Abstract
The membrane-trafficking system underpins cellular trafficking of material in eukaryotes and its evolution would have been a watershed in eukaryogenesis. Evolutionary cell biological studies have been unraveling the history of proteins responsible for vesicle transport and organelle identity revealing both highly conserved components and lineage-specific innovations. Recently, endomembrane components with a broad, but patchy, distribution have been observed as well, pieces that are missing from our cell biological and evolutionary models of membrane trafficking. These data together allow for new insights into the history and forces that shape the evolution of this critical cell biological system.
The eukaryotic membrane-trafficking system affects virtually every cellular component. Genome sequencing, molecular phylogenetics, and cell biology have advanced our understanding of its evolution.
A major feature of eukaryotic cells is subcompartmentalization. Specific components are concentrated within restricted regions of the cell, necessitating the presence of one or more targeting mechanisms. The eukaryotic membrane-trafficking system facilitates intracellular transport of proteins and lipids between organelles and further acts to build the interface between the cell and external environment. This system touches, at some level, virtually every cellular compartment and component; its proper function is crucial for modern eukaryotes.
The establishment of the membrane-trafficking system represented a tremendous milestone in the restructuring that took place during the transition from the prokaryotic to eukaryotic cellular configuration. As it does today, a membrane-trafficking system would have enhanced the ability of even the earliest eukaryotes to remodel their cell surface, export proteins to modify their external environment by exocytosis, as well as acquire nutrients by endocytosis. Subcompartmentalization of the cell and the ability to direct material to specific compartments would have allowed for intracellular specializations, for example, the sequestration of metabolic processes. Membrane trafficking also likely served to integrate fledgling endosymbiotic interactions (Flinner et al. 2013; Wideman et al. 2013), regardless of the precise timing of the mitochondrial endosymbiotic event with respect to the evolution of endogenously derived organelles (Martin and Muller 1998; Cavalier-Smith 2002; Martin and Koonin 2006; Forterre 2011). Finally, trafficking could have also facilitated a size increase for the proto-eukaryotic organisms and enabled their colonization of novel ecological niches; for example, phagocytosis is a critical function that would have been made possible by this change in morphology.
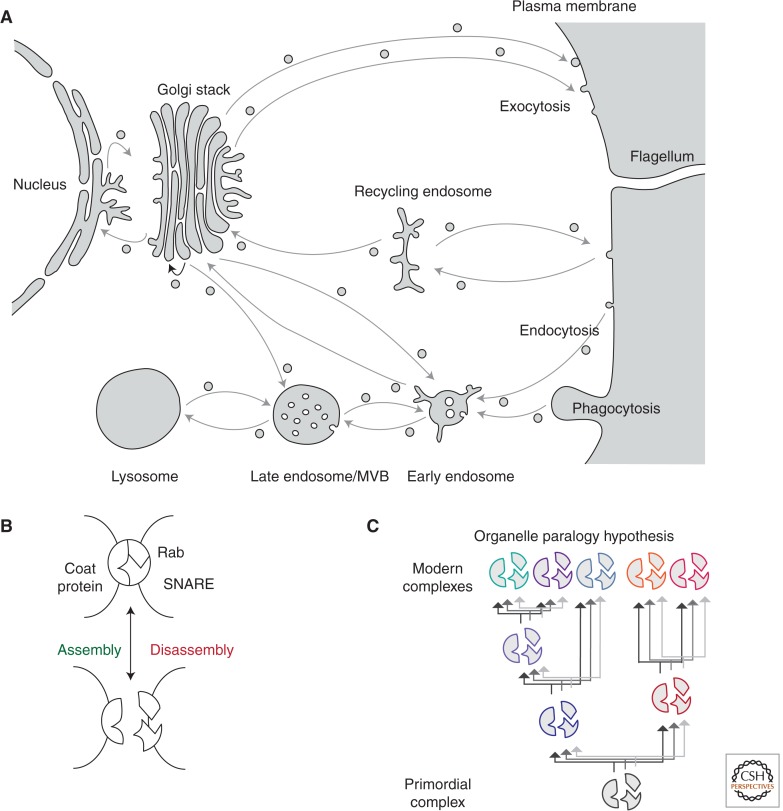
In the textbook definition (e.g., Alberts 2002), the membrane-trafficking system consists of the endoplasmic reticulum, the Golgi body, trans-Golgi network (TGN), various types of endolysosomal organelles (early, recycling, and late endosomes and lysosomes/vacuoles), as well as the plasma membrane (Fig. 1A). However, recent work has uncovered greater integration between these classical membrane-trafficking compartments and other organelles including the nucleus (Dokudovskaya et al. 2009), peroxisomes (Agrawal and Subramani 2013), and even the endosymbiotic organelles, particularly the mitochondria (Braschi et al. 2010; Michel and Kornmann 2012; Sandoval and Simmen 2012). Although the molecular details of the latter are still being unearthed, much insight has been gained into the processes of transport between membrane-trafficking organelles by vesicle formation and the subsequent delivery and fusion of the transport vesicle with a target organelle.
Figure 1.
Eukaryotic endomembrane organelles and evolution. (A) A eukaryotic cell depicting the major endomembrane organelles and trafficking pathways (denoted by arrows). Figure created from data in Wideman et al. (2013). (B) Depiction of specificity machinery encoded by multiple components of the vesicle formation and fusion machinery. For diagrammatic simplicity only the Coats, Rabs, and SNAREs are shown. (C) The organelle paralogy hypothesis for the evolution of novel endomembrane organelles by duplication and coevolution of identity-encoding genes.
The core molecular machinery for transport between endomembrane organelles consists of proteins and lipids that must, in a combinatorial manner, encode the information required for transport specificity (Cai et al. 2007). The generally accepted model for packaging of material into vesicles at a given organelle involves GTPases of the Arf/Sar family, along with a number of activating and effector proteins (Bonifacino and Glick 2004). Further to this is a requirement for cargo selection, membrane deformation, and scission involving one or more coat protein complexes (COPI, COPII, clathrin/adaptins, ESCRTs, retromer) to generate the transport carriers. Delivery of the carrier initially involves a tethering step involving Rab GTPases, and their modulating GTPase-activating proteins (GAPs) and guanine nucleotide exchange factors, as well as multisubunit tethering complexes (MTCs). The final fusion between the transport carrier and target organelle involves additional protein families such as SNAREs and SM proteins (Bonifacino and Glick 2004). Increasingly, the lines between these various sets of machineries have been blurring, with complexes being identified composed of a mixture of proteins initially identified as involved in either vesicle formation or fusion (e.g., Miller et al. 2007; Pryor et al. 2008). To add a level of complexity, many of the aforementioned proteins are, in fact, protein families in which each paralog performs the same mechanistic role, but at defined organelles or transport pathways within the cell (Bonifacino and Glick 2004). With the number of individual components involved in the membrane-trafficking process, the interconnectivity between the machineries and organelles, and with the diversity of eukaryotic organisms possessing membrane-trafficking machinery, understanding the processes of transport specificity and organelle identity benefits from a more holistic view.
Evolutionary cell biology, one aspect of which is the application of comparative molecular evolutionary analysis to cell biology (Brodsky et al. 2012), is particularly valuable in addressing such sweeping questions. Using a toolkit comprising comparative genomics, molecular phylogenetics, and, more recently, mathematical modeling, it has been possible to reconstruct the characteristics and complements of the membrane-trafficking machinery in early eukaryotic ancestors. Importantly, it has been possible to validate some of these in silico predictions of function and behavior of protein components through molecular cell biological characterization in model eukaryotes beyond mammals and yeast. This provides increased confidence in predictions of ancient membrane-trafficking systems, rather than being solely reliant on deduced histories of protein families. Furthermore, by considering the evolutionary histories of trafficking components as an integrated set or cohort, it has been possible to begin deriving mechanistic models of how nonendosymbiotic organelles may evolve. Interestingly, as surveys have advanced in scope, some unexpected patterns of conservation have begun to emerge in the machinery of membrane trafficking that have shed light on the evolution of the system, but also raised questions as to the processes that have shaped it.
A SOPHISTICATED ANCIENT MEMBRANE-TRAFFICKING MACHINERY AND AN EVOLUTIONARY MECHANISM OF NONENDOSYMBIOTIC ORGANELLE EVOLUTION
The availability of genome sequences from diverse eukaryotic organisms, and the tools to sensitively identify genes common between genomes, have allowed evolutionary investigations into the history of the membrane-trafficking system back to more than two billion years ago. The most tractable point of reconstruction is the theoretical ancestor of extant eukaryotes, the last eukaryotic common ancestor (LECA). Broadbrush surveys of membrane-trafficking machinery, at the level of the major protein families (Dacks and Doolittle 2001; Dacks and Field 2004), showed that essentially all of the major players, as defined in mammals and yeast, are likely common to most eukaryotes and thus predicted to be present in their ancestor. More specific investigations into entire trafficking pathways or specific sets of machineries (e.g., Koumandou et al. 2013) also showed the presence of near complete complements for many of these systems, as defined in animals and fungi, in the LECA. For example, the major coat proteins (COPI, COPII, clathrin, adaptins, retromer), Arf GAPs, the small GTPases (Arfs, Sar, Rabs), Syntaxins, EpsinR, and ESCRTs are all found in diverse eukaryotes, indicating once again that the LECA possessed a membrane-trafficking system at least as complex as that of most living eukaryotes. Phylogenetic analysis of paralogous protein families such as Syntaxins, Longins, Adaptins, Rabs, Arf GAPs, and TBCs has shown broad conservation of organelle-specific paralogs (Dacks and Doolittle 2002, 2004; Vedovato et al. 2009; Hirst et al. 2011; Elias et al. 2012; Gabernet-Castello et al. 2013) suggesting that these are a part of the plesiomorphic state of eukaryotes, adding depth to the deduced level of reconstructed complexity of a membrane-trafficking system in the LECA.
These analyses also permitted the postulation of a mechanism (Dacks and Field 2007; Dacks et al. 2008) to explain the evolution of nonendosymbiotic organelles, the organelle paralogy hypothesis (OPH). The OPH stems from the observation that organelle identity is the product of combinatorial interaction of the trafficking proteins found at a particular organelle (Fig. 1B) and many of these proteins belong to paralogous families. The OPH proposes that novel autogenous organelles arose as the result of gene duplication and neofunctionalization of the preexisting trafficking machinery (Fig. 1C). The strongest evidence in favor of this mechanism was the observation that, whereas many of the organelle-specific subfamilies of SNAREs, Rabs, and Adaptins had duplicated pre-LECA, a few of the paralogs associated with endocytic organelles had emerged in lineage-specific fashion independently, but with parallel functions implying a general process for organelle evolution acting on the system (Dacks et al. 2008). Recent computer simulations have confirmed that such a mechanism could indeed produce an organelle-generating mechanism based on purely theoretical calculations of protein–protein interactions and the evolution of specificity within paralogs (Ramadas and Thattai 2013).
The OPH predicts that, because organelle-specific paralogs are inferred to track the evolution of organelles, if the order of paralog duplication can be resolved, then this sequence would provide the order of emergence of the organelles as well. Because many of the identity-encoding genes, or the homologous regions within the genes, are themselves relatively short and the number of paralogs is rather high, phylogenetic resolution has been elusive until recently. However, several studies have now provided new data that can serve to test the OPH and construct hypotheses for the evolution of endomembrane organelles. ScrollSaw, a new phylogenetic pipeline for handling these difficult-to-analyze paralogous gene families, was used successfully to reconstruct the presence of up to 23 ancient Rab paralogs in the LECA (Elias et al. 2012). A separate, nearly concurrent, study using distinct methodology (Diekmann et al. 2011) derived similarly large numbers of pre-LECA Rab paralogs. Interestingly, ScrollSaw also resolved around half of the LECA Rab paralogs into two large clades corresponding to broadly endocytic and exocytic functions (Elias et al. 2012) in the case of Rab proteins in which the function has been described for extant organisms. This implies that one of the earliest functional differentiations in the trafficking system was into “in” and “out” pathways, and this may have predated the emergence of many of the individual organelles. Furthermore, a concatenated phylogeny of adaptin subunits resolved an order of paralog emergence suggesting that initial separation of COPI and adaptin subunits may have served to bridge the secretory system with the existing phagocytic system with subsequent emergence of a TGN-like organelle (Hirst et al. 2011). Thus far, the phylogenies are consistent in the predictions they make, an important prediction of the OPH. As additional resolution is obtained for additional membrane-trafficking protein families, it will be exciting to see how these data integrate, and thus extend, validate, or refute the predictions made by the OPH.
PATTERNS AND PROCESSES: CONSERVED AND LINEAGE-SPECIFIC PROTEINS
The phylogenetics of membrane-trafficking proteins gave insight into the possible mechanisms of nonendosymbiotic organelle evolution, and together with the comparative genomic analyses helped to establish the complexity of the reconstructed LECA. From these analyses, the membrane-trafficking machinery seems to fall into three gross categories of conservation: nearly ubiquitous, narrowly restricted, and broadly, but patchily distributed. The first two patterns are naïvely predicted a priori and parsed in a straightforward manner as suggestive of a drive toward complexity. The third pattern is somewhat less obvious and its implications are still not entirely clear.
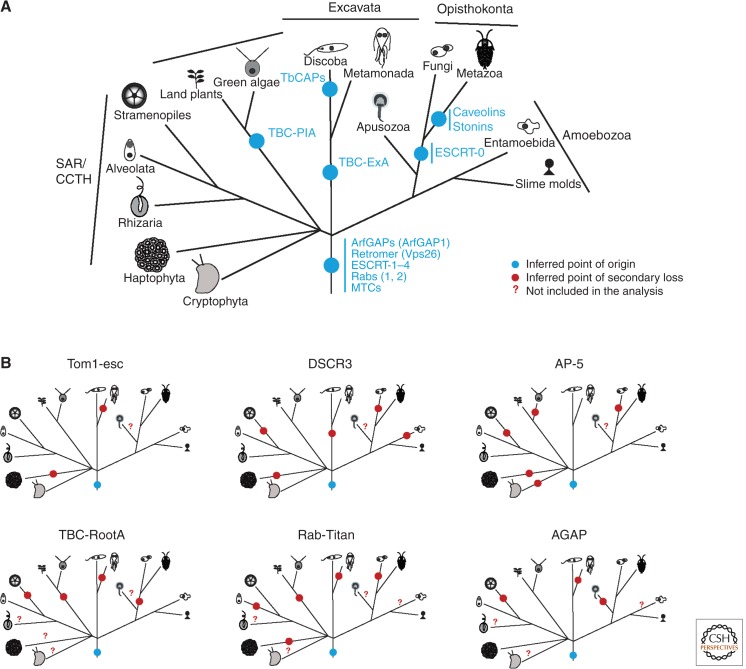
Perhaps the most expected and easy to interpret of these three classes of protein conservation are those that are completely conserved across eukaryote diversity. Such proteins are found in most, if not all, eukaryotes regardless of habitat or lifestyle and are considered necessary to the basic functions and survival of the organism. Their presence gives us confidence in many aspects of the cell biological models for membrane trafficking, with the presence of core sets of machinery in the diversity of taxa-possessing endomembrane organelles. Also, given their wide distribution and near ubiquity across the diversity of eukaryotes, they are considered to have been present in the LECA simply on the basis of parsimony. This type of conservation was the overwhelming observation at the level of the protein families that initially suggested a sophisticated LECA (Dacks and Doolittle 2001; Dacks and Field 2004). It is seen when investigating many key trafficking complexes (Fig. 2A) such as the late endosome ESCRTs (Leung et al. 2008), and the MTCs (Koumandou et al. 2007; Klinger et al. 2013), as well as much of the membrane deformation machinery, for example, COP and clathrin coat complexes (Dacks and Field 2004; Neumann et al. 2010).
Figure 2.
Examples of membrane-trafficking proteins mapped on a schematic tree of eukaryotes. (A) Examples of pre-LECA and lineage-specific membrane-trafficking proteins mapped on a schematic tree of eukaryotes. Relationships are based on the sum of molecular and morphological evidence (e.g., Walker et al. 2011; Adl et al. 2012; Burki et al. 2012). (B) The patterns of loss deduced under a hypothesis of ancient origin for six membrane-trafficking proteins. Distribution data based on Herman et al. (2011) for Tom1-esc, Koumandou et al. (2011) for DSCR3, Hirst et al. (2011) for AP-5, Schlacht et al. (2013) for AGAP, Gabernet-Castello et al. (2013) for TBC-Root A, and Elias et al. (2012) for RabTitan.
Perhaps even more instructive for understanding the evolutionary forces shaping the membrane-trafficking machinery are the cases of paralogous proteins families that show all three of the observed patterns of conservation. One excellent example is the Arf GAP family of proteins. Arf GAPs are GTPase-activating proteins for the Arf small GTPases and, more recently, have also been shown to act as Arf effectors, that is, transducing information to downstream processes (East and Kahn 2011). Originally, 10 subfamilies had been defined in humans based on the domain architectures and primary structures of the ArfGAP domain (Kahn et al. 2008). More recently, comparative genomic analysis was undertaken to assess whether or not all 10 subfamilies were conserved across euakaryotes (Schlacht et al. 2013). Of the 10 defined subfamilies ArfGAP1, ArfGAP2/3, ACAP, and SMAP were very well conserved. These families are involved in a broad range of trafficking steps, with experimental evidence from animals and yeast that both ArfGAP1 and ArfGAP2/3 are involved in retrograde traffic from the Golgi-to-ER, ACAP is involved in general endocytosis, and SMAP is involved in transport within the endosomal system (e.g., Kahn et al. 2008). A second example returns to the Rab phylogeny discussed earlier. Of the identified 23 rab paralogs likely present in the LECA (Elias et al. 2012), nine (Rab 1, 2, 4, 5, 6, 7, 8, 11, and 18) are highly conserved with only occasional losses. The majority of these (Rab 1, 2, 4, 8, 11, and 18) are involved in exocytosis, whereas the others are involved in endocytosis (Rab 5), intra-Golgi transport (Rab 6), and degradation/phagocytosis (Rab 7) (Stenmark 2009). The conservation of machinery across a breadth of processes, whether paralogous protein families or individual complexes such as the MTCs, is further suggestive of the functional complexity in the trafficking system in the LECA.
The second major pattern of conservation is lineage specificity, that is, genes are found with a restricted taxonomic distribution, suggestive of more recent paralogous expansions. Such components are therefore not validly included in generalized models of eukaryotic membrane-trafficking and, furthermore, are more recently derived (Fig. 2A). Examples of such gene products identified in the earliest surveys of endomembrane evolution occur for individual components (caveolin, stonins, GGAs) and novel paralogs of well-conserved proteins families (Boehm and Bonifacino 2001; Field et al. 2007; Kirkham et al. 2008; Diekmann et al. 2011). Two examples from the Arf GAP are ARAP and GIT, which are only found in the Filozoa (metazoa + choanoflagellates + Capsaspora owczarzaki) (Schlacht et al. 2013). Both are involved in cell adhesion through regulation of focal adhesions and cell movement (e.g., Kahn et al. 2008). Although the roles of these proteins in multicelluarity have primarily been studied in animal systems, homologs of these proteins have been identified in nonmetazoan systems. For example, the choanoflagellate Monosiga brevicollis displays the ability to attach to substrates through extracellular matrix proteins that are relatives of adhesive proteins in humans (King et al. 2008), suggesting that substrate adhesion is important to these organisms and possibly predisposes Monosiga and its relatives toward multicellularity.
Much of our understanding of how the membrane-trafficking system functions is derived from work in opisthokont (animal and fungal) model organisms and so there is a seemingly disproportionate wealth of opisthokont-specific machinery to cite. This is essentially a problem of asymmetry. There may be unidentified components in other eukaryotes, but because evolutionary studies have been biased toward searching for the functionally characterized opisthokont machinery, nonopisthokont machinery is viewed as undetected, missing pieces. Improved methodology and recognition of the bias is allowing headway. Earlier phylogenetic analyses allowed for the identification of the independent duplications giving rise to the beta subunits of adaptins 1 and 2 in plants and kinetoplastids (Dacks et al. 2008), whereas the many expansions of Rabs in vascular plants are well established (Rutherford and Moore 2002). Furthermore, the ScrollSaw methodology (Elias et al. 2012) allows the identification of paralogs absent from opisthokonts, either ancient but lost in our line, or lineage specific. Examples here (Fig. 2A) include the Rab GAP TBC-ExA in excavates, TBC-PlA and TBC-PlB in plants, and many additional lineage-specific Rab paralogs (Elias et al. 2012; Gabernet-Castello et al. 2013). Moreover, as molecular cell biological investigations in nonopisthokont models become more sophisticated and depart from the simple validation of opisthokont models, exciting examples are being found in the other supergroups as well. For example, trypanosomes are pathogens of the supergroup Excavata and responsible for a variety of diseases including African sleeping sickness and Chagas’ disease (Barrett and Croft 2012). To maintain infection, trypanosomes constantly recycle surface antigens to evade the host immune system (Allen et al. 2003). They, therefore, depend greatly on endocytosis so much so that its inhibition is lethal. Multiple adaptations have now been reported for the trypanosome endocytic system, including loss of the AP-2 complex (Manna et al. 2013) and presence of apparently trypanosome-specific proteins that associate with clathrin and regulate the budding of clathrin-coated pits from the plasma membrane (Adung’a et al. 2013). Although the function of these novel factors is not yet well characterized, this finding raises the possibility of new aspects of endocytic regulation that are not found in other eukaryotes.
Both the highly conserved and lineage-specific proteins are important for what they tell us functionally and evolutionarily. They provide context for what machinery, which has been defined in the well-characterized model systems, can be generalized to the cell biological process in all eukaryotes. Finding many of the known protein families in other eukaryotes suggests to us that many of the basic cell biological features present in animals and fungi are likely present in other organisms. Moreover, and perhaps paradoxically, these similarities can provide a platform from which we can begin to study differences between organisms to understand how natural selection affects different organisms. For proteins with a restricted distribution, the opposite is true; because these proteins are not found everywhere, they are immensely informative of the cell biology of the organisms in question and show us how they diverge from the general model.
PATTERNS AND PROCESS: PATCHY PROTEINS
More recently, a third intermediate pattern of taxonomic conservation in membrane-trafficking proteins has been reported. A hybrid of the two classes described above, these are proteins with broad retention across eukaryotes, but which are present in only a limited number of extant taxa, resulting in a “patchy” distribution. Even more significant, in some cases, these are proteins that are lost from animals and fungi and therefore at risk of omission from general models of cellular function.
Unlike the fundamental importance of highly conserved proteins or the novelty encoded by lineage-specific expansions, patchy proteins represent more of a challenge to understand. Initially, this type of distribution could be explained by sampling error or as an anomaly. However, there is now a sufficient weight of examples that this distribution needs to be considered more seriously. One example lies within the ESCRT endosomal system responsible for internal budding of vesicles within the multivesicular body (Henne et al. 2011). Subcomplexes I–IV are well conserved, whereas the ESCRT-0 subcomplex is restricted to opisthokonts (Leung et al. 2008; Herman et al. 2011). Intriguingly, a separate protein Tom1-esc has been suggested as serving some overlapping functions, binding ubiquitin, and interacting with components of the ESCRT-I subcomplex (Puertollano 2005; Blanc et al. 2009). Although not present in all taxa (Herman et al. 2011), Tom1-esc has much broader distribution than ESCRT-0 (Fig. 2B). Still within the endosomal system, DSCR3 is a second duplicate of the retromer subunit Vps26 (Hu et al. 2006). Although the retromer complex is involved in recycling of vacuolar receptors from the early endosome back to the TGN (Seaman 2012), the function of DSCR3 is unclear beyond an association with Down’s syndrome. Vps26 is a highly conserved protein, whereas DSCR3 is found widely, but not frequently (Fig. 2B) (Koumandou et al. 2011).
Examples of proteins with a patchy distribution also occur within large paralogous gene families. Adaptin proteins are cargo regulators in the late secretory and endocytic pathways. Adaptin-1 is highly conserved, whereas adaptins 2 to 5 show decreasing frequency (Hirst et al. 2011), with the newly discovered AP-5 complex, involved in trafficking from the late to the early endosome, being the least frequent (Fig. 2B). In the ARF GAP family, AGAP, which functions with AP-1 and AP-3 within the endocytic system (Nie et al. 2005), is found in at least three supergroups (Kahn et al. 2008; Schlacht et al. 2013), but apparently has been lost from some archaeplastids, stramenopiles, metamonads, and apusomonads (Fig. 2B). Additional examples include members of the Rab GTPase GAPs, the TBC family. Similar to AGAP, many of the TBC subfamilies (i.e., TBC-F through TBC-N) are found in multiple supergroups, but also seem to be absent from the majority of taxa (Elias et al. 2012; Gabernet-Castello et al. 2013). More strikingly, there are Rab GAPs and Rab GTPases, for example, TBC-RootA and RabTitan, respectively (Fig. 2B), that are present in multiple lineages but absent from humans and presumably possess roles in other eukaryotes, but are lost from our biology. There is frequently no functional data available for many of these paralogs and it will be intriguing to see whether these proteins have novel functionality that does not exist in their human counterparts or are in some manner redundant or convergent with other cellular factors.
COMPLEXITY OR THE APPEARANCE THEREOF?
The overall interpretation of the data from comparative genomics of intracellular transport, as well as other systems (Koumandou et al. 2013), is of high complexity in the LECA with losses in some lineages and continued expansion in others. Following this interpretation, on diversification, the complex cell biology of the LECA was either retained or trimmed back to a core depending on drift or selection for various niches. Complexity may have also been replenished in many cases by lineage-specific expansions. An implication is that the LECA was more complex than some prominent modern eukaryotes, and that the “patchy” proteins are remnants, essentially echoes resonating down the ages.
The pattern of patchy proteins is further consistent with the idea that the individual components of a complex assemblage may not be selective, but that complexity itself may be (Lukes et al. 2011). Conventionally the cost of retaining a specific system, or of elaborating one, is used as an argument to support secondary loss, essentially a “use it or lose it” mantra in which energetic cost is the major driver. However, under weakly selective environments, it may be favorable to retain redundant machinery, thus reducing the impact of mutations disrupting a complicated system. Under these circumstances, the predicted outcome is a system with complexity and redundancy, but that gets trimmed back and newly expanded by birth and death processes in the machinery, that is, the overall pattern of conservation that is observed with membrane-trafficking machinery, both at the broad-scale or with in-depth analyses of specific components (e.g., Koumandou et al. 2013) and more narrow taxonomic breadth (Pereira-Leal 2008). Further, as Lynch has shown (Lynch 2007; Sung et al. 2012), neutral changes can rapidly become fixed in small populations and many organisms, for example, parasites, experience frequent and extreme bottlenecks so that losses or retention may arise more from stochastic processes rather than true selection. Of course, natural selection still exerts an influence on the resultant genotype.
Nonetheless, a second interpretation for the observed pattern of patchily conserved proteins would be horizontal gene transfer (HGT) between modern eukaryotes, implying that the LECA was actually much less complex than commonly reconstructed. This latter interpretation is generally not embraced by the field, but the arguments in favor of one interpretation or the other have never been formally rallied. To do so, we can ask what aspects of the evidence are consistent with a complex LECA versus a simple LECA with HGT, and what predictions might be made by each hypothesis.
First, it is useful to recall that it is not just reconstructions of membrane transport that predict a complex LECA, but also metabolic processes and the cytoskeleton, as well as interactions and integration with the mitochondrion (e.g., Koumandou et al. 2013). Second, many of the cohorts of membrane-trafficking proteins show a spectrum of conservation from highly retained to patchily distributed, even within the same protein family. These paralogs perform the same task, sometimes redundantly with one another. The adaptin family is perhaps the best example (Fig. 2) with AP-1 never being lost and AP-5, which is infrequently found (Hirst et al. 2011). Although it is possible to invoke HGT to explain the distribution of patchy proteins, this becomes unwieldy as a hypothesis when you also consider highly conserved paralogs. Third, the pattern of organisms possessing patchily distributed proteins is incongruent with other proposed examples of HGT observed in eukaryotes. Many examples of HGT within eukaryotes enable novel metabolic functions and are found in organisms that have moved to a specific new niche, whether pathogenesis, anaerobiasis, or photosynthesis (Andersson 2009). In contrast, the membrane-trafficking proteins with a patchy distribution tend to be absent from parasites or obligate phototrophs, but present in free-living heterotrophic generalist taxa (Fig. 2). Although the latter lineages might be expected to also have high levels of HGT caused by phagocytosis (Doolittle 1998; Archibald et al. 2003; Andersson 2009), the patchy proteins of interest are also frequently found in multicellular lineages such as higher plants and animals, which tend to have very low levels of HGT. These distributions are far more consistent with a complex ancestor and a configuration that is retained if the cell faces complex and changing environments, but is pared down in cases of specialization. Again, the population size here may be significant as, once more, the specializations are associated with parasitism and bottlenecks whereas other examples are of lineages that exist in the environment and, hence, have a very large effective population size (Lynch 2007; Sung et al. 2012).
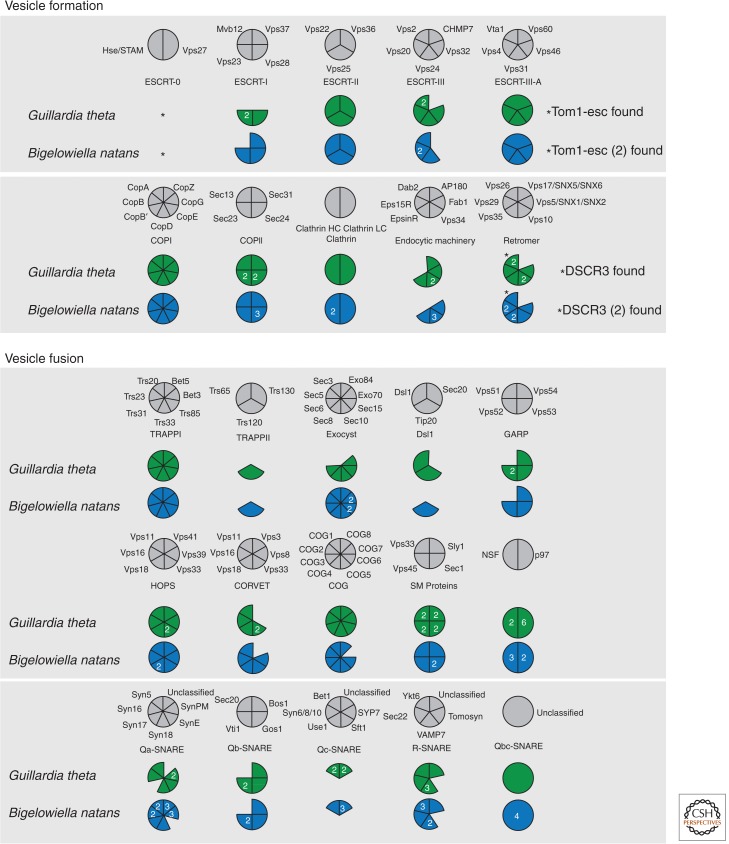
A hypothesis directly stemming from the final point predicts that if a model of ancient complexity, but with subsequent sculpting, is accurate, then as we improve sampling generalist, free-living, eukaryote genomes, we should encounter a larger set of cellular machinery and importantly increased retention of the patchy protein complement. As the first nonparasitic excavate, the Naegleria genome provided a powerful initial example (Fritz-Laylin et al. 2010; Koonin 2010). However, the best example thus far (Fig. 3) has been the membrane-trafficking machinery encoded in the genomes of two algal species, the rhizarian Bigelowiella natans and cryptophyte Guillardia theta (Curtis et al. 2012). Both organisms are free-living marine algae, performing photosynthesis using their unique secondary endosymbiotic organelles that retain the nuclear genome of the red or green algal endosymbiont, in addition to the plastid genome itself. Importantly, however, at least B. natans also ingests prey as a heterotroph and so has a complex lifestyle and exists within to a changeable environment. G. theta possesses a cytostome-like feature (“gullet”) also consistent with a capacity for heterotrophy (Lee et al. 2002). Analysis of the B. natans and G. theta membrane-trafficking complements (Fig. 3) confirmed the presence of the major families involved in vesicle formation and fusion deduced as present in the LECA. In contrast to many other microbial eukaryotic genomes in which significant reductions, for example, Giardia, Cyanydioschyzon or expansions, for example, Trichomonas, of membrane-trafficking machinery have been observed (Matsuzaki et al. 2004; Carlton et al. 2007; Morrison et al. 2007), there was little evidence of modulation of paralog numbers of the “core machinery.” Additionally, both B. natans and G. theta encode many of the proteins described above to have patchy distributions, including Tom1-esc, DSCR3, Vps10 (Fig. 3), and for B. natans, AP-5 (Curtis et al. 2012). The presence of nearly all of these membrane-trafficking proteins with a patchy distribution in B. natans and G. theta is consistent with the retained complexity interpretation.
Figure 3.
Encoded membrane-trafficking machinery in B. natans and G. theta genomes (This figure is based on data from Curtis et al. 2012 and Klinger et al. 2013.)
Although none of these arguments definitively exclude HGT as an explanation for a patchy distribution, we offer that it is more likely that we are recovering ancient complexity, with loss and sculpting. Loss therefore may well make a larger contribution to evolution of eukaryotic cells than previous models would imply.
CONCLUSIONS
In the last decade, molecular evolutionary studies have established a membrane-trafficking protein machinery core that is present in living eukaryotes, and further has deduced a likely quite complex, common ancestor. These studies also lead to a proposed mechanism for how the endomembrane organelles evolved, which is currently being extended and expanded by computational methods and increased taxonomic sampling of organisms for full genome sequencing. Lineage-specific components have also been identified, suggesting ongoing adaptations in the cellular machinery.
More recently, components with a “patchy” distribution have become more commonly observed. These proteins are not only absent from many eukaryotes, but they have been omitted from our understanding of eukaryotic membrane-trafficking evolution and function making them missing pieces of both our cell biological and evolutionary pictures. An obvious question is why this class of proteins was not immediately apparent. In some cases, this can be explained by the machinery examined. Initial evolutionary studies of membrane-trafficking proteins focused on the presence and absence of entire families and then, later on, the most functionally well-studied (and often functionally important) paralogs. These fell into the categories of broadly distributed and retained or more narrowly distributed. As work progressed, more sensitive methods were used to explore the broader scope of all paralogs within protein families, and machinery that was less functionally studied (e.g., Tom1-esc, DSCR3) was included in evolutionary analyses. In other cases, it may be a matter of improved genome sequence availability. What may have initially appeared as lineage-specific machinery because of restricted distribution is revealed as more ancient, as the early obtained genomes of parasitic and strictly autotrophic eukaryotes have been complemented by the genome sequences of free-living generalists.
Generally, proteins with a patchy distribution are often interpreted as having arisen by HGT, and it is certainly possible that some of the “patchy proteins” discussed above may have been transferred between genomes. However, when considered together with patterns of conservation of other membrane-trafficking components, a model of a highly complex LECA cellular system subsequently sculpted by gene loss as the descendant lineages moved to novel ecological niches emerges as a more likely explanation for the majority of examples.
Pairing molecular evolutionary analyses with the rapidly improving capacity for molecular cell biology in nontraditional model organisms results in a powerful toolkit for studying the evolution and basic cell biology of membrane trafficking. With these computational and genomic approaches providing detailed and robust molecular complements, experimental characterization in organisms from multiple and taxonomically diverse lineages can test assumptions of functional homology, and establish both the common and unique features of membrane trafficking in organisms of agricultural, environmental, and medical relevance, as well as enable reconstructions of ancient cell biology. This work will be particularly important in the case of patchy proteins that have previously been overlooked because of their absence or sequence divergence in the key opisthokont organisms, or simply ignored because of emphasis on the higher profile membrane-trafficking families. The more detail that we obtain through higher resolution maps of gene distribution and examining the functions of these newly identified trafficking components, the closer we can come to appreciating both ancient cellular forms and the forces that have shaped the diversity of living eukaryotes.
ACKNOWLEDGMENTS
Work is supported, and gratefully acknowledged, in the authors’ laboratories by the following agencies: Alberta Innovates Technology Futures (New Faculty Award to J.B.D.) and the Wellcome Trust (082813 to M.C.F.), and the Medical Research Council (MR/K008749/1 to M.C.F. A.S. is supported by a Natural Sciences and Engineering Research Council of Canada Post Graduate Scholarship, E.K.H. by an Alberta Innovates Health Solutions Fulltime Studentship, and M.J.K. by a Queen Elizabeth II Graduate Scholarship and a Faculty of Medicine and Dentistry and Alberta Health Services (FoMD/AHS) Recruitment Studentship.
Footnotes
Editors: Patrick J. Keeling and Eugene V. Koonin
Additional Perspectives on The Origin and Evolution of Eukaryotes available at www.cshperspectives.org
REFERENCES
- Adl SM, Simpson AG, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol 59: 429–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adung’a VO, Gadelha C, Field MC 2013. Proteomic analysis of clathrin interactions in trypanosomes reveals dynamic evolution of endocytosis. Traffic 14: 440–457 [DOI] [PubMed] [Google Scholar]
- Agrawal G, Subramani S 2013. Emerging role of the endoplasmic reticulum in peroxisome biogenesis. Front Physiol 4: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B 2002. Molecular biology of the cell. Garland Science, New York [Google Scholar]
- Allen CL, Goulding C, Field MC 2003. Clathrin-mediated endocytosis is essential in Trypanosoma brucei. EMBO J 22: 4991–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JO 2009. Gene transfer and diversification of microbial eukaryotes. Annu Rev Microbiol 63: 177–193 [DOI] [PubMed] [Google Scholar]
- Archibald JM, Rogers MB, Toop M, Ishida K, Keeling PJ 2003. Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc Natl Acad Sci 100: 7678–7683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett MP, Croft SL 2012. Management of trypanosomiasis and leishmaniasis. Br Med Bull 104: 175–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc C, Charette SJ, Mattei S, Aubry L, Smith EW, Cosson P, Letourneur F 2009. Dictyostelium Tom1 participates to an ancestral ESCRT-0 complex. Traffic 10: 161–171 [DOI] [PubMed] [Google Scholar]
- Boehm M, Bonifacino JS 2001. Adaptins: The final recount. Mol Biol Cell 12: 2907–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS 2004. The mechanisms of vesicle budding and fusion. Cell 116: 153–166 [DOI] [PubMed] [Google Scholar]
- Braschi E, Goyon V, Zunino R, Mohanty A, Xu L, McBride HM 2010. Vps35 mediates vesicle transport between the mitochondria and peroxisomes. Curr Biol 20: 1310–1315 [DOI] [PubMed] [Google Scholar]
- Brodsky FM, Thattai M, Mayor S 2012. Evolutionary cell biology: Lessons from diversity. Nat Cell Biol 14: 651. [DOI] [PubMed] [Google Scholar]
- Burki F, Okamoto N, Pombert JF, Keeling PJ 2012. The evolutionary history of haptophytes and cryptophytes: Phylogenomic evidence for separate origins. Proc Biol Sci 279: 2246–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Reinisch K, Ferro-Novick S 2007. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell 12: 671–682 [DOI] [PubMed] [Google Scholar]
- Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, et al. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315: 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol 52: 297–354 [DOI] [PubMed] [Google Scholar]
- Curtis BA, Tanifuji G, Burki F, Gruber A, Irimia M, Maruyama S, Arias MC, Ball SG, Gile GH, Hirakawa Y, et al. 2012. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492: 59–65 [DOI] [PubMed] [Google Scholar]
- Dacks JB, Doolittle WF 2001. Reconstructing/deconstructing the earliest eukaryotes: How comparative genomics can help. Cell 107: 419–425 [DOI] [PubMed] [Google Scholar]
- Dacks JB, Doolittle WF 2002. Novel syntaxin gene sequences from Giardia, Trypanosoma and algae: Implications for the ancient evolution of the eukaryotic endomembrane system. J Cell Sci 115: 1635–1642 [DOI] [PubMed] [Google Scholar]
- Dacks JB, Doolittle WF 2004. Molecular and phylogenetic characterization of syntaxin genes from parasitic protozoa. Mol Biochem Parasitol 136: 123–136 [DOI] [PubMed] [Google Scholar]
- Dacks JB, Field MC 2004. Eukaryotic cell evolution from a genomic perspective: The endomembrane system. In Organelles, genomes and eukaryote phylogeny: An evolutionary synthesis in the age of genomics (ed. Hirt RP, Horner DS), pp. 309–334 CRC, Boca Raton [Google Scholar]
- Dacks JB, Field MC 2007. Evolution of the eukaryotic membrane-trafficking system: Origin, tempo and mode. J Cell Sci 120: 2977–2985 [DOI] [PubMed] [Google Scholar]
- Dacks JB, Poon PP, Field MC 2008. Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc Natl Acad Sci 105: 588–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann Y, Seixas E, Gouw M, Tavares-Cadete F, Seabra MC, Pereira-Leal JB 2011. Thousands of rab GTPases for the cell biologist. PLoS Comput Biol 7: e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokudovskaya S, Waharte F, Schlessinger A, Pieper U, Devos DP, Cristea IM, Williams R, Salamero J, Chait BT, Sali A, et al. 2009. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol Cell Proteomics 10: M110 006478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF 1998. You are what you eat: A gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet 14: 307–311 [DOI] [PubMed] [Google Scholar]
- East MP, Kahn RA 2011. Models for the functions of Arf GAPs. Semin Cell Dev Biol 22: 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias M, Brighouse A, Gabernet-Castello C, Field MC, Dacks JB 2012. Sculpting the endomembrane system in deep time: High resolution phylogenetics of Rab GTPases. J Cell Sci 125: 2500–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Gabernet-Castello C, Dacks JB 2007. Reconstructing the evolution of the endocytic system: Insights from genomics and molecular cell biology. Adv Exp Med Biol 607: 84–96 [DOI] [PubMed] [Google Scholar]
- Flinner N, Ellenrieder L, Stiller SB, Becker T, Schleiff E, Mirus O 2013. Mdm10 is an ancient eukaryotic porin co-occurring with the ERMES complex. Biochim Biophys Acta 1833: 3314–3325 [DOI] [PubMed] [Google Scholar]
- Forterre P 2011. A new fusion hypothesis for the origin of Eukarya: Better than previous ones, but probably also wrong. Res Microbiol 162: 77–91 [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, et al. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140: 631–642 [DOI] [PubMed] [Google Scholar]
- Gabernet-Castello C, O’Reilly AJ, Dacks JB, Field MC 2013. Evolution of Tre-2/Bub2/Cdc16 (TBC) Rab GTPase-activating proteins. Mol Biol Cell 24: 1574–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD 2011. The ESCRT pathway. Dev Cell 21: 77–91 [DOI] [PubMed] [Google Scholar]
- Herman EK, Walker G, van der Giezen M, Dacks JB 2011. Multivesicular bodies in the enigmatic flagellate Breviata anathema and the evolution of ESCRT 0. J Cell Sci 124: 613–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Barlow LD, Francisco GC, Sahlender DA, Seaman MN, Dacks JB, Robinson MS 2011. The fifth adaptor protein complex. PLoS Biol 9: e1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YH, Warnatz HJ, Vanhecke D, Wagner F, Fiebitz A, Thamm S, Kahlem P, Lehrach H, Yaspo ML, Janitz M 2006. Cell array-based intracellular localization screening reveals novel functional features of human chromosome 21 proteins. BMC Genomics 7: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Bruford E, Inoue H, Logsdon JM Jr, Nie Z, Premont RT, Randazzo PA, Satake M, Theibert AB, Zapp ML, et al. 2008. Consensus nomenclature for the human ArfGAP domain-containing proteins. J Cell Biol 182: 1039–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. 2008. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451: 783–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M, Nixon SJ, Howes MT, Abi-Rached L, Wakeham DE, Hanzal-Bayer M, Ferguson C, Hill MM, Fernandez-Rojo M, Brown DA, et al. 2008. Evolutionary analysis and molecular dissection of caveola biogenesis. J Cell Sci 121: 2075–2086 [DOI] [PubMed] [Google Scholar]
- Klinger CM, Klute MJ, Dacks JB 2013. Comparative genomic analysis of multi-subunit tethering complexes demonstrates an ancient pan-eukaryotic complement and sculpting in apicomplexa. PLoS ONE 8: e76278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV 2010. Preview. The incredible expanding ancestor of eukaryotes. Cell 140: 606–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumandou VL, Dacks JB, Coulson RM, Field MC 2007. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol 7: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumandou VL, Klute MJ, Herman EK, Nunez-Miguel R, Dacks JB, Field MC 2011. Evolutionary reconstruction of the retromer complex and its function in Trypanosoma brucei. J Cell Sci 124: 1496–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumandou VL, Wickstead B, Ginger ML, van der Giezen M, Dacks JB, Field MC 2013. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit Rev Biochem Mol Biol 48: 373–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Leedale GF, Bradbury P Eds. 2002. The illustrated guide to the protozoa. Society of Protozoologists, Lawrence, KS [Google Scholar]
- Leung KF, Dacks JB, Field MC 2008. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic 9: 1698–1716 [DOI] [PubMed] [Google Scholar]
- Lukes J, Archibald JM, Keeling PJ, Doolittle WF, Gray MW 2011. How a neutral evolutionary ratchet can build cellular complexity. IUBMB Life 63: 528–537 [DOI] [PubMed] [Google Scholar]
- Lynch M 2007. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci 104: 8597–8604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna PT, Kelly S, Field MC 2013. Adaptin evolution in kinetoplastids and emergence of the variant surface glycoprotein coat in African trypanosomatids. Mol Phylogenet Evol 67: 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Koonin EV 2006. Introns and the origin of nucleus-cytosol compartmentalization. Nature 440: 41–45 [DOI] [PubMed] [Google Scholar]
- Martin W, Muller M 1998. The hydrogen hypothesis for the first eukaryote. Nature 392: 37–41 [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Nishida K, et al. 2004. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428: 653–657 [DOI] [PubMed] [Google Scholar]
- Michel AH, Kornmann B 2012. The ERMES complex and ER-mitochondria connections. Biochem Soc Trans 40: 445–450 [DOI] [PubMed] [Google Scholar]
- Miller SE, Collins BM, McCoy AJ, Robinson MS, Owen DJ 2007. A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles. Nature 450: 570–574 [DOI] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, et al. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317: 1921–1926 [DOI] [PubMed] [Google Scholar]
- Neumann N, Lundin D, Poole AM 2010. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS ONE 5: e13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Fei J, Premont RT, Randazzo PA 2005. The Arf GAPs AGAP1 and AGAP2 distinguish between the adaptor protein complexes AP-1 and AP-3. J Cell Sci 118: 3555–3566 [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB 2008. The Ypt/Rab family and the evolution of trafficking in fungi. Traffic 9: 27–38 [DOI] [PubMed] [Google Scholar]
- Pryor PR, Jackson L, Gray SR, Edeling MA, Thompson A, Sanderson CM, Evans PR, Owen DJ, Luzio JP 2008. Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell 134: 817–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R 2005. Interactions of TOM1L1 with the multivesicular body sorting machinery. J Biol Chem 280: 9258–9264 [DOI] [PubMed] [Google Scholar]
- Ramadas R, Thattai M 2013. New organelles by gene duplication in a biophysical model of eukaryote endomembrane evolution. Biophys J 104: 2553–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S, Moore I 2002. The Arabidopsis Rab GTPase family: Another enigma variation. Curr Opin Plant Biol 5: 518–528 [DOI] [PubMed] [Google Scholar]
- Sandoval CO, Simmen T 2012. Rab proteins of the endoplasmic reticulum: Functions and interactors. Biochem Soc Trans 40: 1426–1432 [DOI] [PubMed] [Google Scholar]
- Schlacht A, Mowbrey K, Elias M, Kahn RA, Dacks JB 2013. Ancient complexity, opisthokont plasticity, and discovery of the 11th subfamily of Arf GAP proteins. Traffic 14: 636–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN 2012. The retromer complex—Endosomal protein recycling and beyond. J Cell Sci 125: 4693–4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H 2009. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10: 513–525 [DOI] [PubMed] [Google Scholar]
- Sung W, Ackerman MS, Miller SF, Doak TG, Lynch M 2012. Drift-barrier hypothesis and mutation-rate evolution. Proc Natl Acad Sci 109: 18488–18492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedovato M, Rossi V, Dacks JB, Filippini F 2009. Comparative analysis of plant genomes allows the definition of the “Phytolongins”: A novel non-SNARE longin domain protein family. BMC Genomics 10: 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G, Dorrell RG, Schlacht A, Dacks JB 2011. Eukaryotic systematics: A user’s guide for cell biologists and parasitologists. Parasitology 138: 1–26 [DOI] [PubMed] [Google Scholar]
- Wideman JG, Gawryluk RM, Gray MW, Dacks JB 2013. The ancient and widespread nature of the ER-mitochondria encounter structure. Mol Biol Evol 30: 2019–2044 [DOI] [PubMed] [Google Scholar]