Abstract
Re-sequencing of target genes is a highly effective approach for identifying mutations in cancers. Mutations, including indels (insertions, deletions, and the combination of the two), play important roles in carcinogenesis. Combining genomic DNA capture using high-density oligonucleotide microarrays (NimbleGen, Inc.) with next-generation high-throughput sequencing, we identified approximately 1600 indels for colorectal cancers in the Chinese population. Among them, 5 indels were localized to exonic regions of genes, including the TFDP1 (transcription factor Dp-1) gene. TFDP1 is an important transcription factor that coordinates with E2F proteins, thereby promoting transcription of E2F target genes and regulating the cell cycle and differentiation. We report here the identification of a recurrent frame-shift indel mutation (named indel84) in the TFDP1 gene in colorectal cancers by next-generation sequencing. We found in a validation set that TFDP1 indel84 is present in 70% of colorectal cancer (CRC) tissues. Wild-type TFDP1 encodes a protein of 410 amino acids with a potential DNA binding site at its N-terminal followed by several functional protein domains. The TFDP1 indel cDNA would generate an alternative TFDP1 protein missing the first 120 amino acids and potentially affecting the DNA binding domain. We further demonstrated that the TFDP1 indel84 mutation generated a gain-of-function phenotype by increasing cell proliferation, migration, and invasion of CRC cells. Our study identified a key molecular event for CRC that might have great diagnostic and therapeutic potentials.
Introduction
Colorectal cancer is one of three most common cancers worldwide, and the incidence has increased annually (Siegel et al., 2013). Many Asian countries, including China, have experienced a 2- to 4-fold increase in the incidence of colorectal cancer during the past few decades (Chen et al., 2013, Hyodo et al., 2010, Sung et al., 2005, Wu et al., 2012).
In recent years, the improved sequencing capacities of next-generation sequencing (NGS) technologies have allowed detailed analysis of cancer genomes at the genomic level (Keller et al., 2011, Sulonen et al., 2011, Wheeler et al., 2008). For colorectal cancers, focused deep sequencing using NGS has been employed as a cost-effective approach to identify common mutations. For example, Pritchard et al. (2012) developed Coloseq to identify pathogenic mutations in MLH1, MSH2, MSH6, PMS2, EPCAM, APC, and MUTYH genes for Lynch syndrome (hereditary nonpolyposis colon cancer) and adenomatous polyposis syndromes. Lipson et al. (2012) analyzed the coding regions of 145 cancer-relevant genes (genes that are associated with cancer-related pathways, targeted therapy, or prognosis) and 37 introns from 14 genes that are frequently rearranged in cancer from 40 formalin-fixed paraffin-embedded (FFPE) specimens of colorectal cancer (CRC) using DNA capture and NGS. The authors identified 125 alterations in 21 genes, including a gene fusion between C2orf44 and ALK (Lipson et al., 2012). We took a similar approach to sequencing and captured whole genomic regions (including promoters, exons, and introns) of 30 important CRC genes, including WNT pathway genes, colon-enriched and CRC-overexpressed genes, and important transcription factors (Supplementary Tables S1 and S2). We have previously summarized the single nucleotide polymorphisms (SNPs) identified in the analysis and reported the association of rs3106189 at the 5′ UTR of TAPBP [TAP binding protein (tapasin)] and rs1052918 at the 3′ UTR of TCF3 (transcription factor 3) with the overall survival of colorectal cancer patients (Shao et al., 2013). We focused in this report on the identification of the indels.
Indels are defined as insertions, deletions, and the combination of both insertions and deletions in DNA sequences (Kondrashov and Rogozin, 2004, Ogurtsov et al., 2004). Among indels, single-nucleotide indels are the most frequent (Iengar, 2012). Many NGS analyses have identified indels as mutations in cancers (Clark et al., 2010), and many have even pinpointed some of the indels as driving mutations using cell-population genetic analysis, as Tao et al. did for hepatocellular carcinoma (Tao et al., 2011). We initially used a pool strategy, similar to the work by Ruark et al. (2012) in their large-scale sequencing of 507 genes for breast and ovarian cancers, to sequence DNA from 30 pairs of CRCs and normal adjacent tissues and then validate the sequences in a new cohort of samples. We report here, for the first time, the identification of an indel, which we named indel84, in the TFDP1 gene (transcription factor Dp-1). TFDP1 is a transcription factor that coordinates with E2F to form E2F/DP complexes, and this complex activates or represses the transcription of E2F target genes (Girling et al., 1993). E2F family members play key roles in the life and death decisions of cells (Iaquinta and Lees, 2007, Polager and Ginsberg, 2009). TFDP1 also interacts with pRB and p53 to regulate the cell cycle and apoptosis (Buchmann et al., 1998, Sorensen et al., 1996). Our laboratory is interested in the role of transcriptional factors in cancer, and the association of TFDP1 with key cellular proteins such as the E2F family transcriptional factors and pRB and p53. Iaquinta and Lees (2007) and Polager and Ginsberg (2009) suggested that TFDP1 is likely a master regulator. Therefore, in this study, we focused on the analysis of indel84 in the TFDP1 gene in colorectal cancer (CRC).
Methods
Ethics statement
There were 60 clinical samples from 30 adult patients involved in this study. Informed written consent was obtained from each patient, and the ethics committee of the Second Affiliated Hospital, College of Medicine, Zhejiang University, approved all aspects of the study. The clinical information of the clinical samples is provided in Supplementary Table S3.
Clinical samples and genomic DNA isolation
The clinical samples and the human colorectal cell lines DLD1, SW480, SW620, RKO, and LoVo were provided by the Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China. The clinical information of the clinical samples is provided in Supplementary Table 3. These cell lines were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The human prostate cell line CL1 and the human glioma cell line U251 were also purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). DLD1, SW480, RKO, and CL1 cells were cultured in RPMI 1640 medium (Invitrogen, Grand Island, NY). SW620 cells were cultured in Leibovitz's L-15 Medium, LoVo cells were cultured in Ham's F12K Medium, and U251 cells were cultured in DMEM medium (Invitrogen). The culture medium was mixed with 10% fetal bovine serum and penicillin/streptomycin (100 μg/mL). Cells were cultured at 37°C and 5% CO2. Genomic DNA was isolated using the E.Z.N.A. ® Tissue DNA Kit (Omega Bio-tek, Norcross, GA) according to the manufacturer's protocol.
NimbleGen capturing and Illumina sequencing
The DNA samples were isolated using the DNeasy Blood and Tissue Kit (Qiagen Inc., Valencia, CA) according to the manufacturer's protocol. Genomic DNA from 30 colon cancer patients was mixed in equal ratios, and the DNA pools were captured on a custom NimbleGen 2.1 M array (Roche/NimbleGen, Madison, WI) that contain probes for the genomic regions of 30 genes, including the following: ETS2, ETV4, FLI1, FUBP1, HMGA1, HSF1, MSX2, SMARCA4, SOX4, STAT5A, TCF3, TFDP1, TRIM28, YBX1, ZNF3, GTF2A2, JUNB, NR3C1, PPARA, VAV1, Axin, LKB1, MDM2, HIPK2, Tip60, Srbc, Hypo2, Tankyrase, Ring25, and APC. The target-enriched DNA was eluted and sequenced as reported previously (Cheng et al., 2010).
Indel analysis
BWA (Li and Durbin, 2009) was used to map the raw reads (one gap allowed) to the genomic regions covering the captured regions, which was referred to as the chip_genes.fasta reference. Samtools (Li et al., 2009) was used to remove the PCR duplicates. The VarScan (Koboldt et al., 2009) was used to call indels (with min coverage 8, p<0.05). Finally, indels were annotated by ANNOVAR (Wang et al., 2010).
Confirmation of indels
To confirm indels identified by the NimbleGen sequencing techniques, we performed allele-specific PCR (AS-PCR) (Wangkumhang et al., 2007, Ye et al., 2001) in a separate cohort of 30 individual colorectal cancer samples and 30 normal adjacent tissues. We used the tetra-primer amplification refractory mutation system-polymerase chain reaction (ARMS-PCR) procedure to confirm the existence of indels. ARMS-PCR is an efficient procedure for genotyping single nucleotide polymorphisms (Wangkumhang et al., 2007, Ye et al., 2001). Each PCR reaction was performed in a total volume of 25 μL containing 10 ng DNA template, 5 U/μL TaKaRa Ex Taq, 1 X TAKARA Ex Taq Buffer (Mg2+ Plus), 1 μL of dNTP Mixture (2.5 mM), and 1 μL of each primer (20 μM). The PCR results were analyzed by 1.5% agarose gel electrophoresis. The outer primers for generating the PCR product at the TFDP1 locus were as follows: outer forward (outer F), 5′-ACCCTCGCCGTGTGGGAGGGGA-3′, and outer reverse (outer R), 5′-TGGGCGGTGCCCAGCCCGACT-3′. The allele-specific inner PCR primers were as follows: inner forward (inner F), 5′-ACCCTGGTGGTAGGAAGCCCACACACCCTCA-3′ and inner reverse (inner R), 5′-CTGGTTCTGAGAGGCAAAGTGAGTGCTGGAGT-3′ (Fig. 1B). Five randomly selected AS-PCR-positive samples for each indel were sequenced by Sanger sequencing (Sangon co. Ltd., Shanghai, China).
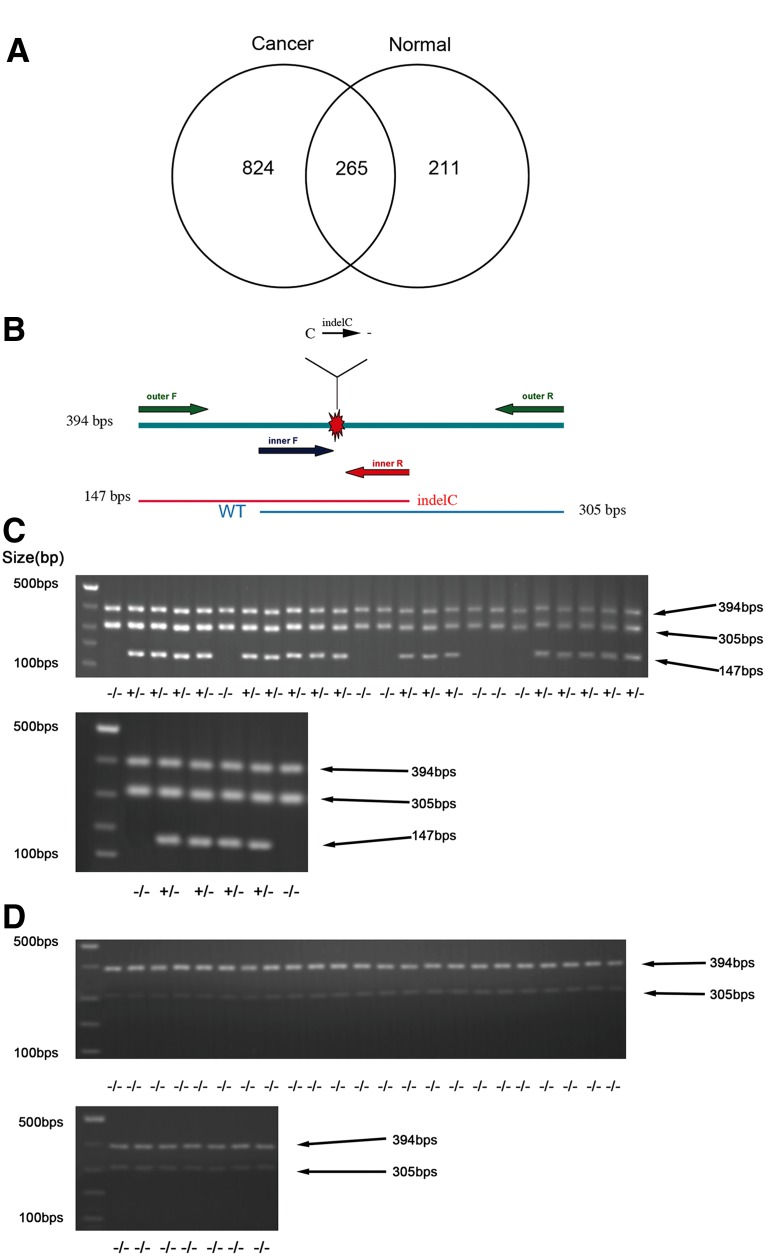
FIG. 1.
The number of indels found in cancer or normal tissues. (A) A Venn diagram showing indels found in cancer tissue, normal tissue, or in both tissues. (B) Schematic illustration of the indel and the inner and outer primers used in the ARMS-PCR. (C) ARMS-PCR data of the TFDP1 indel of CRC cancer tissues demonstrating the detection of the TFDP1 indel mutations. * indicates the PCR product of the outer primers; ** indicates the wild-type TFDP1 PCR product; *** indicates the PCR product of the TFDP1 indel. (D) ARMS-PCR data of the TFDP1 indel of normal adjacent tissues demonstrating no TFDP1 indel mutation detected. * and ** annotations are the same as above.
Western blot analysis
Cells were grown to 70% confluency, washed with cold PBS buffer, and lysed on ice for 30 min in RIPA buffer [25 mmol/L Tris-HCl (pH 7.6), 150 mmol/L NaCl, 0.01 g/mL NP-40, 0.01 g/mL sodium deoxycholate, 1 g/L sodium dodecyl sulfate (SDS)] containing a protease inhibitor cocktail (Pierce, Rockford, IL). Protein concentrations were estimated using Pierce BCA protein Assay Kit (Thermo Scientific). Approximately 20 mg of protein was denatured at 95°C with loading buffer for 5 min and separated by electrophoresis in 12% SDS-PAGE gels. The proteins were transferred onto a PVDF membrane and blocked 2 h in 5% skim milk in TBST buffer. Primary antibodies were diluted and incubated with the PVDF membrane for 3 h at 37°C. The primary antibodies and their dilutions are as follows: anti-TFDP1 (5151-1, Epitomics), 1:1000; anti-APC (1701-1, Epitomics), 1:2000; anti-TNKS2 (ab103781, Abcam), 1:1000; and anti-GAPDH (ab9483, Abcam), 1:5000]. After washing, the membranes were incubated for 2 h at 37°C with an HRP-linked secondary antibody [anti-rabbit (1:5000)], and the signals were detected using ECL reagents (Thermo Scientific).
RNA isolation and Q-PCR
Total RNA was extracted using the protocol for Trizol reagent (Invitrogen), followed by DNase treatment. One microgram of RNA was used for cDNA synthesis using M-MLV Reverse Transcriptase (Promega) in a 20 μL volume, and 0.2 μL was used for Q-PCR. Quantitative real-time PCR reaction was performed in a final volume of 25 μL containing 1 μL of cDNA (1:10 dilution) and 400 nM of primers and SYBRH Pre-mix Ex Taq TM (Takara), and then amplified by Real Time PCR (Bio-RAD CFX96 Rea-Time System). The primers are as follows: QPCR Forward 5′-AAACACCCTGGTGGTAGGAA-3′ and QPCR Reverse 5′-ACGATGATGGGTGAGTGTGA-3′. All samples were amplified in triplicate using the following cycle conditions: 95°C for 2 min, followed by 38 cycles of 95°C for 20 sec,, 60°C for 30 sec, and 72°C for 30 sec. Final RNA levels were normalized to the GAPDH levels.
Plasmid construction and transfection
The full-length human InDel-mutation and wildtype TFDP1 cDNA were generated using gene-on-demand synthesis technology (http://www.genscript.com/gene_synthesis.html) (Genscript, Nanjing, China). The constructs with Kozak sequences and start and stop codons were ligated into pcDNA3.1(+) (Invitrogen). The constructs of pcDNA3.1(+)-InDel TFDP1, pcDNA3.1(+)-wild type TFDP1, and the empty vector control were transfected into SW480 cells by EndofectinTM-Plus (GeneCopoeia, Rockville, MD) according to the manufacturer's protocol. Antibiotic selection medium contained 100 μg/mL ampicillin. The surviving colonies were then subcloned by limiting dilution and amplified to establish sublines overexpressing Indel84 TFDP1, wild type TFDP1, or empty vector controls. Approximately 48 h after transfection, the expression of TFDP1 was detected by Q-PCR and Western blot analysis.
Small interfering RNA transfection
TFDP1 siRNA [TFDP1 siRNA (Q000007027, RiboBio, Guangzhou, China) and negative control siRNA (siN05815122147, RiboBio)] were used for transient knockdown of TFDP1. SW480 cells were cultured in 6-well plates overnight and transfected with siRNAs at a final concentration of 100 nM using Lipofectamine (Invitrogen) according to the manufacturer's instructions. Approximately 48 h after transfection, cells were harvested for Q-PCR and Western blot analysis.
Cell proliferation assays
Approximately 4000 cells of each sample were seeded in 96-well plates in five replicates with 200 μL complete culture medium. After 72-h incubation, cells were counted using the MTT assay (Millipore) according to the manufacturer's protocol.
Wound healing assay
One million SW480 cells were seeded on 6-well plates and incubated until confluent. An artificial homogeneous wound was created in the monolayer cells with a sterile plastic micropipette tip in each well at the same time. After wounding, the debris was removed by washing the cells with PBS buffer, and the cells were grown in normal medium. The healing process was examined 48 h later and recorded with a Nikon Eclipse TS100 microscope (100X).
Cell migration and invasion assays
Cell migration and invasion assays were performed using transwell assays (Corning). For the migration assay, 20,000 cells in 200 μL serum-free RPMI 1640 medium were plated in the upper compartments, whereas the lower compartments were loaded with 500 μL medium containing 10% FBS. After incubation for 36 h, the cells inside the upper chamber were removed with cotton swabs, and the cells on the lower membrane surface were fixed in methanol and stained with 0.1% crystal violet. Photographs of transwells were taken by a Nikon Eclipse TS100 microscope (under 100X). Six visual fields were randomly selected for counting. The invasion assay was performed using transwells that were preloaded with Matrigel (Sigma-Aldrich) on the upper surface. Invading cells were counted as described in the migration assays 60 h after transfection.
Results
Identification of indels in colorectal cancer with targeted genomic sequencing
We selected genes in the WNT signaling pathway as well as important transcription factors and colon-specific or enriched genes that are overexpressed in CRC for targeted capture of their genomic regions, including the 10-kb upstream regions, exons, introns, and the 5-kb downstream regions. After capture by NimbleGen, the DNA was subjected to sequencing with the Illumina platform (GAII) with single-end sequencing of 36 nucleotides. In the end, we obtained 22,190,884 and 12,869,275 raw reads for a pool of 30 cancer tissues and a pool of 30 adjacent normal tissues for the captured target regions, respectively. The raw sequencing data were deposited in the NCBI sequence read archive (SRA) under accession number SRX277359. After BWA (Li and Durbin, 2009) mapping (1 mismatch allowed) to the targeted regions on the reference human genomic sequences, we obtained 6,198,574 and 3,671,898 unique reads that mapped to our targeted genes, respectively. A report summarizing the identification of SNPs was recently published (Shao et al., 2013) and we focus here on the identification and characterization of the indels in this article.
We used Samtools (Li et al., 2009) to remove the PCR duplicates and used VarScan (Koboldt et al., 2009) to identify indels with a minimum coverage of eight and a p value <0.05. In the end, we identified 476 indels for the normal adjacent tissue pool and 1089 indels for the cancer pool (Supplementary Tables 1 and 2, Fig. 1A). Two hundred sixty-five indels are common between the cancer and the adjacent tissue pool (Fig. 1A). Comparing these indels with dbSNP 130, 121 indels in the cancer pool and 69 indels in the adjacent tissue pool were previously identified indels. In the end, we identified 968 and 407 novel indels, respectively, for the CRC and normal adjacent tissues.
We used ANNOVAR (Wang et al., 2010) to annotate indels (Supplementary Tables S1 and S2). Most of the indels were identified in intronic or intergenic regions (Table 1). Three indels were identified in exonic regions in both the cancer and the normal pools, respectively. Fourteen indels in the cancer pool and eight indels in the normal pool were identified in the upstream promoter regions of the genes (Table 1), suggesting the advantages of our targeted captured approach compared with exome sequencing, by which only exonic mutations could be identified.
Table 1.
Numbers of Indels Found at Different Genomic Locations in the Cancer and the Normal Tissue Pools after Targeted Capture and Sequencing
| Genomic locations | Cancer | Normal |
|---|---|---|
| Upstream | 14 | 5 |
| 5′ UTR | 4 | 3 |
| Exonic | 3 (TFDP1, HIPK2, TNKS2) | 3 (TNKS2, 2 in APC) |
| Intronic | 823 | 359 |
| 3′ UTR | 44 | 31 |
| Downstream | 19 | 8 |
| Intergenic | 182 | 67 |
Identification of the recurrent indel84 in the TFDP1 gene in colorectal cancers
We are particularly interested in identifying cancer-specific indels in exonic regions and identified three indels in three genes: TFDP1 (transcription factor DP-1), HIPK2 (homeodomain interacting protein kinase 2), and TNKS2 (tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2) (Table 1). The indels of TFDP1 and HIPK2 were found in the cancer tissue pool but not in the normal tissue pool, whereas the indel of TNKS2 was found in both the cancer and the normal tissue pools. To further identify those indels with obvious functional consequences, we used the cell proliferation assay as a quick way to assess the functional consequences of the three indels that we identified above. We generated constructs containing the indels for three genes. From the cell proliferation assays, we found that only the indel of TFDP1 affected cell proliferation, the indels of other two genes did not (data not shown). We have therefore chosen to the indel of TFDP1 for further validation and analysis.
For validation, we used allele-specific PCR (AS-PCR) (Wangkumhang et al., 2007, Ye et al., 2001). All of the indels for these three genes were confirmed by AS-PCR, but we present here only the TFDP1 data as an example. We used a specific type of AS-PCR called tetra-primer amplification refractory mutation system-polymerase chain reaction (ARMS-PCR) to confirm the existence of the indels. ARMS-PCR is an efficient procedure for genotyping single nucleotide polymorphisms (Wangkumhang et al., 2007, Ye et al., 2001). The procedure combines two inner SNP-specific primers and two outer primers in a single reaction and encompasses deliberate mismatches at position -2 from the 3′ end of inner primers. The primers were designed using the following website: http://primer1.soton.ac.uk/primer1.html (Collins and Ke, 2012). The alignments of the primers with the genomic region of TFDP1 are presented shown in Supplementary Fig. S1, and a representative drawing for the primer design is presented in Fig. 1B. The outer primers for generating the PCR product at the TFDP1 locus are as follows: outer forward (outer F), 5′-ACCCTCGCCGTGTGGGAGGGGA-3′; and outer reverse (outer R), 5′-TGGGCGGTGCCCAGCCCGACT-3′. The allele-specific inner PCR primers are as follows: inner forward (inner F), 5′-ACCCTGGTGGTAGGAAGCCCACACACCCTCA-3′; and inner reverse (inner R), 5′-CTGGTTCTGAGAGGCAAAGTGAGTGCTGGAGT-3′ (Fig. 1B and Supplementary Fig. S1).
We performed AS-PCR on a separate cohort of 30 CRC cancer tissues and 30 normal adjacent tissues for the TFDP1 indel mutations. We found that 21 of the 30 CRC tissues (70.0%) contain the TFDP1 indel84 mutation (manifested as PCR bands of 147 bp) but that none of the 30 adjacent tissues contained the mutation (manifested as PCR bands of 305 bp) (Fig. 1C and D), suggesting that the TFDP1 indel84 mutation is a recurrent mutation specific to cancer tissues.
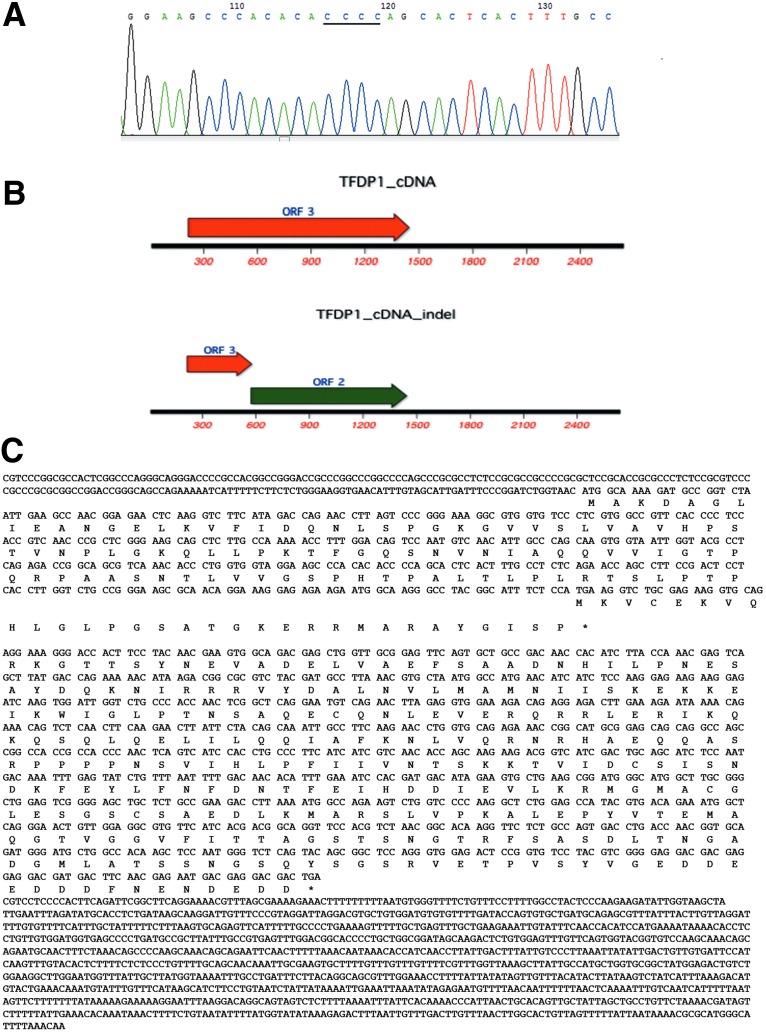
To further confirm the TFDP1 indel84 at the sequence level, we cloned the PCR products from five cancer samples and picked the clone harboring the indel84 by AS-PCR for Sanger sequencing. All five sequencing results confirmed the presence of Indel84 in TFDP1 (Fig. 2A). The TFDP1 Indel84 changes CCCCC to CCCC in the sequence around chr13:114285986-114286018 (GGAAGCCCACACACCCCCAGCACTCACTTTGCC) (underlined sequences are the location of the indel84 deletion). In addition, to confirm that the indel was not a cloning artifact, we performed direct PCR-sequencing of the cancer sample with indel84. From the degenerate sequencing chromatographs after the indel84 position (Supplementary Fig. S2), we notice that the wild-type allele represented the majority of the alleles (70%–80%), whereas the indel84 allele represented approximately 20%–30% of the alleles.
FIG. 2.
(A) Sanger sequencing of the TFDP1 indel. A chromatograph depicting the sequences in the region of chr13:114285986–114286018. The underlined sequences are the locations of the indel. (B) The predicted ORFs of indel84 (labeled TFDP1_cDNA_indel) compared with the ORF of the wild-type TFDP1 cDNA. (C) The alignment of TFDP1 indel84 cDNA and the translation of predicted proteins.
We also further tested the specificity of the ARMS-PCR in identifying the indel using SW480 cells and SW480 cells with the indel or wild-type construct (Supplementary Fig. S3). The indel construct exhibits an additional PCR band of 329 bp resulting from PCR amplification of the outer primer with the inner R primer, whereas the wild-type cells exhibit an additional band of 116 bp resulted from amplification with the outer primer and the inner F primer in additional to the PCR product of 385 bp from the outer primers (Supplementary Fig. S3). This suggests that the ARMS-PCR has the ability to identify indel84.
Based on the Uniprot annotation for TFDP1 (http://www.uniprot.org/uniprot/Q14186), the functional domains for the TFDP1 protein include a potential DNA binding site at amino acid (AA) position 113–195, a potential dimerization domain at AA204–277, a region enhancing the binding of the RB protein to E2F at AA 211–327, a DCB1 domain at 214–246, a DCB2 domain at 259–315, and a DEF box motif at AA 161–195 (Supplementary Table S4). Wild-type TFDP1 encodes a protein of 410 amino acids. However, an open frame search suggested that the TFDP1 indel cDNA could be translated from an internal ATG start codon at nucleotide 572 (Fig. 2B and C), generating an alternative TFDP1 protein only missing the first 120 amino acids and potentially only affecting the DNA binding domain at AA 113–195.
TFDP1 indel84 increased cell proliferation in CRC cells
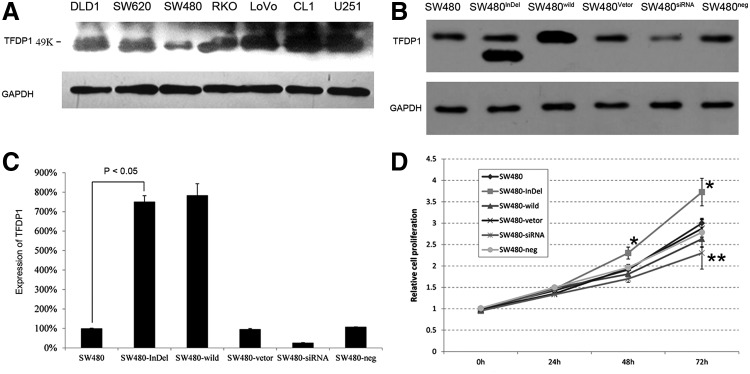
Using Western blot analysis, we measured the expression levels of TFDP1 protein in five CRC cancer cell lines, DLD1, SW480, SW620, RKO, and LoVo, one prostate cancer cell line, CL1, and one glioma cell line, U251 (Fig. 3A). The CRC cell line SW480 exhibited the lowest expression (Fig. 3A) of wild-type TFDP1 and was therefore chosen as a cell line to study the functional consequence of TFDP1 indel84 mutation.
FIG. 3.
In vitro functional analysis of TFDP1. (A) Western blot analysis of different cell lines revealed different TFDP1 expression levels in different cells, with SW480 cells exhibiting the lowest levels. (B) Western blot analysis of the TFDP1 and TFDP1 indel after transfection. Transfected cells SW480-InDel and SW480-wild strongly expressed TFDP1. SiRNA knockdown of TFDP1 (SW480-siRNA) resulted in lower expression compared with the negative siRNA controls (SW480-neg). (C) Quantitative-PCR analysis of the TFDP1 and TFDP1 indel after transfection. SW480-InDel and SW480-wild transfection resulted in approximately 8-fold increase in its protein expression compared with the vector alone. SiRNA knockdown of TFDP1 (SW480-siRNA) resulted in approximately 70% lower expression compared with the negative siRNA controls (SW480-neg). (D) SW480-InDel transfection enhanced cell proliferation, but the siRNA knockdown of TFDP1 inhibited cell proliferation. *p<0.05; **p<0.01.
We constructed the full-length human TFDP1 with the indel84 mutation and the wild-type TFDP1 gene using the pcDNA3.1 vector (Invitrogen) to generate pcDNA3.1(+)-InDel TFDP1 and pcDNA3.1(+)-wild-type TFDP1 constructs. A sequencing analysis revealed that the pcDNA3.1(+)-InDel TFDP1 indeed harbors the indel84 mutation (data not shown). To assess the functional consequences of the TFDP1 indel, we transfected CRC SW480 cells with the constructs for wild-type TFDP1 (SW480-wild), TFDP1 indel84 (SW480-InDel), or the empty vector (SW480-Vector). Western blot analysis (Fig. 3B) showed that the indel84 was translated into a protein around 33 Kd in size, the predicted size of the translated 290 AA indel84 protein. Western blot analysis (Fig. 3B) and quantitative PCR (Fig. 3C) indicated that both the wild-type and the TFDP1 indel84 protein were expressed at high levels (approximately 8-fold increase) after transfection compared with the parental SW480 cells or the cells transfected with the empty vector, SW480-Vector (Fig. 3B and C). In a cell proliferation assay, the cells transfected with TFDP1 indel84 (SW480-InDel) exhibited a significant increase in cell numbers by 48 and 72 hours compared with the SW480-vector control (Fig. 3D). In contrast, the SW480 cells overexpressing the wild-type TFDP1 did not exhibit any growth advantages (Fig. 3D).
Additionally, we included knockdown experiment of TFDP1 using siRNAs to confirm the findings from the overexpression of wild-type TFDP1. Comparing the TFDP1 siRNA knockdown cells (SW480-siRNA) with the scramble controls (SW480-neg), the knockdown efficiency was approximately 70% (Fig. 3B and C). Knockdown of TFDP1 by siRNA resulted in significantly inhibited cell proliferation compared with the siRNA scramble control (SW480-neg) at 72 hours (Fig. 3D). This is consistent with the report by Castillo et al. (2010) who demonstrated that siRNA knockdown of TFDP1 in a lung cancer cell line (HCC33) with a TFDP1 amplification and subsequent protein overexpression reduced cell viability by 50% (Castillo et al., 2010).
The above data demonstrated that the cells overexpressing TFDP1 indel84 exhibited a unique phenotype compared with the TFDP1 knockdown by siRNA, suggesting that the TFDP1 indel84 protein is a gain-of-function mutation.
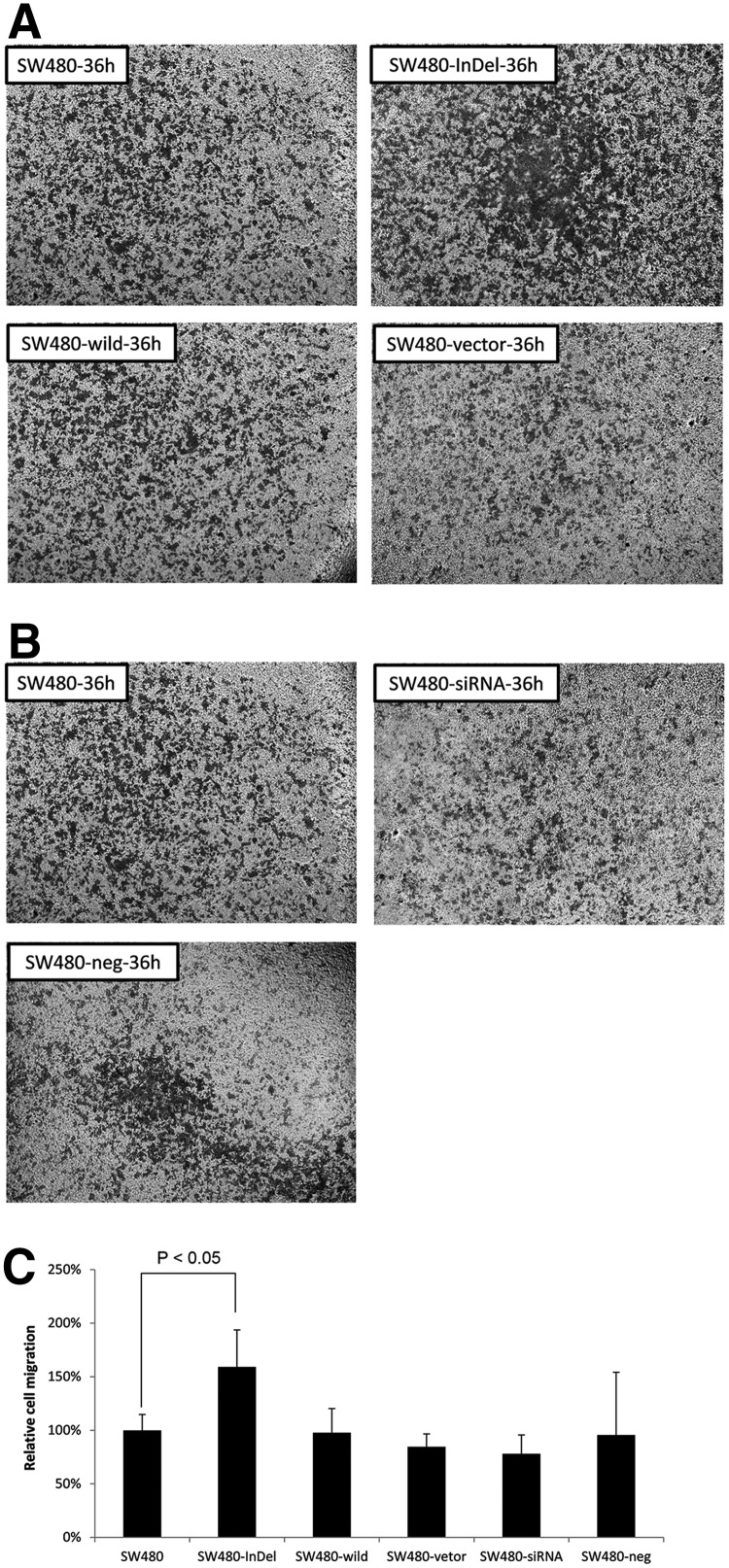
TFDP1 indel84 increased cell migration and invasion in CRC cells
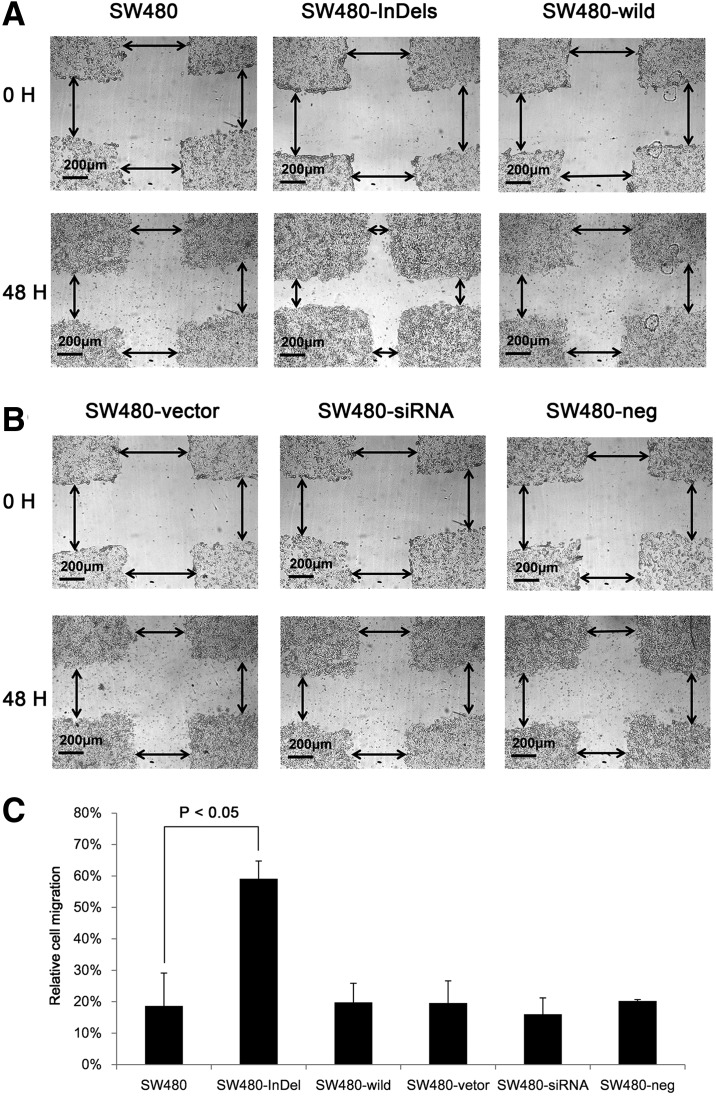
We next examined the effect of TFDP1 indel84 on cell migration and invasion using a wound-healing assay (Dhawan et al., 2005). We observed that the SW480-InDel cells migrated across a wounded area faster than did the parental cells or the wild-type TFDP1-transfected cells (Fig. 4A). However, no difference in cell migration was observed after TFDP1 knockdown in SW480 cells (Fig. 4B). These results again suggest that the TFDP1 indel84 mutation is a gain-of–function mutation that does not mimic the phenotype of the TFDP1 knockdown cells. We further quantified the distances migrated in the wound healing assays and observed that the distance traveled by the SW480-InDel cells was approximately three times that of other cells (Fig. 4C). There was a slight decline in cell migration for the cells with TFDP1 knockdown (SW480-siRNA) compared with the scramble negative controls (Fig. 4C).
FIG. 4.
SW480-InDel transfection increased the migration of CRC cells in a wound-healing assay. (A) Typical images of wound healing assays demonstrating the migrated distances of SW480-, SW480-InDel-l, and SW480-wild-transfected cells 48 hours after transfection. (B) Typical images of wound healing assays demonstrating the migrated distances of cells transfected with SW480-vector, SW480-siRNA, and SW480-neg constructs 48 hours after transfection. (C) Quantification of the images in (A) and (B). The distances indicated by arrows in (A) and (B) were measured and compared.
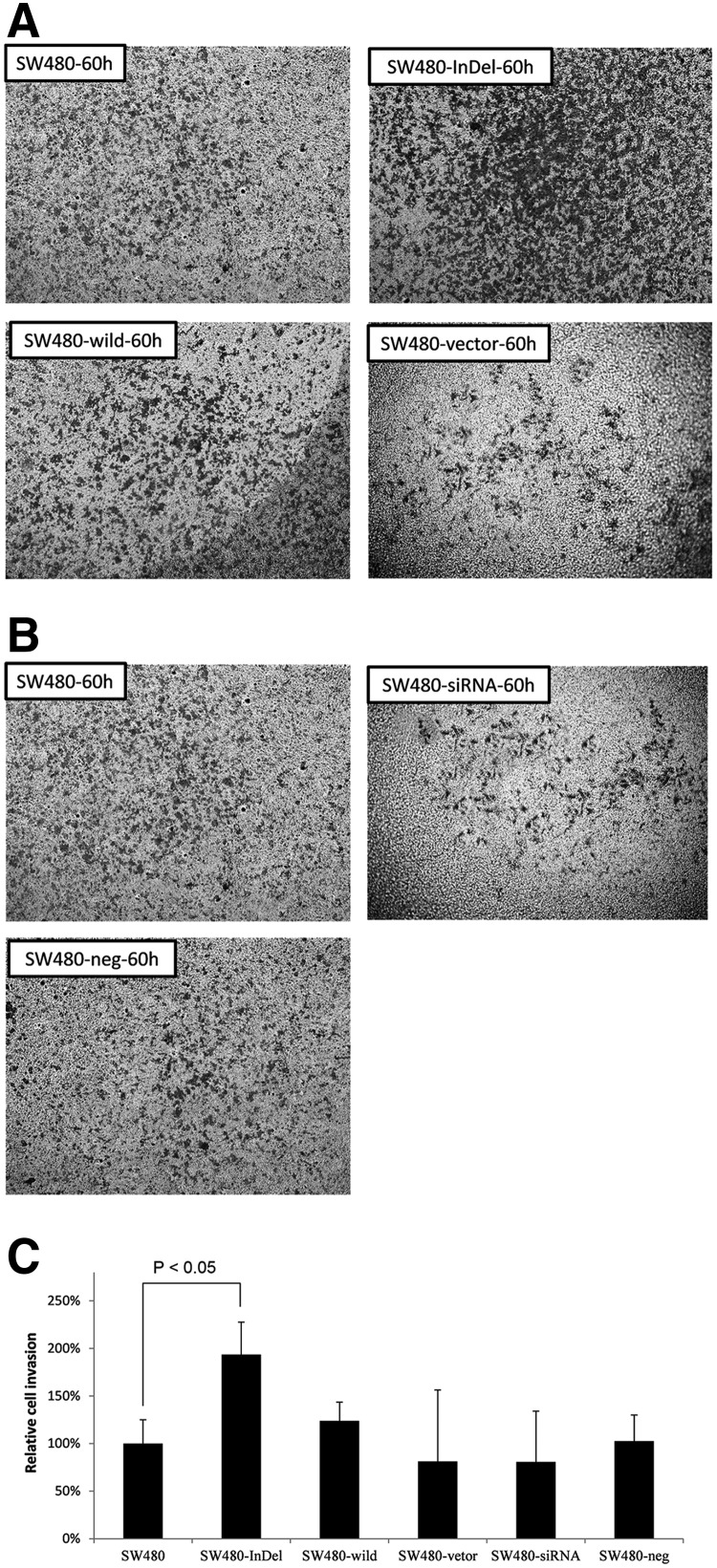
We additionally used transwell migration assays to assess the various constructs on cell migration. Figure 5A–C demonstrates a 1.5-fold increase in cell migration in the SW480 cells with the TFDP1 indel84. A transwell invasion assay also demonstrated that the number of invading cells increased approximately 2-fold in the TFDP1-indel84-transfected cells, compared with the parental cells or the other control groups (Fig. 6A–C).
FIG. 5.
SW480-InDel transfection increased the migration of CRC cells in a transwell migration assay. (A) Images of the migrating cells after 36 hours for cells transfected with the SW480, SW480-InDel, SW480-wild, or SW480 vectors. (B) Images of the migrating cells 36 hours for cells transfected with the SW480, SW480-siRNA, or SW480-neg constructs. (C) Quantification of the cellular migration. Six visual fields for each analysis were randomly selected, and the cells were counted.
FIG. 6.
SW480-InDel transfection increased the migration of CRC cells in a transwell invasion assay. (A) Images of the cells invading through the matrix after 60 hours for cells transfected with the SW480, SW480-InDel, SW480-wild, or SW480 vector. (B) Images of the cells invading through the matrix after 60 hours for cells transfected with the SW480, SW480-siRNA, or SW480-neg constructs. (C) Quantification of the invading cells. Six visual fields for each analysis were randomly selected, and the cells were counted.
Discussion
We identified an indel in the TFDP1 gene and validated the presence of this indel in 21 of 30 CRC tissue samples (70.0%). We further demonstrated that the indel84 was translated into a protein of about 33 Kd (Fig. 3B), suggesting that nonsense-mediated decay (NMD), a pathway is well known to recognize and degrade aberrant mRNAs with truncated open reading frames (ORF) due to the presence of a premature termination codon (PTC) (Palacios, 2012), seemed not playing a role here.
We performed direct PCR-sequencing of cancer samples with indel84. The clinical information of these cancer samples is provided in Supplementary Table S5. We noticed that the wild-type allele consisted of the majority of the alleles, representing approximately 70% of the alleles. This could be due to contamination of normal cells in the cancer samples or due to intratumor heterogeneity (i.e., the existence of tumor cells with or without indel84), or a combination of both factors. Govindan et al. (2012) performed whole-genome and transcriptome sequencing of tumor and adjacent normal tissue samples from 17 patients with non-small cell lung carcinoma (NSCLC). From the variant allele frequencies (VAFs) for somatic mutations identified in each tumor sample, the authors were able to estimate the tumor purity and clonality status (e.g., monoclonal, biclonal, or multiclonal) of the tumors (Govindan et al., 2012). However, we were not able to differentiate contamination of normal cells in the tumor samples from intratumor heterogeneity in our samples because we did not perform whole genome or whole exome sequencing for our samples. Future experimentation using laser capture microdissection (LCM) to isolate pure tumor cells will help to address this issue.
Insertions/deletions (indels) are common mutations for cancers. In CRC, microsatellite instability (MSI), which is the expansion or contraction of DNA repeat tracts caused by the loss of DNA mismatch repair activity, are detected in approximately 15% of all colorectal cancers (Vilar and Gruber, 2010). Govindan et al. (2012) identified 90 indels in 17 NSCLC samples. Mononucleotide runs (≥4 bp) were identified as hotspots for microdeletions/microinsertions (Ivanov et al., 2011). The indel84 that we identified here is also localized to a hotspot consisting of mononucleotide runs of 5 Cs (Supplementary Fig. S2).
Gain-of-function mutations have been reported in many human diseases. For example, Barcia et al. (2012) reported that de novo gain-of-function KCNT1 channel mutations caused malignant migrating partial seizures of infancy. Sanada et al. (2009) identified a gain-of-function mutated C-CBL tumor suppressor in myeloid neoplasm. The authors identified point mutations or stretches of amino acid deletions in the linker-RING finger domain that is central to the E3 ubiquitin ligase activity of the C-CBL gene, as well as mutations that would generate truncated proteins, which would result in a loss-of-function (Sanada et al., 2009). As C-CBL is a tumor suppressor, loss-of-function could be a mechanism for the oncogenicity of these C-CBL mutants. The authors demonstrated that the C-CBL mutants enhanced colony formation in soft agar and tumor generation in nude mice (Sanada et al., 2009). The authors also argued that the gain-of-function could be mediated by a mechanism other than the loss-of-function of the wild-type gene because C-CBL mutants gained several motifs that interacted with numerous signal-transducing molecules (Sanada et al., 2009).
Conclusions
In summary, we identified a recurrent frame-shift indel mutation in the TFDP1 gene in colorectal cancers by next-generation sequencing and validated this mutation to be present in 70% of CRC tissues. We further demonstrated that the TFDP1 indel84 mutation generated a gain-of-function phenotype that increased cell proliferation, migration, and invasion of CRC cells. Our study identified a key molecular event for CRC diagnosis and therapy. Future studies are warranted to explore the diagnostic and therapeutic potential of our discovery. These studies might include in vivo mouse studies to study the in vivo function of the TFDP1 indel84, clinical correlations to study whether indel84 is associated with clinical parameters or patient prognosis, and diagnostic assays to determine whether indel84 is present in stool samples or in circulating tumor cells as a noninvasive diagnostic tool.
Supplementary Material
Acknowledgment
This work was supported by grants from the Ministry of Science and Technology, China (2006AA02Z4A2, 2006AA02A303, 2007DFC30360, 2008DFA11320, and 2006DFA2950), China Postdoctoral Science Foundation (No. 2013M531451), Grant 2012R10021 from Zhejiang Province, and Grant 20110101120153 from the Ministry of Education, China.
Author Disclosure Statement
The authors declare that there are no conflicting financial interests.
References
- Barcia G, Fleming MR, Deligniere A, et al. (2012). De novo gain-of-function KCNT1 channel mutations cause malignant migrating partial seizures of infancy. Nat Genet 44, 1255–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmann AM, Swaminathan S, and Thimmapaya B. (1998). Regulation of cellular genes in a chromosomal context by the retinoblastoma tumor suppressor protein. Mol Cell Biol 18, 4565–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo SD, Angulo B, Suarez-Gauthier A, et al. (2010). Gene amplification of the transcription factor DP1 and CTNND1 in human lung cancer. J Pathol 222, 89–98 [DOI] [PubMed] [Google Scholar]
- Chen W, Zheng R, Zhang S, et al. (2013). Report of incidence and mortality in China cancer registries, 2009. Chin J Cancer Res 25, 10–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Wang J, Shao J, et al. (2010). Identification of novel SNPs by next-generation sequencing of the genomic region containing the APC gene in colorectal cancer patients in China. OMICS 14, 315–325 [DOI] [PubMed] [Google Scholar]
- Clark MJ, Homer N, O'Connor BD, et al. (2010). U87MG decoded: The genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet 6, e1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A, and Ke X. (2012). Primer1: Primer design web service for tetra-primer ARMS-PCR. Open Bioinformat J 6, 55–58 [Google Scholar]
- Dhawan P, Singh AB, Deane NG, et al. (2005). Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest 115, 1765–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girling R, Partridge JF, Bandara LR, et al. (1993). A new component of the transcription factor DRTF1/E2F. Nature 365, 468. [DOI] [PubMed] [Google Scholar]
- Govindan R, Ding L, Griffith M, et al. (2012). Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 150, 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyodo I, Suzuki H, Takahashi K, et al. (2010). Present status and perspectives of colorectal cancer in Asia: Colorectal Cancer Working Group report in 30th Asia-Pacific Cancer Conference. Jpn J Clin Oncol 40Suppl 1, i38–43 [DOI] [PubMed] [Google Scholar]
- Iaquinta PJ, and Lees JA. (2007). Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol 19, 649–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iengar P. (2012). An analysis of substitution, deletion and insertion mutations in cancer genes. Nucleic Acids Res 40, 6401–6413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Hamby SE, Stenson PD, et al. (2011). Comparative analysis of germline and somatic microlesion mutational spectra in 17 human tumor suppressor genes. Hum Mutat 32, 620–632 [DOI] [PubMed] [Google Scholar]
- Keller A, Harz C, Matzas M, et al. (2011). Identification of novel SNPs in glioblastoma using targeted resequencing. PLoS One 6, e18158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Chen K, Wylie T, et al. (2009). VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov AS, and Rogozin IB. (2004). Context of deletions and insertions in human coding sequences. Hum Mutat 23, 177–185 [DOI] [PubMed] [Google Scholar]
- Li H, and Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson D, Capelletti M, Yelensky R, et al. (2012). Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 18, 382–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogurtsov AY, Sunyaev S, and Kondrashov AS. (2004). Indel-based evolutionary distance and mouse-human divergence. Genome Res 14, 1610–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM. (2012). Nonsense-mediated mRNA decay: From mechanistic insights to impacts on human health. Brief Funct Genomics 12, 25–36 [DOI] [PubMed] [Google Scholar]
- Polager S, and Ginsberg D. (2009). p53 and E2f: Partners in life and death. Nat Rev Cancer 9, 738–748 [DOI] [PubMed] [Google Scholar]
- Pritchard CC, Smith C, Salipante SJ, et al. (2012). ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J Mol Diagn 14, 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruark E, Snape K, Humburg P, et al. (2012). Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature 493, 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada M, Suzuki T, Shih LY, et al. (2009). Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature 460, 904–908 [DOI] [PubMed] [Google Scholar]
- Shao J, Lou X, Wang J, et al. (2013). Targeted re-sequencing identified rs3106189 at the 5′ UTR of TAPBP and rs1052918 at the 3′ UTR of TCF3 to be associated with the overall survival of colorectal cancer patients. PLoS One 8, e70307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, and Jemal A. (2013). Cancer statistics, 2013. CA Cancer J Clin 63, 11–30 [DOI] [PubMed] [Google Scholar]
- Sorensen TS, Girling R, Lee CW, Gannon J, Bandara LR, and La Thangue NB. (1996). Functional interaction between DP-1 and p53. Mol Cell Biol 16, 5888–5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulonen AM, Ellonen P, Almusa H, et al. (2011). Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol 12, R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung JJ, Lau JY, Goh KL, and Leung WK. (2005). Increasing incidence of colorectal cancer in Asia: Implications for screening. Lancet Oncol 6, 871–876 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ruan J, Yeh SH, et al. (2011). Rapid growth of a hepatocellular carcinoma and the driving mutations revealed by cell-population genetic analysis of whole-genome data. Proc Natl Acad Sci USA 108, 12042–12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar E, and Gruber SB. (2010). Microsatellite instability in colorectal cancer. The stable evidence. Nat Rev Clin Oncol 7, 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, and Hakonarson H. (2010). ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangkumhang P, Chaichoompu K, Ngamphiw C, et al. (2007). WASP: A Web-based allele-specific PCR assay designing tool for detecting SNPs and mutations. BMC Genomics 8, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, et al. (2008). The complete genome of an individual by massively parallel DNA sequencing. Nature 452, 872–876 [DOI] [PubMed] [Google Scholar]
- Wu QJ, Vogtmann E, Zhang W, et al. (2012). Cancer incidence among adolescents and young adults in urban Shanghai, 1973–2005. PLoS One 7, e42607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Dhillon S, Ke X, Collins AR, and Day IN. (2001). An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res 29, E88–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.