Abstract
Post-transcriptional regulatory mechanisms of several complex and simple retroviruses and retroelements have been elucidated, with the exception of the gammaretrovirus family. We found that, similar to the other retroviruses, gag gene expression of MuLV and XMRV depends on post-transcriptional regulation mediated via an RNA sequence overlapping the pro-pol open reading frame, termed the Post-Transcriptional Element (PTE). PTE function can be replaced by heterologous RNA export elements, e.g. CTE of simian type D retroviruses. Alternatively, Gag particle production is achieved using an RNA/codon optimized gag gene. PTE function is transferable and can replace HIV Rev-RRE-regulated expression of HIV gag. Analysis of PTE by SHAPE revealed a highly structured RNA comprising seven stem-loop structures, with the 5′ and 3′ stem-loops forming an essential bipartite signal. MuLV and XMRV PTE share 98% identity and have highly similar RNA structures, with changes mostly located to single-stranded regions. PTE identification strongly suggests that all retroviruses and retroelements share common strategies of post-transcriptional gene regulation to produce Gag. Expression depends on complex RNA structures embedded within retroviral mRNA, in coding regions or the 3′ untranslated region. These specific structures serve as recognition signals for either cellular or viral proteins.
INTRODUCTION
Post-transcriptional regulation is critical for the expression of cellular and viral mRNAs. While splicing has been associated with marking transcripts as ‘export-ready’, retroviral structural proteins are expressed from unspliced and/or partially or terminally spliced transcripts (1–3). Discovery of the Rev-responsive element (RRE) regulatory mechanism that governs HIV gag/pol and env expression pioneered our understanding of post-transcriptional regulation as an essential step for virus production (4–8). Similar mechanisms were reported to control expression of other complex retroviruses, including those of the lentivirus family [for review see (2,3)], the HTLV family (9–12), the endogenous human HTDV/HERV-K (13), mouse mammary tumor virus (MMTV) (14,15) and the betaretrovirus Jaagsiekte sheep retrovirus (JSRV) (16,17). These viruses share with HIV the feature that the full-length transcript serves not only as genomic RNA, but also as mRNA for gag-pol and as template for several spliced variants encoding env and viral regulatory factors. In addition to the RRE-like RNA export elements, retroviruses also encode post-transcriptional regulatory trans-factors that mediate export and expression of transcripts containing the export elements. Such viral factors share a leucine-rich export signal that provides the interaction site for the nuclear receptor CRM1, defining a distinct export route.
In contrast to the complex retroviruses, simple retroviruses and retroelements produce only two mRNAs, encoding gag/pol and env, respectively. Studies on the Simian retrovirus D (SRV/D) family (Mason Pfizer Monkey virus MPMV, SRV-1 and SRV-2) provided the first evidence of post-transcriptional regulation mediated by the constitutive transport element (CTE) as an essential step in virus production for simple retroviruses (18–20). Expression of Rous sarcoma virus (RSV) (21,22), intracisternal A-type particle retroelements (IAP) (23), murine MusD retroelements (24,25) and the human LINE-1 retrotransposons (26) depend on specific post-transcriptional regulatory mechanisms. Interaction of nuclear export factor 1 (NXF1) with the CTE of the SRV/D retroviruses (27,28) defined the second nuclear export pathway, which was also shown to be essential for the export of cellular RNAs (29) as well as RNAs from the simple retroviruses and retroelements. One exception to the comprehensive analysis of retroviruses and retroelements has been the gammaretrovirus family, exemplified by Moloney murine leukemia virus (MuLV). Here, we investigated the post-transcriptional mechanisms governing expression of MuLV and the closely related xenotropic murine leukemia virus-related virus (XMRV).
MATERIALS AND METHODS
Plasmids
The MuLV plasmid, PRR390, a gift from Dr Alan Rein, is derived from PRR88 and contained full-length proviral genome (Genbank accession number J02255) (30). The XMRV plasmid, pVP62, was a gift from Dr Frank Ruscetti (31,32), and contained the full-length XMRV with the revised nucleotide sequence (Genbank accession number FN692043). Deletions within the MuLV and XMRV proviral clones were obtained by restriction enzyme digestion or sequence replacements using PCR fragments. All constructs were confirmed by sequencing.
pXMRV gag has a deletion of AgeI to PmlI (nt 2431–7683), removing pro-env. pXMRVgag/pol_5869 has env deleted (SexA1, nt 5869–7683). pXMRVenv has a deletion of the HpaI fragment (nt 430–5453 spanning gag/pol). MTE (24), RTEm26-CTE (33) elements were inserted into the SbfI site in a multiple cloning site added 3′ to the XMRV gag gene, between AgeI and PmlI (nt 2431–7683).
pMuLVgag has a deletion of the AgeI-SapI fragment (nt 2445–7658, spanning pol and env); pMuLVgag/pol_5818 has a deletion of the HpaI-ClaI fragment (nt 5818–7675) spanning env; pMuLVgag/pol_4270 has a deletion of the BlpI-ClaI fragment (nt 4270–7675) spanning part of pol and env.
The HIV-1 gag reporter plasmid pNLgag (5) comprises the HIV-1 gag gene flanked by HIV-1 5′ and 3′LTRs. Putative RNA export elements were cloned into the XhoI site 3′ to gag. pNLgag-CTE contains the CTE RNA export element from SRV-1 (20). pNLgag-MTE contains the MTE RNA export element from musD retroelement (24,34). pNLgag-RTE contains the RTE RNA export element from IAP (23), using the stronger RTEm4 (35). pNLgag-RRE contains HIV-1 RRE RNA export element 3′ to the gag gene (5) and was cotransfected with the Rev expression plasmid pBsRev. pDM138 contains the cat gene embedded within HIV env (36,37) and has post-transcriptional regulatory element (PTE) inserted into the ClaI site.
pCMVgag and pCMVgagOPT (a gift from Dr Jeremy Luban) contain the wild-type and RNA-/codon-optimized XMRV gag, respectively, cloned into NheI and NotI sites of pcDNA3.1.
Cells and transfection
HeLa-derived cells (HLtat cells) (38) produce Tat protein that is essential to activate expression from the HIV LTR promoter used in the pNLgag reporter plasmid. Cells were seeded at 0.5 × 106 cells per 60 mm plate and transfected the next day with 200 ng of the NLgag plasmid or 200 ng of the cat reporter DM138 together with 50 ng of GFP plasmid pFRED143 (39) as internal control using the Superfect transfection protocol (Qiagen, Hilden, Germany). Three days later, cells were lysed in 1 ml of 1x Tris-Triton buffer and supernatants were harvested. Total Gag protein production was measured by HIV-1 p24gag ELISA (Zeptometrix, Buffalo, New York). CAT production was measured by ELISA (Roche, Penzberg, Germany) in cell extracts. GFP levels from the cell extracts were measured using the SpectraMax Gemini EM fluorimeter (MolecularDevices, LLC, Sunnyvale, CA, USA).
Expression of MuLV and XMRV plasmids was measured upon transfection of HEK293T cells, seeded at 1 × 106 cells per 60-mm plate. Cells were transfected the next day via calcium chloride coprecipitation and the medium was replaced 24 h later. Cells were transfected with 3 μg DNA, except for the RNA-/codon-optimized gag plasmid, which was transfected at 100 ng per plate and 50 ng of pFRED143 was cotransfected as a transfection control. Three days later, supernatants from two plates were pooled, and the particles were pelleted by centrifugation at 25 000 rpm for 1.5 h at 4°C. The supernatant was removed and the viral particle pellet was resuspended in 100 μl 2x Laemmli sample buffer supplemented with β-mercaptoethanol (Bio-Rad, Inc., CA, USA). Cells were lysed in 1 ml of hypertonic N1 lysis buffer (20 mM HEPES pH7.9, 10% glycerol, 1 mM MgCl2, 400 mM NaCl, 0.5 mM DTT, 0.5% Triton X-100), sonicated briefly for 2 × 6 s and centrifuged at 14 000 rpm for 15 min at 4°C.
Immunological analysis
Twenty-five microliters of the supernatants and 3 μl of the cell extracts (equivalent fractions of total volume) were added to 5 μl of 5x loading dye, brought up to 30 μl with PBS, boiled for 7 min and chilled. For viral particle preparations, 30 μl of the resuspended pellets were boiled and chilled prior to loading on the gel. Proteins were separated on 10% Tris-Glycine polyacrylamide gels (Bio-Rad, CA, USA), transferred overnight at 30 mA to nitrocellulose membranes (Bio-Rad, CA, USA). Membranes were incubated in 5% Blotting Grade Blocker (Bio-Rad, CA, USA). Gammaretroviral proteins were detected by anti-MLV gp30 Gag rabbit serum (a gift from Dr Alan Rein) and anti-RMLV Env gp70 rabbit serum (a gift from Dr Sandra Ruscetti). Proteins were visualized using ECL Prime (GE Life Sciences, PA, USA). Western immunoblot images were captured on a ChemiDoc XRS + imager and analyzed with Image Lab software (Bio-Rad, CA, USA).
Northern blot
Total RNA was isolated using Trizol Reagent (Life Technologies, CA, USA). Cytoplasmic and nuclear RNA fractions were prepared using Ambion PARIS kit (Ambion, Austin, TX, USA), followed by elimination of ribosomal RNA using the RiboMinus kit (Life Technologies, CA, USA). RNA samples were separated on a 1% formaldehyde agarose gel, transferred to nitrocellulose membrane and hybridized with random-labeled probes to detect XMRV gag and HIV gag, or a radiolabeled PCR probe to detect the gag genes expressed from pcDNA3.1 using the shared 3′ untranslated region (UTR) spanning the bovine growth hormone (BGH) pA region. As fractionation controls, the cytoplasmic GAPDH and nuclear lncRNA MALAT1 were probed. Blots were incubated overnight at 42°C, washed twice for 5 min with 2x SSC, 0.1% SDS, then twice for 15 min with 0.1x SSC, 0.1% SDS. The signal was captured by PhosphoImager Storm 860 system and analyzed by ImageQuant software (GE Life Sciences, PA, USA).
In vitro structural RNA analyses
For RNA modification, 2 pmol of RNA were refolded in 100 μl of buffer containing 10 mM Tris-HCl, pH 8.0, 100 mM KCl and 0.1 mM EDTA by heating 3 min at 95°C, placing on ice, adding 50 μl of 3x folding buffer (120 mM Tris-HCl, pH 8.0, 600 mM KCl, 1.5 mM EDTA, 15 mM MgCl2), and incubating for 30 min at 37°C. Folded RNA was divided equally into two tubes and treated with either 1M7 (in DMSO) or DMSO alone (8 μl) and allowed to react for 5 min at 37°C. For Pb2+ probing, Pb(OAc)2 to a final concentration of 0.05 mM or water were added, followed by incubation for 10 min at room temperature. RNA was recovered by ethanol precipitation and resuspended in 10 μl of 5 mM Tris-HCl, pH 7.0, 0.5 mM EDTA.
Detection of sites of cleavage or modification and data processing
Fluorescently labeled primer (1 μl) was added to 0.5 pmol of RNA [Cy5 (+) and Cy5.5 (−); 6 and 4 μM, respectively], and 12 μl of primer-template solutions were incubated at 95°C for 3 min, 37°C for 10 min and 55°C for 2 min in a buffer containing 50 mM Tris-HCl, pH 8.3, 75 mM KCl, 5 mM DTT and 3 mM MgCl2. RNA was reverse transcribed at 50°C for 45 min (Invitrogen Superscript III, Life Technologies, CA, USA). Sequencing ladders were prepared using primers labeled with WellRed D2 and LicorIR-800 and a Thermo Sequenase Cycle Sequencing kit (Affymetrix, CA, USA) according to the manufacturer's protocol. Samples and sequencing ladders were ethanol-precipitated, washed twice with 70% ethanol, dried and resuspended in deionized formamide. Primer extension products were analyzed on a Beckman CEQ8000 Genetic Analysis System (Beckman-Coulter, CA, USA) and electropherograms were processed using SHAPEfinder software (40).
Reactivities were normalized as described (40). Reactivity information was obtained from five overlapping reads (for 1468 nt RNA) of ∼300 nt each and at least three repetitions were obtained for each read. The resulting data were introduced to the RNAstructure 4.6 software as pseudo-energy constraints with default slope and intercept values (41). Different values of the pairing distance were tested during structure development but maximum pairing distance was limited to 600 for the final structure. Tertiary interactions were introduced manually. Secondary structures were visualized using PseudoViewer web application (http://pseudoviewer.inha.ac.kr/) and modified using Adobe Illustrator CS5.1.
RESULTS
A novel element within MuLV and XMRV pro-pol is required for Gag expression
A series of deletions within both the MuLV and XMRV retroviral genomes was constructed to assess their effect on gag gene expression. Mutant clones contained gag/pol or only gag (Figure 1A). HEK293T cells were transfected with the wild-type proviral clones and deletion mutants derived from MuLV (Figure 1B, lanes 1–4) and XMRV (Figure 1C, lanes 1–3). Gag particle production was assessed in supernatants by immunological analysis.
Figure 1.
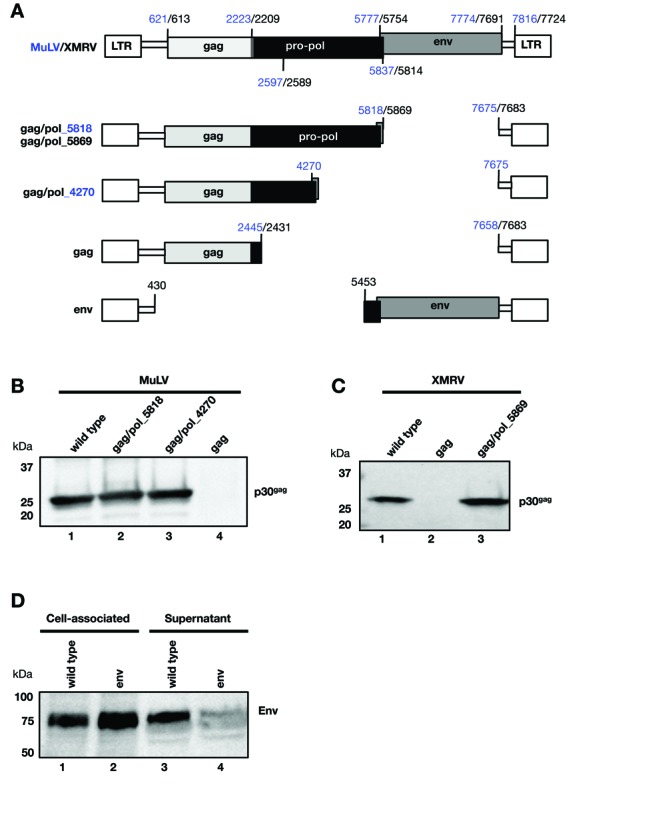
pro-pol is required for Gag particle production. (A) Schematic representation of the MuLV and XMRV genomic structure and the internal deletion mutants are shown. The numbering of the nucleotides follows Genbank accession number J02255 for MuLV (indicated in blue) and Genbank accession number FN692043 for XMRV (indicated in black). (B, C) Western immunoblot analysis of Gag particles produced from transfected HEK293T and detected with anti-MLV-gp30 serum. (B) MuLV particle production was analyzed from the wild-type proviral clone (lane 1), gag/pol_5818 (lane 2), gag/pol_4270 (lane 3) and the empty gag plasmid (lanes 4). (C) XMRV particle production was analyzed from the wild-type proviral clone (lane 1) and plasmids containing only gag (lane 2) and gag/pol_5869 (lane 3). (D) Expression of Env was monitored from the wild-type and the env plasmid upon transfection of HEK293T cells and analyzed by western immunoblot assay. The membrane containing proteins from the cell-associated fractions and the supernatants was probed with anti-RMLV-gp70 antiserum detecting Env (gPr80).
While the wild-type MuLV proviral clone produced Gag particles as expected (Figure 1B, lane 1), no particles were produced from the plasmid lacking the pro-pol and env genes and containing only gag (lane 4). Analysis of the cell-associated fraction confirmed the lack of Gag expression (data not shown), thus excluding a defect in particle release. Plasmids that lacked env and contained the complete gag/pol open reading frame, ending at nt 5818 (lane 2), or a shorter gag/pol sequence ending in RT at nt 4270 (lane 3), produced similar levels of Gag particles as the intact proviral clone (lane 1). Similar data were obtained for XMRV (Figure 1C), i.e. the proviral clone (lane 1) and the gag/pol plasmid (lane 3) produced viral particles, but not the plasmid containing only gag (lane 2). These data collectively suggest that MuLV and XMRV Gag production is severely impaired in the absence of the ∼2 kb region overlapping prt and a portion of pol. This finding is reminiscent of previous reports on the regulated gag expression of complex and simple retroviruses and retroelements (for review see (1–3)). Since both MuLV and XMRV gag gene expression depend on their respective pro/pol and since their sequences share a high degree of similarity, we used both sequences throughout this work. Finally, as Env is produced from a spliced mRNA, we investigated its expression from a plasmid with a deletion of the gag-pro-pol region (Figure 1A). Env was efficiently expressed from this plasmid (Figure 1D, lanes 2 and 4) at levels comparable to the wild-type proviral clone (lanes 1 and 3). In the absence of Gag, Env was mainly found in the cell-associated fraction (lane 2). Together, these experiments demonstrate that only gag gene expression is impaired in the absence of the pro-pol sequence.
gag gene expression depends on post-transcriptional regulation
The dependence of gag RNA on post-transcriptional regulation was interrogated by (i) adding a cis-acting RNA export element (18–20,23,24,33,34) and (ii) altering the nucleotide sequence of the gag gene to remove any cis-acting negative-acting signals (42–45).
We first examined whether gag expression could be achieved in the presence of heterologous cis-acting RNA regulatory elements (Figure 2A–C). We inserted either the MTE RNA export element of the MusD retroelement (24,34) or the RTE-CTE RNA export combination (23,33), between the 3′ terminus of the XMRV gag gene and the 3′LTR (Figure 2A). These elements represent previously reported RNA export elements required for the export of their respective natural RNA targets (23,24,33,34). Specifically, MTE from the MusD retroelement was shown to function similarly to the MPMV CTE (34), while RTE-CTE (a combination of the RTE element from intracisternal A type particles and the CTE from Simian type D retrovirus 1) was shown to provide the strongest export function of all the elements tested (33) (GRP, personal communication). Gag particle production from transfected cells was monitored immunologically (Figure 2B). Supplying MTE (lane 4) or RTE-CTE (lane 6) in cis efficiently promoted Gag expression, whereas no particles were produced from the empty gag plasmid (lanes 3 and 5, respectively). The level of particle production in the presence of the cis-acting MTE element was similar to that obtained from the wild-type proviral clone (lane 1) or a plasmid that contains the complete gag/pol region (lane 2). Note that Gag protein is produced in its unprocessed form (Pr65gag) from plasmids that contain MTE (lane 4) or RTE-CTE (lane 6), since these vectors lack the viral protease. In contrast, Gag produced from the wild-type proviral clone and the plasmid encoding gag/pol is the processed (p30gag) form (lanes 1 and 2). We analyzed total RNA produced from these plasmids and hybridized northern blots with a probe spanning the gag gene (Figure 2C). Full-length gag containing mRNA was efficiently produced from the wild-type proviral clone (lane 1) and the gag/pol plasmid (lane 3). We noted that the empty gag plasmid also produced significant RNA levels, despite the lack of Gag protein production (lane 2). In the presence of MTE (lane 4), higher gag mRNA levels accumulated. Northern blots were re-probed with GAPDH to assure equal extraction and loading of the RNA samples. Thus, we conclude that poor protein production from the gag plasmid reflected post-transcriptional restriction of this mRNA, which could be alleviated in the presence of cis-acting heterologous RNA export elements (MTE, RTE-CTE) resulting in efficient particle production. This finding suggested the MuLV and XMRV pro-pol regions play an analogous role in promoting particle production.
Figure 2.
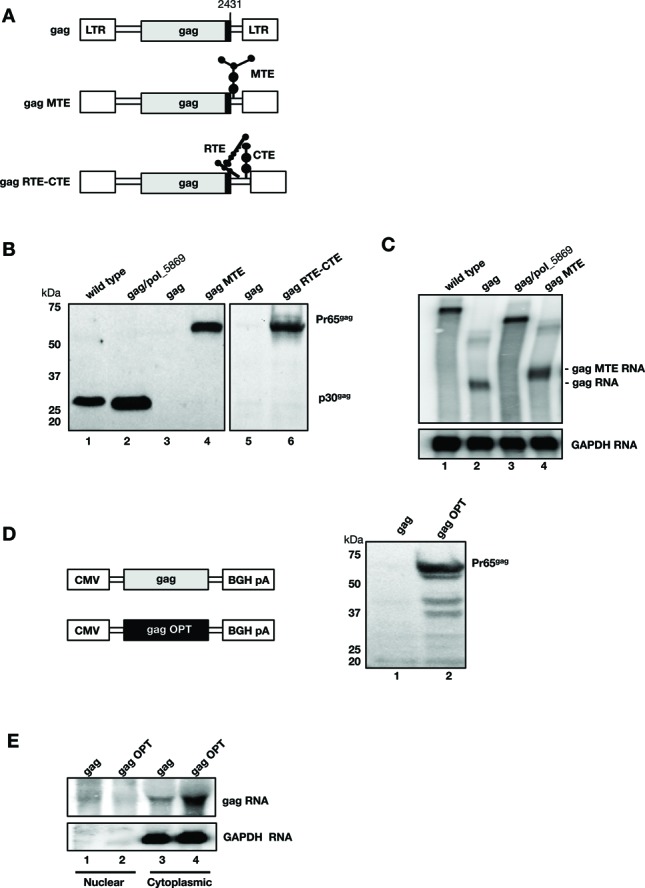
gag expression is controlled at the post-transcriptional level. (A) Schematic representation of the XMRV gag plasmid upon insertion of heterologous RNA export MTE from the musD retrovirus or RTE-CTE element combination. (B) Gag particle production was monitored from the supernatants of HEK293T cells transfected with wild-type clone (lane 1), gag/pol_5869 (lane 2), the empty gag plasmid (lane 3 and 5), the gag plasmid that contains MTE (lane 4) or RTE-CTE (lane 6). The data are from two independent experiments (lanes 1–4 and lanes 5–6, respectively). The blots were probed with anti-MLVgp30 antibody serum, detecting the processed p30gag and the unprocessed Pr65gag proteins. (C) Northern blot containing total RNA from cells transfected with the XMRV-derived plasmids as described in panel B, was probed for gag and GAPDH RNA as a loading control. (D, E) Wild-type and RNA-/codon-optimized gag genes from XMRV were expressed in transfected HEK293T cells from the CMV promoter using plasmid pcDNA3.1. (D) Western immunoblot analysis of viral particles was performed as described for panel B. (E) Northern blot analysis of fractionated RNA from HEK293T cells transfected with plasmids expressing the wild-type and RNA-/codon-optimized gag genes. Nuclear and cytoplasmic gag RNA was probed for the shared 3′UTR region, and GAPDH RNA was used as a marker of cytoplasmic RNA.
We next used a complementary method to understand restriction of gag gene expression, comparing the wild-type XMRV gag gene to that of a RNA-/codon-optimized gene that had 25% of the 1608 nt changed without altering coding potential (Figure 2D, E). Both genes were expressed from the human cytomegalovirus (CMV) promoter and contained the BGH polyA signal. Western blot data revealed that only the expression-optimized gag gene was efficiently produced (Figure 2D, lane 2). The wild-type gag gene failed to produce Gag, even when expressed from the strong CMV promoter (lane 1). We fractionated the RNA produced by these two constructs to determine their subcellular localization (Figure 2E). The transcript containing the optimized gag gene was found to abundantly accumulate in the cytoplasm (lane 4), since the RNA/codon optimization process removes any post-transcriptional inhibitory elements. In contrast, only low cytoplasmic levels of the non-optimized wild-type gag RNA were found (lane 3). Neither of the transcripts accumulated in the nucleus (lanes 1 and 2). Thus, by analogy to HIV-1, altering the nucleotide sequence of the poorly expressed gag gene was sufficient to achieve high levels of expression independent of the viral post-transcriptional regulation (42–45). Therefore, gammaretroviruses and lentiviruses share the feature of poor expression of their gag genes in the absence of a post-transcriptional regulatory mechanism.
Transferring the post-transcriptional RNA regulatory function promotes HIV gag gene expression
We next asked whether the gammaretroviral post-transcriptional regulatory function of the pro-pol region is transferable and can mediate expression from heterologous, poorly expressed genes known to depend on post-transcriptional regulation. Figure 3A illustrates the assays employed, namely, (i) the pNLgag reporter containing the HIV gag gene (5,8) and (ii) pDM138 reporter containing the cat gene (36,37). The gag and cat reporter genes are embedded within intronic HIV sequences flanked by splice sites, and their expression depends on exporting the unspliced transcript under post-transcriptional control (i.e. HIV Rev-RRE; CTE). Expression from the cotransfected GFP plasmid was monitored to control for transfection efficiency.
Figure 3.
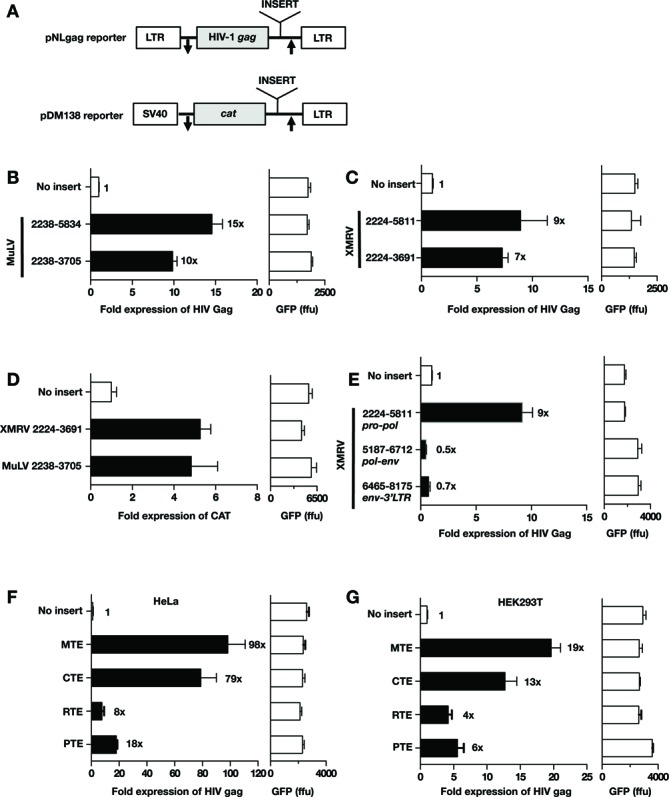
pro-pol promotes export-dependent reporter gene expression. (A) Schematic representation of the HIV pNLgag and the pDM138 reporter plasmids. pNLgag contains the HIV gag gene located between the HIV LTRs. pDM138 contains the cat gene embedded within HIV env sequence and is expressed from the SV40 promoter. The gag and cat genes are flanked by splice sites, indicated by arrows. The expression of gag and cat gene from the unspliced RNA depends on post-transcriptional regulation. Putative RNA export elements were inserted 3′ to gag and cat genes, respectively. (B and C) The pNLgag reporter was used to test the ability of the pro-pol sequences from MuLV (B) and XMRV (C) to promote HIV-1 gag RNA export and expression. Total Gag production was measured upon transfection of HLtat cells. Gag expression from the empty pNLgag reporter was normalized to 1. Raw GFP values are given. (D) The DM138 cat reporter was used to test the ability of the pro-pol sequences from MuLV (2238–3705) and XMRV (nt 2224–3691) to promote cat RNA export and expression. Raw GFP values are given. (E) Testing of the 3′ portion of XMRV for export function. The indicated sequences were inserted into pNLgag reporter and assessed for their ability to promote Gag production as described in panel B. (F, G) Comparison of PTE activity to that of other known RNA export elements. The indicated RNA sequences were cloned into the HIV gag reporter and expression was tested in the HeLa-derived HLtat (F) and HEK293T (G) cells.
Using the pNLgag reporter, we tested whether pro-pol from MuLV (Figure 3B) and XMRV (Figure 3C) could replace the HIV post-transcriptional control. pro-pol sequences were inserted 3′ to the HIV gag gene (Figure 3A). Total Gag production from transfected HLtat cells was quantified by HIV p24gag ELISA. Expression of gag from the empty plasmid was normalized to 1, and expression in the presence of the different pro-pol sequences was assessed. MuLV sequences spanning the complete pro-pol (nt 2238–5834) or the 3′ deletion mutant (nt 2238–3705) activated HIV gag expression (Figure 3B). Similarly, pro-pol sequences from XMRV (nt 2224–5811 and nt 2224–3691, respectively) augmented HIV gag expression (Figure 3C). In both cases, the shorter ∼1.4 kb regions provided ∼70–80% of function and subsequent studies focused on these regions.
Next, we employed the DM138 cat reporter to evaluate post-transcriptional function of pro/pol in an independent reporter RNA. The ∼1.4 kb pro/pol region of MuLV and XMRV was inserted 3′ to the cat gene (Figure 3A). CAT production from transfected HLtat cells was quantified using a CAT ELISA assay. Expression of the cat gene from the empty plasmid was normalized to 1, and expression in the presence of the different pro-pol sequences was assessed. Both MuLV and XMRV sequences augmented CAT production (Figure 3D), demonstrating that they share, with previously identified retroviral elements (Rev-RRE and CTE), the ability to promote post-transcriptional regulation. We therefore designated this gammaretroviral region the PTE. Together, these data showed that the post-transcriptional regulatory function of MuLV and XMRV pro/pol is transferable and that this activity is independent of other viral sequences (5′ or 3′ LTR or gag).
To investigate whether additional sequences outside of pro-pol could mediate post-transcriptional function, we tested different overlapping regions from the 3′ portion of the provirus. With the exception of pro-pol, no other region [spanning 3′ end of pol and env region (nt 5187–6712); env and 3′ LTR (nt 6465–8175)] promoted HIV Gag production (Figure 3E). These data demonstrate that the XMRV and MuLV PTE is contained within ∼1.4 kb of pro-pol. Interestingly, this regulatory element is not located in the untranslated region between env and the 3′LTR, as found in other retroviruses and retroelements, but overlaps a critical viral RNA sequence encoding protease and part of RT.
We further compared PTE activity to that of several previously reported RNA export elements including CTE of SRV-1, MTE of musD retroelement and RTE of an intracisternal A particle retroelement (Figure 3F and G). Comparison of PTE activity was performed in HeLa-derived HLtat cells (Figure 3F) and HEK293T cells (Figure 3G). These data showed that CTE and MTE have comparable activities, whereas PTE activity is ∼4-fold lower in HLtat cells (Figure 3F) and ∼2-fold lower in HEK293T cells (Figure 3G) and is comparable to that of RTE. In summary, these data showed that PTE function falls within the range of other previously reported retroviral RNA export elements.
SHAPE analysis of the PTE reveals a highly complex RNA structure
To derive the structure of the PTE RNA, we first employed selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE, 1M7) (46). Using the ∼3.6 kb XMRV RNA encompassing nt 2224–5811 as a template, modification with 1M7 showed good reactivity toward all four RNA nucleotides, but with slightly lower reactivity toward C residues, which could complicate experimentally supported structure predictions (47). Thus, to complement SHAPE, we performed Pb2+ probing (48). Pb2+ cleavage causes a transesterification reaction that breaks the 5′, 3′-phosphodiester backbone of RNA at single-stranded nucleotides. These complementary methods allowed us to develop a structural model of the 1468 nt 5′- terminal fragment of XMRV pro-pol RNA spanning the PTE (nt 2224–3691) (Figure 4) which was interrogated as the isolated ∼1.4 kb RNA as well as within the ∼3.6 kb RNA. The Supplementary Table S1 shows the normalized SHAPE reactivities for each nucleotide and the PTE structure (Figure 4) indicates the reactivities using a color code.
Figure 4.
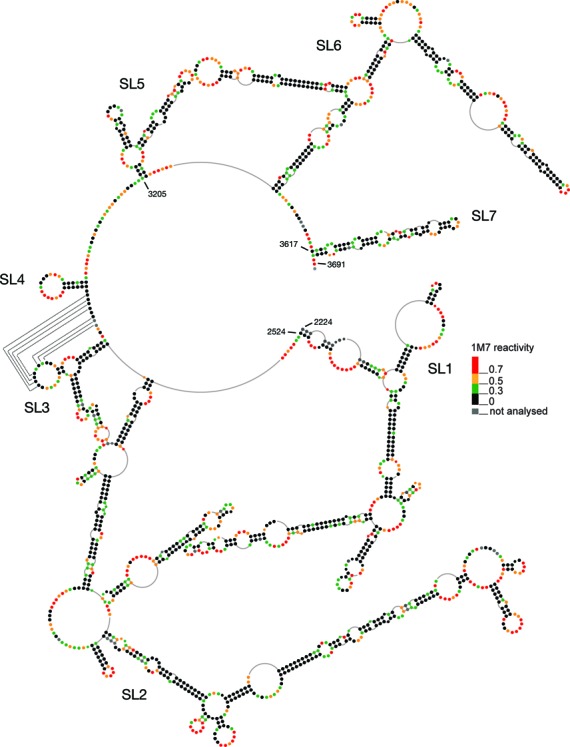
SHAPE analysis of the PTE RNA reveals a highly complex structure. Secondary structure model of the XMRV PTE (nt 2224–3691) based on SHAPE-constrained folding. The stem-loops SL1 through SL7 are indicated. The nucleotides are represented as colored dots, with the respective color of each dot indicating the 1M7 reactivities according to the accompanying scale.
The +2224–3691 RNA folded into a highly structured molecule comprising domains with substantial base-paired secondary structure, which we designate stem-loop (SL) 1 to 7. Reactive nucleotides were mostly located in loops (terminal, internal or junctions) indicating that the model is in good agreement with experimental data. In some cases, reactive residues were predicted to be base-paired, but are located close to single stranded regions, bulges or involve G-U wobble pairing. Such structural elements could cause flexibility in a helical segment (41). In contrast, large loops and junctions are often stabilized by non-canonical base pairs or stacking interactions which could explain low flexibility for nucleotides predicted by RNAstructure to be single stranded. The +2224–3691 RNA showed a median reactivity of 0.24, indicative of a highly structured molecule. It has been suggested that highly structured regions within viral RNAs are associated with regulatory functions. In HIV and SIV, such regions are characterized by median SHAPE reactivities below 0.4 (49), thus the PTE fulfills this criterion.
To confirm correct folding, the +2224–3691 RNA was also probed in the context of the longer, ∼3.6 Kb XMRV pro-pol RNA (nt 2224–5811), to provide a more ‘natural’ structural context (Supplementary Figure S1). SHAPE signatures of the longer and shorter pro-pol-containing RNAs were similar, supporting our contention that +2224–3691 RNA comprised structural elements which were found to be functionally relevant.
Predicted MuLV PTE RNA structure
Comparing the XMRV and MuLV pro-pol sequences showed that despite sequence divergence within the 3′ end of their pol genes, the 5′ ∼1500 nt show little variability, with only 32 nt differences among XMRV (+2224–3691) and the MuLV (+2238–3705) sequences. We therefore used homology modeling to investigate the structure of the MuLV RNA. We noted that nucleotide differences (marked in blue in Supplementary Figure S2) mapped mostly to single-stranded regions bordering bulges, or changed G:C base pairs to G:U, indicating an invariant RNA structure in these regions. Thus, our proposed structure for the MuLV PTE differed minimally from the experimentally supported model of the XMRV counterpart.
SL1 and SL7, but not SL2, are necessary for PTE function
SHAPE analysis suggested an RNA structure, which, although highly complex, could be subdivided into seven distinct stem-loop domains (SL1–7, Figure 4). To define the minimal PTE, we introduced deletions into the 5′ and 3′ stem-loop regions and the internal SL2 and compared their activity in the HIV gag reporter to that of the full-length PTE (nt 2224–3691) (Figure 5A).
Figure 5.
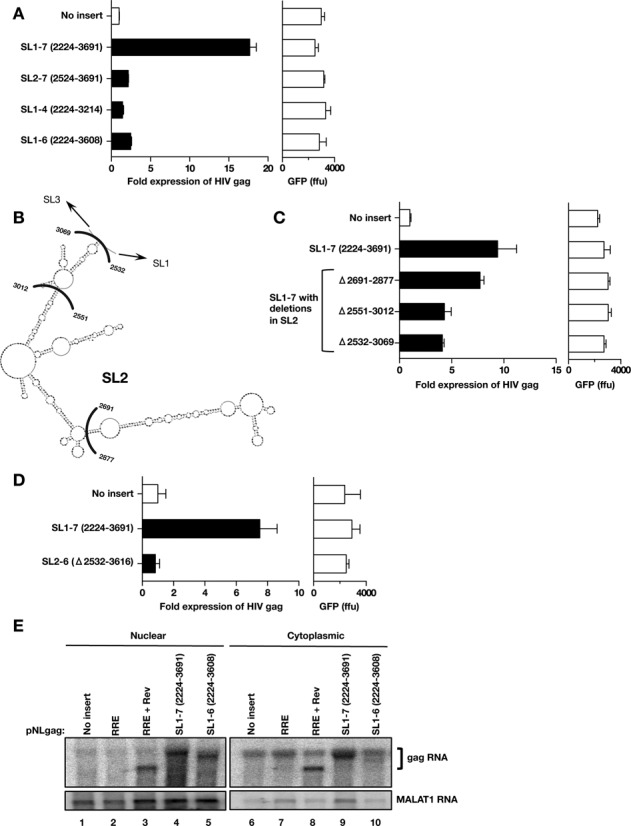
Role of SL1 and SL7 for PTE function. (A) Functional test of PTE mutants with 5′ and 3′ deletion. The following mutants were inserted into pNLgag reporter: SL1–7 (nt 2224–3691), SL1–6 (nt 2224–3608), SL2–7 (nt 2524–3691) and SL1–4 (nt 2224–3214) and Gag production was measured as described in Figure 3. Expression of the cotransfected GFP plasmid was measured. (B) The SL2 domain of the XMRV PTE (from Figure 4) is shown. The indicated nt marks the end points of the internal deletions analyzed in panel C. (C) The wild-type XMRV PTE SL1–7 (nt 2224–3691), and the indicated deletions within SL2 (nt 2691–2877, nt 2551–3012 and nt 2532–3069) were cloned into the pNLgag plasmid 3′ to HIV-1 gag. Gag and GFP production was assessed from transfected HLtat cells as described in Figure 3. Gag expression from the empty pNLgag reporter was normalized to 1 and the fold expression of the plasmids is shown. Raw GFP values are given. (D) The wild-type XMRV PTE SL1–7 (nt 2224–3691), and the PTE mutant deleted for SL2 to SL6 (nt 2532–3616) were cloned into the pNLgag plasmid 3′ to HIV-1 gag. Gag and GFP production was assessed from transfected HLtat cells as described in Figure 3. Gag expression from the empty pNLgag reporter was normalized to 1 and the fold expression of the plasmids is shown. Raw GFP values are given. (E) Northern blot analysis of fractionated RNA from HLtat cells transfected with the pNLgag reporter (lanes 1 and 6) and reporter plasmid containing the XMRV PTE elements SL1–7 (lanes 4 and 9) and SL1–6 (lanes 5 and 10), respectively. The HIV gag-RRE plasmid was transfected in the absence or presence of the Rev expression plasmid (lanes 2, 3 and lanes 7, 8). HLtat cells were transfected with the indicated plasmids, and nuclear and cytoplasmic RNA was probed for HIV-1 gag. The blots were probed for lncRNA MALAT1 as a marker of nuclear RNA.
Loss of PTE function with mutant SL2–7 (nt 2524–3691) demonstrated that SL1 was essential for function. Deleting SL5 to SL7 (mutant SL1–4, nt 2224–3214) or only SL7 (mutant SL1–6, nt 2224–3608) significantly reduced PTE function, supporting a requirement for SL7. Together these data indicated that PTE function derives from SL1 and SL7.
To address a concern that deletions may affect the RNA structure and, consequently impair their function, SHAPE profiles of deletion mutants SL1–4 (nt 2224–3219) (Supplementary Figure S3A) and SL5–7 (nt 3219–3691) (Supplementary Figure S3B) were compared to that of the full-length PTE. RNAs with these 5′ and 3′ deletions showed modification patterns similar to the longer transcript (nt 2224–3691). These data indicated that the 1468 nt (SL1-SL7) RNA structure was stable, and that individual domains folded correctly even when 5′ (SL5–7) or 3′ sequences (SL1–4) were removed. In addition, these data showed that domains of RNAs spanning SL1–4 and SL5–7 folded independently into the structures depicted in Figure 4.
To address the role of SL2, several structure-based internal deletions were constructed in the context of the full-length PTE (nt 2224–3691 Figure 5B). Deleting the tip of SL2 (nt 2691–2877) did not appreciably affect function (Figure 5C), while removing most of SL2 (nt 2551–3012) or the complete SL2 (nt 2532–3069) resulted in ∼50% loss of function. Thus, our data indicate that SL2, in contrast to SL1 and SL7, was not absolutely essential for PTE function. Furthermore, three MuLV genomic clones have been reported that lack active protease but were able to produce immature Gag particles. These mutants contain deletions affecting the 3′ end of SL1 and 5′ end of SL2 including deletion of nt 2421–2546 (50), nt 2519–2567 and nt 2487–2582 (51). Since we showed that SL2 is not essential for PTE function, we tested the sequence of one of the prt− mutants that affected most of SL1. We tested the internal deletion of nt 2421–2546 (50) in the pNLgag reporter assay and found only a 25% loss of activity (data not shown). RNA structure prediction indicated that despite this 183 nt deletion, the 5′- terminal hairpin is preserved (data not shown). Thus, these data support the finding that this prt− mutant is indeed active and can produce particles.
Since our data indicated that the SL1 and SL7 regions are necessary for PTE function, we tested a construct containing solely these regions cloned into the HIV-1 gag reporter (SL2–6 Δ2532–3616; Figure 5D). This minimal PTE was unable to promote Gag expression when compared to the full-length PTE [SL1–7 (2224–3691)], suggesting that while SL1 and SL7 are necessary for expression within the full-length PTE, on their own they are insufficient to provide this function and require the intervening sequence of the PTE for full activity. We cannot rule out a role of the intervening sequence in the proper positioning, i.e. spacing, of SL1 and SL7. Together, these data suggested that a bipartite signal requiring, at least the 5′ 198 nt of SL1 and the 83 nt of SL7, is essential for PTE function.
To further elucidate the PTE mechanism, we analyzed the HIV-1 gag reporter RNA in the absence or presence of wild-type PTE (nt 2224–3691) and an inactive mutant PTE lacking SL7 (nt 2224–3608) (Figure 5E). As a control for the export of the reporter RNA, the pNLgag reporter containing the HIV RRE, was included. As expected, the gag reporter RNA with no insert (lanes 1 and 6) and with RRE in cis (lanes 2 and 7) showed very poor expression. Upon cotransfection of HIV Rev (lanes 3 and 8), elevated levels of the gag-RRE transcript were found both in nuclear and cytoplasmic fraction, resulting in ∼90x increase in HIV Gag production. gag RNA containing the full-length PTE SL1–7 (nt 2224–3691) in cis showed an increase in nuclear RNA (lane 4) and a robust cytoplasmic accumulation (lane 9), in agreement with its ability to promote Gag protein production. The Gag levels were ∼4x lower than those observed by the gag-RRE RNA in the presence of Rev. In contrast, insertion of the inactive PTE SL1–6 (nt 2224–3608) showed a nuclear signal (lane 5), but very poor cytoplasmic accumulation (lane 10), in agreement with the lack of Gag protein production. We showed that PTE function can be replaced by bona fide RNA export elements (MTE, RTE-CTE; Figure 2B) and that PTE in cis functions analogous to other heterologous RNA export elements (CTE, MTE, RTE) in different reporter systems (Figure 3). Together, these data demonstrate that PTE provides an essential role in the export and expression of MuLV/XMRV gag RNA.
DISCUSSION
In this report, we have identified and characterized a novel cis-acting RNA post-transcriptional regulation element, designated PTE, overlapping pro and the 5′ part of pol in MuLV and XMRV. Interestingly, while XMRV and MuLV are 81% identical, their PTE regions show 98% identity, suggesting that this critical region has diverged very little between the two gammaretroviruses.
We demonstrated that the MuLV and XMRV PTE function post-transcriptionally, since (i) no Gag was produced from a plasmid that only contained the gag gene flanked by the 5′ and 3′ LTRs or from a plasmid that expressed gag from the CMV promoter (ii) use of an RNA-/codon-optimized gene, which removed the embedded inhibitory signals without altering the coding potential resulted in robust Gag particle production; (iii) the expression defect of the gag RNA could be overcome by the presence of the cis-acting PTE or heterologous elements (i.e. RTE-CTE and MTE) and (iv) PTE promoted production of the heterologous HIV-1 gag and cat genes from the pNLgag and DM138 reporters, both of which depend on post-transcriptional regulation. These data support the conclusion that gag expression depended on post-transcriptional regulation and that PTE provides a critical function to overcome such restriction of the transcript.
A recent paper by Volkova et al. (52) reported that the U3 region of MuLV contains a cis-acting sequence involved in 3′ processing and the nuclear export of a hybrid reporter transcript comprising the 5′UTR and extended packaging signal of MuLV, neo/GFP reporter genes and the MuLV 3′UTR. The U3 region was shown to exert its function in conjunction with the 5′UTR including the packaging signal but played no role in the expression of internally initiated transcripts. In contrast to the reporter RNAs tested by Volkova et al. (52), the U3 region was unable to promote export and expression of the MuLV/XMRV gag transcript comprising the 5′ and 3′ UTR including the packaging signal and gag ORF (Figures 1 and 2), and the U3 region was also unable to export the unspliced HIV gag transcript containing the MuLV env-3′LTR (Figure 3E). These data indicate that the U3 region and the PTE exert distinct functions and only the presence of the PTE in cis is able to potently augment MuLV/XMRV Gag expression.
Others reported a contribution of the MuLV 5′UTR including R and the extended packaging region on the expression of different reporter plasmids (53–57). Analogous regions from spleen necrosis virus and MPMV were also reported to facilitate export and translation (58–61). Although a sequence spanning R and the extended packaging signal of several viruses showed some benefit on expression of reporter genes, our data demonstrate that this sequence is not sufficient to mediate MuLV/XMRV gag expression. We found that the 5′UTR region was unable to promote MuLV/XMRV gag expression when tested from an RNA that contains an intact 5′UTR and gag ORF (Figure 1), a transcript that is more closely related to the viral unspliced RNA than a heterologous reporter, and solely depends on the presence of the prt region containing PTE in cis for expression. By analogy to MuLV, we and others previously demonstrated that MPMV expression, despite the reported 5′UTR function in promoting expression (60), depends on the presence of CTE that is essential for export and expression of gag/pol RNA and virus production (18,19). Similarly, deletion of RTE rendered the IAP element retrotransposition incompetent (62). Thus, these viruses depend on distinct sequences (PTE, CTE, RTE) that are essential for Gag production.
In addition to a role for the extended packaging sequence, other regions within MuLV gag and/or pol that are distinct from the PTE described herein were previously reported to play a role at post-transcriptional control, in particular the production of the spliced transcript (53,63). An effect on splicing was also reported for the intronic gag/pol sequences of reticuloendotheliosis virus (64). Thus, distinct signals within an MuLV RNA contribute to the control of virus expression at different steps including 5′UTR with the packaging signal, PTE that overlaps the prt region and promotes export and expression of the unspliced RNA, regions in gag and/or pol that control splicing, splice sites and the 3′UTR which controls 3′ processing.
The dependence of Gag production on post-transcriptional regulation is shared among the many retroviruses [reviewed in (1–3)] including simple retroviruses such as SRV/D family (MPMV and SRV), IAP, murine MusD retroelements and Rous sarcoma virus. This can be extended to complex retroviruses including lentiviruses with HIV as prototype, the HTLV family, the endogenous human HTDV/HERV-K; the mouse MMTV and the betaretrovirus JSRV (4,7,9,10,13–26,65). Like MuLV and XMRV, these viruses contain distinct RNA export signals embedded within complex and essential secondary and tertiary structure. Except for the CTE, the prototypic element identified in SRV/D retroviruses that folds into a single stem-loop structure, all the other elements span up to 500 nt and are composed of complex stem-loop structures. Complex interactions among stem-loops were demonstrated for MTE (24) and RRE (66). Thus, the complex stem-loop structure of the PTE is not unique in this context. Although the total length of PTE (∼1.4 kb) is longer than that of other elements, its function is mediated via a bipartite signal (SL1 and SL7) and the internal domains are less critical for function, though they might serve to place SL1 and SL7 in the correct orientation for function.
Post-transcriptional regulatory elements have typically been found in the 3′UTR of the genomes of simple retroviruses and retroelements; some of which are shown in Figure 6. An in-depth comparison of retroviral elements suggests that the localization of the PTE, overlapping pro and 5′ part of pol, is unique. Similar to retroviruses, pararetroviruses from the Hepadnaviridae, such as human hepatitis B virus and woodchuck hepatitis virus, have also evolved post-transcriptional regulation elements to export their mRNAs (36,37,67). Interestingly, these elements are typically bi- or tripartite stem-loop structures, which work in tandem and are required for the cytoplasmic accumulation and expression of a viral RNA (36).
Figure 6.
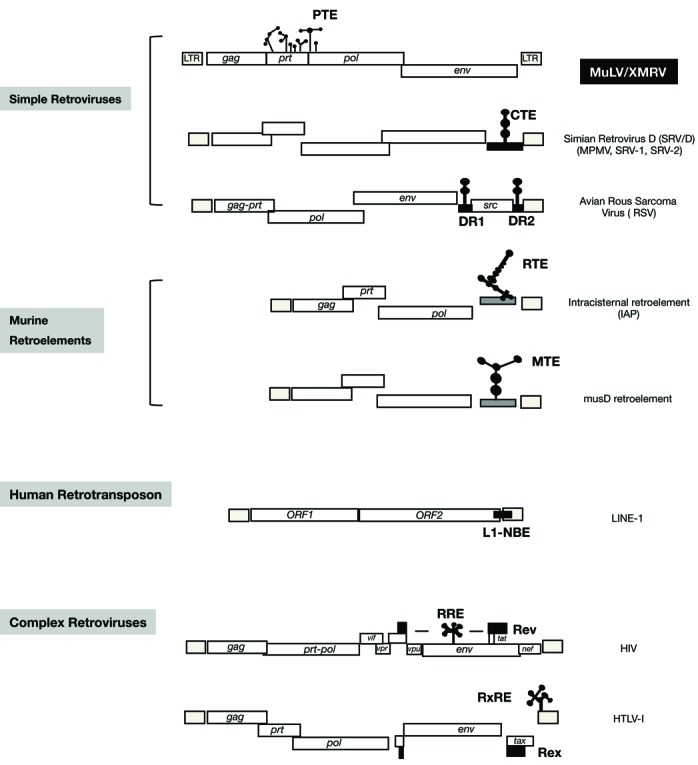
Localization of RNA export elements among simple retroviruses. Cartoon depiction of the genomic structure of MuLV/XMRV and selected retroviruses and retroelements. The location of the essential RNA export elements is shown: Simian retrovirus D (SRV/D) family (MPMV, SRV-1, SRV-2) has the CTE (18–20,65); the murine retroelements (intracisternal A particle retroelement and mus D retroelement) have the RTE (23) and the MTE (24,25), respectively located next to the 3′ LTR. Rous sarcoma virus has the DR elements flanking the src gene (21,22). The human LINE-1 retrotransposons contain the L1-NXF1-binding element overlapping ORF-2 and the 3′ LTR (26). Lentiviruses with HIV as prototype have RRE overlapping env (4,7). The HTLV family has the RXRE overlapping the 3′LTR (9,10). Not depicted are the endogenous human HTDV/HERV-K with the RcRE within the 3′LTR (13), the mouse MMTV with the RmRE overlapping env-3′LTR (14,15) and the betaretrovirus JSRV with the RejRE overlapping env (16,17).
While complex retroviruses also encode the viral post-transcriptional regulatory trans-factors to mediate export and expression of the transcript upon interaction with the nuclear receptor CRM1, simple retroviruses and retroelements depend solely on cellular factors and interaction with the NXF1 nuclear receptor, which was also shown to be essential for the export of cellular RNAs (29). Thus, NXF1 is also the key export receptor shared among simple retroviruses and retroelements. Recent reports have shown that MuLV uses NXF1, and not CRM1, for nuclear export of spliced and unspliced viral transcripts as expected (68–70). In agreement with these studies, we found that expression of pNLgag-PTE reporter was not affected by the addition of fungicidal antibiotic Leptomycin B (LMB), which blocks the export via CRM1, and, by inference, implicates NXF1 nuclear receptor in the export of MuLV (data not shown). The question then arises whether PTE function requires other factors. Potential candidates include RBM15 and RBM15b, which were shown to promote RTE (IAP retroelement) RNA expression and to interact with NXF1 (71–73). Neither factor affected PTE-mediated expression (data not shown). Thus, additional experimentation will be necessary to identify PTE cofactors.
In summary, we have identified a novel post-transcriptional regulatory element, PTE, which is essential for MuLV and XMRV expression. With the addition of the PTE element from gammaretroviruses, RNA regulatory elements have now been identified in every member of the retrovirus family. While typically spanning a couple of hundred nucleotides and being highly structured, they surprisingly share no conservation in sequence or location. Despite this, all are essential for providing efficient post-transcriptional regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors wish to thank J. Luban, S. Ruscetti, F. Ruscetti, A. Rein and G.N. Pavlakis for plasmids, antisera and discussions, the interns K. Apostolakis, P. Martin, N. Coleman, M. Nguyen and H. Kim for outstanding technical help and T. Jones for editorial assistance.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors.
FUNDING
Intramural Research Program of the National Cancer Institute, National Institutes of Health (NCI/NIH) [to S.F.J.L., B.K.F.].; HOMING [PLUS/2012-6/12 to K.J.P.]. Funding for open access charge: Intramural Research Program NIH/NCI.
Conflict of interest statement. None declared.
REFERENCES
- 1.Felber B.K., Valentin A., Rosati M., Bergamaschi C., Pavlakis G.N. HIV DNA vaccine: stepwise improvements make a difference. Vaccines. 2014;2:354–379. doi: 10.3390/vaccines2020354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felber B.K., Zolotukhin A.S., Pavlakis G.N. Posttranscriptional control of HIV-1 and other retroviruses and its practical applications. Adv. Pharmacol. 2007;55:161–197. doi: 10.1016/S1054-3589(07)55005-2. [DOI] [PubMed] [Google Scholar]
- 3.Cochrane A.W., McNally M.T., Mouland A.J. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology. 2006;3:18. doi: 10.1186/1742-4690-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hadzopoulou-Cladaras M., Felber B.K., Cladaras C., Athanassopoulos A., Tse A., Pavlakis G.N. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J. Virol. 1989;63:1265–1274. doi: 10.1128/jvi.63.3.1265-1274.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felber B.K., Hadzopoulou-Cladaras M., Cladaras C., Copeland T., Pavlakis G.N. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl Acad. Sci. U.S.A. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malim M.H., Bohnlein S., Hauber J., Cullen B.R. Functional dissection of the HIV-1 Rev trans-activator–derivation of a trans-dominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 7.Malim M.H., Hauber J., Le S.Y., Maizel J.V., Cullen B.R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 8.Hammarskjold M.L., Heimer J., Hammarskjold B., Sangwan I., Albert L., Rekosh D. Regulation of human immunodeficiency virus env expression by the rev gene product. J. Virol. 1989;63:1959–1966. doi: 10.1128/jvi.63.5.1959-1966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogerd H.P., Huckaby G.L., Ahmed Y.F., Hanly S.M., Greene W.C. The type I human T-cell leukemia virus (HTLV-I) Rex trans-activator binds directly to the HTLV-I Rex and the type 1 human immunodeficiency virus Rev RNA responsive element. Proc. Natl Acad. Sci. U.S.A. 1991;88:5704–5708. doi: 10.1073/pnas.88.13.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogerd H.P., Tiley L.S., Cullen B.R. Specific binding of the human T-cell leukemia virus type I Rex protein to a short RNA sequence located within the Rex-response element. J. Virol. 1992;66:7572–7575. doi: 10.1128/jvi.66.12.7572-7575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solomin L., Felber B.K., Pavlakis G.N. Different sites of interaction for Rev, Tev, and Rex proteins within the Rev-responsive element of human immunodeficiency virus type 1. J. Virol. 1990;64:6010–6017. doi: 10.1128/jvi.64.12.6010-6017.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed Y.F., Hanly S.M., Malim M.H., Cullen B.R., Greene W.C. Structure-function analyses of the HTLV-I Rex and HIV-1 Rev RNA response elements: insights into the mechanism of Rex and Rev action. Genes Dev. 1990;4:1014–1022. doi: 10.1101/gad.4.6.1014. [DOI] [PubMed] [Google Scholar]
- 13.Magin C., Lower R., Lower J. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J. Virol. 1999;73:9496–9507. doi: 10.1128/jvi.73.11.9496-9507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Indik S., Gunzburg W.H., Salmons B., Rouault F. A novel, mouse mammary tumor virus encoded protein with Rev-like properties. Virology. 2005;337:1–6. doi: 10.1016/j.virol.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 15.Mullner M., Salmons B., Gunzburg W.H., Indik S. Identification of the Rem-responsive element of mouse mammary tumor virus. Nucleic Acids Res. 2008;36:6284–6294. doi: 10.1093/nar/gkn608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nitta T., Hofacre A., Hull S., Fan H. Identification and mutational analysis of a Rej response element in Jaagsiekte sheep retrovirus RNA. J Virol. 2009;83:12499–12511. doi: 10.1128/JVI.01754-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofacre A., Nitta T., Fan H. Jaagsiekte sheep retrovirus encodes a regulatory factor, Rej, required for synthesis of Gag protein. J. Virol. 2009;83:12483–12498. doi: 10.1128/JVI.01747-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bray M., Prasad S., Dubay J.W., Hunter E., Jeang K.-T., Rekosh D., Hammarskjold M.-L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl Acad. Sci. U.S.A. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabernero C., Zolotukhin A.S., Valentin A., Pavlakis G.N., Felber B.K. The posttranscriptional control element of the simian retrovirus type 1 forms an extensive RNA secondary structure necessary for its function. J. Virol. 1996;70:5998–6011. doi: 10.1128/jvi.70.9.5998-6011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zolotukhin A.S., Valentin A., Pavlakis G.N., Felber B.K. Continuous propagation of RRE(-) and Rev(-)RRE(-) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J. Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogert R.A., Beemon K.L. Mutational analysis of the rous sarcoma virus DR posttranscriptional control element. J. Virol. 1998;72:3407–3411. doi: 10.1128/jvi.72.4.3407-3411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paca R.E., Ogert R.A., Hibbert C.S., Izaurralde E., Beemon K.L. Rous sarcoma virus DR posttranscriptional elements use a novel RNA export pathway. J. Virol. 2000;74:9507–9514. doi: 10.1128/jvi.74.20.9507-9514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nappi F., Schneider R., Zolotukhin A., Smulevitch S., Michalowski D., Bear J., Felber B.K., Pavlakis G.N. Identification of a novel posttranscriptional regulatory element by using a rev- and RRE-mutated human immunodeficiency virus type 1 DNA proviral clone as a molecular trap. J. Virol. 2001;75:4558–4569. doi: 10.1128/JVI.75.10.4558-4569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Legiewicz M., Zolotukhin A.S., Pilkington G.R., Purzycka K.J., Mitchell M., Uranishi H., Bear J., Pavlakis G.N., Le Grice S.F., Felber B.K. The RNA transport element of the murine musD retrotransposon requires long-range intramolecular interactions for function. J. Biol. Chem. 2010;285:42097–42104. doi: 10.1074/jbc.M110.182840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ribet D., Harper F., Dupressoir A., Dewannieux M., Pierron G., Heidmann T. An infectious progenitor for the murine IAP retrotransposon: emergence of an intracellular genetic parasite from an ancient retrovirus. Genome Res. 2008;18:597–609. doi: 10.1101/gr.073486.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindtner S., Felber B.K., Kjems J. An element in the 3′ untranslated region of human LINE-1 retrotransposon mRNA binds NXF1(TAP) and can function as a nuclear export element. RNA. 2002;8:345–356. doi: 10.1017/s1355838202027759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braun I.C., Herold A., Rode M., Conti E., Izaurralde E. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J. Biol. Chem. 2001;276:20536–20543. doi: 10.1074/jbc.M100400200. [DOI] [PubMed] [Google Scholar]
- 28.Gruter P., Tabernero C., von Kobbe C., Schmitt C., Saavedra C., Bachi A., Wilm M., Felber B.K., Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 29.Tan W., Zolotukhin A.S., Bear J., Patenaude D.J., Felber B.K. The mRNA export in C. elegans is mediated by Ce-NXF-1, an ortholog of human TAP and S cerevisiae Mex67p. RNA. 2000;6:1762–1772. doi: 10.1017/s1355838200000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shinnick T.M., Lerner R.A., Sutcliffe J.G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 31.Lombardi V.C., Ruscetti F.W., Das Gupta J., Pfost M.A., Hagen K.S., Peterson D.L., Ruscetti S.K., Bagni R.K., Petrow-Sadowski C., Gold B., et al. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science. 2009;326:585–589. doi: 10.1126/science.1179052. [DOI] [PubMed] [Google Scholar]
- 32.Paprotka T., Delviks-Frankenberry K.A., Cingoz O., Martinez A., Kung H.J., Tepper C.G., Hu W.S., Fivash M.J., Jr, Coffin J.M., Pathak V.K. Recombinant origin of the retrovirus XMRV. Science. 2011;333:97–101. doi: 10.1126/science.1205292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smulevitch S., Bear J., Alicea C., Rosati M., Jalah R., Zolotukhin A.S., von Gegerfelt A., Michalowski D., Moroni C., Pavlakis G.N., et al. RTE and CTE mRNA export elements synergistically increase expression of unstable, Rev-dependent HIV and SIV mRNAs. Retrovirology. 2006;3:6. doi: 10.1186/1742-4690-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ribet D., Harper F., Dewannieux M., Pierron G., Heidmann T. Murine MusD retrotransposon: structure and molecular evolution of an ‘intracellularized’ retrovirus. J. Virol. 2007;81:1888–1898. doi: 10.1128/JVI.02051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smulevitch S., Michalowski D., Zolotukhin A.S., Schneider R., Bear J., Roth P., Pavlakis G.N., Felber B.K. Structural and functional analysis of the RNA transport element, a member of an extensive family present in the mouse genome. J. Virol. 2005;79:2356–2365. doi: 10.1128/JVI.79.4.2356-2365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donello J.E., Loeb J.E., Hope T.J. Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J. Virol. 1998;72:5085–5092. doi: 10.1128/jvi.72.6.5085-5092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith G.J., 3rd, Donello J.E., Luck R., Steger G., Hope T.J. The hepatitis B virus post-transcriptional regulatory element contains two conserved RNA stem-loops which are required for function. Nucleic Acids Res. 1998;26:4818–4827. doi: 10.1093/nar/26.21.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz S., Felber B.K., Benko D.M., Fenyo E.M., Pavlakis G.N. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 1990;64:2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stauber R.H., Horie K., Carney P., Hudson E.A., Tarasova N.I., Gaitanaris G.A., Pavlakis G.N. Development and applications of enhanced green fluorescent protein mutants. Biotechniques. 1998;24:462–466. doi: 10.2144/98243rr01. 468–471. [DOI] [PubMed] [Google Scholar]
- 40.Vasa S.M., Guex N., Wilkinson K.A., Weeks K.M., Giddings M.C. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deigan K.E., Li T.W., Mathews D.H., Weeks K.M. Accurate SHAPE-directed RNA structure determination. Proc. Natl Acad. Sci. U.S.A. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nasioulas G., Zolotukhin A.S., Tabernero C., Solomin L., Cunningham C.P., Pavlakis G.N., Felber B.K. Elements distinct from human immunodeficiency virus type 1 splice sites are responsible for the Rev dependence of env mRNA. J. Virol. 1994;68:2986–2993. doi: 10.1128/jvi.68.5.2986-2993.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider R., Campbell M., Nasioulas G., Felber B.K., Pavlakis G.N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz S., Campbell M., Nasioulas G., Harrison J., Felber B.K., Pavlakis G.N. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J. Virol. 1992;66:7176–7182. doi: 10.1128/jvi.66.12.7176-7182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz S., Felber B.K., Pavlakis G.N. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J. Virol. 1992;66:150–159. doi: 10.1128/jvi.66.1.150-159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilkinson K.A., Gorelick R.J., Vasa S.M., Guex N., Rein A., Mathews D.H., Giddings M.C., Weeks K.M. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkinson K.A., Vasa S.M., Deigan K.E., Mortimer S.A., Giddings M.C., Weeks K.M. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA. 2009;15:1314–1321. doi: 10.1261/rna.1536209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krzyzosiak W.J., Marciniec T., Wiewiorowski M., Romby P., Ebel J.P., Giege R. Characterization of the lead(II)-induced cleavages in tRNAs in solution and effect of the Y-base removal in yeast tRNAPhe. Biochemistry. 1988;27:5771–5777. doi: 10.1021/bi00415a056. [DOI] [PubMed] [Google Scholar]
- 49.Pollom E., Dang K.K., Potter E.L., Gorelick R.J., Burch C.L., Weeks K.M., Swanstrom R. Comparison of SIV and HIV-1 genomic RNA structures reveals impact of sequence evolution on conserved and non-conserved structural motifs. PLoS Pathog. 2013;9:e1003294. doi: 10.1371/journal.ppat.1003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crawford S., Goff S.P. A deletion mutation in the 5′ part of the pol gene of Moloney murine leukemia virus blocks proteolytic processing of the gag and pol polyproteins. J. Virol. 1985;53:899–907. doi: 10.1128/jvi.53.3.899-907.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh I., Yoshinaka Y., Rein A., Shibuya M., Odaka T., Oroszlan S. Murine leukemia virus maturation: protease region required for conversion from ‘immature’ to ‘mature’ core form and for virus infectivity. Virology. 1985;145:280–292. doi: 10.1016/0042-6822(85)90161-8. [DOI] [PubMed] [Google Scholar]
- 52.Volkova N.A., Fomina E.G., Smolnikova V.V., Zinovieva N.A., Fomin I.K. The U3 region of Moloney murine leukemia virus contains position-independent Cis-acting sequences involved in the nuclear export of full-length viral transcripts. J. Biol. Chem. 2014;289:20158–20169. doi: 10.1074/jbc.M113.545855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Armentano D., Yu S.F., Kantoff P.W., von Ruden T., Anderson W.F., Gilboa E. Effect of internal viral sequences on the utility of retroviral vectors. J Virol. 1987;61:1647–1650. doi: 10.1128/jvi.61.5.1647-1650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smagulova F., Maurel S., Morichaud Z., Devaux C., Mougel M., Houzet L. The highly structured encapsidation signal of MuLV RNA is involved in the nuclear export of its unspliced RNA. J. Mol. Biol. 2005;354:1118–1128. doi: 10.1016/j.jmb.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 55.Cupelli L., Okenquist S.A., Trubetskoy A., Lenz J. The secondary structure of the R region of a murine leukemia virus is important for stimulation of long terminal repeat-driven gene expression. J. Virol. 1998;72:7807–7814. doi: 10.1128/jvi.72.10.7807-7814.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trubetskoy A.M., Okenquist S.A., Lenz J. R region sequences in the long terminal repeat of a murine retrovirus specifically increase expression of unspliced RNAs. J. Virol. 1999;73:3477–3483. doi: 10.1128/jvi.73.4.3477-3483.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.King J.A., Bridger J.M., Gounari F., Lichter P., Schulz T.F., Schirrmacher V., Khazaie K. The extended packaging sequence of MoMLV contains a constitutive mRNA nuclear export function. FEBS Lett. 1998;434:367–371. doi: 10.1016/s0014-5793(98)00948-x. [DOI] [PubMed] [Google Scholar]
- 58.Butsch M., Hull S., Wang Y., Roberts T.M., Boris-Lawrie K. The 5′ RNA terminus of spleen necrosis virus contains a novel posttranscriptional control element that facilitates human immunodeficiency virus Rev/RRE-independent Gag production. J. Virol. 1999;73:4847–4855. doi: 10.1128/jvi.73.6.4847-4855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dangel A.W., Hull S., Roberts T.M., Boris-Lawrie K. Nuclear interactions are necessary for translational enhancement by spleen necrosis virus RU5. J. Virol. 2002;76:3292–3300. doi: 10.1128/JVI.76.7.3292-3300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hull S., Boris-Lawrie K. RU5 of Mason-Pfizer monkey virus 5′ long terminal repeat enhances cytoplasmic expression of human immunodeficiency virus type 1 gag-pol and nonviral reporter RNA. J. Virol. 2002;76:10211–10218. doi: 10.1128/JVI.76.20.10211-10218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roberts T.M., Boris-Lawrie K. The 5′ RNA terminus of spleen necrosis virus stimulates translation of nonviral mRNA. J. Virol. 2000;74:8111–8118. doi: 10.1128/jvi.74.17.8111-8118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zolotukhin A.S., Schneider R., Uranishi H., Bear J., Tretyakova I., Michalowski D., Smulevitch S., O'Keeffe S., Pavlakis G.N., Felber B.K. The RNA transport element RTE is essential for IAP LTR-retrotransposon mobility. Virology. 2008;377:88–99. doi: 10.1016/j.virol.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 63.Hwang L.S., Park J., Gilboa E. Role of intron-contained sequences in formation of moloney murine leukemia virus env mRNA. Mol. Cell. Biol. 1984;4:2289–2297. doi: 10.1128/mcb.4.11.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller C.K., Temin H.M. Insertion of several different DNAs in reticuloendotheliosis virus strain T suppresses transformation by reducing the amount of subgenomic mRNA. J. Virol. 1986;58:75–80. doi: 10.1128/jvi.58.1.75-80.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tabernero C., Zolotukhin A.S., Bear J., Schneider R., Karsenty G., Felber B.K. Identification of an RNA sequence within an intracisternal-A particle element able to replace Rev-mediated posttranscriptional regulation of human immunodeficiency virus type 1. J. Virol. 1997;71:95–101. doi: 10.1128/jvi.71.1.95-101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fang X., Wang J., O'Carroll I.P., Mitchell M., Zuo X., Wang Y., Yu P., Liu Y., Rausch J.W., Dyba M.A., et al. An unusual topological structure of the HIV-1 Rev response element. Cell. 2013;155:594–605. doi: 10.1016/j.cell.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang J., Liang T.J. A novel hepatitis B virus (HBV) genetic element with Rev response element-like properties that is essential for expression of HBV gene products. Mol. Cell. Biol. 1993;13:7476–7486. doi: 10.1128/mcb.13.12.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pessel-Vivares L., Ferrer M., Laine S., Mougel M. MLV requires Tap/NXF1-dependent pathway to export its unspliced RNA to the cytoplasm and to express both spliced and unspliced RNAs. Retrovirology. 2014;11:21. doi: 10.1186/1742-4690-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakuma T., Davila J.I., Malcolm J.A., Kocher J.P., Tonne J.M., Ikeda Y. Murine leukemia virus uses NXF1 for nuclear export of spliced and unspliced viral transcripts. J. Virol. 2014;88:4069–4082. doi: 10.1128/JVI.03584-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sakuma T., Tonne J.M., Ikeda Y. Murine leukemia virus uses TREX components for efficient nuclear export of unspliced viral transcripts. Viruses. 2014;6:1135–1148. doi: 10.3390/v6031135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lindtner S., Zolotukhin A.S., Uranishi H., Bear J., Kulkarni V., Smulevitch S., Samiotaki M., Panayotou G., Felber B.K., Pavlakis G.N. RNA-binding motif protein 15 binds to the RNA transport element RTE and provides a direct link to the NXF1 export pathway. J. Biol. Chem. 2006;281:36915–36928. doi: 10.1074/jbc.M608745200. [DOI] [PubMed] [Google Scholar]
- 72.Zolotukhin A.S., Uranishi H., Lindtner S., Bear J., Pavlakis G.N., Felber B.K. Nuclear export factor RBM15 facilitates the access of DBP5 to mRNA. Nucleic Acids Res. 2009;37:7151–7162. doi: 10.1093/nar/gkp782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uranishi H., Zolotukhin A.S., Lindtner S., Warming S., Zhang G.M., Bear J., Copeland N.G., Jenkins N.A., Pavlakis G.N., Felber B.K. The RNA-binding motif protein 15B (RBM15B/OTT3) acts as cofactor of the nuclear export receptor NXF1. J. Biol. Chem. 2009;284:26106–26116. doi: 10.1074/jbc.M109.040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


