Abstract
Brucella melitensis Rev 1 is the best vaccine available for the prophylaxis of small ruminant brucellosis and, indirectly, for reducing human brucellosis. However, Rev 1 shows anomalously high rates of spontaneous dissociation from smooth (S) to rough (R) bacteria, the latter being inefficacious as vaccines. This S-R instability results from the loss of the O-polysaccharide. To overcome this problem, we investigated whether some recently described mechanisms promoting mutations in O-polysaccharide genes were involved in Rev 1 S-R dissociation. We found that a proportion of Rev 1 R mutants result from genome rearrangements affecting the wbo O-polysaccharide loci of genomic island GI-2 and the wbkA O-polysaccharide glycosyltransferase gene of the wbk region. Accordingly, we mutated the GI-2 int gene and the wbk IS transposase involved in those arrangements, and found that these Rev 1 mutants maintained the S phenotype and showed lower dissociation levels. Combining these two mutations resulted in a strain (Rev 2) displaying a 95% decrease in dissociation with respect to parental Rev 1 under conditions promoting dissociation. Rev 2 did not differ from Rev 1 in the characteristics used in Rev 1 typing (growth rate, colonial size, reactivity with O-polysaccharide antibodies, phage, dye and antibiotic susceptibility). Moreover, Rev 2 and Rev 1 showed similar attenuation and afforded similar protection in the mouse model of brucellosis vaccines. We conclude that mutations targeting genes and DNA sequences involved in spontaneous O-polysaccharide loss enhance the stability of a critical vaccine phenotype and complement the empirical stabilization precautions taken during S Brucella vaccine production.
Introduction
The members of the genus Brucella are gram-negative bacteria that cause brucellosis, an infection affecting domestic and wild animals as well as human beings. The genus includes several species among which B. melitensis, B. abortus and B. suis are the main cause of brucellosis in domestic livestock. B. melitensis preferentially infects sheep and goats and represents the most common cause of human brucellosis, a severe and debilitating disease endemic in large areas of Africa, the Middle East, Asia and Latin America [1]. This distribution reflects the problems associated with the control and eradication of a zoonotic disease in areas where extensive breeding and infrastructure weaknesses are combined. Accordingly, the World Health Organization has classified brucellosis as one of the top neglected zoonosis, a group of diseases that hamper development and contribute to the perpetuation of poverty [2].
The most effective measure to control brucellosis is the vaccination of the host ruminants [3]. Several vaccines have been developed for this purpose but the smooth (S) attenuated B. abortus S19 and B. melitensis Rev 1 strains are superior in controlled experiments [4-6] and the only ones that have proved their usefulness in successful eradication programs. Human-to-human contagion is only anecdotal, and there is abundant evidence showing that the correct use of vaccine Rev 1 results in a parallel decrease of human brucellosis, as illustrated by the experience in Greece [7]. Indeed, the use of a vaccine of good quality is essential and, in this regard, Rev 1 is not lacking problems. It is well know that, as compared to virulent B. melitensis, Rev 1 has a marked tendency to undergo the smooth-rough (S-R) dissociation that results in a dominancy of non-immunogenic R mutants that make the vaccine ineffective [8]. Indeed, procedures that minimize the S-R dissociation and controls to exclude batches containing R bacteria are critical in Rev 1 production [8].
In the brucellae, the S-R dissociation is caused by the spontaneous mutations leading to the loss of the O-polysaccharide (O-PS) of the outer membrane lipopolysaccharide (LPS). The LPS O-PS genes are concentrated in two genetic regions: wbk (which includes genes of glycosiltransferases, enzymes for the synthesis of precursors and bactoprenol priming, and several insertion sequences [IS]) and wbo (carrying the wboA and wboB glycosyltransferase genes), the latter being part of genomic island 2 (GI-2) [9-12]. Other genes coding for glycosyltransferases and synthesis of precursors are scattered in the genome [9,13]. Although the causes of the S-R dissociation have not been completely elucidated, there have been recent advances in our understanding of the main genetic mechanisms involved. In wild-type B. abortus, the spontaneous excision of 15.1 kb GI-2 island (which removes the wboA and wboB loci) by recombination promoted by the GI-2-encoded integrase (int) is one of the main sources of S-R dissociation, and identical dissociation mechanisms have been proposed for B. melitensis and B. suis (Figure 1A) [10]. A similar spontaneous recombination and excision affects wbkA (a wbk glycosyltransferase gene), in this case mediated by RecA, which recognizes the flanking wbkA transposases as substrates for homologous recombination [14] (Figure 1B). In both cases, the resulting R mutants carry a chromosomal scar that results from the release of a circular intermediate (Figure 1A and 1B) that is lost in subsequent rounds of replication. In addition, a proportion of R mutants result from random mutations affecting the manBcore gene and possibly other LPS genes related to the synthesis of O-PS and core oligosaccharide precursors and their assembly [15]. The R phenotype of the B. abortus RB51 vaccine is also known to relate in part to the disruption of gene wboA of GI-2 by IS711 (Figure 1D), which indicates the mobility of IS711 as a source of R mutations [16]. Examination of the LPS genes of B. canis and B. ovis, two naturally R species, confirms some of the above mechanisms and reveals additional possibilities for the generation of defective LPS. B. canis carries a deletion encompassing genes wbkD and wbkF (Figure 1C) probably generated by a slipped mispairing mechanism [17]. B. ovis lacks GI-2 (and bears the corresponding genomic scar) and carries mutations affecting manBcore, wbkF and other wbk genes (Figure 1E) [10-12,15,17].
Figure 1.
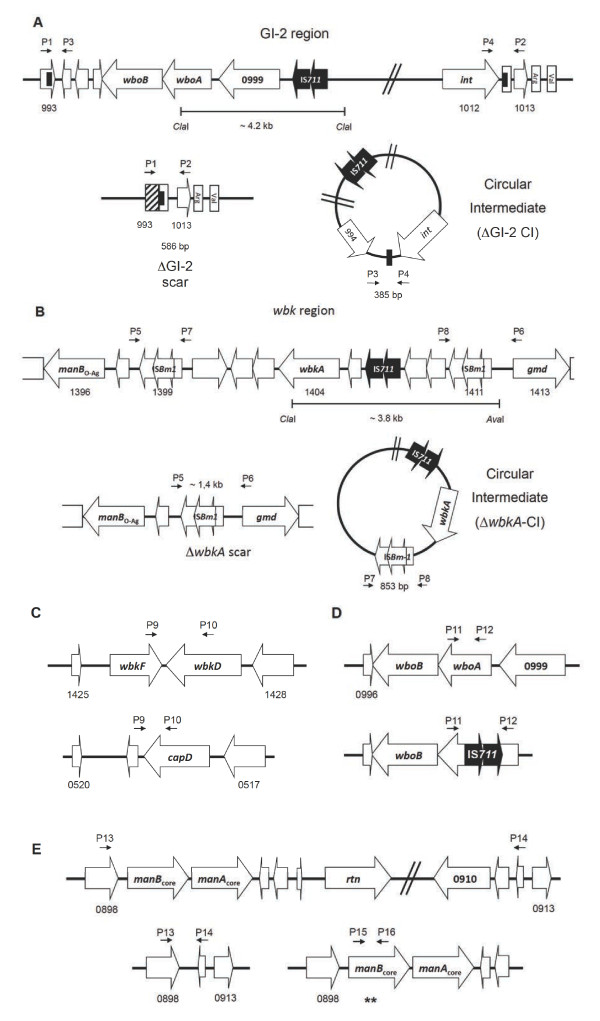
Genetic organization of regions encoding LPS genes that are susceptible to mutation with a subsequent generation of R mutants. (A) GI-2 region of B. melitensis and the scar and circular intermediate (wbo genes are omitted in the circular intermediate) resulting from the int- promoted excision; (B)wbkA region and the corresponding scar and circular intermediate resulting from homologous recombination between ISBm1 transposases; (C) the wbkFD region of B. melitensis and B. canis (wbkD is annotated as capD in B. canis); (D)wbo locus in B. melitensis and B. abortus RB51 and (E)manBAcore locus in B. melitensis chromosome II with its corresponding mutants showing the deletion and point mutations in manBcore gene (asterisks). The position of the primers used in the PCR analyses is indicated as P-number of the primer. The ORF annotations are based on the B. melitensis 16 M genome [GenBank: AE008917 and AE008918] and B. canis chromosome I [GenBank: CP000872].
Since GI-2 and region wbk are conserved in S brucellae, we hypothesized that GI-2 and wbkA excisions could be involved in the S-R dissociation of vaccine Rev 1, and that the stability of this vaccine could be improved. Here we show that these two mechanisms account for the majority of the mutations involved in the S-R dissociation of this vaccine. We also show that mutation of the int gene in GI-2 and the IS-sequences flanking wbkA in Rev 1 stabilizes the S phenotype without altering the biologically relevant characteristics of the vaccine. Accordingly, we propose the corresponding double GI-2 int and wbkA IS mutant of Rev 1 as an improved, more stable vaccine against B. melitensis infection in small ruminants.
Materials and methods
Bacterial strains and culture conditions
The Brucella strains used are listed in Table 1, and the E. coli strains and plasmids are in Additional file 1. For S-R dissociation experiments, three lots of commercial Rev 1 vaccine showing 0%, 5% and 100% of dissociation (determined by the crystal violet exclusion method, following the standard protocol for vaccine quality control [8]) were selected from a collection kept in the Centro de Investigación y Tecnología Agroalimentaria of Aragón (CITA), Zaragoza, Spain. Bacteria were grown either on trypticase soy agar (TSA; Becton Dickinson, Madrid, Spain), Blood Agar Base number 2 (BAB; Oxoid, Hampshire, UK) plates (37 °C, 1–5 days) or in trypticase soy broth (TSB; Becton Dickinson, Madrid, Spain). For cloning, E. coli was grown in Luria-Bertani broth (LB; Becton Dickinson, Madrid, Spain) supplemented with kanamycin (Km; 50 μg/mL) or chloramphenicol (Ch; 20 μg/mL). Subcultures in BAB supplemented with nalidixic acid (Nx; 25 μg/mL) were used for obtaining the B. melitensis H38-NxR spontaneous variant used as challenge strain in vaccine experiments in mice.
Table 1.
Brucella strains used.
| Strain | Characteristics | Source/ reference |
|---|---|---|
|
B. melitensis H38 |
B. melitensis smooth virulent strain; S-LPS |
CITA collection |
| H38-NxR |
B. melitensis H38 spontaneous mutant resistant to nalidixic acid; challenge studies in mice; S-LPS |
This work |
| Rev 1 |
B. melitensis smooth attenuated reference vaccine strain; S-LPS |
CITA collection |
| Rev 1ΔwbkA |
Rev 1 spontaneous deletion mutant in wbkA (ORF BMEI1398 and BME1412); R-LPS |
This work |
| Rev 1ΔGI-2 |
Rev 1 spontaneous deletion mutant in GI-2; R-LPS |
This work |
| Rev 1ΔISBm1 |
Rev 1 in-frame deletion mutant in ISBm-1 transposase gene (ORF BMEI1398 and BME1400); S-LPS |
This work |
| Rev 1Δint |
Rev 1 in-frame deletion mutant in GI-2 phage-integrase gene (nucleotides intΔ53–286); S-LPS |
This work |
| Rev 2(ΔISBm1Δint) |
Rev 1 double in-frame deletion mutant in ISBm-1 transposase and GI-2 phage-integrase; S-LPS |
This work |
| Rev 1ΔwbkA/c |
Rev 1ΔwbkA rough spontaneous mutant complemented with plasmid pMM14; S-LPS |
This work |
| Rev 1ΔGI-2/wboAB |
Rev 1ΔGI-2 rough spontaneous mutant containing the plasmid pMM76; R-LPS |
This work |
| Rev 1ΔGI-2/c | Rev 1ΔGI-2 rough spontaneous mutant complemented with plasmid pBGI-997-99c; S-LPS | This work |
S-LPS: smooth lipopolysaccharide; CITA: Centro de Investigación y Tecnología Agroalimentaria of Aragón (Zaragoza, Spain); R-LPS: rough lipopolysaccharide; GI-2: Genomic Island 2.
Sequence analyses
Confirmation of constructed mutant genotypes and analysis of manBcore locus in spontaneous R mutants was carried out on PCR fragments by DNA sequencing, using the dideoxy method at the Sequencing Unit of Centro de Investigación Médica Aplicada (CIMA, Universidad de Navarra, Spain). In silico mutants were designed using the complete sequence of chromosome I of B. melitensis 16 M as template [GenBank: AE008917] [18]. Similarity searches were performed by BLAST [19] and sequence alignments by Clustal Omega [20]. In silico restriction analyses of wbo and wbk regions were performed with Vector NTI software (Invitrogen, USA).
PCR assays
The primers designed to identify mutations known to affect the O-PS generating R phenotypes are indicated in Figure 1 and listed in Table 2. In addition, primers BMEI0993F and BMEI1013R (GI-2 deletion) or BMEI1398F and BMEI1413R (wbkA deletion) were used for detecting chromosomal scars, and primers BMEI0994R and BMEI1012bF (GI-2 region) or BMEI1400R and BMEI1409F (wbkA gene) were used for detecting the circular intermediates. Primers were generated with Primer3 software [21]. Genomic DNA (gDNA) was obtained by using standard protocols [22] or boiling (95 °C, 10 min) either directly from bacterial colonies grown in TSA plates or from Rev 1 commercial vaccine vials rehydrated in saline. PCR assays were performed in a final volume of 25 μL containing 0.2 μg of gDNA, 12.5 pmol of the correspondent primer, 0.2 mM of dNTPs, 2 mM of MgCl2 and 1U of Immolase DNA polymerase (Bioline, London, UK). The mixture was pre-incubated at 95 °C for 5 min, followed by 30 cycles (95 °C for 20 s; 60 °C for 30 s; and 72 °C for 1 min) with a final extension at 72 °C for 5 min. All PCR rounds were carried out in a Mastercycler gradient PCR machine (Eppendorf, Hamburg, Germany). Amplicons and restriction fragments were resolved by electrophoresis in 1.0-2.0% TBE (45 mM Tris-borate, 1 mM EDTA pH 8.0) agarose gels. All PCR experiments included DNA from S Rev 1 as a control.
Table 2.
List of oligonucleotides used.
| Name | Sequence (5′→ 3′) | Name in Figure1 | Reference |
|---|---|---|---|
| 711u |
cacaagactgcgttgccgacaga |
|
[23] |
| 711d |
catatgatgggaccaaacacctaggg |
|
[23] |
| BMEI0999F |
cacgatcaaaacgatgccct |
|
This study |
| BMEI0999R |
ccaaaatgtcctgagcttgg |
|
This study |
| BMEI1404F |
aagggctggaacctaggaga |
|
This study |
| BMEI1404R |
aatgacttccgctgccatag |
|
This study |
| BMEI0993F |
caacatcgcaaagcctgaaa |
P1 |
[10] |
| BMEI1013R |
cgcaatccagccaatacctg |
P2 |
[10] |
| BMEI0994R |
atcgtcggcattgtctctct |
P3 |
[10] |
| BMEI1012bF |
attatccggcggtatgtgag |
P4 |
[10] |
| BMEI1398F |
gatcttggtatcggcctgtc |
P5 |
[14] |
| BMEI1413R |
tgcgactttcttcacgattg |
P6 |
[14] |
| BMEI1400R |
cgctttaatatctcgcgttcc |
P7 |
[14] |
| BMEI1409F |
ggtcccatcggcatatctt |
P8 |
[14] |
| BMEI1426F |
ctggagtgtgccgaaagtg |
P9 |
This study |
| BMEI1427R |
gctgatctcttccgacaagg |
P10 |
This study |
| BMEI0998F |
ttaagcgctgatgccatttccttcac |
P11 |
This study |
| BMEI0998R |
gccaaccaacccaaatgctcacaa |
P12 |
This study |
| BMEII0898F |
tcggcacagcaagctataaa |
P13 |
This study |
| BMEII0912R |
ggtgtggatattgcgctttc |
P14 |
This study |
| BMEII0899F |
ccgcctatgcctatacgatg |
P15 |
This study |
| BMEII0899R |
gcctcatcatccttgtcgat |
P16 |
This study |
| BMEI1401_F1 |
ttctcgagagcctgaagagc |
|
This study |
| BMEI1401_R2 |
gccttcgtcgagaaaatgag |
|
This study |
| BMEI1397_F3 |
ctcattttctcgacgaaggccgtttgcatcaatcagttcg |
|
This study |
| BMEI1397_R4 |
ctcggctggcagtatctttc |
|
This study |
| BMEI1012_F1 |
caaagagctaagggcattcg |
|
This study |
| BMEI1012_R2 |
cgcgaaactttgaagcatct |
|
This study |
| BMEI1012_F3 |
agatgcttcaaagtttcgcgtctatatcgccggtctgtcc |
|
This study |
| BMEI1012_R4 | tttcagtgctttatgacgaaaat | This study |
F, forward; R, reverse.
Southern blot
The distribution of IS711 sequences was assessed by Southern blot using 1–2 μg of AvaI-ClaI double digested gDNA from B. melitensis Rev 1 related-strains as previously described [23]. Restriction fragments were resolved in 1% agarose-TBE at 25 mA for 10 h and probed with ECL direct-labeled system (GE Healthcare, Waukesha, WI, USA). IS711 fragment was generated by PCR with primers 711u and 711d as well as the corresponding GI-2 (~1.4 kb, BMEI0999 primers) and wbkA (332 bp, BMEI1404 primers) specific DNA probes (Table 2). Chemiluminiscent detection of the hybridized product was performed using a commercial kit (GE Healthcare, Madrid, Spain) and enhanced ECL Films for developing by conventional photographic methods. Blots were stripped with 1% SDS at 80 °C for 30 min and EDTA 20 mM at room temperature for 15 min, and then analyzed for the presence of GI-2 and wbkA with specific DNA probes.
Mutagenesis
The Rev 1 strain used in these experiments was a vaccine seed lot of proved efficacy. It was generously provided by Dr J.M. Blasco (CITA of Aragón, Spain). For mutagenesis, a parental Rev 1 clone was selected directly from colonies grown on agar according to morphology, size and agglutination criteria previously described [24,25]. A first Rev 1 (ΔISBm1 strain) mutant carrying a deletion of 872 bp spanning ORFs BMEI1398-1400 (ISBm1 complete transposase) was constructed by allelic exchange [26]. Briefly, PCR fragments produced with primers pairs BMEI1401_F1, R2 and BMEI1397_F3, R4 (Table 2) were ligated by PCR overlap, and the resulting fragment was cloned into pCR2.1 TOPO (Invitrogen, USA) to produce plasmid pMM20. The BamHI-NotI fragment of pMM20 was subcloned into plasmid pJQK [27] to generate the pMM22 suicide vector, which was transferred to Rev 1 by conjugation using E. coli S17-1λpir[28]. Double crossover transconjugants were selected on polymyxin 1.5 μg/mL and sucrose 5% plates, and the mutants were identified by PCR. The same strategy was used to construct a Rev 1Δint (BMEI1012) non-polar mutant. Two fragments generated with primer pairs BMEI1012_F1, R2 and BMEI1012_F3, R4 (Table 2) were ligated by PCR. The resulting fragment containing a copy of int lacking the nucleotides encoding amino acids 53–286 was cloned into pCR2.1 TOPO to produce the plasmid pMM55. Then, the BamHI-XbaI fragment of pMM55 was subcloned into the plasmid pJQK to generate the pMM56 suicide vector, which in turn was transferred to Rev 1 by conjugation, and the mutants were identified by PCR. Mutant Rev 1 ΔISBm1 was used to construct the double mutant ΔISBm1Δint (hereafter Rev 2) using the mutator plasmid pMM56. Maintenance of the reading frame was verified by sequencing of suicide vectors and of PCR amplified fragments of the final constructs [26].
Complementation
To confirm the origin of the mutation, spontaneous Rev 1 R strains lacking wbkA were complemented with plasmid pMM14, which carries a copy of wbkA[14]. In the case of spontaneous ΔGI-2 Rev1 R mutants, plasmids pMM76 (carrying wboA-wboB) or pBGI-997-99c (carrying wboA-wboB plus ORF BMEI0999) were used [29].
S-R dissociation assessment
A loop of bacteria previously grown on TSA plates for 72–96 h at 37 °C was transferred to a flask containing 10 mL of TSB, and the flask was incubated until bacteria reached the stationary phase (37 °C, 72–96 h). An aliquot of this culture was adjusted to 0.109 of optical density at 750 nm (equivalent to 108 CFU/mL) using sterile TSB and then diluted 1:100 in the same broth. After incubation for 3–4 days at 37 °C, serial ten-fold dilutions were plated on TSA, plates were incubated (37 °C, 5 days), and S-R dissociation was assessed by the crystal violet exclusion method and the R phenotypes confirmed by agglutination with 0.1% acriflavine [5,8]. Dissociation rate was calculated as the ratio between the number of R colonies and the total colony counts. In addition, commercial Rev 1 vaccine lots were analyzed for dissociation by PCR amplification (see above) of ∆GI-2 and ∆wbkA chromosomal scars using DNA obtained directly from vaccine batches reconstituted as recommended by the manufacturers.
Bacteriological characterization
Brucella spp. and Rev 1-specific phenotypic characteristics were confirmed or assessed by established procedures [24,25]. In addition, growth curves were obtained in a BioScreen C [30] apparatus. For this, the parental Rev 1, the Rev 2 double mutant ΔISBm1Δint and the virulent B. melitensis H38 (as a control) were inoculated (5 × 106 CFU/well) by triplicate in Bioscreen multi-well plates containing 250 μL/well of TSB, and the optical density was read at 600 nm wavelength (O.D.600) at 10 min intervals, until to reach the stationary phase.
LPS characterization
The S or R nature of LPS of the mutant and complemented strains was analyzed by Western blot of SDS-proteinase K extracts using the serum of a rabbit immunized with B. melitensis 16 M [9,31]. Purified S and R-LPS obtained as described previously [9,31] were used as controls.
Experiments in mice
The biological properties of Rev 1ΔISBm1Δint mutant (Rev 2) determined in comparison with the reference Rev 1 vaccine were virulence and protective efficacy in the mouse model. Female BALB/c mice 7–8 weeks old were purchased from Charles River International (France) and accommodated in the animal facilities of CITA (registration code ES/50-2970-12005) or Universidad Pública de Navarra (UPNA; registration code ES/31-2016-000002 CR-SU-US) for 1–2 weeks before starting and during the assays, with water and food ad libitum. Animal handling was performed in compliance with current European and national (RD 53/2013) regulations, following the FELASA and ARRIVE guidelines, and with the approval of the CITA or UPNA Animal Experimentation Committees and local Government authorization (approval reference number R130/2012). Mice inoculations were carried out with 0.1 mL of bacterial suspension previously adjusted to an optical density at 600 nm of 0.170 (approximately 1 × 109 CFU/mL) and then diluted to the appropriate dose (see below) in sterile phosphate buffered saline [32]. For virulence, groups of 30 mice each were inoculated intraperitoneally with 1 × 105 CFU/mouse of Rev 2 or Rev 1 attenuated strains. As a control, an additional group of 30 mice were inoculated similarly with the virulent B. melitensis H38 reference strain. Spleen weights and viable counts (n = 5) were determined at 1, 3, 6, 9, 12 and 15 weeks post-inoculation as described previously [32]. The identity of the spleen isolates was confirmed by PCR, and the constancy of the typical Rev 1 phenotypic features confirmed throughout the experiments. Spleen weights were expressed as the mean and SD (n = 5) of grams/spleen and the level of spleen infections as mean ± SD (n = 5) of individual log10 CFU/spleen at the indicated times.
Efficacy studies were carried out in groups of 5 BALB/c mice each vaccinated subcutaneously with 1 × 105 CFU/mouse of Rev 2 or Rev 1 (parental control), or with 0.1 mL of phosphate buffered saline (pH 6.85) as the placebo vaccinated control. Four weeks after vaccination, all mice were challenged intraperitoneally with 1 × 104 CFU/mouse of B. melitensis H38-NxR, and the number of challenge bacteria in spleens was determined 2 weeks afterwards [32]. Differentiation between challenge and residual vaccine bacteria was performed by double plating on BAB and BAB supplemented with Nx 25 μg/mL. The results were expressed as the mean and SD (n = 5) of the log10 of B. melitensis H38-NxR/spleen. In a previous work, the virulence in mice of the B. melitensis H38-NxR challenge strain used in this work was found to be indistinguishable from that of the reference H38 and 16 M strains [9].
In both virulence and protection experiments, statistical comparisons of means were performed by a one-way ANOVA followed by the Fisher’s Protected Least Significant Differences tests [32].
Results
Deletions involving GI-2 and wbkA are the major causes of S-R dissociation of B. melitensis Rev 1
As a first approach to determine the dissociation mechanisms operating in Rev 1, we searched for ∆GI-2 and ∆wbkA chromosomal scars, for deletions in wbkF and wbkD, insertion of IS elements on wboA and manBcore mutations (Figure 1) in Rev 1 spontaneous R mutants. To this end, we studied a collection of 32 R Rev 1 isolates (previously obtained by repeated plating on TSA and crystal violet and acriflavine agglutination screening) and characterized them by PCR using the primers described in Table 1. We found 8 ΔGI-2 and 3 ΔwbkA mutants, but we did not detect redundancy or other mutations known to generate R phenotypes in Brucella, including point mutations in manBcore in the remaining 21 mutants.
To confirm that the above observations corresponded in fact to the expected GI-2 or wbkA deletions and O-PS defects, we first characterized Rev 1 R mutants by IS711-fingerprinting using DNA probes specific for the wbkA and GI-2 loci. We found that, as predicted, ΔGI-2 and ΔwbkA Rev 1 mutants lacked the IS711 bands of 4.2 and 3.8 kb, respectively (Figure 2A). Second, we examined the LPS phenotype by Western blot. Figure 2B shows that the LPS of the Rev 1 mutants lacked the O-PS. Third, we performed complementation experiments in eleven R mutants using plasmids encoding the wbkA (pMM14) or the tandem wboA-wboB of GI-2 (pMM76). We found that the 3 spontaneous Rev 1 ΔwbkA R isolates were complemented with plasmid pMM14 (a representative result can be seen in Figure 2B, lane ΔwbkA/c). However, we repeatedly failed to complement the Rev 1 ΔGI-2 R isolates with plasmid pMM76 (not shown). Rajashekara et al. [29] demonstrated that artificially constructed B. melitensis ΔGI-2 R mutants could be reverted to S phenotype with a plasmid (pBGI-997-99c) bearing wboA and wboB plus ORF BMEI0999. Using this approach, we could revert 8 spontaneous Rev 1 ΔGI-2 R isolates (a representative result can be seen in Figure 2B, lane ΔGI-2/c). These results show that none of these genetically well-characterized R isolates carried more than one R-linked mutation and, in addition, support the hypothesis that the protein encoded by BMEI0999 plays a role in O-PS synthesis. In summary, the results showed that two previously described dissociation mechanisms, int-dependent (for GI-2 excision) and wbk IS-RecA dependent (for wbkA), are actually active in Rev 1.
Figure 2.
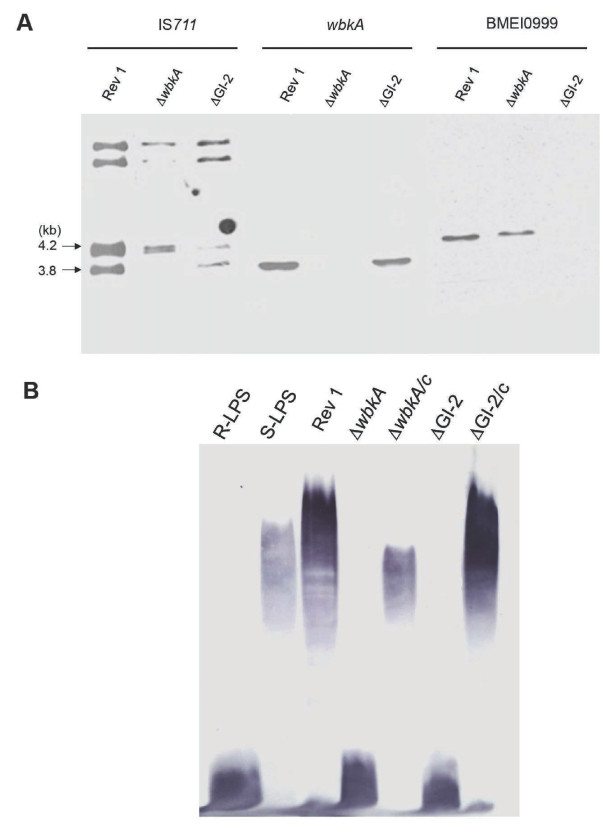
Genetic and phenotypic characterization of spontaneous Rev 1 R mutants. (A) Southern blot analysis of B. melitensis Rev 1 spontaneous R variants with IS711 (left panel), wbkA (center panel) and GI2 BMEI0999 (right panel) as probes. Note that, for the IS711 probe and B. melitensis Rev 1 and ∆wbkA but not ∆GI-2, the signal at 4.2 kb corresponds to two bands (predicted MW 4.2 and 4.14 kb). For simplicity, fragments with molecular masses lower than 3.8 kb were omitted. (B) Western blot of purified R-LPS and S-LPS, Rev 1, wbkA and GI-2 R mutants and the corresponding complemented strains probed with anti-LPS antibodies.
Finally, to study whether ∆GI-2 and ∆wbkA excisions occurred during vaccine manufacturing, we examined three commercial Rev 1 lots with different levels of dissociation (see Material and methods). We identified the GI-2 and wbkA chromosomal scars in these commercial preparations (not shown) and, interestingly, the lot with the highest dissociation rate (almost 100%) presented both chromosomal scars. Considering these results and those of the above-described experiments, we concluded that GI-2 and wbkA deletions occur repeatedly in batches during manufacturing despite the precautions taken to minimize S-R dissociation.
The S-R dissociation of B. melitensis Rev 1 is reduced by mutation of GI-2 int and ISBm1 transposase flanking wbkA
We have previously shown that disruption of GI-2 phage-related integrase int gene reduces the dissociation rate of B. abortus by stabilization of GI-2 [10]. To test whether this also occurs in B. melitensis Rev 1, we constructed mutant Rev 1Δint by in-frame deletion of GI-2 int (see Material and Methods). Similarly, we have previously shown that the excision of the 5.5 kb fragment carrying wbkA is caused by homologous recombination mediated by RecA and the wbkA-flanking IS [14]. Thus, we also constructed mutant Rev 1ΔISBm1, which lacks one of the wbkA-flanking ISBm1 copies. Characterization by PCR demonstrated only the scar and circular intermediate corresponding to GI-2 excision in Rev 1ΔISBm1 (Figure 3A), showing the stabilization of wbkA. Conversely, Rev 1Δint displayed the scar and circular intermediate corresponding to an excision of the wbkA (Figure 3A). To stabilize both genetic regions, we constructed the Rev 1ΔISBm1Δint double mutant (Rev 2) on the Rev 1ΔISBm1 background. As expected, Rev 2 did not show evidence (scars or circular intermediates) of GI-2 or wbkA excision events by PCR (Figure 3A) even in experiments with high amounts of template DNA.
Figure 3.
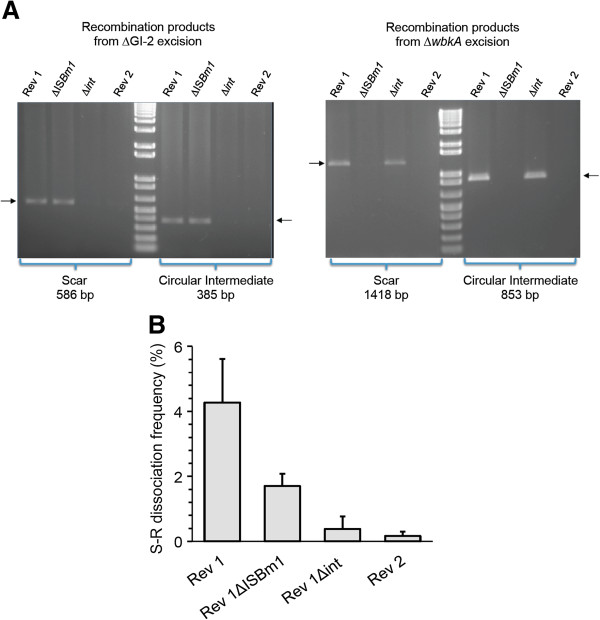
Stabilization of regions GI-2 and wbkA by deletion of appropriate section of GI-2 phage-related integrase (Δint), wbkA-flanking IS (ΔISBm1) and both (Rev 2). (A) PCR detection of the chromosomal scar and circular intermediate resulting from (GI-2 excision (left panel) and wbkA excision (right panel). (B) S-R dissociation rates.
Using the above mutants and the parental Rev 1 and B. melitensis H38 as references, we assessed the importance of the corresponding genetic mechanisms in the frequency of the S-R dissociation of Rev 1. To this end, we used conditions that favor the growth of R mutants and counted the proportions of R bacteria using the crystal violet exclusion method (see Material and methods, S-R dissociation assessment). While dissociation of strain H38 was beyond detection under the experimental conditions, Rev 1 showed dissociation rates of 4.27 ± 1.34% (Figure 3B). In contrast, the single mutants displayed lower (p < 0.0001) dissociation rates than Rev 1. These dissociation rates were significantly (p < 0.01) lower for Rev 1∆int (0.38 ± 0.33) than for Rev 1ΔISBm1 (1.70 ± 0.38). Consistent with the prediction, the double mutant Rev 2 showed the lowest dissociation rates (0.17 ± 0.13%). Therefore, under these experimental conditions, GI-2 and wbkA stabilization resulted in an approximately 25-fold (i.e. about 95%) decrease in dissociation.
Stabilization of the S phenotype does not alter the properties of Rev 1 in vitro or in mice
Standard bacteriological characterization demonstrated that Rev 1ΔISBm1, Rev 1Δint and Rev 2 conserved the phenotypic markers that are characteristic of Rev 1, namely: S phenotype (demonstrated by reactivity with anti-S antibodies and by the crystal violet and acriflavine tests), inhibition by 5 IU/mL penicillin, resistance to 2.5 μg/mL of streptomycin, and comparatively small colony size (1–1.2 mm diameter after 5 days of incubation at 37 °C) [24,25]. Since the latter feature (i.e. slow growth) is a particularly important Rev 1 property ([33]; see also below), we compared the growth curves of Rev 1, Rev 2 and the wild-type B. melitensis H38 reference strain. As can be seen in Figure 4A, Rev 1 and Rev 2 produced almost identical growth curves and both were retarded with respect to B. melitensis H38.
Figure 4.
Genetic stabilization in Rev 2 does not alter the in vitro growth and vaccine properties of Rev 1 reference vaccine. (A) In vitro growth kinetics; (B) Residual virulence (log CFU/spleen); (C) Spleen weights; and (D) Efficacy against a virulent B. melitensis virulent challenge, in BALB/c mice. Statistical comparison of means was performed by one-way ANOVA and Fisher’s PLSD test: aP < 0.005 vs. unvaccinated control (PBS group).
We completed the characterization of Rev 2 by studying whether the critical biological properties of Rev 1 (i.e. residual virulence and efficacy against virulent infection) were maintained in Rev 2. For this purpose, we used the established mouse model for brucellosis vaccine control [34], which is useful in discriminating Rev 1 vaccine lots of inappropriate residual virulence and immunogenicity [25]. Rev 2 showed a behavior almost identical to that of Rev 1 in this model. In virulence assays, both strains persisted similarly and less than the virulent B. melitensis H38 strain (Figure 4B). Likewise, Rev 1 and Rev 2 induced a similarly reduced splenomegaly (Figure 4C). Finally, Rev 2 was as protective as Rev 1 against a challenge infection with the full-virulent B. melitensis H38-NxR strain (Figure 4D).
Discussion
A major practical aspect of vaccine production is quality control. Because of the well-known genetic instability of all brucellae in vitro [34], control is particularly critical in brucellosis vaccines. Therefore, a manufacturing seed-lot system [33] has been adopted to reduce genetic drifts including the appearance of R types. In addition, control of every single batch includes the assessment of colony S phase and morphology [24]. As confirmed here by comparison with B. melitensis H38, dissociation of B. melitensis Rev 1 into S and R forms is particularly troublesome and, although the reasons for this difference between the virulent bacteria and the vaccine are not known, it is possible to indicate probable causes. Our results indicate that the genetic mechanisms operating in Rev 1 dissociation are those described before for other S strains and, therefore, it seems unlikely that this Rev 1 trait follows particular genetic characteristics of this vaccine. A more probable possibility is suggested by the well-known fact that oxygen limitation and other conditions causing energy shortage facilitate the establishment of Brucella R variants [35]. Accordingly, we have proposed [14] that mutants not synthesizing the costly O-PS are more competitive when energy shortage and no selective pressure to maintain the O-PS concur and that, under these circumstances, the excision of GI-2 promoted by the phage-related integrase int gene and the deletion of wbkA by homologous recombination become prominent. In the case of Rev 1, it is known that this vaccine carries a defect in ribosomal protein S12 that relates to its streptomycin resistance [36]. In Salmonella Typhimurium, a similar S12 mutation causes streptomycin resistance, increased rates of ribosomal proofreading and, as a result, decreased rates of protein synthesis, reduced bacterial growth and attenuation [37]. Therefore, it seems likely that the S12 defect in Rev 1 is a major cause of reduced fitness and growth rates (and attenuation), both of which could be compensated for by the reduction in energy expenditure caused by at least (see below) the loss of O-PS, thereby favoring the growth of R mutants. In fact, Rev 1 is characterized by a small colony size and the S-R dissociation is often accompanied by the appearance of larger colonies not appropriate for vaccine use [25].
Although we investigated other known R-generating mutations, we only detected the GI-2 and wbkA deletions in Rev 1. Turse et al. [15] have shown that point mutations on manBcore, a gene required for the complete synthesis of O-PS [9], are a cause of spontaneous appearance of R variants, which have been isolated from infected macrophages and mice [15]. We failed to reveal any point mutations in this locus by sequencing manBcore in the 21 R strains that did not show either wbkA or GI-2 deletions. Since Turse et al. performed their experiments with B. abortus 2308 and B. melitensis 16 M and recovered the R mutants using agar plates saturated with Bk2 phage (specific for the S-phase), it seems that the discrepancies could be explained by the use of different experimental conditions. Concerning the genetic mechanisms, we observed that inactivation of the GI-2 integrase had a more significant effect on O-PS stability. This may reflect a higher instability of this region related to the activity of the integrase and/or to the concomitant loss of Omp25b (a major outer membrane protein encoded in GI-2 [11]), since the ensuing reduction in biosynthetic expenditures would increase competitiveness in vitro. Another possibility is suggested by the fact that recombinases like the one coded for by GI-2 int are able to identify secondary target repeat sequences promoting the excision of unrelated fragments [38]. Therefore, the GI-2 integrase could theoretically take part in other events leading to the R phenotype. This work also shows that mutations other than those described thus far also cause the appearance of R types. This is shown by the fact that dissociation was not completely prevented in Rev 2 strain. Further investigations are necessary for a full understanding of the mutation paths that affect the O-PS in Brucella.
Because the underlying mechanisms were known, we could abrogate the GI-2 and wbkA deletions and generate a more stable Rev 2 strain. Indeed, as shown by the experimental results presented here, the absence of large sections of the DNA sequences involved in this deletion events make highly unlikely the possibility that Rev 2 could revert to an anomalously high S-R dissociation rate. Thus, the genetic strategy described here represents a complement to the empirical methods currently used in the production of smooth Brucella vaccines for the stabilization of smooth phenotype. Although the comparison between the virulent H38 strain and Rev 1 showed differences in both growth and S-R dissociation rates, our results demonstrate that the increased S-R dissociation is not linked to attenuation of Rev 1 in the mouse model, as it might be expected from the characteristic attenuation of Brucella R mutants. As discussed above, the reduced fitness probably caused by the S12 mutation that is manifested by the lower growth rates is a more likely cause of attenuation. Rev 2 conserved this characteristic and also the relevant biological properties of Rev 1 in the mouse model. Although this model represents the best laboratory control for the in vivo properties of Rev 1 [33], experiments in the natural hosts are clearly necessary to prove that Rev 2 retains all the properties that have made Rev 1 a succesful vaccine.
Competing interests
The authors declared that they have no competing interests.
Authors’ contributions
MM designed and performed the molecular biology experiments. MJdM, AZB and BSR carried out the bacteriological studies and experiments in mice. ILG provided financial support. IM, MJG and MM designed the work and wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
E. coli strains and plasmids used. List, characteristics and source/reference of E. coli strains and plasmids used.
Contributor Information
Marcos Mancilla, Email: mancilla.marcos@gmail.com.
María-Jesús Grilló, Email: mariajesus.grillo@unavarra.es.
María-Jesús de Miguel, Email: mjmiguel@aragon.es.
Ignacio López-Goñi, Email: ilgoni@unav.es.
Beatriz San-Román, Email: beatriz.sanroman@unavarra.es.
Ana Zabalza-Baranguá, Email: ana.zabalzab@unavarra.es.
Ignacio Moriyón, Email: imoriyon@unav.es.
Acknowledgements
This work was funded by MINECO (reference project AGL2011-30453-C04) of Spain, the FIMA foundation and the European Union’s FP7/2007-2013 (grant agreement n° 221948, ICONZ - Integrated control of Neglected Zoonoses) and CSIC JAE-Doc program (FSE). We thank to Prof. G. Splitter (Animal Health and Biomedical Sciences, University of Wisconsin-Madison, USA) for providing plasmid pBGI-99-97c, Dr J.M. Blasco (Sanidad Animal, CITA of Aragón, Spain) for the vaccine Rev 1 seed lot and strains, and A. Delgado for skillfull technical assistance.
References
- Grace D, Mutua F, Ocungo P, Kruska R, Jones K, Brierley L, Lapar L, Said M, Herrero M, Duc Phuc P, Elich Tao N, Akuku I, Ogutu F. Mapping of poverty and likely zoonoses hotspots. Zoonoses Project. 2012;4:1–119. [Google Scholar]
- Anonymous. The control of Neglected Zoonoses Diseases: A route to poverty alleviation. Geneva: WHO Press; 2006. [Google Scholar]
- Blasco JM. A review of the use of B. melitensis Rev 1 vaccine in adult sheep and goats. Prev Vet Med. 1997;31:275–283. doi: 10.1016/S0167-5877(96)01110-5. [DOI] [PubMed] [Google Scholar]
- Moriyón I, Grilló MJ, Monreal D, González D, Marín C, Lopez-Goñi I, Mainar-Jaime RC, Moreno E, Blasco JM. Rough vaccines in animal brucellosis: structural and genetic basis and present status. Vet Res. 2004;35:1–38. doi: 10.1051/vetres:2003037. [DOI] [PubMed] [Google Scholar]
- Barrio MB, Grilló MJ, Muñoz PM, Jacques I, González D, De-Miguel MJ, Marín CM, Barberan M, Letesson JJ, Gorvel JP, Moriyón I, Blasco JM, Zygmunt MS. Rough mutants defective in core and O-polysaccharide synthesis and export induce antibodies reacting in an indirect ELISA with smooth lipopolysaccharide and are less effective than Rev 1 vaccine against Brucella melitensis infection of sheep. Vaccine. 2009;27:1741–1749. doi: 10.1016/j.vaccine.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Godfroid J, Scholz HC, Barbier T, Nicolas C, Wattiau P, Fretin D, Whatmore AM, Cloeckaert A, Blasco JM, Moriyón I, Saegerman C, Muma JB, Al-Dahouk S, Neubauer H, Letesson JJ. Brucellosis at the animal/ecosystem/human interface at the beginning of the 21st century. Prev Vet Med. 2011;102:118–131. doi: 10.1016/j.prevetmed.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Minas A, Minas M, Stournara A, Tselepidis S. The “effects” of Rev-1 vaccination of sheep and goats on human brucellosis in Greece. Prev Vet Med. 2004;64:41–47. doi: 10.1016/j.prevetmed.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Alton GG, Jones LM, Angus RD, Verger JM. Techniques for the brucellosis laboratory. Paris, France: INRA; 1988. [Google Scholar]
- González D, Grilló MJ, De-Miguel MJ, Ali T, Arce-Gorvel V, Delrue RM, Conde-Álvarez R, Muñoz PM, Lopez-Goñi I, Iriarte M, Marín CM, Weintraub A, Widmalm G, Zygmunt M, Letesson JJ, Gorvel JP, Blasco JM, Moriyón I. Brucellosis vaccines: assessment of Brucella melitensis lipopolysaccharide rough mutants defective in core and O-polysaccharide synthesis and export. PLoS One. 2008;3:e2760. doi: 10.1371/journal.pone.0002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancilla M, Lopez-Goñi I, Moriyón I, Zarraga AM. Genomic Island 2 is an unstable genetic element contributing to Brucella lipopolysaccharide spontaneous smooth-to-rough dissociation. J Bacteriol. 2010;192:6346–6351. doi: 10.1128/JB.00838-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno N, Caro-Hernandez P, Cloeckaert A, Fernandez-Lago L. DNA polymorphism in the omp25/omp31 family of Brucella spp.: identification of a 1.7-kb inversion in Brucella cetaceae and of a 15.1-kb genomic island, absent from Brucella ovis, related to the synthesis of smooth lipopolysaccharide. Microbes Infect. 2004;6:821–834. doi: 10.1016/j.micinf.2004.04.009. [DOI] [PubMed] [Google Scholar]
- García-Yoldi D, Marín CM, Lopez-Goñi I. Restriction site polymorphisms in the genes encoding new members of group 3 outer membrane protein family of Brucella spp. FEMS Microbiol Lett. 2005;245:79–84. doi: 10.1016/j.femsle.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Conde-Álvarez R, Arce-Gorvel V, Iriarte M, Mancek-Keber M, Barquero-Calvo E, Palacios-Chaves L, Chacón-Díaz C, Chaves-Olarte E, Martirosyan A, Von-Bargen K, Grilló MJ, Jerala R, Brandenburg K, Llobet E, Bengoechea JA, Moreno E, Moriyón I, Gorvel JP. The lipopolysaccharide core of Brucella abortus acts as a shield against innate immunity recognition. PLoS Pathog. 2012;8:e1002675. doi: 10.1371/journal.ppat.1002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancilla M, Marín CM, Blasco JM, Zarraga AM, Lopez-Goñi I, Moriyón I. Spontaneous excision of the O-polysaccharide wbkA glycosyltranferase gene is a cause of dissociation of smooth to rough Brucella colonies. J Bacteriol. 2012;194:1860–1867. doi: 10.1128/JB.06561-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turse JE, Pei J, Ficht TA. Lipopolysaccharide-deficient Brucella variants arise spontaneously during infection. Front Microbiol. 2011;2:54. doi: 10.3389/fmicb.2011.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemulapalli R, McQuiston JR, Schurig GG, Sriranganathan N, Halling SM, Boyle SM. Identification of an IS711 element interrupting the wboA gene of Brucella abortus vaccine strain RB51 and a PCR assay to distinguish strain RB51 from other Brucella species and strains. Clin Diagn Lab Immunol. 1999;6:760–764. doi: 10.1128/cdli.6.5.760-764.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zygmunt MS, Blasco JM, Letesson JJ, Cloeckaert A, Moriyón I. DNA polymorphism analysis of Brucella lipopolysaccharide genes reveals marked differences in O-polysaccharide biosynthetic genes between smooth and rough Brucella species and novel species-specific markers. BMC Microbiol. 2009;9:92. doi: 10.1186/1471-2180-9-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelVecchio VG, Kapatral V, Redkar RJ, Patra G, Mujer C, Los T, Ivanova N, Anderson I, Bhattacharyya A, Lykidis A, Reznik G, Jablonski L, Larsen N, D’Souza M, Bernal A, Mazur M, Goltsman E, Selkov E, Elzer PH, Hagius S, O’Callaghan D, Letesson JJ, Haselkorn R, Kyrpides N, Overbeek R. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc Natl Acad Sci USA. 2002;99:443–448. doi: 10.1073/pnas.221575398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information/Basic Local Aligment Search Tool (NCBI/BLAST) [ http://www.ncbi.nlm.nih.gov/BLAST]
- European Molecular Biology Laboratory-European Bioinformatics Institute (EMBI-EBI) [ http://www.ebi.ac.uk/Tools/clustalo]
- National Center for Biotechnology Information/Primer-BLAST (NCBI/Primer-BLAST) [ http://www.ncbi.nlm.nih.gov/tools/primer-blast/]
- Wilson K. Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol. 1997. pp. 2.4.1–2.4.5. [DOI] [PubMed]
- Ocampo-Sosa AA, Aguero-Balbin J, Garcia-Lobo JM. Development of a new PCR assay to identify Brucella abortus biovars 5, 6 and 9 and the new subgroup 3b of biovar 3. Vet Microbiol. 2005;110:41–51. doi: 10.1016/j.vetmic.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Bosseray N. Brucella melitensis Rev.1 living attenuated vaccine: stability of markers, residual virulence and immunogenicity in mice. Biologicals. 1991;19:355–363. doi: 10.1016/S1045-1056(05)80025-9. [DOI] [PubMed] [Google Scholar]
- Grilló MJ, Bosseray N, Blasco JM. In vitro markers and biological activity in mice of seed lot strains and commercial Brucella melitensis Rev 1 and Brucella abortus B19 vaccines. Biologicals. 2000;28:119–127. doi: 10.1006/biol.2000.0249. [DOI] [PubMed] [Google Scholar]
- Conde-Álvarez R, Grilló MJ, Salcedo SP, De-Miguel MJ, Fugier E, Gorvel JP, Moriyón I, Iriarte M. Synthesis of phosphatidylcholine, a typical eukaryotic phospholipid, is necessary for full virulence of the intracellular bacterial parasite Brucella abortus. Cell Microbiol. 2006;8:1322–1335. doi: 10.1111/j.1462-5822.2006.00712.x. [DOI] [PubMed] [Google Scholar]
- Quandt J, Hynes MF. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pehle A. A broad host range mobilization system for the in vitro genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology. 1983;1:784–890. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- Rajashekara G, Covert J, Petersen E, Eskra L, Splitter G. Genomic island 2 of Brucella melitensis is a major virulence determinant: Functional analyses of genomic islands. J Bacteriol. 2008;190:6243–6252. doi: 10.1128/JB.00520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oy Growth Curves Ab Ltd. [ http://www.bioscreen.fi]
- Aragón V, Díaz R, Moreno E, Moriyón I. Characterization of Brucella abortus and Brucella melitensis native haptens as outer membrane O-type polysaccharides independent from the smooth lipopolysaccharide. J Bacteriol. 1996;178:1070–1079. doi: 10.1128/jb.178.4.1070-1079.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grilló MJ, Manterola L, De-Miguel MJ, Muñoz PM, Blasco JM, Moriyón I, Lopez-Goñi I. Increases of efficacy as vaccine against Brucella abortus infection in mice by simultaneous inoculation with avirulent smooth bvrS/bvrR and rough wbkA mutants. Vaccine. 2006;24:2910–2916. doi: 10.1016/j.vaccine.2005.12.038. [DOI] [PubMed] [Google Scholar]
- OIE. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. OIE; 2009. Bovine Brucellosis; pp. 1–35. Chapter 2.4.3. [Google Scholar]
- Grilló MJ, Blasco JM, Gorvel JP, Moriyón I, Moreno E. What have we learned from brucellosis in the mouse model? Vet Res. 2012;43:29. doi: 10.1186/1297-9716-43-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt P. The nutrition of Brucellae. Bacteriol Rev. 1958;22:91–98. doi: 10.1128/br.22.2.81-98.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloeckaert A, Grayon M, Grepinet O. Identification of Brucella melitensis vaccine strain Rev.1 by PCR-RFLP based on a mutation in the rpsL gene. Vaccine. 2002;20:2546–2550. doi: 10.1016/S0264-410X(02)00159-7. [DOI] [PubMed] [Google Scholar]
- Maisnier-Patin S, Berg OG, Liljas L, Andersson DI. Compensatory adaptation to the deleterious effect of antibiotic resistance in Salmonella Typhimurium. Mol Microbiol. 2002;46:355–366. doi: 10.1046/j.1365-2958.2002.03173.x. [DOI] [PubMed] [Google Scholar]
- Manson JM, Gilmore MS. Pathogenicity island integrase cross-talk: a potential new tool for virulence modulation. Mol Microbiol. 2006;61:555–559. doi: 10.1111/j.1365-2958.2006.05262.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
E. coli strains and plasmids used. List, characteristics and source/reference of E. coli strains and plasmids used.


