Abstract
Monitoring gene expression is crucial for studying the responses of gene therapy and clarifying the function of certain genes in various environments. Molecular imaging is a powerful tool for noninvasive visualization of gene expression. This chapter will summarize the current status of fluorescence and bioluminescence imaging (BLI) of gene expression in live cells and tissues, and the emphasis will be mainly on the early studies that pioneered the field. First, we will describe fluorescence imaging of gene expression with a wide variety of fluorescent proteins. Next, we will discuss the strategies for BLI of gene expression. Besides incorporating the reporter gene into the host DNA, mRNA-based BLI of gene expression will also be briefly mentioned. Lastly, the construction of dual and triple fusion reporter genes will be presented. Since no single imaging modality is perfect and sufficient to obtain all of the necessary information for a given question, a combination of multiple molecular imaging modalities can offer synergistic advantages over any modality alone. Noninvasive optical imaging of gene expression has revolutionized biomedical research and the progress made over the last decade should allow molecular imaging to play a major role in the field of gene therapy. For basic and preclinical research, optical imaging is certainly indispensable for imaging gene expression. However, for clinical imaging of gene expression, positron emission tomography (PET) holds the greatest promise.
INTRODUCTION
With the ability to engineer genes, create “knock-in” and “knock-out” models of human diseases, it has become clear that gene therapy can play an important role in disease management. Over the last four decades, gene therapy has moved from preclinical to clinical studies for many diseases ranging from monogenic recessive disorders (e.g. hemophilia) to more complex diseases such as cancer, cardiovascular disorders, and human immunodeficiency virus (HIV) infection (Serganova et al. 2008; Gillet et al. 2009). To choose the appropriate gene therapy strategy and optimize the therapeutic efficacy, an effective monitoring system is needed. One of the most economical and practical approach is to adopt certain molecular imaging techniques for monitoring gene expression in vivo.
In recent years, noninvasive molecular imaging has emerged as a powerful tool for monitoring cellular and molecular events in vivo (Cai and Chen 2007; Cai et al. 2008a; Cai et al. 2008b). A variety of imaging technologies are being investigated as tools for evaluating gene therapy efficiency and studying gene expression in living subjects. Noninvasive, longitudinal, and quantitative imaging of gene expression can not only help in human gene therapy trials but also facilitate preclinical experimental studies in animal models. Radionuclide-based imaging techniques, i.e. single-photon emission computed tomography (SPECT) and positron emission tomography (PET), have the highest clinical potential and offer many advantages for noninvasive imaging of gene expression.
Besides imaging gene vectors to indirectly visualize the gene expression efficiency, PET and SPECT can play a significant role in imaging gene expression using diverse reporter genes and reporter probes (Kang and Chung 2008). If transcription of a reporter gene is induced, translation of the reporter gene mRNA will lead to a protein product which can interact with the imaging reporter probe (administered in trace amount for PET/SPECT applications, hence the term “tracer”). This interaction may be based on the intracellular enzymatic conversion of the reporter probe with retention of the metabolite(s), or a receptor-ligand based interaction.
Examples of intracellular reporters include the herpes simplex virus type 1 thymidine kinase (HSV1-tk) and its mutant gene (HSV1-sr39tk) (Gambhir et al. 1999; Gambhir et al. 2000; Najjar et al. 2009). Substrates that have been studied to date as PET reporter probes for HSV1-tk can be classified into two main categories: pyrimidine nucleoside derivatives (e.g. 2'-Fluoro-2'-deoxy-5'-[124I]iodo-1β-D-arabinofuranosyluracil; 124I-FIAU) and acycloguanosine derivatives (e.g. 9-[4-[18F]fluoro-3-(hydroxymethyl)butyl]guanine; 18F-FHBG). A few examples of reporters on the cell surface include the dopamine 2 receptor (D2R), receptors for human type 2 somatostatin receptor (hSSTr2), and the sodium iodide symporter (NIS).
Although magnetic resonance imaging (MRI) has a relatively low sensitivity, it can also be used to image gene expression in vivo (Gilad et al. 2007; Gilad et al. 2008). Due to the fact that the generation of images must be related to certain chemical shifts, MRI can be applied for the imaging of iron-related gene products (e.g. transferrin), some special enzymes (e.g. β-galactosidase), and chemical exchange saturation transfer (CEST)-related products (e.g. polylysine-containing proteins) (Gilad et al. 2008).
Despite the advantages and greater clinical potential of PET/SPECT and MRI, optical imaging techniques (e.g. fluorescence and bioluminescence) also have their own benefits (e.g. more economical, easier to handle, no radioisotopes needed, and quite sensitive in certain scenarios). Therefore, optical imaging has been widely used for monitoring gene expression in live cells and tissues. In this chapter, we will summarize the current state-of-the-art in optical imaging of gene expression.
FLUORESCENCE IMAGING OF GENE EXPRESSION
For fluorescence imaging of gene expression, green fluorescent protein (GFP, emission peak at 509 nm) and its variants are the mainstay and have been widely used over the last decade (Figure 1). Because light penetrates tissues better at longer wavelength (red or near-infrared), one of the major research areas in fluorescent proteins over the last decade has been to engineer them so that they can emit in the red or near-infrared.
Figure 1.
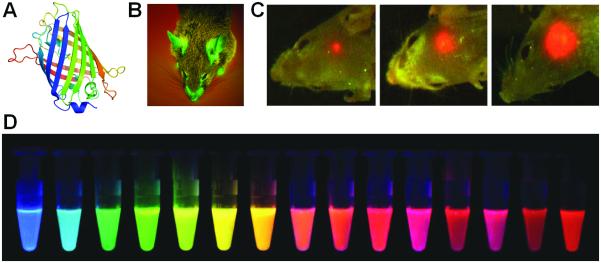
Fluorescence imaging of gene expression. A. The structure of GFP. B. A transgenic GFP mouse. C. Real-time whole-body imaging of a GFP-expressing human glioma growing in the brain of a nude mouse at 1 week (left), 3 weeks (center), and 5 weeks (right) after surgical orthotopic implantation. D. Monomeric and tandem dimeric fluorescent proteins derived from Aequorea GFP or Discosoma RFP, expressed in bacteria, and purified. Adapted with permission from (Shaner et al. 2004; Hoffman and Yang 2006).
GFP-Based Imaging
From the early 1990s, GFP and its variants have become invaluable markers for monitoring protein localization and gene expression in vivo (Heim and Tsien 1996; Jakobs et al. 2000). One early example of imaging gene expression with GFP was the demonstration of vascular endothelial growth factor (VEGF) promoter activity (Fukumura et al. 1998). The VEGF promoter was linked to the GFP gene and expression of GFP was imaged by intravital microscopy. For such intravital studies where limited tissue penetration capability of the reporter protein is needed, GFP was found to be readily detectable, confined to the cell, and stable (with a half-life of up to 24 hours in cells). Although GFP has poor tissue penetration capability, its expression can be visualized noninvasively in intact animals (Figure 1) (Yang et al. 2000; Hoffman and Yang 2006). GFP has also been frequently used in stem cell-related research to monitor the expression of certain genes (Meyer et al. 2000; Niyibizi et al. 2004). Modifications of GFP to increase its signal intensity and thermostability, as well as to alter its emission spectrum, have facilitated the use of GFP in a variety of gene transfer studies (Welsh and Kay 1997).
GFP is highly useful for imaging tumor-related gene expression. Mouse models of metastatic cancer were developed with genetically fluorescent tumors, where the GFP gene was cloned into cancer cell lines and selected for stable GFP expression, which can be imaged in fresh tissue, in situ, as well as externally (Hoffman 2001). With such a model system, tumor location and metastasis was detected and visualized in situ in host organs down to the single-cell level. GFP-transfected tumor cells enabled a fundamental advance in the visualization of tumor growth and metastasis in real time in vivo, and tumor/metastasis growth and their inhibition by certain drugs can be imaged and quantified for rapid anti-cancer drug screening. Subsequently, research on imaging tumor-related gene expression has flourished, with examples such as a reporter gene system consisted of enhanced GFP (EGFP) and wild-type HSV1-tk for both fluorescence and PET imaging (Luker et al. 2002).
GFP can serve as a powerful tool for imaging other diseases such as HIV infection. For example, the function of the HIV-1 Tat protein (capable of traversing cell membranes) has been elucidated by fluorescence imaging (Ferrari et al. 2003). With exogenously added Tat-GFP fusion protein to live HeLa and CHO cells, it was found that the internalization process of full-length Tat, as well as heterologous proteins fused to the transduction domain of Tat, exploits a caveolar-mediated pathway. With fluorescence imaging, the dynamic movement of individual GFP-tagged, Tat-filled caveolae toward the nucleus was observed directly.
RFP-Based Imaging
In 1999, a new red fluorescent protein (RFP) was isolated from tropical corals which was termed “dsRed” (Matz et al. 1999). With emission maxima at 509 and 583 nm respectively, EGFP and DsRed are well suited for virtually crossover-free, dual-color imaging upon simultaneous excitation. Mixed populations of Escherichia coli expressing either EGFP or DsRed were imaged by one-photon and two-photon microscopy. Both excitation modes were found to be suitable for imaging cells expressing either of the fluorescent proteins.
The predominant use of dsRed is for multicolor imaging in plants, together with the GFP variants, since its red-shifted excitation/emission spectra can avoid damaging the cells and tissues by the excitation light (Heikal et al. 2000; Dietrich and Maiss 2002). Dual gene expression has also been imaged in animal models with GFP and dsRed-2, after transduction with a dual-promoter lentiviral vector (Chen et al. 2004). Stem cell research is also a hot-spot of RFP-based gene expression imaging. For example, the whole-body biodistribution and persistence of multipotent adult progenitor cells, transfected with dsRed-2, was visualized in vivo and in tissue sections (Tolar et al. 2005). Another important field for RFP-based research is cancer cell imaging. For example, fluorescent pancreatic cancer cells expressing a high level of the dsRed-2 gene were used to establish an orthotopic, metastatic pancreatic cancer model (Zhou et al. 2008). The high-level expression of dsRed-2 enabled noninvasive imaging of distant micrometastases in their target organs, even in deep tissue such as the lung.
Wild type dsRed has several drawbacks including inefficient folding of the protein, extremely slow maturation of the chromophore, and tetramerization even in dilute solutions. Therefore, stepwise evolution of DsRed to a dimer and then to a true monomer, designated as mRFP1, was carried out (Campbell et al. 2002). The mRFP1 exhibited similar brightness in living cells as dsRed. In addition, the excitation and emission peaks of mRFP1, 584 and 607 nm, are approximately 25 nm red-shifted from DsRed, which can confer greater tissue penetration and spectral separation from autofluorescence and other fluorescent proteins. Subsequently, a series of monomeric fluorescent proteins have been reported which further expanded the toolbox for fluorescence imaging of gene expression (Figure 1) (Shaner et al. 2004; Wang et al. 2004).
Infrared Fluorescent Protein (IFP)
Generally speaking, GFP is still the main choice for fluorescence imaging of gene expression, due to its ease to be fused with other genes and enhanced fluorescence characteristics than RFP. However, for optimal performance in in vivo imaging, near-infrared fluorescent (NIRF, 700-900 nm) proteins will be superior. NIRF signal can pass through deeper tissues and the background fluorescence in this area is also very low.
Recently, it was reported that a bacteriophytochrome from Deinococcus radiodurans, incorporating biliverdin as the chromophore, can be engineered into monomeric, infrared-fluorescent proteins (IFPs) with excitation and emission maxima of 684 and 708 nm, respectively (Shu et al. 2009). The IFPs express well in mammalian cells and mice and spontaneously incorporate biliverdin, which is ubiquitous as the initial intermediate in heme catabolism but has negligible fluorescence by itself. These IFPs provided the base for further engineering of IFPs with better performance characteristics, which is expected to find broad use in future biomedical research, in particular those related to gene expression imaging.
β-Galactosidase-Based Imaging
Besides the fusion of fluorescent proteins with target gene products, several other strategies can also be adopted for fluorescence imaging of gene expression. The use of fluorogenic substrates for certain gene translation products (e.g. enzymes and proteins) is one choice. For example, the bacterial lacZ gene, which encodes for β-galactosidase (β-gal), is a common reporter gene used in transgenic mice (Watson et al. 2008). However, the absence of fluorogenic substrates usable in live animals greatly hampered the applications of this reporter gene. In 2007, a far-red fluorescent substrate, 9H-(1,3-dichloro-9,9-dimethylacridin-2-one-7-yl) β-D-galactopyranoside, was developed for imaging β-gal expression (Josserand et al. 2007). With β-gal as a reporter of tumor growth, as low as 103 β-gal-expressing tumor cells located under the skin were detectable with this substrate.
Aside from the various strategies to image gene expression in live cells and tissues by fluorescence, as mentioned above, there have also been significant advances in the imaging instrumentation. To give the fluorescence readout in a more quantitative manner, fluorescence diffuse tomography (FDT) has been developed recently for small animal imaging (Turchin et al. 2008). The animal is scanned in the trans-illuminative configuration by a single source and detector pair, and a reconstruction algorithm was developed to estimate the fluorophore (e.g. stably transfected fluorescent protein) distribution. Besides FDT, other techniques such as fluorescence-mediated tomography (FMT) can also provide important insights for fluorescence imaging in vivo (Ntziachristos et al. 2003).
With the wide variety of tools for fluorescence imaging of gene expression, multiplexing is highly desirable and certainly possible. Using an imaging system capable of “spectral unmixing”, where fluorophores with different emission spectra can be readily separated and analyzed simultaneously in the same sample (Cai et al. 2006; Cai and Chen 2008), multiple genes fused to different fluorescent proteins can be imaged at the same time which can shed new light onto the biology/mechanism of various diseases. With newly developed fluorescent proteins that emit in longer wavelength and the constantly evolving imaging systems, which can be useful in the clinic in certain sites of the human body (e.g. tissues close to the skin, accessible by endoscopy, and/or during surgery), fluorescence imaging of gene expression can not only serve as invaluable research tools for cell- and animal-based research but also have certain potential clinical applications. Another optical imaging technique, bioluminescence imaging (BLI), is an attractive alternative for optical imaging of gene expression. Although BLI has little to no clinical potential, the virtual absence of background signal is highly desirable for noninvasive and sensitive imaging of gene expression in cells and small animals.
BLI OF GENE EXPRESSION
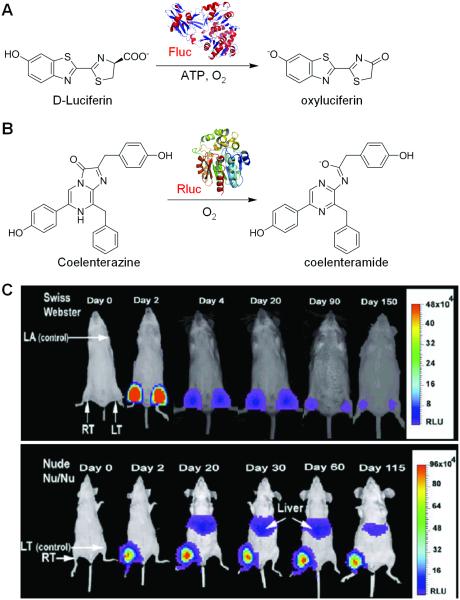
Bioluminescence is the emission of light from biochemical reactions that occur within a living organism. Luciferase is a family of photo-proteins that can be isolated from a large variety of insects, marine organisms, and prokaryotes (Hastings 1996). Luciferase has been used as a reporter gene in transgenic mice but, until the instrumentation was implemented for BLI, the detection of luciferase activity required either sectioning of the animal or excision of tissue and homogenization to measure the luciferase activity in a luminometer (Sadikot and Blackwell 2008). BLI has proven to be a very powerful methodology to detect luciferase activity in intact animal models. It is noninvasive, convenient, and relatively low-cost, thus possessing enormous potential to elucidate the pathobiology of various diseases (e.g. inflammation/injury, infection, and cancer) in animal models. To date, firefly luciferase (Fluc, Figure 2A) and renilla luciferase (Rluc, Figure 2B) are the most widely used reporter proteins for BLI of gene expression in living animals. Another type of luciferase, the bacterial luciferase, is primarily used in the study of infection since it is limited to bacteria that express the reporter gene and also produce the substrate (Contag et al. 1995).
Figure 2.
BLI of gene expression. A. The bioluminescent reaction catalyzed by Fluc. B. The bioluminescent reaction catalyzed by Rluc. C. The magnitude and duration of Fluc gene expression in different host immune systems can be monitored noninvasively and repetitively using the cooled CCD camera. For Swiss Webster mice, the Fluc activity was highest at day 2 and significantly dropped over time. For nude mice, a considerable amount of Fluc activity was seen throughout the study period. Adapted with permission from (Wu et al. 2001).
Fluc-Based BLI
Light emission (yellow-green color, 557 nm) from the firefly Photinus Pyralis, which is generally believed to be the most efficient bioluminescence system known to date, makes Fluc an excellent tool for monitoring gene expression (Branchini et al. 2005). Applications of Fluc in the imaging of gene expression in cells dated back to the early 1990s (Hooper et al. 1990). In 2001, a method for repetitively tracking in vivo gene expression of Fluc in skeletal muscles of mice was described with a cooled charged-coupled device (CCD) camera (Figure 2C) (Wu et al. 2001). The in vivo bioluminescence signals correlated well with the result from in vitro luciferase enzyme assays, which demonstrated the ability of BLI to sensitively and noninvasively track the location, magnitude, and persistence of Fluc-related gene expression.
Fluc can have broad applications in monitoring multiple disease-related gene expression. For example, the cyclooxygenase-2 (COX-2) gene plays a role in a wide variety of physiologic pathways and is a major target for therapeutic intervention in many pathophysiologic contexts such as pain, fever, inflammation, and cancer. Expression of the COX-2 gene can be induced in a wide range of cells, in response to an ever-increasing number of stimuli. BLI was successfully carried out to image the expression of Fluc gene in tumor xenografts, stably transfected with a chimeric gene containing the first kilobase of the murine COX-2 promoter (Nguyen et al. 2003). The imaging data suggested that gene expression from the COX-2 promoter can be easily analyzed in a variety of disease models in which the COX-2 gene is upregulated. BLI of gene expression with Fluc was also tested in larger animals such as rabbits (Li et al. 2005). It was reported that the BLI signal was capable of passing through at least 1 cm of muscle tissue.
BLI with Fluc is also applicable for the evaluation of DNA vaccines. Administration of naked DNA into animals has been used as a research tool to develop DNA vaccines. To monitor the distribution and duration of gene expression of a DNA vaccine in living subjects, the naked DNA encoding Fluc was used as an imaging reporter gene in a mouse model (Jeon et al. 2006). It was concluded that BLI with Fluc could be useful for monitoring the location, intensity and duration of gene expression of naked DNA vaccines in living animals, both noninvasively and repetitively. Similar to the study of DNA vaccines, BLI with Fluc (which serves as a model gene) can also be used to evaluate the gene delivery efficiency of certain gene delivery systems, especially in tumor models (Hildebrandt et al. 2003; Liang et al. 2004).
One major limitation of FLuc is that the light generated is in the yellow-green range which has poor tissue penetration. In 2005, a set of red- and green-emitting luciferase mutants were reported (Branchini et al. 2005). The bioluminescence properties of these mutants are suitable for expanding the use of the P. pyralis system in dual-color reporter assays, biosensor measurements with internal controls, and imaging. Using a combination of mutagenesis methods, a red-emitting luciferase with a bioluminescence maximum of 615 nm was created which also has a narrow emission bandwidth and favorable kinetic properties. Studies in animal models demonstrated that these luciferases could detect gene expression at the attomole level, many orders of magnitude higher than Fluc.
Rluc-Based BLI
Rluc is another promising and common bioluminescence reporter besides the Fluc. Distinct from Fluc in terms of its origin, enzyme structure, and substrate requirements (Inouye and Shimomura 1997), Rluc was isolated from sea pansy (Renilla reniformis) which displays blue-green bioluminescence upon stimulation. It can catalyze the oxidation of coelenterazine, which leads to bioluminescence signal (Figure 2B). Rluc was cloned and sequenced in 1991 (Lorenz et al. 1991) and has been used as a marker for gene expression in bacteria, yeast, plant, and mammalian cells (Lorenz et al. 1996).
BLI of gene expression with Rluc dated back to the early 2000s. In one pioneering report, Rluc was applied for noninvasive BLI of advanced human prostate cancer lesions in living mice by a targeted gene transfer vector (Adams et al. 2002). In another early study, the ability to image gene expression based on Rluc bioluminescence was validated by injecting the substrate coelenterazine in living mice (Bhaumik and Gambhir 2002). Cells transiently expressing the Rluc gene, located in the peritoneum, subcutaneous layer, as well as in the liver and lungs of living mice, were imaged after tail-vein injection of coelenterazine. Most importantly, both Rluc and Fluc expression can be imaged in the same living mouse, however the kinetics of light production is distinct for the two enzymes. The imaging strategy validated in this study can have direct applications in various studies where two molecular events need to be tracked, such as the trafficking of two cell populations, two gene therapy vectors, and indirect monitoring of two endogenous genes.
In a follow-up study, the expression of a novel synthetic Rluc reporter gene (hRluc) in living mice was explored, which has previously been reported to be a more sensitive reporter than the native Rluc in mammalian cells (Bhaumik et al. 2004a). It was found that hRluc:coelenterazine can yield a higher signal than the Fluc:D-luciferin in both cell culture and live animal studies, indicating that hRluc can be a useful primary reporter gene with high sensitivity.
During the prognosis of cancer treatment, early detection of the tumor and its metastases is crucial. In animal models, Rluc can be used for detecting certain gene expression in different stages of tumor development, thus delineating the location of primary and metastatic tumors (Yu et al. 2003). To achieve noninvasive measurement of chemotherapy-induced changes in the expression of genes related to tumor growth, BLI was used to image the alteration in human telomerase reverse transcriptase (hTERT) gene expression in tumor cells before and after 5-fluorouracil treatment (Padmanabhan et al. 2006). Several fusion reporters directed by the hTERT promoter fragments were investigated, which integrate the hRluc (for BLI), mRFP1 (for fluorescence imaging), and a truncated thymidine kinase (for PET imaging with radiolabeled acycloguanosines), respectively. In vitro studies demonstrated that although all three reporter systems can visualize the hTERT promoter activity, BLI was the most powerful one and it could also provide assistance in choosing the optimal tracers for PET imaging (Wang et al. 2006).
BLI has also been widely adopted for stem cell-based studies. The discovery of human embryonic stem cells (hESCs) has dramatically expanded the tools available to scientists/clinicians in the field of regenerative medicine (Thomson et al. 1998). However, direct injection of hESCs, and cells differentiated from hESCs, into living organisms was hampered by significant cell death, teratoma formation, and host immune rejection. Understanding the hESC behavior in vivo after transplantation requires noninvasive imaging techniques, such as BLI, to longitudinally monitor the hESC localization, proliferation, and viability (Wilson et al. 2008). Bioluminescence reporter genes (e.g. Rluc) can be transcribed either constitutively or only under specific biological or cellular conditions, depending on the type of promoter used. Stably transduced cells that carry the reporter construct within their chromosomal DNA will pass the reporter construct DNA to daughter cells, allowing for longitudinal monitoring of hESC survival and proliferation in vivo with BLI. Because expression of the reporter gene product is required for signal generation, only viable parent and daughter cells will generate BLI signal while the apoptotic or dead cells can not, which is the key advantage of report gene-based cell tracking over direct cell labeling (Zhang and Wu 2007).
BLI of mRNA
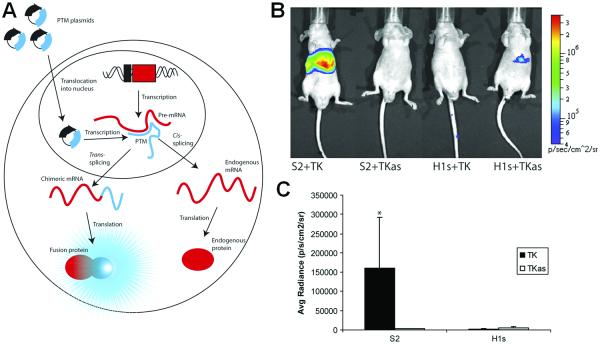
Aside from the abovementioned BLI of gene expression, in which the Fluc or Rluc was incorporated into the DNA, a strategy for imaging mRNA with BLI was designed using spliceosome-mediated RNA trans-splicing (SMaRT) (Bhaumik et al. 2004b; Walls et al. 2008). SMaRT provides an effective means to reprogram mRNAs and the proteins they encode. It can have a broad range of applications, including RNA repair and molecular imaging, each governed by the nature of the sequences delivered by the pre-trans-splicing molecule (PTM). In one ground-breaking study, the ability of SMaRT to optically image the expression of an exogenous gene at the level of pre-mRNA splicing was demonstrated in both cells and living animals (Bhaumik et al. 2004b). Because of the modular design of PTMs, there is potential for SMaRT in imaging the expression of any arbitrary gene of interest in living subjects.
Recently, this strategy was further optimized (Figure 3A) (Walls et al. 2008). A class of generalizable probes that can image pre-mRNA in a sequence-specific manner, using signal amplification and a facile method of delivery, was developed. Incorporating a modular binding domain that confers specificity by base-pair complementarity to the target pre-mRNA, the PTMs were designed to target a chimeric target mini gene and trans-splice the Rluc gene onto the end of the target. After hydrodynamic delivery of the PTMs and target genes in mice, the efficiency and specificity of the trans-splicing reaction was found to vary depending on the binding domain length and structure (Figure 3). Nonetheless, specific trans-splicing was observed in living animals which demonstrated a proof-of-principle for a generalizable imaging probe against RNA, which can amplify the signal on detection and be delivered with existing gene delivery methodology.
Figure 3.
BLI of mRNA. A. A schematic of the SMaRT imaging strategy. B. Representative images of nude mice injected hydrodynamically with combination of PTM and target plasmids. S2+TK: PTM and the target gene. The remaining three mice are negative controls. C. Average radiance for each mouse (mean ± SEM) in photons/s/cm2/steradian. Adapted with permission from (Walls et al. 2008).
BLI of gene expression has been applied in a widely variety of disease models over the last decade and the abovementioned studies are only a representative few. Theoretically, Fluc or Rluc can be fused to almost any gene of interest and the expression of the gene can be noninvasively monitored with BLI. One major advantage of BLI is that it is quite quantitative, therefore the changes in the bioluminescence signal can reflect the differences in gene expression level, provided that all other related variables remain constant (e.g. the depth of the tissue). Over the years, applications of BLI have spanned an enormous range of model systems from stem cell trafficking to cancer therapy to drug screening.
However, one has to bear in mind that BLI is only a research tool in preclinical studies and it can not be used in humans. Among all of the molecular imaging modalities, no single modality is perfect and sufficient to obtain all of the necessary information for a given question. For example, it is difficult to accurately quantify fluorescence signals in living subjects with fluorescence imaging alone, particularly in deep tissues; BLI is quantitative yet it has little to no clinical potential; radionuclide-based imaging techniques (e.g. PET) are very sensitive but have relatively poor spatial resolution. A combination of multiple molecular imaging modalities can offer synergistic advantages over any modality alone. Various double fusion and triple fusion reporter genes have been reported for multimodality imaging of gene expression.
MULTIMODALITY IMAGING OF GENE EXPRESSION
In one early study, a reporter vector encoding a mutant HSV1-sr39tk and Rluc was constructed (Ray et al. 2003). The two genes were joined by a 20 amino acid spacer sequence. Both PET and BLI were able to delineate tumors stably expressing the fusion gene in live, xenograft-bearing mice. Subsequently, several triple fusion reporter genes which can be used for bioluminescence, fluorescence, and PET imaging have been constructed. A triple fusion reporter vector consisting of Rluc, RFP, and HSV1-sr39tk (ttk) was found to confer activity of every protein composition in cell culture (Ray et al. 2004). In xenograft mouse models, the lentiviral vector encoding the triple fusion reporter gene also showed a good correlation of signals from different imaging modalities. Further, the vector expression could last for 40-50 days in living mice.
To exploit the combined strengths of each imaging technique and facilitate multimodality imaging, a dual-reporter construct was established in which Fluc was fused in frame to the N-terminus of a mutant HSV-tk kinetically enhanced for PET (Kesarwala et al. 2006). In addition, a triple reporter construct was developed in which monster GFP was introduced into the fusion vector downstream of an internal ribosome entry site to allow analysis by fluorescence microscopy or flow cytometry without compromising the specific activities of the upstream fusion components. In living mice, somatic gene transfer of a ubiquitin promoter-driven triple fusion plasmid showed a > 1,000-fold increase in liver photon flux and a > 2-fold increase in liver retention of 18F-FHBG by microPET, when compared with mice treated with a control plasmid.
Construction and validation of several triple fusion genes, each composed of a bioluminescent, a fluorescent, and a PET reporter gene, in cell culture and in living subjects was further explored (Ray et al. 2007). A mutant of a thermostable Fluc, bearing the peroxisome localization signal, was designed to have greater cytoplasmic localization and improved access for its substrate, D-luciferin. This mutant exhibited several fold higher activity than Fluc both in vitro and in vivo. Further, the improved version of the triple fusion vector showed significantly higher bioluminescence signal than the previous triple fusion vectors. As the third reporter component of this triple fusion vector for PET applications, a truncated version of wild-type HSV1-tk also retained a higher expression level than the truncated mutant HSV1-sr39tk. It was suggested that this improved triple fusion reporter vector will enable high sensitivity detection of lower numbers of cells from living animals using the combined bioluminescence, fluorescence, and PET imaging techniques.
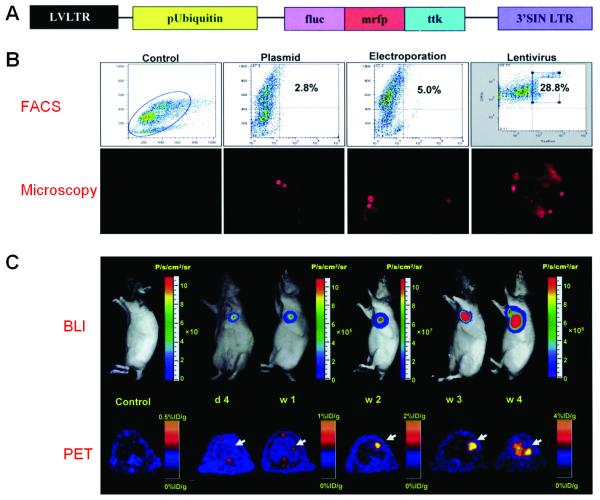
A proof-of-principle study for lentivirus-transduced murine embryonic stem cells (mESCs) that stably express this triple fusion reporter gene was carried out (Cao et al. 2006). It was demonstrated that this molecular imaging platform can be used to monitor the kinetics of stem cell survival, proliferation, migration, and ablation of teratoma sites. The fluorescence feature of the reporter gene was used for cell sorting and microscopy studies while both BLI and PET were used for longitudinal noninvasive monitoring of the transplanted mESCs (Figure 4). Further development of novel imaging techniques will contribute important insights into the biology and physiology of transplanted stem cells, leading to significant potential clinical applications for years to come.
Figure 4.
Multimodality imaging of mESC survival, proliferation, and migration after cardiac delivery with a triple fusion reporter gene. A. Schema of the triple fusion reporter gene. B. FACS histograms of mESCs at 48 hours after transduction with plasmid lipofectamine, electroporation, and lentivirus carrying the triple fusion reporter gene. C. Noninvasive imaging of transplanted mESCs with BLI and PET. Adapted with permission from (Cao et al. 2006).
Radiolabeled luciferase substrates have been explored for multimodality imaging of certain gene expression. In one report, a few 11C-labeled D-luciferin analogs were synthesized (Wang et al. 2006). PET studies showed a low retention of the 11C label at 45 min post-injection in luciferase-expressing tumors, while BLI with unlabeled substrate D-luciferin and the radiolabeled analogs gave tumor signal within a few minutes of photon counting. Since D-luciferin and the radiolabeled analogs are substrates of Fluc with a relatively high turnover rate, this is likely responsible for the poor retention of the tracers in the tumor. In addition, the required injected doses for these two imaging modalities also differ by several orders of magnitude. Therefore, although this is an interesting venue for exploring multimodality imaging, implementing it for optimal performance in animal studies is very challenging.
CONCLUSION
Recent advances in molecular imaging technologies have provided the potential to identify changes at the genetic or molecular level long before they are detectable by conventional diagnostic techniques such as computer tomography or MRI. More importantly, these noninvasive imaging techniques can allow us to interrogate certain biological events (e.g. gene expression) in intact animals that previously could only be determined from in vitro assays of biopsied tissues or body fluids. To date, the vast majority of gene expression imaging studies has been in the preclinical stage. Although there are many gene therapy clinical trials currently ongoing, few of them have incorporated noninvasive imaging of gene expression in the trial yet.
The progress made over the last decade in the development of noninvasive imaging technologies for monitoring gene expression should allow molecular imaging to play a major role in the field of gene therapy (Min and Gambhir 2004). These tools have been validated in gene therapy models for longitudinal and quantitative monitoring of the location(s), magnitude, and time variation of gene delivery and/or expression. This chapter on optical imaging of gene expression is primarily applicable to preclinical research and the emphasis has been mainly on the early studies which pioneered the field. The studies published to date clearly indicated that noninvasive imaging can accelerate the validation of preclinical models, which can give important insight towards clinical monitoring of human gene therapy. For clinical imaging of gene expression, PET holds the greatest promise (Yaghoubi et al. 2009).
Noninvasive optical imaging of gene expression has revolutionized biomedical research. It is no surprise that the 2008 Nobel Prize in Chemistry was awarded to three scientists for “the discovery and development of the GFP”. Although optical imaging (both fluorescence and bioluminescence) has certain inherent disadvantages such as poor tissue penetration and difficult to quantitate, the low cost, convenience, constantly evolving new imaging instrumentation, as well as emerging fluorescent proteins (Shaner et al. 2004; Shu et al. 2009) and luciferases (Loening et al. 2006; Loening et al. 2007) with better optical/physical/biological characteristics, makes optical imaging irreplaceable in basic and preclinical research far beyond the field of imaging gene expression.
ACKNOWLEDGEMENTS
The authors acknowledge financial support from the UW School of Medicine and Public Health’s Medical Education and Research Committee through the Wisconsin Partnership Program, the University of Wisconsin Carbone Cancer Center, NCRR 1UL1RR025011, and a Susan G. Komen Postdoctoral Fellowship (to H. Hong).
REFERENCES
- Adams JY, Johnson M, Sato M, Berger F, Gambhir SS, Carey M, Iruela-Arispe ML, Wu L. Visualization of advanced human prostate cancer lesions in living mice by a targeted gene transfer vector and optical imaging. Nat Med. 2002;8:891–897. doi: 10.1038/nm743. [DOI] [PubMed] [Google Scholar]
- Bhaumik S, Gambhir SS. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc Natl Acad Sci USA. 2002;99:377–382. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S, Lewis XZ, Gambhir SS. Optical imaging of Renilla luciferase, synthetic Renilla luciferase, and firefly luciferase reporter gene expression in living mice. J Biomed Opt. 2004a;9:578–586. doi: 10.1117/1.1647546. [DOI] [PubMed] [Google Scholar]
- Bhaumik S, Walls Z, Puttaraju M, Mitchell LG, Gambhir SS. Molecular imaging of gene expression in living subjects by spliceosome-mediated RNA trans-splicing. Proc Natl Acad Sci USA. 2004b;101:8693–8698. doi: 10.1073/pnas.0402772101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branchini BR, Southworth TL, Khattak NF, Michelini E, Roda A. Red- and green-emitting firefly luciferase mutants for bioluminescent reporter applications. Anal Biochem. 2005;345:140–148. doi: 10.1016/j.ab.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Cai W, Chen X. Nanoplatforms for targeted molecular imaging in living subjects. Small. 2007;3:1840–1854. doi: 10.1002/smll.200700351. [DOI] [PubMed] [Google Scholar]
- Cai W, Chen X. Preparation of peptide conjugated quantum dots for tumour vasculature targeted imaging. Nat Protoc. 2008;3:89–96. doi: 10.1038/nprot.2007.478. [DOI] [PubMed] [Google Scholar]
- Cai W, Niu G, Chen X. Imaging of integrins as biomarkers for tumor angiogenesis. Curr Pharm Des. 2008a;14:2943–2973. doi: 10.2174/138161208786404308. [DOI] [PubMed] [Google Scholar]
- Cai W, Niu G, Chen X. Multimodality imaging of the HER-kinase axis in cancer. Eur J Nucl Med Mol Imaging. 2008b;35:186–208. doi: 10.1007/s00259-007-0560-9. [DOI] [PubMed] [Google Scholar]
- Cai W, Shin DW, Chen K, Gheysens O, Cao Q, Wang SX, Gambhir SS, Chen X. Peptide-labeled near-infrared quantum dots for imaging tumor vasculature in living subjects. Nano Lett. 2006;6:669–676. doi: 10.1021/nl052405t. [DOI] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Lin S, Xie X, Ray P, Patel M, Zhang X, Drukker M, Dylla SJ, Connolly AJ, Chen X, Weissman IL, Gambhir SS, Wu JC. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation. 2006;113:1005–1014. doi: 10.1161/CIRCULATIONAHA.105.588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Zhan X, Kumar A, Du X, Hammond H, Cheng L, Yang X. Detection of dual-gene expression in arteries using an optical imaging method. J Biomed Opt. 2004;9:1223–1229. doi: 10.1117/1.1803842. [DOI] [PubMed] [Google Scholar]
- Contag CH, Contag PR, Mullins JI, Spilman SD, Stevenson DK, Benaron DA. Photonic detection of bacterial pathogens in living hosts. Mol Microbiol. 1995;18:593–603. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x. [DOI] [PubMed] [Google Scholar]
- Dietrich C, Maiss E. Red fluorescent protein DsRed from Discosoma sp. as a reporter protein in higher plants. Biotechniques. 2002;32:286. doi: 10.2144/02322st02. 288-290, 292-283. [DOI] [PubMed] [Google Scholar]
- Ferrari A, Pellegrini V, Arcangeli C, Fittipaldi A, Giacca M, Beltram F. Caveolae-mediated internalization of extracellular HIV-1 tat fusion proteins visualized in real time. Mol Ther. 2003;8:284–294. doi: 10.1016/s1525-0016(03)00122-9. [DOI] [PubMed] [Google Scholar]
- Fukumura D, Xavier R, Sugiura T, Chen Y, Park EC, Lu N, Selig M, Nielsen G, Taksir T, Jain RK, Seed B. Tumor induction of VEGF promoter activity in stromal cells. Cell. 1998;94:715–725. doi: 10.1016/s0092-8674(00)81731-6. [DOI] [PubMed] [Google Scholar]
- Gambhir SS, Barrio JR, Phelps ME, Iyer M, Namavari M, Satyamurthy N, Wu L, Green LA, Bauer E, MacLaren DC, Nguyen K, Berk AJ, Cherry SR, Herschman HR. Imaging adenoviral-directed reporter gene expression in living animals with positron emission tomography. Proc Natl Acad Sci USA. 1999;96:2333–2338. doi: 10.1073/pnas.96.5.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambhir SS, Herschman HR, Cherry SR, Barrio JR, Satyamurthy N, Toyokuni T, Phelps ME, Larson SM, Balatoni J, Finn R, Sadelain M, Tjuvajev J, Blasberg R. Imaging transgene expression with radionuclide imaging technologies. Neoplasia. 2000;2:118–138. doi: 10.1038/sj.neo.7900083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad AA, Winnard PT, Jr., van Zijl PC, Bulte JW. Developing MR reporter genes: promises and pitfalls. NMR Biomed. 2007;20:275–290. doi: 10.1002/nbm.1134. [DOI] [PubMed] [Google Scholar]
- Gilad AA, Ziv K, McMahon MT, van Zijl PC, Neeman M, Bulte JW. MRI reporter genes. J Nucl Med. 2008;49:1905–1908. doi: 10.2967/jnumed.108.053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet JP, Macadangdang B, Fathke RL, Gottesman MM, Kimchi-Sarfaty C. The development of gene therapy: from monogenic recessive disorders to complex diseases such as cancer. Methods Mol Biol. 2009;542:5–54. doi: 10.1007/978-1-59745-561-9_1. [DOI] [PubMed] [Google Scholar]
- Hastings JW. Chemistries and colors of bioluminescent reactions: a review. Gene. 1996;173:5–11. doi: 10.1016/0378-1119(95)00676-1. [DOI] [PubMed] [Google Scholar]
- Heikal AA, Hess ST, Baird GS, Tsien RY, Webb WW. Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: coral red (dsRed) and yellow (Citrine) Proc Natl Acad Sci USA. 2000;97:11996–12001. doi: 10.1073/pnas.97.22.11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- Hildebrandt IJ, Iyer M, Wagner E, Gambhir SS. Optical imaging of transferrin targeted PEI/DNA complexes in living subjects. Gene Ther. 2003;10:758–764. doi: 10.1038/sj.gt.3301939. [DOI] [PubMed] [Google Scholar]
- Hoffman RM. Visualization of GFP-expressing tumors and metastasis in vivo. Biotechniques. 2001;30:1016–1022. doi: 10.2144/01305bi01. 1024-1016. [DOI] [PubMed] [Google Scholar]
- Hoffman RM, Yang M. Whole-body imaging with fluorescent proteins. Nat Protoc. 2006;1:1429–1438. doi: 10.1038/nprot.2006.223. [DOI] [PubMed] [Google Scholar]
- Hooper CE, Ansorge RE, Browne HM, Tomkins P. CCD imaging of luciferase gene expression in single mammalian cells. J Biolumin Chemilumin. 1990;5:123–130. doi: 10.1002/bio.1170050208. [DOI] [PubMed] [Google Scholar]
- Inouye S, Shimomura O. The use of Renilla luciferase, Oplophorus luciferase, and apoaequorin as bioluminescent reporter protein in the presence of coelenterazine analogues as substrate. Biochem Biophys Res Commun. 1997;233:349–353. doi: 10.1006/bbrc.1997.6452. [DOI] [PubMed] [Google Scholar]
- Jakobs S, Subramaniam V, Schonle A, Jovin TM, Hell SW. EFGP and DsRed expressing cultures of Escherichia coli imaged by confocal, two-photon and fluorescence lifetime microscopy. FEBS Lett. 2000;479:131–135. doi: 10.1016/s0014-5793(00)01896-2. [DOI] [PubMed] [Google Scholar]
- Jeon YH, Choi Y, Kang JH, Chung JK, Lee YJ, Kim CW, Jeong JM, Lee DS, Lee MC. In vivo monitoring of DNA vaccine gene expression using firefly luciferase as a naked DNA. Vaccine. 2006;24:3057–3062. doi: 10.1016/j.vaccine.2006.01.033. [DOI] [PubMed] [Google Scholar]
- Josserand V, Texier-Nogues I, Huber P, Favrot MC, Coll JL. Non-invasive in vivo optical imaging of the lacZ and luc gene expression in mice. Gene Ther. 2007;14:1587–1593. doi: 10.1038/sj.gt.3303028. [DOI] [PubMed] [Google Scholar]
- Kang JH, Chung JK. Molecular-genetic imaging based on reporter gene expression. J Nucl Med. 2008;49(Suppl 2):164S–179S. doi: 10.2967/jnumed.107.045955. [DOI] [PubMed] [Google Scholar]
- Kesarwala AH, Prior JL, Sun J, Harpstrite SE, Sharma V, Piwnica-Worms D. Second-generation triple reporter for bioluminescence, micro-positron emission tomography, and fluorescence imaging. Mol Imaging. 2006;5:465–474. [PubMed] [Google Scholar]
- Li JZ, Holman D, Li H, Liu AH, Beres B, Hankins GR, Helm GA. Long-term tracing of adenoviral expression in rat and rabbit using luciferase imaging. J Gene Med. 2005;7:792–802. doi: 10.1002/jgm.720. [DOI] [PubMed] [Google Scholar]
- Liang Q, Yamamoto M, Curiel DT, Herschman HR. Noninvasive imaging of transcriptionally restricted transgene expression following intratumoral injection of an adenovirus in which the COX-2 promoter drives a reporter gene. Mol Imaging Biol. 2004;6:395–404. doi: 10.1016/j.mibio.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Loening AM, Fenn TD, Wu AM, Gambhir SS. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng Des Sel. 2006;19:391–400. doi: 10.1093/protein/gzl023. [DOI] [PubMed] [Google Scholar]
- Loening AM, Wu AM, Gambhir SS. Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat Methods. 2007;4:641–643. doi: 10.1038/nmeth1070. [DOI] [PubMed] [Google Scholar]
- Lorenz WW, Cormier MJ, O'Kane DJ, Hua D, Escher AA, Szalay AA. Expression of the Renilla reniformis luciferase gene in mammalian cells. J Biolumin Chemilumin. 1996;11:31–37. doi: 10.1002/(SICI)1099-1271(199601)11:1<31::AID-BIO398>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Lorenz WW, McCann RO, Longiaru M, Cormier MJ. Isolation and expression of a cDNA encoding Renilla reniformis luciferase. Proc Natl Acad Sci USA. 1991;88:4438–4442. doi: 10.1073/pnas.88.10.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker GD, Luker KE, Sharma V, Pica CM, Dahlheimer JL, Ocheskey JA, Fahrner TJ, Milbrandt J, Piwnica-Worms D. In vitro and in vivo characterization of a dual-function green fluorescent protein--HSV1-thymidine kinase reporter gene driven by the human elongation factor 1 alpha promoter. Mol Imaging. 2002;1:65–73. doi: 10.1162/15353500200201118. [DOI] [PubMed] [Google Scholar]
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- Meyer N, Jaconi M, Landopoulou A, Fort P, Puceat M. A fluorescent reporter gene as a marker for ventricular specification in ES-derived cardiac cells. FEBS Lett. 2000;478:151–158. doi: 10.1016/s0014-5793(00)01839-1. [DOI] [PubMed] [Google Scholar]
- Min JJ, Gambhir SS. Gene therapy progress and prospects: noninvasive imaging of gene therapy in living subjects. Gene Ther. 2004;11:115–125. doi: 10.1038/sj.gt.3302191. [DOI] [PubMed] [Google Scholar]
- Najjar AM, Nishii R, Maxwell DS, Volgin A, Mukhopadhyay U, Bornmann WG, Tong W, Alauddin M, Gelovani JG. Molecular-genetic PET imaging using an HSV1-tk mutant reporter gene with enhanced specificity to acycloguanosine nucleoside analogs. J Nucl Med. 2009;50:409–416. doi: 10.2967/jnumed.108.058735. [DOI] [PubMed] [Google Scholar]
- Nguyen JT, Machado H, Herschman HR. Repetitive, noninvasive imaging of cyclooxygenase-2 gene expression in living mice. Mol Imaging Biol. 2003;5:248–256. doi: 10.1016/s1536-1632(03)00105-7. [DOI] [PubMed] [Google Scholar]
- Niyibizi C, Wang S, Mi Z, Robbins PD. The fate of mesenchymal stem cells transplanted into immunocompetent neonatal mice: implications for skeletal gene therapy via stem cells. Mol Ther. 2004;9:955–963. doi: 10.1016/j.ymthe.2004.02.022. [DOI] [PubMed] [Google Scholar]
- Ntziachristos V, Bremer C, Weissleder R. Fluorescence imaging with near-infrared light: new technological advances that enable in vivo molecular imaging. Eur Radiol. 2003;13:195–208. doi: 10.1007/s00330-002-1524-x. [DOI] [PubMed] [Google Scholar]
- Padmanabhan P, Otero J, Ray P, Paulmurugan R, Hoffman AR, Gambhir SS, Biswal S, Ulaner GA. Visualization of telomerase reverse transcriptase (hTERT) promoter activity using a trimodality fusion reporter construct. J Nucl Med. 2006;47:270–277. [PMC free article] [PubMed] [Google Scholar]
- Ray P, De A, Min JJ, Tsien RY, Gambhir SS. Imaging tri-fusion multimodality reporter gene expression in living subjects. Cancer Res. 2004;64:1323–1330. doi: 10.1158/0008-5472.can-03-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P, Tsien R, Gambhir SS. Construction and validation of improved triple fusion reporter gene vectors for molecular imaging of living subjects. Cancer Res. 2007;67:3085–3093. doi: 10.1158/0008-5472.CAN-06-2402. [DOI] [PubMed] [Google Scholar]
- Ray P, Wu AM, Gambhir SS. Optical bioluminescence and positron emission tomography imaging of a novel fusion reporter gene in tumor xenografts of living mice. Cancer Res. 2003;63:1160–1165. [PubMed] [Google Scholar]
- Sadikot RT, Blackwell TS. Bioluminescence: imaging modality for in vitro and in vivo gene expression. Methods Mol Biol. 2008;477:383–394. doi: 10.1007/978-1-60327-517-0_29. [DOI] [PubMed] [Google Scholar]
- Serganova I, Mayer-Kukuck P, Huang R, Blasberg R. Molecular imaging: reporter gene imaging. Handb Exp Pharmacol. 2008:167–223. doi: 10.1007/978-3-540-77496-9_8. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tolar J, Osborn M, Bell S, McElmurry R, Xia L, Riddle M, Panoskaltsis-Mortari A, Jiang Y, McIvor RS, Contag CH, Yant SR, Kay MA, Verfaillie CM, Blazar BR. Real-time in vivo imaging of stem cells following transgenesis by transposition. Mol Ther. 2005;12:42–48. doi: 10.1016/j.ymthe.2005.02.023. [DOI] [PubMed] [Google Scholar]
- Turchin IV, Kamensky VA, Plehanov VI, Orlova AG, Kleshnin MS, Fiks II, Shirmanova MV, Meerovich IG, Arslanbaeva LR, Jerdeva VV, Savitsky AP. Fluorescence diffuse tomography for detection of red fluorescent protein expressed tumors in small animals. J Biomed Opt. 2008;13:041310. doi: 10.1117/1.2953528. [DOI] [PubMed] [Google Scholar]
- Walls ZF, Puttaraju M, Temple GF, Gambhir SS. A generalizable strategy for imaging pre-mRNA levels in living subjects using spliceosome-mediated RNA trans-splicing. J Nucl Med. 2008;49:1146–1154. doi: 10.2967/jnumed.107.047662. [DOI] [PubMed] [Google Scholar]
- Wang JQ, Pollok KE, Cai S, Stantz KM, Hutchins GD, Zheng QH. PET imaging and optical imaging with D-luciferin [11C]methyl ester and D-luciferin [11C]methyl ether of luciferase gene expression in tumor xenografts of living mice. Bioorg Med Chem Lett. 2006;16:331–337. doi: 10.1016/j.bmcl.2005.09.082. [DOI] [PubMed] [Google Scholar]
- Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci USA. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CM, Trainor PA, Radziewic T, Pelka GJ, Zhou SX, Parameswaran M, Quinlan GA, Gordon M, Sturm K, Tam PP. Application of lacZ transgenic mice to cell lineage studies. Methods Mol Biol. 2008;461:149–164. doi: 10.1007/978-1-60327-483-8_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh S, Kay SA. Reporter gene expression for monitoring gene transfer. Curr Opin Biotechnol. 1997;8:617–622. doi: 10.1016/s0958-1669(97)80038-9. [DOI] [PubMed] [Google Scholar]
- Wilson K, Yu J, Lee A, Wu JC. In vitro and in vivo bioluminescence reporter gene imaging of human embryonic stem cells. J Vis Exp. 2008 doi: 10.3791/740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JC, Sundaresan G, Iyer M, Gambhir SS. Noninvasive optical imaging of firefly luciferase reporter gene expression in skeletal muscles of living mice. Mol Ther. 2001;4:297–306. doi: 10.1006/mthe.2001.0460. [DOI] [PubMed] [Google Scholar]
- Yaghoubi SS, Jensen MC, Satyamurthy N, Budhiraja S, Paik D, Czernin J, Gambhir SS. Noninvasive detection of therapeutic cytolytic T cells with 18F-FHBG PET in a patient with glioma. Nat Clin Pract Oncol. 2009;6:53–58. doi: 10.1038/ncponc1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Baranov E, Moossa AR, Penman S, Hoffman RM. Visualizing gene expression by whole-body fluorescence imaging. Proc Natl Acad Sci USA. 2000;97:12278–12282. doi: 10.1073/pnas.97.22.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YA, Timiryasova T, Zhang Q, Beltz R, Szalay AA. Optical imaging: bacteria, viruses, and mammalian cells encoding light-emitting proteins reveal the locations of primary tumors and metastases in animals. Anal Bioanal Chem. 2003;377:964–972. doi: 10.1007/s00216-003-2065-0. [DOI] [PubMed] [Google Scholar]
- Zhang SJ, Wu JC. Comparison of imaging techniques for tracking cardiac stem cell therapy. J Nucl Med. 2007;48:1916–1919. doi: 10.2967/jnumed.107.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Yu Z, Zhao S, Hu L, Zheng J, Yang D, Bouvet M, Hoffman RM. Lentivirus-Based DsRed-2-Transfected Pancreatic Cancer Cells for Deep In Vivo Imaging of Metastatic Disease. J Surg Res. 2008 doi: 10.1016/j.jss.2008.08.027. [DOI] [PubMed] [Google Scholar]