Abstract
Here we report that RNase P is required for the initial separation of all seven valine tRNAs from three distinct polycistronic transcripts (valV valW, valU valX valY lysY and lysT valT lysW valZ lysY lysZ lysQ). Particularly significant is the mechanism by which RNase P processes the valU and lysT polycistronic transcripts. Specifically, the enzyme initiates processing by first removing the Rho-independent transcription terminators from the primary valU and lysT transcripts. Subsequently, it proceeds in the 3′ → 5′ direction generating one pre-tRNA at a time. Based on the absolute requirement for RNase P processing of all three primary transcripts, inactivation of the enzyme leads to a >4-fold decrease in the levels of both type I and type II valine tRNAs. The ability of RNase P to initiate tRNA processing at the 3′ ends of long primary transcripts by endonucleolytically removing the Rho-independent transcription terminator represents a previously unidentified function for the enzyme, which is responsible for generating the mature 5’ termini of all 86 E. coli tRNAs. RNase E only plays a very minor role in the processing of all three valine polycistronic transcripts.
INTRODUCTION
The Escherichia coli genome contains 86 transfer RNA (tRNA) genes that are organized as either monocistronic transcripts or complex operons, containing other tRNAs, messenger RNAs (mRNA) or ribosomal RNA (rRNA) genes (1,2). Every transcribed tRNA precursor requires subsequent processing at both ends to generate the mature species that will be charged by their cognate aminoacyl tRNA synthetases. In addition, tRNAs that are part of polycistronic transcripts require initial endonucleolytic cleavages to generate the pre-tRNAs that undergo further processing at their 5′ and 3′ termini. The generally accepted model involves endonucleolytic cleavages of polycistronic transcripts by RNase E to generate pre-tRNAs (3–5). Subsequently, the ribozyme RNase P endonucleolytically removes the extra nucleotides at the 5′ terminus, while the 3′ terminus is processed exonucleolytically by a combination of RNase T, RNase PH, RNase D, RNase BN, RNase II and PNPase (6,7).
E. coli RNase P is an essential enzyme consisting of the C5 protein (encoded by rnpA) and the catalytic M1 RNA (encoded by rnpB) subunits (8,9). Although the enzyme has been shown to be involved in the maturation of 4.5S RNA (a component of the essential signal recognition particle) (10), other small RNAs (11), and in the processing of a few polycistronic mRNAs (12,13), the removal of the extra nucleotides at the 5′ end of tRNAs by RNase P has been described as an essential function in the biogenesis of mature tRNA species that can subsequently be aminoacylated (14). However, recent reports suggest that some tRNAs with unprocessed 5′ ends can still be aminoacylated both in vitro and in vivo (15,16).
More importantly, it has recently been demonstrated that RNase P is required for the endonucleolytic separation of certain polycistronic tRNA transcripts such as valV valW, leuQ leuP leuV and secG leuU (15,17). Thus, it was hypothesized that the essential function of RNase P might be related to the complete absence of a particular tRNA that was dependent on the enzyme for initial separation from polycistronic transcripts. In such a scenario, loss of cell viability would result from the shortage of chargeable tRNAs due to immature 3′ ends rather than an inhibition of tRNA charging due to unprocessed 5′ ends.
Here we have examined the 86 tRNA genes in E. coli in silico to determine if perhaps all of the tRNA genes for one specific amino acid were initially processed by RNase P. Based on previous studies (3,7,15,17), many of the tRNAs including those for His, Leu, Pro, Trp, Arg, Asn, Met, Cys, Ala, Gln, Ser, Phe, Thr, Ile and Tyr were ruled out. However, both Val and Lys were possibilities, since we have already shown that the two of the seven valine tRNAs (valV and valW, isotype 2) require RNase P for their initial processing (17) and the processing of the six lysine tRNA genes has not been previously studied. In fact, one lysine tRNA was part of a polycistronic transcript including three valine tRNAs (valU valX valY lysV), while the other five lysine tRNA genes were clustered together with two valine tRNAs (lysT valT lysW valZ lysY lysZ lysQ). However, it has been indicated that the lysT gene cluster is synthesized as four separate transcripts (18).
We now show that the lysT promoter drives the synthesis of a large polycistronic transcript that includes all seven tRNAs. More importantly, all seven valine tRNAs require RNase P for their initial processing. Interestingly, RNase P processing occurs in the 3’→ 5’ direction, generating one pre-tRNA at a time. Thus, functional valine tRNAs, either isotype 1 or 2, are no longer generated, from their primary transcripts upon inactivation of RNase P, leading to a >4-fold drop in the levels of mature valine tRNAs. However, exogenous expression of both valine tRNA isotypes processed independently of RNase P did not complement an rnpA49 mutant at the non-permissive temperature. We also show that the 3’ → 5’ exonuclease, RNase PH, plays a major role in the 3′ end maturation of type I valine isotype tRNAs.
MATERIALS AND METHODS
Bacterial strains
All the E. coli strains used in this study were derivatives of MG1693 (thyA715 rph-1) and are listed in Table 1. The rne-1 and rnpA49 mutations, encoding temperature-sensitive RNase E and RNase P enzymes, respectively, do not support cell viability at either 42 or 44°C and have been previously described (19,20). A P1 phage lysate grown on SK2525 (rnpA49 rph-1) was used to transduce both SK10153 (thyA715) and SK9797 (rne-1 rnzΔ500 rph-1) to generate SK10521 and SK10300, respectively. SK5166 (rnpA49 rph-1) was transduced with a P1 lysate grown on SK4455 (rnc-14 rph-1) to construct SK10525. A P1 lysate grown on SK3170 (rnlA2 rph-1) was used to transduce SK2534 (rne-1 rnpA49 rph-1) to construct SK10523. SK10460 [rneΔ1018 rnpA49/pDH28(rng-219/KmR)] was constructed by the plasmid displacement from SK2539 [rneΔ1018 rnpA49/pMOK15(rneΔ610/CmR)] (Ow and Kushner, unpublished results).
Table 1. List of bacterial strains and plasmids used in this study.
| Strains | Genotype | Reference/source |
|---|---|---|
| MG1693 | rph-1 thyA715 | E. coli Genetic Stock Center |
| SK2525 | rnpA49 rph-1 thyA715 rbsD296::Tn10 TcR | (4) |
| SK2534 | rne-1 rnpA49 rph-1 thyA715 rbsD296::Tn10 TcR | (4) |
| SK2549 | rne-1 rnpA49 rng::cat rph-1 thyA715 rbsD296::Tn10 CmR TcR | (43) |
| SK3170 | rnlA2::kan rph-1 thyA715 KmR | Perwez & Kushner (unpublished results) |
| SK4455 | rnc-14 rph-1 thyA715 TcR | (44) |
| SK4484 | rne-1 rnpA49 rng::cat rnzΔ500::apr rnlA2::kan rph-1 rbsD296::Tn10 CmR KmR AprR TcR | Maples and Kushner (unpublished results) |
| SK4668 | rne-1 rnpA49 rnb-500 pnp-7 rph-1 thyA715 rbsD296::Tn10 TcR | Maples and Kushner (unpublished results) |
| SK5166 | rnpA49 rph-1 thyA715 rbsD3163::Tn10::kan KmR | Stead & Kushner (unpublished results) |
| SK5374 | ΔybeY::cat rnpA49 rph-1 thyA715 rbsD296::Tn10 CmR TcR | Stead & Kushner (unpublished results) |
| SK5665 | rne-1 rph-1 thyA715 | (45) |
| SK5704 | rne-1 pnp-7 rnb-500 rph-1 thyA715 | (45) |
| SK9797 | rne-1 rph-1 rnzΔ500::kan thyA715 KmR | (46) |
| SK10153 | thyA715 | (28) |
| SK10300 | rne-1 rnpA49 rnzΔ500::kan rph-1 thyA715 rbsD296::Tn10 TcR KmR | This study |
| SK10451 | rnpA49 pnp-7 rnb-500 rph-1 thyA715 rbsD296::Tn10 TcR | (17) |
| SK10460 | Δrne-1018::bla/pDHK28(rng-219/KmR) rnpA49 rph-1 thyA715 rbsD296::Tn10 SmR/SpR ApR KmR TcR | This study |
| SK10523 | rne-1 rnpA49 rnlA2::kan rph-1 thyA715 rbsD296::Tn10 TcR KmR | This study |
| SK10525 | rnc-14 rnpA49 rph-1 thyA715 rbsD296::Tn10 TcR KmR | This study |
| SK10530 | rnpA49 rph-1 thyA715 rbsD296::Tn10 TcR/pAAK11 | This study |
| SK10531 | rnpA49 thyA715 rbsD296::Tn10 TcR/pAAK11 | This study |
| SK10533 | rnpA49 rph-1 thyA715 rbsD296::Tn10 TcR/pAAK13 | This study |
| SK10534 | rnpA49 thyA715 rbsD296::Tn10 TcR/pAAK13 | This study |
| SK10537 | rnpA49 rph-1 thyA715 rbsD296::Tn10 TcR /pAAK15 | This study |
| SK10538 | rnpA49 thyA715 rbsD296::Tn10 TcR/pAAK15 | This study |
| SK10539 | rnpA49 rph-1 thyA715 rbsD296::Tn10 TcR/pAAK17 | This study |
| SK10540 | rnpA49 thyA715 rbsD296::Tn10 TcR/pAAK17 | This study |
| SK10541 | rnpA49 rph-1 thyA715 rbsD296::Tn10 TcR/pAAK11 pAAK13 | This study |
| SK10542 | rnpA49 thyA715 rbsD296::Tn10 TcR/pAAK11 pAAK13 | This study |
| Plasmids | ||
| pMS421 | lacIq+, SmR | (22) |
| pBMK11 | pcnB+, CmR | (21) |
| pAAK11 | pBMK11, valU+,CmR | This study |
| pAAK13 | pMS421, valW+,SmR | This study |
| pAAK15 | pBMK11, argX+,CmR | This study |
| pAAK17 | pBMK11, rnpB+,CmR | This study |
| pMOK15 | rneΔ610 CmR | (47) |
| pDHK28 | rng-219 KmR | (32) |
Plasmid constructions
The plasmids pAAK11 (valU+/ CmR), pAAK15 (argX+/CmR) and pAAK17 (rnpB+/CmR) all contain the p15A origin of DNA replication (15–20 copies/cell) and express either valU, argX or the M1 RNA (rnpB), respectively, under the control of the lac promoter. Plasmid pAAK13 (valW+ /SmR) is a 6–8 copy plasmid with a pSC101 origin of DNA replication expressing valW under the control of the lac promoter. A PCR (polymerase chain reaction) fragment containing a lac promoter followed by a gene of interest (either valU, valW, argX or rnpB) and a Rho-independent transcription terminator [derived from leuU, (15)] was generated employing an overlapping PCR technique using Phusion® High-Fidelity DNA Polymerase (NEB). The resulting PCR products were cloned into the BamHI/HindIII sites of either pBMK11 (21) to construct pAAK11 (valU+/ CmR), pAAK15 (argX+/CmR) and pAAK17 (rnpB+/CmR) or pMS421 (22) to construct pAAK13 (valW+/SmR).
All the plasmid constructions were confirmed by DNA sequencing of the cloned fragments (Eurofins MWG Operon). In addition, Northern analysis was used to confirm that the isopropyl-β-D-1-thiosgalactopyranoside (IPTG) induction of plasmids pAAK11, pAAK13, pAAK15 and pAAK17 led to increased intracellular levels of tRNAVal1, tRNAVal2, tRNAArg and M1 RNA, respectively. Plasmid transformations were carried out as described previously (23).
Bacterial strains were typically grown with shaking in Luria broth (24) supplemented with thymine (50 μg/ml). When appropriate, the medium also contained tetracycline (20 μg/ml), kanamycin (25 μg/ml), chloramphenicol (20 μg/ml) or streptomycin (20 μg/ml). Culture growth was monitored using a Klett-Summerson Colorimeter (No.42 Green filter). For temperature-sensitive mutant strains, cultures were initially grown at 30ºC until they reached 50 Klett units above background and then shifted to 44°C, unless noted otherwise. All the cultures were maintained in exponential growth by periodic dilutions with pre-warmed medium. To collect samples for RNA extraction, RNase P (rnpA49 allele) and RNase E (rne-1 allele) mutant strains were shifted to 44°C for 1 and 2 h, respectively. The wild-type control strains were also shifted to 44°C for 1 h.
Isolation of total RNA
Total RNA for steady-state analysis was extracted using the method described by Mohanty et al. (25), with the following modifications. Cell pellets from 3.5 ml of cells were resuspended in 510 μl of Lysis buffer. After lysis, 71 μl of 20 mM acetic acid were added. Following resuspension in 1 ml of 2 M LiCl, each sample was centrifuged at 16 000 x g for 10 min. The resulting pellets were washed with 500 μl of chilled 70% ethanol, followed by centrifugation at 10 000 x g for 10 min. The supernatants were removed and the pellets were spun again at 10 000 x g for 1 min to remove any residual ethanol.
Total RNA for half-life analyses was extracted using the RNAsnapTM method (26). Rifampicin (500 μg/ml) and nalidixic acid (20 μg/ml) were added to the cell cultures at 50 Klett units above background and the samples for 0-min time-point were collected after 70 s (25). All RNA samples were quantified using a NanoDrop (Model 2000c; Thermo Scientific) apparatus. In addition, 500 ng of each RNA sample was separated on a 1% agarose gel and visualized by ethidium bromide staining to check for RNA integrity and to ensure equal loading.
Northern analysis and determination of in vivo aminoacylation levels
Northern analysis was carried out as described by Mohanty and Kushner (17). Briefly, 12 μg of total RNA from each strain were separated on either 6 or 8% polyacrylamide gels containing 8.3 M urea in TBE buffer or 1.2% agarose gels and were subsequently transferred on to a positively charged nylon membrane (NytranTMSPC, WhatmanTM). 32P-labeled DNA oligonucleotides were used as probes. The blots were scanned with a PhosphorImager (StormTM 840; GE Healthcare). Each membrane was often probed with different probes after stripping as previously described (25). Band intensities were determined using ImageQuant TL 5.2 software (GE Healthcare). The percentage of tRNA aminoacylation level was determined as previously described (27,28).
Reverse transcription-polymerase chain reaction (RT-PCR) cloning and sequencing of 5′ → 3′ ligated transcripts
The 5′ and 3′ ends of various processing intermediates derived from the valU and lysT polycistronic transcripts were identified by cloning and sequencing RT-PCR products obtained from 5′ → 3′ end-ligated circular RNAs using the method described by Mohanty and Kushner (15,28,29). In brief, 3 μg of total RNA from rph-1, rnpA49 rph-1 and rne-1 rnpA49 rph-1 strains were initially treated with tobacco acid pyrophosphatase (Epicentre Technologies) to convert any 5′ triphosphates to phosphomonoesters in order to facilitate the self-ligation step. The dephosphorylated RNAs were self-ligated using T4 RNA ligase (NEB) and were subsequently reverse transcribed followed by PCR amplification using several pairs of gene-specific primers.
Primers and probes
The nucleotide sequences of all primers and probes used in this study are shown in Supplementary Table S1 (Supplementary Material). Table S1 RESULTS
Processing of the valU polycistronic transcript is completely dependent on RNase P
The seven valine tRNAs in the E. coli genome are organized into three separate gene clusters, valV valW (17), the valU and lysT operons (Figures 1A and 2A). The initial separation of the valV valW transcript, containing two copies of valine isotype 2, has been shown to be completely dependent on RNase P cleavages at the mature 5′ termini of both tRNAs (17). The remaining five valine tRNAs, belong to the valine isotype 1. Interestingly, the intergenic spacer regions between valY and lysV (CUUC), valT and lysW (CC) and valZ and lysY (CUC) (Figures 1A and 2A) were very similar to the UCCU spacer between valV and valW. This observation suggested that RNase P might be involved in the initial processing of additional valine tRNAs, since these short spacer regions are not canonical RNase E cleavage sites (30,31).
Figure 1.
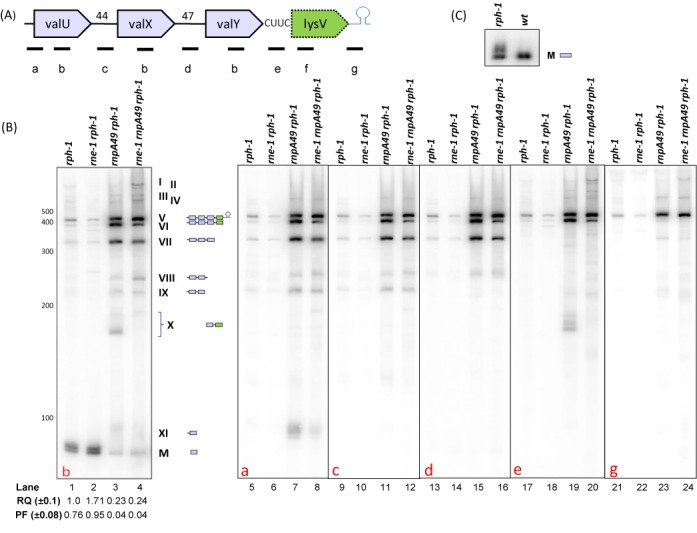
Analysis of valU operon processing. (A) Schematic diagram of the valU operon (not drawn to scale). Relative positions of the oligonucleotide probes (a: valU-UP, b: valUXYTZ, c: valU-X, d: valX-Y, e: valY-lysV, f: lysmature, g:lysV-TER) used in Northern analyses are shown. Probe sequences are listed in Supplementary Table S1 (Supplementary Materials). Numbers indicate the length of the intergenic spacers. (B) Northern analysis of valU operon. The specific oligonucleotide probe used for each blot is indicated in the bottom left corner of each blot. The genotypes of the strains used are listed on the top of each autoradiogram. The RNA molecular size standards (nucleotides) (Fermentas) are shown to the left of the first image. The numerical designations of the different species along with their graphical structures are shown to the right of the first blot. Relative quantity (RQ) of mature tRNAVal (M) in various genetic backgrounds was calculated by setting a level of one in the rph-1 strain. Processed fraction (PF) represents the fraction of mature tRNA relative to the total amount of processed and unprocessed species of that tRNA. Each value represents the average of at least three independent determinations. (C) Analysis of the role of RNase PH in the final maturation of the five valine type I tRNAs.
Figure 2.
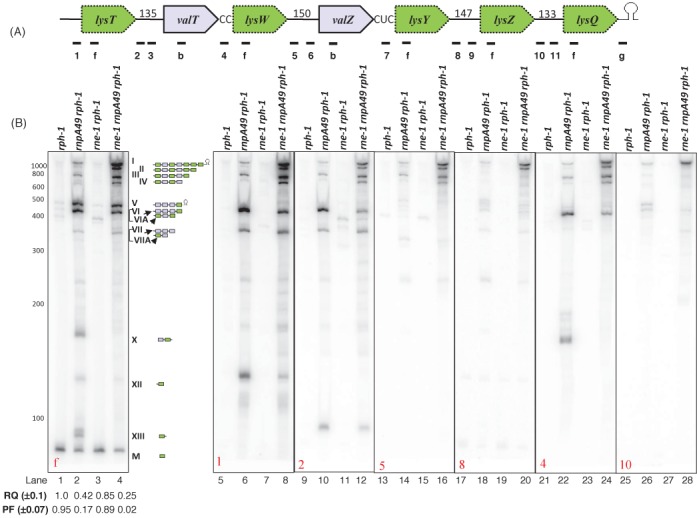
Analysis of the lysT operon processing. (A) Schematic diagram of the operon (not drawn to scale). Relative positions of the oligonucleotide probes (1: lysT-UP, f: lysmature, 2: lysT-valT1, 3: lysT-valT3, b: valUXYTZ, 4: valT-lysW, 5: lysW-valZ1, 6: valZ-UP2, 7: valZ-lysY, 8: lysY-lysZ1, 9: lysY-lysZ2, 10: lysZ-lysQ1, 11: lysZ-lysQ2, g: lysV-TER). Probe sequences are listed in Supplementary Table S1 (Supplementary Materials). Numbers indicate the length of the intergenic spacers. (B) Northern analysis of lysT operon. The specific oligonucleotide probe used in each blot is indicated in the bottom left corner of the blot. The genotypes of the strains used are indicated on the top of the each autoradiogram. The RNA molecular size standards (nucleotides) (Fermentas) are shown to the left of the first blot. The numerical designations of the different species along with their graphical structures are shown to the right of the blot. Bands VI and VII arise from the valU operon and comigrate with bands VIA and VIIA, respectively. Two independent Northern blots were run (one was probed with f, 1, 2, 5 and 8; second was probed with 4 and 10). The RQ and PF values were calculated as described in the legend to Figure 1.
Accordingly, we examined the processing of the valU operon in the rph-1, rne-1 rph-1, rnpA49 rph-1 and rne-1 rnpA49 rph-1 isogenic strains by Northern analysis using operon-specific oligonucleotide probes (Figure 1A). Since all five isotype I valine tRNAs have identical sequences, we first probed with an oligonucleotide that was complementary to the mature valine type I coding sequence (probe b, Figure 1A). No significant differences in processing intermediates were observed between the rph-1 and rne-1 rph-1 strains (Figure 1B, lanes 1–2). The most prominent species were the mature tRNA (M) and a slightly larger band. This larger band was due to the RNase PH deficiency (rph-1) in all the strains resulting in unprocessed 3′ ends (See below). There were also three high molecular weight species (V, VI and VII) that were weakly visible (lanes 1, 2).
The intensity of species V, VI and VII increased dramatically in the absence of RNase P (rnpA49 rph-1) along with an almost 4-fold decrease in the amount of the mature species (M) (lane 3). In addition, new species (VIII, IX and X) appeared in the absence of RNase P. When both RNase P and RNase E (rne-1 rnpA49 rph-1) were inactivated, species X disappeared along with slightly increased intensities of species V and VIII and small decreases in the levels of species VI and VII (lane 4), suggesting a very minor role for RNase E in the absence of RNase P. The level of mature species (M) remained unchanged compared to the rnpA49 rph-1 double mutant. In fact, the processed fraction (PF) for the mature tRNAVal1 decreased from 0.76 and 0.95 in the rph-1 and rne-1 rph-1 strains, respectively, to 0.04 in the rnpA49 rph-1 and rne-1 rnpA49 rph-1 strains (lanes 1–4).
The composition of all the various processing intermediates seen in Figure 1B (lanes 1–4) was subsequently determined by probing the blot with oligonucleotides that were complementary to various regions of the operon (Figure 1A). For example, probe a (complementary to the upstream region of valU) hybridized to all the processing intermediates (species V-IX, XI) with the exception of species X (lanes 5–8). In contrast, probe g (complementary to the Rho-independent transcription terminator, Figure 1A) only hybridized to species V (lanes 21–24) demonstrating that it was the full-length valU operon transcript (predicted size ∼438 nt).
In addition, probes c, d and e hybridized to species V and VI (lanes 9–20). Since species VI also hybridized to probe a (lanes 5–8), but not to probe g (lanes 21–24), it was identified as the complete transcript minus the Rho-independent transcription terminator. Our data also suggested that the Rho-independent transcription terminator associated with species V was removed primarily by RNase P, because its level increased significantly in the rnpA49 rph-1 strain compared to the rne-1 rph-1 mutant (lanes 2, 3).
Probe e, which was complementary to the intergenic region between valY and lysV (Figure 1A), did not hybridize to either mature tRNAVal or tRNALys and also did not hybridize to species VII (Figure 1B, lanes 17–20). However, species VII was detected with probe d (lanes 13–16), indicating that it arose from a cleavage between valY and lysV tRNAs. In contrast, species VIII appeared to arise from a cleavage downstream of valX. The multiple bands labeled as species X were observed with probes b and e in the rnpA49 rph-1 strain (lanes 3, 19), but disappeared in the rne-1 rnpA49 rph-1 strain (lanes 4, 20). This result suggested that this group of processing intermediates containing valine coding sequences arose from occasional RNase E cleavages at different sites within the valU operon transcript in the rnpA49 rph-1 strain.
Species XI was detected by probes b and a in the rnpA49 rph-1 (lanes 3, 7), but with reduced intensity in the rne-1 rnpA49 rph-1 strain (lanes 4, 8). However, this band was not detected with probe c (complementary to the intergenic region between valU and valX), even in the rnpA49 rph-1 strain (lane 11), indicating that in the absence of RNase P, RNase E cleaved inefficiently in the intergenic region between valU and valX, resulting in a fragment containing valU with the 5′ leader sequence and perhaps some portion of the intergenic region at its 3′ end.
The detection of bands I-IV with probes b, e and g (Figure 1B, lanes 4, 20, 24) was initially surprising, since their sizes were much larger than the full-length valU polycistronic transcript. However, upon examining the sequences of the valU operon and lysT gene cluster (18), we realized that the intergenic region between valY and lysV (CUUC, Figure 1A) was similar to the intergenic regions between valT and lysW (CC, Figure 2A) and valZ and lysY (CUC, Figure 2A). Since all the five valine tRNAs and the six lysine tRNAs present in the valU operon and lysT gene cluster are identical to each other, respectively, we hypothesized that probes b and e (Figure 1A) were likely hybridizing to RNA fragments originating from both the valU operon and the lysT gene cluster. Additionally, we observed that the Rho-independent transcription terminators downstream of the lysV and lysQ genes were identical in sequence (data not shown). Thus, the large species detected with probes b, e and g (lanes 4, 20, 24) probably originated from transcripts of the lysT gene cluster (see below).
RNase PH is required for the final 3′ end maturation of the valine type 1 tRNAs
It should be noted that our experiments were carried out in a parental strain (MG1693) that was defective in RNase PH (rph-1). Sequence analysis of 5′ → 3′ ligated tRNAs from rph-1 strains identified val tRNAs that had either one or two extra nucleotides downstream of the CCA determinant (data not shown). These results were consistent with the species that was slightly larger than the mature valine tRNA that was observed in the rph-1 and rne-1 rph-1 strains (Figure 1B, lanes 1, 2). In fact, the larger species disappeared in SK10153 an isogenic rph+derivative of MG1693 (Figure 1C).
ThelysT gene cluster is transcribed as a polycistronic transcript
Based on our observations with probes b, e and g (Figure 1B), we hypothesized that the larger species (I-IV) were derived from the lysT gene cluster, even though it is indicated in the EcoCyc database that the seven genes are part of four separate transcripts (18). If all seven tRNAs in the lysT gene cluster were synthesized as a polycistronic transcript, a full-length species of over 1100 nt would be generated. In fact, a species greater than 1100 nt in length (I) was observed in the rnpA49 rph-1 and rne-1 rnpA49 rph-1 strains (Figure 2B, lanes 2, 4) along with multiple lower molecular weight species (II, III, IV, V, VI, VIA, VII, VIIA, X, XII, XIII and M). However, only species I hybridized to both probe 1 (Figure 2A and B, lanes 6, 8) and probe g (Figures 1A and 2B, lane 24), demonstrating that it constituted the full-length lysT transcript. The larger species (I-IV) were barely visible in the rph-1 and rne-1 rph-1 strains (Figure 2B, lanes 1, 3), suggesting a very minimal role for RNase E in processing the lysT transcript if RNase P were present.
Processing of the lysT polycistronic operon requires RNase P but is stimulated by RNase E
Unlike what was seen with the analysis of the valU operon, there was a single mature lysine tRNA species (M) in both the rph-1 and rne-1 rph-1 strains and surprisingly few other processing intermediates (Figure 2B, lanes 1, 3). In contrast, the amount of the mature tRNALys species (M) was significantly reduced in the rnpA49 rph-1 and rne-1 rnpA49 rph-1 mutants (lanes 2, 4) along with a concomitant increase in larger species (I-V, VIA, VIIA, X, XII and XIII). In fact, the PF of mature tRNALys decreased from 0.95 and 0.89 in the rph-1 and rne-1 rph-1 strains to 0.17 and 0.02 in the rnpA49 rph-1 and rne-1 rnpA49 rph-1 mutants, respectively.
To ascertain the composition of the larger species (I-IV), we probed the blot with oligonucleotides that were complementary to the 5′ leader region (Figure 2A, probe 1) as well as intergenic regions between lysT and valT (Figure 2A, probes 2, 3), lysW and valZ (probes 5, 6), lysY and lysZ ( probes 8, 9), and lysZ and lysQ (probes 10, 11). With probes 10 and 11, species I, but not species II-IV, was observed in both rnpA49 rph-1 and rne-1 rnpA49 rph-1 strains (lanes 25–28; data not shown). It should be noted that the additional bands present in Figure 2B, lanes 26 and 28, arose from incomplete stripping of earlier probes. With probes 8 and 9, species I (full-length lysT operon) was detected in the rnpA49 rph-1 strain, while species I and II were present in the rne-1 rnpA49 rph-1 strain (lanes 17–20; data not shown). These results suggested that species II was generated by RNase E cleavage between the lysZ and lysQ tRNAs, since it hybridized with probes 8 and 9, but not 10 or 11 (lanes 20, 28).
Similarly, species III arose from a cleavage between lysY and lysZ tRNAs based on its hybridization to probes 5 and 6, but not the probes 8 and 9 (lanes 13–20, data not shown). Species IV was most likely a lysT valT lysW valZ processing intermediate because of its molecular weight, but we could not unequivocally make this assignment.
Based on the data shown in Figure 1, the hybridization of probe f to species V in both the rnpA49 rph-1 and rne-1 rnpA49 rph-1 mutants (Figure 2B, lanes 2 and 4), represented the detection of the full-length valU polycistronic transcript. The failure of probe 1 (Figure 2A) to hybridize to species V (Figure 2B, lanes 6 and 8) confirmed this identification. However, probe 1 hybridized to the two most prominent species of ∼430 and ∼140 nt (VIA and XII), respectively, in the rnpA49 rph-1 double mutant (Figure 2B, lane 6). Since species VIA also hybridized to probes 2 and 4 (Figure 2A, B, lanes 10, 22), but not to probe 5 (Figure 2B, lane 14), this species was identified as lysT valT lysW processing intermediate that retained its 5′ leader region.
Species XII was identified as a lysT intermediate with the 5′ leader region, since it hybridized to probes f and 1 (Figure 2A and B, lanes 2, 6). The intensities of VIA and XII decreased significantly in the rne-1 rnpA49 rph-1 triple mutant (Figure 2B, lane 8 versus lane 6) along with concomitant increases in the intensities of species I-IV, suggesting these processing intermediates arose from RNase E cleavages within the larger lysT polycistronic transcript. It should be noted that species VIA (lysT valT lysW) is similar in size to species VI (Figure 1B, valU valX valY lysV).
Species VIIA was observed with probes that were complementary to the spacer region between lysT and valT (probes 2, 3) (Figure 2B, lanes 10, 12; data not shown), but was not detected with a probe complementary to the intergenic region between valT and lysW (probe 4) (Figure 2B, lanes 22, 24). Thus, species VIIA was a lysT valT processing intermediate that retained its 5′ terminal sequence. It was designated as VIIA to distinguish it from species VII that was observed in Figure 1B, but was a valU valX valY processing intermediate that was almost identical in size. Species XIII was only observed with probe f (Figure 2B, lane 2) and probe 9 (data not shown), suggesting it was a lysZ intermediate that retained some 3′ sequences.
Species X hybridized to probe 4 (Figure 2A and B, lanes 2, 22) as well as probe 7 (data not shown). However, it failed to hybridize with probes 2, 5 and 8 (Figure 2B, lanes 10, 14 and 18). Thus, this species seemed to be a mixture of valT lysW and valZ lysY intermediates. The absence of species X in the rne-1 rnpA49 rph-1 triple mutant indicated that these intermediates were generated by RNase E cleavages upstream of valT, valZ and lysZ in the absence of RNase P.
Identification of cleavage sites using RNA circularization
Although the data in Figures 1 and 2 clearly identified the composition of the various tRNA processing intermediates from the valU and lysT operons, the Northern analyses did not provide insights into the precise cleavage sites. Since primer extension analysis was not technically feasible for these two polycistronic transcripts, we employed cloning and sequencing of cDNAs derived from reverse transcription of self-ligated tRNAs (29) to identify the 5′ and 3′ termini of various species. This method has been successfully employed previously to simultaneously obtain the 5′ and 3′ termini of specific individual tRNA species (15,28).
Accordingly, clones were obtained from rph-1, rnpA49 rph-1 and rne-1 rnpA49 strains and sequenced (see Materials and Methods). The data presented in Figure 3, represent the analysis of over 150 independent isolates obtained from the three genetic backgrounds. Although it was not possible to obtain sequence information for every cleavage site, we were able to identify RNase P cleavages at the mature 5′ termini of the valU, valX, valY, lysV and lysW transcripts (Figure 3, P1–5), since there are no known 5′ → 3′ exonucleases in E. coli. Furthermore, it appeared that RNase P and perhaps RNase E cleaved two nucleotides downstream of the CCA of lysV to remove the Rho-independent transcription terminator from the polycistronic valU primary transcript (T5, Figure 3). It should also be noted that the transcription start-site (TSS) of the valU operon is 9 nt upstream of the mature 5′ terminus of valU (Figure 3), not 7 nt as indicated at the EcoCyc Website. The identification of various 5′ and 3′ ends (Figure 3) was consistent with several processing intermediates (M, VI, VII, VIII, XI and X) that had been characterized by Northern analysis (Figures 1 and 2).
Figure 3.
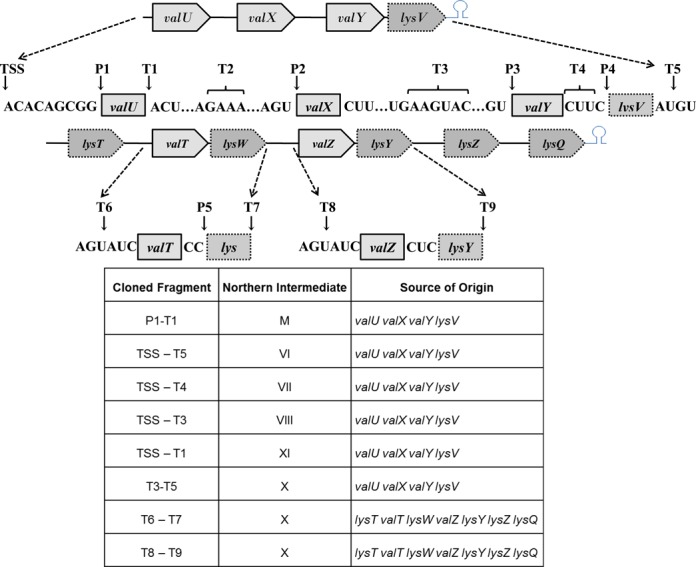
Identification of cleavage sites within the valU and lysT operons by cloning and sequencing of processing intermediates. cDNAs of self-ligated RNA molecules were amplified, cloned and sequenced to identify the 5′ and 3′ ends as described in ‘Materials and Methods’. Each arrow represents either a 5′ or 3′ end based on the DNA sequence. Multiple ends were obtained at the T2, T3 and T4 locations. The transcription, start-site (TSS), RNase P cleavage sites (P1–5), 3′ cleavage sites [either at the mature 3’ end or in the intergenic region for respective upstream tRNAs (T1–5, 7 and 9)] and 5′ cleavage sites (in the intergenic regions) for respective downstream tRNAS (T6, 8) are indicated. The 3′ and 5′ ends of some of the major processing intermediates observed in the Northern analyses (Figures 1 and 2) are shown below the diagram.
RNase P is the only enzyme that generates valine pre-tRNAs
In order to further confirm that RNase P was both essential and sufficient for the generation of the valine pre-tRNAs, we carried out half-life measurements on the three large valU operon species (Figure 1B, V, VI and VII). We predicted that if other endoribonucleases were involved in processing this polycistronic transcript, even inefficiently, we would see the decay of one or more of these species in an rnpA49 rph-1 double mutant. In the rph-1 control, both species V (the full-length transcript) and VII (valU valX valY) had identical half-lives of ∼1 min (Figure 4, Table 2), while the half-life of species VI was estimated at <0.5 min (Figure 4, Table 2). In the rne-1 rph-1 mutant, all three species had half-lives of <0.5 min (Figure 4, Table 2), explaining the higher RQ and PF values of steady-state valU tRNAs compared to the rph-1 strain (Figure 1B, lanes 1, 2). Although the exact reason for faster processing of the valU transcript in the rne-1 rph-1 double mutant is not clear, it might simply be due to better access of RNase P to its processing sites in the absence of RNase E.
Figure 4.
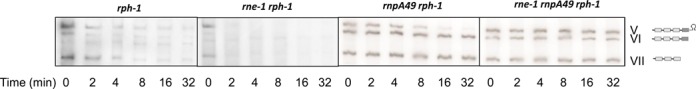
Northern analysis of the decay of valU operon. The genotypes of the strains are indicated on the top and time-points are shown at the bottom of the autoradiograms. The blots were probed with probe b (Figure 1A). The numerical designations of the different species with their graphical structures are shown to the right and are identical to those shown in Figure 1.
Table 2. Half-lives of valU and lysT operon transcripts in various strains.
| (A) valU | ||||
|---|---|---|---|---|
| Species | Half-life (min) | |||
| rph-1 | rnpA49 rph-1 | rne-1 rph-1 | rne-1 rnpA49 rph-1 | |
| V | 1.0 ± 0.4 | 7.7 ± 1.4 | <0.5 | >30 |
| VI | <0.5 | >30 | <0.5 | >30 |
| VII | 1.1 ± 0.1 | >30 | <0.5 | >30 |
| (B) lysT | ||||
| Species | Half-life (min) | |||
| rph-1 | rne-1 rnpA49 rph-1 | rnpA49 rneΔ1019 rng-219 rph-1 | rne-1 rnpA49 Δrnz Δrng rnlA2 rph-1 | |
| I | <0.5 | >30 | >30 | >30 |
In contrast, in the absence of RNase P, the half-life of species V increased over 7-fold, while that of species VI and VII increased over 30-fold (Figure 4, Table 2). In the absence of both RNase E and RNase P, all three species had half-lives of >30 min. These data provided further support that normally RNase P catalyzes the initial removal of the Rho-independent transcription terminator from the valU transcript. However, in the absence of RNase P, RNase E can substitute inefficiently to carry out this reaction.
Since the lysT polycistronic transcript is over 1100 nt in length, we carried out half-life experiments for this operon using 1.2% agarose gels. Unlike what was observed with the full-length valU transcript, we could not detect the full-length lysT transcript in either the rph-1 or rne-1 rph-1 strains even at time zero, indicating a half-life of <0.5 min in these genetic backgrounds (data not shown). In contrast, the half-life of species I was >30 min in an rne-1 rnpA49 rph-1 triple mutant (Table 2).
Although these data strongly suggested that RNase P was primarily responsible for the initial processing of all five valine type 1 tRNAs, we wanted to directly rule out the possibility that other known ribonuclease(s) besides RNase E might be involved in their processing. Accordingly, we tested a variety of multiple mutants that were deficient in RNase III (rnpA49 rnc-14 rph-1), RNase G (rne-1 rnpA49 Δrng rph-1), RNase LS (rne-1 rnpA49 rnlA2 rph-1), YbeY (rnpA49 ΔybeY rph-1) and RNase Z (rne-1 rnpA49 Δrnz rph-1) as well as either RNase P or RNase E. None of the additional endonucleases tested affected the processing of the valU operon (data not shown). We also eliminated the possibility of residual RNase E activity in strains carrying the rne-1 allele (15) by examining valU operon processing in an RNase E deletion strain [rnpA49 Δrne-1018/ rng-219 rph-1, data not shown, see (32)].
Since species VII (valU valX valY, Figure 1) was present in both the rnpA49 rph-1 and rne-1rnpA49 rph-1 strains, we also tested the possibility that the lysV tRNA was removed by exonucleolytic processing from the 3′ terminus. However, the processing of the valU primary transcript in multiple mutant strains deficient in PNPase (pnp) and RNase II (rnb) (rne-1 pnp-7 rnb-500 rph-1; rnpA49 pnp-7 rnb-500 rph-1; and rne-1 rnpA49 rnb-500 pnp-7,rph-1), was unchanged compared to a rnpA49 rph-1 strain (data not shown).
Ectopic expression of mature valine tRNAs does not suppress the inviability associated with an rnpA49 mutant
Because the loss of RNase P results in tRNAs with immature 5′ ends, it has been assumed that such pre-tRNAs cannot be aminoacylated leading to an inhibition of protein synthesis and a subsequent loss of cell viability. However, recent studies suggest that tRNAs with immature 5′ but mature 3′ ends can be functional both in vivo and in vitro (15). Alternatively, the loss of RNase P activity could be lethal if all of a particular species of tRNA(s) remained as unprocessed primary transcripts. From the analysis of the processing of the valU and lysT operons (Figures 1 and 2), it was evident that the amount of mature tRNAVal(UAC) was reduced >4-fold in an rnpA49 rph-1 strain. Since the processing of tRNAVal(GAC) is also completely dependent on RNase P (17), we hypothesized that the loss of cell viability in the absence of RNase P might arise from the significantly reduced levels of functional valine tRNAs [Figure 1B, lane 3; (17)].
In order to test this hypothesis directly, we generated a set of compatible plasmids from which the two valine isotypes, tRNAval(GAC) (valW) and tRNAval(UAC) (valU) were ectopically expressed in a way that did not require RNase P processing at either the 5′ or 3′ terminus. In both cases, the valW and valU genes were constructed such that transcription termination was controlled by the leuU Rho-independent transcription terminator, which is known to be removed efficiently by RNase E (15). Furthermore, to eliminate the need for RNase P processing of the tRNAs synthesized from the plasmids, they were designed such that transcription initiated at the mature 5′ terminus of each tRNA.
To ensure that the tRNA species being expressed from the two plasmids were mature and functional, we measured the in vivo aminoacylation levels of the two valine tRNA isotypes. Surprisingly, the percentage of aminoacylation of both the valine tRNA isotypes was less than 40% in an rph-1 strain (Table 3). This result was unexpected since other tRNAs have charging efficiencies ranging between 60 and 90% (33,34) in the same genetic background. Surprisingly, the level of aminoacylation improved only marginally in the rph+ strain in spite of the significant improvement in 3′ end maturation (Figure 1C). As expected, the extent of aminoacylation of both valine tRNAs decreased between 1.9 and 2.3-fold (Table 3) in the absence of RNase P, but the aminoacylation levels of both tRNAs returned to the level observed in the rph-1 control upon ectopic expression of both tRNAs from plasmids in an rnpA49 rph-1 strain (Table 3).
Table 3. Aminoacylation levels of mature valine tRNAs.
| tRNA isotype/genes | Percentage of aminoacylated tRNA | |||
|---|---|---|---|---|
| rph-1 | rnpA49 rph-1 | rnpA49 rph-1/valU+ valW+ | rph+ | |
| Val1/valU, X,Y, T, Z | 39 ± 2 | 17 ± 6 | 37 ± 3 | 41 ± 5 |
| Val2/valV, W | 38 ± 4 | 20 ± 3 | 37 ± 3 | 47 ± 5 |
The percentage of each aminoacylated tRNA was determined by the method of Varshney (27) as previously described (33). valU+ and valW+ represent tRNA species transcribed from plasmids pAAK11 and pAAK13, respectively as 5′-mature molecules with a leuU transcription terminator known to be removed efficiently by RNase E (15) (See Materials and Methods). Data shown are the average of at least two independent determinations.
Subsequently, we measured the growth and cell viability of an rnpA49 rph-1 strain in which both valine tRNAs were ectopically expressed. As a positive control we also overexpressed the M1 RNA component of RNase P, since increased levels of the M1 RNA have been shown to partially complement the rnpA49 allele at 42°C (35,36). As shown in Figure 5, the simultaneous expression of tRNAval(GAC) and tRNAval(UAC) did not suppress the temperature sensitivity of the rnpA49 strain. In contrast, overproduction of the rnpB gene did lead to a small improvement in the growth of the rnpA49 rph-1 strain at 42°C. In addition, there was no change in cell viability in the strain ectopically expressing both mature valine tRNAs (data not shown). It should be noted that identical results were obtained in an rnpA49 rph+ strain (data not shown).
Figure 5.
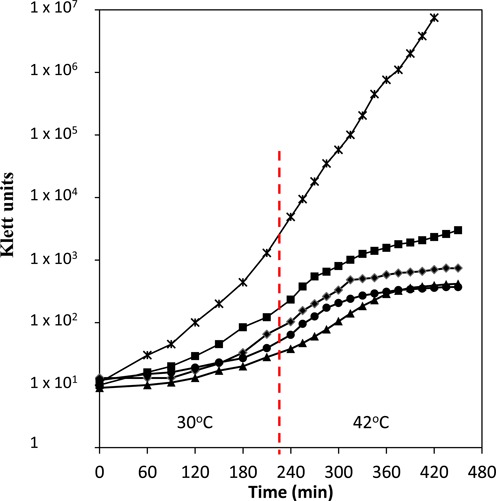
Growth curve of various strains. All strains were initially grown at 30°C until 50 Klett units above background when IPTG was added to a final concentration of 350 μM. The cultures were diluted with pre-warmed Luria broth (LB)/thymine or LB/thymine/IPTG to maintain them in exponential growth (Klett 50–70). For this experiment, all the cultures were shifted to 42°C when the slowest growing culture reached Klett 50 above background (vertical dashed line), since it has been previously shown that overproduction of rnpB does not complement at 44°C (36). The optical densities (OD) were measured with a Klett-Summerson Colorimeter (No. 42 Green filter). x: MG1693 (rph-1); ♦: SK2525 (rnpA49 rph-1); ▴: SK10537 (rnpA49 rph-1 argX+); •: SK10541 (rnpA49 rph-1 valU+valW+); ▪: SK10539 (rnpA49 rph-1 rnpB+).
Ecotopic expression of the mature 4.5S RNA or tRNAArg(CCG) does not suppress the inviability associated with the rnpA49 allele
It has previously been suggested that overexpression of tRNAArg(CCG) can complement an rnpA49 mutation (35). However, as shown in Figure 5, ectopic expression of this tRNA had no effect on the growth of an rnpA49 rph-1 mutant at the elevated temperature. It is known that RNase P is required to generate the mature 5′ end of the essential 4.5S RNA (8). In fact, it has been suggested that this processing reaction could be the essential function of RNase P in E. coli (37). Accordingly, we expressed a 4.5S RNA species from a plasmid that did not require RNase P processing at it 5’ terminus. Increased levels of this species also did not suppress the conditional lethality of the rnpA49 allele (data not shown).
DISCUSSION
The data presented here demonstrate that all five of the valine isotype 1 pre-tRNAs derived from the valU and lysT operons are initially generated by RNase P cleavages (Figures 1 and 2). When coupled with our previous work showing that the two valine isotype 2 tRNAs (valV and valW) also exclusively use RNase P for their initial processing (17), it is clear that all seven valine tRNA species in E. coli require RNase P for separation from their primary polycistronic transcripts. The more than 4-fold decrease in the level of mature valine tRNA after shift to the non-permissive temperature was consistent with the absolute requirement for initial RNase P cleavages [Figure 1, (17)].
What is most striking about the results presented here relates to how RNase P processes both the valU and lysT polycistronic transcripts. There are three possible mechanisms by which RNase P could process the primary transcripts: (i) initiate cleavage at the 5′ terminus of the first tRNA in the operon and proceed in the 5′ → 3′ direction to the end of the transcript; (ii) initiate processing by first removing the Rho-independent transcription terminator and proceeding in the 3′→ 5′ direction to the 5′ proximal end of the transcript; or (iii) cleave the transcripts randomly with some sites being preferred over others.
These three possibilities can be distinguished by examining the processing intermediates obtained in the various rnpA49 mutants (Figures 1 and 2). For example, in the case of the valU operon, all of the major processing intermediates observed after inactivation of RNase P retained their immature 5′ termini (Figure 1B, lanes 3, 7). These data clearly rule out RNase P initiating cleavage at the 5′ proximal terminus and proceeding in the 5′ → 3′ direction. In addition, both Northern analysis and cDNA sequencing of processing intermediates suggested that each of the intermediates detected in the rnpA49 rph-1 mutant was shortened by one tRNA (Figure 1B, species VI, VII, VIII and XI: Figure 3). It would thus appear that the separation of the valU polycistronic transcript is not random but rather proceeds in the 3′ → 5′ direction, as outlined in Figure 6, with the first cleavage reaction being the removal of the Rho-independent transcription terminator. This is in contrast to the processing of the valV valW dicistronic transcript, where it was suggested that RNase P cleavage at the 5′ terminus of valW tRNA might occur cotranscriptionally, since computational models of the dicistronic transcript indicated that this RNase P site might be occluded (17).
Figure 6.
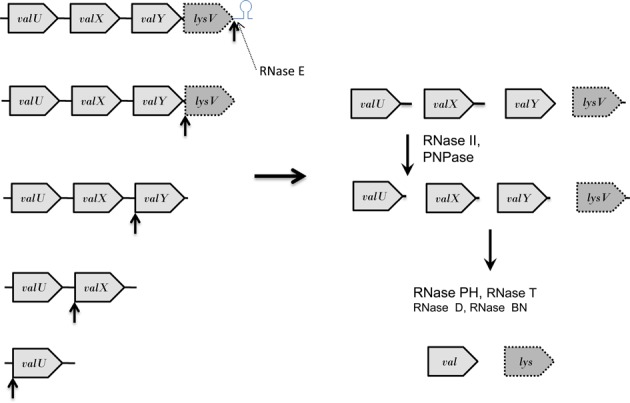
Processing pathway for the valU polycistronic transcript. Similar to what has been observed with the valV valW and secG leuU primary transcripts (15,17), RNase P (upward black arrows) is the primary processing enzyme for the valU transcript. Initially, RNase P efficiently removes the Rho-independent transcription terminator (upward black arrow). Subsequent sequential cleavages occur in the 3′ → 5′ direction at the mature 5′ termini of lysV, valY, valX and valU (upward black arrows). The valU and valX pre-tRNAs will have long 3′ termini of 44 and 47 nt, respectively (Figure 1A) that will be initially processed by a combination of RNase II and PNPase (17,48). Subsequently, the final processing of the valine pre-tRNAs will be carried out primarily by RNase PH because of the presence of C residues downstream of the CCA determinant (42). The final processing of the lysV pre-tRNA employs RNase T. In the absence of RNase P, RNase E can inefficiently remove (dashed arrow) the Rho-independent transcription terminator (Figure 1B). Transcripts are not drawn to scale.
Furthermore, the data shown in Figures 1 and 4 demonstrate that the only role RNase E plays in the processing of the valU operon is to inefficiently remove the Rho-independent transcription terminator in the absence of RNase P, since the processing pattern of the full-length transcript remained unchanged in the rne-1 rph-1 mutant compared to the rph-1 single mutant except for the slightly reduced signal intensity of species V (Figure 1B, lanes 1, 2). The half-life data (Figure 4) and our analysis of other multiple mutants lacking RNase III, RNase G, YbeY, RNase Z and RNase LS (data not shown) also confirmed that RNase P was the only endonuclease that processed the full-length valU transcript and thus was primarily responsible for the removal of the Rho-independent transcription terminator. These results were also very different from what was observed with the metT operon in which inactivation of RNase E by itself led to a significant change in the observed processing intermediates (15).
The processing of the lysT operon is somewhat more complex. On the one hand, RNase E does not play a significant role in the separation of any of the seven tRNAs in the presence of RNase P as judged by the lack of any new decay intermediates in the rne-1 rph-1 strain (Figure 2B, lane 3). Furthermore, as seen with the valU operon, the vast majority of the processing intermediates in both the rnpA49 rph-1 and rne-1 rnpA49 rph-1 strains retained their 5′ termini, again suggesting that RNase P initiates the processing of the large 1100 nt transcript at the 3′ and proceeds in the 3′→ 5′ direction. The increase in the amounts of species I-IV in the rne-1 rnpA49 rph-1 triple mutant can be ascribed to the ability of RNase E to cleave within the large spacer regions between lysT and valT (135 nt), lysW and valZ (150 nt), lysY and lysZ (147 nt), and lysZ and lysQ (133 nt) (Figure 2A) in the absence of RNase P due to stabilization of the primary transcript. The identification of valT lysW and valZ lysY intermediates with 5′ unprocessed ends (Figure 3) is consistent with this analysis. These cleavages do not occur in the presence of RNase P (Figure 2B, lane 3), since each of the tRNAs is processed at the 5′ mature terminus (Figure 7). Thus, we propose a processing pathway for the lysT operon (Figure 7), which is similar to that described for the valU operon (Figure 6).
Figure 7.
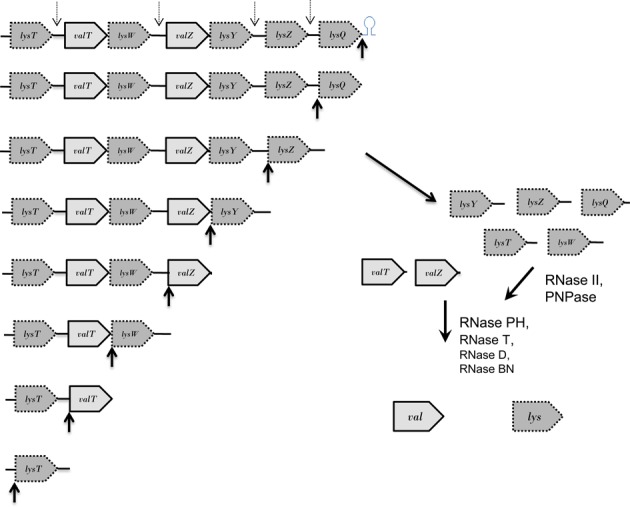
Processing pathway for the lysT polycistronic operon. Similar to what was described in Figure 6, after RNase P removes the Rho-independent transcription terminator (upward black arrow), subsequent sequential cleavages will occur in the 3′ → 5′ direction at the mature 5′ termini of lysQ, lysZ, lysZ, lysW, valT and lysT (upward black arrows) to generate pre-tRNAs containing between 2 and150 nt extra nucleotides at their 3′ ends (Figure 2A). The long 3′ ends of the five lys pre-tRNAs will be initially processed by a combination of RNase II and PNPase (17,48). Final maturation of the lys tRNAs primarily is carried out by RNase T, while the final processing of the two valine species is done by RNase PH. In the absence of RNase P, RNase E (dashed downward arrows) can cleave within the long spacer regions between lysT and valT, lysW and valZ, lysY and lysZ, and lysZ and lysQ. However, RNase E does not separate valT and lysW or valZ and lysY. Transcripts are not drawn to scale.
The data presented above suggest that transcription must be complete before any processing occurs, indicating that nascent transcripts are initially protected from degradation. Although the basis of this protection is not clear, it is possible that either the secondary structure of the transcript itself or RNA chaperones like Hfq (38) prevent processing until the entire transcript is completed. Taken together, our data indicate that RNase P can efficiently process large transcripts starting from their 3′ termini in vivo. In fact, based on the half-life data (Table 2), the enzyme works more efficiently on the 1100 nt lysT transcript than the 440 nt valU transcript. These results are consistent with the fact that RNase P cleaves the 5′ end of a tRNA poorly in the presence of a long 3′ trailer (39).
It should also be noted that Hansen et al. (40) showed that RNase P could cleave single-stranded oligonucleotides in vitro particularly between U and G. Moreover, Mohanty and Kushner (7) demonstrated that the Rho-independent transcription terminator associated with the leuX transcript was removed in vivo by an RNase P cleavage reaction between a U and G residue seven nucleotides downstream of the CCA determinant. It is thus not surprising that the Rho-independent transcription terminator associated with the valU operon was also removed endonucleolytically by an RNase P cleavage between a U and G residue three nucleotides downstream of the CCA (Figure 3). Furthermore, since the terminators of both the valU and lysT operons have identical sequences, it is presumed that the Rho-independent transcription terminator associated with the lysT operon is removed in a similar fashion.
However, the model shown in Figure 6 does not completely explain the origin of species VII (valU valX valY), which was present in all the genetic backgrounds (Figure 1B, lanes 1–4). Since there is only a four nucleotide spacer between valY and lysV (CUUC), this sequence should not be cleaved by RNase E (30,31). Yet there is an almost 2-fold decrease in the amount of this species in the rne-1 rnpA49 rph-1 triple mutant and the rne-1 rnpA49 Δrnz Δrng rnlA2 rph-1 hextuple mutant (data not shown) compared to the rnpA49 rph-1 double mutant (Figure 1B, lanes 3, 4). Thus, it is possible that RNase E is cleaving poorly in between valY and lysV (ACCACUUC). In addition, we could not demonstrate any exonucleolytic removal of lysV tRNA from the full-length transcript, similar to the removal of the leuX Rho-independent transcription terminator by PNPase (7). Thus, another possibility is that this species arises from premature transcription termination at a site immediately downstream of valY. Alternatively, it is possible that there is another endonuclease, as yet unidentified.
Based on the absolute dependence on RNase P for the initial separation of all seven valine tRNAs [Figures 1 and 2, (17)], we hypothesized that ectopic expression of both valine isotypes in a fashion that did not require RNase P to generate mature species might suppress the lethality associated with the rnpA49 mutation. To our surprise, even though ectopic expression of the two valine tRNAs led to a significant increase in the amount of aminoacylated valine tRNAs (Table 3), no suppression of the growth defect was observed (Figure 5). In fact, when both valine tRNAs were simultaneously expressed, the strain grew more slowly than the rnpA49 mutant on its own (Figure 5). The only improvement in growth in the rnpA49 mutant after shift to 42°C occurred in the presence of increased expression of the M1 RNA, in agreement with previously reported results (36). Furthermore, we were not able to reproduce the work of Kim et al. (35), who suggested that overproduction of an arginine tRNA could suppress the rnpA49 allele. In addition, we also ruled out the possibility that the processing of the 5′ end of the 4.5S RNA was the essential function of RNase P (data not shown).
Since it has been reported that the dephosphorylation of the 5′ triphosphate by the RppH enzyme from Bacillus subtilis requires a single-stranded region of at least two nucleotides (41), it is possible that the ectopically expressed RNAs retained their 5′ triphosphates. Although the data in Table 3 demonstrate that the potential presence of a triphosphate did not interfere with aminoacylation, we cannot rule out the possibility that the triphosphate moiety at the 5′ terminus of the charged tRNA interfered with its binding to the ribosome.
Finally, it should be noted that the final maturation of the 3′ termini of the valine type 1 tRNAs required RNase PH (Figure 1C), while the type II valine tRNAs (17) and the lysine tRNAs (Figure 2) did not. These observations are in agreement with the results of Zuo and Deutscher (42) who showed that C nucleotides immediately downstream of the CCA determinant will inhibit RNase T activity. In the case of the type I valine tRNAs, four of the five species have at least one C immediately downstream of the CCA (CCAACUAC, CCACUUUC, CCACUUC, CCACC, CCACUC). In contrast, this is not the case with the valV and valW transcripts (CCAUCCU, CCAGAUU) or all six lys transcripts (data not shown). It is also worth noting that even though the presence of wild-type RNase PH activity led to a single mature val tRNA species (Figure 1C), the fraction of aminoacylated type 1 tRNA did not significantly increase (Table 3, data not shown). Thus, the relatively low levels of valine aminoacylation are not related to 3′ end processing but rather to inefficient charging.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online. Supplementary Table S1
Acknowledgments
We thank Mark Stead, Valerie Maples, Maria Ow and Khailee Marischuk for constructing some of the strains used in this study.
FUNDING
National Institutes of Health [GM57220 and GM81554 to S.R.K.]. Funding for open access charge: National Institutes of Health Research Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Blattner F.R., Plunkett G., III, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., et al. The complete sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 2.Berlyn M.K.B. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol. Mol. Biol. Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Z., Deutscher M.P. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ow M.C., Kushner S.R. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev. 2002;16:1102–1115. doi: 10.1101/gad.983502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deutscher M.P. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z., Deutscher M.P. Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell. 1996;86:503–512. doi: 10.1016/s0092-8674(00)80123-3. [DOI] [PubMed] [Google Scholar]
- 7.Mohanty B.K., Kushner S.R. Processing of the Escherichia coli leuX tRNA transcript, encoding tRNAleu5, requires either the 3′−5′ exoribonuclease polynucleotide phosphorylase or RNase P to remove the Rho-independent transcription terminator. Nucleic Acids Res. 2010;38:597–607. doi: 10.1093/nar/gkp997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakano H., Yamada S., Ikemura T., Shimura Y., Ozeki H. Temperature sensitive mutants of Escherichia coli for tRNA synthesis. Nucleic Acids Res. 1974;1:355–371. doi: 10.1093/nar/1.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stark B.C., Kole R., Bowman E.J., Altman S. Ribonuclease P: an enzyme with an essential RNA component. Proc. Natl Acad. Sci. U.S.A. 1978;75:3717–3721. doi: 10.1073/pnas.75.8.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown S., Fournier M.J. The 4.5 S RNA gene of Escherichia coli is essential for cell growth. J. Mol. Biol. 1984;178:533–550. doi: 10.1016/0022-2836(84)90237-7. [DOI] [PubMed] [Google Scholar]
- 11.Bothwell A.L., Stark B.C., Altman S. Ribonuclease P substrate specificity: cleavage of a bacteriophage phi80-induced RNA. Proc. Natl Acad. Sci. U.S.A. 1976;73:1912–1916. doi: 10.1073/pnas.73.6.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alifano P., Rivellini F., Piscitelli C., Arraiano C.M., Bruni C.B., Carlomagno M.S. Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes Dev. 1994;8:3021–3031. doi: 10.1101/gad.8.24.3021. [DOI] [PubMed] [Google Scholar]
- 13.Li Y., Cole K., Altman S. The effect of a single, temperature-sensitive mutation on global gene expression in Escherichia coli. RNA. 2003;9:518–532. doi: 10.1261/rna.2198203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phizicky E.M., Hopper A.K. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohanty B.K., Kushner S.R. Rho-independent transcription terminators inhibit RNase P processing of the secG leuU and metT tRNA polycistronic transcripts in Escherichia coli. Nucleic Acids Res. 2008;36:364–375. doi: 10.1093/nar/gkm991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perret V., Florentz C., Giege R. Efficient aminoacylation of a yeast transfer RNAAsp transcript with a 5′ extension. FEBS Lett. 1990;270:4–8. doi: 10.1016/0014-5793(90)81221-9. [DOI] [PubMed] [Google Scholar]
- 17.Mohanty B.K., Kushner S.R. Ribonuclease P processes polycistronic tRNA transcripts in Escherichia coli independent of ribonuclease E. Nucleic Acids Res. 2007;35:7614–7625. doi: 10.1093/nar/gkm917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keseler I.M., Collado-Vides J., Santos-Zavaleta A., Peralta-Gil M., Gama-Castro S., Muniz-Rascado L., Bonavides-Martinez C., Paley S., Krummenacker M., Altman T., et al. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39:D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schedl P., Primakoff P. Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proc. Natl Acad. Sci. U.S.A. 1973;70:2091–2095. doi: 10.1073/pnas.70.7.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ono M., Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of mRNA. J. Mol. Biol. 1979;129:343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- 21.Mohanty B.K., Kushner S.R. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol. 1999;34:1094–1108. doi: 10.1046/j.1365-2958.1999.01673.x. [DOI] [PubMed] [Google Scholar]
- 22.Grana D., Gardella T., Susskind M.M. The effects of mutations in the ant promoter of phage P22 depend on context. Genetics. 1988;120:319–327. doi: 10.1093/genetics/120.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kushner S.R. In: Genetic Engineering. Boyer HW, Nicosia S, editors. Amsterdam: Elsevier/North-Holland Biomedical Press; 1978. pp. 17–23. [Google Scholar]
- 24.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohanty B.K., Giladi H., Maples V.F., Kushner S.R. Analysis of RNA decay, processing, and polyadenylation in Escherichia coli and other prokaryotes. Methods Enzymol. 2008;447:3–29. doi: 10.1016/S0076-6879(08)02201-5. [DOI] [PubMed] [Google Scholar]
- 26.Stead M.B., Agarwal A., Bowden K.D., Nasir R., Meagher R.B., Mohanty B.K., Kushner S.R. RNAsnap™ : a rapid, quantitative, and inexpensive, method for isolating total RNA from bacteria. Nucleic Acids Res. 2012;40:e156. doi: 10.1093/nar/gks680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varshney U., Lee C.P., RajBhandary U.L. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 28.Mohanty B.K., Maples V.F., Kushner S.R. Polyadenylation helps regulate functional tRNA levels in Escherichia coli. Nucleic Acids Res. 2012;40:4589–4603. doi: 10.1093/nar/gks006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohanty B.K., Kushner S.R. In vivo analysis of polyadenylation in prokaryotes. Methods Mol. Biol. 2014;1125:229–249. doi: 10.1007/978-1-62703-971-0_19. [DOI] [PubMed] [Google Scholar]
- 30.McDowall K.J., Lin-Chao S., Cohen S.N. A + U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J. Biol. Chem. 1994;269:10790–10796. [PubMed] [Google Scholar]
- 31.Kaberdin V.R. Probing the substrate specificity of Escherichia coli RNase E using a novel oligonucleotide-based assay. Nucleic Acids Res. 2003;31:4710–4716. doi: 10.1093/nar/gkg690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung D.-H., Min Z., Wang B.-C., Kushner S.R. Single amino acid changes in the predicted RNase H domain of E. coli RNase G lead to the complementation of RNase E mutants. RNA. 2010;16:1371–1385. doi: 10.1261/rna.2104810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohanty B.K., Kushner S.R. Deregulation of poly(A) polymerase I in Escherichia coli inhibits protein synthesis and leads to cell death. Nucleic Acids Res. 2013;41:1757–1766. doi: 10.1093/nar/gks1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grunberg-Manago M. In: Escherichia coli and Salmonella. Neidhardt FC, Curtiss RI, Ingraham J, Lin EC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HD, editors. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1432–1457. [Google Scholar]
- 35.Kim M.S., Park B.H., Kim S., Lee Y.J., Chung J.H., Lee Y. Complementation of the growth defect of an rnpA49 mutant of Escherichia coli by overexpression of arginine tRNA(CCG) Biochem. Mol. Biol. Int. 1998;46:1153–1160. doi: 10.1080/15216549800204712. [DOI] [PubMed] [Google Scholar]
- 36.Jain S.K., Gurevitz M., Apirion D. A small RNA that complements mutants in the RNA processing enzyme ribonuclease P. J. Mol. Biol. 1982;162:515–533. doi: 10.1016/0022-2836(82)90386-2. [DOI] [PubMed] [Google Scholar]
- 37.Altman S. Ribonuclease P: an enzyme with a catalytic RNA subunit. Adv. Enzymol. Relat. Areas Mol. Biol. 1989;62:1–36. doi: 10.1002/9780470123089.ch1. [DOI] [PubMed] [Google Scholar]
- 38.Moll I., Leitsch D., Steinhauser T., Blasi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altman S., Baer M.F., Gold H., Guerrier-Takada C., Lawrence N., Lumelsky N., Vioque A. In: Cleavage of RNA by RNase P. Inouye M, Dudock BS, editors. Orlando: Academic Press; 1987. pp. 3–15. [Google Scholar]
- 40.Hansen A., Pfeiffer T., Zuleeg T., Limmer S., Ciesiolka J., Feltens R., Hartmann R.K. Exploring the minimal substrate requirements for trans-cleavage by RNase P holoenzymes from Escherichia coli and Bacillus subtilis. Mol. Microbiol. 2001;41:131–143. doi: 10.1046/j.1365-2958.2001.02467.x. [DOI] [PubMed] [Google Scholar]
- 41.Hsieh P.K., Richards J., Liu Q., Belasco J.G. Specificity of RppH-dependent RNA degradation in Bacillus subtilis. Proc. Natl Acad. Sci. U.S.A. 2013;110:8864–8869. doi: 10.1073/pnas.1222670110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuo Y., Deutscher M.P. The physiological role of RNase T can be explained by its unusual substrate specificity. J. Biol. Chem. 2002;277:29654–29661. doi: 10.1074/jbc.M204252200. [DOI] [PubMed] [Google Scholar]
- 43.Ow M.C., Perwez T., Kushner S.R. RNase G of Escherichia coli exhibits only limited functional overlap with its essential homologue, RNase E. Mol. Microbiol. 2003;49:607–622. doi: 10.1046/j.1365-2958.2003.03587.x. [DOI] [PubMed] [Google Scholar]
- 44.Stead M.B., Marshburn S., Mohanty B.K., Mitra J.P., Ray C.L.D., Hughes T., Kushner S.R. Analysis of E. coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res. 2010;39:3188–3203. doi: 10.1093/nar/gkq1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arraiano C.M., Yancey S.D., Kushner S.R. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J. Bacteriol. 1988;170:4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perwez T., Kushner S.R. RNase Z in Escherichia coli plays a significant role in mRNA decay. Mol. Microbiol. 2006;60:723–737. doi: 10.1111/j.1365-2958.2006.05124.x. [DOI] [PubMed] [Google Scholar]
- 47.Ow M.C., Liu Q., Kushner S.R. Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNase E-based degradosome assembly. Mol. Microbiol. 2000;38:854–866. doi: 10.1046/j.1365-2958.2000.02186.x. [DOI] [PubMed] [Google Scholar]
- 48.Li Z., Deutscher M.P. The role of individual exoribonucleases in processing at the 3’ end of Escherichia coli tRNA precursors. J. Biol. Chem. 1994;269:6064–6071. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


