Abstract
Introduction
A few studies have reported an association between NADP(H): quinine oxidoreductase 1 (NQO1) C609T polymorphism and susceptibility to colorectal cancer (CRC). However, the results were inconsistent rather than conclusive. We performed a meta-analysis to examine this association in various populations.
Material and methods
Eligible articles were identified by a search of several databases up until June 30, 2013. Summary odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to assess the strength of the association.
Results
Overall, 14 case-control studies with 4,461 cases and 5,474 controls were included in this meta-analysis. The results indicated that the NQO1 C609T polymorphism was significantly associated with CRC susceptibility (summary ORs (95% CIs): 1.30 (1.07–1.59) for CT vs. CC, 1.64 (1.15–2.33) for TT vs. CC, 1.34 (1.10–1.64) for TT/CT vs. CC, and 1.43 (1.10–1.87) for TT vs. CT/CC). Subgroup analyses indicated that the T allele was significantly associated with CRC susceptibility in both Asians and Caucasians, and was also observed in high quality studies and hospital-based case-control studies. Specifically, we found a positive association between the NQO1 C609T polymorphism and CRC susceptibility in smokers, but not in non-smokers.
Conclusions
The results of this meta-analysis suggest that the NQO1 C609T polymorphism significantly contributes to increased susceptibility to CRC in both Asians and Caucasians.
Keywords: colorectal cancer, NQO1, polymorphism, meta-analysis
Introduction
The incidence of colorectal cancer (CRC) is differently shaped over time: the incidence is stabilizing or even gradually decreasing in some countries of Northern and Western Europe [1], while it is increasing in others [2]. It is reported that both environmental and genetic factors contribute to colorectal carcinogenesis [3, 4]. In the last decade, great efforts have been focused on unraveling the genetic underpinnings of CRC [5]; however, its driving genes and genetic determinants that contribute to the development of CRC so far remain elusive.
NADP(H): quinine oxidoreductase 1 (NQO1), also named diphtheria toxin diaphorase, is located on chromosome 16q22. NQO1 has been reported to play a crucial role in the detoxification of potentially mutagenic and carcinogenic quinones (derived from tobacco or the diet) and catalyzing the two-electron reduction of quinoid compounds into hydroquinones, which can alleviate cancer development [6]. In addition, NQO1 is implicated in protection of cells against oxidative stress and carcinogenesis by maintaining antioxidant forms of ubiquinone [7].
Several polymorphisms have been discovered in the NQO1 gene [8]. The most commonly studied polymorphism in this gene is the C609T polymorphism (dbSNP: rs1800566) at exon 6 of the gene, which leads to a proline to serine substitution in the protein sequence. This polymorphism occurs at a frequency of 50% in the human population, with 10% being homozygous for T alleles [9]. Phenotyping studies suggested that the homozygosity for the NQO1 protein has little or no activity (2–4% activity of the wild type). In contrast, the heterozygous (CT) genotype showed threefold decreased enzymatic activity compared with the wild-type allele.
The NQO1 C609T polymorphism has been widely evaluated in relation to susceptibility of CRC across various ethnicities, yet with inconsistent results [10–14]. A case-control study carried out by Van der Logt et al. [13] showed that, compared with the CC genotype, the TT genotype was significantly associated with the risk of CRC, with the adjusted odds ratio (OR) (95% confidence interval (CI)) of 1.6 (1.03–2.4). Specifically, a recent study from China found that, in addition to the positive relationship with CRC susceptibility, an interaction between NQO1 polymorphism and smoking was also observed [15]. However, other studies showed that there was no relationship between NQO1 polymorphism and CRC susceptibility [10, 11, 14]. Recently, several meta-analyses [16–19] have evaluated the association between the NQO1 C609T polymorphism and risk of colorectal neoplasm. The most recent analysis by Wang et al. [18] found an obvious association between NQO1 C609T polymorphism and colorectal cancer risk in both Caucasians and Asians. In another meta-analysis, Zhu et al. [19] found that the NQO1 C609T polymorphism might have a significantly increased risk of upper digest tract cancer, but not risk of CRC. That study included 9 studies on CRC involving 4,461 cases and 4,825 controls.
In the current study, we aimed to conduct a comprehensive meta-analysis of the association between only invasive colorectal neoplasm and the risk of NQO1 C609T polymorphism in both Caucasians and Asians. We also explored the interaction between NQO1 genotype and smoking status.
Material and methods
Data sources and searches
Data searches were conducted by two independent investigators (C.R. and Z.B.A.). A computerized literature search was conducted in several databases for all published reports on the association between NQO1 polymorphisms and the risk of CRC from the indexing to 30 June, 2013. For English articles, MEDLINE and EMBASE databases were searched; and for Chinese articles, the CNKI database, the China WanFang database, and the China Weipu database were searched. We applied the following algorithm to both the Medical Subject Heading (MeSH) and the full text: 1) “quinone oxidoreductase” OR “DT-diaphorase” OR “quinone reductase” OR “NAD(P)H: quinone oxidoreductase 1” OR “NQO1” OR “DTD”; 2) “colorectal” OR “colon” OR “rectal”; 3) “cancer” OR “carcinoma” OR “adenocarcinoma” or “neoplasm”; AND 4) “polymorphism” OR “allele” OR “genotype” OR “variant” OR “variation”. We also reviewed the reference lists of the relevant articles to identify additional studies. Unpublished studies were not considered.
Selection and exclusion criteria
Studies included in the meta-analysis must meet all the following inclusion criteria: 1) being an independent case-control, nested case-control, or cohort study; 2) evaluating the association between NQO1 C609T polymorphism and the risk of colorectal cancer; 3) having sufficient data for calculating an OR with 95% CI; and 4) reported in English or in Chinese. Exclusion criteria were: 1) duplicate data; 2) abstract, case report, comment, review and editorial; 3) no sufficient genotyping data; 4) the outcome was benign tumors, precancerous lesions, and adenomas; and 5) family-based study.
Data extraction
The following information was collected from all eligible publications according to the criteria listed above: first author's last name, year of publication, countries or region of origin, ethnicity, sources of controls (population-based or hospital-based), numbers of cases and controls, and Hardy-Weinberg equilibrium (HWE) for the control group. Two of us (C.R. and Z.B.A.) assessed and extracted the data in a standardized data extraction form each publication. When discrepancies were found, a third investigator would make the definitive decision for study eligibility and data extraction. To retrieve the missing data, we also contacted the authors of primary studies. Only one study provided the relevant data, although communication with the authors had taken place [15].
Quality score assessment
Two reviewers (C.R. and Z.B.A.) assessed the quality of each selected study using the quality assessment criteria, which were modified from a previously published meta-analysis of molecular association studies [20, 21]. Any discrepancies were resolved by consultation with the third authors. We included the following factors related to both traditional epidemiological considerations and cancer genetic issues in terms of quality of the studies: representativeness of the cases, representativeness of the controls, ascertainment of outcome, matching of case and control participants, genotyping examination, and total sample size. The criteria are described in detail in Supplementary Table I, and the scores were defined as 0 to 2 points given to each component. A numerical score ranging from 0 to 12 was assigned as a quantitative measure of literature quality. Studies were categorized as “high quality” if the quality score was ≥ 7; otherwise, studies were categorized as “low quality”.
Table I.
Main characteristics of all studies included in the meta-analysis
| First author | Year | Recruitment period | Country | Ethnicity | Control source | Cases/controls(n) | Adjustments/matching | Quality score | Value of p*† |
|---|---|---|---|---|---|---|---|---|---|
| Harth et al. [41] | 2000 | 1996–1999 | Germany | White | PB | 323/205 | Area | 6 | 0.792 |
| Lafuente et al. [24] | 2000 | 1996–1997 | Spain | White | HB | 247/296 | Age, sex, smoking | 6 | – |
| Mitrou et al. [42] | 2002 | 1998–2000 | UK | White | HB | 206/345 | None | 8 | 0.932 |
| Sachse et al. [14] | 2002 | 1997–2001 | UK | White | PB | 490/593 | Age, sex, study centre and general practitioner | 9 | 0.555 |
| Hamajima et al. [43] | 2002 | 1999–2000 | Japan | Asian | HB | 146/640 | Age, sex | 5 | 0.076 |
| Dai et al. [44] | 2004 | 2001–2002 | China | Asian | HB | 101/103 | None | 8 | 0.749 |
| Van der Logt et al. [13] | 2006 | 2002–2004 | Netherlands | White | PB | 369/415 | Age, sex | 7 | 0.947 |
| Begleiter et al. [9] | 2006 | – | Canada | White | PB | 280/327 | Age, sex | 8 | 0.452 |
| Hlavata et al. [10] | 2010 | 2004–2006 | Czech | White | HB | 495/495 | Age, sex, smoking, education, living area | 8 | 0.849 |
| Nisa et al. [11] | 2010 | 2000–2003 | Japan | Asian | PB | 684/777 | Age, sex, residence area, smoking, alcohol, BMI, type of job, physical activity | 11 | 0.068 |
| Sameer et al. [12] | 2010 | 2008–2009 | India | Asian | HB | 86/160 | Age, sex, smoking, dwelling | 4 | 0.447 |
| Xiane et al. [45] | 2010 | 2005–2006 | China | Asian | HB | 286/286 | Age, sex, race, dwelling | 7 | 0.316 |
| Su et al. [25] | 2012 | 2008–2010 | China | Asian | HB | 76/160 | Age, sex | 5 | 0.017 |
| Peng et al. [15] | 2013 | 2006–2010 | China | Asian | HB | 672/672 | Age, sex, income, marriage, job, education, smoking, alcohol intake, family history, and diet | 10 | 0.241 |
PB – population-based, HB – hospital-based, BMI – body mass index.
p-value of the χ 2 goodness of fit test for Hardy-Weinberg equilibrium (HWE) in controls
The HWE test cannot be conducted because only the total number of genotypes (TT vs. CT/CC) was available, and the HWE test was not mentioned in this study [24]
Statistical analysis
All statistical analyses were performed using STATA, version 11.0 (STATA, College Station, TX, USA), and a p value < 0.05 was considered significant. We assessed the departure from the HWE for the control group in each study using Pearson's goodness-of-fit χ2 test with 1 degree of freedom. Summary ORs and corresponding 95% CIs were used to estimate the association between the NQO1 C609T polymorphism and CRC risk, which were calculated by several comparisons, that is, a homozygote model (TT vs. CC), a heterozygote model (CT vs. CC), a dominant model (CT/TT vs. CC), and a recessive model (TT vs. CT/CC).
Heterogeneity among studies was evaluated by the Cochran Q and I 2 statistics. For the Q statistic, a p value < 0.10 was considered statistically significant; for I 2, a value > 50% was considered a measure of severe heterogeneity. To summarize the risk estimation, we used the method of a random-effects model, which accounts for heterogeneity among studies [22]. The potential source of heterogeneity across studies was explored by stratified analyses, which were conducted by several study characteristics, including ethnicity, sources of controls, smoking status and the quality score of studies (quality score, < 7 and ≥ 7).
Sensitivity analyses were conducted by removing one study at a time to assess the stability of the results. Asymmetry funnel plots were inspected to assess potential publication bias. Both Begg's test and Egger's test [23] were used to assess publication bias. A p value of less than 0.10 was considered to indicate statistically significant publication bias.
Results
Characteristics of selected studies
Based on our search strategy, a total of 14 case-control studies with 4,461 CRC patients and 5,474 controls met the inclusion criteria. The detailed baseline characteristics of qualified studies are presented in Table I. Of these 14 studies, 9 studies were based on a hospital-based design and 5 studies on a population-based design. Twelve studies were published in English and 2 in Chinese. Of these 14 studies, 7 studies were conducted in Caucasian populations, 7 in Asian populations (4 in Chinese, 2 in Japanese and 1 in Indians). The polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) method was used to determine the genotype in all the included studies.
The genotype distributions of the NQO1 gene C609T polymorphism were in agreement with the HWE among control groups of all but two studies [24, 25]. The HWE test in the study by Lafuente et al. was not mentioned [24]; we also could not perform the HWE test for the subjects (either cases or controls) in that study, because only the total number of the combined genotypes (TT vs. CT/CC) was available. Quality scores for the individual studies ranged from 4 to 11, with 64.3% (9 of 14) of the studies being classified as high quality (≥ 7).
Overall analyses
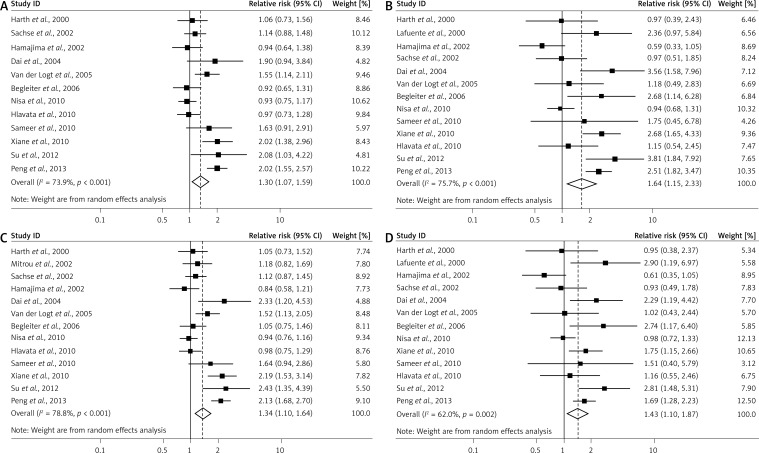
As shown in Table II, compared to the wild-type CC homozygous genotype, the TT homozygous and CT heterozygous genotype were significantly associated with an increased risk for CRC (CT vs. CC: summary ORs = 1.30, 95% CIs: 1.07–1.59, Figure 1A; TT vs. CC: summary ORs = 1.64, 95% CIs: 1.15–2.33, Figure 1B). A main effect was significant in the dominant model (CT/TT vs. CC: summary ORs = 1.34, 95% CIs: 1.10– 1.64, Figure 1C) and the recessive model (TT vs. CT/CC: summary ORs = 1.43, 95% CIs: 1.10–1.87, Figure 1D).
Table II.
Subgroup analyses of NQO1 gene C609T polymorphism with risk of colorectal cancer
| Variables | N | No. of cases/controls | Heterozygote | Homozygote | Dominant model | Recessive model | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CT vs. CC | TT vs. CC | CT/TT vs. CC | TT vs. CT/CC | |||||||||||
| OR (95% CI) | P het | I 2 (%) | OR (95% CI) | P het | I 2 (%) | OR (95% CI) | P het | I 2 (%) | OR (95% CI) | P het | I 2 (%) | |||
| All | 14 | 4461/5474 | 1.30 (1.07–1.59) | < 0.001 | 73.9 | 1.64 (1.15–2.33) | < 0.001 | 75.7 | 1.34 (1.10–1.64) | 0.001 | 78.8 | 1.43 (1.10–1.87) | 0.002 | 62.0 |
| Design: | ||||||||||||||
| HB | 9 | 2315/3157 | 1.45 (1.11–1.90) | < 0.001 | 74.6 | 1.83 (1.19–2.82) | 0.001 | 75.3 | 1.53 (1.13–2.08) | 0.001 | 82.8 | 1.53 (1.11–2.12) | 0.011 | 62.6 |
| PB | 5 | 2146/2317 | 1.09 (0.85–1.40) | 0.048 | 62.0 | 1.20 (0.77–1.89) | 0.163 | 41.4 | 1.12 (0.94–1.34) | 0.139 | 42.3 | 1.19 (0.77–1.85) | 0.165 | 41.1 |
| Ethnicity: | ||||||||||||||
| White | 7 | 2410/2676 | 1.23 (1.03–1.39) | 0.159 | 39.4 | 1.20 (1.01–1.47) | 0.331 | 13.0 | 1.14 (1.00–1.29) | 0.385 | 4.9 | 1.38 (1.01–1.79) | 0.159 | 37.1 |
| Asian | 7 | 2051/2798 | 1.51 (1.08–2.12) | < 0.001 | 80.6 | 1.84 (1.16–2.89) | < 0.001 | 81.9 | 1.40 (1.11–2.34) | < 0.001 | 86.6 | 1.46 (1.02–2.10) | < 0.001 | 74.5 |
| Quality of study: | ||||||||||||||
| Low (< 7) | 5 | 917/1451 | 1.41 (1.00–1.99) | 0.025 | 64.0 | 1.59 (0.74–3.45) | 0.001 | 82.1 | 1.47 (0.96–2.24) | 0.001 | 79.4 | 1.34 (0.74–2.42) | 0.004 | 73.6 |
| High (≥ 7) | 9 | 3544/4023 | 1.25 (0.96–1.61) | < 0.001 | 80.1 | 1.65 (1.09–2.51) | 0.001 | 74.0 | 1.29 (1.01–1.63) | < 0.001 | 80.7 | 1.47 (1.08–2.00) | 0.022 | 57.4 |
| Smoking status: | ||||||||||||||
| Nonsmokers | 2 | 952/999 | 1.05 (0.78–1.42) | 0.559 | 0 | 1.41 (0.92–2.15) | 0.216 | 34.6 | 1.12 (0.85–1.49) | 0.673 | 0 | 1.30 (0.90–1.87) | 0.184 | 43.4 |
| Smokers | 2 | 952/999 | 2.22 (1.69–2.92) | 0.01 | 83.1 | 5.05 (3.22–7.92) | 0.045 | 75.0 | 2.57 (0.98–3.33) | 0.001 | 83.4 | 2.83 (1.86–4.31) | 0.583 | 0 |
PB – population-based, HB – hospital-based
Figure 1.
Study specific and summary odds ratios with 95% confidence intervals describing the association of NQO1 C609T polymorphism with risk of colorectal cancer. The NQO1 C609T polymorphism was associated with a modestly increased risk of colorectal cancer in heterozygous (CT vs. CC; A), homozygous (CT/TT vs. CC; B), dominant (CT/TT vs. CC; C) and recessive models (TT vs. CT/CC; D)
Subgroup analyses and sensitivity analyses
Ethnicity
Subgroup analysis by ethnicity showed that the association was significant in both Caucasians (CT vs. CC: ORs = 1.23, 95% CIs: 1.03–1.39; TT vs. CC: ORs = 1.20, 95% CIs: 1.01–1.47; CT/TT vs. CC: ORs = 1.14, 95% CIs: 1.00–1.29; TT vs. CT/CC: ORs = 1.38, 95% CIs: 1.01–1.79) and Asians (CT vs. CC: ORs = 1.51, 95% CIs: 1.08–2.12; TT vs. CC: ORs = 1.84, 95% CIs: 1.16–2.89; CT/TT vs. CC: ORs = 1.40, 95% CIs: 1.11–2.34; TT vs. CT/CC: ORs = 1.46, 95% CIs: 1.02–2.10; Table II).
Sources of controls
Additional stratification by sources of controls showed significant associations between the NQO1 C609T polymorphism and CRC risk in these four genetic models for hospital-based subgroups, but not for population-based controls.
Study quality
After an analysis according to the quality score of studies, we found a significantly positive association of CRC for the four genetic models in the studies with a high quality score (≥ 7) (summary ORs (95% CIs), 1.41 (1.00–1.99) for TC vs. CC; 1.65 (1.09–2.51) for TT vs. CC; 1.29 (1.01–1.63) for CT + TT vs. CC; 1.47 (1.08–2.00) for TT vs. CT + CC), whereas no significant associations were found in these four genetic models based on a meta-analysis of low quality studies.
Smoking status
To determine the effect of smoking on the association between NQO1 C609T polymorphism and the risk of CRC, we performed a stratified analysis by tobacco smoking. Based on two studies which reported these data [9, 15], we found a modified effect of smoking on this association: a positive association was found in smokers, but not in non-smokers (Table II). In addition, compared with nonsmokers carrying the CC genotype, smokers carrying CT/TT genotypes had a significantly increased risk of colorectal cancer (summary OR = 2.74, 95% CI = 2.08–3.62).
Sensitivity analysis
We also conducted a sensitivity analysis by omitting one study at a time and calculating the pooled ORs for the remainder of studies, and found that there were no changes in the direction of effect when any one study was excluded. This analysis confirmed the stability of the positive association between the NQO1 C609T polymorphism and the risk of CRC in these four genetic models.
Publication bias
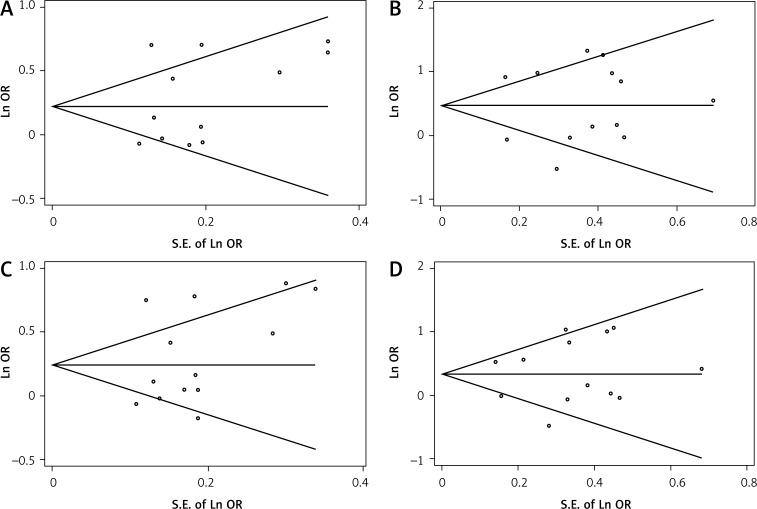
The shape of the funnel plots did not reveal any evidence of the obvious asymmetry for all genetic models in the overall meta-analysis. Begg's test and Egger's test did not reveal any significant evidence of publication bias for any of the genetic models (CT vs. CC: P Begg = 0.340 and P Egger = 0.824 (Figure 2A); TT vs. CC: P Begg = 0.855 and P Egger = 0.380 (Figure 2B); CT/TT vs. CC: P Begg = 0.428 and P Egger = 0.771 (Figure 2C); and TT vs. CT/CC: P Begg = 0.855 and P Egger = 0.434 (Figure 2D).
Figure 2.
Funnel plot analysis to detect publication bias. No evident publication bias was detected for heterozygous (CT vs. CC; A), homozygous (CT/TT vs. CC; B), dominant (CT/TT vs. CC; C) or recessive models (TT vs. CT/CC; D)
Discussion
The results of this meta-analysis suggested that the NQO1 C609T polymorphism was associated with increased risk of CRC. Statistical significance was shown in the heterozygote, homozygote, dominant and recessive models. Subgroup analyses indicated that the C609T polymorphism was associated with increased risk of CRC in both Asians and Caucasians. In addition, an increased risk of CRC associated with the NQO1 C609T polymorphism was only observed in smokers, but not in nonsmokers.
It has been reported that the NQO1 C609T polymorphism is associated with the risk of many types of malignancies, such as cancers of the stomach [26], breast [27] and esophagus [28]. However, a meta-analysis by Guo et al. [29] involving 7,286 patients and 9,167 controls suggested that the NQO1 C609T polymorphism was not associated with the risk of lung cancer for both Caucasians and Asians. Therefore, different primary sites of cancers may have different risk relationships between NQO1 C609T polymorphism and malignancies. In terms of the association between NQO1 C609T polymorphism and CRC risk, the most recent meta-analysis by Wang et al. [18] included 12 studies involving 4,026 cases and 4,855 controls, and found that the NQO1 C609T polymorphism was positively associated with risk of CRC in both Caucasians and Asians. However, this study did not perform a study quality assessment and did not evaluate the modified role of smoking on this association. In addition, Zhu et al. [19] included 21 case-control studies and found that the NQO1 C609T polymorphism might be associated with a significantly increased risk of upper digestive tract cancer, but not risk of CRC. The further subgroup analysis by ethnicity showed significant associations for colorectal cancer in Caucasians, but not in Asians [19]. The null association between the NQO1 C609T polymorphism and CRC risk in Asians may be due to the limited number of studies included (n = 2 studies). In the current comprehensive meta-analysis, we identified a total of 14 case-control studies with 4,461 CRC patients and 5,474 controls and found a significant association between the NQO1 C609T polymorphism and CRC risk in both Caucasians (n = 7 studies) and Asians (n = 7 studies). Moreover, we found that smoking had a modified role in this association: the increased risk of CRC associated with NQO1 C609T polymorphism was only observed in smokers, but not in nonsmokers.
In terms of the biological functions of the NQO1 protein and gene, the results of our meta-analysis are biologically plausible and reliable. In vitro studies have indicated that NQO1 C609T polymorphism is associated with decreased enzyme activity of NQO1 protein: homozygous variant cells have little or no NQO1 enzyme activity, and heterozygous variant cells have approximately half of the NQO1 enzyme activity of wild-type cells [30]. Wild-type NQO1 has been shown to sensitize cells to undergo apoptosis and also to stabilize the p53 tumor suppressor protein, whereas heterozygous variant does not [31, 32]. Furthermore, in vivo studies have suggested that wild-type NQO1 could inhibit colon carcinogenesis at both the initiation and post-initiation stages by directly detoxifying colon carcinogens and tumor promoters [33]. Alternately, NQO1 knockout mice were reported to exhibit a significantly increased prevalence rate of skin carcinogenesis induced by 7,12-dimethylbenz(a)anthracene (DMBA) and benzo(a)pyrene (BP) [34].
Tobacco smoking is a source of carcinogenic compounds such as nitrosamines, benzene, BP, and vinyl chloride [35], which is an established risk factor in developing CRC [36]. In the current meta-analysis, we observed an interaction between smoking and NQO1 C609T polymorphism in cancer risk: the risk with NQO1 C609T polymorphism was elevated in smokers, but absent in nonsmokers. Similar interactions were also observed in previous studies on colorectal adenoma risk, though these did not reach statistical significance due to the small sample size [37, 38]. Hou et al. [37] recruited 725 Caucasian cases with advanced colorectal adenoma and 729 gender- and ethnicity-matched controls, and found that subjects carrying NQO1 Ser187 alleles were weakly associated with risk of colorectal adenoma; however, a higher risk of CRC was observed among recent (including current) (OR = 2.2, 95% CI: 1.5–3.2) and heavy cigarette smokers (> 20 cigarettes/day) (OR = 1.8, 95% CI: 1.2–2.7) compared with non-smokers who did not carry this variant. This genotype was unassociated with risk in non-smokers [37]. Similar results were also reported in the UKFSS study [38], which recruited 946 polyp-free controls and 894 colorectal adenoma cases. The mechanism by which tobacco smoking has a modified effect on the association between NQO1 C609T variant and colorectal cancer risk may involve decreased NQO1 enzyme activity, leading to increased levels of BP metabolites in smokers and thus leading to an increased risk of CRC. Animal and human studies have suggested that dietary BP increased the risk of CRC [39, 40].
Our meta-analysis has some advantages. First, this is to date the largest analysis exploring the association of the NQO1 gene C609T polymorphism with CRC risk, which included 14 studies; thus, the results of our study are more reliable. Second, the present meta-analysis comprehensively evaluated the quality score of the included studies according to the quality score criteria, and restricting analyses to high quality studies generated similar findings. Third, our meta-analysis explored the interactive role of tobacco smoking in this association, and we found a positive association between NQO1 C609T polymorphism and the risk of CRC in smokers, but not in non-smokers.
Our meta-analysis has some limitations that may affect the interpretation of the results. First, the current meta-analysis included only 14 retrospective studies. Because of the limited data, we could not determine a relationship between NQO1 C609T polymorphism and the risk of CRC in Africans and in Latin Americans. Second, high between-study heterogeneity was detected, which would throw some doubt on the reliability of the summary OR estimates. Significant heterogeneity may exist in terms of ethnicity, sources of controls and study quality. By using sub-group analyses with random-effect models, we found the high heterogeneity may exist among Asian studies and hospital-based case-control studies; thus, our results should be interpreted with caution and further larger studies are needed. Third, an effect of the modified role of smoking in the association between the NQO1 C609T polymorphism and the risk of CRC is likely to be present. Unfortunately, the information of smoking status was unavailable in the majority of the included studies, although we made our best efforts to contact the authors of primary studies. Fourth, as with all meta-analyses, publication bias might have occurred, although an appropriate search strategy was used to identify eligible studies. Because our analyses were based entirely on published studies from English- and Chinese-language journals, it is possible that some relevant unpublished studies, which may have met the inclusion criteria, were missed. However, neither the funnel plots nor standard statistical tests indicated remarkable publication bias in the meta-analysis.
In conclusion, this meta-analysis indicates that the NQO1 C609T polymorphism is significantly associated with risk of CRC in both Caucasians and Asians, and in smokers but not in nonsmokers. However, because of the high heterogeneity and potential bias, the results should be interpreted with caution, and further well-designed studies with large sample sizes are warranted to confirm our findings.
Acknowledgments
The authors thank Dr Peng Xian'e for her kindness and willingness to provide additional information on the article.
References
- 1.Jemal A, Thun MJ, Ries LA, et al. Annual report to the nation on the status of cancer, 1975-2005, featuring trends in lung cancer, tobacco use, and tobacco control. J Natl Cancer Inst. 2008;100:1672–94. doi: 10.1093/jnci/djn389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klimczak A, Kempinska-Miroslawska B, Mik M, Dziki L, Dziki A. Incidence of colorectal cancer in Poland in 1999-2008. Arch Med Sci. 2011;7:673–8. doi: 10.5114/aoms.2011.24138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vrieling A, Kampman E. The role of body mass index, physical activity, and diet in colorectal cancer recurrence and survival: a review of the literature. Am J Clin Nutr. 2010;92:471–90. doi: 10.3945/ajcn.2010.29005. [DOI] [PubMed] [Google Scholar]
- 4.Smolinska K, Paluszkiewicz P. Risk of colorectal cancer in relation to frequency and total amount of red meat consumption. Systematic review and meta-analysis. Arch Med Sci. 2010;6:605–10. doi: 10.5114/aoms.2010.14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X, Shan Y, Xue B. Int7G24A polymorphism (rs334354) and cancer risk. Arch Med Sci. 2013;9:3–7. doi: 10.5114/aoms.2013.33341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross D, Kepa JK, Winski SL, Beall HD, Anwar A, Siegel D. NAD(P)H:quinone oxidoreductase 1 (NQO1): chemoprotection, bioactivation, gene regulation and genetic polymorphisms. Chem Biol Interact. 2000;129:77–97. doi: 10.1016/s0009-2797(00)00199-x. [DOI] [PubMed] [Google Scholar]
- 7.Siegel D, Gustafson DL, Dehn DL, et al. NAD(P)H:quinone oxidoreductase 1: role as a superoxide scavenger. Mol Pharmacol. 2004;65:1238–47. doi: 10.1124/mol.65.5.1238. [DOI] [PubMed] [Google Scholar]
- 8.Nebert DW, Roe AL, Vandale SE, Bingham E, Oakley GG. NAD(P)H:quinone oxidoreductase (NQO1) polymorphism, exposure to benzene, and predisposition to disease: a HuGE review. Genet Med. 2002;4:62–70. doi: 10.1097/00125817-200203000-00003. [DOI] [PubMed] [Google Scholar]
- 9.Begleiter A, Hewitt D, Maksymiuk AW, Ross DA, Bird RP. A NAD(P)H:quinone oxidoreductase 1 polymorphism is a risk factor for human colon cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:2422–6. doi: 10.1158/1055-9965.EPI-06-0661. [DOI] [PubMed] [Google Scholar]
- 10.Hlavata I, Vrana D, Smerhovsky Z, et al. Association between exposure-relevant polymorphisms in CYP1B1, EPHX1, NQO1, GSTM1, GSTP1 and GSTT1 and risk of colorectal cancer in a Czech population. Oncol Rep. 2010;24:1347–53. doi: 10.3892/or_00000992. [DOI] [PubMed] [Google Scholar]
- 11.Nisa H, Kono S, Yin G, et al. Cigarette smoking, genetic polymorphisms and colorectal cancer risk: the Fukuoka Colorectal Cancer Study. BMC Cancer. 2010;10:274. doi: 10.1186/1471-2407-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sameer AS, Shah ZA, Syeed N, Rasool R, Afroze D, Siddiqi MA. NAD(P)H:quinone oxidoreductase 1 (NQO1) Pro187Ser polymorphism and colorectal cancer predisposition in the ethnic Kashmiri population. Asian Pac J Cancer Prev. 2010;11:209–13. [PubMed] [Google Scholar]
- 13.van der Logt EM, Bergevoet SM, Roelofs HM, et al. Role of epoxide hydrolase, NAD(P)H:quinone oxidoreductase, cytochrome P450 2E1 or alcohol dehydrogenase genotypes in susceptibility to colorectal cancer. Mutat Res. 2006;593:39–49. doi: 10.1016/j.mrfmmm.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 14.Sachse C, Smith G, Wilkie MJ, et al. A pharmacogenetic study to investigate the role of dietary carcinogens in the etiology of colorectal cancer. Carcinogenesis. 2002;23:1839–49. doi: 10.1093/carcin/23.11.1839. [DOI] [PubMed] [Google Scholar]
- 15.Peng XE, Jiang YY, Shi XS, Hu ZJ. NQO1 609C > T polymorphism interaction with tobacco smoking and alcohol drinking increases colorectal cancer risk in a Chinese population. Gene. 2013;521:105–10. doi: 10.1016/j.gene.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 16.Ding R, Lin S, Chen D. Association of NQO1 rs1800566 polymorphism and the risk of colorectal cancer: a meta-analysis. Int J Colorectal Dis. 2012;27:885–92. doi: 10.1007/s00384-011-1396-0. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Lin Y, Zhang R, Huang ZJ, Pan XG. Contribution of NAD(P)H quinone oxidoreductase 1 (NQO1) Pro187Ser polymorphism and risk of colorectal adenoma and colorectal cancer in Caucasians: a meta-analysis. Arch Med Res. 2012;43:58–66. doi: 10.1016/j.arcmed.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Zhang G, Luo Y. Association between NQO1 C609T polymorphism and colorectal cancer risk. Tumor Biol. 2013;34:4027–32. doi: 10.1007/s13277-013-0993-7. [DOI] [PubMed] [Google Scholar]
- 19.Zhu CL, Huang Q, Liu CH, Lin XS, Xie F, Shao F. NAD(P)H: quinone oxidoreductase 1 (NQO1) C609T gene polymorphism association with digestive tract cancer: a meta-analysis. Asian Pac J Cancer Prev. 2013;14:2349–54. doi: 10.7314/apjcp.2013.14.4.2349. [DOI] [PubMed] [Google Scholar]
- 20.Thakkinstian A, McKay GJ, McEvoy M, et al. Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: a HuGE review and meta-analysis. Am J Epidemiol. 2011;173:1365–79. doi: 10.1093/aje/kwr025. [DOI] [PubMed] [Google Scholar]
- 21.Yu H, Liu H, Wang LE, Wei Q. A functional NQO1 609C > T polymorphism and risk of gastrointestinal cancers: a meta-analysis. PLoS One. 2012;7:e30566. doi: 10.1371/journal.pone.0030566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 23.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lafuente MJ, Casterad X, Trias M, et al. NAD(P)H:quinone oxidoreductase-dependent risk for colorectal cancer and its association with the presence of K-ras mutations in tumors. Carcinogenesis. 2000;21:1813–9. doi: 10.1093/carcin/21.10.1813. [DOI] [PubMed] [Google Scholar]
- 25.Su XL, Yan MR, Yang L. NQO1 C609T polymorphism correlated to colon cancer risk in farmers from western region of Inner Mongolia. Chin J Cancer Res. 2012;24:317–22. doi: 10.3978/j.issn.1000-9604.2012.08.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Wang ZT, Zhong J. Meta-analysis demonstrates that the NAD(P)H: quinone oxidoreductase 1 (NQO1) gene 609 C > T polymorphism is associated with increased gastric cancer risk in Asians. Genet Mol Res. 2012;11:2328–37. doi: 10.4238/2012.August.13.6. [DOI] [PubMed] [Google Scholar]
- 27.Yuan W, Xu L, Chen W, et al. Evidence on the association between NQO1 Pro187Ser polymorphism and breast cancer risk in the current studies: a meta-analysis. Breast Cancer Res Treat. 2011;125:467–72. doi: 10.1007/s10549-010-0966-0. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z, Hu J, Zhong J. Meta-analysis of the NAD(P)H: quinine oxidoreductase 1 gene 609 C > T polymorphism with esophageal cancer risk. DNA Cell Biol. 2012;31:560–7. doi: 10.1089/dna.2011.1332. [DOI] [PubMed] [Google Scholar]
- 29.Guo S, Gao M, Li X, et al. Lack of Association between NADPH quinone oxidoreductase 1 (NQO1) gene C609T polymorphism and lung cancer: a case-control study and a meta-analysis. PLoS One. 2012;7:e47939. doi: 10.1371/journal.pone.0047939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Begleiterabc A, Leith MK, Doherty GP, Digbya TJ, Pan S. Factors influencing the induction of DT-diaphorase activity by 1,2-dithiole-3-thione in human tumor cell lines. Biochem Pharmacol. 2001;61:955–64. doi: 10.1016/s0006-2952(01)00537-8. [DOI] [PubMed] [Google Scholar]
- 31.Siemankowski LM, Morreale J, Butts BD, Briehl MM. Increased tumor necrosis factor-alpha sensitivity of MCF-7 cells transfected with NAD(P)H:quinone reductase. Cancer Res. 2000;60:3638–44. [PubMed] [Google Scholar]
- 32.Asher G, Lotem J, Kama R, Sachs L, Shaul Y. NQO1 stabilizes p53 through a distinct pathway. Proc Natl Acad Sci U S A. 2002;99:3099–104. doi: 10.1073/pnas.052706799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Begleiter A, Sivananthan K, Lefas GM, Maksymiuk AW, Bird RP. Inhibition of colon carcinogenesis by post-initiation induction of NQO1 in Sprague-Dawley rats. Oncol Rep. 2009;21:1559–65. doi: 10.3892/or_00000388. [DOI] [PubMed] [Google Scholar]
- 34.Long DJ, 2nd, Waikel RL, Wang XJ, Roop DR, Jaiswal AK. NAD(P)H:quinone oxidoreductase 1 deficiency and increased susceptibility to 7,12-dimethylbenz[a]-anthracene-induced carcinogenesis in mouse skin. J Natl Cancer Inst. 2001;93:1166–70. doi: 10.1093/jnci/93.15.1166. [DOI] [PubMed] [Google Scholar]
- 35.Nioi P, Hayes JD. Contribution of NAD(P)H:quinone oxidoreductase 1 to protection against carcinogenesis, and regulation of its gene by the Nrf2 basic-region leucine zipper and the arylhydrocarbon receptor basic helix-loop-helix transcription factors. Mutat Res. 2004;555:149–71. doi: 10.1016/j.mrfmmm.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 36.Botteri E, Iodice S, Bagnardi V, Raimondi S, Lowenfels AB, Maisonneuve P. Smoking and colorectal cancer: a meta-analysis. JAMA. 2008;300:2765–78. doi: 10.1001/jama.2008.839. [DOI] [PubMed] [Google Scholar]
- 37.Hou L, Chatterjee N, Huang WY, et al. CYP1A1 Val462 and NQO1 Ser187 polymorphisms, cigarette use, and risk for colorectal adenoma. Carcinogenesis. 2005;26:1122–8. doi: 10.1093/carcin/bgi054. [DOI] [PubMed] [Google Scholar]
- 38.Mitrou PN, Watson MA, Loktionov AS, et al. Role of NQO1C609T and EPHX1 gene polymorphisms in the association of smoking and alcohol with sporadic distal colorectal adenomas: results from the UKFSS Study. Carcinogenesis. 2007;28:875–82. doi: 10.1093/carcin/bgl194. [DOI] [PubMed] [Google Scholar]
- 39.Hakura A, Seki Y, Sonoda J, et al. Rapid induction of colonic adenocarcinoma in mice exposed to benzo[a] pyrene and dextran sulfate sodium. Food Chem Toxicol. 2011;49:2997–3001. doi: 10.1016/j.fct.2011.07.057. [DOI] [PubMed] [Google Scholar]
- 40.Sinha R, Kulldorff M, Gunter MJ, Strickland P, Rothman N. Dietary benzo[a]pyrene intake and risk of colorectal adenoma. Cancer Epidemiol Biomarkers Prev. 2005;14:2030–4. doi: 10.1158/1055-9965.EPI-04-0854. [DOI] [PubMed] [Google Scholar]
- 41.Harth V, Donat S, Ko Y, Abel J, Vetter H, Bruning T. NAD(P)H quinone oxidoreductase 1 codon 609 polymorphism and its association to colorectal cancer. Arch Toxicol. 2000;73:528–31. doi: 10.1007/s002040050004. [DOI] [PubMed] [Google Scholar]
- 42.Mitrou P, Watson M, Bingham S, Stebbings WS, Speakman CT, Loktionov A. NQO1 and mEH exon 4 (mEH4) gene polymorphisms, smoking and colorectal cancer risk. IARC Sci Publ. 2002;156:495–7. [PubMed] [Google Scholar]
- 43.Hamajima N, Matsuo K, Iwata H, et al. NAD(P)H: quinone oxidoreductase 1 (NQO1) C609T polymorphism and the risk of eight cancers for Japanese. Int J Clin Oncol. 2002;7:103–8. doi: 10.1007/s101470200013. [DOI] [PubMed] [Google Scholar]
- 44.Enyong D, Zhenxia L, Jieping S, Yaqin Y, Jing Z. NAD(P)H: quinone oxidoreductase gene polymorphism and its association with colorectal cancer. Chinese J Clin Oncol. 2004;31:89–91. [Google Scholar]
- 45.Peng X, Jiang Y, Shi X, et al. Relationship between NQO1 C609T gene polymorphism and risk of colorectal cancer. Chin J Public Health. 2010;26:415–6. [Google Scholar]