1 immunoprecipitates from OV2008 and C13* cells were immune blotted with anti-phospho- p53 (serine 15 and serine 20). CDDP increased the interaction of phospho-p53 (serine 15) and Phb1 in OV2008 cells but not in C13* cells (p < 0.001, Fig. 6A). In contrast, phospho-p53 (serine 20) did not interact with Phb1 irrespective of the presence of CDDP (Fig. 6A). Interestingly, Opa1 was also detected in both OV2008 and C13* cells without CDDP treatment, and CDDP decreased the interaction of Opa1 and Phb1 in OV2008 cells but not in C13* cells (p < 0.001, Fig. 6A).
FIGURE 6.
Phosphorylated p53 (serine 15) dissociates Phb1 from Opa1-Phb1 complex. A, OV2008 and C13* cells were treated with CDDP (0–10 μm, 6 h). Contents of Phb1, p-p53 (Ser-15), p-p53 (Ser-20), Opa1, and GAPDH were examined by Western blot. CDDP increased the content of both phospho-p53 (serine 15 and 20) in the chemosensitive cells but not in their resistant counterpart (n = 3). Cell lysates were immunoprecipitated with IgG (control (ctl); lane 1) or Phb1 antibody. Protein-protein interaction was determined by Western immunoprecipitation. Phb1 immunoprecipitates were immunoblotted (immunoprecipitation, Phb1; Western blot, Opa1, phosphorylated p53 (serine 15 and serine 20)). CDDP increased the interaction of phospho-p53 (serine 15, not serine 20) and Phb1 in OV2008 cells, not in C13* cells (***, p < 0.001, versus DMSO, n = 3). Results show representative images from three independent experiments. B, OV2008 cells were transfected with wild type p53, serine 15 mutant p53, serine 20 mutant p53, serine 15 plus serine 20 mutant p53 plasmid, and treated with CDDP (10 μm, 6 h). The overexpression was confirmed by probing HA tag in the whole cell lysates. Protein-protein interaction was determined by Western immunoprecipitation. Phb1 immunoprecipitates were immunoblotted (immunoprecipitation, Phb1; Western blot, Opa1, phosphorylated p53 (serine 15 and serine 20)). Serine 15 mutant p53 and serine 15 plus serine 20 mutant p53 significantly enhanced the interaction of Opa1 and Phb1 and decreased that of Phb1 and phospho-p53 (serine 15) (***, p < 0.001, versus wild type p53, n = 3). Results show representative images from three independent experiments.
To further test whether phosphorylated p53 (serine 15 and serine 20) is required for the CDDP-induced dissociation of Opa1 from the Opa1-Phb1 complex, we transfected OV2008 cells with wild type p53, serine 15 mutant p53, serine 20 mutant p53, and serine 15 plus serine 20 mutant p53 plasmid, and treated the cells with CDDP (10 μm, 6 h). Overexpression was confirmed by probing HA tag in the whole cell lysates. Phb1 immunoprecipitates were immunoblotted, and interactions between Phb1 and Opa1 and Phb1 and phospho-p53 (serine 15 and serine 20) were examined. The interaction of Phb1 with phospho-p53 (serine 15) was significantly reduced (p < 0.001, Fig. 6B), whereas that between Opa1 and Phb1 was significantly enhanced (p < 0.001, Fig. 6B) by serine 15 mutation and serine 15 plus 20 mutation. In contrast, mutation of p53 at serine 20 alone had no significant effect on p-p53 (serine 15)-Phb1 and Phb1-Opa1 interactions (p > 0.05, Fig. 6B).
DISCUSSION
L-Opa1 processing is essential for mitochondrial fission and subsequent cell death (27). Therefore, it is important to understand the molecular mechanisms by which L-Opa1 expression and processing is regulated. In the present study, we have shown for the first time dysregulated mitochondrial dynamics in chemoresistant cancer cells and the involvement of Oma1 and Opa1 in the regulation of mitochondrial dynamics in chemosensitive cancer cells. We have also demonstrated for the first time that activation p53 at the serine 15 but not serine 20 is required for CDDP-induced Oma1-mediated L-Opa1 processing and mitochondrial fragmentation in these chemosensitive gynecologic cancer cells. The action of phospho-Ser-15 p53 in these processes involves its binding to and displacement of Opa1 from the Phb1-Opa1 complex.
Mitochondrial fission and fusion influence nearly all aspects of mitochondrial function, including respiration, calcium buffering, and apoptosis (28–31). However, to our knowledge, whether mitochondrial dynamics are dysregulated in chemoresistant cells has not been reported. Here, for the first time, our results indicate a much higher level of mitochondrial fusion in chemoresistant gynecologic cancer cells than in chemosensitive counterparts, as evident by a higher percentage of cells with tubular mitochondria, suggesting mitochondria fusion may be a determinant in chemoresistance. Fused mitochondria in chemoresistant cells appeared incapable of undergoing fragmentation in response to CDDP treatment. Dysregulated mitochondrial dynamics may be the key reason for chemoresistance. Interestingly, Tondera et al. (32) reported mitochondria hyperfusion could be induced by selective stresses and conferred the cells resistance to stress. Together with our present observations, these findings indicate that fused mitochondria promote cell survival, which could be due to better mitochondria function such as ATP production and transportation.
Mitochondria dynamics are controlled by the action of key regulating proteins, which include fission proteins Drp1 and Fis1, and fusion proteins Mfn1, Mfn2, and Opa1. However, with the exception of Opa1, these regulatory molecules in both sensitive and resistant CECA cells appeared not to be affected by CDDP treatment. Our findings do not exclude the possible involvement of these key mitochondrial fission/fusion proteins in the regulation of mitochondrial dynamics and CDDP responsiveness. Whether these molecules regulate mitochondrial dynamics and chemoresistance by post-translational modifications (i.e. phosphorylation) needs to be further investigated. Interestingly, the chemoresistant cells (C13*) have lower expression of Opa1, whereas exhibit higher activity of fusion and resistant to CDDP-induced mitochondrial fragmentation, suggesting that Opa1 level is not a “rate-limiting” or determinant of mitochondrial fusion and, that other events, such as the balance of long and short forms of Opa1, could be more important.
In the present studies, five Opa1 forms have been recognized, and L-Opa1 isoforms are processed in chemosensitive CECA cells during CDDP-induced apoptosis, which is consistent with other reported observations (9). Interestingly, we have shown for the first time chemoresistant cells (C13*) do not exhibit two isoforms (88 and 81 kDa), suggesting that either certain proteases involved in the processing of long forms to short forms could be absent or the expression of the short forms were too low to be detectable by the current assay technique. These possibilities require further investigation. Although the function of individual Opa1 form remains unclear, the three isoforms in chemoresistant cells might be more essential for hyper-fused mitochondria. CDDP induces L-Opa1 processing, mitochondrial fragmentation, and apoptosis in chemosensitive CECA cells (OV2008) but not in its resistant variant (C13*), confirming that stabilized L-Opa1 is needed for mitochondrial fusion and cells survival. Interestingly, our results also suggest that the abundance of different Opa1 splice forms is associated with chemosensitivity of the CECA cells, and CDDP decreases L-Opa1 contents partly by down-regulating the abundance of Opa1 mRNA.
Although different reports and our present study suggest that Oma1 is responsible for proteolytic inactivation of Opa1, certain issues remain unclear. We found Oma1 exists as a 55- and a 40-kDa forms in chemosensitive cancer cells, the latter evident only with CDDP treatment, although at a much lower level compared with the 55-kDa form. Interestingly, the 40-kDa form of Oma1 increased with CDDP treatment, a phenomenon absent in chemoresistant cells irrespective of the presence of CDDP. We thus propose that the 40-kDa form is functional form of Oma1 and is derived from the 55-kDa form. This notion is contrary to the opinion of the van der Bliek group (11) who, by using Oma1-HA construct in Hela cells, showed that Oma1 exists as a 60- and a 40-kDa form and that the 60-kDa form increased during mitochondrial fragmentation. The relationship and function of two Oma1 forms remain unclear. Whether the observed difference of the Oma1 form pattern during mitochondrial fragmentation is due to differences in cell types examined requires further investigations. Consistent with Opa1 mRNA results, knockdown of Oma1 by siRNA failed to completely abolish the effect of CDDP in inducing L-Opa1 loss, suggesting CDDP-induced Opa1 mRNA abundance decrease may be involved in L-Opa1 loss. Another possible mechanism is that other proteases are activated or still functioning when Oma1 is knocked down such as the matrix AAA protease AFG3L1 and -2, and this needs to be further investigated (10). Interestingly, Oma1 knockdown not only prevents CDDP-induced Opa1 processing but also increased the level of L-Opa1. These findings are consistent with this notion that Oma1 is involved in the processing of L-Opa1. Whether Oma1 is directly involved in the degradation of Opa1 requires further investigations.
Although it is known that p53 could mediate apoptosis in a transcription-independent manner by targeting mitochondria and regulates outer mitochondrial membrane potential (33–35), very little evidence indicates that p53 is involved in the regulation of mitochondrial dynamics. Our present study strongly suggests that p53 is involved in the regulation of mitochondrial dynamics by controlling the production of Oma1 (40 kDa) and L-Opa1 processing. We thus propose that p53 targets mitochondria and activates a yet to be determined protease, which converts full-length Oma1 (55 kDa) to functional Oma1 (40 kDa), and the latter processes L-Opa1 and induces mitochondrial fission in chemosensitive cells. Our previous study has showed that p53 fails to translocate to the mitochondria and to induce Smac release in chemoresistant cells when challenged with CDDP (3, 19). This may in part explain the absence of the functional Oma1 form, stabilized L-Opa1, and fused mitochondria in chemoresistant cells. A requirement for p53 in the regulation of mitochondrial dynamics is further supported by the observation that p53 reconstitution not only induced mitochondrial fragmentation in p53 mutant or null chemoresistant OVCA cells but also sensitized the cells to CDDP-induced mitochondrial fragmentation.
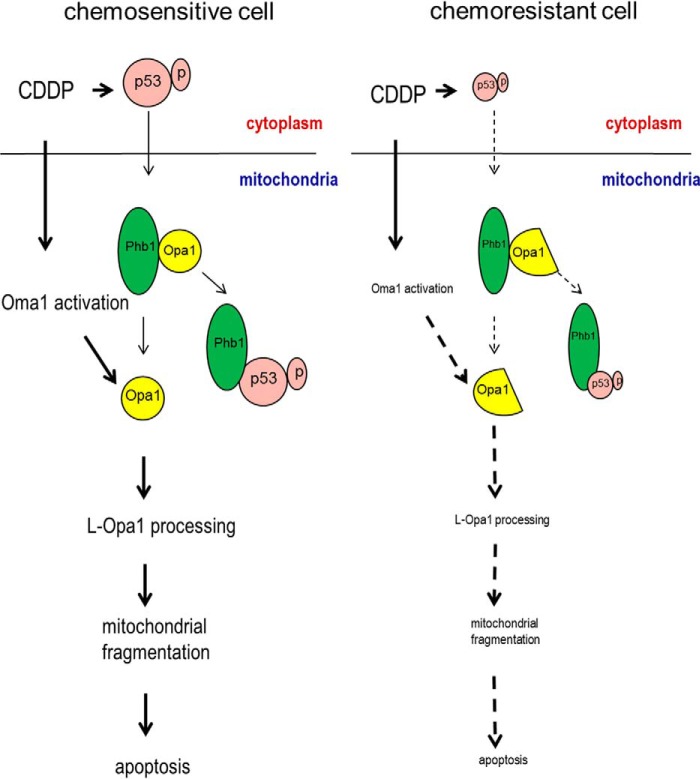
Phb1 is localized in mitochondria (14). Accumulated evidence suggests that it is involved in the regulation of L-Opa1 processing (13, 14), although the underlying mechanism is not clear. Our results strongly suggest Phb1 and activated p53 are critical in the regulation of L-Opa1 processing by L-Opa1 sequestration and protection from proteolysis. We have showed that phospho- p53 (serine 15) targeting mitochondria is required for apoptosis induced by CDDP in chemosensitive cells (24). This study suggests that phosphorylated p53 (serine 15 but not serine 20) induce apoptosis by regulating mitochondrial dynamics through competitive binding to Phb1, therefore exposing L-Opa1 to its protease (Fig. 7). To our knowledge, this is the first study showing the mechanism by which p53 regulates mitochondrial dynamics.
FIGURE 7.
A hypothetical model illustrating the involvement of p53, Phb1, Oma1, and Opa1 in the regulation of mitochondrial fragmentation, apoptosis, and chemosensitivity in gynecologic cells. In chemosensitive cells, CDDP induces p53 phosphorylation (serine 15), which translocates to the mitochondria and targets the complex of Phb1 and Opa1 (five forms). Phospho-p53 (serine 15) binds to Phb1 and releases Opa1. CDDP also induces Oma1 activation, which processes L-Opa1 and induces mitochondrial fragmentation and subsequent apoptosis. In chemoresistant cells, CDDP-induced p53 phosphorylation is minimal or inhibited and therefore stabilizes the complex of Phb1 and Opa1 (three forms). CDDP-induced Oma1 activation is also inhibited. L-Opa1 is protected from being processed, leading to the failure of CDDP to induce mitochondrial fragmentation and apoptosis.
In summary, we have demonstrated that CDDP-induced Oma1 (40 kDa) increase, L-Opa1 processing as well as mitochondrial fragmentation are differentially regulated in chemosensitive and chemoresistant gynecologic cancer cells. Mitochondrial dynamics is regulated by p53- and Oma1-mediated Opa1 processing. The binding of phosphorylated 53 (serine 15) to Phb1 is needed for consequent L-Opa1 release. Determining the molecular mechanisms by which p53 controls Oma1-mediated L-Opa1 processing may contribute to the current understanding of mitochondrial dynamics and apoptosis and, ultimately, of the mechanisms of chemoresistance in human gynecologic cancer.
This work was supported by Canadian Institutes of Health Research Grant MOP-126144.
- L-Opa1
- long form of Opa1
- OVCA
- ovarian cancer
- CECA
- cervical cancer
- CDDP
- cisplatin
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1. Li J., Feng Q., Kim J. M., Schneiderman D., Liston P., Li M., Vanderhyden B., Faught W., Fung M. F., Senterman M., Korneluk R. G., Tsang B. K. (2001) Human ovarian cancer and cisplatin resistance: possible role of inhibitor of apoptosis proteins. Endocrinology 142, 370–380 [DOI] [PubMed] [Google Scholar]
- 2. Kigawa J., Sato S., Shimada M., Takahashi M., Itamochi H., Kanamori Y., Terakawa N. (2001) p53 gene status and chemosensitivity in ovarian cancer. Hum. Cell 14, 165–171 [PubMed] [Google Scholar]
- 3. Yang X., Fraser M., Moll U. M., Basak A., Tsang B. K. (2006) Akt-mediated cisplatin resistance in ovarian cancer: modulation of p53 action on caspase-dependent mitochondrial death pathway. Cancer Res. 66, 3126–3136 [DOI] [PubMed] [Google Scholar]
- 4. Otera H., Ishihara N., Mihara K. (2013) New insights into the function and regulation of mitochondrial fission. Biochim. Biophys. Acta 1833, 1256–1268 [DOI] [PubMed] [Google Scholar]
- 5. Shutt T. E., McBride H. M. (2013) Staying cool in difficult times: mitochondrial dynamics, quality control and the stress response. Biochim. Biophys. Acta 1833, 417–424 [DOI] [PubMed] [Google Scholar]
- 6. Chen H., Chan D. C. (2009) Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum. Mol. Genet. 18, R169–R176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Twig G., Elorza A., Molina A. J., Mohamed H., Wikstrom J. D., Walzer G., Stiles L., Haigh S. E., Katz S., Las G., Alroy J., Wu M., Py B. F., Yuan J., Deeney J. T., Corkey B. E., Shirihai O. S. (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Griparic L., Kanazawa T., van der Bliek A. M. (2007) Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J. Cell Biol. 178, 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guillery O., Malka F., Landes T., Guillou E., Blackstone C., Lombès A., Belenguer P., Arnoult D., Rojo M. (2008) Metalloprotease-mediated OPA1 processing is modulated by the mitochondrial membrane potential. Biol. Cell 100, 315–325 [DOI] [PubMed] [Google Scholar]
- 10. Ehses S., Raschke I., Mancuso G., Bernacchia A., Geimer S., Tondera D., Martinou J. C., Westermann B., Rugarli E. I., Langer T. (2009) Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 187, 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Head B., Griparic L., Amiri M., Gandre-Babbe S., van der Bliek A. M. (2009) Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 187, 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Quirós P. M., Ramsay A. J., Sala D., Fernández-Vizarra E., Rodríguez F., Peinado J. R., Fernández-García M. S., Vega J. A., Enríquez J. A., Zorzano A., López-Otín C. (2012) Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. EMBO J. 31, 2117–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Merkwirth C., Dargazanli S., Tatsuta T., Geimer S., Löwer B., Wunderlich F. T., von Kleist-Retzow J. C., Waisman A., Westermann B., Langer T. (2008) Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 22, 476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Osman C., Merkwirth C., Langer T. (2009) Prohibitins and the functional compartmentalization of mitochondrial membranes. J. Cell Sci. 122, 3823–3830 [DOI] [PubMed] [Google Scholar]
- 15. Chander H., Halpern M., Resnick-Silverman L., Manfredi J. J., Germain D. (2010) Skp2B attenuates p53 function by inhibiting prohibitin. EMBO Rep. 11, 220–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shaw T. J., Senterman M. K., Dawson K., Crane C. A., Vanderhyden B. C. (2004) Characterization of intraperitoneal, orthotopic, and metastatic xenograft models of human ovarian cancer. Mol. Ther. 10, 1032–1042 [DOI] [PubMed] [Google Scholar]
- 17. Abedini M. R., Muller E. J., Bergeron R., Gray D. A., Tsang B. K. (2010) Akt promotes chemoresistance in human ovarian cancer cells by modulating cisplatin-induced, p53-dependent ubiquitination of FLICE-like inhibitory protein. Oncogene 29, 11–25 [DOI] [PubMed] [Google Scholar]
- 18. Abedini M. R., Qiu Q., Yan X., Tsang B. K. (2004) Possible role of FLICE-like inhibitory protein (FLIP) in chemoresistant ovarian cancer cells in vitro. Oncogene 23, 6997–7004 [DOI] [PubMed] [Google Scholar]
- 19. Woo M. G., Xue K., Liu J., McBride H., Tsang B. K. (2012) Calpain-mediated processing of p53-associated parkin-like cytoplasmic protein (PARC) affects chemosensitivity of human ovarian cancer cells by promoting p53 subcellular trafficking. J. Biol. Chem. 287, 3963–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fraser M., Leung B. M., Yan X., Dan H. C., Cheng J. Q., Tsang B. K. (2003) p53 is a determinant of X-linked inhibitor of apoptosis protein/Akt-mediated chemoresistance in human ovarian cancer cells. Cancer Res. 63, 7081–7088 [PubMed] [Google Scholar]
- 21. Rambold A. S., Kostelecky B., Elia N., Lippincott-Schwartz J. (2011) Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. U.S.A. 108, 10190–10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taguchi N., Ishihara N., Jofuku A., Oka T., Mihara K. (2007) Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 282, 11521–11529 [DOI] [PubMed] [Google Scholar]
- 23. Mai S., Klinkenberg M., Auburger G., Bereiter-Hahn J., Jendrach M. (2010) Decreased expression of Drp1 and Fis1 mediates mitochondrial elongation in senescent cells and enhances resistance to oxidative stress through PINK1. J. Cell Sci. 123, 917–926 [DOI] [PubMed] [Google Scholar]
- 24. Fraser M., Bai T., Tsang B. K. (2008) Akt promotes cisplatin resistance in human ovarian cancer cells through inhibition of p53 phosphorylation and nuclear function. Int. J. Cancer 122, 534–546 [DOI] [PubMed] [Google Scholar]
- 25. Olichon A., Elachouri G., Baricault L., Delettre C., Belenguer P., Lenaers G. (2007) OPA1 alternate splicing uncouples an evolutionary conserved function in mitochondrial fusion from a vertebrate restricted function in apoptosis. Cell Death Differ. 14, 682–692 [DOI] [PubMed] [Google Scholar]
- 26. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 27. Yamaguchi R., Perkins G. (2009) Dynamics of mitochondrial structure during apoptosis and the enigma of Opa1. Biochim. Biophys. Acta 1787, 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aon M. A., Cortassa S., O'Rourke B. (2004) Percolation and criticality in a mitochondrial network. Proc. Natl. Acad. Sci. U.S.A. 101, 4447–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frieden M., James D., Castelbou C., Danckaert A., Martinou J. C., Demaurex N. (2004) Ca2+ homeostasis during mitochondrial fragmentation and perinuclear clustering induced by hFis1. J. Biol. Chem. 279, 22704–22714 [DOI] [PubMed] [Google Scholar]
- 30. Lee Y. J., Jeong S. Y., Karbowski M., Smith C. L., Youle R. J. (2004) Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol. Biol. Cell 15, 5001–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Skulachev V. P. (2001) Mitochondrial filaments and clusters as intracellular power-transmitting cables. Trends Biochem. Sci. 26, 23–29 [DOI] [PubMed] [Google Scholar]
- 32. Tondera D., Grandemange S., Jourdain A., Karbowski M., Mattenberger Y., Herzig S., Da Cruz S., Clerc P., Raschke I., Merkwirth C., Ehses S., Krause F., Chan D. C., Alexander C., Bauer C., Youle R., Langer T., Martinou J. C. (2009) SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 28, 1589–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shibue T., Suzuki S., Okamoto H., Yoshida H., Ohba Y., Takaoka A., Taniguchi T. (2006) Differential contribution of Puma and Noxa in dual regulation of p53-mediated apoptotic pathways. EMBO J. 25, 4952–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wolff S., Erster S., Palacios G., Moll U. M. (2008) p53's mitochondrial translocation and MOMP action is independent of Puma and Bax and severely disrupts mitochondrial membrane integrity. Cell Res. 18, 733–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moll U. M., Marchenko N., Zhang X. K. (2006) p53 and Nur77/TR3-transcription factors that directly target mitochondria for cell death induction. Oncogene 25, 4725–4743 [DOI] [PubMed] [Google Scholar]