Abstract
Increasing evidence is implicating mitochondrial dysfunction as a central factor in the etiology of Alzheimer's disease (AD). The most significant risk factor in AD is advanced age and an important neuropathological correlate of AD is the deposition of amyloid-β peptide (Aβ40 and Aβ42) in the brain. An AD-like dementia is also common in older individuals with Down syndrome (DS), though with a much earlier onset. We have shown that somatic mitochondrial DNA (mtDNA) control region (CR) mutations accumulate with age in post-mitotic tissues including the brain and that the level of mtDNA mutations is markedly elevated in the brains of AD patients. The elevated mtDNA CR mutations in AD brains are associated with a reduction in the mtDNA copy number and in the mtDNA L-strand transcript levels. We now show that mtDNA CR mutations increase with age in control brains; that they are markedly elevated in the brains of AD and DS and dementia (DSAD) patients; and that the increased mtDNA CR mutation rate in DSAD brains is associated with reduced mtDNA copy number and L-strand transcripts. The increased mtDNA CR mutation rate is also seen in peripheral blood DNA and in lymphoblastoid cell DNAs of AD and DSAD patients, and distinctive somatic mtDNA mutations, often at high heteroplasmy levels, are seen in AD and DSAD brain and blood cells DNA. In aging, DS, and DSAD, the mtDNA mutation level is positively correlated with β-secretase activity and mtDNA copy number is inversely correlated with insoluble Aβ40 and Aβ42 levels. Therefore, mtDNA alterations may be responsible for both age-related dementia and the associated neuropathological changes observed in AD and DSAD.
Keywords: Alzheimer's disease, amyloid-β, AβPP, control region, dementia, Down syndrome, mitochondria, mitochondrial dysfunction, mtDNA
INTRODUCTION
Alzheimer's disease (AD) is the major cause of dementia in the elderly, yet despite two decades of research, not only is there no cure but its etiology also remains unclear. Age is the greatest risk factor for AD, and AD is associated with the accumulation of amyloid-β peptide (Aβ40 and Aβ42) containing senile plaques and intraneuronal neurofibrillary tangles in the brain. While early-onset familial AD has been linked to amyloid-β protein precursor (AβPP), presenillin1 (PSEN1), and presenillin2 (PSEN2) gene mutations, mutations in these genes account for less than 5% of AD cases [1–7].
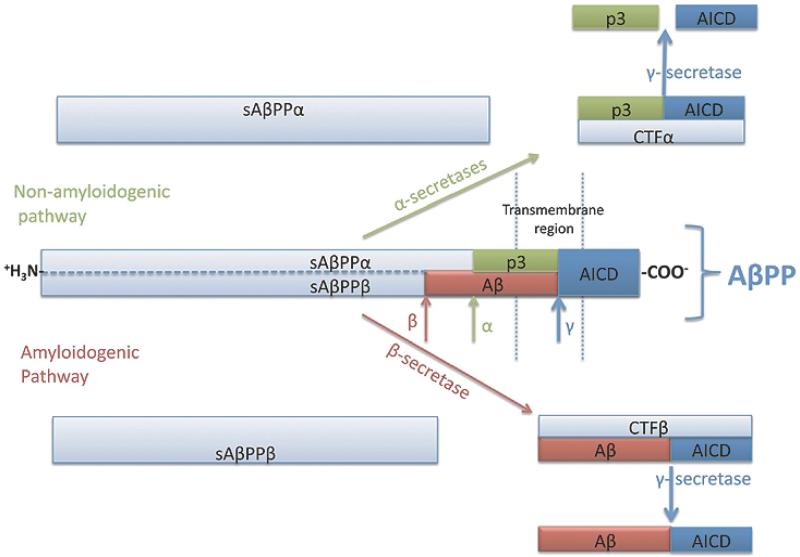
Since Alois Alzheimer's [8] original report of a demented patient with amyloid deposition in the brain, it has been assumed that the accumulation of Aβ is the cause of AD dementia. Aβ peptide is the result of the processing of the AβPP, an integral membrane protein that is processed proteolytically by two pathways: amyloidogenic and non-amyloidogenic (Fig. 1). The amyloidogenic pathway is initiated by β-secretase cleavage of AβPP to yield the N-terminal soluble AβPPβ (sAβPPβ) plus a C-terminal fragment β (CTFβ). CTFβ can then be processed by γ-secretase to generate Aβ and a free AβPP intracellular domain (AICD). The non-amyloidogenic pathway is initiated by α-secretase cleavage of AβPP that produces the N-terminal soluble AβPPα (sAβPPα) and the remaining C-terminal fragment α (CTFα). CTFα can then be processed by γ-secretase to yield the small peptide p3 and AICD. Since the α-secretase cleavage of AβPP occurs within the Aβ sequence, the generation of Aβ is precluded. Hence, increased β-secretase activity and Aβ peptide are indicative of an increase in the amyloidogenic pathway [9,10].
Fig. 1.
Schematic representation of AβPP processing for amyloidogenic and non-amyloidogenic pathways.
Aβ deposition in the brains of Down syndrome (DS) patients is also associated with the development of dementia after the 4 to 5th decade of their life [11]. DS is the result of trisomy of all or part of chromosome 21, which includes the AβPP gene as well as β-site AβPP cleaving enzyme 2 (BACE2) [12–14]. In DS, AβPP is overexpressed in brain and peripheral lymphocytes throughout life [15] yet Aβ levels are similar between young DS patients and controls under 30 years. However, after 40 years, Aβ plaques begin to accumulate in AD brains [16,17]. As a result, it has been hypothesized that the rise in Aβ in DS after 40 years is due to a change in the proteolytic cleavage of AβPP toward the amyloidogenic pathway [18].
While progressive dementia and amyloid plaque deposition correlate with each other in AD, this does not necessarily indicate that amyloid plaque deposition causes the dementia. It is possible that some third factor causes both the dementia and the accumulation of Aβ. Evidence that this might be the case is seen for late onset “sporadic” AD. Late onset AD accounts for more than 90% of AD cases and has not been linked to mutations in AβPP or presenilins. Rather, the strongest genetic associations are with the ε4 allele of the apolipoprotein E gene (ApoE ε4) and closely linked short allele of the TOM40 protein gene [7,19,20]. The ApoE protein is a lipid carrier, and lipids are required for the assembly of the mitochondria and are oxidized by the mitochondrion to generate energy. TOM40 is the central channel protein for the import of cytosolicly synthesized proteins through the mitochondrial outer membrane and thus into the mitochondrion [21]. Hence, late-onset AD appears to be associated to mitochondrial function.
Mitochondrial dysfunction and oxidative stress are commonly observed in AD patients [22,23]. The brain tissue, platelets, and cell lines from AD patients have been reported to manifest partial defects in mitochondrial oxidative phosphorylation (OXPHOS), specifically respiratory complex IV (cytochrome c oxidase or COX) [24–32]. Moreover, the Aβ peptide has been reported to enter the mitochondrion and inhibit COX and L-3-hydroxyacyl coenzyme Q dehydrogenase (ABAD). Inhibition of these enzymes in turn contributes to mitochondrial reactive oxygen species (ROS) production [33–38]. Similarly, the ApoE ε4 protein has been reported to be cleaved by a neuron-specific protease, with the resulting peptides also entering the mitochondrion, inhibiting OXPHOS, and increasing ROS production [39,40].
In the brains of mutant mice overexpressing AβPP, tau, and PSEN1, mitochondrial dysfunction has been detected as one of the earliest events in AD [33,41–43]. Mitochondrial defects include reduced membrane potential, an OXPHOS defect, reduced ATP production, and increased ROS production. Furthermore, one-third of the proteins that are deregulated in the triple transgenic mice are mitochondrial including subunits of complex I and complex IV [41].
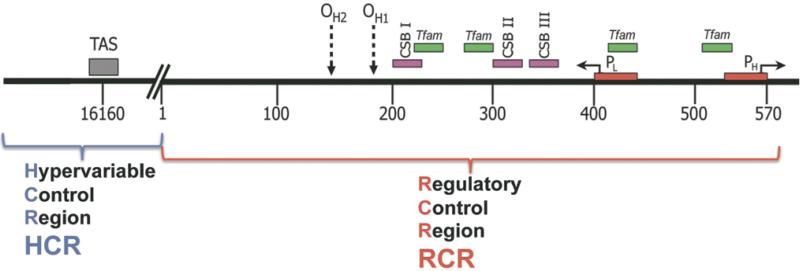
Mitochondria generate the majority of cellular energy by OXPHOS, are a major generator of cellular ROS, regulate cytosolic Ca++, and control cell death through activation of the mitochondrial permeability transition pore (mtPTP) [21]. Every cell contains hundreds of mitochondria and mtDNAs, with each mtDNA encoding 13 essential OXPHOS polypeptides, the rRNAs and tRNAs for mitochondrial translation, and a regulatory control region (CR) spanning nucleotides (nt) 16024 through 16569/0 to 576 (Fig. 2). The CR can be divided into the regulatory control region (RCR) and the hypervariable control region (HCR). The RCR is located between nt 1 and 576 and encompasses the origin of the heavy (H)-stand DNA replication (OH); the promoters for the H-strand (HSP) and the light (L)-strand (LSP); transcription factor A mitochondrial, TFAM, binding sites; conserved sequence blocks (CSBs) I, II, and III. The HCR is located between nt 16024 to 16569, and has a much higher sequence diversity across the human populationthan do other regions of the mtDNA, and encompasses the termination associated sequence (TAS) and the transcription termination factor 3 (mTERF3) binding site [21,44] (Fig. 2).
Fig. 2.
Schematic representation of human mtDNA control region. Complete mtDNA control region is divided into the regulatory control region (RCR, located between nt 1 and 576) and the hypervariable control region (HCR, located between nt 16024 to 16569). OH , the origin of the heavy (H)-stand DNA replication; HSP, the promoter for the H-strand; LSP, the promoter for the light (L)-strand; TFAM, transcription factor A mitochondrial binding sites; CSBs, conserved sequence blocks I, II and III; TAS, termination associated sequence.
The human mtDNA sequence variation is much greater than that seen in comparable nuclear DNA (nDNA) genes [45,46]. Furthermore, the accumulation of mtDNA mutations in post-mitotic tissues has been proposed to erode cellular energetics and thus cause the progressive decline of function perceived as aging [47, 48]. Clinical observations suggest that mtDNA variation may be important in the etiology of late onset AD comes from the observations that AD is 1.7 to 3.6 times more likely to be observed in a patient's mother than father [49,50], and from Aβ imaging studies that indicate the offspring from AD mothers have higher brain Aβ loads than the brains of offspring from AD father [51]. Certain germline mtDNA mutations and haplotypes are also associated with AD. The mtDNA haplogroup H5a, which harbors the tRNAGln nt 4336G variant, has been repeatedly associated with late onset AD [52–61], while mtDNA haplogroups J, T, and K have been reported to be protective of AD [62–64].
Somatic mtDNA mutations have also been found to be increased in AD brains. The common mtDNA 5 kilobase (kb) deletion was found to be elevated 15 fold in AD brains [65] and somatic mtDNA CR mutations were found to be increased by 73% in AD brains. Many of the AD somatic mtDNA variants alter known CR functional elements, while CR mutations found in normal brains generally do not. The mtDNA CR T414G mutation, which accumulates with age in skin fibroblasts [66], can be detected at a low level of heteroplasmy in 65% of AD brains [67] but cannot be detected in control brains [68]. Somatic mtDNA mutation levels in AD brains also correlate with a reduction in mtDNA/nuclear DNA (nDNA) ratio and with the L-strand ND6 transcript/H-strand ND2 transcript mRNA ratios [67]. This mutation alters the binding of TFAM at its binding site associated with the LSP.
Mitochondrial dysfunction has also been implicated in DS. The triplicated region of chromosome 21 encodes not only AβPP and BACE2, but also important antioxidant genes including Cu/Zn SOD and mitochondrial genes such as NDUFV3, MRPS6 and MRPL39, HLCS, ES1, ATP5J and ATP5O [69]. Mitochondrial dysfunction and elevated oxidative stress have been observed in DS primary neuronal cultures, brains, fibroblasts, erythrocytes, and urine [70–74]. Specific DS mitochondrial alterations include decreased mitochondrial COX, complex I proteins, and heat shock proteins [75–81], and elevated mitochondrial aconitase and isocitrate dehydrogenase levels [82].
Based on these observations, we can now propose that the etiology of premature dementia is the result of an underlying mitochondrial dysfunction resulting in energetic deficiency, increased oxidative stress, altered calcium regulation, and predilection to cell death through activation of the mtPTP [21]. A systemic mitochondrial defect would preferentially affect the brain because of its very high energy demand [83]. High levels of Aβ and the ApoE ε4 degradation products can also cause dementia because they also inhibit mitochondrial function. Regardless of whether mitochondrial OXPHOS is inhibited by an inherited defect or due to a toxic peptide, inhibition of mitochondrial function will result in increased ROS production which will cause the accumulation of somatic mtDNA mutations. The resulting decline in mitochondrial function ultimately results in mtPTP activation and loss of neuronal processes and neurons. In cases where the initiating defect is a systemic mitochondrial defect, as would commonly be the case for late-onset AD, then we would expect that somatic mtDNA mutation levels would also be increased in non-brain tissues. Furthermore, the AD-like dementia of older DS patients should also be associated with increased mitochondrial dysfunction and the accumulation of somatic mtDNA mutations. In the current study, we demonstrate that these predictions are correct. Therefore, mitochondrial dysfunction appears to be a causal variable resulting in premature dementia [21].
MATERIALS AND METHODS
Sample collection
Frontal cortex samples were collected from equal proportions of male and female DSAD, DS, AD, and control subjects from the Alzheimer Disease Research Center (ADRC) at UCI and Emory University, Atlanta. These included 14 DSAD (age range 40 to 62 years old), 11 DS (newborn to 45 years old), 13 AD (55 to 90 years), and 25 Controls (newborn to 95 years). All brain samples were characterized for the presence and frequency of senile plaques and neurofibrillary tangles. This AD-like neuropathology was identified in the AD and DSAD brains but not in the DS and control samples.
Because of the influence of age on the level of accumulated somatic mtDNA CR mutations, comparisons were made between AD and DSAD samples and age-matched controls when possible. Since virtually all DS over 45 develop dementia, it was not possible to obtain older DS subjects without dementia. Studies of DS without dementia included subjects and controls from < 1 to 45 years. The DSAD cases were also compared to control brains, ages 40 to 64 years of age. None of the AD cases had known AD-related nuclear gene mutations, and thus all would be classified as sporadic AD cases.
The probability of developing AD doubles every five years after the age of 65 [84]. Therefore 2–3% of individuals between ages 64–69 exhibit signs of AD, while 25–50% of people over 85 have evidence of AD. To correct for this age effect in comparisons using our AD cohort, we subdivided our AD and controls into two cohorts: young AD ages 55 to 70 years and old AD ages 81 to 90 years, with comparable age-matched controls.
Peripheral tissues from DS, DSAD, AD, and control subjects were analyzed for mtDNA somatic mutation levels using the same approaches as applied to the brain samples of these 4 groups. For six each of the AD and control cases, ages 81 to 95 years, from which brain samples were analyzed, clotted serum samples were available from which DNA was isolated, called cell free DNA samples (CF-DNA). These were analyzed for mtDNA mutation levels. Furthermore, lymphoblastoid cell lines (LCLs) were analyzed from 4 different groups of 5 to 7 subjects each of DS, DSAD, AD, and controls, all in the range of ages 45 to 62 years of age.
mtDNA CR sequence and haplotyping
The mtDNA haplogroup of each sample was determined by sequencing the CR (np-16024-570) and confirmed by testing for haplogroup specific coding region single nucleotide polymorphisms (http://www.mitomap.org). All nucleotide variants were reported relative to the modified Cambridge mtDNA Reference Sequence. To be assured that variants analyzed were not the result of amplification of nuclear DNA pseudogenes, each PCR protocol was applied to genomic DNA isolated from cells lacking mtDNA (ρo cells). If no PCR band was generated, then the primer set was assumed to avoid pseudogene amplification.
Detection of the T414G mutation
PNA-clamping PCR was used to detect the T414G mutation in frontal cortex DNAs. This procedure can detect one mutant mtDNA in 1000 wild-type molecules. By this procedure, the T414G mutation was detected by PCR amplification of a 334 bp PCR using a primer that overlaps with nt 414 in the presence of the wild type blocking PNA. The amplification of the mutant product was confirmed by digestion with the FokI restriction enzyme [85].
Identification of heteroplasmic mtDNA CR mutations by cloning and sequencing
The full array of heteroplasmic mtDNA CR mutations was identified by sequencing between 35 to 52 mtDNA CR clones in each individual case. Genomic DNA was extracted from frontal cortex using the Pure-Gene kit (Gentra system) and the CR amplified with the high fidelity PFU Taq DNA polymerase (iStrate-gene UltraPFU) using primers nt 15978–15998 and nt 616–636. Purified PCR fragments were cloned into the pCR 4Blunt-TOPO vector (Invitrogen), the cloned DNA directionally amplified with universal m13 reverse and forward primers, the products cyclically amplified using BigDye dideoxy chain terminator chemistry (Applied Biosystem), and the sequence determined using an ABI 3130 capillary sequencer and analyzed using “Sequencer v4.8” (Gene Codes Corporation).
By cloning and sequencing the full CR from nt 15978 through 16569/1 to 636, each clone included both the “regulatory control region (RCR)” (nt 1 to 600) and the “hypervariable control region (HCR)” (nt 16000to 16569). The HCR is hypervariable when comparing the mtDNAs from different individuals; however, the HCR was much less variable than the RCR when comparing mtDNAs isolated from a tissue of a single individual. Hence, in the current study, the HCR sequence provided the haplogroup signature nucleotides, thus guaranteeing that each clone is from the correct subject, while the RCR sequence permitted the detection of heteroplasmic somatic mtDNA mutations.
The sequences were aligned and deviant nucleotides identified. Unless otherwise stated, only the heteroplasmic mutations of the RCR were reported, which were assumed to have arisen within the tissue and thus be somatic in origin.
Quantification of mitochondrial transcript efficiency and mtDNA copy number
Mitochondrial L-strand/H-strand gene expression ratio was quantified from total cortical RNA extracted using TRIZOL (Gibco-BRL system). mtDNA L-Strand encodes ND6 and 8 tRNAs H-Strand encodes rest of the mRNAs and tRNAs and rRNAs. The ratio of mtDNA L-strand to H-strand transcripts was evaluated as mitochondrial L-strand/H-strand gene expression ratio. ND6 gene expression represents the L-stand transcription, and ND2 gene expression represents the H-Strand. mRNAs were quantified using quantitative reverse transcription-PCR (qRT-PCR) using Roche Light Cycler S480 Real-Time PCR system. The gene expression of ND2 and ND6 determined by absolute quantification using a reference plasmid which was derived from the ND2 and ND6 amplicones cloned in the pCR 4Blunt-TOP® vector (Invitrogen).
The mtDNA copy number was determined in the total DNA extracted from the frontal cortices. The mtDNA ND2 gene copy number was quantified to indicate the mtDNA copy number and the 18S rDNA nuclear DNA (nDNA) copy number was estimated to determine the nDNA copy number. The absolute copy number values were determined by comparison with quantification of reference plasmids derived with the ND2 and 18S rDNA amplicones cloned in pCR® 4Blunt-TOP vector (Invitrogen). The relative mtDNA copy number was then reported as a ratio of ND2 to 18S rDNA gene copy numbers. The primers are listed in a previous report [67].
Quantitative analysis of β-secretase activity, AβPP, and Aβ Levels
The β-secretase activity and AβPP and Aβ levels for DS, DSAD, and control brains have been reported previously [86].
Data analysis and statistics
Statistical analyses were performed using GraphPad Prism 5 software (GraphPad Software, Inc.).
RESULTS
Frequency of mtDNA RCR mutations in frontal cortices
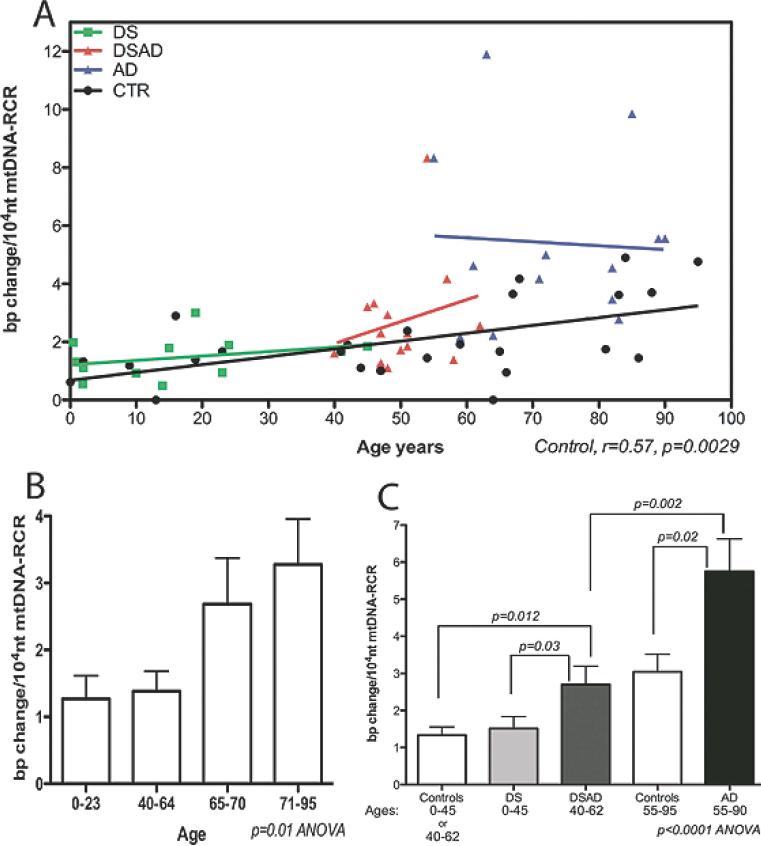
The frequency of somatic mutations in the mtDNA RCR increases with age in normal human brain (r = 0.57, p = 0.0029) (Fig. 3A). This increase rises markedly in the majority of samples after age 65. The mean RCR mutation frequency at 0 to 23 year-old brains was 1.3 × 10−4/nt, in 40 to 64 year old brains was 1.4 × 10−4/nt, in 65 to 70 year-old brains was 2.7 × 10−4/nt, and in 71 to 95 year-old brains was 3.3 × 10−4/nt (p = 0.01 ANOVA) (Fig. 3B).
Fig. 3.
mtDNA RCR mutation frequency in the frontal cortices of control, DS, DSAD and AD patients. A) The correlation of mtDNA RCR mutation frequency with aging in all four groups. B) The comparison of mtDNA RCR mutation frequency in four control brain groups for the age ranges: 0–23, 40–64, 65–70 and 71–95 years. C) The comparison of mtDNA RCR mutation frequencies of DSAD and AD brains to age-matched controls and DS brains. Because the control groups for DS (ages 0–40) and DSAD (ages 40–62) were not significantly different (see panel 3b), DS and DSAD control groups were grouped together.
In AD brains, the frequency of the mtDNA RCR mutations was significantly higher at 5.8 × 10−4/nt than in control brains at 2.3 × 10−4/nt, in the age range of 55 to 90 years (p = 0.002, t-test) (Fig. 3A,C). This confirms and extends our previous observation that mtDNA CR mutations are increased in AD brains [67].
The mean frequency of mtDNA RCR mutations in DSAD brains, ages 40 to 62 years, was 2.7 × 10−4/nt, which is also significantly higher than the 1.4 × 10−4/nt age-matched control brains (p = 0.012) (Fig. 3C), though two-fold less than AD brains (2.7 × 10−4 versus 5.8 × 10−4/nt). By contrast, the mean RCR mutation frequency of 0 to 45 year-old DS brains was 1.5 × 10−4/nt, the same as age-matched controls at 1.3 × 10−4/nt (Fig. 3C). Therefore, the mean mtDNA RCR somatic mutation frequency is elevated in both AD and DSAD brains, relative to age-matched control and DS brains.
Frequency of mtDNA RCR mutations in the peripheral tissues
To determine if the increased mtDNA mutation rate seen in AD brains was brain-specific or systemic, we analyzed the mtDNA RCR mutation frequency in DNA extracted from serum samples (cell free DNA, CF-DNA) collected from some of the same patients and controls as were analyzed for brain somatic mtDNA mutations. This revealed that the frequency RCR mutations in AD serum samples (ages 81 to 95 years) was 3.7 × 10−4/nt, 2.5 times more than that found in age-matched control serum samples (1.5 × 10−4/nt) (p = 0.02 Two-way ANOVA) (Fig. 4A).
Fig. 4.
mtDNA RCR mutation frequency in the peripheral tissue samples from control and dementia patient. A) The analysis of mtDNA RCR mutation frequency in the frontal cortex and serum DNA (CF-DNA) from the same AD and control cases (n = 6, between the ages of 81 to 95 years). B) The analysis of lymphoblastoid cell lines (LCLs) from DS, DSAD, AD, and controls (n = 5–7, between the ages of 45 to 62 years).
The elevation of somatic mtDNA mutations in AD blood cells was confirmed by analyzing the steady state mtDNA RCR mutation frequency in lymphoblastoid cell lines derived from AD, DSAD, DS, and control subjects, derived from 45 to 62 year-old donors. The mtDNA RCR levels revealed that both the AD (3.9 × 10−4/nt) and DSAD (3.6 × 10−4/nt) lymphoblastoid cell lines have approximately 2.5 fold more somatic mtDNA RCR mutations than control (1.7 × 10−4/nt) and DS (1.4 × 10−4/nt) lymphoblastoid cell lines (p < 0.001 ANOVA) (Fig. 4B). Therefore, the somatic mtDNA mutation levels are consistently elevated in peripheral blood cells of AD and DSAD patients, confirming that the elevated mtDNA mutation frequency is a systemic phenomenon in patients with dementia. Since lymphoblastoid cell lines are continually replicating, the somatic mtDNA mutation levels must be maintained at a steady state level in both patients and controls. Therefore, the increased frequency of somatic mtDNA mutations in AD and DSAD lymphoblastoid cells must reflect an increased mtDNA mutation rate in the lymphoblasts from demented patients.
The nature of mtDNA somatic mutations in the brains and bloods
We previously observed that specific mtDNA RCR mutations were found in AD brains that were either rare or absent in control brains, the most striking of these being the mtDNA T414G mutation [67]. To confirm this result and determine if the T414G mutation was also found in DSAD brains, the PNA-clamping PCR procedure was used to detect the T414G mutation in AD, DS, DSAD, and control brains. The T414G mutation was found in 65% of AD brain samples but not in controls, and was also detected in 57% of DSAD brains but not in DS brains. Therefore, the accumulation of the T414G mutation is specific for brains with dementia.
In addition to the T414G mutation, other RCR somatic mutations were found to accumulate in AD, DS, DSAD, and control brains, documented using the mtDNA CR cloning and sequencing procedure. We considered a mutation to be somatic if different clones from the same sample had different bases at the same nucleotide position indicating that the site is heteroplasmic and thus that the nucleotide change arose recently. All variants are reported relative to the Revised Cambridge Reference Sequence. In AD brains, a heteroplasmic, somatic C mutation nt T414 was observed, suggesting that the T to C variant at nt 414 is more prevalent than the T to G variant. Additional heteroplasmic mutations that were more common in AD brains than control brains included G68A, G70A, T72C, G185A, G207A, G228A, 309delC, 309insC, T408C, T414C, C418T, and 466.2insCC (Table 1). Only the A189G/C, 309.3in CCC, A415G, and 456delC mutations were more prevalent in control brains than AD brains (Table 1).
Table 1.
Specific mtDNA RCR heteroplasmic nucleotide changes reported as the percentage of the total individuals that harbor the heteroplasmic mutation
| Tissue type → |
BRAIN |
LCL |
CELL FREE DNA |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of cases → | 24 | 13 | 14 | 12 | 5 | 6 | 6 | 7 | 6 | 6 | ||
| np | rCRS | Change | Control | AD | DSAD | DS | Control | AD | DSAD | DS | Control | AD |
| 68 | G | A | O | 15% | 14% | O | O | O | 17% | O | O | O |
| 70 | G | A | 4% | 31% | 14% | 8% | O | O | 17% | O | O | O |
| 72 | T | C | 4% | 8% | 14% | O | O | O | O | O | O | 17% |
| 73 | A | G | 4% | O | O | O | 60% | O | O | 71% | O | O |
| 93 | A | G | 4% | O | O | O | O | O | O | O | O | 17% |
| 103 | G | A | O | 8% | 7% | O | O | O | O | O | O | O |
| 114 | C | T | O | 8% | O | 8% | O | O | 17% | O | O | O |
| 150 | C | T | O | O | O | 17% | 40% | O | O | O | O | O |
| 152 | T | C | O | O | 7% | O | O | O | O | O | O | O |
| 185 | G | A | O | 8% | 7% | 17% | O | O | O | O | O | 17% |
| 189 | A | G/C | 38% | 23% | 36% | O | O | 17% | 17% | O | O | 17% |
| 195 | T | C | 4% | O | 14% | O | O | O | O | O | O | O |
| 199 | T | C | O | O | O | O | 20% | O | O | O | O | O |
| 204 | T | C | 4% | O | 7% | O | O | O | 33% | O | 33% | 33% |
| 207 | G | A | 4% | 23% | 7% | O | O | O | O | O | O | 17% |
| 228 | G | A | O | 8% | O | O | O | O | O | O | O | O |
| 234 | A | G | O | O | O | O | O | O | 17% | O | 17% | O |
| 293 | T | C | O | O | O | O | O | O | O | O | O | 17% |
| 309 | C | Δ | O | 23% | 7% | O | O | O | O | O | O | O |
| 309.1 | : | C | 38% | 85% | 29% | 58% | 20% | 100% | 83% | 43% | 50% | 50% |
| 309.2 | : | C | 25% | 38% | 29% | 58% | 20% | 50% | 83% | 29% | 50% | 50% |
| 309.3 | : | C | 17% | 8% | 14% | 33% | 20% | 17% | 33% | O | 17% | 17% |
| 310 | T | C | O | O | O | 8% | 20% | O | O | O | O | 17% |
| 357 | A | G | O | O | O | O | O | O | O | 14% | O | O |
| 377 | C | T | O | O | O | O | O | 33% | 17% | O | O | O |
| 408 | T | A | O | 31% | O | O | O | O | O | O | O | O |
| 414 | T | C | O | 8% | O | O | O | 17% | O | O | O | O |
| 415 | A | G | 4% | O | O | O | O | O | O | O | O | O |
| 418 | C | T | 4% | 8% | O | O | O | O | O | O | O | O |
| 456 | C | A | 4% | O | O | O | O | O | 17% | O | O | O |
| 466.2 | : | CC | O | 8% | O | O | O | O | O | O | O | 17% |
| 489 | T | C | O | O | O | O | O | 17% | O | O | O | O |
| 499 | G | A | O | O | O | O | O | O | O | O | O | O |
| 522–23.1 | : | CA | 20% | 23% | 29% | 8% | O | 17% | 17% | O | O | O |
| 522–23 | CA | Δ | O | O | O | O | O | 17% | O | 14% | O | O |
| 537 | C | T | O | O | 7% | O | O | O | 17% | O | O | O |
Heteroplasmic mutations were preferentially found in DSAD brains relative to controls. These included G68A, G70A, T72C, G103A, T125C, G185A, T195C, and C537T. Some of these were also elevated in AD brains (Table 1). Finally, heteroplasmic mutations that were more common in DS brains relative to controls included G70A, C114T, C150T, G185A, and 309.1-3insC (1–3).
Certain mtDNA RCR mutations were more prevalent in AD serum than control serum DNAs. These included T72C, A93G, G185A, A189G/C, G207A, T293C, T310C, and 466.2insCC. The only RCR mtDNA mutation preferentially found in control serum DNA was A234G.
Among the lymphoblastoid cell lines, six heteroplasmic variants were commonly found in AD cell lines, than control lines: A189G/C, 309.1inC, 309.2inCC, C377T, T414C, T489C, 522-523inCA, and 522-523delCA (Table 1). Three RCR mutations were elevated in control lymphoblastoid cell lines: A73G, C150T, and T199C. Several additional variants were preferentially found in DSAD lymphoblasts versus controls: G68A, G70A, C114T, A189G/C, T204C, A324G, 309.1-3insC(1–3), 377T, 456delC, 522-523inCA, and C537T. DS lymphoblasts share the A73G variant with control lymphoblasts while they more commonly have the 309.1inC, A357G, and 522–523delCA (Table 1).
Heteroplasmic somatic mutations shared by AD and DSAD brains in excess of controls were C68A, G70A, T72C, G103A, G185A, G207A, and C309del. Mutations shared by AD and DSAD lymphoblasts included A189G/C, 309.1, 309.2, C377T, and 522–523.
In addition to the actual location and proportion of cases that harbored heteroplasmic mutations, there was a striking elevation in the percentage of mutant mtDNAs (heteroplasmy level) in AD and DSAD tissues or cell types (Table 2). Surveying all casess in which the percentage heteroplasmy was greater than 10%, every case but three was observed in either AD or DSAD. The only exceptions were one DS brain which was 22% for the G70A variant, one DS brain which was 29% for the A214G variant, and one control brain that was 97% for the 522–523.1 CA insertion. Several AD and DSAD cases had mutations at remarkably high heteroplasmy levels (Table 2). Of the AD brains, individual cases with greater than 50% heteroplasmy included three cases with high G207A mutations (90%, 88%, 75%), two cases with a high proportion of 315.1 C insertion mtDNAs (68%, 84%), and three cases with the 522–523.1 CA insertion (86%, 61%, 97%). Similarly, of the DSAD brains, one case had the G68A mutation at 80%, one case of the A189G/C at 96%, one case of the A200G at 90%, and one with the G207A at 92%.
Table 2.
Number of cases that have ≥ 10% heteroplasmy rate. Right site of the table (under Brain, LCL and Cell free DNA (CF DNA) columns) shows all the cases – including cases reported in column 4 – that have the particular nucleotide position (np) full range of heteroplasmy in each tissue type
| Brain |
LCL |
Cell free DNA |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of cases → | 24 | 13 | 14 | 12 | 5 | 6 | 6 | 7 | 6 | 6 | ||||||
| np | rCRS | Change | # of cases (% heteroplasmy) | Condition | Tissue | Ctr | AD | DSAD | DS | Ctr | AD | DSAD | DS | Ctr | AD | |
| 7 | A | G | 2 (12 & 80%) | DSAD | brain | 1 | 0 | 2 | 0 | |||||||
| 68 | G | A | 1 (11%) | AD | brain | 0 | 2 | 2 | 0 | |||||||
| ” | 1 (80%) | DSAD | brain | |||||||||||||
| ” | 1 (13%) | DSAD | LCL | 0 | 0 | 1 | 0 | |||||||||
| 70 | G | A | 1 (11%) | AD | brain | 1 | 4 | 2 | 1 | |||||||
| ” | 2 (12 & 80%) | DSAD | brain | |||||||||||||
| ” | 1 (22%) | DS | brain | |||||||||||||
| ” | 1 (13%) | DSAD | LCL | 0 | 0 | 1 | 0 | |||||||||
| 71 | G | Δ | 1 (20%) | AD | brain | 6 | 3 | 4 | 6 | |||||||
| 72 | T | C | 1 (14%) | ctr | brain | 1 | 1 | 2 | 0 | |||||||
| 93 | A | G | 1 (39%) | ctr | brain | 1 | ||||||||||
| 185 | G | A | 1 (91%) | AD | CF DNA | 0 | 1 | |||||||||
| 189 | A | G/C | 1 (10%) | AD | brain | 10 | 3 | 5 | 0 | |||||||
| ” | 1 (13 & 96%) | DSAD | brain | |||||||||||||
| 200 | A | G | 1 (90%) | DSAD | brain | 0 | 0 | 1 | 0 | |||||||
| 204 | T | C | 1 (26%) | DSAD | LCL | 0 | 0 | 2 | 0 | |||||||
| 207 | G | A | 3 (90 & 88 & 75%) | AD | brain | 1 | 3 | 1 | 0 | |||||||
| ” | 1 (92%) | DSAD | brain | |||||||||||||
| ” | 1 (94%) | AD | CF DNA | 0 | 1 | |||||||||||
| 214 | A | G | 1 (29%) | DS | brain | 0 | 0 | 1 | 1 | |||||||
| 234 | A | G | 1 (12%) | DSAD | LCL | 0 | 0 | 1 | 0 | |||||||
| 241 | A | G | 1 (10%) | DSAD | brain | 0 | 0 | 1 | 0 | |||||||
| 263 | G | A | 1 (63%) | DSAD | LCL | 0 | 0 | 1 | 0 | |||||||
| 490 | A | G | 1 (14%) | AD | brain | 0 | 1 | 0 | ||||||||
| 537 | C | T | 1 (10%) | DSAD | brain | 0 | 0 | 1 | 0 | |||||||
| 586 | A | G | 1 (11%) | AD | brain | 0 | 1 | 0 | ||||||||
| 309 | C | Δ | 2 (29 & 14%) | AD | brain | 0 | 3 | 1 | 0 | |||||||
| 315.1 | : | C | 2 (68 & 84%) | AD | brain | 0 | 2 | 0 | 0 | |||||||
| 368 1.8 | : | ins | 1 (30%) | AD | CF DNA | 0 | 1 | |||||||||
| 522–523.1 | : | CA | 1 (97%) | ctr | brain | 5 | 3 | 4 | 1 | |||||||
| ” | 3 (86 & 61 & 97%) | AD | brain | |||||||||||||
| ” | 1 (96%) | AD | LCL | 0 | 1 | 1 | 0 | |||||||||
The level of heteroplasmy in lymphoblasts was generally low. However, one AD case had the 522–523.1 CA insertion in 96% of the mtDNAs.
For the serum samples, over 50% heteroplasmy levels were seen for one case at G185A (91%), and one case at G207A (94%). Remarkably, the brains from these same two patients had high levels of the same CR mutation observed in the blood. The patient with the 91% G185A mutation in blood was found to be approximately 100% mutant in the brain. This high percentage mutant was not of a germline transmission of the G185A mutation since this patient's mtDNA haplogroup was European U5a1 and U5a1 is not associated with the G185A variant. Indeed, the G185A variant is only rarely found in European mtDNAs, being more common in Asian haplogroups A and D and occasionally being seen in African L mtDNAs. Similarly, in the patient whose serum DNA was 94% for the G207A mutation, his brain mtDNA was 75% mutant. Interestingly, the G207A mutation was also present in 88% of the mtDNAs in the brain of the patient which harbored the G185A mutation in both serum and brain mtDNAs. These results indicate that certain somatic mutations arise before the separation of the neurological and blood cell lineages and become broadly distributed in cells throughout these organ systems. Subsequently selective amplification of these mutant mtDNAs would then result in the predominance of the mutation in the different tissues.
Correlation of CR mutations with functional elements
To better understand the potential significance of the AD and DSAD somatic mutations, we also compared the position of the somatic mutations relative to the location of known functional elements of the mtDNA RCR in AD, DSAD, DS, and control samples (Table 3). The most significant observation is that the frequency of somatic mutations seen in the CSBII element was significantly higher in AD brains than controls, with 10.7% of the total AD brain clones having a CSBII site mutation as compared to 4.8% of the age-matched controls (p = 0.01 Fisher's exact test). The increase in CSBII mutations in AD was also seen for lymphoblast and cell free DNAs. The next most significant difference for brain samples was seen at the LSP site where 2.3% of all the AD clones had mutations versus 1.2% of age-matched control clones. A comparable increase was seen in lymphoblast when the data from all dementia subject (AD + DSAD) clones were compared with the data from all non-dementia subject (control + DS) clones. In brain, the proportion of clones with mutations in the CSBII and LSP regions was 6.3% versus 4% for the CSBII site (p = 0.036), and 1.4% versus 0.4% for the LSP site, (p = 0.01 Fisher's test) (Table 4). More mutant clones were also observed between nts 1–212 in dementia subjects than controls, 7.4% versus 3.6% (p = 0.0009 Fisher's test), a trend also seen in lymphoblast and cell free DNA samples (Table 3). The nt 1 to 212 RCR region contains multiple H-strand origin replication start sites [87]. Therefore, somatic mtDNA mutations in specific CR regulatory elements are significantly increased in AD and DSAD relative to controls and DS subjects (Table 4).
Table 3.
The percentage of heteroplasmic clones that are distributed on the regulatory elements of mtDNA RCR in AD, DS, DSAD, and control brains and peripheral tissues
| Brains |
Lymphoblastoid cell lines |
Cell free DNA |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All CTR | CTR | AD | CTR | DSAD | CTR | DS | CTR | AD | DSAD | DS | CTR | AD | ||
| Total clone → | 658 | 250 | 262 | 226 | 377 | 182 | 470 | 112 | 164 | 137 | 172 | 177 | 135 | |
| Locus | nt span | |||||||||||||
| 1–212 | 4.7% | 4.4% | 8.4% | 2.2% | 6.6% | 0.5% | 2.1% | 2.7% | 5.0% | 3.6% | 0.6% | 1.7% | 3.7% | |
| CSBI | 213–35 | 0% | 0.8% | 0.4% | 0% | 0.3% | 0% | 0.2% | 0% | 1.0% | 1.5% | 0.0% | 1.1% | 0% |
| TFX | 233–60 | 0% | 0% | 0.8% | 0% | 0.3% | 0% | 0% | 0% | 1.0% | 0.7% | 0% | 0% | 0% |
| TFY | 276–303 | 1.0% | 1.2% | 0.8% | 0.4% | 0% | 0% | 0.2% | 0% | 0.0% | 0% | 0% | 0.6% | 0.7% |
| CSBII | 299–315 | 4.0% | 4.8% | 10.7%* | 2.2% | 3.2% | 3.8% | 4.5% | 3.6% | 11.0% | 8.8% | 3.5% | 4.5% | 7.4% |
| CSBIII | 346–63 | 0% | 0% | 0% | 0% | 0.3% | 0% | 0.2% | 0% | 1.0% | 0.0% | 0.6% | 0.6% | 0% |
| mt4H | 371–91 | 0% | 0.8% | 0% | 0% | 0% | 0% | 0% | 0% | 2.0% | 0.7% | 0% | 0% | 0% |
| LSP | 392–445 | 1.0% | 1.2% | 2.3% | 0% | 0.8% | 0.5% | 0% | 1.8% | 3.0% | 0.7% | 0.6% | 0.6% | 0.7% |
| TFL | 418–45 | 0% | 0.4% | 0.4% | 0% | 0.3% | 1.0% | 0% | 0% | 2.0% | 0.7% | 0% | 0.6% | 0% |
| TFH | 523–50 | 1.0% | 1.2% | 1.9% | 0.9% | 1.3% | 0.5% | 0.6% | 0% | 2.0% | 1.5% | 1.2% | 0% | 0% |
| HSP1 | 545–67 | 0% | 0% | 0% | 0% | 0.3% | 0% | 0% | 0.9% | 1.0% | 0% | 0% | 0% | 0% |
p = 0.01 Fisher test when AD CSBII values compared to controls. CSBI, II, III: Conserved Sequence Blocks 1, 2, 3; TFX and TFY: mitochondrial transcription factor A (TFAM) binding sites other than promoters; TFL and TFH: TFAM binding site for L-Strand Promoter (LSP) and H-Strand promoter (HSP); mt4H: H-strand control element, CTR: controls, nt: nucleotide.
Table 4.
The percentage and the significance of mtDNA mutation distribution on the regulatory elements of RCR in the dementia versus non-dementia groups
| Locus | nt span | Brains |
Lymphoblastoid cell lines |
||||
|---|---|---|---|---|---|---|---|
| Non-D | D | p | Non-D | D | p | ||
| 1–212 | 3.6% | 7.4% | 0.0009 | 1.8% | 4.7% | 0.06 | |
| CSBII | 299–315 | 4.0% | 6.3% | 0.013 | 3.5% | 7.6% | 0.03 |
| LSP | 392–445 | 0.4% | 1.4% | 0.01 | 1.0% | 1.3% | NS |
Non-demented values are combination of controls and DS; demented values are AD and DSAD. Non-D: Non-Demented, D: demented, NS: non-significant, nt: nucleotide.
The copy number of mtDNA in brains
Mutations that alter the regulatory elements of the mtDNA RCR might be expected to inhibit mtDNA replication (Fig. 2), resulting in a reduction in the ratio of mtDNA to nDNA copy number. This was examined using quantitative PCR (qPCR) of the mtDNA ND2 gene versus the nDNA 18S rRNA gene in the brains of controls, AD, DS, and DSAD subjects (Fig. 5A). Overall, the mtDNA to nDNA copy number declined in control brains with age. The average copy numbers of brain samples of 0 to 23 year-old subjects was 282 mtDNAs/nDNA, that for 41 to 65 year-old subjects was 290 mtDNAs/nDNA, and that for 66 to 88 year old subjects was 148 mtDNAs/nDNA) (ANOVA p = 0.01). Hence, the mtDNA copy number declined after age 65 years in parallel with the increased mtDNA mutation rate.
Fig. 5.
mtDNA amounts in the brain samples of control, DS, DSAD and AD patients represented as ND2 DNA amount normalized to nuclear 18S rDNA. A) The correlation of mtDNA amount with age in all four groups. The control brain mtDNA amount declined with aging (p = 0.02). The AD and DSAD mtDNA levels were below the control brains while DS mtDNA levels were above. B-D) The comparison of AD (B), DSAD (C) and DS (D) mtDNA amounts to age-matched controls (∗p < 0.05).
The mtDNA copy number of AD brains was significantly lower than age-matched controls for 66 to 88 year-old individuals, 110 mtDNA/nDNA versus 197 mtDNA/nDNA (p = 0.013) (Fig. 5B). The mtDNA copy number of DSAD brains was also significantly lower than age-matched controls, 145 mtDNA/nDNA in DSAD versus 290 mtDNA/nDNA (p = 0.016) (Fig. 5C). By contrast, there is a trend toward increased mtDNA copy number of DS brains relative to age-matched controls, 388 mtDNA/nDNA in DS versus 282 mtDNA/nDNA in controls (p = 0.1) (Fig. 5D). Therefore, the brain mtDNA copy number is strikingly reduced in association with dementia.
mtDNA strand dependent gene expression ratio
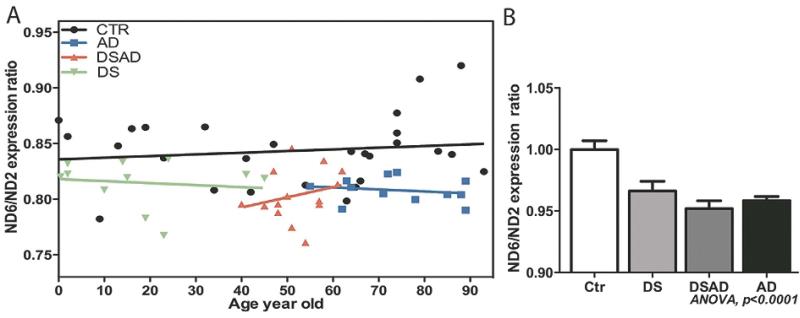
Mutations in the mtDNA control region, particularly in LSP and the associated TFAM binding sites might be expected to reduce mitochondrial L-strand transcription relative to H-strand transcription. To test this prediction, we compared the relative levels of the mtDNA ND6 L-strand mRNA to that of the mtDNA H-strand ND2 mRNA by qRT-PCR for control, AD, DSAD, and DS brain samples (Fig. 6A). The brain ND6 to ND2 mRNA ratio did not change significantly with age in control brains. However, the ND6 to ND2 transcript ratio was significantly lower in both AD and DSAD brains relative to control values. Moreover, the ND6 to ND2 levels were also reduced in the DS brains (ANOVA, p = 0.001) (Fig. 6A,B). Hence, mental retardation and dementia were both associated with reduced mtDNA L-strand transcript levels.
Fig. 6.
The ratio of mtDNA L-strand to H-strand transcripts in control, DS, DSAD, and AD brains. A) The correlation of mitochondrial L-strand/ H-strand gene expression ratio with age in all four groups. ND6 gene expression represents the L-stand transcription, and ND2 gene expression represents the H-Strand. B) The L-strand/ H-strand gene expression ratio was reduced in DS, DSAD, and AD brains relative to controls.
Relation between mtDNA alterations and AβPP processing in DS and DSAD
AD brains typically have heightened levels of Aβ40 and Aβ42 as well as increased levels of somatic mtDNA deletions and base substitution mutations [21]. In DS and DSAD brains, the levels of AβPP protein are consistently elevated relative to controls (p = 0.0004 t-test) (Fig. 7A), as expected for the triplication of the AβPP gene. We then compared the mtDNA alterations found with our published data on β-secretase activity and Aβ peptide levels in DS and DSAD brains.
Fig. 7.
The comparison of AβPP protein and processing products to mtDNA mutation frequency and mtDNA amount in DS, DSAD, and control groups. A) AβPP protein is overexpressed in DS and DSAD brains compared to controls (p = 0.0004, and the values are shown as mean plus SD). B) The correlation of β-secretase activity with mtDNA mutation frequency. The dashed lines show the linear regression of individual groups, whereas the solid black line shows the combination of all three groups. C) The correlation of β-secretase activity with mtDNA amount. The dashed lines show the linear regression of individual groups, whereas the solid black line shows the combination of all three groups. D) The correlation of insoluble Aβ40 levels with mtDNA amount. The only significant correlation was found with DSAD group (r = −0.65, p = 0.02, red dashed lines). E) The correlation of the percent of insoluble Aβ42 over total Aβ (i.e., Aβ40 and Aβ42) with mtDNA amount. The proportion of Aβ42 increased with mtDNA levels in the DSAD group (r = +0.72, p = 0.008, red dashed lines).
The β-secretase activity increased in parallel with the mtDNA somatic mutation frequency in the aggregate data from all DS, DSAD, and control brains (r = +0.55, p = 0.0005) (Fig. 7B). In addition, increased β-secretase activity was correlated with decreased mtDNA amount in DSAD brains (r = –0.57, p = 0.05) (Fig. 7C). An inverse relationship was seen between mtDNA amount and Aβ levels such that the lower the mtDNA copy number, the higher Aβ40 in DSAD brains (r = −0.61, p = 0.02) (Fig. 7D). The proportion of Aβ42 also increased relative to the mtDNA content (r = +0.72, p = 0.008). Hence at low mtDNA levels, Aβ40 predominates (Fig. 7E). Therefore, for DSAD the decline in mtDNA function is associated with increased Aβ production.
DISCUSSION
The current study supports the hypothesis that mitochondrial dysfunction is a major factor in the age-related dementia associated with AD and advanced age DS patients. The frequency of mtDNA RCR mutations was shown to increase with age in control brains, and to be significantly elevated in AD and DSAD brains, relative to age-matched control and DS brains. The increase mtDNA somatic mutation frequency observed in AD brains was also observed in serum DNA samples collected from the same individuals and the frequency of somatic mtDNA RCR mutations was significantly elevated in lymphoblastoid cell lines from AD and DSAD patients relative to lymphoblastoid cell lines derived from age-matched controls. Therefore, for late-onset AD and DSAD individuals, the elevated brain mtDNA RCR mutation frequency is the result of a systemic elevation in the mtDNA mutation frequency.
The elevated somatic mtDNA mutation frequency in AD and DSAD brains is associated with the accumulation of specific mtDNA RCR mutations, a number of which are rarely if ever found in normal brains. These mutations include the T414G mutation, which accumulates in skin fibroblasts but not the brain during normal aging. While we could not detect this variant in non-demented brains, it was present in 65% of AD brains and 57% of DSAD brains. Since this mutation alters an LSP TFAM binding site [66], it should reduce L-strand transcription. Since L-strand transcription is thought to create the primer for H-strand replication, the T414G mutation should also reduce mtDNA copy number [88]. The T414C mutation was also found in AD.
Other tissue-specific somatic mtDNA mutations were also identified in AD and DSAD brains. Two somatic mtDNA mutations, T408A and A189G, have been found to accumulate in skeletal muscle [89]. Strikingly, somatic mtDNA mutations at nt 408 were found in 31% of AD brains, but not in control, DS, or DSAD brains, blood DNAs or lymphoblasts. Variation in the 408T nt would be expected to alter the mtDNA H-strand replication origins and L-strand transcription [89]. Further evidence that the somatic mtDNA RCR mutations that accumulate in AD and DSAD are functionally relevant come from the statistically significant increase in somatic mtDNA mutations found in the CSBII, LSP, and the nt 1–212 region. The CSBII directs transcriptional termination for generation of the primer for H-strand replication [90], the LSP initiates L-strand transcription which generates the L-strand encoded ND6 mRNA and multiple tRNAs but also is terminated at CBSII to generate the H-strand replication primer [88], and the 1 to 212 nt region encompasses multiple H-strand replication origins [87].
Assuming that post conception somatic mtDNA mutations are central to the pathophysiology of AD, then a high mtDNA mutation load in the brain could result from either a chronically increased mtDNA mutation rate or from the appearance of a single deleterious mutation early in development with subsequent distribution and amplification in multiple organs. The first scenario would account for the diverse array of somatic mtDNA mutations found in most AD and DSAD brains and lymphoblasts, while the second scenario would account for finding the same mutation at high heteroplasmy levels in both brain and blood.
The somatic mtDNA RCR mutations that accumulate in AD and DSAD brains would be expected to affect mtDNA replication and L-strand transcription. This prediction is supported by the fact that both the mtDNA copy number and the mtDNA L-strand transcription levels are significantly depressed in both AD and DSAD brains, relative to age-matched control brains. A reduction in the mtDNA copy number would result in an overall decline in OXPHOS and a disproportionate decrease in the L-stand ND6 transcript would specifically inhibit respiratory complex I. These reductions would result in reduced mitochondrial energy output, increased mitochondrial ROS production, reduced mitochondrial membrane potential and thus altered calcium regulation, and an increased probability for the activation of the mtPTP, predisposing neurons to loss of processes and ultimately death.
The accumulation of Aβ peptides in AD brains is well-documentedand has also recently been documented in DSAD [86]. Since we have shown that mtDNA somatic RCR mutations are increased in AD and DSAD and that mtDNA copy number and L-transcripts are reduced in AD and DSAD, then the increase in mtDNA mutations and decline in mtDNA function should correlate with increased Aβ. This was confirmed by comparing the brain β-secretase activity and Aβ levels determined in DS, DSAD, and control brains [86] with mtDNA parameters, which we determined from the brains of the same subjects. This revealed a direct correlation between increased mtDNA somatic mutations and β-secretase activity in all samples examined. In addition, as mtDNA copy number declined in DSAD brains, the β-secretase and the Aβ levels increased. Hence, mtDNA alterations are directly related to Aβ production. Given that increased mtDNA somatic mutation levels are systemic, this implies that they precede the development of brain dementia and Aβ accumulation. It then follows that the accumulation of Aβ may be a consequence of mitochondrial dysfunction. This would be logical if the function of the Aβ monomer is to protect neurons from mitochondrially generated oxidative stress. However, after prolonged induction, the concentration of Aβ becomes sufficiently high to form oligomers which are the toxic form of Aβ [21]. The elevated Aβ might then drive Aβ into the mitochondrion where it inhibits OXPHOS, ultimately activating the mtPTP and destroying the energetically compromised neuron.
This mitochondrial model for age-related dementia provides an explanation for a number of previously unexplained observations on AD patients. Multiple studies have been published in which the mtDNA from peripheral blood cells such as platelets of AD patients have been transferred into cultured human cells that lack mtDNA (ρo cells) by cell fusion. These AD mtDNA cybrids have been shown to exhibit mitochondrial defects and have increased Aβ production [91–96]. If AD were specifically due to the accumulation of Aβ in the brains of patients, the AD patients’ blood cell mtDNAs should be no different from the mtDNAs of control subjects. However, our discovery is that the increased somatic mtDNA mutation rate is systemic and is reflected in the mtDNA of serum DNA and lymphoblastoid cells, which is consistent with mtDNA defects being transferred from AD blood cells to somatic cell cybrids.
In conclusion, we have shown that somatic mtDNA RCR mutations accumulate systemically in patients with AD and DSAD dementia, that this results in a decline in mtDNA copy number and mtDNA L-strand transcription, and that the decline in mitochondrial function correlates with increased β-secretase activity and the accumulation of Aβ peptide. Since inherited mtDNA variants such as the tRNAGln nt 4336 mutation has been repeatedly demonstrated to be associated with late-onset AD [52] and a variety of mtDNA mutations have been shown to cause a wide range neurological diseases including dementia [88], it follows logically that the underlying etiology of AD and DSAD is progressive mitochondrial decline. This mitochondrial decline can be initiated in a variety of ways: inherited germline mtDNA ancient or recent variants and mutations, genetic or environmental factors that increase the somatic mtDNA mutation rate, environmental factors that chronically inhibit mitochondrial function, or aberrant over-expression of Aβ leading to oligomerization and inhibition of OXPHOS.
ACKNOWLEDGMENTS
This work was supported by an ADRC Grant (AG-16573), with sub-projects awarded to I.T.L. and D.C.W. Additional funding came from in part by National Institutes of Health grants R01 NS211328, AG24373, DK73691, AG16573, and NS41850; CIRM Comprehensive Grant RC1-00353-1 awarded to D.C.W.
Footnotes
Authors’ disclosures available online (http://www.jalz.com/disclosures/view.php?id=427).
REFERENCES
- 1.Selkoe DJ. Alzheimer's disease. Missense on the membrane. Nature. 1995;375:734–735. doi: 10.1038/375734a0. [DOI] [PubMed] [Google Scholar]
- 2.Chartier-Harlin MC, Crawford F, Houlden H, Warren A, Hughes D, Fidani L, Goate A, Rossor M, Roques P, Hardy J, Mullan M. Early-onset Alzheimer's disease caused by mutations at codon 717 of the beta-amyloid precursor protein gene. Nature. 1991;353:844–846. doi: 10.1038/353844a0. [DOI] [PubMed] [Google Scholar]
- 3.Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, Mant R, Newton P, Rooke K, Roques P, Talbot C, Pericak-Vance M, Roses A, Williamson R, Rossor M, Owen M, Hardy J. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 4.Murrell J, Farlow M, Ghetti B, Benson MD. A mutation in the amyloid precursor protein associated with hereditary Alzheimer's disease. Science. 1991;254:97–99. doi: 10.1126/science.1925564. [DOI] [PubMed] [Google Scholar]
- 5.Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, Tsuda T, Mar L, Foncin J-F, Bruni AC, Montesi MP, Sorbi S, Rainero I, Pinessi L, Nee L, Chumakov I, Pollen D, Brookes A, Sanseau P, Polinsky RJ, Wasco W, Da Silva HAR, Haines JL, Pericak-Vance MA, Tanzi RE, Roses AD, Fraser PE, Rommens JM, St. George-Hyslop PH. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 6.Pericak-Vance MA, Bebout JL, Gaskell PC, Jr., Yamaoka LH, Hung W-Y, Alberts MJ, Walker AP, Bartlett RJ, Haynes CA, Welsh KA, Earl NL, Heyman A, Clark CM, Roses AD. Linkage studies in familial Alzheimer disease: evidence for chromosome 19 linkage. Am J Hum Genet. 1991;48:1034–1050. [PMC free article] [PubMed] [Google Scholar]
- 7.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 8.Jarvik L, Green-son H. About a peculiar disease of the cerebral cortex. By Alois Alzheimer, 1907. Alzheimer Dis Assoc Disord. 1987;1:3–8. [PubMed] [Google Scholar]
- 9.Fukumoto H, Rosene DL, Moss MB, Raju S, Hyman BT, Irizarry MC. Beta-secretase activity increases with aging in human, monkey, and mouse brain. Am J Pathol. 2004;164:719–725. doi: 10.1016/s0002-9440(10)63159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyler SJ, Dawbarn D, Wilcock GK, Allen SJ. alpha-and beta-secretase: profound changes in Alzheimer's disease. Biochem Biophys Res Commun. 2002;299:373–376. doi: 10.1016/s0006-291x(02)02635-9. [DOI] [PubMed] [Google Scholar]
- 11.Lott IT, Head E. Down syndrome and Alzheimer's disease: a link between development and aging. Ment Retard Dev Disabil Res Rev. 2001;7:172–178. doi: 10.1002/mrdd.1025. [DOI] [PubMed] [Google Scholar]
- 12.Capone GT. Down syndrome: advances in molecular biology and the neurosciences. J Dev Behav Pediatr. 2001;22:40–59. doi: 10.1097/00004703-200102000-00007. [DOI] [PubMed] [Google Scholar]
- 13.Head E, Lott IT. Down syndrome and beta-amyloid deposition. Curr Opin Neurol. 2004;17:95–100. doi: 10.1097/00019052-200404000-00003. [DOI] [PubMed] [Google Scholar]
- 14.Lott IT, Head E. Alzheimer disease and Down syndrome: factors in pathogenesis. Neurobiol Aging. 2005;26:383–389. doi: 10.1016/j.neurobiolaging.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Rumble B, Retallack R, Hilbich C, Simms G, Multhaup G, Martins R, Hockey A, Montgomery P, Beyreuther K, Masters CL. Amyloid A4 protein and its precursor in Down's syndrome and Alzheimer's disease. N Engl J Med. 1989;320:1446–1452. doi: 10.1056/NEJM198906013202203. [DOI] [PubMed] [Google Scholar]
- 16.Teller JK, Russo C, DeBusk LM, Angelini G, Zaccheo D, Dagna-Bricarelli F, Scartezzini P, Bertolini S, Mann DM, Tabaton M, Gambetti P. Presence of soluble amyloid beta-peptide precedes amyloid plaque formation in Down's syndrome. Nat Med. 1996;2:93–95. doi: 10.1038/nm0196-93. [DOI] [PubMed] [Google Scholar]
- 17.Hirayama A, Horikoshi Y, Maeda M, Ito M, Takashima S. Characteristic developmental expression of amyloid beta40, 42 and 43 in patients with Down syndrome. Brain Dev. 2003;25:180–185. doi: 10.1016/s0387-7604(02)00209-7. [DOI] [PubMed] [Google Scholar]
- 18.Selkoe DJ. Normal and abnormal biology of the beta-amyloid precursor protein. Annu Rev Neurosci. 1994;17:489–517. doi: 10.1146/annurev.ne.17.030194.002421. [DOI] [PubMed] [Google Scholar]
- 19.Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, Saykin AJ, Orro A, Lupoli S, Salvi E, Weiner M, Macciardi F. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer's disease. PLoS One. 2009;4:e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roses AD, Lutz MW, Amrine-Madsen H, Saunders AM, Crenshaw DG, Sundseth SS, Huentelman MJ, Welsh-Bohmer KA, Reiman EM. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer's disease. Pharmacogenomics J. 2009 doi: 10.1038/tpj.2009.69. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Ann Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aliev G, Gasimov E, Obrenovich ME, Fischbach K, Shenk JC, Smith MA, Perry G. Atherosclerotic lesions and mitochondria DNA deletions in brain microvessels: implication in the pathogenesis of Alzheimer's disease. Vasc Health Risk Manag. 2008;4:721–730. doi: 10.2147/vhrm.s2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdul HM, Sultana R, St Clair DK, Markesbery WR, Butterfield DA. Oxidative damage in brain from human mutant APP/PS-1 double knock-in mice as a function of age. Free Radic Biol Med. 2008;45:1420–1425. doi: 10.1016/j.freeradbiomed.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker WD, Jr., Filley CM, Parks JK. Cytochrome oxidase deficiency in Alzheimer's disease. Neurology. 1990;40:1302–1303. doi: 10.1212/wnl.40.8.1302. [DOI] [PubMed] [Google Scholar]
- 25.Parker WD, Jr., Parks J, Filley CM, Kleinschmidt-DeMasters BK. Electron transport chain defects in Alzheimer's disease brain. Neurology. 1994;44:1090–1096. doi: 10.1212/wnl.44.6.1090. [DOI] [PubMed] [Google Scholar]
- 26.Maurer I, Zierz S, Moller H. A selective defect of cytochrome c oxidase is present in brain of Alzheimer disease patients. Neurobiol Aging. 2000;21:455–462. doi: 10.1016/s0197-4580(00)00112-3. [DOI] [PubMed] [Google Scholar]
- 27.Kish SJ, Bergeron C, Rajput A, Dozic S, Mastrogiacomo F, Chang LJ, Wilson JM, DiStefano LM, Nobrega JN. Brain cytochrome oxidase in Alzheimer's disease. J Neurochem. 1992;59:776–779. doi: 10.1111/j.1471-4159.1992.tb09439.x. [DOI] [PubMed] [Google Scholar]
- 28.Mutisya EM, Bowling AC, Beal MF. Cortical cytochrome oxidase activity is reduced in Alzheimer's disease. J Neurochem. 1994;63:2179–2184. doi: 10.1046/j.1471-4159.1994.63062179.x. [DOI] [PubMed] [Google Scholar]
- 29.Reddy PH, Beal MF. Are mitochondria critical in the pathogenesis of Alzheimer's disease? Brain Res Brain Res Rev. 2005;49:618–632. doi: 10.1016/j.brainresrev.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan PG, Brown MR. Mitochondrial aging and dys-function in Alzheimer's disease. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29:407–410. doi: 10.1016/j.pnpbp.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Teng FY, Tang BL. Widespread gamma-secretase activity in the cell, but do we need it at the mitochondria? Biochem Biophys Res Commun. 2005;328:1–5. doi: 10.1016/j.bbrc.2004.12.131. [DOI] [PubMed] [Google Scholar]
- 32.Bubber P, Haroutunian V, Fisch G, Blass JP, Gibson GE. Mitochondrial abnormalities in Alzheimer brain: mechanistic implications. Ann Neurol. 2005;57:695–703. doi: 10.1002/ana.20474. [DOI] [PubMed] [Google Scholar]
- 33.Caspersen C, Wang N, Yao J, Sosunov A, Chen X, Lustbader JW, Xu HW, Stern D, McKhann G, Yan SD. Mitochondrial Abeta: a potential focal point for neuronal metabolic dysfunction in Alzheimer's disease. FASEB J. 2005;19:2040–2041. doi: 10.1096/fj.05-3735fje. [DOI] [PubMed] [Google Scholar]
- 34.Canevari L, Clark JB, Bates TE. beta-Amyloid fragment 25-35 selectively decreases complex IV activity in isolated mitochondria. FEBS Lett. 1999;457:131–134. doi: 10.1016/s0014-5793(99)01028-5. [DOI] [PubMed] [Google Scholar]
- 35.Lustbader JW, Cirilli M, Lin C, Xu HW, Takuma K, Wang N, Caspersen C, Chen X, Pollak S, Chaney M, Trinchese F, Liu S, Gunn-Moore F, Lue LF, Walker DG, Kuppusamy P, Zewier ZL, Arancio O, Stern D, Yan SS, Wu H. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease. Science. 2004;304:448–452. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- 36.Takuma K, Yao J, Huang J, Xu H, Chen X, Luddy J, Trillat AC, Stern DM, Arancio O, Yan SS. ABAD enhances Abeta-induced cell stress via mitochondrial dysfunction. FASEB J. 2005;19:597–598. doi: 10.1096/fj.04-2582fje. [DOI] [PubMed] [Google Scholar]
- 37.Yan SD, Stern DM. Mitochondrial dysfunction and Alzheimer's disease: role of amyloid-beta peptide alcohol dehydrogenase (ABAD). Int J Exp Pathol. 2005;86:161–171. doi: 10.1111/j.0959-9673.2005.00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibson GE, Karuppagounder SS, Shi Q. Oxidant-induced changes in mitochondria and calcium dynamics in the pathophysiology of Alzheimer's disease. Ann N Y Acad Sci. 2008;1147:221–232. doi: 10.1196/annals.1427.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang S, ran Ma T, Miranda RD, Balestra ME, Mahley RW, Huang Y. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc Natl Acad Sci U S A. 2005;102:18694–18699. doi: 10.1073/pnas.0508254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer's disease. Proc Natl Acad Sci U S A. 2006;103:5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhein V, Song X, Wiesner A, Ittner LM, Baysang G, Meier F, Ozmen L, Bluethmann H, Drose S, Brandt U, Savaskan E, Czech C, Gotz J, Eckert A. Amyloid-beta and tau synergistically impair the oxidative phosphorylation system in triple transgenic Alzheimer's disease mice. Proc Natl Acad Sci U S A. 2009;106:20057–20062. doi: 10.1073/pnas.0905529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao J, Irwin RW, Zhao L, Nilsen J, Hamilton RT, Brinton RD. Mitochondrial bioenergetic deficit precedes Alzheimer's pathology in female mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 2009;106:14670–14675. doi: 10.1073/pnas.0903563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manczak M, Jung Y, Park BS, Partovi D, Reddy PH. Time-course of mitochondrial gene expressions in mice brains: implications for mitochondrial dysfunction, oxidative damage, and cytochrome c in aging. J Neurochem. 2005;92:494–504. doi: 10.1111/j.1471-4159.2004.02884.x. [DOI] [PubMed] [Google Scholar]
- 44.Pellegrini M, Asin-Cayuela J, Erdjument-Bromage H, Tempst P, Larsson NG, Gustafsson CM. MTERF2 is a nucleoid component in mammalian mitochondria. Biochim Biophys Acta. 2009;1787:296–302. doi: 10.1016/j.bbabio.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 45.Wallace DC, Ye JH, Neckelmann SN, Singh G, Webster KA, Greenberg BD. Sequence analysis of cDNAs for the human and bovine ATP synthase b-subunit: mitochondrial DNA genes sustain seventeen times more mutations. Current Genetics. 1987;12:81–90. doi: 10.1007/BF00434661. [DOI] [PubMed] [Google Scholar]
- 46.Neckelmann N, Li K, Wade RP, Shuster R, Wallace DC. cDNA sequence of a human skeletal muscle ADP/ATP translocator: lack of a leader peptide, divergence from a fibroblast translocator cDNA, and coevolution with mitochondrial DNA genes. Proc Natl Acad Sci U S A. 1987;84:7580–7584. doi: 10.1073/pnas.84.21.7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Linnane AW, Marzuki S, Ozawa T, Tanaka M. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet. 1989;1:642–645. doi: 10.1016/s0140-6736(89)92145-4. [DOI] [PubMed] [Google Scholar]
- 48.Wei YH, Wu SB, Ma YS, Lee HC. Respiratory function decline and DNA mutation in mitochondria, oxidative stress and altered gene expression during aging. Chang Gung Med J. 2009;32:113–132. [PubMed] [Google Scholar]
- 49.Duara R, Lopez-Alberola RF, Barker WW, Loewenstein DA, Zatinsky M, Eisdorfer CE, Weinberg GB. A comparison of familial and sporadic Alzheimer's disease. Neurology. 1993;43:1377–1384. doi: 10.1212/wnl.43.7.1377. [DOI] [PubMed] [Google Scholar]
- 50.Edland SD, Silverman JM, Peskind ER, Tsuang D, Wijsman E, Morris JC. Increased risk of dementia in mothers of Alzheimer's disease cases: evidence for maternal inheritance. Neurology. 1996;47:254–256. doi: 10.1212/wnl.47.1.254. [DOI] [PubMed] [Google Scholar]
- 51.Mosconi L, Rinne JO, Tsui WH, Berti V, Li Y, Wang H, Murray J, Scheinin N, Nagren K, Williams S, Glodzik L, De Santi S, Vallabhajosula S, de Leon MJ. Increased fibrillar amyloid-{beta} burden in normal individuals with a family history of late-onset Alzheimer's. Proc Natl Acad Sci U S A. 2010;107:5949–5954. doi: 10.1073/pnas.0914141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shoffner JM, Brown MD, Torroni A, Lott MT, Cabell MR, Mirra SS, Beal MF, Yang C, Gearing M, Salvo R, Watts RL, Juncos JL, Hansen LA, Crain BJ, Fayad M, Reckord CL, Wallace DC. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17:171–184. doi: 10.1006/geno.1993.1299. [DOI] [PubMed] [Google Scholar]
- 53.Hutchin T, Cortopassi G. A mitochondrial DNA clone is associated with increased risk for Alzheimer disease. Proc Natl Acad Sci U S A. 1995;92:6892–6895. doi: 10.1073/pnas.92.15.6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cortopassi GA, Hutchin TP. Germline inheritance of a rare mtDNA variant leads to greatly increased risk for Alzheimer's disease. Am J Hum Genet. 1994;55(Suppl):A149. [Google Scholar]
- 55.Egensperger R, Kosel S, Schnopp NM, Mehraein P, Graeber MB. Association of the mitochondrial tRNA(A4336G) mutation with Alzheimer's and Parkinson's diseases. Neuropathol Appl Neurobiol. 1997;23:315–321. [PubMed] [Google Scholar]
- 56.Mayr-Wohlfart U, Rodel G, Henneberg A. Mitochondrial tRNA(Gln) and tRNA(Thr) gene variants in Parkinson's disease. Eur J Med Res. 1997;2:111–113. [PubMed] [Google Scholar]
- 57.Tanno Y, Okuizumi K, Tsuji S. mtDNA polymorphisms in Japanese sporadic Alzheimer's disease. Neurobiol Aging. 1998;19:S47–51. doi: 10.1016/s0197-4580(98)00028-1. [DOI] [PubMed] [Google Scholar]
- 58.Hutchin TP, Heath PR, Pearson RC, Sinclair AJ. Mitochondrial DNA mutations in Alzheimer's disease. Biochem Biophys Res Commun. 1997;241:221–225. doi: 10.1006/bbrc.1997.7793. [DOI] [PubMed] [Google Scholar]
- 59.Chinnery PF, Thorburn DR, Samuels DC, White SL, Dahl HM, Turnbull DM, Lightowlers RN, Howell N. The inheritance of mitochondrial DNA heteroplasmy: random drift, selection or both? Trends Genet. 2000;16:500–505. doi: 10.1016/s0168-9525(00)02120-x. [DOI] [PubMed] [Google Scholar]
- 60.Chinnery PF, Taylor GA, Howell N, Andrews RM, Morris CM, Taylor RW, McKeith IG, Perry RH, Edwardson JA, Turn-bull DM. Mitochondrial DNA haplogroups and susceptibility to AD and dementia with Lewy bodies. Neurology. 2000;55:302–304. doi: 10.1212/wnl.55.2.302. [DOI] [PubMed] [Google Scholar]
- 61.Garcia-Lozano JR, Aguilera I, Bautista J, Nunez-Roldan A. A new mitochondrial DNA mutation in the tRNA leucine 1 gene (C3275A) in a patient with Leber's hereditary optic neuropathy. Hum Mut. 2000;15:120–121. doi: 10.1002/(SICI)1098-1004(200001)15:1<120::AID-HUMU33>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 62.Chagnon P, Gee M, Filion M, Robitaille Y, Belouchi M, Gauvreau D. Phylogenetic analysis of the mitochondrial genome indicates significant differences between patients with Alzheimer disease and controls in a French-Canadian founder population. Am J Med Genet. 1999;85:20–30. doi: 10.1002/(sici)1096-8628(19990702)85:1<20::aid-ajmg6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 63.Carrieri G, Bonafe M, De Luca M, Rose G, Varcasia O, Bruni A, Maletta R, Nacmias B, Sorbi S, Corsonello F, Feraco E, Andreev KF, Yashin AI, Franceschi C, De Benedictis G. Mitochondrial DNA haplogroups and APOE4 allele are non-independent variables in sporadic Alzheimer's disease. Hum Genet. 2001;108:194–198. doi: 10.1007/s004390100463. [DOI] [PubMed] [Google Scholar]
- 64.van der Walt JM, Dementieva YA, Martin ER, Scott WK, Nicodemus KK, Kroner CC, Welsh-Bohmer KA, Saunders AM, Roses AD, Small GW, Schmechel DE, Murali Doraiswamy P, Gilbert JR, Haines JL, Vance JM, Pericak-Vance MA. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett. 2004;365:28–32. doi: 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 65.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Mc-Kee AC, Beal MF, Graham BH, Wallace DC. Marked changes in mitochondrial DNA deletion levels in Alzheimer brains. Genomics. 1994;23:471–476. doi: 10.1006/geno.1994.1525. [DOI] [PubMed] [Google Scholar]
- 66.Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science. 1999;286:774–779. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- 67.Coskun PE, Beal MF, Wallace DC. Alzheimer's brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci U S A. 2004;101:10726–10731. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murdock DG, Christacos NC, Wallace DC. The age-related accumulation of a mitochondrial DNA control region mutation in muscle, but not brain, detected by a sensitive PNA-directed PCR clamping based method. Nucleic Acids Res. 2000;28:4350–4355. doi: 10.1093/nar/28.21.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gardiner K, Costa AC. The proteins of human chromosome 21. Am J Med Genet C Semin Med Genet. 2006;142C:196–205. doi: 10.1002/ajmg.c.30098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anneren KG, Epstein CJ. Lipid peroxidation and super-oxide dismutase-1 and glutathione peroxidase activities in trisomy 16 fetal mice and human trisomy 21 fibroblasts. Pediatr Res. 1987;21:88–92. doi: 10.1203/00006450-198701000-00019. [DOI] [PubMed] [Google Scholar]
- 71.Brooksbank BW, Balazs R. Superoxide dismutase, glutathione peroxidase and lipoperoxidation in Down's syndrome fetal brain. Brain Res. 1984;318:37–44. doi: 10.1016/0165-3806(84)90060-9. [DOI] [PubMed] [Google Scholar]
- 72.Bras A, Monteiro C, Rueff J. Oxidative stress in trisomy 21. A possible role in cataractogenesis. Ophthalmic Paediatr Genet. 1989;10:271–277. doi: 10.3109/13816818909009882. [DOI] [PubMed] [Google Scholar]
- 73.Jovanovic SV, Clements D, MacLeod K. Biomarkers of oxidative stress are significantly elevated in Down syndrome. Free Radic Biol Med. 1998;25:1044–1048. doi: 10.1016/s0891-5849(98)00137-3. [DOI] [PubMed] [Google Scholar]
- 74.Busciglio J, Yankner BA. Apoptosis and increased generation of reactive oxygen species in Down's syndrome neurons in vitro. Nature. 1995;378:776–779. doi: 10.1038/378776a0. [DOI] [PubMed] [Google Scholar]
- 75.Castellani R, Hirai K, Aliev G, Drew KL, Nunomura A, Takeda A, Cash AD, Obrenovich ME, Perry G, Smith MA. Role of mitochondrial dysfunction in Alzheimer's disease. J Neurosci Res. 2002;70:357–360. doi: 10.1002/jnr.10389. [DOI] [PubMed] [Google Scholar]
- 76.Nagy Z, Esiri MM, LeGris M, Matthews PM. Mitochondrial enzyme expression in the hippocampus in relation to Alzheimer-type pathology. Acta Neuropathol (Berl) 1999;97:346–354. doi: 10.1007/s004010050997. [DOI] [PubMed] [Google Scholar]
- 77.Kim SH, Vlkolinsky R, Cairns N, Fountoulakis M, Lubec G. The reduction of NADH ubiquinone oxidoreductase 24- and 75-kDa subunits in brains of patients with Down syndrome and Alzheimer's disease. Life Sci. 2001;68:2741–2750. doi: 10.1016/s0024-3205(01)01074-8. [DOI] [PubMed] [Google Scholar]
- 78.Kim SH, Vlkolinsky R, Cairns N, Lubec G. Decreased levels of complex III core protein 1 and complex V beta chain in brains from patients with Alzheimer's disease and Down syndrome. Cell Mol Life Sci. 2000;57:1810–1816. doi: 10.1007/PL00000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee SH, Lee S, Jun HS, Jeong HJ, Cha WT, Cho YS, Kim JH, Ku SY, Cha KY. Expression of the mitochondrial ATPase6 gene and Tfam in Down syndrome. Mol Cell. 2003;15:181–185. [PubMed] [Google Scholar]
- 80.Bozner P, Wilson GL, Druzhyna NM, Bryant-Thomas TK, LeDoux SP, Pappolla MA. Deficiency of chaperonin 60 in Down's syndrome. J Alzheimers Dis. 2002;4:479–486. doi: 10.3233/jad-2002-4604. [DOI] [PubMed] [Google Scholar]
- 81.Martin J, Horwich AL, Hartl FU. Prevention of protein denaturation under heat stress by the chaperonin Hsp60. Science. 1992;258:995–998. doi: 10.1126/science.1359644. [DOI] [PubMed] [Google Scholar]
- 82.Bajo M, Fruehauf J, Kim SH, Fountoulakis M, Lubec G. Proteomic evaluation of intermediary metabolism enzyme proteins in fetal Down's syndrome cerebral cortex. Proteomics. 2002;2:1539–1546. doi: 10.1002/1615-9861(200211)2:11<1539::AID-PROT1539>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 83.Wallace DC. Why do we have a maternally inherited mitochondrial DNA? Insights from Evolutionary Medicine. Ann Rev Biochem. 2007 Jul;76 doi: 10.1146/annurev.biochem.76.081205.150955. 2007. [DOI] [PubMed] [Google Scholar]
- 84.Schutte DL. Alzheimer disease and genetics: anticipating the questions. Am J Nurs. 2006;106:40–47. doi: 10.1097/00000446-200612000-00018. quiz 47-48. [DOI] [PubMed] [Google Scholar]
- 85.Murdock DG, Wallace DC. PNA-mediated PCR clamping. Applications and methods. Meth Mol Biol. 2002;208:145–164. doi: 10.1385/1-59259-290-2:145. [DOI] [PubMed] [Google Scholar]
- 86.Nistor M, Don M, Parekh M, Sarsoza F, Goodus M, Lopez GE, Kawas C, Leverenz J, Doran E, Lott IT, Hill M, Head E. Alpha- and beta-secretase activity as a function of age and beta-amyloid in Down syndrome and normal brain. Neurobiol Aging. 2007;28:1493–1506. doi: 10.1016/j.neurobiolaging.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci U S A. 1985;82:351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wallace DC, Lott MT, Procaccio V. Mitochondrial Genes in Degenerative Diseases, Cancer and Aging. In: Rimoin DL, Connor JM, Pyeritz RE, Korf BR, editors. Emery and Rimoin's Principles and Practice of Medical Genetics. 5th Edition Churchill Livingstone Elsevier; Philadelphia, PA: 2007. pp. 194–298. [Google Scholar]
- 89.Wang Y, Michikawa Y, Mallidis C, Bai Y, Woodhouse L, Yarasheski KE, Miller CA, Askanas V, Engel WK, Bhasin S, Attardi G. Muscle-specific mutations accumulate with aging in critical human mtDNA control sites for replication. Proc Natl Acad Sci U S A. 2001;98:4022–4027. doi: 10.1073/pnas.061013598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pham XH, Farge G, Shi Y, Gaspari M, Gustafsson CM, Falkenberg M. Conserved sequence box II directs transcription termination and primer formation in mitochondria. J Biol Chem. 2006;281:24647–24652. doi: 10.1074/jbc.M602429200. [DOI] [PubMed] [Google Scholar]
- 91.Ghosh SS, Swerdlow RH, Miller SW, Sheeman B, Parker WD, Jr., Davis RE. Use of cytoplasmic hybrid cell lines for elucidating the role of mitochondrial dysfunction in Alzheimer's disease and Parkinson's disease. Ann N Y Acad Sci. 1999;893:176–191. doi: 10.1111/j.1749-6632.1999.tb07825.x. [DOI] [PubMed] [Google Scholar]
- 92.Sherer TB, Trimmer PA, Parks JK, Tuttle JB. Mitochondrial DNA-depleted neuroblastoma (rho-zero) cells exhibit altered calcium signaling. Biochim Biophys Acta. 2000;1496:341–355. doi: 10.1016/s0167-4889(00)00027-6. [DOI] [PubMed] [Google Scholar]
- 93.Bijur GN, Davis RE, Jope RS. Rapid activation of heat shock factor-1 DNA binding by H2O2 and modulation by glutathione in human neuroblastoma and Alzheimer's disease cybrid cells. Brain Res Mol Brain Res. 1999;71:69–77. doi: 10.1016/s0169-328x(99)00168-0. [DOI] [PubMed] [Google Scholar]
- 94.Sheehan JP, Swerdlow RH, Miller SW, Davis RE, Parks JK, Parker WD, Tuttle JB. Calcium homeostasis and reactive oxygen species production in cells transformed by mitochondria from individuals with sporadic Alzheimer's disease. J Neurosci. 1997;17:4612–4622. doi: 10.1523/JNEUROSCI.17-12-04612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Trimmer PA, Borland MK. Differentiated Alzheimer's disease transmitochondrial cybrid cell lines exhibit reduced organelle movement. Antioxid Redox Signal. 2005;7:1101–1109. doi: 10.1089/ars.2005.7.1101. [DOI] [PubMed] [Google Scholar]
- 96.Thiffault C, Bennett JP., Jr. Cyclical mitochondrial deltapsiM fluctuations linked to electron transport, F0F1 ATP-synthase and mitochondrial Na+/Ca+2 exchange are reduced in Alzheimer's disease cybrids. Mitochondrion. 2005;5:109–119. doi: 10.1016/j.mito.2004.12.002. [DOI] [PubMed] [Google Scholar]