Abstract
The origin of the eukaryotic cell can be regarded as one of the hallmarks in the history of life on our planet. The apparent genomic chimerism in eukaryotic genomes is currently best explained by invoking a cellular fusion at the root of the eukaryotes that involves one archaeal and one or more bacterial components. Here, we use a phylogenomics approach to reevaluate the evolutionary affiliation between Archaea and eukaryotes, and provide further support for scenarios in which the nuclear lineage in eukaryotes emerged from within the archaeal radiation, displaying a strong phylogenetic affiliation with, or even within, the archaeal TACK superphylum. Further taxonomic sampling of archaeal genomes in this superphylum will certainly provide a better resolution in the events that have been instrumental for the emergence of the eukaryotic lineage.
Phylogenomic studies suggest that eukaryotes may have emerged from the archaeal TACK superphylum. Orthologs of several eukaryotic signature proteins (e.g., actin) have recently been identified in genomes of TACK Archaea.
The origin of the eukaryotic cell can be regarded as one of the hallmarks in the history of life on our planet. Yet, despite its evolutionary significance, the emergence of eukaryotes remains poorly understood (Embley and Martin 2006). At the cellular level, the gap between prokaryotes (Bacteria and Archaea) and Eukarya is immense, with the latter cell types being exceedingly compartmentalized with organelles and cell structures such as mitochondria, peroxisomes, Golgi complex, and endoplasmic reticulum, and the central nexus of the eukaryotic cell, the nucleus. Although cellular compartmentalization is also observed in several prokaryotic lineages (e.g., in Cyanobacteria, Planctomycetes, and Crenarchaeota), these compartments do not seem evolutionarily related to those observed in eukaryotes (e.g., see McInerney et al. 2011). The absence of such missing links, or intermediate stages of eukaryogenesis, significantly hampers the delineation of more sophisticated models for the emergence of the eukaryotic cell (Martijn and Ettema 2013).
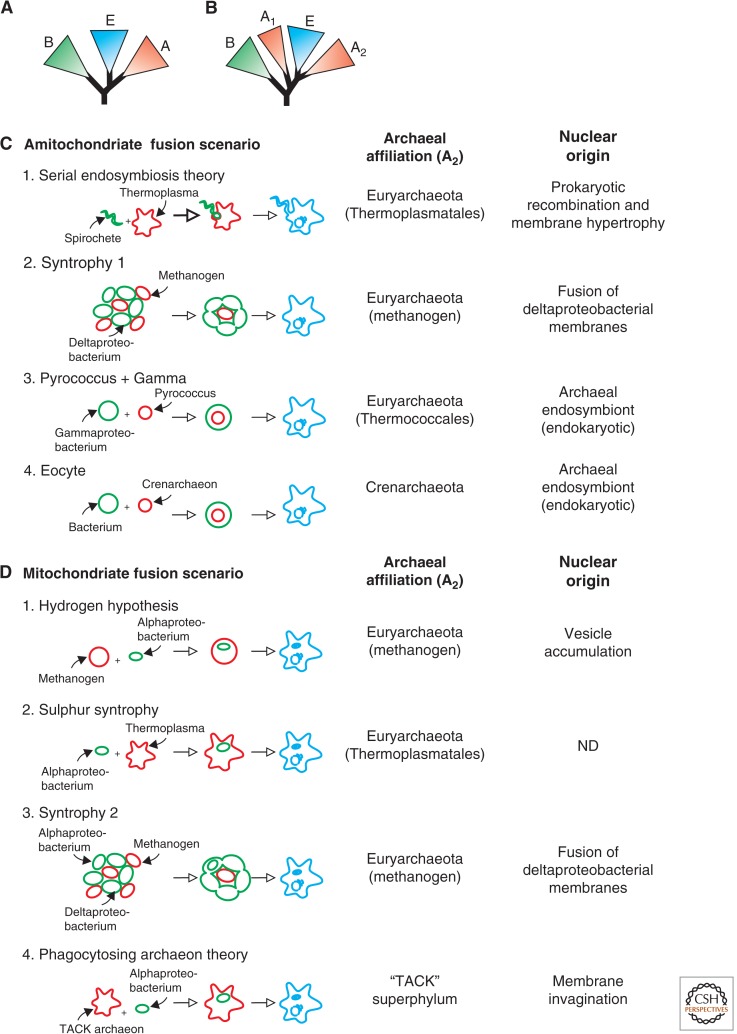
The lack of understanding of how eukaryotes emerged echoes back in another central dilemma. How are the domains of cellular life related to each other? Ever since Carl Woese and George Fox (1977) discovered the “third domain of life” in the late 1970s, the Archaea, followed by their subsequent proposal for the classical three domains of life model (Fig. 1A) (Woese 1987; Woese et al. 1990), scientists have had heated debates on their evolutionary affiliation. In particular, they have failed to reach consensus regarding the placement of the eukaryotic branch in the tree of life. Early attempts to resolve the placement of eukaryotes in the tree of life, including these of Carl Woese himself, placed eukaryotes as a sister clade with respect to the Archaea (Fig. 1A), whereas others proposed an alternative topology in which eukaryotes grouped within the Archaea (Fig. 1B), as a sister to the Crenarchaeota (“eocytes”) (Henderson et al. 1984; Lake et al. 1984).
Figure 1.
Overview of scenarios for the origin of the eukaryotic cell. Schematic depiction of the classical three-domain tree of life (A) and a tree that supports fusion hypotheses in which the eukaryotic nuclear lineage evolved from within the archaeal radiation (B). Of the latter category, a number of hypotheses have been proposed that can be classified as amitochondriate fusion scenarios (i.e., the fusion event leads to a mitochondrion-lacking proto-eukaryotic lineage). The following scenarios have been outlined schematically: (1) The Serial Endosymbiosis Theory (Margulis et al. 2006), which involves a fusion between a Spirochete and a Thermoplasma-like archaeon; (2) “Syntrophy 1” representing the original syntrophic hypothesis proposed by Moreira and Lopez-Garcia (1998), involving a fusion between a syntrophic community comprising hydrogen producing deltaproteobacterial cells and hydrogen consuming methanogens; (3) “Pyrococcus + Gamma,” depicting the endokaryotic model proposed by Horiike et al. (2004) in which the eukaryotic lineage emerges via a Pyrococcus-related archaeal endosymbiont in a gammaproteobacterial host; (4) The eocyte model proposed by Lake (1988), which suggests that the eukaryotic nucleus evolved from a crenarchaeal lineage. Another class of fusion models involves scenarios in which the origin of the proto-eukaryotic lineage coincides with that of the mitochondrial origin (D), and include the following examples: (1) The Hydrogen hypothesis, involving the endosymbiosis of a hydrogen-producing alphaproteobacterium in a methanogen (Martin and Muller 1998); (2) Sulfur Syntrophy, in which eukaryotes evolved from a sulfur-dependent syntrophy between a Thermoplasma-like archaeaon and an alphaproteobacterium (Searcy and Hixon 1991; Pisani et al. 2007); (3) “Syntrophy 2,” which involves a refined version of the original Syntrophic hypothesis, which now also includes anaerobic methane oxidizing alphaproteobacterial cells from which the mitochondria supposedly emerged (Lopez-Garcia and Moreira 1999); (4) Phagocytosing Archaeon Theory (PhAT), which involves the engulfment of an alphaproteobacterium by a phagocytic archaeon belonging to the TACK superphylum (Martijn and Ettema 2013). Archaeal, bacterial, and (proto)eukaryotic cells are depicted in red, green and blue, respectively. A, Archaea; B, Bacteria; E, Eukarya; ND, not determined. (C,D, Inspired by Table 1 in Martin 2005.)
The phylogenetic incongruence regarding the placement of eukaryotes in the tree of life has triggered the emergence of a wide variety of evolutionary models to explain the origin of eukaryotes, each of which having its own unique angle to it. Whereas some of these models are essentially compatible with the classical three domains view of life, such as the Archezoa hypothesis (Cavalier-Smith 1989), other models propose an autonomous origin of eukaryotes, such as the Neomuran hypothesis (Cavalier-Smith 2002a,b), which proposes that Archaea and Eukaryotes emerged from a common ancestor related to Gram-positive bacteria, and the PVC hypothesis, which suggests that eukaryotes evolved from an ancestor of the PVC superphylum (Devos and Reynaud 2010; Forterre and Gribaldo 2010; Forterre 2011; Reynaud and Devos 2011). Yet another class of hypotheses imply an important role for viruses in the prokaryote-to-eukaryote transition (Bell 2001; Takemura 2001; Forterre 2005, 2006), and some suggest that Bacteria, Archaea, and eukaryotes have evolved from a complex, eukaryote-like ancestor (Kurland et al. 2006). However, in-depth analyses of the genomic data that became available during the last decade of the previous century deemed many of these hypotheses implausible. Importantly, these analyses pointed out two other important issues. First, they confirmed that Archaea had highly distinct gene repertoires, compatible with them evolving independently from the bacterial domain (Bult et al. 1996; Brown and Doolittle 1997; Olsen and Woese 1997; Rivera et al. 1998; Makarova et al. 1999; Ettema et al. 2005), and second, they pointed out that eukaryotic genomes are chimeric in nature, comprising two distinct gene sets (Rivera et al. 1998); genes for information storage and processing are Archaea related (see, for example, Yutin et al. 2008), and genes for metabolic or “operational” processes are mostly bacterial in nature. These observations suggest that at the root of the eukaryotic domain of life, some sort of fusion event has taken place that involved an archaeal partner, and one or more bacterial partners (Koonin 2010).
The idea of a cellular fusion event at the basis of the eukaryotic lineage is not new. During the late 1980s, Zillig and colleagues interpreted the myriad of characters shared between Archaea, Bacteria, and Eukarya as evidence of chimerism in the eukaryotic domain of life (Zillig et al. 1985, 1989a,b). The fusion hypothesis postulated by Zillig and coworkers fitted in nicely with the Serial Endosymbiosis Theory (repopularized by Lynn Margulis), which placed fusion events as a central avenue in cellular evolution, in particular, that of eukaryotes (Sagan 1967). Altogether, both concepts stood at the basis of a new wave of hypotheses for the origin of the eukaryotic cell in which fusion events played a central role. Although the identity, nature, and amount of fusion partners differed markedly between different hypotheses, we can roughly enumerate two classes based on whether the end product of the fusion represents an amitochondriate (mitochondrion-lacking) (Fig. 1C) or a mitochondriate (mitochondrion-bearing) (Fig. 1D) cellular entity. The former “amitochondriate category” is compliant with the Archezoa theory (Cavalier-Smith 1989), and other “Woesean” models, in the sense that they implement the origin of the eukaryotic cell and mitochondrion as two independent evolutionary events. Several studies have suggested, however, that these two events most likely co-occurred, based on several lines of evidence. For example, we now know that previously assumed amitochondriate eukaryotes do harbor remnants of the mitochondriate past in the form of hydrogenosomes, mitosomes, and possibly other mitochondria-like organelles. In addition, phylogenetic analyses have argued against the existence of a deeply rooting Archezoan clade (see Embley 2006 for a detailed argument).
Significant heterogeneity exists among the fusion models with respect to the envisioned archaeal fusion partner (Fig. 1C,D). Most of these models, especially those that were delineated before the genomic age, are based on biological (metabolic or cytological) considerations. For example, several models implement a syntrophic interaction between a methanogenic archaeon and hydrogen-producing bacterial partner (e.g., the hydrogen and other syntrophic hypotheses; see Fig. 1C,D). Yet another group of models envision that the nucleus emerged from an archaeal endosymbiont (so-called endokaryotic models) (Martin 2005), such as the eocyte (Lake 1988) and pyrococcus-gammaproteobacteria fusion models (Fig. 1C) (Horiike et al. 2004).
Obviously, obtaining information regarding the identity and nature of the archaeal fusion partner holds the key to solving the evolutionary puzzle of the origin of the eukaryotic cell. Yet, the plethora of phylogenetic studies that has tried to resolve this conundrum has thus far failed to reach consensus, prompting some to even speak of a “phylogenomic impasse” (Gribaldo et al. 2010). Clearly, the phylogenomic approaches taken to resolve these deep evolutionary relationships have to be performed with great care as they are prone to all sorts of biases and artifacts (Delsuc et al. 2005; Gribaldo et al. 2010). Some of those, such as compositional bias and varying evolutionary rates (heterotachy), are inherent in the nature of biological sequence data. Yet, apart from biological issues, the outcome of phylogenomic studies are strongly influenced by data selection (genes and taxa), site filtering, choice of sequence alignment and phylogenetic algorithms, evolutionary model, etc. (see Delsuc et al. 2005 for an overview). Delsuc et al. (2005) hit the nail on the head by summarizing these issues wittily: “garbage in, garbage out.” Therefore, the outcome of the earliest genome-scale phylogenetic studies, in which such issues were not yet properly addressed, should be regarded in light of such artifacts. With this in mind, it is interesting to note that the first phylogenomic studies that aimed to depict the tree of life generally supported the iconic “Woesean” three-domains tree (e.g., Snel et al. 1999; Tekaia et al. 1999; Ciccarelli et al. 2006). However, recent studies that analyzed more extensive genomic data sets with more sophisticated phylogenetic models and algorithms embedded the eukaryotic branch within the archaeal domain, creating a paraphyletic archaeal domain. More specifically, they seem to support a scenario in which eukaryotes emerged from the “TACK superphylum” (Cox et al. 2008; Foster et al. 2009; Guy and Ettema 2011; Williams et al. 2012), which, apart from Cren-, Kor-, and Thaumarchaeota, also comprises the recently proposed Aigarchaeota phylum (Nunoura et al. 2011). These latter findings thus lend support to an extended version of the eocyte hypothesis (Lake 1988), or the recently proposed phagocytosing archaeon theory (PhAT), which poses that eukaryotes evolved from a phagocytic TACK archaeon (Martijn and Ettema 2013). It should, however, be noted that whereas the eocyte hypothesis represents a so-called endokaryotic fusion model (Lake 1988; Martin 2005) with an amitochondriate, nucleated eukaryote as result, the PhAT hypothesis, as an extension of previous scenarios (e.g., Yutin et al. 2009), proposes a cellular fusion that results in a mitochondrion-containing but nucleus-lacking cell (Fig. 1). Apart from phylogenetic studies, the evolutionary affiliation between eukaryotes and the TACK superphylum is seemingly supported by the recent identification of several previously presumed eukaryotic signature proteins (ESPs) in genomes of TACK Archaea (Guy and Ettema 2011). These ESPs are generally involved in pivotal processes in eukaryotes and, among others, include bona fide archaeal orthologs of actin (Yutin et al. 2009; Bernander et al. 2011; Ettema et al. 2011), tubulin (Yutin and Koonin 2012), ESCRT proteins (Lindås et al. 2008; Ettema and Bernander 2009; Makarova et al. 2010), as well as several proteins that are involved in transcription and translation (Guy and Ettema 2011).
Here, we present a reassessment of the phylogenetic affiliation between the archaeal domain and eukaryotes by performing phylogenomic analyses in which we have investigated the impact of gene and taxon selection and compositional bias removal while using a variety of phylogenetic algorithms and evolutionary models. Our analyses indicate that a broadened taxon sampling and increased gene selection unequivocally support a scenario in which the eukaryotic branch emerged from within or at the base of the TACK superphylum.
RESULTS AND DISCUSSION
An Unbiased Phylogenomics Approach to Study the Evolutionary Affiliation between Archaea and Eukaryotes
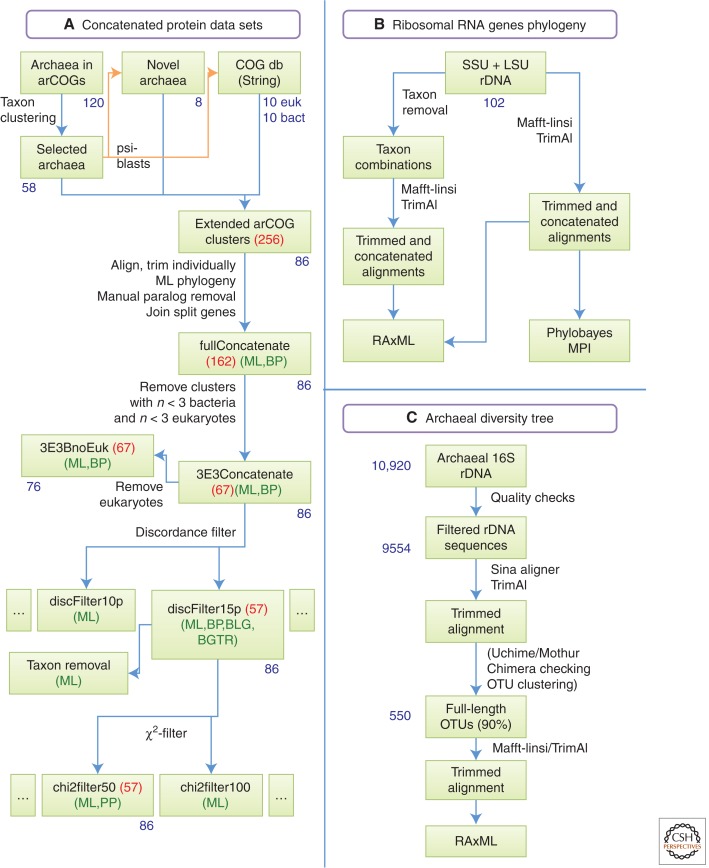
A dual strategy was designed to investigate the evolutionary affiliation between Archaea and eukaryotes. One analysis included a phylogenomic analysis of highly conserved, universal protein coding genes, and a second analysis focused on the phylogenetic analysis of 16S/18S and 23S/28S ribosomal DNA (rDNA) genes (Fig. 2). Because it was anticipated that the eukaryotic lineage might branch within the archaeal domain, care was taken to include an unbiased sample of all main archaeal groups in all data sets that were analyzed. The latter was achieved by clustering concatenated protein sequences comprising 55 panorthologs present in 120 archaeal species using data extracted from the arCOG database (Fig. 2; see supplemental Fig. 1 online) (Wolf et al. 2012). Using a clustering cutoff of 70% sequence identity, a set of 58 taxa was obtained that represented all major archaeal groups. This set was completed by 10 bacterial and 10 eukaryotic genomes, as well as eight recently published archaeal genomes, including the genome of the Geoarchaeote NAG1, which was proposed to represent a novel archaeal phylum (Kozubal et al. 2013) (see supplemental Table 1 online for a detailed list). Assignment of proteins encoded by each of these genomes to the arCOG clusters was achieved by performing PSI-BLAST (position-specific iterative basic local alignment search tool) analysis followed by phylogenetic evaluation of each updated arCOG to remove paralogous sequences, as well as genes that were obviously horizontally transferred (see the Material and Methods section for details). Eventually, a data set was obtained that comprised 67 protein clusters (see supplemental Table 2 online), each of which contained at least 52 archaeal sequences, as well as at least three bacterial and three eukaryotic protein sequences. This data set represented the starting point of a number of analyses that aimed to gain insight into the phylogenetic position of eukaryotes in the tree of life, focusing on effects of gene selection, taxon selection, and compositional bias. In parallel, 16S/18S and 23S/28S rDNA gene sequences were obtained from the same taxa present in the protein data set, followed by addition of 16S and 23S sequences from further archaeal taxa representing deeply diverging lineages. These sequences were also used to infer the phylogenetic position of eukaryotes in the rDNA-based tree of life and investigate any potential taxon sampling effects on this placement (see Fig. 2).
Figure 2.
Flowcharts of the data selection processes. Numbers in blue outside boxes represent the number of sequences or organisms included. Numbers in red represent the number of genes or clusters. Abbreviations in green show the phylogeny algorithms applied to the data set: ML, maximum-likelihood (RAxML); BP, BLG, and BGTR, Bayesian under CAT-Poisson, CAT-LG, and CAT-GTR model, respectively (Phylobayes). (A) Protein concatenated data sets. (B) Ribosomal RNA genes phylogeny. (C) Archaeal diversity tree.
Robust Support for the Archaeal TACK Superphylum
Recent studies have reported that the eukaryotic lineage emerged from within the so-called TACK superphylum, comprising the Thaum-, Aig-, Cren-, and Korarchaeota (Guy and Ettema 2011; Williams et al. 2012). To reevaluate these findings, we decided to first investigate whether the existence of this proposed superphylum is supported by means of phylogenetic analysis of the concatenated protein and rDNA data sets. Bayesian and maximum-likelihood (ML) analyses were performed on the data set of 67 conserved proteins clusters (see above and Fig. 2), but leaving out eukaryotic sequences, hence, comprising sequences up to 66 archaeal and 10 bacterial taxa. Likewise, Bayesian and ML analyses were performed on concatenated data sets of archaeal and bacterial 16S and 23S rDNA sequences. Bayesian and ML analyses of the concatenated protein data set, as well as the concatenated rDNA data set, provided very strong support of the existence of the TACK superphylum with BS = 100 and PP = 1.00 for both data sets (Table 1; see supplemental Fig. 2 online).
Table 1.
Summary of the phylogenies inferred from concatenated protein alignments
| Data set | Software | Model | Topologya | TACK + E supportb | See supplemental figures online |
|---|---|---|---|---|---|
| Full concatenate | RAxML | PROTCATLG | TAC,KE:74 | 74 | 3A |
| Phylobayes | CAT-Poisson | TACK:1,E | 1 | 3B | |
| 3E3B | RAxML | PROTCATLG | TAC,KE:85 | 85 | 3C |
| Phylobayes | CAT-Poisson | TACK:0.92,E | 1 | 3D | |
| 3E3BnoEuk | RAxML | PROTCATLG | (K,TA),C | 100c | 3A |
| Phylobayes | CAT-Poisson | (K,TA),C | 1c | 3B | |
| discFilter15p | RAxML | PROTCATLG | TAC,KE:100 | 100 | 3E |
| Phylobayes | CAT-Poisson | TAC,KE:0.99 | 1 | 3F | |
| CAT-GTR | TAC,KE:1 | 1 | 3G | ||
| CAT-LG | TAC,KE:0.5 | 1 | 3H | ||
| chi2filter50sd | RAxML | PROTCATLG | (TA,KE:100),C | 100 | 3I |
| Phylobayes | CAT-Poisson | (TA,KE:0.88),C | 1 | 3J |
Trees corresponding to each row of the table are available in supplemental Figure 3 online, except for the trees built on the 3E3BnoEuk data set, which are shown in supplemental Figure 2 online.
T, Thaumarchaeota; A, Aigarchaeota; C, Crenarchaeota; K, Korarchaeota; E, eukaryotes.
aTopology inside the TACK + E superphylum, in a pseudo-Newick format.
bSupport is given in percent of bootstrap for RAxML runs, in posterior probability (PP) for Bayesian trees.
cThese two values represent the support for the TACK superphylum, without eukaryotes.
The strong support obtained here agrees with previous phylogenetic analysis of 26 conserved protein-coding genes (Guy and Ettema 2011) and universally conserved ribosomal protein sequences (Wolf et al. 2012; Yutin et al. 2012). Moreover, the branching order within TACK observed in the present analysis corresponds with the branching order observed in both studies, with Korarchaeota representing the deepest branch, and Aig- and Thaumarchaeota representing sister-phyla, subseeded by Crenarchaeota—(((Thaum,Aig)Cren)Kor). It should be noted that the analysis by Yutin et al. (2012) did not include sequence data from Aigarchaeota.
Phylogenomic Analysis of Concatenated Protein and rDNA Data Sets Supports the Affiliation between TACK and Eukaryotes
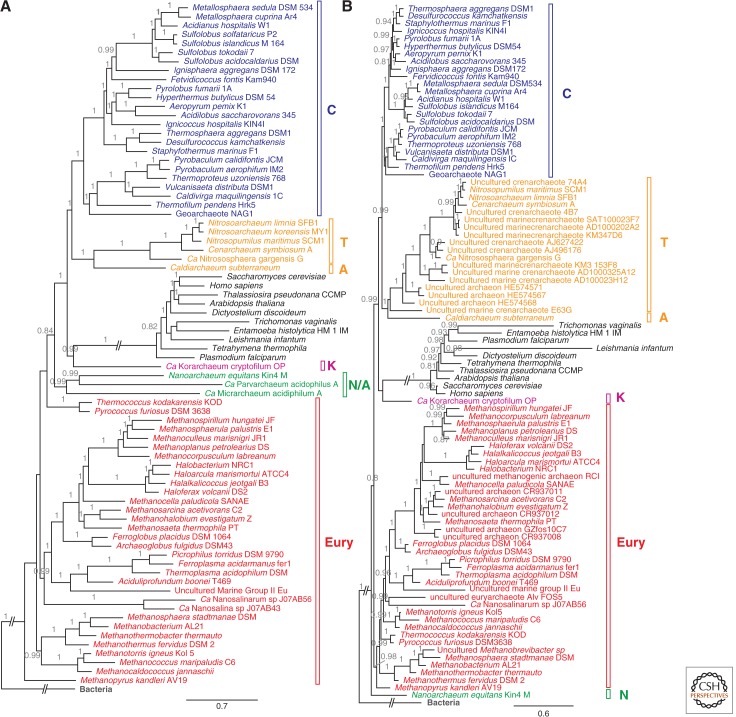
Having established strong phylogenetic support for the existence of a TACK superphylum in the archaeal domain, we sought to address the phylogenetic relationship between this group of Archaea and the eukaryotic lineage. We performed Bayesian and ML analyses on the same data set of 67 conserved proteins, as well as on the concatenated data set of 16S and 23S rDNA sequences, but now including protein, 18S, and 28S data from up to 10 eukaryotic taxa. The results of the protein data set analyses provide strong support for a phylogenetic affiliation between the TACK superphylum and eukaryotes, with bootstrap support (BS) of 85 and posterior probability (PP) of 1.00 (Table 1; see supplemental Fig. 3C,D online). The analysis of the concatenated 16S/18S and 23S/28S rDNA data sets also supported the TACK-eukaryotes affiliation, albeit with slightly lower support values in the ML analysis (BS = 79, PP = 1.00) (Table 1; see supplemental Fig. 4A online; Fig. 4B). The same analyses performed on a larger data set (162 clusters) (see supplemental Table 2 online) in which more missing data was tolerated (presence of at least 90% of archaeal sequences and one eukaryotic sequence) yielded similar results, with the TACK superphylum supported with BS = 74 and PP = 1.00 (Table 1; see supplemental Fig. 3A,B online). Altogether, these results corroborate those of a recent study performed by Williams et al. (2012), which rejected the three-domain tree of life by providing strong support of a phylogenetic affiliation of TACK and eukaryotes in both concatenated protein and rDNA gene data sets.
Figure 4.
Bayesian trees obtained from concatenated amino acid sequence alignments (A) and the concatenated sequences of the small and large ribosomal subunits (SSU and LSU, respectively) (B). In A, the tree was obtained from the alignment of the 85% least-divergent proteins (data set discFilter15p), running four chains of Phylobayes under a CAT-Poisson model. In B, the sequences for both ribosomal subunits were concatenated and a phylogeny was similarly inferred. Eukaryotes are shown in black, Bacteria in gray, Euryarchaeota in red, Nanoarchaeota and ARMAN in green, Korachaeota in pink, Thaumarchaeota and Aigarchaeota in orange, and Crenarchaeota in blue. PPs are shown on the branches. PP support values lower than 0.8 are not displayed. Branches leading to eukaryotes and Bacteria have been shortened for readability. Full figures are available as supplemental Figure 3F and 3B online, respectively.
The Phylogenetic Affiliation between TACK and Eukaryotes Is Improved by Removal of Discordant Protein Clusters
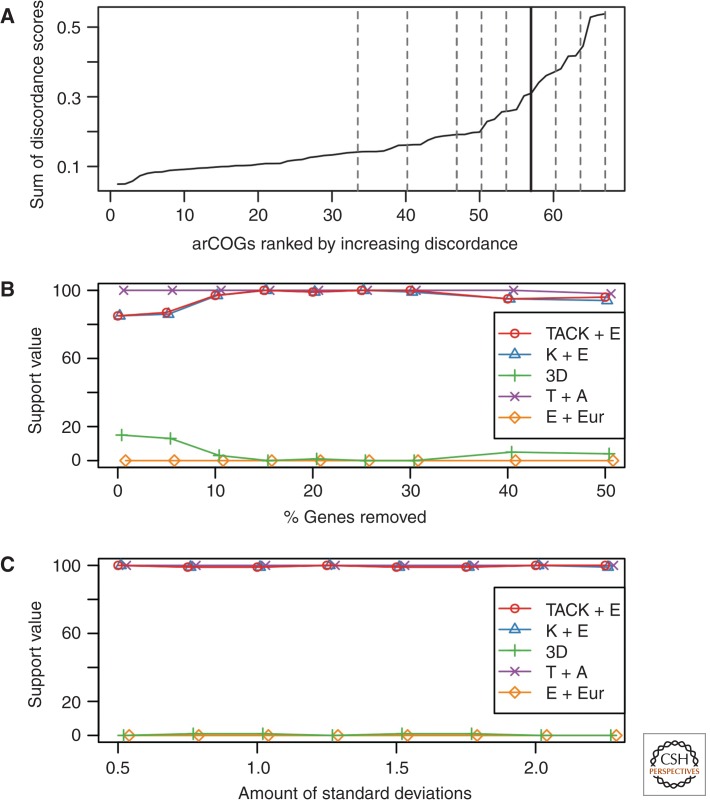
To mitigate any conflicting effect of horizontal gene transfers or fast-evolving eukaryotic sequences on the inferred TACK-eukaryotes relationship, protein clusters displaying the most divergent phylogenetic signals were removed using a discordance filtering procedure similar to the one used by Williams et al. (2010). After ranking all 67 protein clusters based on their discordance score (Fig. 3A), different fractions of the most discordant clusters were gradually removed and the remaining data sets were analyzed using an ML analysis. Removal of the most discordant clusters resulted in an increased support for the TACK-eukaryotes affiliation (red circles in Fig. 3B), and any remaining support for the three-domain tree topology (green crosses in Fig. 3B) was completely diminished after removal of 15% of most discordant clusters. These results indicate that the discordant clusters mainly were supporting the three-domain tree topology, which might point at the presence of fast-evolving eukaryotic sequences and traces of chimerism in composite microbial genomes that resided in the protein clusters (see supplemental Fig. 5 online).
Figure 3.
Evaluation of the discordance and χ2 filters. (A) Discordance score (y-axis) for all clusters (x-axis) ranked by increasing score. Dashed lines are shown at 50%, 40%, 30%, 20%, 15%, 10%, and 5% of the data, which correspond at the fractions of genes removed and further tested; see B. A thick line is shown at 15%, which is the fraction chosen for subsequent χ2 filter (C). (B) Effect of the discordance filter on selected bipartitions: TACK superphylum and eukaryotes monophyletic (TACK + E); Korarchaeota grouping with eukaryotes (K + E); each of all three domains monophyletic, corresponding to the three-domain hypothesis (3D); Thaumarchaeota and Aigarchaeota monophyletic (T + A); Crenarchaeota and Geoarchaeota (NAG1) monophyletic (C + NAG1); and Crenarchaeota not including Geoarchaeota (NAG1) monophyletic, that is, Geoarchaeota branching at the root or within the Crenarchaeota (C-NAG1). Colors and symbols, see C. The x-axis values represent the fraction of genes removed, slightly scattered to improve readability, and the y value represents the number of times a specific bipartition was found in 100 ML bootstrap trees. (C) Effect of the χ2 filter on the same bipartitions as in B. Sites diverging from the average amino acid composition by an increasing amount of standard deviations (x-axis) were removed and the support for specific bipartitions was inferred from the number of times the bipartition was found in 100 ML bootstraps (y-axis).
Removing the 15% most discordant protein clusters further substantiated the support for the eukaryotic clade to nest within the TACK superphylum, in addition to the ML analysis (BS = 100), Bayesian phylogenies run under three different models (CAT-Poisson, CAT-GTR, and CAT-LG) unanimously confirm it with posterior probabilities of 1.00. Interestingly, removing discordant protein clusters also increasingly associated eukaryotes with Korarchaeota (blue triangles, Fig. 3B). Stepwise removal of the most discordant clusters caused the BS for this affiliation to increase from 85 (no removal) to 100 (15% removal), and robust support was observed even when removing up to half of all data (BS = 94) (see Fig. 3B and supplemental Fig. 6 online). Given that this affiliation has also previously been observed by Williams et al. (2012, see their Fig. 2A), we decided to analyze the data set in which 15% of the most discordant clusters were removed using ML and Bayesian methods under the CAT-Poisson, CAT-LG, and CAT-GTR models. Intriguingly, only CAT-LG failed to retrieve the eukaryotes-Korarchaeota affiliation with significant support (PP = 0.50) (see supplemental Fig. 3H online). The ML analysis very strongly supported (BS = 100) (see supplemental Fig. 3E online) the eukaryotes-Korarchaeota affiliation, and the Bayesian analyses of this data set with the CAT-Poisson and CAT-GTR models, which are regarded to be significantly more robust against long-branch attraction artifacts compared to all other models (Lartillot et al. 2009), confidently retrieved this grouping as well (PP of 0.99 and 1.00 for CAT-Poisson and CAT-GTR, respectively) (see Fig. 4A, Table 1, and supplemental Fig. 3F,G online). The eukaryotes-Korarchaeota clade is also supported in the ML analyses of both larger data sets (BS = 74 and 85 for the 162 and 67 protein data sets, respectively) (see supplemental Fig. 3A,C online), but not in the Bayesian analyses performed on the same set in which eukaryotes are a sister clade to the TACK superphylum (PP = 1.00 and 0.92 for the 162 and 67 protein data sets, respectively) (see supplemental Fig. 3B,D online). However, given that the Korarchaeota are represented by only a single taxon, ‘Candidatus Korarchaeum cryptofilum’ (Elkins et al. 2008), it will be interesting to see whether the eukaryotes-Korarchaeota affiliation will remain stable with the inclusion of more genomic data from Korarchaeota and related archaeal species whenever this becomes available.
The Phylogenetic Affiliation between TACK and Eukaryotes Is Not Caused by Compositional Bias
Compositional bias in biological sequence data has been shown to be a major source of incorrect inference of phylogenetic relationships (Delsuc et al. 2005). To assess whether such bias is also affecting the abovementioned TACK-eukaryotes relation and nested affiliation of eukaryotes with Korarchaeota within TACK, we performed an analysis in which we gradually removed sites that were potentially compositionally biased. To do so, a χ2 filter was used that determines the relative contribution to the global amino acid composition heterogeneity for each site in a given alignment (Viklund et al. 2012). ML analyses of protein data sets in which those sites that contributed most to the composition heterogeneity were removed in a stepwise manner revealed that both the TACK-eukaryotes affiliation, as well as the nested affiliation of eukaryotes with Korarchaeota, remained strongly supported, even when removing sites more than half a standard deviation away from the average amino acid heterogeneity, that is, in total 28.4% of the most heterogeneous sites of the alignment (Fig. 3C; see supplemental Fig. 7 online). These findings indicate that the phylogenetic signal underlying the observed TACK-eukaryotes affiliation is not caused by compositional bias.
Taxon Sampling Effects Do Not Affect the Affiliation of the TACK Superphylum and Eukaryotes
In addition to the availability of informative characters in phylogenetic analyses, empirical studies underline the impact that taxon sampling can exert on the reliability of phylogenetic inference (Townsend and Lopez-Giraldez 2010). Archaeal taxa are generally sampled sparsely with respect to amount, diversity, and available genomic sequences and, moreover, several deeply rooting phyla comprise of only a single representative (e.g., Korarchaeota, Aigarchaeota, and Nanoarchaeota). Therefore, taxon-sampling artifacts might have a significant impact on the phylogenomic reconstruction of the archaeal species tree itself and placement of the eukaryotic root, in particular. To assess such effects on the placement of eukaryotes in the archaeal tree in concatenated protein and rDNA data sets, we decided to perform phylogenetic analysis on data sets from which predefined (combinations of) archaeal clades were omitted (Table 2).
Table 2.
Effect of taxon removal on the position and phylogeny of the TACK superphylum and eukaryotes
| Removed taxa | Remaining taxa | Proteins | SSU + LSU | ||||
|---|---|---|---|---|---|---|---|
| TACK + E support | Topology inside TACKa | See supplemental figures online | TACK + E support | Topology inside TACKa | See supplemental figures online | ||
| – | TACK + E | 100 | KE:100,TAC:74 | 3E | 79 | TACK:51,E | 4 |
| E | TACK | 100b | K,TAC:59 | 8A | 100b | K,TAC:86 | 9A |
| ACK | T | 100 | N.A. | 8B | 57 | N.A. | 9B |
| TCK | A | 100 | N.A. | 8C | NA | N.A. | 9C |
| TAK | C | 72 | N.A. | 8D | 60 | N.A. | 9D |
| TAC | K | 100 | N.A. | 8E | 69 | N.A. | 9E |
| CK | TA | 99 | E,TA:100 | 8F | 68 | E,TA:100 | 9F |
| AK | TC | 100 | ET:99,C | 8G | 67 | E,TC:97 | 9G |
| AC | TK | 95 | ET:36,K | 8H | 80 | E,TK:79 | 9H |
| TC | AK | 100 | E,AK:79 | 8I | 82 | E,AK:85 | 9I |
| TK | AC | 78 | E,AC:71 | 8J | 76 | E,AC:100 | 9J |
| TA | CK | 99 | EK:99,C | 8K | NA | CK:52 | 9K |
| T | ACK | 100 | EK:85,A,C | 8L | 69 | EK:37,AC:93 | 9L |
| A | TCK | 100 | EK:94,T,C | 8M | 70 | E,TCK:72 | 9M |
| C | TAK | 100 | EK:98,TA | 8N | 75 | E,TAK:83 | 9N |
| K | TAC | 91 | ETA:65,C | 8O | 73 | E,TAC:99 | 9O |
All phylogenies were inferred with RAxML under PROTCATLG, running 100 bootstraps. T, Thaumarchaeota; A, Aigarchaeota; C, Crenarchaeota; K, Korarchaeota; E, eukaryotes; B, bacteria; NA, not applicable; SSU, small ribosomal subunits; LSU, large ribosomal subunits.
aTopology inside the TACK + E superphylum using a pseudo-Newick format.
bThese two values represent support for the TACK superphylum, without eukaryotes.
The analyses of the concatenated protein data sets using ML methods indicated that removal of individual archaeal TACK phyla, as well as removal of any permutations thereof, had only very little affect on the TACK-eukaryotes affiliation. Even if the data set included only a single TACK phylum, all bootstrap values were above 70 (Table 2; see supplemental Fig. 8 online). Interestingly, in the case in which only a single phylum was removed, only removal of Korarchaeota (represented by only a single taxon) decreased support for the TACK-eukaryotes affiliation (BS = 92 vs. BS = 100 for separate removal of Thaum-, Aig-, or Crenarchaeota). The latter observation could indicate that the affiliation between Korarchaeota and eukaryotes that was observed before (see above) is genuine rather than the result of an artifact.
Support for TACK superphylum remains strong in phylogenetic analyses of concatenated rDNA sequences with a combination of excluded taxa within the TACK-eukaryotes affiliation (Table 2; see supplemental Fig. 9 online). In trees in which a single TACK phylum was removed, BS was equal or higher than 70 except for those from which Thaumarchaeota was removed. In trees where two different clades were removed (CK, AK, AC, TC, TK, or TA; for abbreviations, see Table 2), TACK + E support was above 70 in three of them (AC, TC, and TK), and in the rest of the combinations, the placement of the eukaryotic root could not be confidently established (Table 2).
In light of the results discussed above and those presented in Table 2, the effect of taxon sampling in terms of removal of any combination of TACK phyla on the phylogenetic affiliation between TACK and eukaryotes seems to be minor, and none of the analyses of concatenated protein and rDNA data sets retrieved significant support for the three-domains tree of life scenario.
CONCLUDING REMARKS
The origin of the eukaryotic cell represents an enigmatic evolutionary puzzle—a puzzle that is currently lacking too many pieces to fully appreciate the process of eukaryogenesis. Phylogenomic and comparative genomic analyses of genomic sequence data that has recently become available have already started to unveil key pieces of this puzzle. Several independent phylogenomic studies (Cox et al. 2008; Foster et al. 2009; Guy and Ettema 2011; Kelly et al. 2011; Williams et al. 2012) have now reported results that refute the three-domains tree of life, supporting scenarios in which the eukaryotic lineage evolved from a fusion event that involved a “garden variety” archaeon (Koonin 2010) as a source for the eukaryotic nuclear lineage.
In the present study, we have reevaluated the phylogenetic affiliation between Archaea and eukaryotes, focusing on effects of marker (gene) selection, taxon sampling, and compositional bias removal. The results in our study unequivocally support a phylogenetic affiliation between eukaryotes and the archaeal TACK superphylum, underscoring the conclusions drawn in recent studies (Guy and Ettema 2011; Williams et al. 2012), and providing further support for fusion-like scenarios at the root of the eukaryotic lineage. Moreover, the apparent affiliation with the TACK superphylum provides evidence against fusion scenarios that involve archaeal members of the Euryarchaeota, such as the hydrogen and syntrophic hypotheses, which involve a methanogenic fusion partner, and the sulfur syntrophy hypothesis and serial endosymbiotic theory, which envision a Thermoplasma-like fusion partner (Fig. 1C,D). Yet, our data is compatible with an extended version of the eocyte theory (Williams et al. 2012) or the recently proposed PhAT (Martijn and Ettema 2013), which both propose TACK-affiliated archaeal fusion partners. It should be noted that, whereas the eocyte hypothesis represents a so-called endokaryotic fusion model (Lake 1988; Martin 2005) with an amitochondriate, nucleated eukaryote as result (Fig. 1C), the PhAT hypothesis proposes a cellular fusion that results in a mitochondrion-containing but nucleus-lacking cell (Fig. 1D).
In several of our analyses we retrieved support for a eukaryotes-Korarchaeota sister relationship (also observed in Williams et al. 2012). Although we cannot rule out that this affiliation is artificial, for example, the result of poor taxon sampling (Korarchaeota are represented only by a single, deeply rooting taxon), it could also indicate that the eukaryotic nuclear lineage evolved from an archaeal lineage that is distantly related to Korarchaeota and which remains to be identified. Possibly, this lineage could belong to the genomically unexplored lineages that are part of the TACK superphylum (Fig. 5), such as archaeal clades with enigmatic names as Deep Sea Archaea Group, Marine Hydrothermal Vent Group , and Ancient Archaea Group (Fig. 5). The genomic exploration of these archaeal lineages, as well as those that thus far have remained undetected in the microbial biosphere, represents a unique opportunity to identify some key pieces of the puzzle of eukaryotic origins, which will hopefully allow us to gain a more profound insight into the evolutionary transition from prokaryotic to eukaryotic life on our planet.
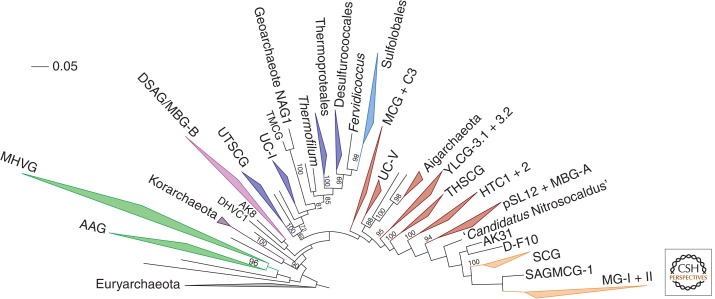
Figure 5.
Phylogenetic diversity of major archaeal clades of the TACK superphylum. The tree was constructed from an alignment of full-length sequences from 459 representative operative taxonomic units, along with 91 guide 16S rRNA sequences used in the 16S + 23S rRNA gene phylogeny, except that Nanoarchaeota was excluded. Known major clades are collapsed and shown as wedges and only bootstrap values above 70 are shown. For viewing clarity, major clades are shown as wedges and each shaded in a different color. AAG, Ancient Archaeal Group; MHVG, Marine Hydrothermal Vent Group; DHVC1, Deep-sea Hydrothermal Vent Crenarchaeotic group 1; DSAG/MBG-B, Deep-Sea Archaeal Group/Marine Benthic Group B; UTSCG, Uncultured Thermoacidic Spring Clone Group; UC-I and V, Uncultured Crenarchaeota groups I and V; TMCG, Terrestrial Miscellaneous Crenarchaeotic Group; MCG, Miscellaneous Crenarchaeal Group; YLCG-3.1 + 3.2, Yellowstone Lake Crenarchaeal Groups 3.1 and 3.2; THSCG, Terrestrial Hot Spring Crenarchaeotic Group; HTC1 + 2, Hot Thaumarchaeota-related Clades 1 and 2; pSL12 + MBG-A, pSL12-related group + Marine Benthic Group A; SCG, Soil Crenarchaeotic Group; SAGMCG-1, South African Gold Mine Crenarchaeotic Group-1; MG-I+II, Marine Groups I and II. AK8, AK31, and D-F10 are clone names. Details regarding methods for the construction of the phylogenetic tree are available in the supplemental methods online.
MATERIALS AND METHODS
A detailed description of the materials and methods is available in the supplemental material online.
ACKNOWLEDGMENTS
We thank Johan Viklund (Uppsala University) for advice and technical support on the computational analyses described here, and the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) for providing computational resources. The work in T.J.G.E.’s laboratory is supported by the Swedish Research Council (Grant No. 621-2009-4813), European Research Council (ERC) (Grant No. 310039-PUZZLE_CELL), a Marie Curie European Reintegration Grant (ERG) (Grant No. 268259-RICKOCHET), and Carl Tryggers Stiftelse (Grant No. CTS11:127). We dedicate this work to the memory of Professor Carl R. Woese, revolutionary and visionary in evolutionary biology and discoverer of the Archaea, the third domain of life.
Footnotes
Editors: Patrick J. Keeling and Eugene V. Koonin
Additional Perspectives on The Origin and Evolution of Eukaryotes available at www.cshperspectives.org
REFERENCES
- Bell PJ 2001. Viral eukaryogenesis: Was the ancestor of the nucleus a complex DNA virus? J Mol Evol 53: 251–256 [DOI] [PubMed] [Google Scholar]
- Bernander R, Lind AE, Ettema TJ 2011. An archaeal origin for the actin cytoskeleton: Implications for eukaryogenesis. Commun Integr Biol 4: 664–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR, Doolittle WF 1997. Archaea and the prokaryote-to-eukaryote transition. Microbiol Mol Biol Rev 61: 456–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, et al. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273: 1058–1073 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 1989. Molecular phylogeny. Archaebacteria and Archezoa. Nature 339: l00–101 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 2002a. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int J Syst Evol Microbiol 52: 7–76 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 2002b. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol 52: 297–354 [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287 [DOI] [PubMed] [Google Scholar]
- Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM 2008. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci 105: 20356–20361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H 2005. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet 6: 361–375 [DOI] [PubMed] [Google Scholar]
- Devos DP, Reynaud EG 2010. Evolution. Intermediate steps. Science 330: 1187–1188 [DOI] [PubMed] [Google Scholar]
- Elkins JG, Podar M, Graham DE, Makarova KS, Wolf Y, Randau L, Hedlund BP, Brochier-Armanet C, Kunin V, Anderson I, et al. 2008. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc Natl Acad Sci 105: 8102–8107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM 2006. Multiple secondary origins of the anaerobic lifestyle in eukaryotes. Philos Trans R Soc Lond B Biol Sci 361: 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM, Martin W 2006. Eukaryotic evolution, changes and challenges. Nature 440: 623–630 [DOI] [PubMed] [Google Scholar]
- Ettema TJ, Bernander R 2009. Cell division and the ESCRT complex: A surprise from the Archaea. Commun Integr Biol 2: 86–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettema TJ, de Vos WM, van der Oost J 2005. Discovering novel biology by in silico archaeology. Nat Rev Microbiol 3: 859–869 [DOI] [PubMed] [Google Scholar]
- Ettema TJ, Lindås AC, Bernander R 2011. An actin-based cytoskeleton in Archaea. Mol Microbiol 80: 1052–1061 [DOI] [PubMed] [Google Scholar]
- Forterre P 2005. The two ages of the RNA world, and the transition to the DNA world: A story of viruses and cells. Biochimie 87: 793–803 [DOI] [PubMed] [Google Scholar]
- Forterre P 2006. Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: A hypothesis for the origin of cellular domain. Proc Natl Acad Sci 103: 3669–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P 2011. A new fusion hypothesis for the origin of Eukarya: Better than previous ones, but probably also wrong. Res Microbiol 162: 77–91 [DOI] [PubMed] [Google Scholar]
- Forterre P, Gribaldo S 2010. Bacteria with a eukaryotic touch: A glimpse of ancient evolution? Proc Natl Acad Sci 107: 12739–12740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PG, Cox CJ, Embley TM 2009. The primary divisions of life: A phylogenomic approach employing composition-heterogeneous methods. Philos Trans R Soc Lond B Biol Sci 364: 2197–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribaldo S, Poole AM, Daubin V, Forterre P, Brochier-Armanet C 2010. The origin of eukaryotes and their relationship with the Archaea: Are we at a phylogenomic impasse? Nat Rev Microbiol 8: 743–752 [DOI] [PubMed] [Google Scholar]
- Guy L, Ettema TJ 2011. The archaeal “TACK” superphylum and the origin of eukaryotes. Trends Microbiol 19: 580–587 [DOI] [PubMed] [Google Scholar]
- Henderson E, Oakes M, Clark MW, Lake JA, Matheson AT, Zillig W 1984. A new ribosome structure. Science 225: 510–512 [DOI] [PubMed] [Google Scholar]
- Horiike T, Hamada K, Miyata D, Shinozawa T 2004. The origin of eukaryotes is suggested as the symbiosis of pyrococcus into γ-proteobacteria by phylogenetic tree based on gene content. J Mol Evol 59: 606–619 [DOI] [PubMed] [Google Scholar]
- Kelly S, Wickstead B, Gull K 2011. Archaeal phylogenomics provides evidence in support of a methanogenic origin of the Archaea and a thaumarchaeal origin for the eukaryotes. Proc Biol Sci 278: 1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV 2010. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol 11: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubal MA, Romine M, Jennings R, Jay ZJ, Tringe SG, Rusch DB, Beam JP, McCue LA, Inskeep WP 2013. Geoarchaeota: A new candidate phylum in the Archaea from high-temperature acidic iron mats in Yellowstone National Park. ISME J 7: 622–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Collins LJ, Penny D 2006. Genomics and the irreducible nature of eukaryote cells. Science 312: 1011–1014 [DOI] [PubMed] [Google Scholar]
- Lake JA 1988. Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences. Nature 331: 184–186 [DOI] [PubMed] [Google Scholar]
- Lake JA, Henderson E, Oakes M, Clark MW 1984. Eocytes: A new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc Natl Acad Sci 81: 3786–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S 2009. PhyloBayes 3: A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25: 2286–2288 [DOI] [PubMed] [Google Scholar]
- Lindås AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R 2008. A unique cell division machinery in the Archaea. Proc Natl Acad Sci 105: 18942–18946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia P, Moreira D 1999. Metabolic symbiosis at the origin of eukaryotes. Trends Biochem Sci 24: 88–93 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI, Koonin EV 1999. Comparative genomics of the Archaea (Euryarchaeota): Evolution of conserved protein families, the stable core, and the variable shell. Genome Res 9: 608–628 [PubMed] [Google Scholar]
- Makarova KS, Yutin N, Bell SD, Koonin EV 2010. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat Rev Microbiol 8: 731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L, Chapman M, Guerrero R, Hall J 2006. The last eukaryotic common ancestor (LECA): Acquisition of cytoskeletal motility from aerotolerant spirochetes in the Proterozoic Eon. Proc Natl Acad Sci 103: 13080–13085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martijn J, Ettema TJ 2013. From archaeon to eukaryote: The evolutionary dark ages of the eukaryotic cell. Biochem Soc Trans 41: 451–457 [DOI] [PubMed] [Google Scholar]
- Martin W 2005. Archaebacteria (Archaea) and the origin of the eukaryotic nucleus. Curr Opin Microbiol 8: 630–637 [DOI] [PubMed] [Google Scholar]
- Martin W, Muller M 1998. The hydrogen hypothesis for the first eukaryote. Nature 392: 37–41 [DOI] [PubMed] [Google Scholar]
- McInerney JO, Martin WF, Koonin EV, Allen JF, Galperin MY, Lane N, Archibald JM, Embley TM 2011. Planctomycetes and eukaryotes: A case of analogy not homology. BioEssays 33: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Lopez-Garcia P 1998. Symbiosis between methanogenic Archaea and δ-proteobacteria as the origin of eukaryotes: The syntrophic hypothesis. J Mol Evol 47: 517–530 [DOI] [PubMed] [Google Scholar]
- Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H, Chee GJ, Hattori M, Kanai A, Atomi H, et al. 2011. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res 39: 3204–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen GJ, Woese CR 1997. Archaeal genomics: An overview. Cell 89: 991–994 [DOI] [PubMed] [Google Scholar]
- Pisani D, Cotton JA, McInerney JO 2007. Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol 24: 1752–1760 [DOI] [PubMed] [Google Scholar]
- Reynaud EG, Devos DP 2011. Transitional forms between the three domains of life and evolutionary implications. Proc Biol Sci 278: 3321–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera MC, Jain R, Moore JE, Lake JA 1998. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci 95: 6239–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan L 1967. On the origin of mitosing cells. J Theor Biol 14: 255–274 [DOI] [PubMed] [Google Scholar]
- Searcy DG, Hixon WG 1991. Cytoskeletal origins in sulfur-metabolizing archaebacteria. Bio Systems 25: 1–11 [DOI] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA 1999. Genome phylogeny based on gene content. Nat Genet 21: 108–110 [DOI] [PubMed] [Google Scholar]
- Takemura M 2001. Poxviruses and the origin of the eukaryotic nucleus. J Mol Evol 52: 419–425 [DOI] [PubMed] [Google Scholar]
- Tekaia F, Lazcano A, Dujon B 1999. The genomic tree as revealed from whole proteome comparisons. Genome Res 9: 550–557 [PMC free article] [PubMed] [Google Scholar]
- Townsend JP, Lopez-Giraldez F 2010. Optimal selection of gene and ingroup taxon sampling for resolving phylogenetic relationships. Syst Biol 59: 446–457 [DOI] [PubMed] [Google Scholar]
- Viklund J, Ettema TJ, Andersson SG 2012. Independent genome reduction and phylogenetic reclassification of the oceanic SAR11 clade. Mol Biol Evol 29: 599–615 [DOI] [PubMed] [Google Scholar]
- Williams KP, Gillespie JJ, Sobral BW, Nordberg EK, Snyder EE, Shallom JM, Dickerman AW 2010. Phylogeny of gammaproteobacteria. J Bacteriol 192: 2305–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Foster PG, Nye TM, Cox CJ, Embley TM 2012. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc Biol Sci 279: 4870–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR 1987. Bacterial evolution. Microbiol Rev 51: 221–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Fox GE 1977. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc Natl Acad Sci 74: 5088–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML 1990. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci 87: 4576–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Makarova KS, Yutin N, Koonin EV 2012. Updated clusters of orthologous genes for Archaea: A complex ancestor of the Archaea and the byways of horizontal gene transfer. Biol Direct 7: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Koonin EV 2012. Archaeal origin of tubulin. Biol Direct 7: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV 2008. The deep archaeal roots of eukaryotes. Mol Biol Evol 25: 1619–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Wolf MY, Wolf YI, Koonin EV 2009. The origins of phagocytosis and eukaryogenesis. Biol Direct 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Puigbo P, Koonin EV, Wolf YI 2012. Phylogenomics of prokaryotic ribosomal proteins. PloS ONE 7: e36972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W, Schnabel R, Stetter KO 1985. Archaebacteria and the origin of the eukaryotic cytoplasm. Curr Topics Microbiol Immunol 114: 1–18 [DOI] [PubMed] [Google Scholar]
- Zillig W, Klenk HP, Palm P, Leffers H, Pühler G, Gropp F, Garrett RA 1989a. Did eukaryotes originate by a fusion event? Endocytobiosis Cell Res 6: 1–25 [Google Scholar]
- Zillig W, Klenk HP, Palm P, Puhler G, Gropp F, Garrett RA, Leffers H 1989b. The phylogenetic relations of DNA-dependent RNA polymerases of archaebacteria, eukaryotes, and eubacteria. Canadian J Microbiol 35: 73–80 [DOI] [PubMed] [Google Scholar]