Abstract
Objective:
We aimed to assess the effect of APOE ε variants on warfarin-related intracerebral hemorrhage (wICH), evaluated their predictive power, and tested for interaction with warfarin in causing wICH.
Methods:
This was a prospective, 2-stage (discovery and replication), case-control study. wICH was classified as lobar or nonlobar based on the location of the hematoma. Controls were sampled from ambulatory clinics (discovery) and random digit dialing (replication). APOE ε variants were directly genotyped. A case-control design and logistic regression analysis were utilized to test for association between APOE ε and wICH. A case-only design and logistic regression analysis were utilized to test for interaction between APOE ε and warfarin. Receiver operating characteristic curves were implemented to evaluate predictive power.
Results:
The discovery stage included 319 wICHs (44% lobar) and 355 controls. APOE ε2 was associated with lobar (odds ratio [OR] 2.46; p < 0.001) and nonlobar wICH (OR 1.67; p = 0.04), whereas ε4 was associated with lobar (OR 2.09; p < 0.001) but not nonlobar wICH (p = 0.35). The replication stage (63 wICHs and 1,030 controls) confirmed the association with ε2 (p = 0.03) and ε4 (p = 0.003) for lobar but not for nonlobar wICH (p > 0.20). Genotyping information on APOE ε variants significantly improved case/control discrimination of lobar wICH (C statistic 0.80). No statistical interaction between warfarin and APOE was found (p > 0.20).
Conclusions:
APOE ε variants constitute strong risk factors for lobar wICH. APOE exerts its effect independently of warfarin, although power limitations render this absence of interaction preliminary. Evaluation of the predictive ability of APOE in cohort studies is warranted.
While reducing the risk of ischemic stroke in the setting of atrial fibrillation,1–3 anticoagulation with warfarin is associated with increased risk of intracerebral hemorrhage (ICH).4 Given the small therapeutic margin of warfarin,5 new insights into the mechanisms of warfarin-related ICH (wICH) may assist clinicians in determining the risk-to-benefit ratio of this therapeutic strategy in a given patient.
An important genetic risk factor for lobar spontaneous ICH (sICH) is the ε variants within the APOE gene.6 This association is mediated by the effect of APOE ε variants on cerebral amyloid angiopathy (CAA),7–9 a cerebral small vessel disease that accounts for 60% of lobar sICHs.10 While wICH is generally considered to be mediated by the same vasculopathies that cause sICH, the specific role of APOE ε variants in warfarin-exposed populations has not been extensively studied.
Genetic contribution is also important in warfarin metabolism.11 Identification of APOE ε as a risk factor for wICH could improve existing pharmacogenetic-based dosing algorithms12,13 for warfarin by adding information on subjects at increased risk of intracranial bleeding. Moreover, this genetic information could enhance existing risk stratification strategies, such as the CHADS2 score,14 that assist clinicians in the decision to start anticoagulation.
We aimed to establish to what extent previous results on APOE ε and sICH are applicable to wICH specifically. We hypothesized that CAA is a strong contributor to lobar wICH, mirroring the pathologic process observed for lobar sICH, and that the combination of high-risk APOE ε variants and warfarin treatment potentiate each other, resulting in higher-than-additive effects.
METHODS
Study design.
The discovery phase included data from the case-control Genetics of Cerebral Hemorrhage on Anticoagulation (GOCHA) Study,15 a multicenter US study that includes the Massachusetts General Hospital, Beth Israel Deaconess Medical Center, Mayo Clinic Jacksonville, and the Universities of Michigan, Virginia, Florida at Jacksonville, and Washington. Independent replication was undertaken in the case-control Genetic and Environmental Risk Factors for Hemorrhagic Stroke (GERFHS) Study,16 at the University of Cincinnati, Ohio.
Standard protocol approvals, registrations, and patient consents.
All studies were approved by the local institutional review board at each participating institution. Informed consent was obtained from all subjects, their legally authorized representatives, or waived via protocol-specific allowance for subjects who died during hospitalization.
Cases.
Cases enrolled in both studies were subjects of self-reported European ancestry older than 55 years in GOCHA and older than 18 years in GERFHS. Cases were ascertained across participating studies using predefined standardized criteria.6,16 wICH was defined as a new and acute neurologic deficit in warfarin-exposed subjects with compatible findings in neuroimaging. Exclusion criteria included brain tumor, trauma, vascular malformation, hemorrhagic transformation of cerebral infarction, and any other cause of secondary ICH.
Imaging.
wICH was confirmed by CT and categorized as lobar or nonlobar by stroke neurologists blinded to genotype data. wICH originating at the cortex and cortical–subcortical junction was categorized as lobar, and wICH selectively involving the basal ganglia, internal capsule, thalamus, deep periventricular white matter, cerebellum, and brainstem was categorized as nonlobar. Patients with multiple simultaneous hemorrhages involving both lobar and nonlobar regions were categorized as mixed ICH and excluded from the study (n = 3). Individuals with CT images of poor quality that precluded location assignment were excluded (n = 5). CAA status was evaluated by means of the Boston criteria.17
Controls.
Unmatched controls enrolled in both studies were subjects of self-reported European ancestry older than 55 years in GOCHA and older than 18 years in GERFHS. In the GOCHA Study, controls were ICH-free, warfarin-exposed subjects randomly sampled from ambulatory clinics at each participating study site. Warfarin-exposed controls were not available in the GERFHS Study, and study controls in this instance were ICH-free subjects sampled using random digit dialing. The distribution of APOE ε variants was confirmed to be similar in warfarin-exposed and warfarin-unexposed controls (see below). In both studies, previous diagnoses of ICH, pre-enrollment dementia, or Alzheimer disease were rule out in controls through interview and revision of medical charts.
Genotyping.
The 2 single nucleotide polymorphisms that define the ε variants in APOE (rs7412 and rs429358) were genotyped utilizing 2 different direct genotyping methods.18 The genotypes of rs7412 and rs429358 were used to determine APOE ε status (ε2ε2, ε3ε3, ε4ε4, ε3ε2, ε3ε4, or ε2ε4). Research staff in charge of genotyping was blinded to neuroimaging and clinical data. Controls were confirmed to be in Hardy-Weinberg equilibrium for APOE genotypes. Differential missingness of APOE ε variants across cases and controls was evaluated through χ2 tests. Genome-wide genotyping was also performed in 379 GOCHA samples using Illumina HumanHap610-Quad (San Diego, CA), with subsequent implementation of standard quality-control procedures.19
Statistical analysis.
Hypothesis testing.
Association analyses between APOE ε genotypes and wICH were completed separately for lobar and nonlobar hemorrhages. Logistic regression models were fitted assuming independent additive genetic effects for ε2 and ε4 (1-df additive trend test for each), and adjusting for age, sex, and hypertension. Secondary analyses comprised inclusion of additional covariates in regression models (antiplatelet medications and vascular risk factors), and restriction of the replication sample to subjects older than 55 years, the latter seeking to mirror the inclusion criteria of the discovery study.
Population stratification.
To account for population structure, principal component analysis20 was implemented in the subset of GOCHA subjects with available genome-wide genotyping. Association testing in this subset of subjects was repeated as described above adding principal components 1 and 2 as covariates in the regression models.
Meta-analysis.
Results from the discovery and replication phases were meta-analyzed using inverse-variance, fixed-effects meta-analysis. Heterogeneity was assessed by computing Cochrane Q (with corresponding p) and I2.
Genetic modeling.
All available data were reanalyzed under dominant and recessive models. Differences in model fit between these modeling strategies and the additive approach were evaluated using likelihood ratio tests.
Predictive power.
The overall contribution of APOE ε genotype data to discrimination of case/control status was evaluated by computing receiver operating characteristic curves and corresponding C statistics for different models containing APOE information, after stratifying by ICH location. Fitted models included (1) age and sex, (2) age, sex, and APOE ε, and (3) age, sex, vascular risk factors, and APOE ε. Comparison of these models was performed using likelihood ratio tests.
Interaction analysis.
Interaction between warfarin exposure and APOE ε variants was assessed using a case-only approach,21 in which the environmental exposure of interest was treated as the dependent variable and the genetic locus believed to interact with the environmental factor was tested as the independent variable.
All statistical analyses were completed using SAS 9.3 (SAS Institute Inc., Cary, NC). Quality-control processing of genome-wide data was implemented using PLINK 1.07.22 Results were considered significant at Bonferroni-corrected p < 0.025 (2 tests for lobar and nonlobar wICH).
RESULTS
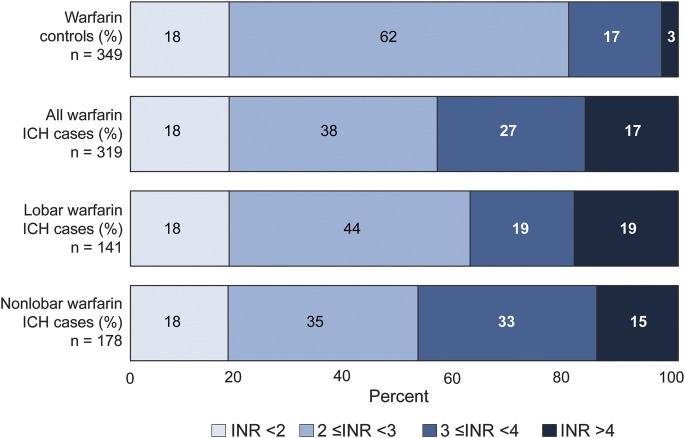
A total of 319 wICH cases (44% lobar and 56% nonlobar) and 355 controls were included in the discovery analysis (table 1). The majority of both cases (n = 232, 73%) and controls (n = 254, 72%) were receiving warfarin due to atrial fibrillation. A similar proportion of cases (18%) and controls (18%) were on infra-therapeutic levels of anticoagulation, as measured by an international normalized ratio (INR) <2, but a significantly higher proportion of cases than controls were in upper levels of anticoagulation as defined by INR ≥3 (p < 0.001, figure 1). The GERFHS Study (replication stage) included 63 wICH cases (43% lobar and 57% nonlobar) and 1,030 controls in the full cohort, and 58 wICH cases (42% lobar and 58% nonlobar) and 699 controls when restricting to subjects older than 55 years (table 1).
Table 1.
Population characteristics
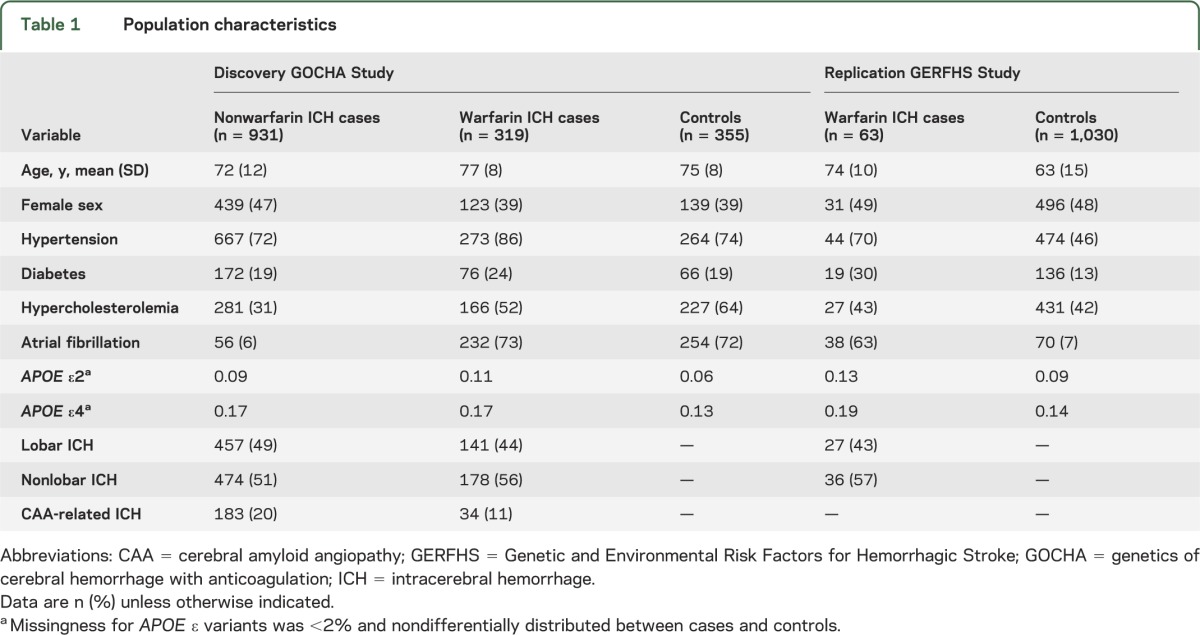
Figure 1. INR distribution by case/control status.
ICH = intracerebral hemorrhage; INR = international normalized ratio.
Lobar wICH.
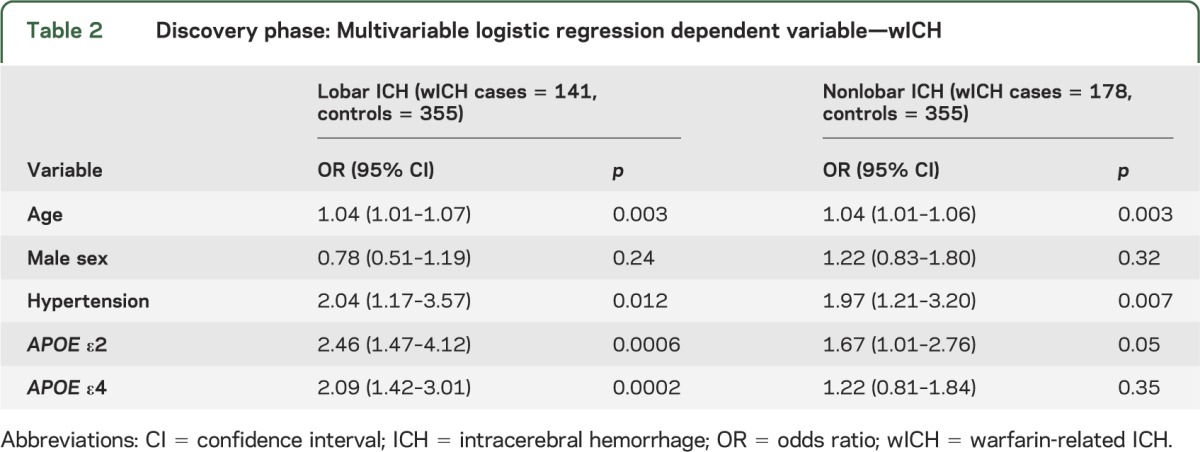
In the discovery stage, APOE ε2 and ε4 were significantly associated with lobar wICH (table 2). After adjusting for age, sex, and hypertension, each additional ε2 allele was associated with a 2.5-fold increase in risk of lobar wICH (odds ratio [OR] 2.46, 95% confidence interval [CI] 1.47–4.12; p < 0.001), whereas each additional ε4 allele was associated with 2-fold increase in the same risk (OR 2.09, 95% CI 1.42–3.01; p < 0.001). These results remained unchanged when adjusting for several additional covariates (table e-1 on the Neurology® Web site at Neurology.org). In this secondary analysis, age (p = 0.007) and hypertension (p = 0.004) were also associated with increased risk of lobar wICH, whereas hypercholesterolemia (p < 0.001) was associated with decreased risk (table e-1).
Table 2.
Discovery phase: Multivariable logistic regression dependent variable—wICH
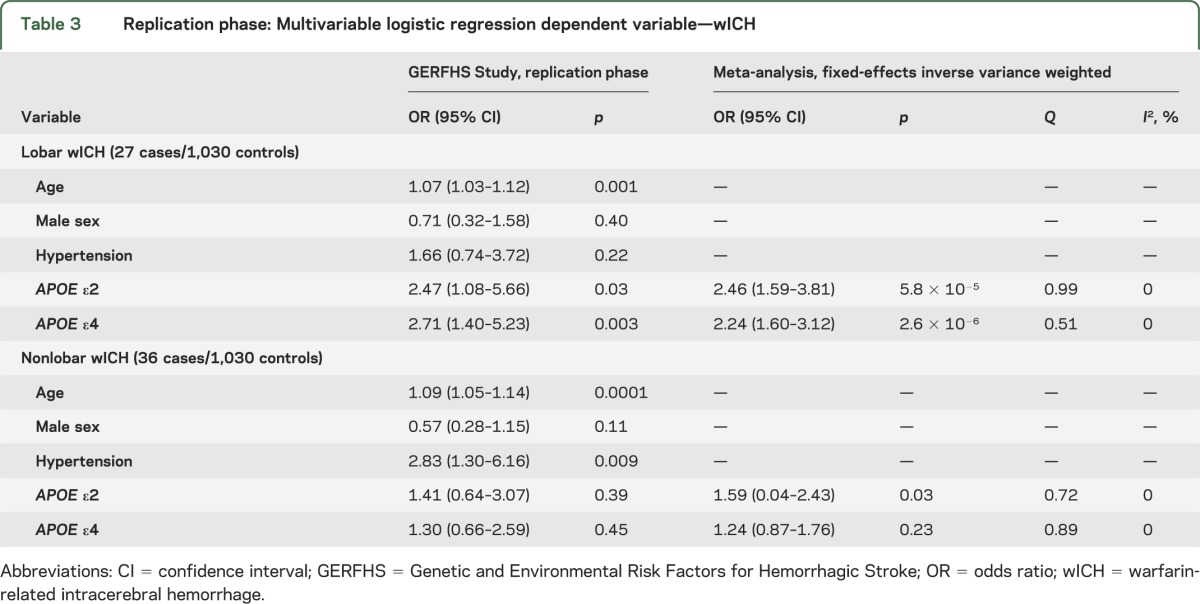
The associations described above were confirmed in the replication stage (table 3) for both ε2 (OR 2.47, 95% CI 1.08–5.66; p = 0.03; meta-analysis p = 5.8 × 10−5) and ε4 (OR 2.71, 95% CI 1.40–5.23; p = 0.003; meta-analysis p = 2.6 × 10−6). Similar results were obtained when restricting the replication sample to subjects older than 55 years (ε2 p = 0.05 and ε4 p = 0.03). The distribution of APOE ε variants was similar in warfarin-exposed controls utilized in the discovery stage and in controls not exposed to warfarin included in the replication stage (ε2 p = 0.81 and ε4 p = 0.88). No heterogeneity was observed when pooling effect estimates across the discovery and replication stages for either ε2 (Q p = 0.99, I2 = 0%) or ε4 (Q p = 0.51, I2 = 0%). Evaluation for different modeling strategies confirmed that the assumption of additive genetic effects offered superior model fit over recessive (p < 0.001) or dominant (p < 0.001) models.
Table 3.
Replication phase: Multivariable logistic regression dependent variable—wICH
CAA-associated lobar wICH.
The effect of ε2 and ε4 was stronger when the discovery sample of lobar wICH was restricted to definite/probable CAA cases, as defined by the Boston criteria.17 Thirty-four of 141 lobar wICH cases (24%) had definite/probable CAA (44% female, mean age 77 years): 7 (5%) were autopsy-confirmed, 4 (3%) had supporting pathology (brain biopsy), and 23 (16%) presented several lobar hemorrhages in neuroimaging. Despite the limited sample size and consequent widening in CIs, associations for ε2 and ε4 remained significant with increases in calculated point estimates for ORs. Each additional ε2 allele was associated with a 2.5-fold increase in risk of CAA-associated lobar wICH (OR 2.44, 95% CI 1.20–5.83; p = 0.045), whereas each additional ε4 allele was associated with a 3-fold increase in the same risk (OR 2.90, 95% CI 1.48–5.67; p = 0.002).
Nonlobar wICH.
For nonlobar wICH, only ε2 was nominally associated with risk in the discovery stage (table 2), but this association could not be reproduced in replication (table 3). After adjusting for age, sex, and hypertension, each additional ε2 allele was associated with a 67% increase in risk of nonlobar wICH (OR 1.67, 95% CI 1.01–2.76; p < 0.05). These results remained unchanged when adjusting for several other covariates (table e-1). In this secondary analysis, age (p = 0.003), hypertension (p = 0.008), and diabetes (p = 0.03) were also associated with increased risk of nonlobar wICH.
Replication results for ε2 were consistent in the direction of the effect estimate but statistically nonsignificant (OR 1.41, 95% CI 0.64–3.07; p = 0.45). Similar results were obtained when restricting the replication sample to subjects older than 55 years (ε2 p = 0.37). Meta-analysis results trended toward the Bonferroni-corrected threshold for significance (OR 1.59, 95% CI 0.04–2.43; p = 0.03; Q p = 0.72, I2 0%). As for lobar wICH, statistical modeling using additive genetics effects offered superior fit to the data when compared with recessive or dominant models (all p < 0.001).
Population stratification.
Association results were reassessed after accounting for population structure. Genome-wide data were available in 190 cases and 189 controls included in the discovery stage. In this subset of subjects, 29 population outliers (21 cases and 8 controls) were identified and removed using principal component analysis (figure e-1). After these exclusions, associations were corroborated for ε2 (OR 3.55, 95% CI 1.47–8.46; p = 0.004) and ε4 (OR 2.25, 95% CI 1.17–4.34; p = 0.01) with lobar wICH, and for ε2 (OR 2.09, 95% CI 1.07–4.18; p = 0.03) with nonlobar wICH (table e-2).
Discrimination of case/control status.
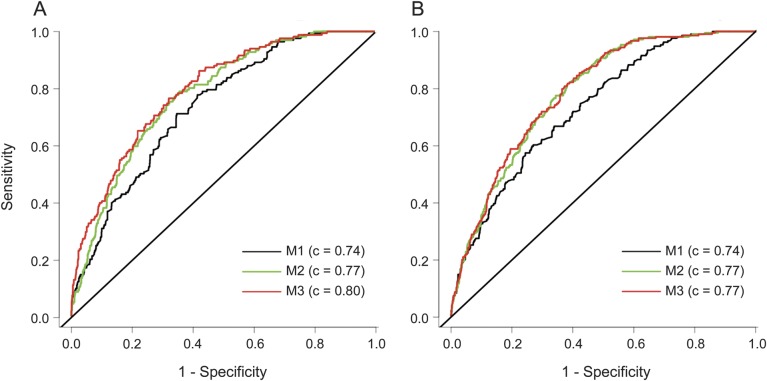
Utilization of ε2 and ε4 genotype data in regression models improved discrimination between cases and controls for lobar but not for nonlobar wICH (figure 2). Taking models that included age and sex as the reference (M1 in figure 2), inclusion of ε2 and ε4 increased the C statistic of models with (M3 in figure 2; p = 0.04) and without (M2 in figure 2; p < 0.001) vascular risk factors.
Figure 2. Receiver operating characteristic curves.
(A) Lobar wICH. (B) Nonlobar wICH. M1 = wICH ∼ age + sex; M2 = wICH ∼ age + sex + hypertension + diabetes mellitus + hypercholesterolemia; M3 = ICH ∼ age + sex + hypertension + diabetes mellitus + hypercholesterolemia + ε2 + ε4. Likelihood ratio tests for lobar wICH: M1 vs M2 p < 0.001; M1 vs M3 p < 0.001; M3 vs M2 p = 0.04. Likelihood for ratio tests nonlobar wICH: M1 vs M2 p < 0.001; M1 vs M3 p < 0.001; M3 vs M2 p = 0.23. ICH = intracerebral hemorrhage; wICH = warfarin-related ICH.
Interaction between warfarin and APOE status.
No evidence of interaction between warfarin exposure and APOE status was found in the present analysis (table e-3). Interaction was assessed utilizing a case-only design that included 931 spontaneous nonwarfarin ICH cases (457 lobar [49%] and 474 nonlobar) plus the 319 wICH cases used in the previous analyses for risk. Results for interaction analysis were null for both lobar (ε2 p = 0.92 and ε4 p = 0.17) and nonlobar hemorrhages (ε2 p = 0.18 and ε4 p = 0.85).
Post hoc power calculations.
For each wICH type, assuming a 2-sided α = 0.025, equal distribution of bleeding locations, and utilizing the allele frequencies and sample sizes described in table 1, the discovery stage had 80% power to detect ORs of 1.91 and 1.74 per each additional ε2 and ε4 allele, respectively. For the case-only interaction analysis, the study had 80% power to detect risk increments of 45% and 39% in subjects exposed to both warfarin and either ε2 or ε4 alleles, respectively.
DISCUSSION
In this large, prospective, 2-stage (discovery and replication), case-control study assessing the role of APOE ε variants in the occurrence of wICH, we demonstrated that APOE ε2 and ε4 represent strong risk factors for lobar wICH. Our results also suggest that this association is mediated by the effect of APOE ε variants on CAA. No evidence of interaction between warfarin treatment and APOE was found, indicating that subjects carrying high-risk variants and receiving warfarin are exposed to additive increases in risk of sustaining wICH, but that these 2 exposures do not potentiate each other. Finally, these data provide preliminary evidence to support a role of APOE ε genotype information in prediction models of wICH aimed to identify individuals at high risk of this condition.
Our results confirm and extend previous reports on the role of APOE in lobar wICH. A previous small (32 lobar and 9 nonlobar wICH cases), unreplicated study found that APOE ε2, but not ε4, was overrepresented in subjects with CAA-related lobar wICH compared with ICH-free controls unexposed to warfarin.23 In that study, ε2 was associated with 3.8-fold increase in risk of lobar wICH. Our results support this previous communication by providing similar effect estimates in a larger sample size that included warfarin-exposed controls, independent replication, and convincing adjustment for population stratification.
We also found evidence for a role of APOE ε4 in risk of lobar wICH. While the study mentioned above did not find a relationship between ε4 and wICH,23 a large multicenter study on sICH found that each additional ε4 allele was associated with a 2.2-fold increase in risk of lobar bleeding.6 By showing location specificity for lobar hemorrhages and a significant association even when restricting the analysis to a small number of probable/possible CAA-related hemorrhages, our results extend the role of ε4 to lobar wICH and highlight that CAA may represent an important biological mechanism for wICH. In general, these results support the notion that similar mechanisms may underlie the occurrence of wICH and sICH.
Our study did not find evidence of effect potentiation in subjects exposed to both warfarin treatment and high-risk APOE ε variants. Interaction analysis involving these 2 risk factors is difficult because of a very low prevalence of warfarin exposure in the general population. To overcome this limitation, we implemented a case-only design,21 in which the environmental exposure of interest becomes the dependent variable and the genetic locus believed to interact with the environmental factor is tested as the independent variable. This approach increases discovery power for gene-environment interactions in a variety of scenarios.24 Further studies assessing the presence of biological interaction with different designs and statistical tools are needed to confirm our findings.
Genetic information on APOE ε variants could have an important predictive role in 2 clinical settings: evaluation of the risk-benefit ratio of warfarin use in newly diagnosed cases of atrial fibrillation and specification of the optimal dosing scheme to achieve effective anticoagulation once warfarin is indicated. We have demonstrated that APOE ε status improves the ability of regression models to discriminate lobar wICH cases and controls. These results must be interpreted with caution because they were derived from a case-control study design with a fixed exposure prevalence. Nonetheless, these data do provide important support to justify further evaluation of the predictive role of APOE ε variants in wICH in properly designed cohort studies.
Several strengths of the present study contribute to the robustness our findings. All ICH cases were confirmed by neuroimaging and classified as lobar or nonlobar by expert neurologists and neuroradiologists, a crucial point given the biological heterogeneity underlying ICH in different locations of the brain.25,26 In addition, controls enrolled in the discovery stage of the study were also receiving warfarin, thus providing a more valid description of the distribution of exposure in the population that gave rise to the cases. Finally, both an independent replication and an appropriate strategy to account for population structure were implemented.
An important limitation of this study is that warfarin-exposed controls were only available in the discovery stage of the study. We address this issue by showing that the distribution of APOE genotypes in controls exposed and nonexposed to warfarin is similar. In addition, the clinic-based sampling technique for controls implemented in the discovery stage could have introduced selection bias because it would only capture warfarin-exposed individuals that are followed in ambulatory clinics. This bias is likely to be minor because the population of warfarin-exposed individuals that originated the cases requires routine follow-up in the ambulatory setting. Third, only one INR measurement per subject was available for analysis, providing only limited information on the overall temporal trend of each individual's response to warfarin. Fourth, the null results described for warfarin-APOE interaction analyses should be considered preliminary because of power limitations. Finally, these results were obtained in subjects of European ancestry and may not be applicable to other ethnicities.
The results of this study indicate that both ε2 and ε4 variants in APOE are associated with increased risk of lobar wICH. Given the known relationship between these alleles and CAA, our results provide suggestive evidence for a role of this specific cerebral small vessel disease in causing lobar intraparenchymal bleeding in warfarin-exposed subjects. APOE genotype information improves case/control discrimination, which could justify future efforts to assess the predictive power of these genetic data. We did not find evidence of potentiation of effects in subjects exposed to both warfarin and high-risk APOE ε variants.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Tammy Gillis and Marcy McDonald for technical assistance in genotyping APOE variants and Miguel Hernan, MD, DrPH, for expert advice on study design and data analysis.
GLOSSARY
- CAA
cerebral amyloid angiopathy
- CI
confidence interval
- GERFHS
Genetic and Environmental Risk Factors for Hemorrhagic Stroke
- GOCHA
Genetics of Cerebral Hemorrhage on Anticoagulation
- ICH
intracerebral hemorrhage
- INR
international normalized ratio
- OR
odds ratio
- sICH
spontaneous intracerebral hemorrhage
- wICH
warfarin-related intracerebral hemorrhage
Footnotes
Supplemental data at Neurology.org
AUTHOR CONTRIBUTIONS
Study design: G.J.F., F.R., H.B.B., A.B., J.R., C.D.A. Data collection: T.W.K.B., W.J.D., V.V., M.R.R., L.P.C., A.M.A., K.S., J.N.G., A.V., S.M.G., M.S., J.F.M., D.L.B., B.B.W., S.L.S., D.L.T., M.L.F., S.R.M., R.D., D.W. Statistical analysis: G.J.F. Manuscript drafting: G.J.F., F.R., H.B.B., J.R., C.D.A. Critical revision of the manuscript: T.W.K.B., W.J.D., V.V., M.R.R., L.P.C., A.M.A., K.S., J.N.G., A.V., S.M.G., M.S., J.F.M., D.L.B., B.B.W., S.L.S., D.L.T., M.L.F., S.R.M., R.D., A.B., P.K., D.W.
STUDY FUNDING
Supported by the NIH and the National Institute for Neurological Disorders and Stroke (NINDS) through grants R01NS059727 and P50NS061343 (GOCHA Study), NS36695 (Genetic and Environmental Risk Factors for Hemorrhagic Stroke), and NS30678 (Hemorrhagic and Ischemic Stroke among Blacks and Whites). Drs. Falcone and Brouwers were supported by the NIH-NINDS SPOTRIAS fellowship grant P50NS061343. Dr. Anderson was supported by the American Brain Foundation, and received support from the Massachusetts General Hospital Institute for Heart, Vascular, and Stroke Care for his cerebrovascular disease research.
DISCLOSURE
G. Falcone, F. Radmanesh, H. Brouwers, T. Battey, W. Devan, V. Valant, M. Raffeld, L. Chitsike, A. Ayres, K. Schwab, J. Goldstein, A. Viswanathan, and S. Greenberg report no disclosures relevant to the manuscript. M. Selim is supported by the NIH/NINDS (U01 NS074425). J. Meschia reports no disclosures relevant to the manuscript. D. Brown serves as an editorial board member of Neurology® and Stroke, is funded by NIH grants R01 NS062675, R01 HL098065, and R01 NS070941, received research support from the Blue Cross Blue Shield of Michigan Foundation and Michigan Department of Community Health, and receives research support from the University of Michigan for stroke-related research. B. Worrall serves as associate editor for Neurology. S. Silliman, D. Tirschwell, M. Flaherty, S. Martini, R. Deka, A. Biffi, P. Kraft, and D. Woo report no disclosures relevant to the manuscript. J. Rosand serves as an editorial board member of Stroke and Lancet Neurology. C. Anderson reports no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Hart RG, Pearce LA, Aguilar MI. Meta-analysis: antithrombotic therapy to prevent stroke in patients who have nonvalvular atrial fibrillation. Ann Intern Med 2007;146:857–867 [DOI] [PubMed] [Google Scholar]
- 2.Aguilar MI, Hart R. Oral anticoagulants for preventing stroke in patients with non-valvular atrial fibrillation and no previous history of stroke or transient ischemic attacks. Cochrane Database Syst Rev 2005;(3):CD001927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hylek EM, Skates SJ, Sheehan MA, Singer DE. An analysis of the lowest effective intensity of prophylactic anticoagulation for patients with nonrheumatic atrial fibrillation. N Engl J Med 1996;335:540–546 [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez-Perez A, Gaist D, Garcia Rodriguez LA. Warfarin and absolute risk of hemorrhagic stroke. CMAJ 2013;185:687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hylek EM, Go AS, Chang Y, et al. Effect of intensity of oral anticoagulation on stroke severity and mortality in atrial fibrillation. N Engl J Med 2003;349:1019–1026 [DOI] [PubMed] [Google Scholar]
- 6.Biffi A, Sonni A, Anderson CD, et al. Variants at APOE influence risk of deep and lobar intracerebral hemorrhage. Ann Neurol 2010;68:934–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bales KR, Verina T, Dodel RC, et al. Lack of apolipoprotein E dramatically reduces amyloid beta-peptide deposition. Nat Genet 1997;17:263–264 [DOI] [PubMed] [Google Scholar]
- 8.Bales KR, Verina T, Cummins DJ, et al. Apolipoprotein E is essential for amyloid deposition in the APP(V717F) transgenic mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA 1999;96:15233–15238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holtzman DM, Fagan AM, Mackey B, et al. Apolipoprotein E facilitates neuritic and cerebrovascular plaque formation in an Alzheimer’s disease model. Ann Neurol 2000;47:739–747 [PubMed] [Google Scholar]
- 10.Greenberg SM. Cerebral amyloid angiopathy: prospects for clinical diagnosis and treatment. Neurology 1998;51:690–694 [DOI] [PubMed] [Google Scholar]
- 11.Cavallari LH, Shin J, Perera MA. Role of pharmacogenomics in the management of traditional and novel oral anticoagulants. Pharmacotherapy 2011;31:1192–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pirmohamed M, Burnside G, Eriksson N, et al. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med 2013;369:2294–2303 [DOI] [PubMed] [Google Scholar]
- 13.Kimmel SE, French B, Kasner SE, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med 2013;369:2283–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gage BF, Waterman AD, Shannon W, Boechler M, Rich MW, Radford MJ. Validation of clinical classification schemes for predicting stroke: results from the National Registry of Atrial Fibrillation. JAMA 2001;285:2864–2870 [DOI] [PubMed] [Google Scholar]
- 15.Genes for Cerebral Hemorrhage on Anticoagulation (GOCHA) Collaborative Group. Exploiting common genetic variation to make anticoagulation safer. Stroke 2009;40:S64–S66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo D, Sauerbeck LR, Kissela BM, et al. Genetic and environmental risk factors for intracerebral hemorrhage: preliminary results of a population-based study. Stroke 2002;33:1190–1195 [DOI] [PubMed] [Google Scholar]
- 17.Knudsen KA, Rosand J, Karluk D, Greenberg SM. Clinical diagnosis of cerebral amyloid angiopathy: validation of the Boston criteria. Neurology 2001;56:537–539 [DOI] [PubMed] [Google Scholar]
- 18.Greenberg SM, Rebeck GW, Vonsattel JP, Gomez-Isla T, Hyman BT. Apolipoprotein E epsilon 4 and cerebral hemorrhage associated with amyloid angiopathy. Ann Neurol 1995;38:254–259 [DOI] [PubMed] [Google Scholar]
- 19.Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc 2010;5:1564–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–909 [DOI] [PubMed] [Google Scholar]
- 21.Dennis J, Hawken S, Krewski D, et al. Bias in the case-only design applied to studies of gene-environment and gene-gene interaction: a systematic review and meta-analysis. Int J Epidemiol 2011;40:1329–1341 [DOI] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosand J, Hylek EM, O’Donnell HC, Greenberg SM. Warfarin-associated hemorrhage and cerebral amyloid angiopathy: a genetic and pathologic study. Neurology 2000;55:947–951 [DOI] [PubMed] [Google Scholar]
- 24.Tchetgen Tchetgen EJ. On the interpretation, robustness, and power of varieties of case-only tests of gene-environment interaction. Am J Epidemiol 2010;172:1335–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher CM. Pathological observations in hypertensive cerebral hemorrhage. J Neuropathol Exp Neurol 1971;30:536–550 [DOI] [PubMed] [Google Scholar]
- 26.Vinters HV. Cerebral amyloid angiopathy: a critical review. Stroke 1987;18:311–324 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.