Abstract
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive cancer with poor survival rates and frequently carries oncogenic KRAS mutation. However, KRAS has thus far not been a viable therapeutic target. We found that the abundance of YAP mRNA, which encodes Yes-associated protein (YAP), a protein regulated by the Hippo pathway during tissue development and homeostasis, was increased in human PDAC tissue compared with that in normal pancreatic epithelia. In genetically engineered KrasG12D and KrasG12D: Trp53R172H mouse models, pancreas-specific deletion of Yap halted the progression of early neoplastic lesions to PDAC without affecting normal pancreatic development and endocrine function. Although Yap was dispensable for acinar to ductal metaplasia (ADM), an initial step in the progression to PDAC, Yap was critically required for the proliferation of mutant Kras or Kras:Trp53 neoplastic pancreatic ductal cells in culture and for their growth and progression to invasive PDAC in mice. Yap functioned as a critical transcriptional switch downstream of the oncogenic KRAS–mitogen-activated protein kinase (MAPK) pathway, promoting the expression of genes encoding secretory factors that cumulatively sustained neoplastic proliferation, a tumorigenic stromal response in the tumor microenvironment, and PDAC progression in Kras and Kras: Trp53 mutant pancreas tissue. Together, our findings identified Yap as a critical oncogenic KRAS effector and a promising therapeutic target for PDAC and possibly other types of KRAS-mutant cancers.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal cancers with a 5-year survival rate of less than 5% (1). PDAC tumors are thought to originate from mature acinar cells that transdifferentiate into ductal-like cells, a process known as acinar to ductal metaplasia (ADM) (2, 3). ADM can be induced by pancreatitis or oncogenic mutations. Only under sustained oncogenic insult will pancreatic cells that have undergone ADM progress through a series of histopathological stages called pancreatic intraepithelial neoplasias (PanINs) to invasive PDAC.
Nearly all PDAC and a high percentage of early PanIN lesions have KRAS mutations (4). The human ADM-to-PanIN-to-PDAC progression has been recapitulated using genetically engineered mouse models (GEMMs) in which endogenous expression of oncogenic Kras is induced in the developing pancreas, either alone or in conjunction with the in-activation of other commonly mutated tumor suppressor genes, such as p16, Trp53, or Smad4 (5–8). When oncogenic Kras is activated alone, ADM and early PanINs readily develop, but progression into late-stage PanINs and eventually PDAC is delayed (8). This process is substantially accelerated by mutation of p16, Trp53, or Smad4, suggesting that the proteins encoded by these genes inhibit the proliferative signals mediated by mutant Kras (5–7).
Despite recent progress, oncogenic KRAS itself remains an extremely challenging therapeutic target (9, 10). Thus, much effort is devoted to identifying critical downstream effectors of oncogenic KRAS. Because the development and progression of PDAC are strongly influenced by the microenvironment, GEMMs represent the most physiologically relevant models. In the past few years, the aforementioned GEMMs have been crossed with mice harboring various loss-of-function mutant alleles, leading to the identification of genes required for the initiation (EGFR, ADM17, PDK1, Glil, and Sox9) or progression (Racl, STAT3, IL-6, NEMO, and IKK2) of PDAC in the context of Kras mutation, either alone or concurrent with p16 deletion (11–22). However, introduction of Trp53 mutations into these models (which mimic the TP53 mutations found in the majority of human PDAC) mitigated the requirement for many of these genes during PDAC initiation and progression (11,17,19,20). In accord with these findings in GEMMs, clinical trials of inhibitors targeting the epidermal growth factor receptor (EGFR), the RAF-mitogen-activated protein kinase (MAPK) pathway, phosphoinositide 3-kinase, or the Hedgehog pathway have been largely unsuccessful (23). Thus, there remains an urgent need to discover the “Achilles’ heel” that governs PDAC development in the presence of KRAS:TP53 mutations.
The Yes-associated protein (YAP), encoded by YAP1, referred to herein simply as Yap, is a bona fide oncoprotein, and its abundance and activity are frequently increased in many types of cancers (24–36). YAP is a transcriptional coactivator that partners with the TEAD family of transcription factors to promote the expression of pro-proliferative and antiapoptotic genes (37–46). Extensive genetic studies that focused primarily on organ size control in Drosophila and mice identified that the Hippo pathway is the canonical regulator of YAP activity (39, 47–50). The Hippo pathway is composed of a kinase cascade in which the MST1 and MST2 (MST1/2) Hippo kinases are facilitated by scaffold protein WW45 to phosphorylate the LATS1 andLATS2 (LATS1/2) kinases and their adaptor protein, MOB1 (43, 45, 51–53). Phosphorylated LATS1/2 kinases in turn phosphorylate YAP, inactivating YAP by causing it to be retained in the cytoplasm and degraded (54, 55). A host of factors [including cell density, extracellular matrix stiffness, G protein (heterotrimeric guanine nucleotide–binding protein)–coupled receptors, protease-activated receptors, EGFR, and leukemia inhibitory factor receptor] influence YAP activity by modulating the Hippo pathway (56–60). Additionally, accumulating evidence indicates that YAP activity can be regulated by noncanonical, Hippo-independent mechanisms (61–67).
GEMMs developed in recent years have revealed critical functions of YAP and the Hippo pathway in the maintenance of tissue homeostasis, the organ size checkpoint, and tumorigenesis. Tissue-specific overexpression of Yap or inactivation of Hippo signaling through the homozygous deletion of Mst1/2 or genes encoding other components of the Hippo pathway resulted in enlargement of the liver, heart, and intestine, as well as tumorigenesis in the liver (47,49,68, 69). In contrast, deletion of Mst1/2 or ectopic expression of Yap in the developing mouse pancreas induces ADM and impairs differentiation of exocrine and endocrine compartments without increasing the size of the pancreas or inducing pancreatic tumor development (70, 71). These studies demonstrate that activation of YAP alone is insufficient to induce PDAC, but have not determined whether YAP is necessary for PDAC development. Here, we examined YAP abundance in primary human PDAC, its role in mutant KRAS– and KRAS:TP53-mediated PDAC initiation and progression, and the molecular mechanisms underlying oncogenic RAS-YAP crosstalk.
RESULTS
YAP is expressed in normal and neoplastic pancreatic ductal cells
YAP abundance is increased in human PDAC (72, 73). Through a metaanalysis of published human PDAC microarray data sets (74–77), we confirmed that overall YAP mRNA abundance was significantly increased in human PDAC when compared to normal pancreatic tissues (fig. S1A). For reference, the genotypes of all GEMMs used in this study are listed in table S1. The KrasG12D/+:p48-Cre (KC) and KrasG12D/+:Trp53R172H/+:p48-Cre (KPC) GEMMs in which p48-Cre (also known as Ptfla-Cre) induces the expression of mutant Kras alone or together with mutant Trp53 in murine pancreatic epithelium fully recapitulate the pathogenesis of human PDAC and are generally regarded as two of the best GEMMs for human PDAC (23). By Western blot analysis, we found that Yap protein abundance was also markedly greater in pancreatic tissue from KPC mice that had developed PDAC compared with that from wild-type mice (fig. S1B).
To explore this association further, we performed immunohisto-chemistry (IHC) analysis for YAP on two human tissue microarrays (TMAs) containing 31 normal pancreata, 64 chronic pancreatitis or tumor adjacent tissues, and 140 PDAC tumor cores, as well as pancreatic sections from multiple wild-type, KC, and KPC mice (figs. S1, C to I, and S2A). Consistent with previous reports (72, 73), we found that YAP abundance was restricted to ductal and terminal-duct centroacinar cells in normal human and mouse pancreas tissue (figs. S1D and S2A and table S2). Variable amounts of cytoplasmic or nuclear YAP were also detected in a significant fraction of neoplastic ductal epithelial cells and stromal cells in almost all of the PDAC tumors and adjacent tissues containing benign lesions and chronic pancreatitis (fig. S1, C and E to G, and table S2). Similar patterns of Yap staining were observed in pancreata from KC and KPC mice that had early PanINs or fully established PDAC (fig. S1, H and I). Although a fraction of human PDAC tumors exhibited intense staining for YAP (particularly in late-stage tumors), the majority of human PDAC tumors appeared to have an abundance of YAP within individual tumor cells that was comparable to that in normal centroacinar and ductal cells (fig. S1, C to G, and table S2). Thus, the overall increase in YAP abundance in PDAC measured by micro-array and Western blot analyses (fig. S1, A and B) likely resulted from the expansion of neoplastic ductal and stromal cell populations, rather than from a net increase in YAP transcript and abundance in individual PDAC cells.
Yap is dispensable for normal pancreatic development and glucose metabolism
Because YAP is present in normal centroacinar and ductal cells, we investigated whether the deletion of Yap might affect the normal development or function of the pancreas by generating Yapflox/flox:p48-Cre (YC) mice. YC mice were born at the Mendelian ratio and had normal weight, physical appearance, and life expectancy. IHC analysis confirmed the complete loss of Yap expression in the pancreatic epithelium of YC mice (fig. S2A). Despite the absence of Yap, YC pancreata were indistinguishable from wild-type pancreata in overall histology by hematoxylin and eosin (H&E), staining patterns for major pancreatic cell lineage markers [including amylase (in acinar cells), cytokeratin 19 (CK19; in ductal cells), insulin (in β cells within the islets), and glucagon (in α cells within the islets)], or organization of ductal networks (fig. S2, B and C). A glucose tolerance test also revealed no apparent difference between YC and wild-type mice in their ability to modulate blood glucose levels (fig. S2D). Collectively, these data suggest that YAP deletion from the pancreatic epithelium is inconsequential to normal pancreatic development, homeostasis, and glucose metabolism.
Inactivation of Yap blocks PDAC development in Kras and Kras:Trp53 compound mutant pancreas
To determine whether Yap is involved in PDAC oncogenesis, we crossed Yapflox/flox with KC and KPC mice to generate KrasG12D/+:Yapflox/+:p48-Cre, KrasG12D/+p53R172H/+:Yapflox/+:p48-Cre,KrasG12D/+:Yapflox/flox:p48-Cre (KYC), and KrasG12D/+:p53R172H/+:Yapflox/flox:p48-Cre (KPYC) mice.
KC (including KmsG12D/+:Yap+/+:p48-Cre and KrasG12D/+:Yapflox/+:p48-Cre) or KPC (including KrasG12D/+:p53R172H/+:Yap+/+:p48-Cre and KrasG12D/+: p53R172H/+:Yapflox/+:p48-Cre) mice with one or two intact Yap alleles developed ADM and early PanINs from 4 to 8 weeks of age, respectively (table S1 and fig. S3). These ADM and early PanINs progressed through late-stage PanINs and eventually to invasive PDAC by 2 to 4 months in KPC mice, or from 6 months to 2 years in KC mice (Fig. 1, A to C, and fig. S3). In contrast, all the KYC and KPYC mice with homozygous Yap deletions completely lacked any late-stage PanINs or PDAC when analyzed at all the tested time points, even at the age of 12 months (for KC mice) or 10 months (for KPC mice) (Fig. 1, A to C, and fig. S3).
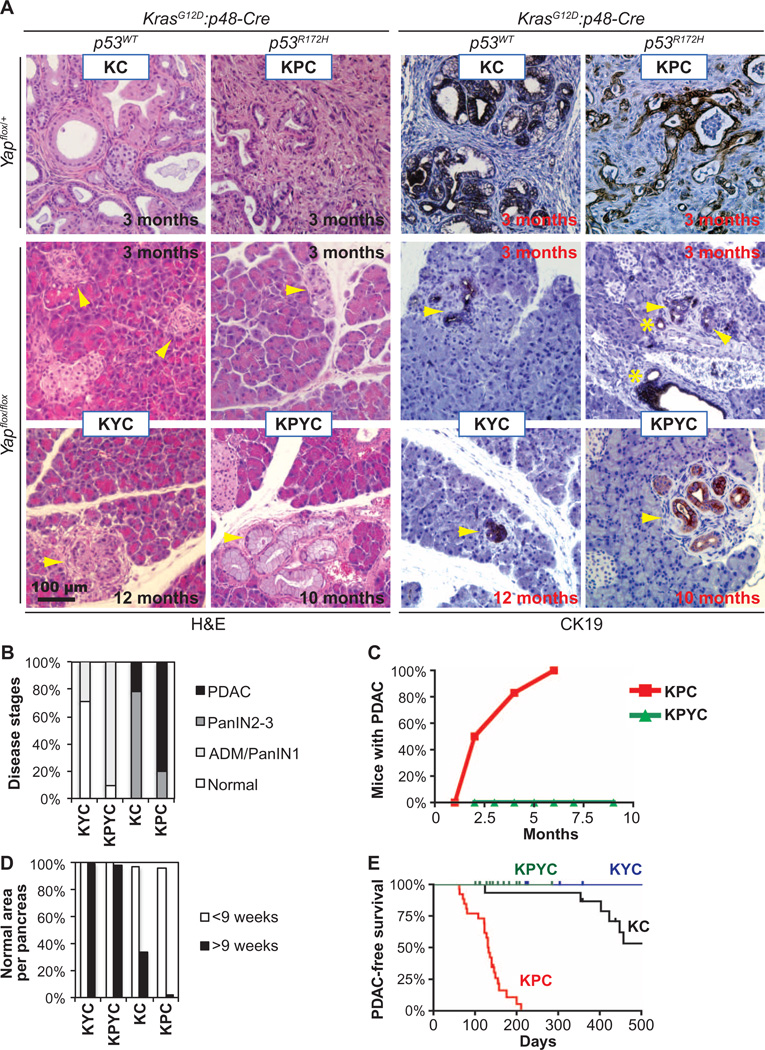
Fig. 1. Deletion of Yap blocks PDAC development in Kras and Kras:Trp53 mutant pancreas.
(A) Representative images of H&E and CK19 IHC staining of pancreatic sections from KC, KPC, KYC, and KPYC mice of indicated ages. Asterisks, normal ducts; arrowheads, ADM or early PanINs. Scale bar, 100 µm. (B) Quantification of the disease stages in KC (n = 11 mice), KPC (n = 10), KYC (n = 7), and KPYC (n = 11) mice older than 9 weeks. (C) Quantification of the percentage of KPC (n = 13) or KPYC (n = 26) mice that developed PDAC over time. (D) Quantification of the mean percentage of histologically normal areas in pancreata from KC (n = 14), KPC (n = 13), KYC (n = 9), and KPYC (n = 16) mice younger or older than 9 weeks of age. (E) PDAC-free survival analysis of KPC (n = 26), KPYC (n = 14), KC (n = 16), and KYC (n = 10) mice.
Although ADM and some PanIN-1 lesions that were CK19-positive and Yap-negative (CK19+Yap−) still developed in KYC and KPYC pancreata (Fig. 1, A and B, and fig. S3, A and B), nearly all pancreatic tissue areas in these mice were histologically normal across all age groups (Fig. 1, A and D, and fig. S3A). In contrast, from the age of 9 weeks, the number of lesions at various stages of progression increased in the pancreata from KC and KPC mice (Fig. 1, A and D, and fig. S3A).
Whereas nearly all KPC and half of KC mice died from PDAC by 6 months or 1.5 years of age, respectively, not a single KPYC or KYC mouse in our experimental cohort succumbed to PDAC within the same periods (Fig. 1E). These results clearly demonstrate that YAP is essential in mutant KRAS– and KRAS:TP53-mediated PDAC development.
Deletion of Yap does not affect ADM induced by oncogenic Kras or pancreatitis
ADM caused by KRAS mutation or pancreatic injury is thought to be the initiating step in PDAC development. Examination of pancreatic tissues from young KPC and KPYC mice (4 to 8 weeks old) showed that pERK+ (phosphorylated extracellular signal–regulated kinase):EGFR+:CK19+ ADM initiated at a similar rate in KPC and KPYC littermates (Fig. 2, A and B). In contrast to KPC pancreata, in which Yap was abundant in both ADMs and a fraction of the surrounding stromal cells, the CK19+:α-SMA− epithelial cells lining the ADM lesions were completely negative for Yap staining in KPYC mice (Fig. 2A and fig. S3B). These results suggest that deletion of YAP does not affect oncogenic KRAS– induced ADM in vivo.
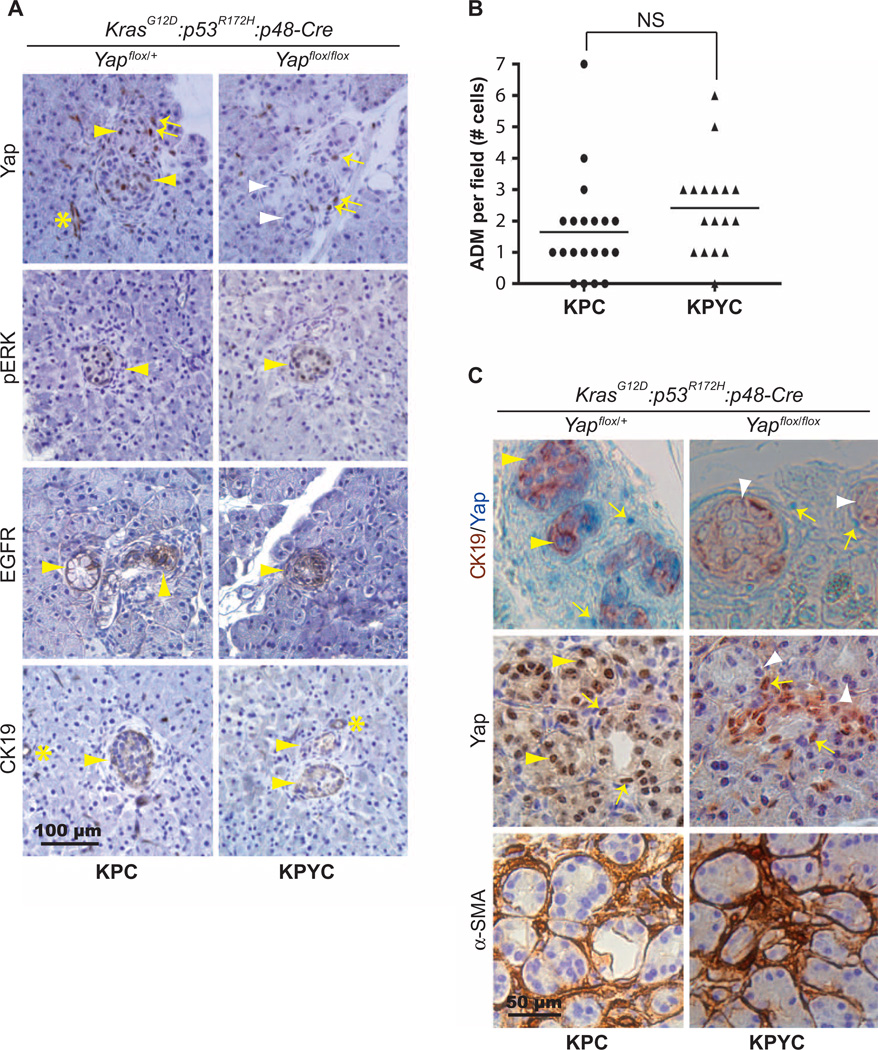
Fig. 2. Deletion of Yap does not affect ADM induced by oncogenic Kras.
(A) Representative images of IHC staining for Yap, phosphorylated ERK(pERK), EGFR, and CK19 in pancreatic sections from 7-week-old KPC and KPYC mice. Scale bar, 100 µm. (B) Number of ADM lesions per 20× microscopic field in pancreata from 7-week-old KPC (n = 20) and KPYC (n = 17) mice. Lines mark the median numbers of ADM lesions per field for each genotype. (C) Representative IHC images of CK19 (brown) and Yap (blue) double staining and staining of Yap and α-SMA in serial pancreatic sections from 7-week-old KPC and KPYC mice. Asterisks, positively stained normal ducts; arrows, positively stained stromal cells; yellow arrowheads, positively stained ADM lesions; white arrowheads, negatively stained ADM lesions. Scale bar, 50 µm.
Previous studies showed that the expression of oncogenic Kras also induces ADM in culture (21, 78). To perform an in vitro ADM assay, we isolated normal pancreatic acinar explants from KrasG12D:Yapflox/+ and KrasG12D:Yapflox/flox mice that did not carry p48-Cre, infected the cells with Ad-GFP and Ad-Cre-GFP viruses, and embedded them in collagen with serum-free medium. Consistent with those previous reports, we found that after several days of culture under this condition, both Ad-Cre-GFP–treated KrasG12D:Yapflox/+ and Kras:Yapflox/flox acinar clusters spontaneously transformed into CK19+ duct-like spherical structures, whereas Ad-GFP (con-trol)–treated cultures retained acinar morphology and remained CK19-negative (fig. S4, A and B). Genomic polymerase chain reaction (PCR) confirmed the complete recombination of KrasG12D and Yapflox alleles in Ad-Cre-GFP–treated KrasG12D: Yapflox/+ and KrasG12D:Yapflox/flox pancreatic epithelial explants (fig. S4C). Thus, YAP is also dispensable for oncogenic KRAS–induced ADM in culture.
To examine the role of Yap in the pancreatic response to injury, we treated wild-type and YC mice with the cholecystokinin analog caerulein. After caerulein treatment, both wild-type and YC pancreata developed acute pancreatitis, indicated by the formation of widespread CK19+:EGFR+ duct-like structures and an increase in the overall abundance of CK19 and EGFR (fig. S5, A and B). These results suggest that the ability of the pancreatic acini to undergo ductal transdifferentiation in response to injury also does not depend on YAP.
Yap is required for the proliferation of Kras and Kras:Trp53 mutant pancreatic ductal cells
To understand how Yap deletion blocked the progression of PanINs to PDAC, we stained pancreatic sections from KC, KYC, KPC, and KPYC mice with markers for proliferation (Ki-67), apoptosis (cleaved caspase-3), and senescence (SA-β-gal). Although there were no general differences in cleaved caspase-3 or SA-β-gal staining in the pancreata from KPC and KPYC mice (fig. S6, A to C), pancreata from KYC and KPYC mice exhibited a markedly reduced number of Ki-67–positive cells compared with those from KC and KPC mice (Fig. 3, A and B). In agreement with previous reports (79), we found that a large fraction of PanINs and surrounding stromal cells were positive for Ki-67 in pancreata from KC and KPC mice (Fig. 3, A and B). In contrast, all epithelial cells in pancreata from KYC and KPYC mice, including those lining ADM and early PanIN lesions, were devoid of Ki-67 staining (Fig. 3, A and B). These data indicate that the loss of YAP may trigger cell cycle arrest in mutant KRAS– and KRAS:TR53-transformed pancreatic ductal cells in vivo.
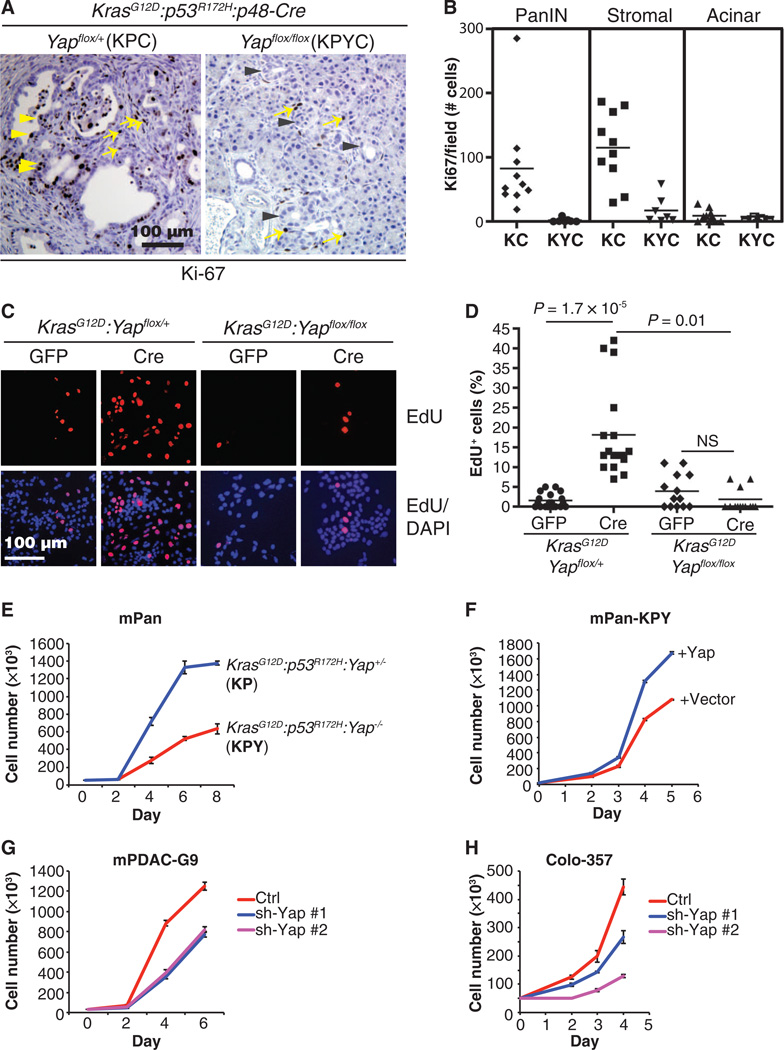
Fig. 3. Deletion of Yap blocks the proliferation of Kras and Kras:Trp53mutant pancreatic ductal cells in vivo and in vitro.
(A) Representative IHC images of Ki-67 in pancreatic sections from KPC and KPYC mice. Yellow arrowheads, Ki-67+ proliferating pancreatic ductal cells in KPC mice; gray arrowheads, Ki-67− ADMs in KPYC mice; yellow arrows, Ki-67+ proliferating stromal cells in both KPC and KPYC mice. Scale bar, 100 µm. (B) Number of Ki-67+ PanIN, stromal, and acinar cells per field from KC (n = 10) and KYC (n = 7) pancreata. Each data point represents the sum of five randomly selected fields from the pancreatic section of a different mouse. (C) Representative images from EdU incorporation assays of KrasG12D:Yapflox/+ and KrasG12D:Yapflox/flox pancreatic epithelial cells infected with Ad-GFP (GFP) or Ad-Cre-GFP (Cre). EdU-incorporated cells are red; nuclei are blue. Scale bar, 100 µm. (D) Quantification of the percentage of EdU+ cells per field from (C). Lines indicate median percentages of EdU+ cells. KrasG12D:Yapflox/+ (GFP), n = 23; KrasG12D:Yapflox/+ (Cre), n = 16; KrasG12D:Yapflox/flox (GFP), n= 13; KrasG12D:Yapflox/flox (Cre), n = 14. NS, not significant; P values were calculated using two-tailed t test. (E to H) Cell proliferation assessed in (E) KP and KPY cells, (F) KPY cells reconstituted with Yap or control vector, (G) mPDAC-G9 cells transfected with control or two independent mYap shRNAs (#1 and #2), and (H) human Colo-357 PDAC cells transfected with control or two independent hYap shRNAs (#1 and #2). Data are means ± SD from three independent experiments.
To gain further insight into the role of YAP in KRAS mutant pancreatic epithelial cell proliferation, we isolated pancreatic epithelium explants from KrasG12D:Yap+/+, KrasG12D:Yapflox, and KrasG12D:Yapflox/flox mice and infected them in vitro with Ad-GFP or Ad-Cre-GFP As expected 5-ethynyl-2′-deoxyuridine (EdU) incorporation assays showed that Ad-Cre-GFP–treated KrasG12D:YapWT (KrasG12D:Yap+/+ or KrasG12D:Yapflox/+) pancreatic epithelium explants exhibited a significant increase in the percentage of replicating cells compared with controls (Fig. 3, C and D). In contrast, homozygous deletion of Yap prevented the mutant Kras–induced increase in proliferation in Ad-Cre-GFP-treated KrasG12D:Yapflox/flox pancreatic epithelial cells (Fig. 3, C and D).
To take a step further, we established Yap-positive (Yap+) or Yap-negative (Yap−) primary Kras:Trp53 mutant mouse pancreatic ductal cell cultures by isolating normal pancreatic acinar cells from KrasG12D/+:p53R172H/+:Yapflox/+(KP) and KrasG12D/+:p53R172H/+:Yapflox/flox (KPY) mice, infecting them with Ad-Cre-GFP, and embedding them in collagen as in the ADM assay. After further passaging in two-dimensional collagen-coated plates, we obtained cell populations composed entirely of CK19+:Sox9+:amylase− ductal cells (fig. S7A). Genomic PCR confirmed the recombination of mutant Kras and Trp53 alleles in both cell lines and the homozygous deletion of Yap in the KPY line treated with Ad-Cre-GFP (fig. S7B). Yap-deficient KPY cells proliferated significantly slower than Yap+ KP cells (Fig. 3E), and the proliferative defect in KPY cells was rescued by ectopic re-expression of Yap (Fig. 3F), further supporting that YAP is critical for the proliferation of pancreatic ductal cells.
Finally, partial knockdown of YAP with two independent YAP-targeted short hairpin RNAs (shRNAs) significantly inhibited the proliferation of mouse PDAC (mPDAC) lines established from KPC mice and KRAS mutant human Colo-357 PDAC cells (Fig. 3, G and H, and fig. S8, A and B).
Collectively, our data demonstrate that YAP is essential in maintaining persistent cell proliferation required for PanIN progression into PDAC in KRAS and KRAS: TP53 mutant pancreata.
Yap drives the expression of multiple secretory factors in KRAS and KRAS:TP53 mutant pancreatic ductal cells
We hypothesized that as a transcriptional regulator, YAP likely controls the proliferation of KRAS and KRAS:TP53 mutant pancreatic ductal cells by regulating the expression of KRAS-induced pro-proliferative genes. We performed an unbiased screen of 54 genes reportedly regulated by KRAS signaling. Using quantitative real-time PCR (qRT-PCR) and Western blot analyses of either human Colo-357 cells or KPY mouse cells in which YAP abundance was experimentally manipulated, we identified only six genes for which the transcript or encoded protein [connective tissue growth factor (CTGF), cysteinerich angiogenic inducer 61 (CYR61), matrix metalloproteinase 7 (MMP7), interleukin-6 (IL-6), IL-1α, and cyclooxygenase 2 (COX2)] positively correlated with YAP abundance across all the pancreatic ductal cell groups examined (Fig. 4, A to C, and fig. S8, C to F). All six genes are implicated in PDAC progression; five of these encode secretory proteins and the sixth encodes COX2, which is responsible for the synthesis of the lipid prostaglandin E2 (PGE2), which promotes inflammation, among other activities (12–14, 80–84). Consistent with the notion that YAP sustains the proliferation of KRAS-mutant pancreatic ductal cells by promoting the expression of pro-proliferative secretory factors, we found that conditioned medium from Yap-reconstituted KPY cells overcame the proliferative defect exhibited by Yap-deficient KPY cells (Fig. 4D). Moreover, combined treatment with recombinant MMP7,I1–6, Il-1α, and PGE2 stimulated the proliferation of Yap-deficient KPY cells, but not Yap-reconstituted KPY cells (Fig. 4E), whereas treatment with individual proteins or various combinations had variable and generally less pronounced effects on the proliferation of either Yap+ or Yap− pancreatic ductal cells. These results suggest that lack of expression of these secreted factors underlies the proliferative defect of Yap null KPY cells and also highlight the complex interactions among these factors in the context of YAP.
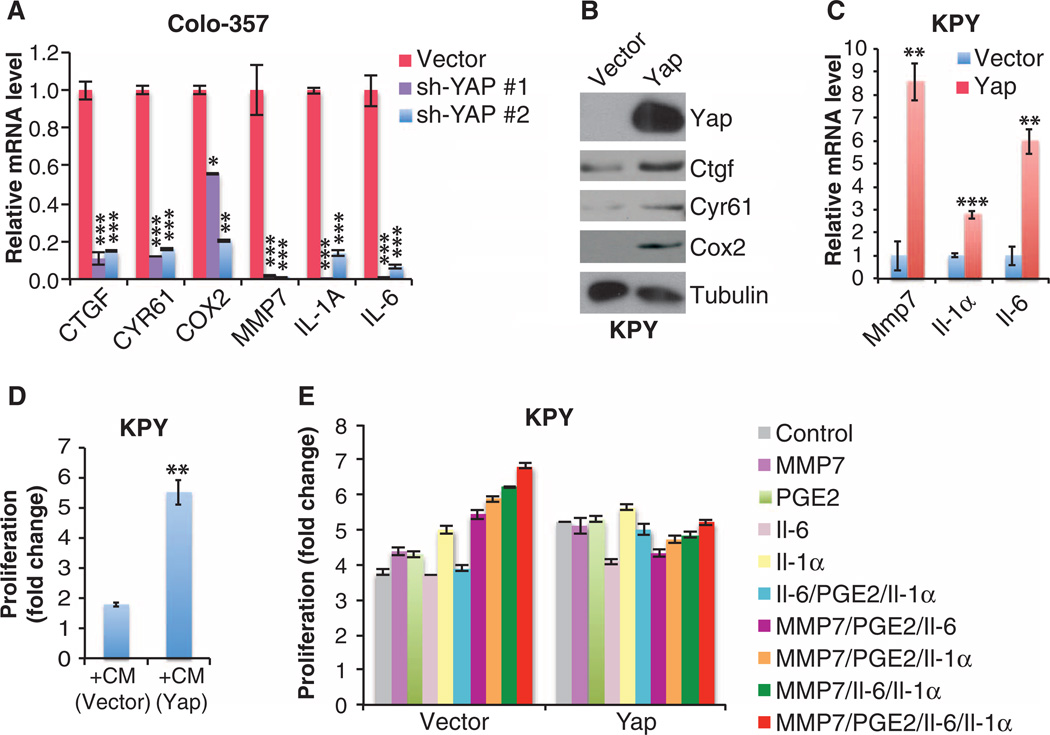
Fig. 4. Yap promotes the proliferation of Kras and Kras:Trp53 mutant pancreatic ductal cells by sustaining the expression of a group of secreted factors.
(A) qRT-PCR analysis of the indicated mRNA amounts in human Colo-357 PDAC cells infected with control or Yap shRNAs. Data are means ± SD of three independent experiments. (B) Western blot analysis in KPY cells infected with control or Yap retroviral vector, representative of three independent experiments. (C) qRT-PCR analysis of the indicated mRNA abundance in the cells as from (B). Data are means ± SD from three independent experiments. (D) Fold proliferation in KPY cells after 2 days in conditioned medium (+CM) from Yap-reconstituted (Yap) or control (vector) KPY cells. Data are means ± SD from four independent experiments. (E) Fold proliferation in Yap-reconstituted or control KPY cells after 3 days of exposure to dimethyl sulfoxide (DMSO) or phosphate-buffered saline (PBS) (control), MMP7 (200 ng/ml), PGE2 (10 µM), II-6 (150 ng/ml), or ll-1α (10 ng/ml), singly or in combination as indicated. Data are means ± SD from three independent experiments. *P< 0.01, **P< 0.001, ***P < 0.0001, two-tailed t test.
COX2 and MMP7 are two novel YAP target genes that contribute to sustaining the proliferation of KRAS:TP53 mutant pancreatic ductal cells
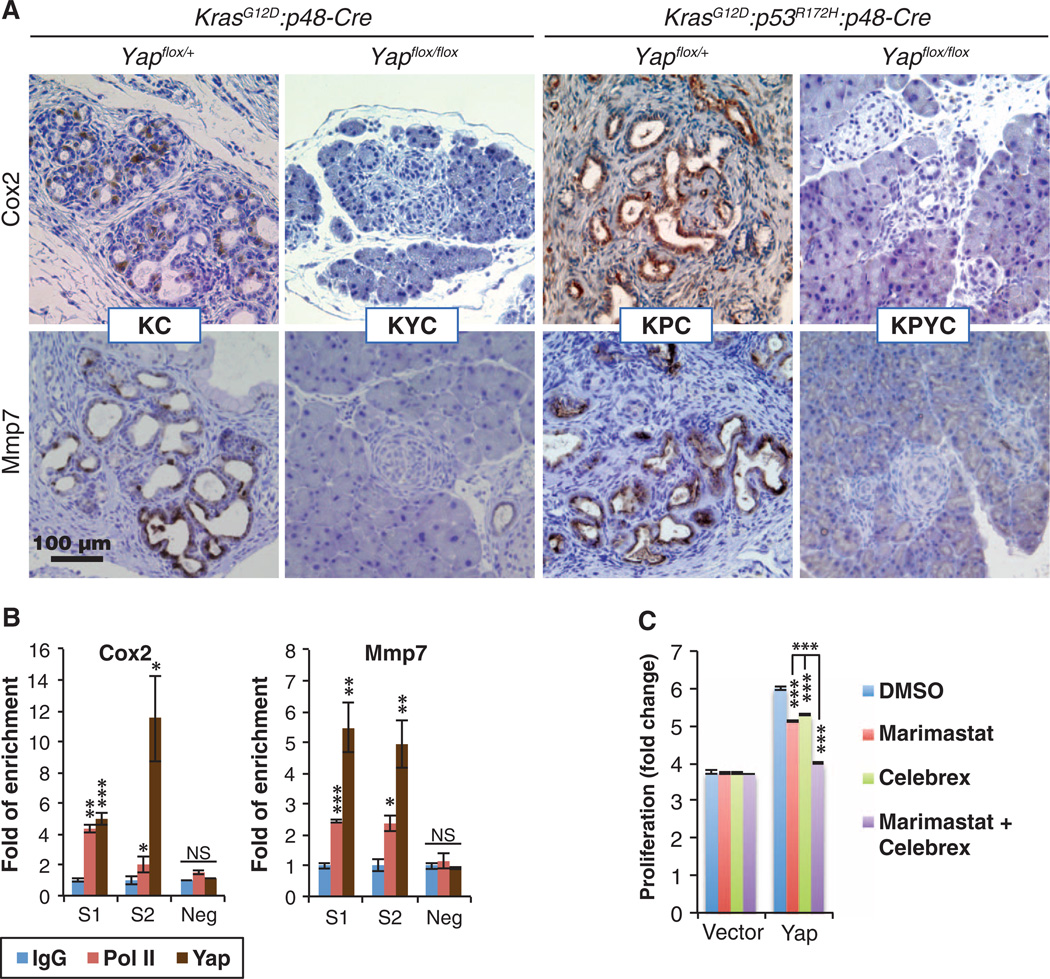
Whereas CTGF and CYR61 are well-established YAP target genes in multiple cell types (85–87), to our knowledge no previous reports link the abundance of COX2, MMP7, IL-6, and IL-1α to YAP. We chose to focus on COX2 and MMP7 for the current study because genetic deletion of either Ptgs2 (encoding Cox2) or Mmp7 delays PanIN progression in KC mice, and inhibitors targeting either human protein have undergone or are undergoing clinical trials for PDAC or other types of cancers (12,13,80). First, we confirmed through IHC analysis of pancreata from KC, KYC, KPC, and KPYC mice that deletion of Yap also blocked the induction of Cox2 and Mmp7 abundance by oncogenic Kras in vivo (Fig. 5A). Similar to the promoters of Ctgf and Cyr61, the promoters of Cox2 and Mmp7 contain multiple putative TEAD-binding sites. Chromatin immunoprecipitation (ChIP) assays confirmed that Yap bound to the promoter regions of Cox2 and Mmp7, but not the coding regions, which do not contain TEAD-binding motifs (Fig. 5B). As expected we also detected a specific association between Yap and the promoters of Ctgf and Cyr61 (fig. S8G).
Fig. 5. Yap controls the expression of Cox2 and Mmp7 in vitro and in vivo.
(A) Representative images of IHC staining for Cox2 and Mmp7 in pancreatic sections from KC, KPC, KYC, and KPYC mice. Scale bar, 100 µm. (B) qRT-PCR analysis of ChIP with antibodies to immunoglobulin G (IgG), polymerase II (Pol II), and Yap on Cox2 and Mmp7 promoter regions that contain (S1 and S2) or do not contain (neg) putative TEAD-binding sites in Yap-reconstituted KPY cells. Data are means ± SD from three independent experiments. (C) Fold proliferation in KPY cells expressing control or Yap vector 3 days after addition of DMSO (control), marimastat (MMP inhibitor, 5 µM), or Celebrex (Cox2 inhibitor, 10 µM). Data are means ± SD from three independent experiments. *P< 0.01, **P< 0.001, ***P< 0.0001, two-tailed t test.
To further investigate how COX2 and MMP7 might contribute to YAP-mediated regulation of pancreatic ductal cell proliferation, we treated KPY cells expressing either the Yap or vector control with marimastat (a clinical MMP inhibitor), Celebrex (a clinical COX2 inhibitor), or their combination. In Yap-reconstituted KPY cells, treatment with either inhibitor alone modestly reduced cell proliferation, whereas combined treatment significantly suppressed the proliferative rate of these cells to near that of control KPY cells (Fig. 5C). In contrast, individual or combined treatment had no effect on the proliferation of Yap-deficient KPY cells that have minimal abundance of Cox2 and Mmp7 (Figs. 4, B and C, and 5C), indicating that the proliferative inhibition by marimastat and Celebrex in Yap-reconstituted KPY cells was unlikely to be due to off-target effects.
Ablation of Yap from Kras and Kras:Trp53 mutant pancreatic epithelial cells dampens their ability to elicit stromal response in vivo
CTGF, CYR61, COX2, MMP7, IL-1α, and IL-6 are all implicated in various aspects of the stromal response that creates a tumor microenvironment that fuels PDAC progression (12, 14, 82, 88–95). In pancreata from KYC and KPYC mice, loss of Yap from pancreatic epithelial cells not only blocked epithelial cell proliferation but also led to a marked reduction in the number of Ki-67–positive stromal cells surrounding the ADM and early PanIN lesions (Fig. 3, A and B). Thus, both autocrine and paracrine mechanisms likely contribute to the profound effects of Yap and the Yap-controlled secretome on PanIN cell proliferation and progression in vivo.
To understand how Yap modulates the tumor microenvironment by controlling the abundance of these secreted factors, we stained pancreatic tissue from 3-month-old and 7-week-old KC, KYC, KPC, and KPYC mice with markers for activated cancer-associated fibroblasts (CAFs), collagen, and infiltrating immune cells, which are main components of the desmoplastic stroma that encapsulate PanINs and PDAC (96).
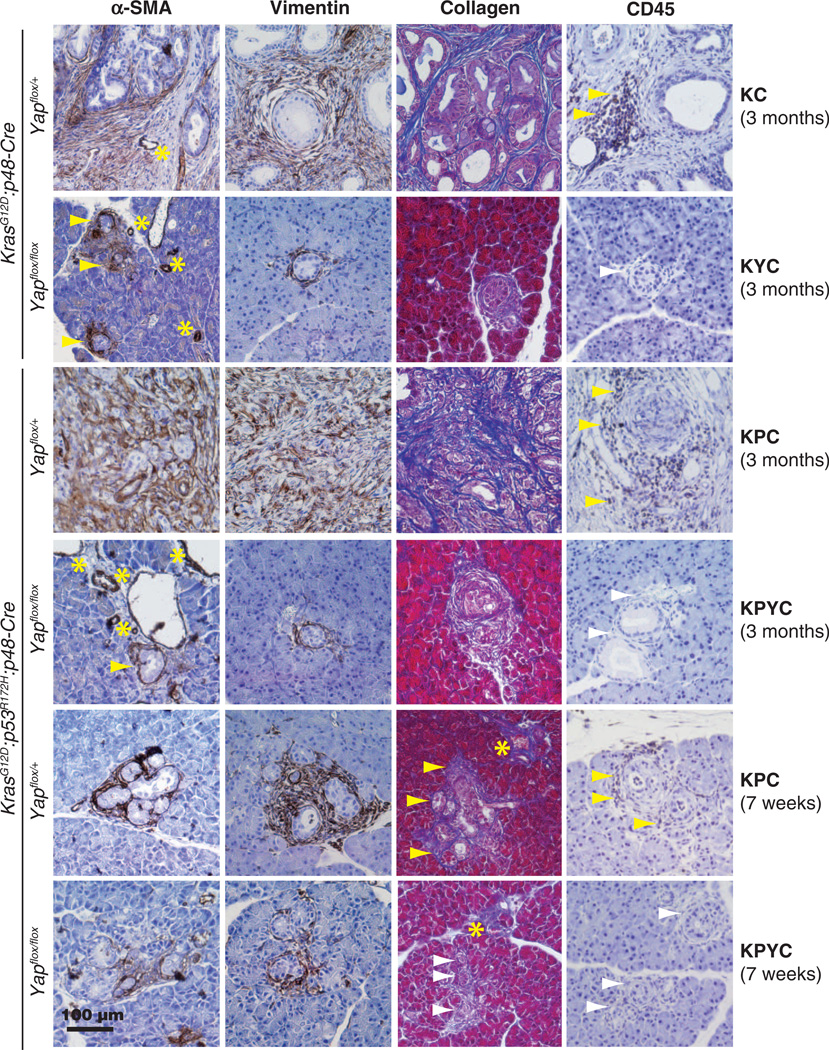
CAFs, a major cellular component and source of collagen in PDAC-associated stroma, express the myofibroblast marker α-SMA and the mesenchymal marker vimentin (97). We found that α-SMA:vimentin+ CAFs accumulated around many of the neoplastic lesions in all of the KC, KYC, KPC, and KPYC pancreata examined regardless of age (Fig. 6). Nevertheless, we did notice a moderate decrease in the intensity and area of α-SMA and vimentin staining around ADM and early PanIN lesions from 7-week-old KPYC mice, which was accompanied by a significant reduction in collagen abundance (Fig. 6). These findings suggest that loss of YAP in KRAS- and TP53-mutant pancreatic epithelial cells compromises the recruitment and activation of stromal fibroblasts and consequently, the production of collagen.
Fig. 6. Deletion of Yap dampens the stromal response in Kras and Kras:Trp53 mutant pancreata.
Representative images of IHC staining for α-SMA, vimentin, collagen, and CD45 on pancreatic sections from 3-month-old KC, KPC, KYC, and KPYC mice and 7-week-old KPC and KPYC mice. Yellow arrowheads, positively stained stroma; white arrowheads, negatively stained stroma; asterisks, blood vessels. Scale bar, 100 µm.
We used an antibody against CD45 (a lymphocyte marker) to detect the presence and localization of infiltrating immune cells in pancreata from KC, KYC, KPC, and KPYC mice. Whereas a large number of CD45 lymphocytes were recruited to the stromal compartments in pancreata from 3-month-old KC and KPC mice, as previously reported (98), we found that CD45+ immune infiltrates were absent in KYC and KPYC pancreata of the same age or older (Fig. 6). To determine whether the differences in CD45 staining in pancreata from adult KC, KYC, KPC, and KPYC mice was due to differences in disease progression, we performed IHC analysis for CD45 on pancreatic sections from 7-week-old KPC and KPYC mice. We detected CD45+ lymphocytes in the vicinity of the ADM and early PanIN lesions from KPC, but not KPYC, mice (Fig. 6). These results illustrate that YAP deficiency in pancreatic neoplastic epithelial cells impedes their ability to elicit a pro-tumor immune response that is critical for PDAC progression (96).
Oncogenic KRAS induces posttranscriptional modification of YAP and augments its transcriptional activity through the MAPK pathway
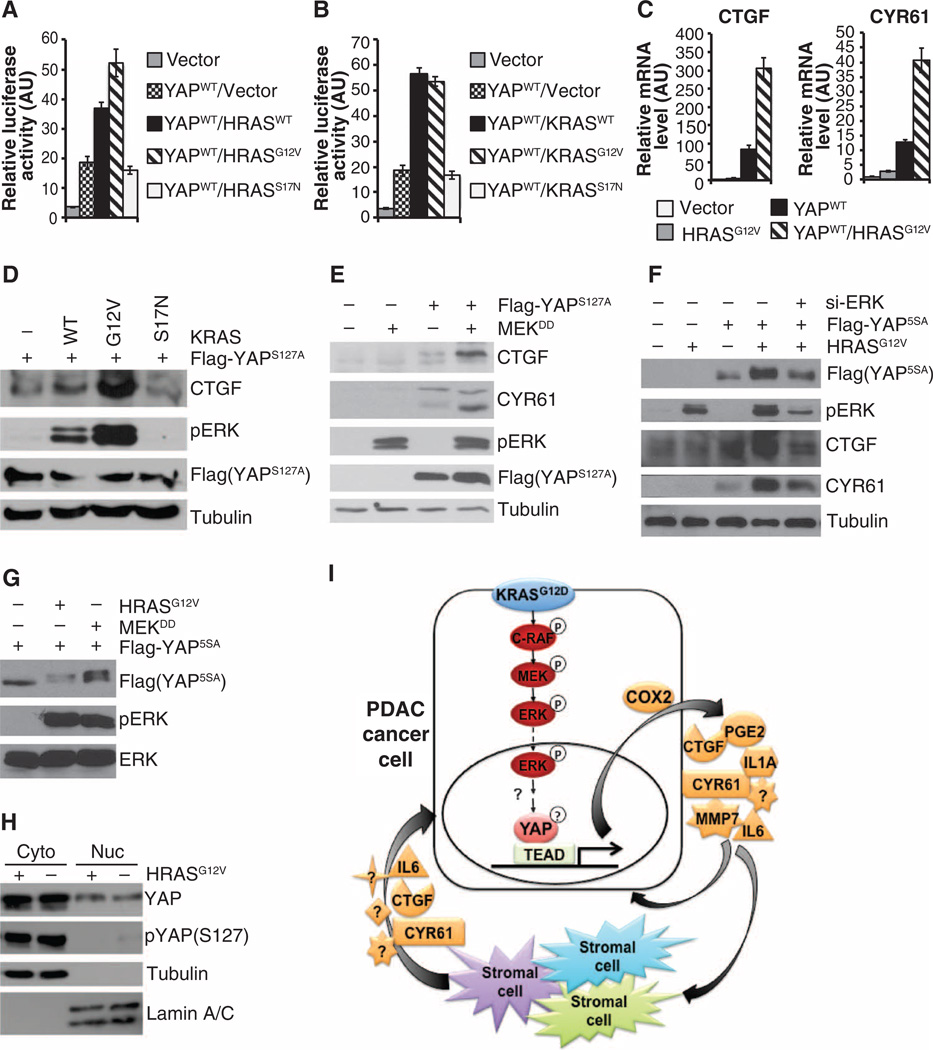
Deletion of Yap did not affect the activation of major oncogenic Kras effector pathways in the mouse pancreas (Fig. 2A), suggesting that YAP most likely acts more downstream in the oncogenic RAS signaling network. Therefore, we focused on examining how oncogenic RAS, its main effector pathways, and YAP coordinate in transcription regulation. First, we examined how expression of wild-type, constitutively active (G12V), or dominant-negative (S17N) Ras (either HRAS or KRAS, herein referred to as H/KRAS) affected YAP/TEAD-mediated transcription using a CTGF luciferase reporter containing TEAD-binding motifs in human embryonic kidney (HEK) 293T cells. H/KRASWT and H/KRASG12V but not H/KRASS17N, promoted the transcriptional activity of YAP (Fig. 7, A and B). By qRT-PCR and Western blot analysis, we found that coexpression of constitutively active RAS (H/KRASG12V) or constitutively active MAPK-ERK kinase (MEKDD) with wild-type YAP, YAPS127A (mutation of the critical serine residue Ser127 to which 14–3–3 binds when phosphorylated by LATS1), or YAP5SA (mutated at one or all five LATS1/2 phosphorylation sites, rendering it insensitive to regulation by the Hippo kinase cascade and thus constitutively localized to the nucleus) also synergistically stimulated CTGF and CYR61 transcript and protein abundance (Fig. 7, B to F). Moreover, expression of HRASG12V or MEKDD induced mobility shifts for YAP5SA on SDS–polyacrylamide gel electrophoresis (SDS-PAGE), and this effect and that on the abundance of CTGF and CYR61 protein were reversed by treatment with small interfering RNAs (siRNAs) targeting ERK1 and ERK2 (Fig. 7, F and G). These results suggest that oncogenic RAS induces posttranscriptional modification of YAP through the MAPK pathway and augments its transcriptional activity. Consistent with oncogenic RAS acting independently of the Hippo pathway, we found that expression of HRASG12V did not affect either the subcellular distribution of endogenous YAP or its phosphorylation at Ser127 (Fig. 7H).
Fig. 7. Oncogenic Ras signals through the MAPK pathway to promote the transcriptional activity of Yap.
(A and B) Dual luciferase reporter assays of HEK293T cells transfected with Yap alone or in combination with wild-type (WT), G12V, or S17N HRAS (A) or KRAS (B). Data are means ± SD of the relative luciferase activity from three independent experiments. (C) qRT-PCR analysis for CTGF and CYR61 in HEK293T cells transfected with control vector, HRASG12V, or wild-type YAP alone or in combination. Data are means ± SD from three independent experiments. (D) Western blot analysis of HEK293T cells transfected with Flag-YAPS127A alone or in combination with wild-type KRAS, KRASG12V, or KRASS17N. (E) Western blot analysis of HEK293T cells transfected with vector control, MEKDD, or Flag-YAPS127A, either alone or in combination. (F) Western blot analysis of HEK293T cells transfected with vector control, HRASG12V, Flag-YAP5SA, and ERK siRNA (si-ERK) alone or in combination. (G) Western blot analysis of lysates from HEK293T cells expressing Flag-YAP5SA in combination with a control vector, HRASG12V, or MEKDD after extended separation on SDS-PAGE. (H) Western blotting for total and phosphorylated YAP (Ser127) in cytoplasmic and nuclear fractions of HEK293T cells transfected with either control vector or HRASG12V. Tubulin, cytoplasmic marker; lamin A/C, nuclear marker. All blots are representative of at least three independent experiments. (I) Schematic of the function of YAP in PDAC cells as a master transcriptional switch of the KRAS secre-tome promoting PDAC cell proliferation.
There are multiple conserved S/TP residues (the minimal ERK phosphorylation motif) within the YAP protein sequence. We mutated a number of these sites and found that only S367A mutation abolished oncogenic KRAS–induced YAP mobility shift (fig. S9A). However, mutation of S367A alone or in combination with several other putative ERK phosphorylation sites did not significantly alter the transcriptional activity of YAP in the presence or absence of oncogenic RAS (fig. S9B), suggesting that additional phosphorylation or other posttranscriptional events are necessary to modulate the transcriptional activity of YAP in response to oncogenic RAS and MAPK signaling. Finally, we found that the ability of KRASG12V to induce YAP5SA-mediated transcription was severely compromised in the TEAD-binding defective YAP5SA/S94A mutant (fig S9C) (99), implying the requirement of intact YAP/ TEAD transcriptional complexes in mediating oncogenic RAS–induced transcription program.
DISCUSSION
KRAS mutations are present in nearly all PDAC (4). Point mutations of the TP53 tumor suppressor have also been described in about 75% of PDAC (7). Here, we identified Yap as a critical partner to mutant Kras and Trp53 in driving PDAC oncogenesis in mice by acting as a critical transcriptional switch in multiple autocrine and paracrine signaling loops required for sustaining not only the proliferation of KRAS-mutant neoplastic ductal cells but also the stromal response and PanIN progression to PDAC (Fig. 7I).
A number of genes, including EGFR, ADM17, PDK1, Gli1, and Sox9, are required for KRAS-induced ADM, commonly thought to be the initiating step in PDAC development (16–22,100). We found that Yap was dispensable for ADM in mice but was necessary for subsequent progression to late-stage PanINs and PDAC in both Kras and Kras:Trp53 mutant mice. The marked effect of Yap deletion on PanIN progression is also in contrast to those of Rac1 STAT3, Gli, IKK2,Il Mmp7, and Cox2, which when knocked out singly in the same GEMMs delayed but did not prevent PDAC development (11–14, 17, 18, 101). Thus, YAP may be the first molecule identified to be absolutely required for PanIN progression into PDAC.
Our data indicate that YAP acts as a critical transcriptional switch by promoting the expression of an oncogenic KRAS secretome that includes CTGF, CYR61, COX2 and PGE, MMP7, IL-6, and IL-1α We also found that in culture, the YAP-controll secretome can act in an autocrine fashion to promote the proliferation of KRAS mutant pancreatic ductal cells. Its in vivo roles are much more profound, observed as epithelial cell proliferation arrest, compromised immune response, and a blockade of PanIN progression resulting from Yap inactivation in Kras or Kras:Trp53 mutant pancreatic neoplastic epithelial cells in mice. Individual inactivation of these secreted factors genetically or pharmacologically only slowed the progression to PDAC in KC or KPC mice (13, 80, 81,101), whereas only when added in combination in culture did these factors effectively rescue the proliferation defect in Yap-deficient mouse pancreatic ductal cells. Thus, the prevention of PDAC development in mice by Yap deletion most probably reflected the cumulative effects of transcriptional blockade of these six—and perhaps additional—Yap target genes. Further studies will identify the rninimum set of YAP targets required for sustaining neoplastic proliferation and PDAC progression in KRAS mutant pancreas.
Although YAP is reportedly broadly expressed in the developing pancreas (72, 73), Yap deletion had no apparent effect on pancreatic development, homeostasis, or function in mice. This is in contrast to the liver, in which Yap knockout impairs the development of bile ducts, the survival of he-patocytes, and normal organ function and fails to prevent oncogenic Kras–induced hepatocellular carcinoma (102). Thus, there are clear tissue-specific differences in the function of YAP during development and tumorigenesis.
Notably, there may be a tissue-specific requirement of certain downstream effectors in mutant KRAS–induced oncogenesis. For example, deletion of Egfr prevents oncogenic KrasG12V-driven PDAC development but does not affect KrasG12V-driven lung and intestinal tumors in mice (19). Similarly, abundance of phosphoinositide-dependent kinase 1 (PDK1) is rate-limiting for KrasG12V-driven PDAC, but not non–small cell lung carcinoma (NSCLC), formation (21). On the other hand deletion of Craf1, which encodes c-Raf, blocks KrasG12D-induced NSCLC but not PDAC development (21, 103, 104). These findings underscore the complexity of oncogenic KRAS–induced carcinogenesis and the necessity for further studies to determine the role of YAP in other KRAS-induced cancer types.
Although we and others have shown that the overall mRNA and protein abundance of YAP is increased in human and mouse PDAC compared with normal pancreatic tissues (72, 73), we found robust cytoplasmic and nuclear localization of YAP in normal ductal and centroacinar cells similar to those in PanINs and PDACs, suggesting that, unlike other cancer types, the overall increase in YAP abundance in PDAC tumors likely reflects the shift in cell composition from a majority of acinar cells (which have low YAP abundance) in the normal pancreas to a majority of neoplastic ductal cells (which have high YAP abundance) in PDAC tumors.
Another interesting finding was that oncogenic KRAS signaled through the MAPK pathway to modulate the transcriptional activity of YAP (including mutant YAPS127A and YAP5SA, which are uncoupled from Hippo pathway regulation), without affecting its subcellular localization. Indeed pancreatic ductal cells (both normal and neoplastic) likely contain minimal Hippo activity, as indicated by the abundance of nuclear YAP in these cells. Our results suggest that oncogenic KRAS may bypass the Hippo pathway and potentiate or direct the transcriptional activity of YAP through phosphorylation or other forms of post-transcriptional modification mediated by ERK and its downstream targets (Fig. 7I). Notably, the primary phosphorylation site causing the mobility shift for YAP in response to oncogenic RAS (fig. S9A), Ser367, is located within the transactivation domain (TA) of YAP. Oncogenic RAS and the MAPK pathway induce phosphorylation within the TA domain of several other transcription factors, leading to phosphorylation-specific recruitment of coactivators and enhanced transcription (105–107). It is tempting to speculate that posttranscriptional modification of YAP at Ser367 and additional sites may recruit coactivators that cooperate with YAP in promoting the transcription of KRAS and YAP co-regulated genes. Further studies will identify these putative YAP transcriptional partners and the specific mechanisms of their recruitment.
Because of the limitation of our GEMMs in which Yap was deleted concurrently with activation of mutant Kras and Trp53, our current study did not directly address whether YAP is required for tumor maintenance in established PDAC. Developing new transgenic mouse models that enable Yap deletion to be temporally separated from activation of mutant Kras and Trp53 are necessary to establish unequivocally the role of YAP in PDAC maintenance and progression. Nonetheless, we did uncover several lines of evidence supporting the continued requirement for YAP in established PDAC. First, we demonstrated that YAP is widely expressed in primary human PDAC. Second we showed that YAP expression is necessary for maintaining the expression of the six noted secreted factors in KRAS mutant human and mouse PDAC lines. Last, we found that even a modest decrease in YAP expression was sufficient to inhibit the proliferation of the aforementioned PDAC lines.
As a transcriptional regulator with no enzymatic pocket, YAP poses substantial challenges as a drug target. Despite this, there has been some recent progress in developing small-molecule YAP inhibitors (108–112). An alternative to directly targeting YAP to treat PDAC may be to combine a cocktail of inhibitors or therapeutic antibodies against multiple components of the KRAS/YAP secretome. Clinical trials have been conducted with COX2 and MMP7 inhibitors as single agents for PDAC, yielding mostly disappointing results (113–115). A clinical trial is currently ongoing for a CTGF-targeted antibody in PDAC and other cancers (116). Clinical agents targeting IL-6 and IL-1α are also available, although they have not been tested in PDAC (117). We found that combination of COX2 and MMP7 inhibitors more efficiently inhibited the proliferation of Yap-positive pancreatic ductal cells than either inhibitor alone, suggesting that combination strategies with available agents may achieve optimum clinical outcome in PDAC without causing unmanageable toxicity.
MATERIALS AND METHODS
Generation of mouse strains
Genetically engineered mouse strains Yapflox/flox, KrasLSL-G12D, Trp53LSL-R172H, and p48-Cre (102, 118–120) were interbred to generate all experimental cohorts (table S1). All animal experiments were conducted according to protocol #11–055 approved by the Institutional Animal Care and Use Committee at Georgetown University.
Cell culture, transfection, and infection
The human pancreatic cancer cell line Colo-357 was cultured in RPMI 1640 (Sigma-Aldrich Corp.) supplemented with 10% fetal bovine serum (FBS). HEK293T, 293 Phoenix-A and mouse pancreatic tumor cell line mPDAC-G9 were cultured in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich) supplemented with 10% FBS. The primary mouse pancreatic cells were cultured in Waymouth’s MB 752/1 (Sigma-Aldrich) supplemented with 10% FBS and soybean trypsin inhibitor (STI; 0.1 mg/ml; AMRESCO, LLC) on collagen-coated plates.
pLKO-shRNA lentiviral constructs targeting human and mouse Yap (TTRCN0000107266: 5′-TTCTTTATCTAGCTTGGTGGC-3′, TRCN0000107268: AAAGGATCTGAGCTATTGGTC, and TRCN0000095864: 5′-TTAACAAAGGAATCTGTCTGC-3′) were purchased from Thermo Scientific Open Biosystems. Lentiviral and retroviral productions were performed as previously described (121).
Mouse acinar explant preparation and in vitro ADM assay
Mouse pancreatic explant cultures were established by modifying previously published protocols (122,123). Briefly, whole pancreata were harvested and digested in collagenase type 4 (4000 U/ml, Worthington Biochemical Corp.) for 50 min at 37°C. After multiple washes with Waymouth’s MB 752/1 supplemented with 5% FBS, collagenase-digested pancreatic tissue was sequentially filtered through 90-µm metal mesh filters. The filtrate was then passed through a 30% FBS cushion by centrifugation for 5 min at 1OOOg. The cell pellet was resuspended in Waymouth’s culture medium [Waymouth’s MB 752/1 Medium supplemented with penicillin/streptomycin and STI (0.1 mg/ml)]. After incubation with adenoviruses carrying GFP or Cre-GFP (Gene Transfer Vector Core of University of Iowa) at 37°C for 1 hour, the cells were pelleted and resuspended in Waymouth’s culture medium. An equal volume of neutralized rat-tail collagen type I (RTC) (BD Biosciences) mixture was added to the cellular suspension. The cellular/RTC suspension was pipetted into culture dish precoated with RTC. After solidification, Waymouth’s culture medium was added. Cultures were maintained at 37°C in a 5% CO2 humidified chamber for up to 3 days, then switched to Waymouth’s culture medium supplemented with 10% FBS.
Cell proliferation and EdU incorporation assays
For cell proliferation assays, human or mouse pancreatic tumor cell lines were seeded in triplicate onto 12-well plates, trypsinized, and counted at the indicated times using Multisizer 3 Coulter Counter (Beckman Coulter Inc.). Colo-357 and mPDAC-G9 cells expressing control or Yap shRNAs were plated at 5 × 104 cells per well. Primary mouse pancreatic cells expressing pBABE vector or pBABE-Yap were plated at 2 × 104 cells per well. For treatment with secreted factors, recombinant human MMP7 (200 ng/ml, Mllipore), murine Il-6 (150 ng/ml, Invitrogen), murine Il-1α (10 ng/ml, Sino Biological Inc.), and PGE2 (10 µM Enzo Life Sciences Inc.) were added singly or in combination to the medium, and cells were incubated for three additional days.
For EdU incorporation assays, freshly isolated pancreatic epithelial cells infected with Ad-GFP or Ad-Cre-GFP were plated onto collagen-coated coverslips in Waymouth’s MB 752/1 Medium supplemented with penicillin G (1000 U/ml), streptomycin (100 µg/ml), STI (0.1 mg/ml), and 10% FBS. After 3 days in culture, EdU was added to the medium, and cells were incubated for an additional 14 hours. EdU-positive cells were detected using the Click-iT EdU Alexa Fluor 594 Imaging Kit according to the manufacturer’s instructions (Invitrogen). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole.
Glucose tolerance test
Three-month-old YC mice with age-matched wild-type littermates were subjected to fasting for 12 hours before the baseline blood glucose level was first measured for each mouse using One Touch UItraMni Blood Glucose Monitoring System according to the manufacturer’s instructions (Life-Scan Inc.). Sterilized D-glucose (200 mg/ml) was then intraperitoneally injected into each mouse (2 mg/g in PBS), and blood glucose was measured again at 30, 60, and 120 min after the injection.
Acute pancreatitis induction
Two-month-old wild-type and YC mice were intraperitoneally injected seven times with caerulein (50 mg/kg) at 1-hour intervals. Mce were euthanized 48 hours after the first injection, and the entire pancreas was dissected and fixed in 10% formalin.
Quantitative real-time RT-PCR
Total mRNA was isolated using TRIzol reagent (Invitrogen). Reverse transcription was performed using iScript cDNA Synthesis Kit according to the manufacturer’s instructions (Bio-Rad Laboratories Headquarters). The resulting complementary DNA (cDNA) products were amplified with iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories). All reactions were performed in triplicate. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (for human) or hypoxanthine-guanine phosphoribosyltransferase (HPRT) (for mouse) was used for normalization. Relative gene expression was calculated as unit value of 2−ΔCt = 2−[Ct(GAPDH or HPRT)– Ct(gene of interest)], where Ct is the threshold cycle value defined as the fractional cycle number at which the target fluorescent signal passes a fixed threshold above baseline. The sequences of the primers used in the study are listed in table S3.
Chromatin immunoprecipitation
ChIP analysis was performed as previously described using normal rabbit IgG (Santa Cruz Biotechnology, sc-2027X) and antibodies against Yap (Santa Cruz Biotechnology, sc-15407X) and Pol II (Santa Cruz Biotechnology sc-899X) (121). Precipitated DNA was eluted and amplified using qRT-PCR with primer pairs flanking different regions of the mouse Cyr61, Ctgf, Cox2, and Mmp7 promoters that contain putative TEAD-binding sites. The primer sequences used in the study are listed in table S4.
Western blot, IHC, and immunofluorescence
Western blot analysis was performed as previously described (124). Human pancreatic TMAs (#PA1001 and #PA207) were purchased from US Biomax Inc. Mouse pancreas was fixed in 10% buffered formalin and embedded in paraffin. For IHC, unstained pancreatic slides were deparaffinized and heated in standard citrate or tris-EDTA retrieval buffer for 30 min at 95°C After incubation overnight with the primary antibodies at 4°C, the slides were incubated with biotinylated secondary antibodies (Vector Laboratories Ltd.) for 1 hour at room temperature. Antibody labeling was visualized using the VECTASTAIN ABC kit (Vector Laboratories Ltd.) followed by staining with 3,3′-diaminobenzidine tetrahydrochloride plus (DAB+) according to the manufacturer’s instructions (Thermo Scientific) or using the Vector Blue Alkaline Phosphatase Substrate Kit (Vector Laboratories Ltd.). The antibodies used for Western blot and IHC analyses are listed in table S5. Collagen staining was performed using the Masson Trichrome Stain Kit according to the manufacturer’s instructions (Sigma).
Subcellular fractionation
For subcellular fractionation experiments, the cytoplasmic fraction was extracted with hypotonic buffer [10 mM Hepes (pH 7.9), 1.5 mM MgCl2, 10 mM KC1, and 0.5 mM fresh dithiothreitol], and pellets containing nuclei were washed twice with hypotonic buffer and subsequently lysed in radio-immunoprecipitation assay buffer.
Luciferase reporter assay
The CTGF luciferase reporter was a gift from K. Lyons [University of California, Los Angeles (UCLA)] (125). Renilla luciferase vector was purchased from Promega. Luciferase reporter activities were determined with the Dual Luciferase Assay Kit (Promega). The reporter’s firefly luciferase activity was normalized to that of the internal control Renilla luciferase before statistical analysis. The annotated relative luciferase activity is the ratio between firefly and Renilla luciferase activities.
Microarray data mining and statistical analysis
Gene Expression Omnibus data sets GSE15471, GSE18670, GSE19650, and GSE16515 containing either normal pancreatic tissue or PDAC tumor cDNA samples hybridized to Human Genome U133 Plus 2.0 GeneChips were included in our analysis. Array data were normalized, and samples having an SE greater than 1.1 were excluded. GC-RMA was used for probe-level normalization of array intensities (126). Batch effects caused by multiple data sources were corrected using ComBat (127). Probes from each probe set with the greatest interquartile range were retained for gene expression analysis. The Linear Models for Mcroarray (LIMMA) package was used for expression calculations (128). Bonferroni-corrected Student’s t tests were used to calculate P values. All analyses were done in the R programming and language software environment using packages available through Bioconductor (129).
Supplementary Material
Acknowledgments
We thank D. J. Pan (Johns Hopkins University) for providing us with Yap conditional knockout mice and K. Lyons (UCLA) for providing us with the CTGF lu-ciferase reporter. We are thankful for the technical assistance from J. Garee in ChIP analysis, from D. L. Berry, E. Permaul, and S. Sen of Histopathology & Tissue Shared Resource (HTSR) in IHC analysis, and from P. Johnson of Lombardi Cancer Center Microscopy & Imaging Shared Resource (MISR) in microscope imaging.
Funding: This work is supported by a Georgetown Lombardi Cancer Center new faculty startup fund to C.Y., National Cancer Institute grant CA71508 to A.W., and the Burroughs Wellcome Clinical Scientist Award in Translational Research and NIH grants R01CA133662 and R01CA138212 to J.T. The Lombardi Cancer Center Shared Resource is supported by a Cancer Center Support Grant, CA051008.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/7/324/ra42/DC1
Fig. S1. YAP is expressed in PDAC.
Fig. S2. Deletion of Yap does not affect normal pancreatic development and glucose metabolism.
Fig. S3. Ablation of Yap blocks PDAC development in Kras and Kras:Trp53 mutant pancreas.
Fig. S4. Yap is not required for oncogenic Kras–induced ADM in vitro.
Fig. S5. Yap is not required for induction of ADM by acute pancreatitis in vivo.
Fig. S6. Deletion of Yap does not cause significant changes in apoptosis or senescence.
Fig. S7. Characterization of Yap+ and Yap− primary Kras:Trp53 mutant pancreatic ductal cells.
Fig. S8. Yap selectively promotes the expression of a group of genes encoding oncogenic Kras–induced secretory factors.
Fig. S9. Ser367 is the primary phosphorylation site responsible for the oncogenic RAS–induced mobility shift of YAP.
Table S1. Abbreviations of all the mouse genotypes used in this study.
Table S2. IHC analysis of YAP in human pancreatic TMAs.
Table S3. Mouse and human primers used in qRT-PCR assay.
Table S4. Mouse primers used in ChIP assay.
Table S5. Antibodies used in IHC and Western blot analyses.
Author contributions: C.Y., W.Z., and N.N. designed and conducted the bulk of the experiments. C.Y., W.Z., and G.G. co-wrote the manuscript. Y.S., S.Z.L., and M.W. carried out the mouse studies. M.M., A.S., M.C., and S.G. contributed to some of the in vitro experiments. G.G. analyzed the microarray data and performed other statistical analysis. E.E.V. generated PDAC cell lines. M.J. and F.W. contributed to IHC analysis. B.K. performed pathological evaluations of all human and mouse pancreatic tissues. A.W. and J.T. provided intellectual input into the design and presentation of the study. C.Y., A.W., and J.T. obtained funding for the project.
Competing interests: Georgetown University is filing a patent application related to technology described in this paper. One or more authors have intellectual property interests in the technology related to this research.
REFERENCES AND NOTES
- 1.Warshaw AL, Fernandezdel Castillo C. Pancreatic carcinoma. N. Engl. J. Med. 1992;326:455–465. doi: 10.1056/NEJM199202133260706. [DOI] [PubMed] [Google Scholar]
- 2.Maitra A, Leach SD. Disputed paternity: The uncertain ancestry of pancreatic ductal neoplasia. Cancer Cell. 2012;22:701–703. doi: 10.1016/j.ccr.2012.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hruban RH, Wilentz RE, Kern SE. Genetic progression in the pancreatic ducts. Am. J. Pathol. 2000;156:1821–1825. doi: 10.1016/S0002-9440(10)65054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Löhr M, Klöppel G, Maisonneuve P, Lowenfels AB, Luttges J. Frequency of K-ras mutations in pancreatic intraductal neoplasias associated with pancreatic ductal adenocarcinoma and chronic pancreatitis: A meta-analysis. Neoplasia. 2005;7:17–23. doi: 10.1593/neo.04445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, Hezel AF, Horner J, Lauwers GY, Hanahan D, DePinho RA. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. 7rp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 8.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Kaiser CE, Frett B, Li HY. Targeting mutant KRAS for anticancer therapeutics: A review of novel small molecule modulators. J. Med. Chem. 2013;56:5219–5230. doi: 10.1021/jm3017706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson H. US National Cancer Institute’s new Ras project targets an old foe. Nat. Med. 2013;19:949–950. doi: 10.1038/nm0813-949. [DOI] [PubMed] [Google Scholar]
- 11.Heid I, Lubeseder-Martellato C, Sipos B, Mazur PK, Lesina M, Schmid RM, Siveke JT. Early requirement of Rac1 in a mouse model of pancreatic cancer. Gastroenterology. 2011;141:719.e7–730.e7. doi: 10.1053/j.gastro.2011.04.043. [DOI] [PubMed] [Google Scholar]
- 12.Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Klöppel G, Yoshimura A, Reindl W, Sipos B, Akira S, Schmid RM, Algul H. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19:456–469. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Fukuda A, Wang SC, Morris IV JP, Folias AE, Liou A, Kim GE, Akira S, Boucher KM, Firpo MA, Mulvihill SJ, Hebrok M. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19:441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling J, Kang Y, Zhao R, Xia Q, Lee DF, Chang Z, Li J, Peng B, Fleming JB, Wang H, Liu J, Lemischka IR, Hung MC, Chiao PJ. KrasG12D-induced IKK2/β/NF-κB activation by IL-1α and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:105–120. doi: 10.1016/j.ccr.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maier HJ, Wagner M, Schips TG, Salem HH, Baumann B, Wirth T. Requirement of NEMO/IKKγ for effective expansion of KRAS-induced precancerous lesions in the pancreas. Oncogene. 2013;32:2690–2695. doi: 10.1038/onc.2012.272. [DOI] [PubMed] [Google Scholar]
- 16.Maniati E, Bossard M, Cook N, Candido JB, Emami-Shahri N, Nedospasov SA, Balkwill FR, Tuveson DA, Hagemann T. Crosstalk between the canonical NF-κB and Notch signaling pathways inhibits Pparγ expression and promotes pancreatic cancer progression in mice. J. Clin. Invest. 2011;121:4685–4699. doi: 10.1172/JCI45797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajurkar M, De Jesus-Monge WE, Driscoll DR, Appleman VA, Huang H, Cotton LJ, Klimstra DS, Zhu LJ, Simin K, L Xu, McMahon AP, Lewis BC, Mao J. The activity of Gli transcription factors is essential for Kras-induced pancreatic tumorigenesis. Proc. Natl. Acad. Sci. USA. 2012;109:E1038–E1047. doi: 10.1073/pnas.1114168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mills LD, Zhang Y, Marier RJ, Herreros-Villanueva M, Zhang L, Almada LL, Couch F, Wetmore C, Pasca di Magliano M, Fernandez-Zapico ME. Loss of the transcription factor GLI1 identifies a signaling network in the tumor microenvironment mediating KRAS oncogene-induced transformation. J. Biol. Chem. 2013;288:11786–11794. doi: 10.1074/jbc.M112.438846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navas C, Hernández-Porras I, Schuhmacher AJ, Sibilia M, Guerra C, Barbacid M. EGF receptor signaling is essential for K-Ras oncogene-driven pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:318–330. doi: 10.1016/j.ccr.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ardito CM, Grüner BM, Takeuchi KK, Lubeseder-Martellato C, Teichmann N, Mazur PK, Delgiorno KE, Carpenter ES, Halbrook CJ, Hall JC, Pal D, Briel T, Herner A, Trajkovio-Arsic M, Sipos B, Liou GY, Storz P, Murray NR, Threadgill DW, Sibilia M, Washington MK, L Wilson C, Schmid RM, Raines EW, Crawford HC, Siveke JT. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell. 2012;22:304–317. doi: 10.1016/j.ccr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eser S, Reiff N, Messer M, Seidler B, Gottschalk K, Dobler M, Hieber M, Arbeiter A, Klein S, Kong B, Michalski CW, Schlitter AM, Esposito I, Kind AJ, Rad L, Schnieke AE, Baccarini M, Alessi DR, Rad R, Schmid RM, Schneider G, Saur D. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell. 2013;23:406–420. doi: 10.1016/j.ccr.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 22.L Kopp J, von Figura G, Mayes E, Liu FF, Dubois CL, Morris JP, 4th, Pan FC, Akiyama H, Wright CV, Jensen K, Hebrok M, Sander M. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pérez-Mancera PA, Guerra C, Barbacid M, Tuveson DA. What we have learned about pancreatic cancer from mouse models. Gastroenterology. 2012;142:1079–1092. doi: 10.1053/j.gastro.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Dong Q, Zhang Q, Li Z, Wang E, Qiu X. Overexpression of yes-associated protein contributes to progression and poor prognosis of non-small-cell lung cancer. Cancer Sci. 2010;101:1279–1285. doi: 10.1111/j.1349-7006.2010.01511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall CA, Wang R, Miao J, Oliva E, Shen X, Wheeler T, Hilsenbeck SG, Orsulic S, Goode S. Hippo pathway effector Yap is an ovarian cancer oncogene. Cancer Res. 2010;70:8517–8525. doi: 10.1158/0008-5472.CAN-10-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, George J, Deb S, Degoutin JL, Takano EA, Fox SB, AOCS Study group. Bowtell DD, Harvey KF. The Hippo pathway transcriptional co-activator YAP is an ovarian cancer oncogene. Oncogene. 2011;30:2810–2822. doi: 10.1038/onc.2011.8. [DOI] [PubMed] [Google Scholar]
- 27.Kang W, Tong JH, Chan AW, Lee TL, Lung RW, Leung PP, So KK, Wu K, Fan D, Yu J, Sung JJ, To KF. Yes-associated protein 1 exhibits oncogenic property in gastric cancer and its nuclear accumulation associates with poor prognosis. Clin. Cancer Res. 2011;17:2130–2139. doi: 10.1158/1078-0432.CCR-10-2467. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Ye DX, Pan HY, Wei KJ, Wang LZ, Wang XD, Shen GF, Y Zhang Z. Yes-associated protein promotes cell proliferation by activating Fos Related Activator-1 in oral squamous cell carcinoma. Oral Oncol. 2011;47:693–697. doi: 10.1016/j.oraloncology.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Helias-Rodzewicz Z, Pérot G, Chibon F, Ferreira C, Lagarde P, Terrier P, Coindre JM, Aurias A. YAP1 VGLL3 encoding two cofactors of TEAD transcription factors are amplified and overexpressed in a subset of soft tissue sarcomas. Genes Chromosomes Cancer. 2010;49:1161–1171. doi: 10.1002/gcc.20825. [DOI] [PubMed] [Google Scholar]
- 30.Orr BA, Bai H, Odia Y, Jain D, Anders RA, Eberhart CG. Yes-associated protein 1 is widely expressed in human brain tumors and promotes glioblastoma growth. J. Neuropathol. Exp. Neurol. 2011;70:568–577. doi: 10.1097/NEN.0b013e31821ff8d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yokoyama T, Osada H, Murakami H, Tatematsu Y, Taniguchi T, Kondo Y, Yatabe Y, Hasegawa Y, Shimokata K, Horio Y, Hida T, Sekido Y. YAP1 is involved in mesothelioma development and negatively regulated by Merlin through phosphorylation. Carcinogenesis. 2008;29:2139–2146. doi: 10.1093/carcin/bgn200. [DOI] [PubMed] [Google Scholar]
- 32.Muramatsu T, Imoto I, Matsui T, Kozaki K, Haruki S, Sudol M, Shimada Y, Tsuda H, Kawano T, Inazawa J. YAP is a candidate oncogene for esophageal squamous cell carcinoma. Carcinogenesis. 2011;32:389–398. doi: 10.1093/carcin/bgq254. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Shi S, Guo Z, Zhang X, Han S, Yang A, Wen W, Zhu Q. Overexpression of YAP and TAZ is an independent predictor of prognosis in colorectal cancer and related to the proliferation and metastasis of colon cancer cells. PLOS One. 2013;8:e65539. doi: 10.1371/journal.pone.0065539. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Liu T, Liu Y, Gao H, Meng F, Yang S, Lou G. Clinical significance of Yes-associated protein overexpression in cervical carcinoma: The differential effects based on histo-types. Int. J. Gynecol. Cancer. 2013;23:735–742. doi: 10.1097/IGC.0b013e31828c8619. [DOI] [PubMed] [Google Scholar]
- 35.Su LL, Ma WX, Yuan JF, Shao Y, Xiao W, Jiang SJ. Expression of Yes-associated protein in non-small cell lung cancer and its relationship with clinical pathological factors. Chin. Med. J. 2012;125:4003–4008. [PubMed] [Google Scholar]
- 36.Zhang J, Xu ZP, Yang YC, Zhu JS, Zhou Z, Chen WX. Expression of Yes-associated protein in gastric adenocarcinoma and inhibitory effects of its knockdown on gastric cancer cell proliferation and metastasis. Int. J. Immunopathol. Pharmacol. 2012;25:583–590. doi: 10.1177/039463201202500304. [DOI] [PubMed] [Google Scholar]
- 37.Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, Huebner K, Lehman D. Characterization of the mammalian YAP (Yes-associated protein) gene its role in defining a novel protein module the WW domain. J. Biol. Chem. 1995;270:14733–14741. doi: 10.1074/jbc.270.24.14733. [DOI] [PubMed] [Google Scholar]
- 38.Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- 39.Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, Yaffe MB. TAZ: A novel transcriptional co-activator regulated by interactions with 14–3–3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 43.Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 44.Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with Salvador and warts . Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 45.Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber D, Hariharan IK. Salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 46.Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation apoptosis by Mob as tumor suppressor Mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 47.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cel1. 2007;30:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, L Li, Zheng P, Ye K, Chinnaiyan A, Haider G, Lai ZC, L Guan K. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 50.Zeng Q, Hong W. The emerging role of the Hippo pathway in cell contact inhibition organ size control and cancer development in mammals. Cancer Cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 51.Harvey K, Tapon N. The Salvador-Warts-Hippo pathway-An emerging tumour-suppressor network. Hat. Rev. Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 52.Udan RS, Kango-Singh M, Nolo R, Tao C, Haider G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 53.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation apoptosis by inactivating Yorkie the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 54.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP . J. Biol. Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 55.Zhao B, L Li, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCFβ-TRCP . Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reddy BV, Irvine KD. Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev. Cell. 2013;24:459–471. doi: 10.1016/j.devcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan R, Kim NG, Gumbiner BM. Regulation of Hippo pathway by mitogenic growth factors via phosphoinositide 3-kinase and phosphoinositide-dependent kinase-1. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2569–2574. doi: 10.1073/pnas.1216462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mo JS, Yu FX, Gong R, Brown JH, Guan KL. Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs) Genes Dev. 2012;26:2138–2143. doi: 10.1101/gad.197582.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piccolo S, Cordenonsi M, Dupont S. Molecular PATHWAYS: YAP and TAZ take center stage in organ growth and tumorigenesis. Clin. Cancer Res. 2013;19:4925–4930. doi: 10.1158/1078-0432.CCR-12-3172. [DOI] [PubMed] [Google Scholar]
- 60.Chen D, Sun Y, Wei Y, Zhang P, Rezaeian AH, Teruya-Feldstein J, Gupta S, Liang H, Lin HK, Hung MC, Ma L. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat. Med. 2012;18:1511–1517. doi: 10.1038/nm.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oudhoff MJ, Freeman SA, Couzens AL, Antignano F, Kuznetsova E, Min PH, Northrop JP, Lehnertz B, Barsyte-Lovejoy D, Vedadi M, Arrowsmith CH, Nishina H, Gold MR, Rossi FM, Gingras AC, Zaph C. Control of the Hippo pathway by Set7-dependent methylation of Yap. Dev. Cell. 2013;26:188–194. doi: 10.1016/j.devcel.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 62.Nguyen HB, Babcock JT, Wells CD, Quilliam LA. LKB1 tumor suppressor regulates AMP kinase/mTOR-independent cell growth and proliferation via the phosphorylation of Yap. Oncogene. 2013;32:4100–4109. doi: 10.1038/onc.2012.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J, Park JS, Wei Y, Rajurkar M, Cotton JL, Fan Q, Lewis BC, Ji H, Mao J. TRIB2 acts downstream of Wnt/TCF in liver cancer cells to regulate YAP and C/EBPα function. Mol. Cell. 2013;51:211–225. doi: 10.1016/j.molcel.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Konsavage WM, Kyler SL, Jr, Rennoll SA, Jin G, Yochum GS. Wnt/β-catenin signaling regulates Yes-associated protein (YAP) gene expression in colorectal carcinoma cells. J. Biol. Chem. 2012;287:11730–11739. doi: 10.1074/jbc.M111.327767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomlinson V, Gudmundsdottir K, Luong P, Leung KY, Knebel A, Basu S. JNK phosphorylates Yes-associated protein (YAP) to regulate apoptosis. Cell Death Dis. 2010;1:e29. doi: 10.1038/cddis.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee KK, Yonehara S. Identification of mechanism that couples multisite phosphorylation of Yes-associated protein (YAP) with transcriptional coactivation and regulation of apoptosis. J. Biol. Chem. 2012;287:9568–9578. doi: 10.1074/jbc.M111.296954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hata S, Hirayama J, Kajiho H, Nakagawa K, Hata Y, Katada T, Furutani-Seiki M, Nishina H. A novel acetylation cycle of transcription co-activator Yes-associated protein that is downstream of Hippo pathway is triggered in response to SN2 alkylating agents. J. Biol. Chem. 2012;287:22089–22098. doi: 10.1074/jbc.M111.334714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Haider G, Finegold MJ, Lee JS, Johnson RL. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, Bardeesy N. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao T, Zhou D, Yang C, Singh T, Penzo-Méndez A, Maddipati R, Tzatsos A, Bardeesy N, Avruch J, Stanger BZ. Hippo signaling regulates differentiation and maintenance in the exocrine pancreas. Gastroenterology. 2013;144:1543.e1–1553.e1. doi: 10.1053/j.gastro.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.George NM, Day CE, Boerner BP, Johnson RL, Sarvetnick NE. Hippo signaling regulates pancreas development through inactivation of Yap. Mol. Cell. Biol. 2012;32:5116–5128. doi: 10.1128/MCB.01034-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo J, Kleeff J, Zhao Y, Li J, Giese T, Esposito I, Büchler MW, Korc M, Friess H. Yes-associated protein (YAP65) in relation to Smad7 expression in human pancreatic ductal adenocarcinoma. Int. J. Mol. Med. 2006;17:761–767. [PubMed] [Google Scholar]
- 73.Diep CH, Zucker KM, Hostetter G, Watanabe A, Hu C, Munoz RM, Von Hoff DD, Han H. Down-regulation of Yes Associated Protein 1 expression reduces cell proliferation and clonogenicity of pancreatic cancer cells. PLOS One. 2012;7:e32783. doi: 10.1371/journal.pone.0032783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 75.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, Petersen G, Lou Z, Wang L. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hiraoka N, Yamazakiltoh R, Ino Y, Mizuguchi Y, Yamada T, Hirohashi S, Kanai Y. CXCL17 and ICAM2 are associated with a potential anti-tumor immune response in early intraepithelial stages of human pancreatic carcinogenesis. Gastroenterology. 2011;140:310–321. doi: 10.1053/j.gastro.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 77.Sergeant G, van Eijsden R, Roskams T, Van Duppen V, Topal B. Pancreatic cancer circulating tumour cells express a cell motility gene signature that predicts survival after surgery. BMC Cancer. 2012;12:527. doi: 10.1186/1471-2407-12-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guerra C, Schuhmacher AJ, Cañamero M, Grippo PJ, Verdaguer L, Pérez-Gallego L, Dubus P, Sandgren EP, Barbacid M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 79.Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, L Mercer K, Grochow R, Hock H, Crowley D, Hingorani SR, Zaks T, King C, Jacobetz MA, Wang L, Branson RT, Orkin SH, DePinho RA, Jacks T. Endogenous oncogenic K-rasG12D stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 80.Hill R, Li Y, Tran LM, Dry S, Calvopina JH, Garcia A, Km C, Wang Y, Donahue TR, Herschman HR, Wu H. Cell intrinsic role of COX-2 in pancreatic cancer development. Mol. Cancer Ther. 2012;11:2127–2137. doi: 10.1158/1535-7163.MCT-12-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Neesse A, Frese KK, Bapiro TE, Nakagawa T, Sternlicht MD, Seeley TW, Pilarsky C, Jodrell DI, Spong SM, Tuveson DA. CTGF antagonism with mAb FG-3019 enhances chemotherapy response without increasing drug delivery in murine ductal pancreas cancer. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12325–12330. doi: 10.1073/pnas.1300415110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haque I, Mehta S, Majumder M, Dhar K, De A, McGregor D, Van Veldhuizen PJ, Banerjee SK, Banerjee S. Cyr61/CCN1 signaling is critical for epithelial-mesenchymal transition and sternness and promotes pancreatic carcinogenesis. Mol. Cancer. 2011;10:8. doi: 10.1186/1476-4598-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Haque I, De A, Majumder M, Mehta S, McGregor D, Banerjee SK, Van Veldhuizen P, Banerjee S. The matricellular protein CCN1/Cyr61 is a critical regulator of Sonic Hedgehog in pancreatic carcinogenesis. J. Biol. Chem. 2012;287:38569–38579. doi: 10.1074/jbc.M112.389064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kirane A, Toombs JE, Ostapoff K, Carbon JG, Zaknoen S, Braunfeld J, Schwarz RE, Burrows FJ, Brekken RA. Apricoxib, a novel inhibitor of COX-2 markedly improves standard therapy response in molecularly defined models of pancreatic cancer. Clin. Cancer Res. 2012;18:5031–5042. doi: 10.1158/1078-0432.CCR-12-0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pan D. The Hippo signaling pathway in development and cancer. Dev. Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lai D, Ho KC, Hao Y, Yang X. Taxol resistance in breast cancer cells is mediated by the Hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF . Cancer Res. 2011;71:2728–2738. doi: 10.1158/0008-5472.CAN-10-2711. [DOI] [PubMed] [Google Scholar]
- 88.L Bennewith K, Huang X, Ham CM, Graves EE, Erier JT, Kambham N, Feazell J, Yang GP, Koong A, Giaccia AJ. The role of tumor cell-derived connective tissue growth factor (CTGF/CCN2) in pancreatic tumor growth. Cancer Res. 2009;69:775–784. doi: 10.1158/0008-5472.CAN-08-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hartel M, Di Mola FF, Gardini A, Zimmermann A, Di Sebastiano P, Guweidhi A, Innocenti P, Giese T, Giese N, Büchler MW, Friess H. Desmoplastic reaction influences pancreatic cancer growth behavior. World J. Surg. 2004;28:818–825. doi: 10.1007/s00268-004-7147-4. [DOI] [PubMed] [Google Scholar]
- 90.lacobuzio-Donahue CA, Ryu B, Hruban RH, Kern SE. Exploring the host desmoplastic response to pancreatic carcinoma: Gene expression of stromal and neoplastic cells at the site of primary invasion. Am. J. Pathol. 2002;160:91–99. doi: 10.1016/S0002-9440(10)64353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Colby JK, Klein RD, McArthur MJ, Conti CJ, Kiguchi K, Kawamoto T, Riggs PK, Pavone AI, Sawicki J, Fischer SM. Progressive metaplastic and dysplastic changes in mouse pancreas induced by cyclooxygenase-2 overexpression. Neoplasia. 2008;10:782–796. doi: 10.1593/neo.08330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Müller-Decker K, Fürstenberger G, Annan N, Kucher D, Pohl-Arnold A, Steinbauer B, Esposito I, Chiblak S, Friess H, Schirmacher P, Berger I. Preinvasive duct-derived neoplasms in pancreas of keratin 5-promoter cyclooxygenase-2 transgenic mice. Gastroenterology. 2006;130:2165–2178. doi: 10.1053/j.gastro.2006.03.053. [DOI] [PubMed] [Google Scholar]
- 93.Groblewska M, Siewko M, Mroczko B, Szmitkowski M. The role of matrix metal-loproteinases (MMPs) and their inhibitors (TIMPs) in the development of esophageal cancer. Folia Histochem. Cytobiol. 2012;50:12–19. doi: 10.2478/18691. [DOI] [PubMed] [Google Scholar]
- 94.Tjomsland V, Spangeus A, Valila J, Sandstrom P, Borch K, Druid H, Falkmer S, Falkmer U, Messmer D, Larsson M. Interleukin 1α sustains the expression of inflammatory factors in human pancreatic cancer microenvironment by targeting cancer-associated fibroblasts. Neoplasia. 2011;13:664–675. doi: 10.1593/neo.11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Feurino WL, Zhang Y, Bharadwaj U, Zhang R, Li F, Fisher WE, Brunicardi FC, Chen C, Yao Q, Min L. IL-6 stimulates Th2 type cytokine secretion and upregulates VEGF and NRP-1 expression in pancreatic cancer cells. Cancer Biol. Ther. 2007;6:1096–1100. doi: 10.4161/cbt.6.7.4328. [DOI] [PubMed] [Google Scholar]
- 96.Feig C, Gopinathan A, Neesse A, Chan DS, Cook N, Tuveson DA. The pancreas cancer microenvironment. Clin. Cancer Res. 2012;18:4266–4276. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hinz B. Formation and function of the myofibroblast during tissue repair. J. Invest. Dermatol. 2007;127:526–537. doi: 10.1038/sj.jid.5700613. [DOI] [PubMed] [Google Scholar]
- 98.Clark CE, Hingorani SR, Mick R, Combs C, Tuveson DA, Vonderheide RH. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res. 2007;67:9518–9527. doi: 10.1158/0008-5472.CAN-07-0175. [DOI] [PubMed] [Google Scholar]
- 99.Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, Guan KL, Xu Y. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24:235–240. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mazur PK, Einwächter H, Lee M, Sipos B, Nakhai H, Rad R, Zimber-Strobl U, Strobl LJ, Radtke F, Klöppel G, Schmid RM, Siveke JT. Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc. Natl. Acad. Sci. U.S.A. 2010;107:13438–13443. doi: 10.1073/pnas.1002423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Funahashi H, Satake M, Dawson D, Huynh NA, Reber HA, Hines OJ, Eibl G. Delayed progression of pancreatic intraepithelial neoplasia in a conditional KrasG12D mouse model by a selective cyclooxygenase-2 inhibitor. Cancer Res. 2007;67:7068–7071. doi: 10.1158/0008-5472.CAN-07-0970. [DOI] [PubMed] [Google Scholar]
- 102.Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Karreth FA, Frese KK, DeNicola GM, Baccarini M, Tuveson DA. C-Raf is required for the initiation of lung cancer by K-RasG12D . Cancer Discov. 2011;1:128–136. doi: 10.1158/2159-8290.CD-10-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Blasco RB, Francoz S, Santamaría D, Cañamero M, Dubus P, Charron J, Baccarini M, Barbacid M. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell. 2011;19:652–663. doi: 10.1016/j.ccr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Foulds CE, Nelson ML, Blaszczak AG, Graves BJ. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol. Cell. Biol. 2004;24:10954–10964. doi: 10.1128/MCB.24.24.10954-10964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Behre G, Singh SM, Liu H, Bortolin LT, Christopeit M, Radomska HS, Rangatia J, Hiddemann W, Friedman AD, Tenen DG. Ras signaling enhances the activity of C/EBPα to induce granulocytic differentiation by phosphorylation of serine 248. J. Biol. Chem. 2002;277:26293–26299. doi: 10.1074/jbc.M202301200. [DOI] [PubMed] [Google Scholar]
- 107.Monje P, Marinissen MJ, Gutkind JS. Phosphorylation of the carboxyl-terminal transactivation domain of c-Fos by extracellular signal-regulated kinase mediates the transcriptional activation of AP-1 and cellular transformation induced by platelet-derived growth factor. Mol. Cell. Biol. 2003;23:7030–7043. doi: 10.1128/MCB.23.19.7030-7043.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, Pan D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sudol M, Shields DC, Farooq A. Structures of YAP protein domains reveal promising targets for development of new cancer drugs. Semin. Cell Dev. Biol. 2012;23:827–833. doi: 10.1016/j.semcdb.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bao Y, Nakagawa K, Yang Z, Ikeda M, Withanage K, Ishigami-Yuasa M, Okuno Y, Hata S, Nishina H, Hata Y. A cell-based assay to screen stimulators of the Hippo pathway reveals the inhibitory effect of dobutamine on the YAP-dependent gene transcription. J. Biochem. 2011;150:199–208. doi: 10.1093/jb/mvr063. [DOI] [PubMed] [Google Scholar]
- 111.Park HW, Guan KL. Regulation of the Hippo pathway and implications for anticancer drug development. Trends Pharmacol. Sci. 2013;34:581–589. doi: 10.1016/j.tips.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jiao S, Wang H, Shi Z, Dong A, Zhang W, Song X, He F, Wang Y, Zhang Z, Wang W, Wang X, Guo T, Li P, Zhao Y, Ji H, Zhang L, Zhou Z. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25:166–180. doi: 10.1016/j.ccr.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 113.Dragovich T, Burris H, III, Loehrer P, Von Hoff DD, Chow S, Stratton S, Green S, Obregon Y, Alvarez I, Gordon M. Gemcitabine plus celecoxib in patients with advanced or metastatic pancreatic adenocarcinoma: Results of a phase II trial. Am. J. Clin. Oncol. 2008;31:157–162. doi: 10.1097/COC.0b013e31815878c9. [DOI] [PubMed] [Google Scholar]
- 114.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: Trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 115.Zucker S, Hymowitz M, Rollo EE, Mann R, Conner CE, Cao J, Foda HD, Tompkins DC, Toole BP. Tumorigenic potential of extracellular matrix metalloproteinase inducer. Am. J. Pathol. 2001;158:1921–1928. doi: 10.1016/S0002-9440(10)64660-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dornhöfer N, Spong S, Bennewith K, Salim A, Klaus S, Kambham N, Wong C, Kaper F, Sutphin P, Nacamuli R, Höckel M, Le Q, Longaker M, Yang G, Koong A, Giaccia A. Connective tissue growth factor-specific monoclonal antibody therapy inhibits pancreatic tumor growth and metastasis. Cancer Res. 2006;66:5816–5827. doi: 10.1158/0008-5472.CAN-06-0081. [DOI] [PubMed] [Google Scholar]
- 117.Jones SA, Scheller J, Rose-John S. Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J. Clin. Invest. 2011;121:3375–3383. doi: 10.1172/JCI57158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Johnson L, Mercer K, Greenbaum D, Branson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 119.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Branson RT, Crowley D, Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 120.Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 121.Yi C, Shen Z, Stemmer-Rachamimov A, Dawany N, Troutman S, Showe LC, Liu Q, Shimono A, Sudol M, Holmgren L, Stanger BZ, Kissil JL. The p130 isoform of angiomotin is required for Yap-mediated hepatic epithelial cell proliferation and tumor-igenesis. Sci. Signal. 2013;6:ra77. doi: 10.1126/scisignal.2004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.De Lisle RC, Logsdon CD. Pancreatic acinar cells in culture: Expression of acinar and ductal antigens in a growth-related manner. Eur. J. Cell Biol. 1990;51:64–75. [PubMed] [Google Scholar]
- 123.Githens S, Patke CL, Schexnayder JA. Isolation and culture of rhesus monkey pancreatic ductules and ductule-like epithelium. Pancreas. 1994;9:20–31. doi: 10.1097/00006676-199401000-00003. [DOI] [PubMed] [Google Scholar]
- 124.Yi C, Troutman S, Fera D, Stemmer-Rachamimov A, Avila JL, Christian N, Persson NL, Shimono A, Speicher DW, Marmorstein R, Holmgren L, Kissil JL. A tight junction-associated Merlin-angiomotin complex mediates Merlin’s regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell. 2011;19:527–540. doi: 10.1016/j.ccr.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huang BL, Brugger SM, Lyons KM. Stage-specific control of connective tissue growth factor (CTGF/CCN2) expression in chondrocytes by Sox9 and β-catenin. J. Biol. Chem. 2010;285:27702–27712. doi: 10.1074/jbc.M110.108498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wu ZJ, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004;99:909–917. [Google Scholar]
- 127.Barrera L, Benner C, Tao YC, Winzeler E, Zhou Y. Leveraging two-way probe-level block design for identifying differential gene expression with high-density oligonucleotide arrays. BMC Bioinformatics. 2004;5:42. doi: 10.1186/1471-2105-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 129.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, lacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.