Abstract
The Hippo-Yap signaling pathway regulates a number of developmental and adult cellular processes, including cell fate determination, tissue growth, and tumorigenesis. Members of the scaffold protein angiomotin (Amot) family interact with several Hippo pathway components, including Yap (Yes-associated protein), and either stimulate or inhibit Yap activity. We used a combination of genetic, biochemical, and transcriptional approaches to assess the functional consequences of the Amot-Yap interaction in mice and in human cells. Mice with a liver-specific Amot knockout exhibited reduced hepatic “oval cell” proliferation and tumorigenesis in response to toxin-induced injury or when crossed with mice lacking the tumor suppressor Nf2. Biochemical examination of the Amot-Yap interaction revealed that the p130 splicing isoform of Amot (Amot-p130) and Yap interacted in both the cytoplasm and nucleus, which involved binding of PPxY and LPxY motifs in Amot-p130 to WW domains of Yap. In the cytoplasm, Amot-p130 prevented the phosphorylation of Yap by blocking access of the WW domains to the kinase Lats1. Within the nucleus, Amot-p130 was associated with the transcriptional complex containing Yap and Teads (TEA domain family members) and contributed to the regulation of a subset of Yap target genes, many of which are associated with tumorigenesis. These findings indicated that Amot acts as a Yap cofactor, preventing Yap phosphorylation and augmenting its activity toward a specific set of genes that facilitate tumorigenesis.
INTRODUCTION
Angiomotin (Amot), angiomotin-like 1 (AmotL1), and angiomotin-like 2 (AmotL2) comprise the Motin family, a group of scaffold proteins that associate with a number of PDZ, WW, and coiled-coil domain–containing proteins through specific binding motifs (1–5). Amot, the first reported member of the family, was originally identified as an angiostatin-binding protein in endothelial cells (6). The protein exists in two major splicing isoforms (p80 and p130), both of which are localized primarily to tight junctions (7). During angiogenesis, Amot is thought to coordinate cell migration and junctional remodeling by promoting trafficking of Syx [synectin-binding Ras homolog gene family, member A (RhoA)–specific guanine exchange factor] together with tight junction proteins Patj [protein associated with lin seven 1 (PALS1)–associated tight junction protein] and Mupp1 (multiple PDZ domain protein 1) to the leading edge of migrating endothelial cells, leading to focal activation of RhoA at the leading edge (8). In vivo studies using transgenic zebrafish and mouse models show that Amot functionally overlaps with AmotL1 and AmotL2 in promoting angiogenesis and is required for normal blood vessel formation during development (9–13). Amot also inhibits the activity of two other Rho family small guanosine triphosphatases (GTPases), Rac1 and Cdc42, by inhibiting the activity of the GTPase-activating protein Rich1 at tight junctions (1, 2).
In addition to its role in regulating the activity of Rho family small GTPases, Amot has been linked to the Hippo-Yap (Yes-associated protein) pathway, an evolutionarily conserved kinase cascade that functions in a context-dependent manner in cell fate determination, cell polarity, organ growth, tissue regeneration, stem cell maintenance, and tumorigenesis (14). In mammalian cells, the core pathway is composed of a kinase cascade, in which the mammalian STE20-like protein kinases 1 and 2 (Mst1/2) phosphorylate and activate the large tumor suppressor homologs 1 and 2 (Lats1/2) kinases. Activated Lats1/2 kinases in turn phosphorylate the transcriptional coactivator Yap, leading to its cytoplasmic retention, ubiquitination, and proteasomal degradation (15, 16). When the Mst and Lats kinases are inactive, hypophos-phorylated Yap translocates into the nucleus where it complexes with Tead (TEA domain family) transcription factors to regulate gene expression (17). In addition to these core components of the pathway, several regulatory components have been identified, including Merlin (moesinezrin-radixin–like protein), Kibra (kidney and brain protein), WW45 (45-kDWW domain protein), and Mob1 [Mps one binder (MOB) kinase activator 1] (14, 18).
Both Amot-p80 and Amot-p130 bind directly to Merlin through their mutual coiled-coil domains (2). Furthermore, Amot-p130, AmotL1, and AmotL2—but not Amot-p80—interact with the WW domains of Yap through PPxY motifs located within a conserved N-terminal glutamine-rich domain that is absent in Amot-p80 (3–5, 19, 20). These results suggest that Amot can interact with, and potentially regulate, Hippo signaling components at both proximal and distal points along the pathway.
This is of critical importance because previous studies have resulted in conflicting findings regarding the roles of the Motin family in Hippo signaling and tumorigenesis. For example, overexpression of Amot in cell lines that do not normally express endogenous Amot results in Yap phosphorylation, its cytoplasmic sequestration, and repression of two known Yap target genes: CTGF (which encodes connective tissue growth factor) and Cyr61 (which encodes cysteine-rich, angiogenic inducer, 61) (3–5, 20). Likewise, knockdown of AmotL2 causes cellular transformation in MDCK canine kidney and MCF10A human breast cell lines in vitro, suggesting a tumor-suppressive role for Motin proteins (4, 5). In contrast to these in vitro findings, results from several in vivo studies suggest that Amot plays a pro-proliferative role during blood vessel development and tumorigenesis. For example, using an orthotopic implantable model, we previously demonstrated that Amot is required for tumorigenesis associated with Merlin in-activation (2). Furthermore, knocking down or deleting Motin family members, either individually or in combination, causes defects in endothelial cell polarization, migration, and proliferation in both mouse and zebrafish embryos (4, 8, 10, 11, 21, 22). Here, we took advantage of several well-defined systems to clarify the role of Amot in tumorigenesis and its relationship with the Hippo-Yap pathway.
RESULTS
Amot deletion impedes injury-induced proliferation of biliary epithelial cells
Previous studies suggested that Amot acts as a Yap antagonist and would therefore be expected to function as a tumor suppressor. To determine whether this is the case, we crossed Amot conditional knockout (KO) mice (Amotflox/flox) with Alb-cre transgenic mice to generate AmotKO (Alb-cre :Amotflox/flox) mice with a liver-specific deletion of Amot. AmotKO mice were born at the expected Mendelian ratio and exhibited a normal life span. Livers from AmotKO mice were indistinguishable from cre-negative littermate controls in overall appearance, histology, and liver weight/body weight ratios. No tumors were observed in any of the AmotKO mouse livers examined at any time point. These results indicate that Amot is dispensable for normal liver development and does not act as a tumor suppressor gene in the liver.
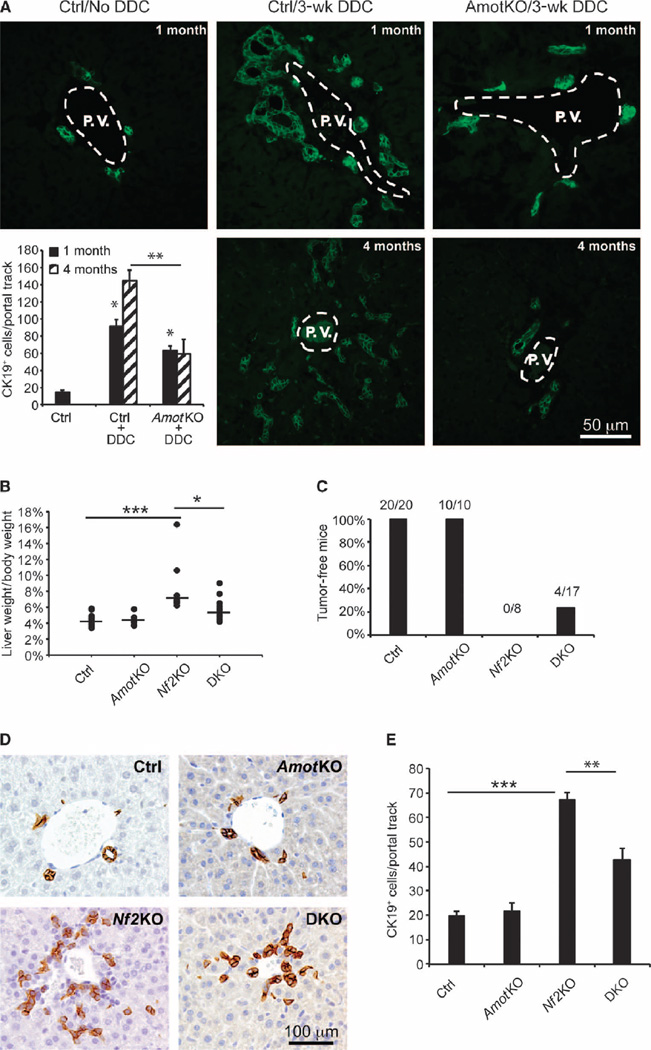
To investigate whether Amot plays a role in a pathological response, we examined whether Amot ablation affects hepatic cell proliferation after liver injury. The diet of AmotKO and control mice was supplemented with 0.2% 3,5-diethoxycarbonyl-l,4-dihydrocollidine (DDC), a porphyrinogenic hepatotoxin that elicits the proliferation of biliary epithelial cells (BECs) in portal tract regions, a histological phenomenon known as the “oval cell response” (23). As expected control mice exhibited a marked increase in the number of cells that stained positive for cytokeratin 19 (CK19+), a BEC marker, 3 weeks after DDC treatment (Fig. 1A). In contrast, the average number of CK19+ cells was significantly reduced in AmotKO mice treated with DDC compared with the treated controls, an effect that was evident in 1-month-old mice and further pronounced in 4-month-old mice (Fig. 1A). These results suggest that Amot is required for injury-induced proliferation of BECs.
Fig. 1. Deletion of Amot attenuates oval cell proliferation and tumorigenesis induced by DDC treatment or Nf2KO in the liver.
(A) Immunofluorescence (IF) analysis of CK19 abundance in liver sections from untreated or DDC-treated wild-type (WT) (Ctrl) or AmotKO mice. Representative images of CK19-stained livers from each experimental group are shown. Oval cell response is shown as mean ± SEM of CK19+ cells per portal track (two portal tracks per animal, n = 5 mice). *P = 0.02; **P = 2 × 10−5, analysis of variance (ANOVA) with Bonferroni correction. One-month-old DDC experimental groups: Ctrl/–DDC (n = 3 mice), Ctrl/+DDC (n = 4), AmotKO/+DDC (n = 6). Four-month-old DDC experimental groups: Ctrl/+DDC (n = 5), AmotKO/+DDC (n = 5). P.V., portal vein. Scale bar, 50 µm (B) Percentage of liver weight to body weight in control (Ctrl; n = 20 mice), AmotKO (n = 10), Nf2KO (n = 8), and DKO (n = 17) mice at 7 weeks of age. *P = 0.02; ***P < 0.001, ANOVA with Bonferroni correction. (C) Percentage and ratio of tumor-free WT (Ctrl; n = 20 mice), AmotKO (n = 10), Nf2KO (n = 8), and DKO (n = 17) mice. (D) Immunohistochemistry (IHC) analysis of CK19 abundance in livers from WT (Ctrl), AmotKO, Nf2KO, and DKO mice. Representative images of CK19-stained livers from each experimental group are shown. Scale bar, 100 µm. (E) Analysis of (D). Data are means ± SEM of CK19+ cells per portal track in the nontumor regions of the livers from each genotype. WT (Ctrl; n = 5 mice), AmotKO (n = 3), Nf2KO (n = 4), and DKO (n = 5). **P = 0.024; ***P < 0.001, two-tailed t test.
Amot deletion inhibits hepatomegaly and tumorigenesis associated with Nf2 deficiency
We have previously shown that short hairpin RNA (shRNA)–mediated silencing of Amot inhibited the tumorigenicity of the Nf2-null Schwann cells in an orthotopic mouse model (2). To test whether this holds true in an endogenous tumor model, we crossed mice bearing floxed alleles of the Nf2 and Amot genes (Nf2flox/flox, Amoflox,flox mice) to Alb-cre mice to generate liver-specific Nf2 knockout (Nf2KO) mice and Nf2:Amot double knockout (DKO) mice. Because the Alb-cre allele mediates recombination at the hepatoblast stage of liver development (24), such mice would be expected to lack Nf2 and/or Amot in both hepatocytes and BECs.
Consistent with previous reports (25, 26), we found that 100% of the Nf2KO mice exhibited an increased number of CK19+ ductal cells in the liver, leading to an increase in the liver weight/body weight ratio from 4.5% in control and AmotKO mice to 7% in Nf2KO mice. In DKO mice, this ratio was reduced to 5.5% (Fig. 1B). Moreover, whereas 100% (8 of 8) of the Nf2KO mice displayed visible hepatic tumors by 7 weeks of age, nearly a quarter (4 of 17) of the DKO mice remained tumor-free at this time point (Fig. 1C). Correspondingly, the increase in the number of CK19+ cells in the DKO livers was also significantly reduced when compared to the Nf2KO livers (Fig. 1, D and E). These results demonstrate that Amot deletion partially prevents the phenotypes associated with Nf2 deficiency in the liver, consistent with our previous finding that Amot is required for the tumorigenicity of Nf2-null Schwann cells (2). Together, these data indicate that Amot is required in vivo for hepatic ductal cell proliferation and tumor formation in response to either Nf2 loss or DDC injury.
The abundance of Amot is increased in Nf2-null liver tumors and human schwannomas
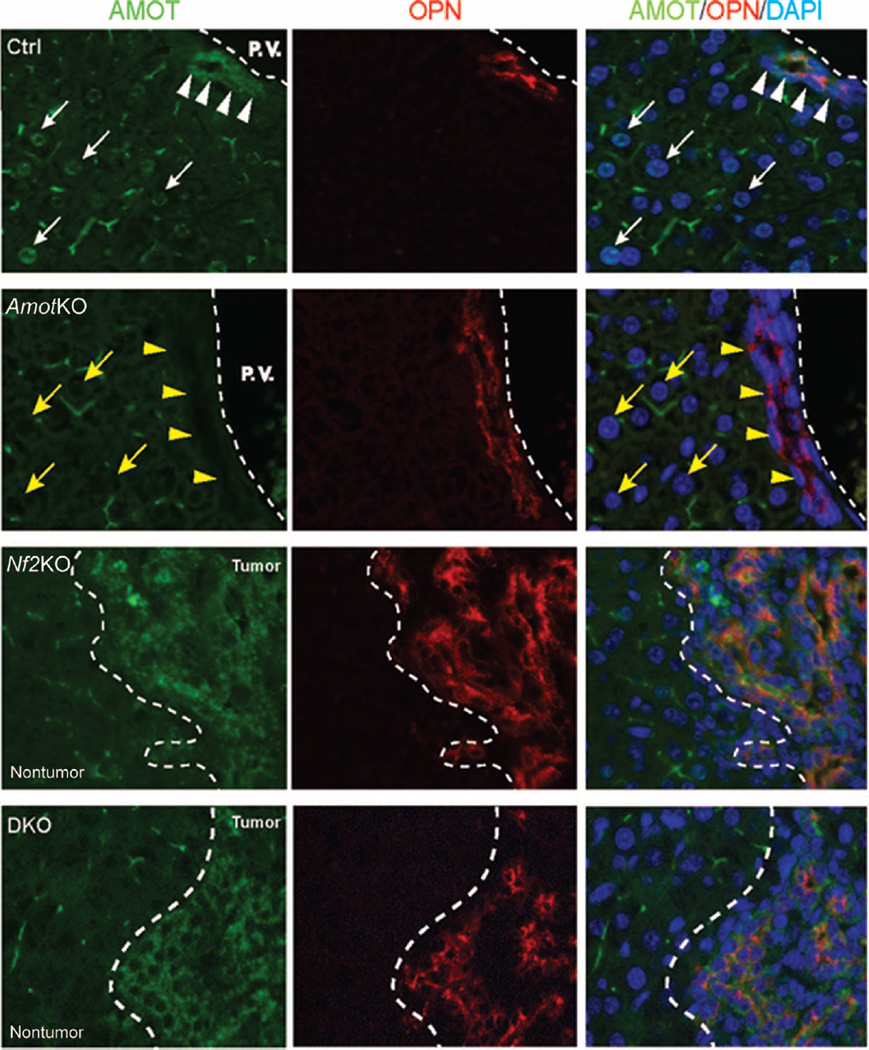
To elucidate how Amot may contribute to tumor development after Nf2 in-activation in the liver, we examined Amot protein expression in the livers from control, AmotKO, Nf2KO, and DKO mice, using immunofluorescence. In control livers, Amot staining was detected in osteopontin (OPN)–positive BECs (Fig. 2, white arrowheads) and within the nuclei of a subpopulation of hepatocytes (Fig. 2, white arrows). Both staining patterns were undetectable in the AmotKO livers (Fig. 2, yellow arrows and arrowheads), confirming the specificity of the immunofluorescent signal.
Fig. 2. Amot abundance is increased in liver tumors from Nf2KO mice.
Immunofluorescence analysis of Amot and OPN in livers from control (Ctrl), AmotKO, Nf2KO, and DKO mice. White arrowheads indicate OPN+ BECs, and white arrows indicate the nuclei of hepatocytes that were positive for Amot. Yellow arrowheads and arrows indicate BECs and hepatocyte nuclei, respectively, in AmotKO livers that did not have detectable Amot. In Nf2KO and DKO livers, tumor and nontumor tissue regions are demarcated. Images are representative of results from three WT mice (Ctrl), three AmotKO mice, three Nf2KO mice, and five DKO mice. DAPI, 4′,6-diamidino-2-phenylindole.
In livers from Nf2KO mice, we observed a substantial increase in Amot abundance within the tumors compared to the adjacent nontumor regions (Fig. 2). Surprisingly, a similar pattern of Amot staining was observed in the DKO livers that did develop tumors (Fig. 2), implying that tumors from DKO livers may have formed from cells that “escaped” recombination of the Amot locus. Consistent with this hypothesis, polymerase chain reaction (PCR) analysis of genomic DNA from DKO livers revealed that, compared to the adjacent nontumor tissue, the tumors retained more of noncombined Amot allele and correspondingly less of the deleted Amot allele (fig. S1).
In humans, loss of Merlin (encoded by NF2) is primarily associated with neurofibromatosis type 2 (NF2) (27). To assess the relevance of Amot to NF2, we analyzed Amot abundance in tissue samples from 10 NF2-mutant human schwannomas and 1 normal human nerve by IHC In contrast to the normal nerve, which showed negligible Amot staining (Fig. 3), 9 of 10 human schwannoma samples exhibited a substantial increase in Amot abundance, with variable staining primarily in the nucleus, as well as in the cytoplasm and along the membrane (Fig. 3).
Fig. 3. Elevated Amot expression in human schwannomas.
A normal human nerve and 10 human schwannoma sections were analyzed by IHC with an Amot antibody. Shown are two representative images from schwannoma sample XT3908 and one from a normal nerve. In each case, the entire section was scored and moderate diffuse staining (upper panel) was scored as +1, strong focal staining (middle panel) was scored as +2, and minimal background staining (bottom panel) was scored as +/−. Right panels show the higher-magnification images of the areas enclosed by the dotted lines. Overall staining scores for all the schwannomas analyzed are listed in the table (bottom right). Scale bars, 50 µm.
Amot-p130 binds to the two WW domains of Yap through LPxY-PPxY motifs
The effects of Amot deletion on oval cell proliferation and liver tumorigenesis induced by injury or Nf2 mutation are reminiscent of those of YapKO mice (26, 28). Given the reported interactions between Motin family members and Yap (3–5), we sought to determine whether Amot modulates liver cell proliferation and tumorigenesis phenotypes through direct interactions with Yap.
Endogenous Amot-p130, but not Amot-p80, specifically bound to endogenous Yap in human embryonic kidney (HEK) 293 cells through reciprocal immunoprecipitation assays with antibodies against Amot or Yap (fig. S2A). Consistent with previous reports, we found that the N-terminal domain of Amot-p130 alone was necessary and sufficient for Yap binding (fig. S2B), and Amot-p130 bound to both WW domains of Yap (fig. S2, C and D). However, upon closer examination of the Amot-p130 primary sequence (fig. S3), we discovered a previously unidentified, conserved, atypical LPxY motif (LPTY, denoted PY1), which lies upstream of the two previously reported PPxY motifs (denoted PY2 and PY3) within the N-terminal domain of Amot-p130 (fig. S2C and S3). This LPxY motif is also conserved in AmotL1 and AmotL2 (fig. S3).
To investigate the functional importance of this LPxY motif, we mutated each of these proline-containing sequences individually or in combination. Whereas single mutation of either the LPxY motif (PY1) or the first PPxY motif (PY2) caused partial or no reduction in Amot-Yap binding, simultaneous mutation of both motifs completely abolished this interaction (fig. S2E). These results demonstrate that the Amot-Yap interaction is mediated by the binding of the LPxY-PPxY motifs of Amot to the two WW domains of Yap.
Amot-p130 promotes Yap nuclear translocation by inhibiting its interaction with Lats1/2
Previous studies examining the impact of Amot on Yap activity relied on overexpression of Amot-p130 in cell lines that do not normally express this protein, resulting in the formation of abnormal cytoplasmic aggregates, which may have confounded interpretations (3–5). To avoid this pitfall, we expressed N-terminal Flag-tagged Amot-p80 and Amot-p130 in HEK293 cells, which have high basal abundance of both Amot isoforms.
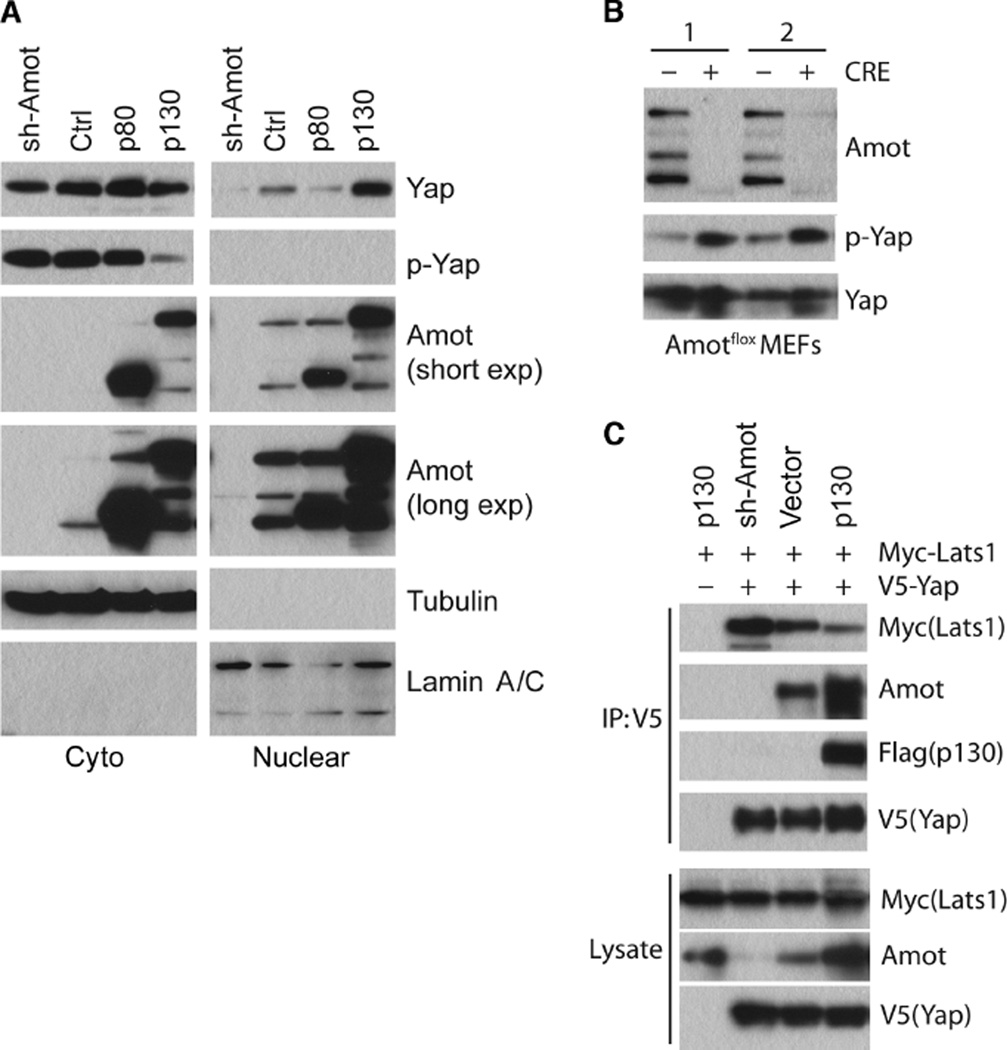
Subcellular fractionation indicated that, similar to silencing Merlin, overexpressing Amot-p130, but not Amot-p80, inhibited Yap phosphorylation at Ser127, which appeared to increase the amount of nuclear Yap and correspondingly decreased cytoplasmic Yap (Fig. 4A and fig. S4A). Conversely, shRNA-mediated knockdown of Amot in HEK293 cells reduced the abundance of nuclear Yap (Fig. 4A and S4A), whereas Cre-mediated deletion of Amot from mouse embryonic fibroblasts (MEFs) increased the amount of phosphorylated Yap (p-Yap) without affecting total Yap protein abundance (Fig. 4B). These results suggest that Amot-p130 functions to maintain the steady-state abundance of active, nuclear Yap at least in part by inhibiting its phosphorylation.
Fig. 4. Amot promotes Yap dephosphorylation and antagonizes Yap-Lats1 interaction.
(A) Western blotting analysis for Yap, p-Yap (at Ser127), or Amot in cytoplasmic and nuclear fractions of HEK293 cells transfected with Amot shRNAs (sh-Amot), a shRNA vector control (Ctrl), Amot-p80, or Amot-p130. Tubulin was used as the cytoplasmic marker and lamin A/C as the nuclear marker. (B) Western blotting analysis of Amotflox MEFs treated with Adeno (−) or Adeno-Cre (+) virus using Amot, p-Yap, and Yap antibodies as indicated. “1” and “2” represent MEFs from two independent embryos. (C) Immunoprecipitation (IP) with the V5-tag antibody and Western blotting analysis to assess the Yap-Lats1 interaction in HEK293 cells cotransfected with V5-Yap and Myc-Lats1 in the presence of vector control, Flag-Amot-p130, or Amot shRNAs. Western blotting data in (A) and (C) are representative of three independent experiments.
Because both Amot-p130 and the Lats1/2 kinases bind to the WW domains of Yap, we hypothesized that Amot-p130 may inhibit Yap phosphorylation and promote Yap nuclear translocation by blocking access of Lats1/2 to Yap. Consistent with this scenario, overexpression of Amot-p130 inhibited the Lats1-Yap interaction in HEK293 cells, and conversely, shRNA-mediated Amot knockdown enhanced this interaction (Fig. 4C). Overexpression of Amot-p130 had no effect on the binding of the WW domain-containing protein Sav1 (also known as WW45) to either Lats1 or Mst2 (fig. S4B), suggesting that Amot-p130 specifically regulates the Lats1-Yap interaction.
Amot-p130 requires binding to Yap to support the proliferation of Nf2-deficient cells
To determine whether the regulation of Yap’s subcellular localization by Amot-p130 requires direct Yap binding, we transfected vector control, Amot-p80, Amot-p130N (unique N-terminal domain of p130), wild-type Amot-p130, or single, dual, or triple PY-mutant Amot-p130 into HEK293 cells and performed subcellular fractionation studies. As expected, overexpression of wild-type Amot-p130 or the PY single mutants caused a localization shift of Yap from the cytoplasm to the nucleus (Fig. 5A). By contrast, expression of Amot-p80, Amot-p130N, or Amot-p130 PY double or triple mutants that are defective in Yap binding appeared to have little effect on the ratio of cytoplasmic to nuclear Yap compared with controls (Fig. 5A). These results demonstrate that Amot-p130 influences the subcellular localization of Yap through direct interactions.
Fig. 5. Amot-p130 promotes the proliferation of Nf2-deficient Schwann cells by promoting Yap nuclear translocation.
(A) Western blotting analysis using Flag and Yap antibodies as indicated of cytoplasmic (labeled with “C”) and nuclear (labeled with “N”) fractions from HEK293 cells transfected with vector or Flag-tagged Amot-p80, Amot-p130N, WT Amot-p130, or Amot-p130 PY mutants. Tubulin was used as a cytoplasmic marker and lamin A/C as a nuclear marker. Asterisk indicates the number of PY mutations. Yellow arrow indicates residual Flag-Amot-p130N bands that were not completely stripped from the membrane before reprobing with tubulin antibody. (B) Immunofluorescence for Yap or Amot in SC4 cells stably expressing control (Ctrl) or Amot-specific shRNA (sh-Amot) infected with retroviruses carrying pBabe (Vector), WT pBabe-Amot-p130, or PY1+2 mutant pBabe-Amot-p130. (C) Fold increase in SC4 cell numbers from the lines described in (B) after 2 days in culture. Data are means ± SEM from three independent experiments.
We have previously generated Nf2-null mouse SC4 Schwann cell lines that stably express Amot shRNAs (SC4/sh-Amot), which exhibit reduced proliferation and tumorigenesis compared with control SC4 cells (2). Consistent with what we observed in HEK293 cells using subcellular fractionation analysis (Fig. 4A and fig. S4A), analysis by immunofluorescence staining indicated that knockdown of Amot in SC4 cells caused a shift of Yap localization from the nucleus to the cytoplasm (Fig. 5B). This effect of Amot knockdown on Yap subcellular localization appeared to be reversed by reintroduction of wild-type, but not LY1/PY2 double mutant, Amot-p130 into SC4/sh-Amot cells (Fig. 5B), again suggesting that Amot-p130 requires direct binding to Yap to promote its nuclear translocation. Additionally only reintroduction of wild-type, but not LY1/PY2 double mutant, Amot-p130 rescued the proliferation defect of SC4/sh-Amot cells (Fig. 5C). Together, these data suggest that Amot-Yap interaction is essential for the pro-proliferative role of Amot in the absence of Merlin.
Amot-p130 is a component of the Yap-Tead transcriptional complex and regulates the transcription of a subset of Yap target genes
We noted from our cell fractionation studies with HEK293 cells that the two isoforms of Amot were present in both the cytoplasmic and the nuclear compartments (Figs. 4A and 5A). Relative to Amot-p80, Amot-p130 appeared to be enriched in the nucleus in these cells (Fig. 4A). This finding prompted us to examine whether the Amot-Yap interaction is present in the nucleus and not restricted to the cytoplasmic compartment, as suggested by previous studies (4, 5).
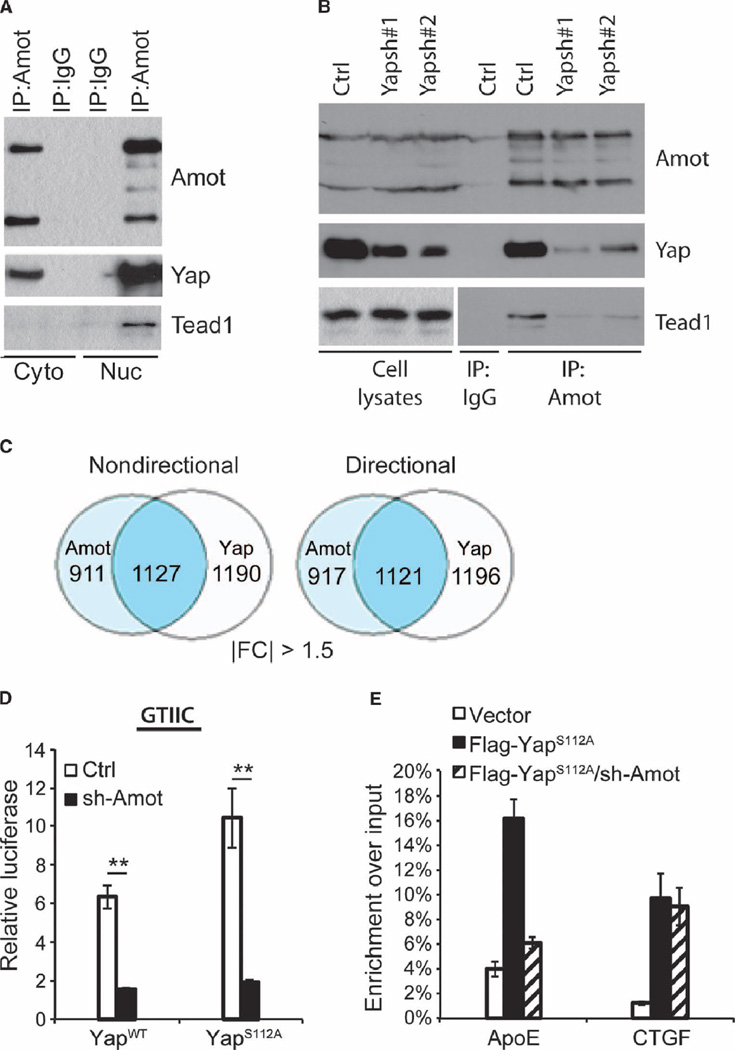
Immunoprecipitation with antibodies against Amot efficiently pulled down endogenous Yap from both cytoplasmic and nuclear lysates of HEK293 cells (Fig. 6A and fig. S4D). Amot immunoprecipitates also pulled down Tead1 exclusively in the nuclear fraction (Fig. 6A). This interaction was dependent on Yap because Yap knockdown using each of two independent shRNAs reduced the amount of Tead1 that coprecipitated with Amot (Fig. 6B). Overexpression of Amot-p130 together with Yap and Tead1 to Tead4 did not substantially alter the amount of Teads that coprecipitated with Yap (fig. S4C). These results suggest that Amot-p130 is recruited by Yap to the Yap-Tead complex within the nucleus.
Fig. 6. Amot is part of a nuclear Yap-Tead complex and required for Yap-Tead–mediated transcription.
(A) Immunoprecipitation and Western blotting of endogenous Amot-Yap and Amot-Yap-Tead complexes from cytoplasmic and nuclear fractions of HEK293 cells. Whole-lysate blot is present in fig. S4D. Image is representative of three independent experiments. (B) Immunoprecipitation and Western analysis of Amot, Yap, and Tead1 in total cell lysates from HEK293 cells stably expressing control (Ctrl) or two independent Yap shRNAs (Yapsh#1 or Yapsh#2). Image is representative of three independent experiments. (C) Microarray-derived Venn diagram of genes for which expression changed more than 1.5-fold by knock-down of Amot (light blue) or Yap (white) compared with control HEK293 cells. The number of genes that changed in both conditions is in the overlap (blue), either regardless of direction (nondirectional) or which either increased in both or decreased in both conditions (directional). IFCI = absolute fold change. (D) Dual-luciferase reporter assays of control and Amot-KD HEK293 cells transfected with WT or S112A, GTIIC-luciferase, and phRL-CMV vectors. Data are means ± SEM of the relative luciferase activity (calculated by normalizing firefly luciferase activity to Renilla luciferase activity from the same well) from three independent experiments. **P < 0.01, two-tailed t test. (E) ChIP analysis for Flag-Yap in lysates from control and Amot-KD HEK293 cells transfected with vector or Flag-Yap S112A. Real-time quantitative PCR (qPCR) analysis was performed with eluted DNA using primers targeting the promoter regions of ApoE and CTGF. Data are means ± SEM from three independent experiments.
To examine the possibility that Amot-p130 directly participates in Yap-Tead-mediated transcription, we carried out a microarray analysis of control, Amot-knockdown (Amot-KD), and Yap-knockdown (Yap-KD) HEK293 cells. As many as 48 to 55% of the genes differentially expressed in Amot-KD cells were also differentially expressed in Yap-KD cells (Fig. 6C and fig. S5A). Among the genes co-regulated by Amot and Yap (table S1), 99.5% or more were regulated in the same direction. Of the 1127 genes whose expression changed by more than 1.5-fold in both Amot-KD cells and Yap-KD cells, 1121 were either increased or decreased in both data sets concordantly (Fig. 6C), whereas this was observed for 100% of genes whose expression changed more than 2-fold (fig. S5A). Gene set enrichment analysis (GSEA) revealed a strong correlation between top-ranked Amot-regulated genes and top-ranked Yap-regulated genes (fig. S5B). In aggregate, our transcriptional analysis suggests that Amot and Yap primarily act in an overlapping fashion regarding their effects on gene transcription.
To assess the role of Amot in Yap-Tead-mediated transcription, we used a GTIIC-luciferase reporter construct that carries eight copies of the minimal Tead-binding sequence (29). As expected, overexpression of either wild-type or a constitutively active S112A (corresponding to S127A in human Yap) mouse Yap (mYap), but not the empty vector, induced GTIIC reporter activity in control HEK293 cells, with the S112A mutant exerting a greater effect (Fig. 6D and fig. S6A). In contrast, neither wild-type nor S112A mYap expression had a significant effect on GTIIC reporter activity in Amot-KD HEK293 cells (Fig. 6D and fig. S6A). Reexpression of Amot-p130, but not of Amot-p80 or PY-mutant Amot-p130, partially rescued the ability of Yap to stimulate GTIIC reporter activity in Amot-KD HEK293T cells (fig. S6C), suggesting that Amot-p130 is necessary for Yap-Tead-mediated transcriptional activity. We further conducted chromatin immunoprecipitation (ChIP) with Flag antibodies in wild-type or Amot-KD HEK293 cells transfected with Flag-tagged Yap S112A. Silencing Amot inhibited the binding of Flag-Yap S112A to the promoter of ApoE, an Amot/Yap negatively co-regulated gene identified from our microarray study (Fig. 6E and fig. S6B). In contrast, the ability of Flag-Yap S112A to bind to the promoter of CTGF, which is positively regulated by Yap, was not significantly affected by knockdown of Amot (Fig. 6E and fig. S6B). Thus, Amot likely modulates Yap-Tead–mediated transcription by affecting the recruitment of Yap to the promoters of a subset of Yap target genes.
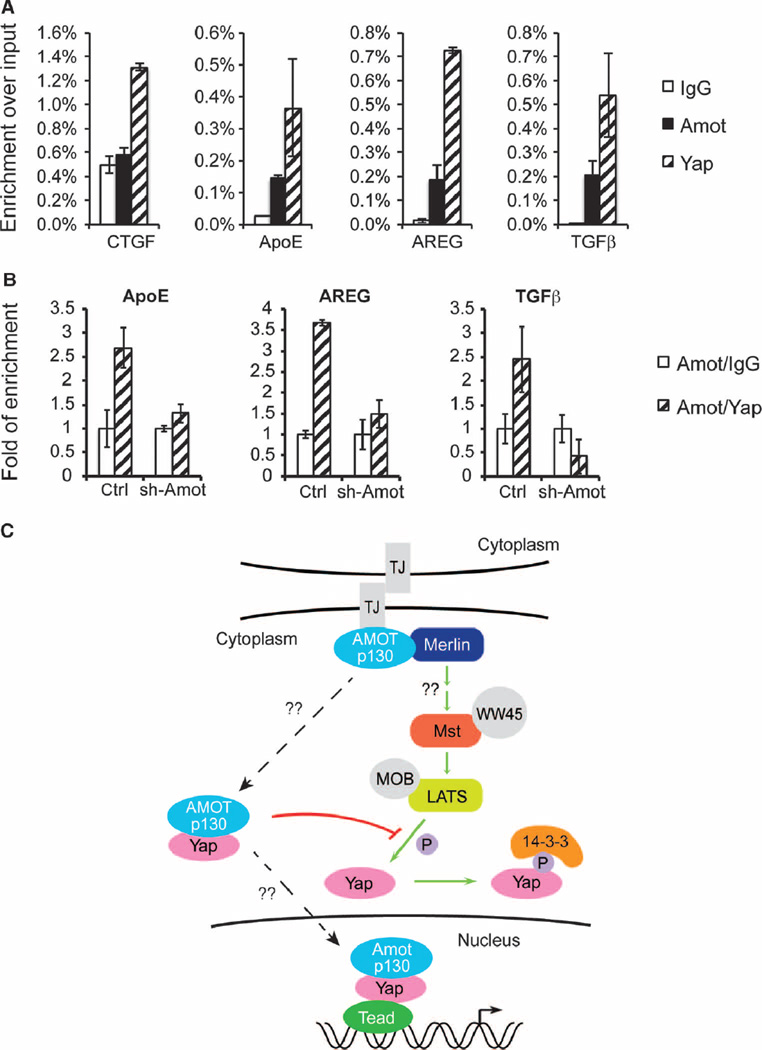
To investigate whether Amot-p130 is part of an endogenous DNA-bound Yap-Tead transcriptional complex, we conducted ChIP analysis using immunoglobulin G (IgG) (control), Amot, and Yap antibodies. Both endogenous Amot and Yap were enriched at the proximal promoter of ApoE as well as two previously reported Yap targets: AREG (which encodes amphiregulin) and TGFB1 [which encodes transforming growth factor–β1 (TGFβ1)] (9, 30, 31) (Fig. 7A). Sequential ChIP first with Amot antibody followed by either IgG control or Yap antibody revealed that Amot and Yap concurrently occupied ApoE, AREG, and TGFB1 in wild-type HEK293 cells, suggesting that Amot and Yap are present within a common regulatory complex on the promoters of these genes (Fig. 7B). No enrichment of these genes was detected in Amot-KD HEK293 cells, thus verifying the specificity of the Amot antibody used in this study (fig. S6D and Fig. 7B). Although we observed association of Yap with the CTGF promoter, we did not obtain significant enrichment of Amot at the same region in wild-type HEK293 cells under the same conditions (Fig. 7A and fig. S6D). Together, these data suggest that Amot-p130 is a component of the nuclear Yap-Tead transcriptional complex and directly participates in the transcription regulation of a subset of Yap target genes.
Fig. 7. Amot associates with a chromatin-bound Yap-Tead transcriptional complex.
(A) ChIP with IgG, Amot, or Yap antibody in HEK293 cell lysates, followed by real-time qPCR analysis of the promoter regions of ApoE, AREG, TGFB1, and CTGF. Enrichment of each product was calculated relative to input. Data are means ± SEM from three independent experiments. (B) Sequential ChIP with Amot antibody followed by IgG or Yap in control and Amot-KD HEK293 cell lysates, followed by real-time qPCR analysis of the promoter regions of ApoE, AREG, and TGFB1 genes. The fold of enrichment of each product with Yap antibody was calculated relative to IgG after Amot ChIP. Data are means ± SEM from three independent experiments. (C) Schematic representation of the multiple interactions of Amot with Merlin-Hippo-Yap signaling in both the cytoplasm and the nucleus. In this model, Amot-p130 prevents Yap-Lats1/2 (LATS) interaction in the cytoplasm and LATS-mediated Yap phosphorylation. Subsequently, the Amot-p130/Yap complex translocates to the nucleus to regulate Tead-target gene transcription. TJ, tight junction.
DISCUSSION
Given the broad context-dependent activities of the Hippo-Yap pathway, crosstalk between the core Hippo kinase cascade and other signaling inputs is likely to influence the nature and/or intensity of the signaling output (32–34). One candidate mediator of such crosstalk is Amot, a scaffold protein that can interact with other proteins through PDZ-binding, WW-binding, and coiled-coiled domains. Although we and others have found that Amot binds to components of the Hippo-Yap pathway, the nature and significance of such interactions remain poorly understood. Here, we used mice with a liver-specific deletion of Amot to determine the functional role of Amot in vivo and conducted a series of biochemical and transcriptional analyses to understand how Amot regulates Yap activity. We conclude that Amot acts to augment, rather than inhibit, Yap function by influencing its phosphorylation, localization, and capacity to transactivate its transcriptional targets (Fig. 7C).
Several groups have reported that Amot is a major Yap-interacting protein (3–5, 20, 35). Indeed, we pursued these studies, in part because we independently identified Amot as a major Yap-interacting protein through unbiased mass spectrometry experiments, a surprising result given that Amot was thought to be a tight junction protein, whereas Yap is a transcriptional coactivator that shuttles between the cytoplasm and nucleus. A similar example of such an interaction is that of the junctional protein ZO2, which interacts with Yap2 in the nucleus (36). We used HEK293 cells, which normally express Yap and Amot, to confirm that this interaction occurs endogenously. We further showed that Yap-Amot-p130 binding is mediated by the two WW domains of Yap and one of the two previously identified PY motifs, as well as a third upstream atypical PY motif located in the N-terminal domain of Amot-p130.
One major mechanism by which the cell regulates Hippo-Yap activity is by controlling Yap localization through phosphorylation-dependent sequestration and degradation (15, 16). Using HEK293 cells, we found that Amot overexpression promotes Yap nuclear localization, whereas Amot knockdown promotes cytoplasmic localization. This effect was associated with changes in Yap phosphorylation status. Moreover, changes in localization were specific to Amot-p130 (and not Amot-p80) and required a direct interaction between Amot and Yap because PY mutations that abrogated the Amot-Yap interaction also abrogated this effect on nuclear localization.
The finding that Amot-p130 promotes nuclear localization of Yap is at odds with recent studies that found that in HeLa, MCF10A, and MDCK cells, Amot-p130 overexpression causes translocation of Yap from the nucleus into the cytoplasm or to tight junctions in the case of MDCK cells (3–5). However, because HeLa and MCF10A do not normally express Amot-p130 [fig. S1D in (5)], and exogenously expressed Amots in these cells form mainly cytoplasmic aggregates, the results obtained using these cell lines must be interpreted with caution. Stable expression of Amot-p130 in MDCK cells, where it is correctly targeted to tight junctions, leads to an increase in both nuclear and total Yap [fig. S3B in (5)]. Loss-of-function studies showed that silencing of AmotL2, but not AmotL1, increased the localization of Yap and Taz to the nucleus and induced cellular transformation in MDCK and MCF10A cells (3–5). Thus, some of the discrepancies between our finding and other studies could also be due to tissue- or cell type–specific functions of different members of the Motin family.
Although the biochemical basis for Amot’s effect on Yap phosphorylation and localization remains to be determined, one possible mechanism involves the interaction between Yap and Lats1. We found that Amot-p130 overexpression disrupts formation of the Yap-Lats1 complex, whereas Amot knockdown enhances it. Thus, we speculate that cytoplasmic Amot acts as a rheostat for the association of Yap and its inhibitory kinase (Fig. 7C). In the setting of limiting levels of Amot, Yap associates with Lats1 and is phosphorylated, leading to its cytoplasmic retention and degradation, whereas high levels of Amot interfere with the Yap-Lats1 interaction, preventing Yap phosphorylation and thereby facilitating its nuclear entry.
Our findings stand in contrast to a report suggesting that the Amots increase Yap phosphorylation by interacting with and activating the Lats kinases (20). One significant caveat of this study is that the experiments were performed primarily by overexpressing C-terminal–tagged Amots. Because Amot requires a free C-terminal PDZ-binding motif to associate with PDZ domain–containing proteins such as Patj/Mupp1, adding a C-terminal tag to Amot would likely interfere with its normal functions/localizations. Indeed despite our repeated attempts, we were unable to detect, any specific association between our N-terminal–tagged Amot constructs and the Lats kinases in HEK293 cells.
In the course of our cellular fractionation experiments, we found that a large fraction of endogenous Amot-p130 is present in the nucleus of HEK293 cells. This result was surprising given previous reports that ectopic expression of Amot-p130 in MCF10A and HeLa cells leads to cytoplasmic retention of Yap or its paralog Taz (3–5). Moreover, we found that immunoprecipitation of Amot from the nuclear fraction pulled down Tead1 in addition to Yap, suggesting that Amot may be part of a transcriptionally active nuclear complex (Fig. 7C). To test this possibility, we examined the effects on global transcription after knocking down either Amot or Yap in the same cell line. This analysis revealed a significant overlap between the transcriptional changes caused by Amot knockdown and those caused by Yap knockdown, with more than 40% of the genes that exhibited a transcriptional change with either Amot or Yap knockdown showing a corresponding change in the other group. Almost all changes in transcript abundance occurred in the same direction, such that most genes whose expression increased after Amot knockdown were also increased after Yap knockdown, and the same was true for genes whose expression decreased.
Notably, the well-characterized Yap target CTGF was among a half-dozen genes whose transcript amounts did not change in the same direction (CTGF transcript abundance decreased after Yap-KD and increased after Amot-KD; fig. S6B). In the aforementioned studies by Chan et al. (3) and Zhao et al. (5), the authors concluded that Amot antagonizes Yap transcriptional activity, in part because knockdown of Amot with shRNAs increased transcript abundance of CTGF. Although our global transcriptional analysis confirms this finding, it also indicates that this effect of Amot knockdown on CTGF is the exception rather than the rule, in that it was one of a very small number of genes whose expression was regulated in an opposite manner by Amot and Yap. At a global gene regulation level, more than 99% of genes that responded to loss of Amot or Yap did so in the same direction, strongly suggesting that the two proteins generally function in a cooperative manner.
Consistent with this notion, we found that Amot-p130 is part of a Tead1-containing nuclear complex that associates with the proximal promoters of several Yap targets, including ApoE, AREG, and TGFβ (but not CTGF). Sequential ChIP demonstrated that Yap and Amot are present within the same transcription factor complex, and luciferase and ChIP assays showed that Amot knockdown inhibits Yap-Tead–dependent transcription of several Yap target genes because of compromised recruitment of Yap to their promoters. Together, these results strongly suggest that Amot-p130 can act as a co-factor in a transcriptionally active Yap-Tead– containing complex at a subset of Yap target genes.
Questions remain as to how Amot-p130 may modulate the specificity of Yap-Tead– mediated transcription. GSEA of the genes regulated by Amot and Yap in HEK293 cells shows substantial enrichment of genes involved in proteasome pathways and several metabolic pathways (table S2). However, future studies in perhaps more disease-relevant cell types will be necessary to cross-validate the significance of these findings and to identify the additional factors or modifications that govern Amot-p130’s selectivity.
Given the discrepancies arising from in vitro readouts for Amot’s effects on Yap localization and function, it was essential to determine whether, and if so how, Amot modulated Yap activity in vivo. In AmotKO livers, we observed a reduction in the biliary ductal cell proliferation that occurs after toxin exposure (the so-called oval cell response), a finding that is quite similar to the blunted response to bile duct ligation observed in yap-null livers (28). Furthermore, our results are consistent with previous observations that conditional deletion of the Hippo-Yap pathway components WW45/Sav1 or Mst1/2 results in an exacerbated oval cell response to DDC, effects that are mediated by increased Yap activity (37–39). Moreover, Amot deficiency inhibited tumorigenesis caused by Nf2 deletion, which requires Yap activity (26). Indeed, the tumors that arose in DKO mice, similar to those in Nf2KO mice, had high levels of Amot expression, indicating that tumors in DKO mice had arisen from cells that escaped Amot deletion. These results, together with our observation of increased Amot abundance in the majority of NF2-associated human schwannomas, raise the possibility that increased abundance of Amot facilitates tumorigenesis after NF2 inactivarion by increasing Yap activity. Contrary to previous reports, our in vivo genetic studies demonstrated that Amot acts in concert with Yap during injury-related bile duct cell proliferation and tumorigenesis.
It is also important to note that although deletion of Yap during liver development leads to bile duct paucity (26), there were no detectable effects on bile duct development in AmotKO mice. Thus, Amot deficiency does not phenocopy Yap deficiency in a developmental context. There are several possible explanations for this difference. First, the appearance of an Amot phenotype during oval cell proliferation and tumorigenesis, rather than embryonic development, may reflect a “threshold” effect whereby Amot is required when a biological response hinges on the “high” activity of Yap signaling, as occurs after biliary injury. Alternatively, Amot may act as a co-regulator at a different set of targets during development compared with adulthood. Finally, functional redundancy could account for the differences in Yap- and Amot-mutant phenotypes because different biological processes may be differentially sensitive to compensation by other Motin family members (AmotL1 and AmotL2). Additional experiments using conditional deletion of multiple Motin family members will help to clarify this question.
In summary, we have shown that Amot facilitated the nuclear entry and transcriptional activity of Yap and was required in the liver for ductal cell proliferation and tumorigenesis in response to liver injury or Nf2 deletion. Questions remain as to how loss of Nf2 leads to increased abundance of Amot in murine livers and human schwannomas and whether overexpression of Amot is sufficient to drive tumorigenesis in the absence of Nf2 deletion. In the future, it will also be critical to understand how Amot itself is regulated. Because Amot functions in other contexts as a tight junction scaffold protein that modulates cell polarity complexes, it is possible that this protein also participates in the well-established connection between the Hippo-Yap pathway and apical-basal polarity (40).
MATERIALS AND METHODS
Animal experiments
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committees of the Wistar Institute and University of Pennsylvania. The strains were bred on a mixed background. The Nf2 and Amot knockout mice were described previously (41,42). For liver tumor studies, mice were fed a regular diet and euthanized at 7 weeks of age. For the liver injury experiments, mouse chow containing 0.2% DDC (Harlan Teklad) was fed to 1- or 4-month-old male mice continuously for 3 weeks until euthanization.
Constructs and shRNAs
pCMV-Flag–tagged Amot-p80 and Amot-p130, V5-tagged Yap, Myc-tagged Lats1 and Lats2, and Flag-tagged WW45 were generated as described previously (2, 43). pCMV-Flag–tagged Amot-p80 and Amot-p130 LPxY/PPxY mutants were generated with Site-Directed Mutagenesis Kit from Stratagene. Wild-type mYap was generated by cloning full-length mYAP complementary DNA (cDNA) (Addgene) into the pCMV-tag-2A vector (Stratagene). The S112A mYAP mutant was generated with the Stratagene mutagenesis kit. Gal4-TEAD3 and Gal4-TFAD4 were gifts from J. Hogenesch (University of Pennsylvania). Gal4-TEAD2 was constructed by cloning the full-length TEAD2 cDNA (Addgene) into the CM-Gal4 vector (44). pLKO-shRNA lead-viral constructs targeting human Yap (clone IDs: TRCN0000107266 and TRCN0000107268) were purchased from Thermo Scientific Open Bio-systems. Two human Amot shRNA vectors were described previously (2).
Antibodies
Purified rabbit polyclonal antibodies against Amot were previously described (2). The following antibodies were commercially purchased: Merlin (c-18), c-Myc (9E10), HA (Y11), HA (F7), Gal4, and GST (Z5) from Santa Cruz Technology; Merlin, Yap, p-Yap (Ser127), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) from Cell Signaling Technology; tubulin, rabbit Flag, and mouse Flag (M2) from Sigma; and OPN from R&D Systems. Antibodies against CK19 were gifts from D Melton (Harvard University).
Cell culture, transfection, and infection conditions
All cells were maintained in low-glucose Dulbecco’s modified Eagle’s medium + 10% fetal calf serum + 1% penicillin-streptomycin. Wild-type and Amot-KD HEK293 cells were transfected with Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen). For retroviral production, retroviral vectors were transfected into 293 Phoenix A packaging cells. For lentiviral production, lentiviral vectors were cotransfected with packaging vectors Δ8.5 and VSVG into HEK293 cells. Viral supernatants were collected at 48 and 72 hours after transfection. To infect cells, we added 2 ml of viral supernatants containing polybrene (8 µg/ml) to cells grown on six-well plates and spun at 3000 rpm at room temperature for 2 hours. Forty-eight hours after infection, cells were selected with puromycin (2 ng/ml). MEFs were isolated from embryonic day 13.5 Amoflox mouse embryos and infected with Adeno or Adeno-Cre viruses as previously described (45).
Immunoprecipitation and subcellular fractionation
Immunoprecipitation was performed as described previously (2). For subcellular fractionation experiments, the cytoplasmic fraction was extracted with hypotonic buffer [10 mM Hepes (pH 7.9), 1.5 mM MgCl2, 10 mM KC1, and 0.5 mM fresh dithiothreitol (DTT)], and pellets containing nuclei were washed twice with hypotonic buffer and subsequently lysed in radio-immunoprecipitation assay buffer.
Immunohistochemistry and immunofluorescence
Tissues were paraffin-embedded and processed. Antigen retrieval was performed by incubating deparaffinized slides with R-Buffer A with 0.1% trypsin in an antigen retrieval chamber (Electron Microscopy Science) for 15 min at 37°C. Amot immunofluorescence was performed with the TSA (tyramide signal amplification) kit (Perkin Elmer). The ABC (avidin/biotin complex) kit was used for IHC (Vector Lab). For both IHC and TSA kits, the endogenous peroxidase activity was quenched with a 30-min incubation in 7.5% hydrogen peroxide (Sigma-Aldrich) immediately after retrieval. Slides are blocked with either 2% normal donkey serum (Jackson ImmunoResearch) for IHC or buffer provided with the TSA kit. The slides are incubated with primary antibodies overnight at 4°C. All secondary antibody steps are done at room temperature for 1 to 1.5 hours.
Luciferase reporter assay
GTIIC-luciferase reporter was a gift from H. Sasaki at RIKEN, Japan (29). Renilla luciferase vector was purchased from Promega. Cells were transfected in 96-well plates with FuGene 6 (Roche R&D) 24 hours before the luciferase assay. The GTIIC-luciferase reporter activity was determined with the Dual Luciferase Assay kit (Promega). The reporter’s firefly luciferase activity was normalized to that of the internal control Renilla luciferase before all statistical analysis. The annotated relative luciferase activity is the ratio between firefly and Renilla luciferase activities.
Chromatin immunoprecipitation
Cells were fixed with 1% formaldehyde for 10 min, followed by incubation in 0.125 M glycine for 5 min. The fixed cells were resuspended in SDS lysis buffer [50 mM tris (pH 8.0), 10 mM EDTA, and 1% SDS] and sonicated for 30 min (high, 10 min per cycle, 30 s on/off) on a Bioruptor (Diagenode). The sonicated lysates were diluted with ChIP dilution buffer [20 mM tris (pH 8.0), 2 mM EDTA, 150 mM NaCl, and 1% Triton X-100] and precleared for 2 hours before incubating with Flag-conjugated agarose beads (Sigma-Aldrich) or the indicated primary antibodies plus protein G–agarose beads with salmon sperm DNA (Millipore) overnight at 4°C. The beads were washed first with low-salt buffer [20 mM tris (pH 8.0), 2 mM EDTA, 150 mM NaCl, 0.1% SDS, and 1% Triton X-100], followed by high-salt buffer [20 mM tris (pH 8.0), 2 mM EDTA, 500 mM NaCl, 0.1% SDS, and 1% Triton X-100], then LiCl buffer [10 mM tris (pH 8.0), 1 mM EDTA, 250 mM LiCl, 1% sodium deoxycholate, and 1% NP-40], and finally twice with TE [10 mM tris (pH 8.0) and 1 mM EDTA] before eluting with freshly prepared elution buffer (1% SDS and 0.1 M NaHCO3). The cross-link reversal was performed at 65°C overnight in the presence of 200 mM NaCl. The eluted chromatins were then incubated with protein digestion buffer [5 µl of 0.5 M EDTA, 10 µl of 1 M tris (pH 6.5), and 1 µl of proteinase K (10 mg/ml)] at 45°C for 4 hours and finally purified with the PCR purification kit (Qiagen). Quantitative analysis of chromatin enrichment was performed by real-time PCR. Primer sequences are provided in table S3.
For sequential ChIP, both rounds of the ChIP were performed as described above. The beads used for the ChIP were protein A/G beads with salmon sperm DNA (Millipore). Instead of using the standard elution buffer at the end of the first ChIP with the Amot antibody, the beads were eluted in tris-EDTA with 10 mM DTT at 37°C for 30 min. The resulting supernatant was diluted with ChIP dilution buffer and subjected to a second round of ChIP with either IgG or Yap antibodies.
Real-time PCR
RNAs were extracted with Qiagen RNeasy kit and then reverse-transcribed into cDNA with either Superscript III kit (Invitrogen) or iScript Advanced cDNA Synthesis kit (Bio-Rad). qPCR was performed with SYBR Green (Applied Biosystems) with ABI7900HT or SsoAdvanced SYBR Green with CFX384 (Bio-Rad). Relative gene expression between control, Amot-KD, and Yap-KD was calculated with the 2−ΔΔCT method (46). Primer sequences are provided in table S3.
Microarray
Microarray analysis was performed to compare gene expression in Amot-KD, Yap-KD, and control HEK293 cells. Samples from three individual experiments for each of the three groups were analyzed. Total RNA was isolated with the modified protocol of Sigma-Aldrich TRI reagent. The Epicentre TargetAmp Nano-g Biotin-aRNA Labeling Kit was used to generate amplified complementary RNA (cRNA). Biotinylated, amplified cRNA at 750 ng was then hybridized to the Illumina HumanHT-12 v4 Expression BeadChip. All arrays were processed in the Wistar Institute Genomics Facility.
Statistical analysis
ANOVA and post hoc Bonferroni procedure were used for the comparisons of the liver weight/body weight ratio and CK19+ cell count among the four mouse strains (control, AmotKO, Nf2KO, and DKO). For each pairwise comparison, Bonferroni’s adjusted P < 0.05 was considered statistically significant. Two-group t test was used for comparing CK19+ cell counts between control and AmotKO tissue at 1 and 4 months of age, and P < 0.05 was considered statistically significant.
For microarray data analysis, raw data were first quantile-normalized and then filtered to remove noninformative probes that were either expressed at background level or showing little variation among samples (maximum fold change <1.2). A t test was used to assess changes in gene expression between the two knockdown groups and the controls (Amot-KD versus controls and Yap-KD versus controls). In both comparisons, significant genes were selected with a cutoff of P < 0.05. Three fold-change cutoffs (FC >1.5, 2, and 3) were applied to select for genes that were most differentially expressed. These two lists were then overlapped to determine the similarity in the effects of the Yap compared with Amot knockdowns. All data preprocessing and analysis were performed in MATLAB R2010a.
Supplementary Material
Acknowledgments
We thank M. Giovannini (House Ear Institute) for providing us with Nf2 and Amot conditional knockout mice. We thank A. Gardini (The Wistar Institute) and J. Li (Georgetown Lombardi Cancer Center) for technical assistance. We thank D. J. Pan (Johns Hopkins), H. Sasaki (RIKEN, Japan), J. Hogenesch (University of Pennsylvania), and D. Melton (Harvard) for providing us with reagents.
Funding: This work was supported in part by the NIH (DK083355 and DK083111 to B.Z.S., CA142295 and NS077952 to J.L.K., and CA0180815 and CA132098 to L.C.S.), the Commonwealth of Pennsylvania (66651-01 to L.C.S.), the PA Breast Cancer Coalition (60707 and 920093 to M.S.), the Abramson Family Cancer Research Institute, the Geisinger Clinic, and the Pew Charitable Trusts. C.Y. was a recipient of a Young Investigator Award from the Children’s Tumor Foundation and is currently supported by a Georgetown Lombardi Cancer Center new faculty startup fund. Z.S. was a recipient of the Cell and Molecular Biology training grant (GM 07229-35). We are thankful for the help from Lombardi Cancer Center tissue culture and microscopy core facilities supported by Cancer Center Support Grant CA051008.
Footnotes
Author contributions: C.Y., J.L.K., and B.Z.S. conceived and obtained funding for the project. C.Y. and Z.S. designed and conducted the bulk of the experiments. C.Y., Z.S., J.L.K., and B.Z.S. co-wrote the manuscript. A.S.-R. conducted Amot IHC analysis on human schwannomas. N.D. and L.C.S. performed microarray data analysis. S.T. generated plasmids that were used in this study. Q.L. performed statistical analysis of animal studies. A.S., M.S., and L.H. provided reagents and intellectual input for this study.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Array data are available on the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE49406.
REFERENCES AND NOTES
- 1.Wells CD, Fawcett JP, Traweger A, Yamanaka Y, Goudreault M, Elder K, Kulkami S, Gish G, Virag C, Lim C, Colwill K, Starostine A, Metalnikov P, Pawson T. A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell. 2006;125:535–548. doi: 10.1016/j.cell.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 2.Yi C, Troutman S, Fera D, Stemmer-Rachamimov A, Avila JL, Christian N, Persson NL, Shimono A, Speicher DW, Marmorstein R, Holmgren L, Kissil JL. A tight junction-associated Merlin-angiomotin complex mediates Merlin’s regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell. 2011;19:527–540. doi: 10.1016/j.ccr.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J. Biol. Chem. 2011;286:7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W, Huang J, Chen J. Angiomotin-like proteins associate with and negatively regulate YAP1. J. Biol. Chem. 2011;286:4364–4370. doi: 10.1074/jbc.C110.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Troyanovsky B, Levchenko T, Mansson G, Matvijenko O, Holmgren L. Angiomotin: An angiostatin binding protein that regulates endothelial cell migration and tube formation. J. Cell Biol. 2001;152:1247–1254. doi: 10.1083/jcb.152.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bratt A, Birot O, Sinha I, Veitonmaki N, Aase K, Ernkvist M, Holmgren L. Angiomotin regulates endothelial cell-cell junctions and cell motility. J. Biol. Chem. 2005;280:34859–34869. doi: 10.1074/jbc.M503915200. [DOI] [PubMed] [Google Scholar]
- 8.Ernkvist M, Luna Persson N, Audebert S, Lecine P, Sinha I, Liu M, Schlueter M, Horowitz A, Aase K, Weide T, Borg JP, Majumdar A, Holmgren L. The Amot/Patj/ Syx signaling complex spatially controls RhoA GTPase activity in migrating endothelial cells. Blood. 2009;113:244–253. doi: 10.1182/blood-2008-04-153874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aase K, Ernkvist M, Ebarasi L, Jakobsson L, Majumdar A, Yi C, Birot O, Ming Y, Kvanta A, Edholm D, Aspenström P, Kissil J, Claesson-Welsh L, Shimono A, Holmgren L. Angiomotin regulates endothelial cell migration during embryonic angiogenesis. Genes Dev. 2007;21:2055–2068. doi: 10.1101/gad.432007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garnaas MK, Moodie KL, Liu ML, Samant GV, Li K, Marx R, Baraban JM, Horowitz A, Ramchandran R. Syx, a RhoA guanine exchange factor, is essential for angiogenesis in vivo. Circ. Res. 2008;103:710–716. doi: 10.1161/CIRCRESAHA.108.181388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du D, Xu F, Yu L, Zhang C, Lu X, Yuan H, Huang Q, Zhang F, Bao H, Jia L, Wu X, Zhu X, Zhang X, Zhang Z, Chen Z. The tight junction protein occludin, regulates the directional migration of epithelial cells. Dev. Cell. 2010;18:52–63. doi: 10.1016/j.devcel.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Li Z, Xu P, Huang L, Tong J, Huang H, Meng A. Angiomotin-like2 gene (amotl2) is required for migration and proliferation of endothelial cells during angiogenesis. J. Biol. Chem. 2011;286:41095–1104. doi: 10.1074/jbc.M111.296806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao B, Tumaneng K, Guan LK. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCFβ-TRCP . Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Haider G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong W, Guan KL. The YAP and TAZ transcription co-activators: Key downstream effectors of the mammalian Hippo pathway. Semin. Cell Dev. Biol. 2012;23:785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tapon N, Harvey KF. The Hippo pathway—From top to bottom and everything in between. Semin. Cell Dev. Biol. 2012;23:768–769. doi: 10.1016/j.semcdb.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Oka T, Schmitt AP, Sudol M. Opposing roles of angiomotin-like-1 and zona occludens-2 on pro-apoptotic function of YAP. Oncogene. 2012;31:128–134. doi: 10.1038/onc.2011.216. [DOI] [PubMed] [Google Scholar]
- 20.Paramasivam M, Sarkeshik A, Yates JR, III, Fernandes MJ, McCollum D. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol. Biol. Cell. 2011;22:3725–3733. doi: 10.1091/mbc.E11-04-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang H, Lu FI, Jia S, Meng S, Cao Y, Wang Y, Ma W, Yin K, Wen Z, Peng J, Thisse C, Thisse B, Meng A. Amotl2 is essential for cell movements in zebrafish embryo and regulates c-Src translocation. Development. 2007;134:979–988. doi: 10.1242/dev.02782. [DOI] [PubMed] [Google Scholar]
- 22.Zheng Y, Vertuani S, Nyström S, Audebert S, Meijer I, Tegnebratt T, Borg JP, Uhlén P, Majumdar A, Holmgren L. Angiomotin-like protein 1 controls endothelial polarity and junction stability during sprouting angiogenesis. Circ. Res. 2009;105:260–270. doi: 10.1161/CIRCRESAHA.109.195156. [DOI] [PubMed] [Google Scholar]
- 23.Preisegger KH, Factor VM, Fuchsbichler A, Stumptner C, Denk H, Thorgeirsson SS. Atypical ductular proliferation and its inhibition by transforming growth factor pi in the 3,5-diethoxycarbonyl-1,4-dihydrocollidine mouse model for chronic alcoholic liver disease. Lab. Invest. 1999;79:103–109. [PubMed] [Google Scholar]
- 24.Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, Stanger BZ. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136:1727–1739. doi: 10.1242/dev.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benhamouche S, Curto M, Saotome I, Gladden AB, Liu CH, Giovannini M, McClatchey AI. NF2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 2010;24:1718–1730. doi: 10.1101/gad.1938710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClatchey AI. Neurofibromatosis. Annu. Rev. Pathol. 2007;2:191–216. doi: 10.1146/annurev.pathol.2.010506.091940. [DOI] [PubMed] [Google Scholar]
- 28.Bai H, Zhang N, Xu Y, Chen Q, Khan M, Potter JJ, Nayar SK, Cornish T, Alpini G, Bronk S, Pan D, Anders RA. Yes-associated protein regulates the hepatic response after bile duct ligation. Hepatology. 2012;56:1097–1107. doi: 10.1002/hep.25769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Ji JY, Yu M, Overholtzer M, Smolen GA, Wang R, Brugge JS, Dyson NJ, Haber DA. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat. Cell Biol. 2009;11:1444–1450. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varelas X, Wrana JL. Coordinating developmental signaling: Novel roles for the Hippo pathway. Trends Cell Biol. 2012;22:88–96. doi: 10.1016/j.tcb.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Bao Y, Hata Y, Ikeda M, Withanage K. Mammalian Hippo pathway: From development to cancer and beyond. J. Biochem. 2011;149:361–379. doi: 10.1093/jb/mvr021. [DOI] [PubMed] [Google Scholar]
- 34.Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, Fu XD, Mills GB, Guan KL. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oka T, Remue E, Meerschaert K, Vanloo B, Boucherie C, Gfeller D, Bader GD, Sidhu SS, Vandekerckhove J, Gettemans J, Sudol M. Functional complexes between YAP2 ZO-2 are PDZ domain-dependent, and regulate YAP2 nuclear localization and signalling. Biochem. J. 2010;432:461–472. doi: 10.1042/BJ20100870. [DOI] [PubMed] [Google Scholar]
- 37.Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, Park O, Chang J, Simpson RM, Wang CY, Gao B, Jiang J, Yang Y. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc. Natl. Acad. Sci. USA. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, Bardeesy N. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Haider G, Finegold MJ, Lee JS, Johnson LR. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Genevet A, Tapon N. The Hippo pathway and apico-basal cell polarity. Biochem. J. 2011;436:213–224. doi: 10.1042/BJ20110217. [DOI] [PubMed] [Google Scholar]
- 41.Giovannini M, Robanus-Maandag E, van der Valk M, Niwa-Kawakita M, Abramowski V, Goutebroze L, Woodruff JM, Berns A, Thomas G. Conditional biallelic Nf2 mutation in the mouse promotes manifestations of human neurofibromatosis type 2. Genes Dev. 2000;14:1617–1630. [PMC free article] [PubMed] [Google Scholar]
- 42.Shimono A, Behringer RR. Angiomotin regulates visceral endoderm movements during mouse embryogenesis. Curr. Biol. 2003;13:613–617. doi: 10.1016/s0960-9822(03)00204-5. [DOI] [PubMed] [Google Scholar]
- 43.Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev. Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amelio LA, Miraglia LJ, Conkright JJ, Mercer BA, Batalov S, Cavett V, Orth AP, Busby J, Hogenesch JB, Conkright MD. A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20314–20319. doi: 10.1073/pnas.0707999105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kissil LJ, Wilker EW, Johnson KC, Eckman MS, Yaffe MB, Jacks T. Merlin the product of the Nf2 tumor suppressor gene, is an inhibitor of the p21-activated kinase, Pak1. Mol. Cell. 2003;12:841–849. doi: 10.1016/s1097-2765(03)00382-4. [DOI] [PubMed] [Google Scholar]
- 46.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.