Abstract
Methylation of the base cytosine in DNA is critical for silencing endogenous retroviruses, regulating gene expression, and establishing cellular identity, and has long been regarded as an indelible epigenetic mark. The recent discovery that the ten eleven translocation (TET) proteins can oxidize 5-methylcytosine (5mC) resulting in the formation of 5-hydroxymethylcytosine (5hmC) and other oxidized cytosine variants in the genome has triggered a paradigm shift in our understanding of how dynamic changes in DNA methylation regulate transcription and cellular differentiation, thus influencing normal development and disease.
5-methylcytosine can be oxidized by the TET family of enzymes, forming 5-hydroxymethylcytosine and other cytosine variants. The roles of these variants in neurogenesis, development, and other processes continue to be explored.
Methylation of the base cytosine (termed 5-methylcytosine or 5mC) is an epigenetic mark often referred to as the fifth base, to underscore its heritability and importance in development. 5mC is considered an epigenetic mark because it directs biological function (i.e., transcriptional repression) without altering the protein coding capacity of the local DNA sequence dictated by the four conventional bases. 5mC is vital for processes including embryogenesis, parental imprinting, X inactivation, the silencing of endogenous retroviruses, and the regulation of gene expression and splicing. Cytosine methylation influences these processes by both modulating protein–DNA interactions and nucleating the formation of repressive heterochromatic structures. In 2009, 5-hydroxymethylcytosine (5hmC) was simultaneously identified by two research groups as a normal constituent of genomic DNA in mammalian neurons and embryonic stem (ES) cells (Kriaucionis and Heintz 2009; Tahiliani et al. 2009). This landmark finding has stimulated a tremendous amount of research focused on understanding how this modification exerts its influence on the regulation of the genome and how this modification ties into a 5mC demethylation pathway that was previously lacking in enzymatic players.
5hmC was serendipitously identified in Nathaniel Heintz’s laboratory when Skirmantas Kriaucionis was elucidating the chromatin make-up of the strikingly euchromatic nuclei of cerebellar Purkinje neurons. Isolating Purkinje cell nuclei in itself was a technical achievement, requiring the use of transgenic mice with an eGFP labeled nucleolus (bacTRAP) and high-capacity fluorescence-activated cell sorting to get enough material for the assays. The goal was to compare 5mC abundance in Purkinje cells with granule cells using the classic “nearest neighbor” DNA composition analysis technique dating back to Kornberg’s classic experiments of 1961 and used in Adrian Bird’s pioneering experiments quantifying global levels of methylated CpGs. Unexpectedly, this sensitive, unbiased, and robust method revealed an additional signal, which was reproducibly enriched in Purkinje neurons and detectable in other neuronal cell types. The most exciting phase of these experiments was identifying the signal as 5hmC, a novel base modification in genomic DNA (Kriaucionis and Heintz 2009).
5hmC was concurrently discovered by Mamta Tahiliani in Anjana Rao’s laboratory when her quest to identify a DNA demethylase took an unexpected twist. The search for such an enzyme was primarily motivated by the demonstration that DNA methylation is actively erased in the paternal genome immediately after fertilization. This seminal finding strongly suggested that resetting methylation patterns might be critical for epigenetic reprogramming (as illustrated in Fig. 3 of Li and Zhang 2014). Mamta’s bioinformatics collaborator L. Aravind predicted that the TET family of proteins were dioxygenases with a specificity for nucleic acids. Distantly related dioxygenases had recently been shown to remove methyl groups from both histones and damaged DNA bases. Therefore, the TET proteins were extremely attractive DNA demethylase candidates. In her initial experiments, Mamta found that overexpression of TET1 diminished levels of 5mC by immunofluorescence, suggesting tantalizingly that TET1 was acting as a true DNA demethylase. However, her attempts to confirm demethylation using thin-layer chromatography yielded puzzling results because the reduction in 5mC was not accompanied by the predicted increase in cytosine. However, when she adjusted the contrast on the scanned image, she noticed that what had appeared to be a faint smear under cytosine took on the shape of an independent spot suggesting that TET1 might be converting 5mC to a novel species. Because many dioxygenases initiate catalysis by hydroxylating their substrates, Mamta hypothesized and then confirmed that this nucleotide was 5hmC. The group also showed that 5hmC was present in the genome of ES cells, and that both TET1 and 5hmC levels decline when ES cells are differentiated. This suggested that 5hmC is a normal constituent of mammalian DNA, and that TET proteins and 5hmC play an important role in regulating gene expression and cell identity in ES cells (Tahiliani et al. 2009). Subsequent studies by multiple laboratories have established that each member of the TET family (TET1/TET2/TET3) is able to convert 5mC to 5hmC (Wu and Zhang 2011). However, studies in mice have shown that Tet3 is the only member of the TET family required in vivo for normal development.
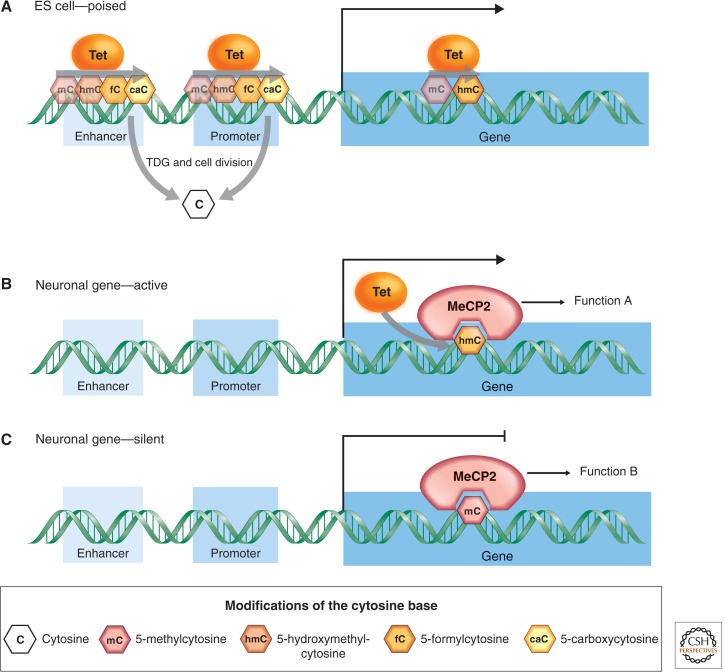
The discovery that TET enzymes can oxidize 5mC to 5hmC led to the question of whether full DNA demethylation from 5hmC to C was passive (i.e., achieved by replication-dependent dilution), or actively catalyzed. TET enzymes have now been shown to successively oxidize 5hmC to 5-formylcytosine (5fC) and 5-carboxylcytosine (5acC); reviewed in Wu and Zhang 2011). The rapid loss of 5mC in the paternal genome coincides with the translocation of TET3 to the nucleus and the large-scale conversion of 5mC to 5hmC, 5fC, and 5caC (Wu and Zhang 2011). Immunostaining of metaphase chromatin further revealed that all three oxidized derivatives of 5mC are largely retained on the original strands of DNA and are passively diluted by replication during the early cleavage cycles, indicating that TET-mediated oxidation of 5mC can stimulate passive loss of 5mC oxidation products through replication. Alternatively or even concurrently, 5fC and 5caC can be removed by thymine DNA glycosylate (TDG) and replaced by cytosine via base excision repair (Fig. 1A) (Wu and Zhang 2011; Fig. 6 of Li and Zhang 2014). When and where in the genome these mechanisms operate remains a topic of active research.
Figure 1.
Distribution and metabolism of cytosine modifications within genes in ES cells and neurons. (A) TET-mediated oxidation of 5mC followed by base excision repair (BER)-mediated removal of 5caC keeps promoters and enhancers free of methylation in ES cells. It is also possible that oxidation of 5mC blocks maintenance methylation at these regions. MeCP2 binds both 5mC (B) and 5hmC (C) in neuronal gene bodies, where the cytosine modification state correlates with the level of expression.
Understanding the biological function of 5hmC has required the development of innovative tools to detect it and distinguish it unequivocally from 5mC and C. It is clear now that bisulfite sequencing cannot distinguish 5hmC from 5mC, and also misinterprets 5fC and 5caC as cytosine (Pastor et al. 2013). Therefore, it is important to note that decades of bisulfite sequencing data must be interpreted with caution, as “methylation” could be either 5mC or 5hmC, whereas positions previously identified as cytosine could actually contain 5fC or 5caC. A number of techniques have now been developed to enrich for 5hmC-containing DNA and most recently to sequence it at single nucleotide resolution (Pastor et al. 2013).
Multiple lines of evidence indicate that 5hmC is not simply a demethylation intermediate, but rather a novel modification in DNA with an effector program of its own. 5hmC is present in a variety of mature cell types in adult organisms, and its levels range from 0.05% of all bases in some immune cells to as high as 0.6% in Purkinje cells. This leads to the question of whether readers of this mark exist to translate the presence of this modification into biological function, much as unmethylated cytosines can be read by CXXC domain-containing proteins (see Blackledge et al. 2013), or methylated CpGs are recognized by MBD proteins. A number of proteins have already been identified that bind to 5hmC including MeCP2, MBD3, and Uhrf2, which are known to regulate transcription. 5fC- and 5caC-bound proteins include a number of DNA repair proteins, consistent with a role for these modifications as demethylation intermediates.
The cell type, developmental stage, and genomic locus specific distribution of 5hmC is beginning to suggest particular functions of this DNA modification. Techniques enriching for 5hmC as well as single nucleotide sequencing techniques have shown that in ES cells 5hmC levels are elevated at enhancers and CpG island (CGI)-containing promoters, which are free of methylation despite their high CpG content (Pastor et al. 2013). In neuronal cells, 5hmC is enriched in gene bodies (Fig. 1B,C) (Mellén et al. 2012; Pastor et al. 2013). Although gene body enrichment was also noted in ES cells, single nucleotide techniques have not verified this finding. It has been proposed that TET proteins and 5hmC play a role in keeping CGIs free of methylation in ES cells, whereas the function of gene body 5hmC in neuronal cells is still unclear.
Future research will need to address the precise function of 5hmC in early development, hematopoiesis and neuronal function. It will be intriguing to know whether a single model can explain 5hmC function in all cell types or whether its function will vary for each cell type examined.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- *.Blackledge NP, Thomson JP, Skene PJ 2013. CpG island chromatin is shaped by recruitment of ZF-CxxC proteins. Cold Spring Harb Perspect Biol 5: a018648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Li E, Zhang Y 2014. DNA methylation in mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N 2012. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151: 1417–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A 2013. TETonic shift: Biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol 14: 341–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y 2011. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev 25: 2436–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]