Abstract
Stress granules (SGs) are formed in response to stress, contain mRNAs, 40S ribosomal subunits, initiation factors, RNA-binding and signaling proteins, and promote cell survival. Our study describes a novel function of the protein heterodimer SRP9/14 and Alu RNA in SG formation and disassembly. In human cells, SRP9/14 exists assembled into SRP, bound to Alu RNA and as a free protein. SRP9/14, but not SRP, localizes to SGs following arsenite or hippuristanol treatment. Depletion of the protein decreases SG size and the number of SG-positive cells. Localization and function of SRP9/14 in SGs depend primarily on its ability to bind directly to the 40S subunit. Binding of SRP9/14 to 40S and Alu RNA is mutually exclusive indicating that the protein alone is bound to 40S in SGs and that Alu RNA might competitively regulate 40S binding. Indeed, by changing the effective Alu RNA concentration in the cell or by expressing an Alu RNA binding-defective protein we were able to influence SG formation and disassembly. Our findings suggest a model in which SRP9/14 binding to 40S promotes SG formation whereas the increase in cytoplasmic Alu RNA following stress promotes disassembly of SGs by disengaging SRP9/14 from 40S.
INTRODUCTION
Eukaryotic cells have evolved elaborate mechanisms to cope with stress. Four cellular kinases (PKR, HRI, PERK and GCN2) are able to integrate different stress signals and to phosphorylate the initiation factor eIF2α. Phosphorylation of eIF2α impairs formation of the eIF2-GTP-tRNAiMet ternary complex causing a rapid decrease of global translation while the synthesis of some proteins such as transcription factors and molecular chaperones, which help cells resist to stress, is favored (1).
Inhibition of translation initiation also results in the formation of stress granules (SGs). These cytoplasmic foci are composed of 40S ribosomal subunits, initiation factors and mRNAs in the form of non-functional initiation complexes, as well as a plethora of other RNA-binding proteins and signaling molecules (2). Formation of SGs is triggered by the oligomerization of low complexity sequences contained in several RNA-binding proteins such as TIA-1 and G3BP (3,4), and SGs are generally considered as pro-survival entities, which prevent apoptosis by sequestering key signaling molecules (2). In addition, since there is a constant and rapid flux of molecules between SGs and other cytoplasmic structures, most notably the polysomes (5,6), SGs were proposed to participate in regulating the composition and functional activity of messenger ribonucleoproteins (mRNPs).
The heterodimeric protein complex SRP9/SRP14 (SRP9/14) is a component of the signal recognition particle (SRP). As part of SRP, the heterodimer binds the 7SL RNA, but it also binds to cytoplasmic Alu RNA to form a complex known as Alu RNP (7,8). In these two forms, the SRP9/14 dimer participates in two different mechanisms of translation regulation: (i) in SRP, it is required to delay polypeptide elongation in order to maintain nascent chains in a translocation-competent state until they reach the membrane of the endoplasmic reticulum (ER) (9,10); (ii) in Alu RNPs, it plays a role in preventing polysome formation and thus, most likely inhibits initiation of protein synthesis (11).
Alu elements are derived from the ancestral 7SL RNA gene (12) and amplified by retrotransposition such that over 1 million copies are now present in the human genome (13). Alu elements are 300 nucleotides long and composed of two arms joined by an A-rich linker (14,15). They function as independent transcription units, which are transcribed into noncoding Alu RNAs by RNA polymerase III (Pol III). Alu RNAs can be further processed into scAlu RNAs that accumulate in the cytoplasm (Supplementary Figure S3A) (16). Rodent species instead contain the B1 repetitive elements, which are also derived from the 7SL RNA gene. They are transcribed into B1 RNA comprising one Alu-like arm of 130 nucleotides followed by an A-rich region, and are processed into scB1 RNAs, which accumulate in the cytoplasm and have been shown to bind the SRP9/14 dimer (Supplementary Figure S3B) (17,18).
While Alu and B1 elements are expressed at a low level under standard growth conditions, their expression is upregulated following heat shock (19–21) and viral infection (22–25). This observation, together with the well-established association of SRP9/14 with Alu RNAs, prompted us to investigate possible functions of these two components in the response to stress. Based on our results, we propose a model in which binding of SRP9/14 to the 40S ribosomal subunit promotes formation of SGs, while the increase of Alu RNA seen in response to stress favors SG disassembly.
MATERIALS AND METHODS
Cell lines, transfections and stress treatments
HeLa, HeLa Kyoto, HEK 293T and NIH 3T3 cells were grown at 37°C in Dulbecco's Modified Eagle Medium (Sigma) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin (PAA). Cells were transfected with calcium phosphate, unless otherwise specified. Experiments including the expression of 14-9VN proteins were performed 48 h post-transfection, while experiments including the expression of Alu RNAs were performed 24 h post-transfection. Different stress treatments with sodium arsenite (Sigma-Aldrich), hippuristanol (a gift from Dr. J. Pelletier, McGill University, Montreal, Canada) or heat shock were applied for different time periods and concentrations as specified in the figure legends. After sodium arsenite treatment, cells were washed twice in medium and incubated in new medium during recovery. After heat shock, cells were allowed to recover at 37°C for the indicated time periods. The final concentration of actinomycin D (Sigma-Aldrich) in the medium was 8 μM.
Plasmids
The plasmid pSP6-U6 (26) was a gift of Dr. Henri Tiedge. The p14-9VN was obtained by inserting the cDNAs of human SRP14 and SRP9 in the pBiFC-VN173 vector (27). The plasmids expressing shLuc, sh14 and sh9 are described in (10,28). Alu and scAlu RNAs were expressed in cells from the plasmids pAluYNF1 and pscAluYNF1. pAluYNF1 was generated from the plasmid Alu neoTet (29) by removing the neoTet reporter gene. The internal A-rich linker and the Alu right arm were deleted from pAluYNF1 to obtain pscAluYFN1. For the expression of 4.5S RNA, the Alu element in the plasmid pAluYNF1 was replaced by the 4.5S RNA gene (18) resulting in pDL4.5S.
Immunofluorescence staining and in situ hybridization
For immunofluorescence, cells grown on glass coverslips were rinsed with phosphate buffered saline (PBS), fixed using cold (−20°C) acetone/methanol (7:3) for 20 min at 4°C, washed twice in Tris-buffered saline (TBS) with 0.2% Tween and blocked with 2% bovine serum albumin for 30 min. Fragile X Mental Retardation Protein (FMRP) was detected using the monoclonal antibody 1C3 (30). Affinity-purified anti-human SRP14, anti-human SRP19, anti-human SRP72 and anti-S15 antibodies were described before (7,28,31). Anti-human SRP9 antibodies were raised in chicken and affinity-purified. Anti-human SRP68 and anti-flag M2 antibodies were purchased from Sigma-Aldrich, anti-eIF3η subunit and anti-GFP antibodies from Santa Cruz and Takara/Clontech, respectively. Secondary antibodies were purchased from Jackson Immunoresearch (Alexa Fluor 488 goat anti-rabbit, Texas Red sheep anti-mouse, Texas Red Donkey anti-chicken) and from Invitrogen (Alexa Fluor 594 donkey anti-goat and Alexa Fluor 488 donkey anti-mouse antibodies), and used following the suppliers’ protocols.
For in situ hybridization, cells were rinsed with PBS, fixed in 4% formaldehyde for 15 min, permeabilized with 0.2% Triton X-100 for 10 min and washed twice in 2x saline sodium citrate (SSC). Pre-hybridization was performed in 2xSSC, 20% formamide, 1 mg/ml tRNA and 1x Denhardt's solution for 2 h in a humid chamber at 37°C. For hybridization, the cells were incubated overnight at 37°C in fresh buffer containing 0.5 ng/μl of a biotinylated 50-mer oligo(dT) probe. Cells were washed twice in 2x SSC at room temperature and twice in 1xSSC at 37°C for 30 min. Immunofluorescence was performed using anti-biotin antibodies (Bethyl Laboratories, Montgomery, TX, USA).
Images were captured on a SP2 laser scanning confocal microscope (63X/1.4 numerical aperture, PlanApo), on a SP5 laser scanning confocal microscope (63X/1.4 numerical aperture, HCX PL APO CS), or on a LSM-710 Laser scanning microscope (Carl Zeiss Microimaging, Inc.) (63X).
Quantification of small and big SGs
SGs were identified by immunostaining of FMRP in control or knock-down HeLa cells. Confocal images were captured on a SP5 laser scanning confocal microscope (63X/1.4 numerical aperture, HCX PL APO CS). SGs with an area <0.78 μm2 (corresponding to a circular SG with a diameter <1 μm) were defined as small SGs while SGs with an area equal or bigger than 0.78 μm2 were defined as big SGs. The ratio between small SGs and big SGs per cell was obtained using the ImageJ software. A total of 235 control or 270 knock-down cells were analyzed from five independent experiments.
Northern blots
Total RNAs were extracted using Tri Reagent (Sigma-Aldrich) and aliquots of the samples displayed on a 6% urea/polyacrylamide gel. Alu and scAlu RNAs were detected using 24 nt-long oligonucleotide probes described in (17). Note that this probe does not cross-hybridize with the Alu domain of the 7SL RNA as indicated by the completely different Northern blot obtained using a polymerase chain reaction (PCR)-probe against the complete S domain of the 7SL RNA (compare Figure 7B and Supplementary Figure S7A). 4.5S and U6 RNAs were detected using PCR probes against the entire RNAs. U2 RNA was detected using the probe: 5′-GGTGCACCGTTCCTGGAGGTAC-3′. Detection was performed using the DIG detection system (Roche Applied Science).
Figure 7.
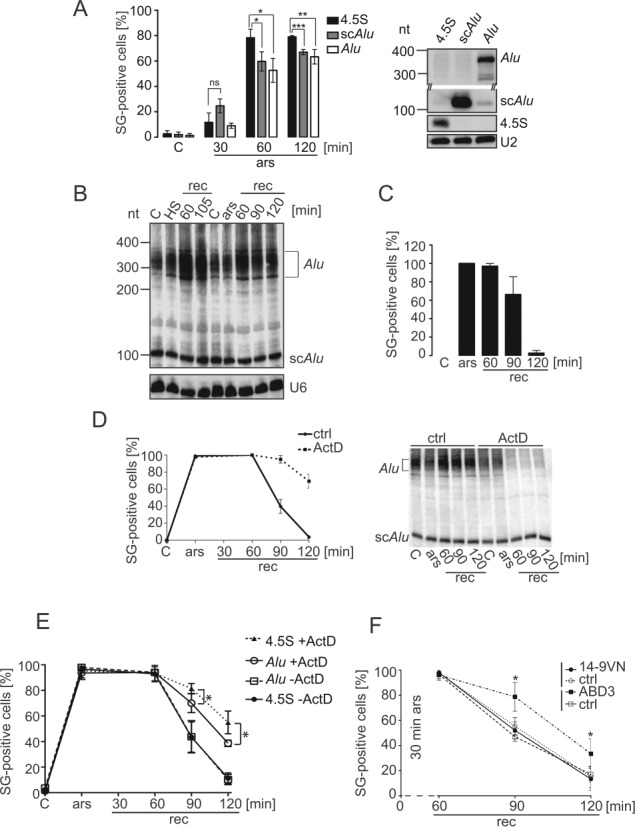
Alu RNA expression affects SG formation and disassembly. (A) Quantification of SG formation upon Alu RNA expression. HeLa cells expressing the flag-VN protein as a reporter for transfection, together with either 4.5S, scAlu or Alu RNA were incubated with 50 μM sodium arsenite for the indicated time periods and processed for double-immunofluorescence staining with anti-flag antibodies to identify transfected cells and with anti-eIF3 antibodies as a marker of SGs. Left panel: quantification. Error bars are shown as SD, n = 3. Right panel: Northern blot showing the expression of the 4.5S, scAlu and Alu RNAs. Expression levels of scAlu and Alu RNAs were significantly increased as compared to the endogenous levels, which were hardly detectable. (B) Expression of Alu RNAs during stress recovery. HeLa Kyoto cells were heat-shocked or arsenite-treated and allowed to recover for the times indicated. RNA samples were displayed on 6% denaturing polyacrylamide gel followed by Northern blot analysis. HS: heated 45°C for 30 min; ars: 500 μM sodium arsenite for 30 min; rec: recovery. (C) Quantification of SG-positive cells during recovery. SGs were revealed with anti-FMRP antibodies in arsenite-treated HeLa Kyoto cells. Error bars are shown as SD, n = 3. (D) Time course of SG-positive cells during recovery with and without actinomycin D. Stress: 30 min of 500 μM sodium arsenite. Left panel: quantification of SG-positive cells during recovery (rec) using FMRP antibodies. Right panel: Northern blot of RNA samples. ActD: 8 μM actinomycin D; ctrl: without actinomycin D. Error bars are shown as SD, n = 3. (E) Arsenite-treated HeLa cells expressing Alu or 4.5S RNA were allowed to recover with and without actinomycin D and the number of SGs counted as indicated in (A) at the time periods indicated. Error bars are shown as SD, n ≥ 3. (F) Arsenite-treated HeLa Kyoto cells expressing 14-9VN or 14-9VNABD3 were allowed to recover for the indicated time periods and processed for immunofluorescence staining using anti-FMRP antibodies. SG-positive cells were counted in 100 cells expressing the fusion proteins. In each sample, untransfected SG-positive cells were also quantified (ctrl). Error bars are shown as SD, n ≥ 2. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
In vitro binding experiments
Expression vectors and purification of the different recombinant human SRP9/14 proteins are described in (10,18). For in vitro binding of SRP9/14 to 40S ribosomal subunits, three pmoles of recombinant SRP9/14 protein were incubated with three pmoles of high-salt purified 40S ribosomal subunit (33) for 10 min on ice, followed by 10 min at 37°C in 30 μl of binding buffer containing 1.5 mM MgOAc; 20 mM Tris–HCl pH 7.5; 150 mM KOAc; 2 mM dithiothreitol (DTT); 0.01% Nikkol. The volume was adjusted to 100 μl with binding buffer, layered on top of a 500 μl 15% w/v sucrose cushion (binding buffer without Nikkol) and centrifuged at 120 000 rpm for 17 min in a Beckman MLA-130 rotor. Supernatant and pellet fractions were precipitated in 10% trichloroacetic acid and analyzed by Western blot using anti-SRP14 and anti-S15 antibodies.
For the protein transfer assay, the scAlu-SRP9/14 and scAlu-SRP9/14A5 RNPs were reconstituted by combining the biotinylated RNA with an excess of human SRP9/14 in a buffer containing 5 mM MgOAc, 350 mM KOAc, 20 mM Hepes-KOH, pH 7.5, 2 mM DTT and 0.01% Nikkol followed by two incubations for 10 min on ice and at 37°C. RNPs were separated from free RNA and protein on a Superdex 200 column, the RNP fractions were pooled and dialyzed in the same buffer supplemented with 10% glycerol. The OD260 was determined and the concentration of the scAlu RNP calculated based on its RNA content (40 μg/OD). Experiments with scB1 RNP were done without purification of the RNP on Superdex 200. Six pmoles of biotinylated scB1 RNA and 12 pmoles of human SRP9/14 were reconstituted as before in a buffer containing 5 mM MgOAc, 150 mM KOAc, 20 mM Tris–HCl pH 7.5, 2 mM DTT and 0.01% Nikkol and the B1-SRP9/14 RNP immobilized on magnetic Streptavidin beads for 30 min at 4°C. Excess protein was removed with two washes in the same buffer. Six pmoles of either 40S, U6 RNA, scAlu RNA or scB1 RNA were then added to immobilized RNPs in 30 μl binding buffer and incubated for 10 min at 4°C, 5 min at 37°C and 5 min on ice. The supernatants were separated from the beads, which were washed twice for 1 min. Aliquots of the supernatant and the bead fractions were analyzed by Western blot using anti-SRP14 and anti-S15 antibodies.
Fractionations of cell extracts on sucrose gradients
HEK 293T cells were lysed 48 h after transfection in a buffer containing 150 mM KOAc; 5 mM MgOAc; 20 mM Tris–HCl pH 7.5; 0.01% Nikkol; 2 mM DTT; AEBSF (Uptima, London, UK); peptide inhibitor cocktail (Sigma-Aldrich) and RNAseOUT (Invitrogen). The cell extracts were centrifuged for 7 min at 3000 rpm followed by 5 min at 10 000 g and 1 mg aliquots of the supernatants loaded on 10–40% sucrose gradients in lysis buffer. Centrifugation was performed for 254 min at 38 000 rpm in a Beckman SW 40 Ti rotor. Thirteen fractions were collected and aliquots analyzed for protein content by Western blot using anti-SRP14 and anti-SRP19 antibodies and for ribosome content by extraction of the RNA followed by its separation on 2% agarose gels and ethidium bromide-staining.
Additional antibodies for Western blots
Anti-Phospho-eIF2α (Ser51) antibodies (Cell Signaling Technology), anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) antibody (Abcam), anti-HSP70 antibody (StressMarq Biosciences) and anti-α-tubulin antibody (Sigma-Aldrich) were used as indicated by the supplier. Anti-L9 antibodies have been described (31).
RESULTS
SRP9/14 localizes to SGs in response to stress
The proteins SRP9 and SRP14 exist as a stable SRP9/14 protein heterodimer and in excess over SRP in human cells (7). In addition, only the protein heterodimer can bind SRP and Alu RNAs (32,34). Immunofluorescence staining of SRP14 and SRP9 in HeLa cells revealed a strong cytoplasmic staining of both proteins (Figure 1A–C). SRP14 could be detected in nucleoli when cells were fixed with acetone/methanol (Figure 1A) but not after formaldehyde fixation (Figure 1B). SRP9 was not detected in nucleoli (Figure 1C). Since mammalian SRP is assembled in nucleoli (35) and SRP9 and SRP14 function as a protein heterodimer, we believe that SRP9 is also present in nucleoli. However, the antibodies might not recognize it. After arsenite treatment, SRP14 and SRP9 accumulated in cytoplasmic granules (Figure 1A–C), which were identified as SGs using the three different SG markers FMRP, mRNAs and eIF3 (36–38). As for mRNAs, eIF3 (Figure 1B and C) and 40S subunits (Supplementary Figure S1) only a fraction of SRP9 and SRP14 localized to SGs, and staining throughout the cytoplasm persisted. We could not directly quantify the fraction of SRP9 and SRP14 present in SGs, since protein detection by antibodies is not linear over a sufficiently large concentration range. In contrast, high-resolution in situ hybridization has previously been used to determine quantitatively in HeLa cells the fractions of mRNAs and of 40S ribosomal subunits present in SGs (39). They were found to be about 15% and 5%, respectively. 40S subunits and the dimer SRP9/14 are about equally abundant in HeLa cells and by comparing S15 staining to SRP14 staining in SGs (compare Supplementary Figure S1 to Figure 1), we estimated that 5–10% of SRP9/14 was localized to SGs.
Figure 1.
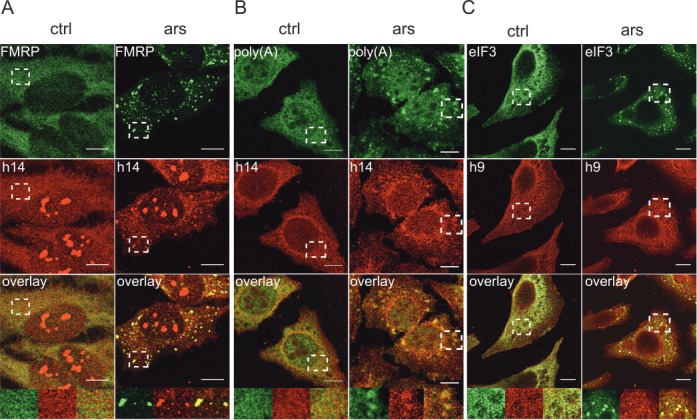
SGs contain SRP9/14. Immunofluorescence images of HeLa cells stained with antibodies against human SRP14 (h14), SRP9 (h9) and against different SG markers. ars: cells treated with 500 μM sodium arsenite for 30 min; ctrl: untreated cells. (A) Antibodies against h14 and FMRP. (B) Anti-h14 antibodies and in situ hybridization of mRNAs with oligo(dT). (C) Anti-h9 and anti-eIF3 antibodies. Anti-h9 antibodies failed to stain nucleoli. (A–C) Images were captured at the SP2 laser scanning confocal microscope (63x/1.4 numerical aperture, PlanApo). Areas denoted by rectangles are shown at higher magnification. Scale bars: 10 μm.
While arsenite interferes with protein synthesis by activation of the kinase HRI, which phosphorylates eIF2α, hippuristanol induces SG formation (40) by inactivating the RNA helicase eIF4A (41). Both subunits of SRP9/14 were present in SGs after hippuristanol treatment (Supplementary Figure S2A and B). This indicated that different mechanisms of translation inhibition caused the protein dimer SRP9/14 to localize to SGs.
SRP is stable during stress and absent from SGs
The SRP9/14 present in SGs might be there as part of SRP. To address this question, we first had to determine whether SRP remains intact in response to arsenite treatment. To this end, postnuclear supernatants were fractionated on glycerol gradients. SRP migrated in fractions 5–7 (Figure 2A, left panel) as indicated by the co-migration of SRP19 and SRP14. As mentioned before, SRP14 was also detected in slower migrating fractions representing SRP9/14 bound to scAlu RNA and free SRP9/14 (7). SRP14 in the bottom fractions might represent ribosome-associated protein and aggregates.
Figure 2.
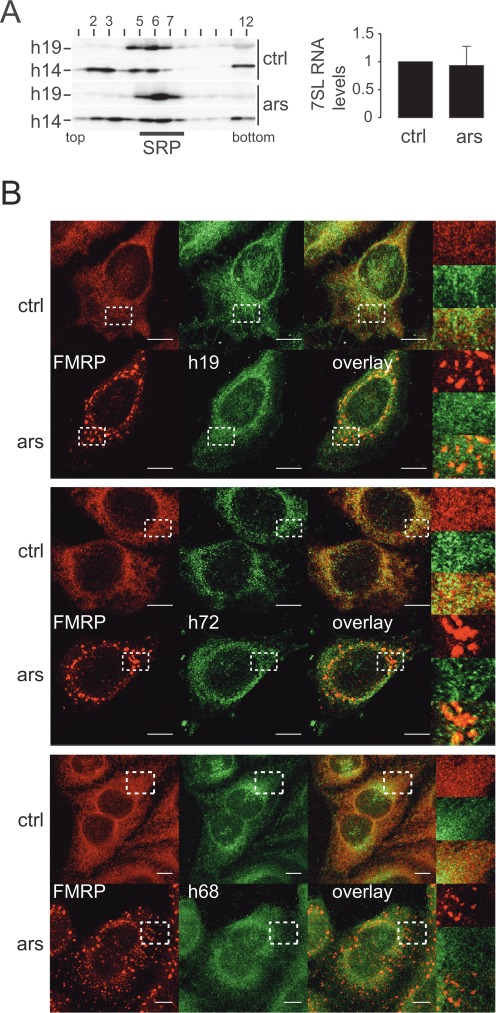
The SRP complex remains intact during stress and does not localize to SGs. (A) Velocity sedimentation fractionations of HeLa cell postnuclear supernatants on 12–30% glycerol gradients. Left panel: Western blots of gradient fractions using anti-h14 and anti-h19 antibodies. Right panel: quantification of 7SL RNA in HeLa cell extracts by qRT-PCR. Levels were standardized to GAPDH mRNA and normalized to control cells. Error bars are shown as SD, n = 3. (B) Immunofluorescence images of HeLa cells stained with antibodies against h19 and FMRP (upper panel) against h72 and FMRP (middle panel) and against h68 and FMRP (lower panel). Images were captured using a 63x lens on LSM-710 Laser scanning microscope. Areas denoted by rectangles are shown at higher magnification. Scale bars: 10 μm. ars: 500 μM sodium arsenite for 30 min; ctrl: untreated cells. h19: human SRP19; h72: human SRP72; h68: human SRP68
The gradient profiles of SRP19 and SRP14 were the same for arsenite-treated and control cells (upper and lower panels) indicating that SRP remained stable during stress. Furthermore, we quantified the total cellular levels of 7SL RNA in stressed and control cells and found that they remained unchanged (Figure 2A, right panel). Since 7SL RNA is not stable when cells are depleted of any of the proteins SRP72, SRP14 and SRP54 (28), this finding independently confirmed that SRP remained stable and SRP9/14 associated with SRP upon arsenite treatment.
We then performed double immunofluorescence staining experiments in HeLa cells with antibodies against one of three other subunits of SRP, SRP72, SRP68 or SRP19, together with antibodies against FMRP. Confocal microscopy revealed a predominantly reticular staining for SRP72, SRP68 and SRP19 in control cells as expected for SRP (Figure 2B). After arsenite treatment, we did not observe granular staining for any of the three SRP subunits (enlarged insets), whereas SGs were clearly detected with the marker FMRP. In the overlay, anti-SRP antibodies did not stain most of the granular structures. The partial overlap that was occasionally observed might be due to a fortuitous colocalization of SGs with SRP. Specificity of the anti-SRP68 and anti-SRP72 antibodies was assessed by Western blot on total cell extract (Supplementary Figure S2E and F).
SRP subunits also failed to be localized to SGs after hippuristanol treatment of HeLa cells (Supplementary Figure S2C and D). Since three SRP subunits failed to co-localize with FMRP in response to stress, we concluded that SRP did not localize to SGs and that therefore SRP9/14, when present in SGs, was not part of SRP.
SRP9/14 also localizes to SGs in rodent cells
Like the human proteins SRP9/14, murine SRP9 and SRP14 form a stable heterodimer, which forms a complex with SRP and scB1 RNAs (18,42). In contrast to human cells, there is no large excess of SRP9/14 over SRP in murine cells. However, scB1 RNA is similarly abundant in murine cells as scAlu RNA in human cells (43) suggesting that different cellular pools of SRP9/14 may also exist in murine cells. To address this question, we fractionated extracts of NIH 3T3 cells on glycerol gradients (Figure 3A). SRP was mostly present in fractions 7–8 as indicated by the co-migration of SRP19 with SRP14. SRP14 was also detected in fractions 3–5 indicating that murine cells contain a small pool of SRP9/14 not assembled into SRP. SRP9/14 in these fractions migrated as expected for the protein dimer bound to scB1 RNA and not as expected for the free protein. Importantly, the gradient profiles did not change in response to arsenite treatment confirming that SRP remained intact.
Figure 3.
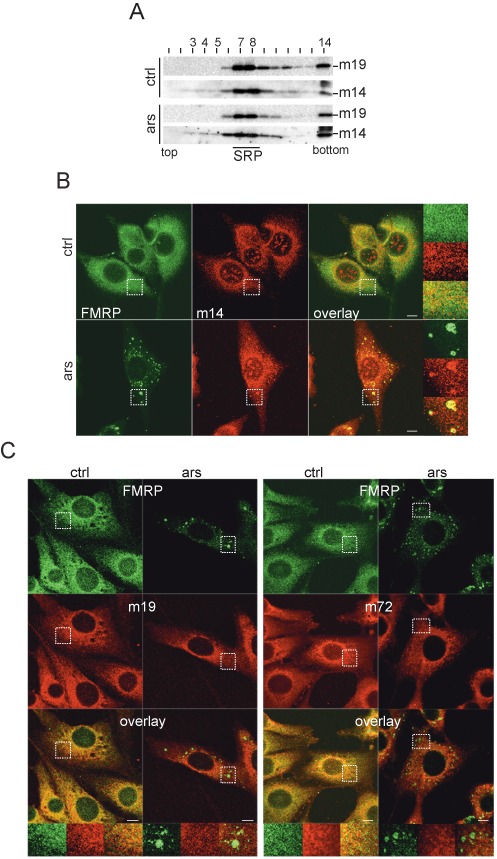
SRP9/14 localizes to SGs following stress in murine cells. (A) Velocity sedimentation fractionation of NIH 3T3 postnuclear supernatants on 12–30% glycerol gradients. Western blots of gradient fractions using anti-h14 and anti-h19 antibodies. (B) Double immunofluorescence staining of NIH 3T3 cells with anti-h14 and anti-FMRP antibodies. (C) Double immunofluorescence staining of NIH 3T3 cells with anti-h19 (left panel) and anti-h72 (right panel) antibodies together with anti-FMRP antibodies. Images were captured using a 63x lens on LSM-710 laser scanning microscope. Areas denoted by rectangles are shown at higher magnification. Scale bars: 10 μm. m14: murine SRP14; m19: murine SRP19; m72: murine SRP72. ars: 500 μM sodium arsenite for 1h; ctrl: untreated cells.
To examine whether SRP9/14, but not SRP, localizes to SGs in mouse cells, we performed double immunofluorescence staining experiments of NIH 3T3 cells using antibodies against human SRP14, SRP19 or SRP72 in combination with anti-FMRP antibodies. Since most of murine SRP9/14 is in a complex with SRP, staining for the three SRP proteins, SRP14, SRP19 and SRP72 was strongly reticular and less diffuse as compared to SRP9/14 in HeLa cells (Figure 3B and C). After arsenite treatment, SRP14 localized to granular structures together with FMRP (Figure 3B, lower panel). In contrast, SRP19 and SRP72 failed to enrich in granular structures after arsenite treatment (Figure 3C). Although we could not detect the murine SRP9 with antibodies against the human SRP9, the fact that the murine and human proteins behave the same in biochemical studies, let us to conclude that it is the protein heterodimer SRP9/14, which is present in SGs and that it is not associated with SRP. The function of SRP9/14 in SGs might therefore be conserved between mice and humans.
Functional domains in SRP9/14 involved in SG localization
As a tool to study mutated proteins, we decided to express the human SRP9/14 as a fusion protein flanked by two tags (14-9VN, Figure 4A). A fusion protein of murine SRP9 and SRP14 has previously been shown to replace functionally the protein dimer in reconstituted SRP (44). 14-9VN expressed in HeLa cells behaved like the endogenous SRP9/14. It had a cytosolic and nucleolar staining in control cells, and a fraction of the protein localized to SGs upon arsenite treatment (Figure 4B).
Figure 4.
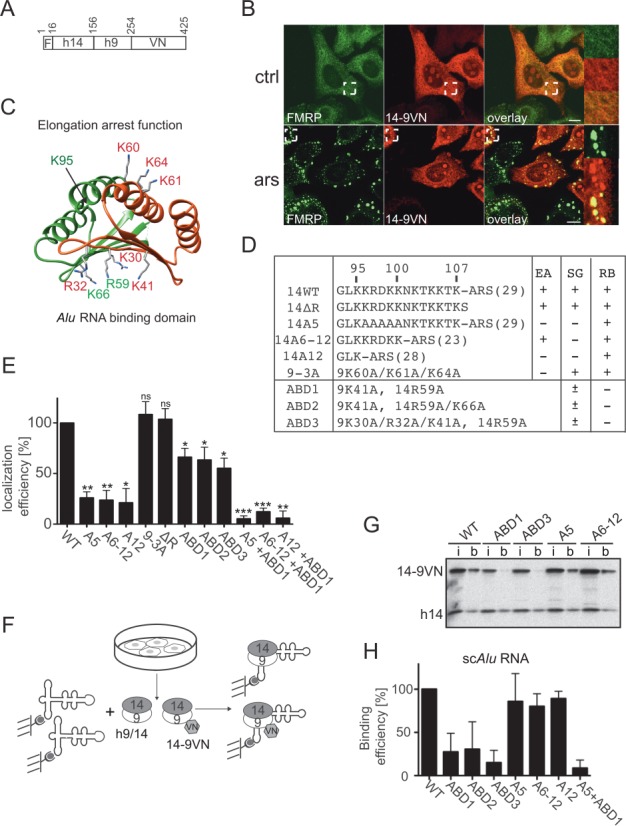
Functional determinants for SG localization in SRP9/14. (A) Schematic representation of the 14-9VN fusion protein. F: flag epitope: VN: 173 N-terminal amino acid residues of the Venus protein. Numbering refers to amino acid residues in 14-9VN. (B) Double immunofluorescence staining of HeLa cells with anti-GFP and anti-FMRP antibodies. ars: 500 μM sodium arsenite for 30 min; ctrl: untreated cells. Images were captured using a 63x lens on the LSM-710 laser scanning microscope. Areas denoted by rectangles are shown at higher magnification. Scale bars: 10 μm. (C) Structure of the protein dimer. h9: red, h14: green. Mutated amino acids are highlighted. The structure of the protein sequence following K95 in h14 could not be solved in the SRP-Alu-h9/14 complex (32). (D) Description of the mutations in the reporter protein 14-9VN. Amino acids are numbered according to the human sequences. ARS: alanine-rich sequence; brackets: number of amino acid residues; EA: elongation arrest activity; SG: SG localization; RB: Alu RNA binding activity. ABD1–3: proteins with mutations in the Alu RNA Binding Domain. (E) Efficiency of SG localization of the mutated proteins. Wild-type (WT) and mutated reporter proteins were expressed in HeLa Kyoto cells. Double immunofluorescence staining of cells with antibodies against GFP and FMRP revealed the fusion proteins and SGs, respectively. The presence of the reporter protein in SGs was counted in 100 transfected and SG-positive cells. In 74 ± 10% (n = 8) of these cells, 14-9VN was present in SGs. Localization efficiencies of the mutated proteins were normalized to 14-9VN, which was arbitrarily set to 100%. Error bars are shown as SD, n = 3. (F) Schematic representation of the RNA binding assay. Synthetic biotinylated scAlu RNA immobilized on magnetic streptavidin beads was incubated with postnuclear supernatants of HeLa Kyoto cells expressing the fusion proteins. (G) Equivalent aliquots of the input (i) and scAlu RNA-bound (b) protein fractions were analyzed by Western blot. Additional Western blots of RNA binding experiments are shown in Supplementary Figure S4. (H) Quantification of the RNA binding efficiencies of all fusion proteins normalized to the WT, which was set to 100%. Error bars are shown as SD, n ≥ 2. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
SGs contain 40S subunits and SRP9/14 in SRP is likely to interact with ribosomal subunits to inhibit nascent chain elongation (10,31). We therefore examined whether sequences important for elongation arrest activity of SRP were also required for SG localization. The domain involved in elongation arrest activity includes two patches of basic amino acid residues, the pentapeptide K96-K100 in SRP14 and the three lysines K60, K61 and K64 in SRP9 (9,10) (Figure 4C and D). Mutations in these residues were introduced into 14-9VN (Figure 4D, 14A5, 14A12, 9-3A). In addition, we mutated residues 96–107 (Figure 4D, 14A6-12) and removed the C-terminal alanine-rich sequence (ARS) in SRP14 (Figure 4D, ΔR). The alanine-rich tail represents a low complexity sequence (45) and might therefore trigger oligomerization of 14-9VN, contributing to SG nucleation as described for TIA-1 (3).
All fusion proteins were expressed at comparable levels in HeLa cells (Figure 4G, Supplementary Figure S4B, input lanes (i)) and did not affect SG formation. We chose to do these experiments in the presence of the endogenous SRP9/14 to avoid the negative effects on cell growth, structure and function caused by certain mutations (9).
The efficiency of SG localization of the mutated proteins was quantified and normalized to the one of 14-9VN (Figure 4E). SG localization of both 14-9VNA5 and 14-9VNA6–12 was strongly reduced. Simultaneous removal of both regions (A12) did not enhance the localization defect suggesting a role for these residues in the same function. In contrast, the protein 14-9VN9-3A was localized efficiently to SGs. Hence, there was only a partial overlap in the amino acid residues involved in SG localization of 14-9VN and in elongation arrest activity of SRP (summarized in Figure 4D). In addition, removal of the ARS did not affect the localization of the fusion protein to SGs (Figure 4E, ΔR). Since murine SRP14 lacks the ARS, this result was in agreement with the localization of murine SRP9/14 to SGs.
Next, we introduced mutations that interfered with the Alu RNA binding function of 14-9VN (Figure 4D, ABD1–3). The RNA binding domain of SRP9/14 involves primarily basic amino acid residues on the six-stranded β-sheet formed by the two polypeptides (Figure 4C, 14R59 and K66, 9K30, R32 and K41) (32,42,46). The RNA binding capacity of the mutated fusion proteins was determined by incubating cell extracts with scAlu RNA immobilized on streptavidin beads (Figure 4F, Supplementary Figure S4A). The proteins ABD1–3 and A5 + ABD1 had a reduced RNA binding capacity whereas it remained unchanged for the proteins 14-9VNA5, 14-9VNA6–12 and 14-9VNA12 (Figure 4G and H, Supplementary Figure S4B). These results were comparable to SRP9/14 binding to the Alu portion of 7SL RNA (9–10,32) and therefore confirmed that protein–RNA interactions were largely conserved between scAlu- and 7SL-SRP9/14 complexes.
All proteins with reduced RNA binding activity still localized to SGs following stress, albeit with reduced efficiency (Figure 4E). Simultaneous mutations of the RNA-binding domain and of the basic C-terminal region of SRP14 moderately, but significantly, decreased the localization efficiency consistent with the interpretation that the two regions have different functions in SG localization of SRP9/14. Together these results indicated that amino acids 96–107 in SRP14 are essential for SG localization whereas an intact RNA-binding function improves its efficiency.
SRP9/14 depletion increases the number of cells without SGs and decreases the size of SGs
In order to assess the role of SRP9/14 in SG formation, we reduced endogenous SRP9 and SRP14 levels by RNAi in HeLa cells (see ‘Materials and Methods’ section). The SRP14 protein levels at 72 h post-transfection were reproducibly lowered to 18% in cells expressing both sh9 and sh14 RNAs (Figure 5A). This level of depletion was chosen to minimize the defects in SRP-mediated protein translocation into the ER, which would result in deleterious protein mislocalization and growth defects (9,28).
Figure 5.
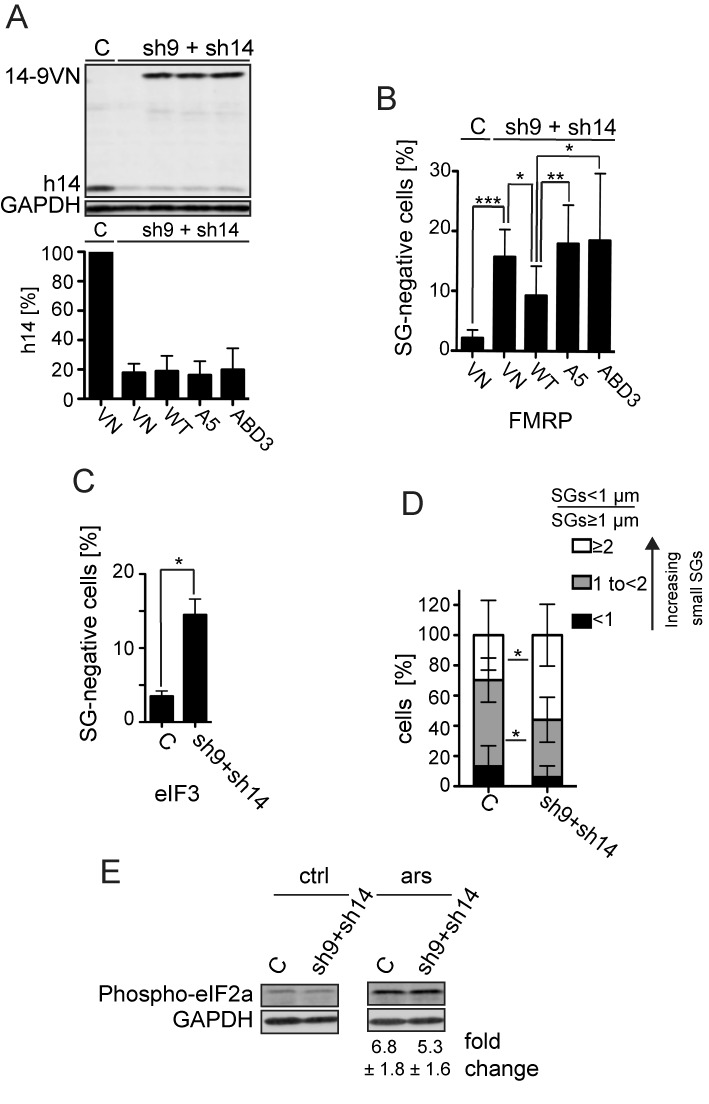
Effect of the SRP9/14 knockdown on SG formation. HeLa cells were transfected simultaneously with plasmids expressing shRNAs and the reporter proteins as indicated. After 24 h, cells were selected with puromycin (3 μg/ml) for 24 h and harvested following 30 min of sodium arsenite (500 μM) treatment at 72 h posttransfection. (A) Western blot (upper panel) and quantification standardized to GAPDH (lower panel) of the h9/14 knockdown. C: shLuc RNA; VN: venus protein; WT: 14-9VN; A5: 14-9VNA5; ABD3: 14-9VNABD3. Values were normalized to C, which was set to 100%. Error bars are shown as SD, n = 7. (B and C) Number of cells without SGs in knockdown cells. Arsenite-treated cells from (A) were subjected to immunofluorescence staining using anti-FMRP antibodies (B) or anti-eIF3 antibodies (C) and the number of cells devoid of SGs was counted in a sample of 100 cells. Error bars are shown as SD, n = 7 (B) or n = 2 (C). Unpaired one-tailed t-test. (D) Cells were categorized into three groups according to the ratio = SGs < 1 μm of diameter/SGs ≥ 1 μm of diameter. The three categories <1, 1 to <2 and ≥2 represent cells with an increasing number of small SGs. Error bars are shown as SD, n = 5 (E) Phosphorylation of eIF2α in response to stress in SRP9/14 knockdown cells. Western blots of cell lysates of arsenite-treated (ars) and untreated (ctrl) cells using anti-phospho-eIF2α antibodies. Phosphorylated protein levels were standardized to GAPDH. Numbers indicate the fold increases of phosphorylated eIF2α in response to arsenite treatment. The difference was not significant between the samples. Error bars are shown as SD; n = 4. *P≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
SGs were stained with the anti-FMRP antibody, and the cells lacking SGs were counted. The percentage of cells devoid of SGs increased from 2 to 16% after partial depletion of SRP9/14 (Figure 5B). A similar increase in SG-negative cells was observed using antibodies to the SG marker eIF3 (Figure 5C). The increase in SG-negative cells was due to the depletion of the protein dimer, since SG-negative cells decreased following the expression of the fusion protein 14-9VN (Figure 5B). Moreover, the expression of the mutated proteins A5 and ABD3 failed to reduce the number of SG-negative cells.
We noticed that SGs were generally smaller in SRP9/14 knockdown cells. We therefore decided to evaluate the changes in SG size as well. SGs have a diameter of about 1 μm following arsenite treatment (39). Since the total number of SGs per cell is quite variable, we decided to determine the ratio between the number of small (diameter < 1 μm) and large (diameter ≥ 1 μm) SGs in each SG-positive cell. In five independent experiments, we analyzed a total of 235 control and 270 knockdown cells and classified cells into three groups with ratios of <1, 1 to <2 and ≥2, representing cells with an increasing number of small SGs (Figure 5D). In control and SRP9/14 knockdown cells, a minor fraction of cells had more large SGs than small SGs (ratio <1, black bar). The majority of cells had more small SGs than large SGs (gray and white bars). Notably, the number of cells with a higher proportion of small SGs (ratio ≥2, white bars) increased significantly whereas cells with a lower proportion of small SGs (ratio 1 to <2, gray bars) decreased significantly in knockdown cells as compared to control cells.
Furthermore, we quantified the phosphorylation of eIF2α by Western blot (Figure 5E). Control and knockdown cells had comparable levels of eIF2α phosphorylation indicating that the depletion of SRP9/14 did not affect early steps in the stress-induced signaling pathway.
In summary, SRP9/14 knockdown interferes with proper SG formation by decreasing the number of cells forming SGs and the size of SGs, consistent with a role for SRP9/14 in the stress response.
The basic region in SRP14 mediates binding to 40S subunits
As mentioned before, the localization of SRP9/14 to SGs could be mediated by its binding to 40S ribosomal subunits. To demonstrate direct binding of SRP9/14 to 40S, we added wild-type (WT) and mutated human recombinant SRP9/14 proteins (h9/14) to 40S subunits purified from rabbit reticulocyte lysate. Free and 40S-bound proteins were separated on sucrose cushions. 40S subunits were always found in the pellet fraction (Figure 6A, P), and in the absence of 40S subunits, h9/14 was largely found in the supernatant fraction (ctrl, S). The WT h9/14 protein bound very efficiently to 40S subunits and was therefore found in the pellet. All h9/14 proteins lacking the region comprising amino acids 96–107 in SRP14 (A5, K100, K95) bound much less well to 40S subunits. In contrast, h9-3A bound well to 40S subunits. Hence, we observed a correlation between SG localization and 40S binding of SRP9/14 strongly suggesting that SRP9/14 is bound to 40S subunits in SGs.
Figure 6.
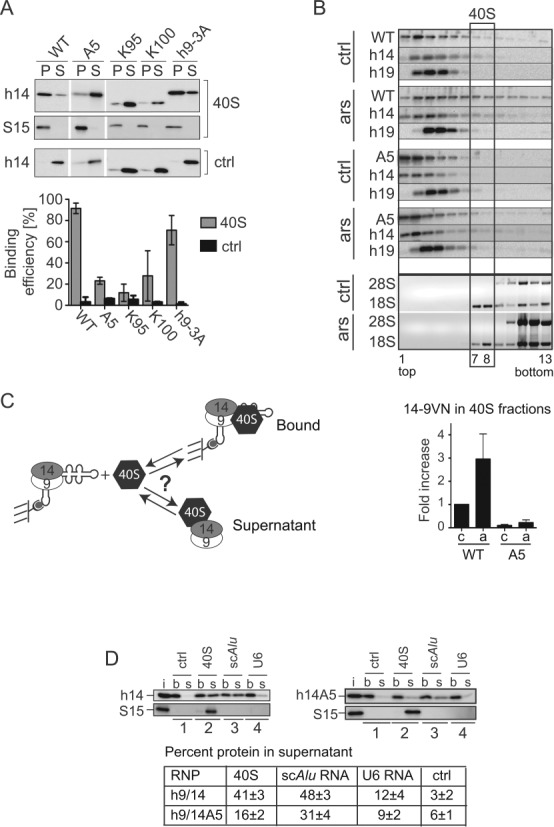
SRP9/14 binds to 40S subunits and cannot bind Alu RNA and 40S subunits simultaneously. (A) Binding of WT and mutated h9/14 to 40S. Equal amounts (100 nM) of purified recombinant proteins and purified rabbit 40S ribosomal subunits were incubated in a buffer containing 150 mM potassium acetate and 1.5 mM magnesium acetate. 40S-bound and free proteins were separated on sucrose cushions. Pellet (P) and supernatant (S) fractions were analyzed by Western blots (upper panel) and the results quantified (lower panel). K100 and K95 proteins were truncated at positions 100 and 95 in h14, respectively (see Figure 4D) (9,10). Ctrl: without 40S. Error bars are shown as SD, n ≥ 2. (B) Velocity sedimentation fractionation on 10–40% sucrose gradients of postnuclear supernatants obtained from HEK 293T cells expressing 14-9VN and 14-9VNA5. Western blots of gradient fractions with anti-h14 and anti-h19 antibodies. Bottom panels: agarose gels of RNA samples stained with ethidium bromide. ars: 500 μM sodium arsenite for 40 min; ctrl: untreated cells. Lower panel: relative amounts of the proteins 14-9VN and 14-9VNA5 present in fractions 7–8 as compared to untreated cells expressing 14-9VN (upper panel). Error bars are shown as SD, n ≥ 3. (C) Schematic representation of the protein transfer assay (PTA). Purified scAlu RNP is captured on magnetic streptavidin beads and purified 40S added in a buffer containing 150 mM potassium acetate and 5 mM magnesium acetate. Immobilized Alu RNP (b) is separated from the supernatant (s) after 5 min. (D) Western blots of the fractions from PTA experiments using anti-S15 and anti-h14 antibodies. Left panel: WT h9/14. Right panel: h9/14A5. Lane 1: mock control; lane 2: 40S; lane 3: nonbiotinylated scAlu RNA; lane 4: nonbiotinylated U6 RNA. (i): aliquots of h9/14 and 40S equivalent to the amounts loaded on the beads. Lower panel: quantification of the results.
The binding of SRP9/14 to 40S subunits might therefore be increased following stress. To address this question, postnuclear supernatants from HEK 293T cells expressing 14-9VN and 14-9VNA5 were fractionated on 10–40% sucrose gradients. 40S subunits were present in fractions 7–8 (Figure 6B). Both exogenous and the endogenous proteins were mostly present in fractions 1–6, which contain SRP, Alu RNPs and free SRP9/14. Interestingly, the percentage of 14-9VN co-migrating with 40S subunits increased about 3-fold following stress and represented about 6% of the total protein (Figure 6B, lower panel). The mutated 14-9VNA5 protein was almost not detectable in fractions 7–8 in control and arsenite-treated cells consistent with its deficiency in 40S binding. In both samples, the endogenous SRP9/14 increased also 2–3-fold in the 40S fractions following stress (not shown).
Alu RNA and 40S subunits compete for SRP9/14 binding
To elucidate a hypothetical function of the Alu RNA in SG localization of SRP9/14, we examined whether SRP9/14 was bound to 40S in complex with Alu RNA. To this end, we developed an assay allowing the rapid separation of 40S-bound and free complexes in vitro referred to as protein transfer assay (PTA, Figure 6C). In short, a complex of synthetic biotinylated scAlu RNA and recombinant h9/14 was purified by size exclusion chromatography and immobilized on streptavidin beads. After a 5 min incubation with 40S subunits, the supernatant (s) and the bead (b) fractions were separated and analyzed.
Notably, 40S subunits never bound to the immobilized scAlu RNA-h9/14 complex (Figure 6D, 40S) or to immobilized scAlu RNA alone (Supplementary Figure S5A) and therefore remained in the supernatant. However, in the presence of 40S, about 41% of h9/14 dissociated from the Alu RNA and was bound to 40S subunits in the supernatant (Figure 6D, 40S). Apparently, the protein bound to 40S without the Alu RNA. Hence, binding of h9/14 to 40S and to scAlu RNA were mutually exclusive indicating that the Alu RNA binding domain of SRP9/14 might be occupied by 18S rRNA in the 40S-SRP9/14 complex. In addition, it indicated that SRP9/14 in SGs is not bound to Alu RNA.
Equimolar amounts of nonbiotinylated scAlu RNA were added as a positive control and the protein also rapidly transferred from the immobilized scAlu RNA to the free scAlu RNA (Figure 6D, scAlu). The transfer efficiencies to scAlu RNA and 40S were comparable suggesting similar affinities of h9/14 for the two ligands. We also replaced scAlu RNA with the mouse scB1 RNA and found that h9/14 transferred from the scB1 RNP to 40S subunits (Supplementary Figure S5B).
Using purified scAlu-h9/14A5 RNP we found that much less protein transferred to 40S subunits (16%) consistent with the impaired 40S binding capacity of h9/14A5 observed in the protein binding experiments (Figure 6A). Surprisingly, we observed a slightly reduced transfer efficiency of h9/14A5 to scAlu RNA. The mutated protein SRP9/14A5 bound well to scAlu RNA (Figure 4G and H) and to the Alu domain of 7SL RNA (9). It is conceivable that the basic pentapeptide, which does not bind Alu RNA (32) accelerates the kinetics of the transfer reactions by initiating contacts with the competing target RNA.
Alu RNA affects SG formation
The competition between Alu RNA and 40S for SRP9/14 binding suggested that a high concentration of Alu RNA might influence the time course and the efficiency of SG formation. To address this question, Alu and scAlu RNAs were expressed in HeLa cells together with a reporter protein, which allowed the identification of transfected cells. As a negative control, we expressed the murine 4.5S RNA, which fails to bind human SRP9/14 (18). The expression of the RNAs was confirmed by Northern blot (Figure 7A, right panel).
At 24 h after transfection, a low, sublethal concentration of sodium arsenite (50 μM) was added to the cells and the cells were fixed at the indicated time points. SG formation was assessed in transfected cells using anti-eIF3 antibodies. Arsenite was present throughout the experiment, since cells are able to establish a long-term resistance to the toxic stress produced at this concentration of arsenite. At 30 min of arsenite treatment, a low percentage of cells have SGs in all samples (Figure 7A, left panel). After 60 and 120 min, the number of SG-positive cells was significantly lower in Alu RNA expressing cells as compared to the negative control cells. In parallel experiments, we used twice the concentration of arsenite (100 μM), which accelerated SG formation (Supplementary Figure S6). The expression of Alu RNAs did not affect SG formation at early time points. We even observed a small enhancement of the formation of SG by scAlu RNA after 20 min. However, after 30 min of arsenite treatment, SG formation was significantly reduced in cells expressing Alu RNA as compared to control cells. At 60 min all cells contained SGs. These results indicated that Alu RNA had the potential to delay the stress response under mild stress conditions compatible with cell survival. Hence, most likely by sequestering SRP9/14, Alu RNA expression interfered with an effective cellular stress response.
Alu RNA expression increases after arsenite treatment
As mentioned before, dimeric Alu RNA levels increase during recovery from heat shock (19). We decided to investigate whether Alu RNA expression was also induced after arsenite treatment. Cells were arsenite-treated or, as a control, heat-treated for 30 min, and cellular Alu RNA levels were determined at different time points after arsenite removal (recovery). After both treatments, dimeric Alu RNA levels were significantly increased at 60 min of recovery (Figure 7B). By contrast and as previously reported (19), scAlu (Figure 7B) and 7SL RNA levels (Supplementary Figure S7A) remained constant during recovery. In parallel samples, we assessed SG-positive cells using an anti-FMRP antibody (Figure 7C). All cells contained SGs after arsenite treatment (ars, 500 μM) and at 60 min of recovery. Subsequently, SGs-positive cells decreased and finally disappeared at 120 min when dimeric Alu RNA levels are still significantly increased.
Dimeric Alu RNA and the Alu RNA binding domain of SRP9/14 are required for efficient SG disassembly
To examine whether Alu RNA plays a role in SG disassembly, we attempted to interfere with Alu RNA expression after stress. α-amanitine, a well-known inhibitor of Pol III was not suitable because transcription inhibition is only observed after several hours of treatment (47). ML-60218, another Pol III inhibitor, could not be used because the solvent dimethyl sulfoxide (DMSO), an anti-oxidant, interfered with the stress response to arsenite. siRNAs against Alu sequences were very toxic for cells, presumably because they targeted Alu sequences in mRNAs causing unwanted side effects. We therefore used actinomycin D, which is rapidly incorporated into cells and has a general inhibitory effect on transcription (47).
Arsenite-treated cells were allowed to recover in media with or without actinomycin D. The presence of actinomycin D completely abolished dimeric Alu RNA expression during stress recovery (Figure 7D, right panel). In contrast, scAlu RNA was not affected, presumably because of a longer half-life (180 min) as compared to Alu RNA (30 min) (48,49). We quantified SG-positive cells over 120 min of recovery (Figure 7D, left panel). Actinomycin D treatment strongly interfered with the disassembly of SGs. The actinomycin D treatment per se did not induce SG formation (not shown) and the levels of an early stress response protein, Hsp70, remained the same during the short time of analysis (Supplementary Figure S7B). This was in agreement with earlier results showing that the expression of Alu RNA following heat shock precedes the expression of ubiquitin and HSP70 mRNAs (19). Moreover, the time course of dephosphorylation of eIF2α during stress recovery was unchanged in the presence of actinomycin D (Supplementary Figure S7C).
To consolidate the link between Alu RNA expression levels and SG disassembly kinetics, we transfected cells 24 h before stress treatment with plasmids expressing either Alu RNA (Supplementary Figure S7D) or, as a negative control, 4.5S RNA. All transfected cells contained SGs after arsenite treatment (ars, 500 μM) and at 60 min of recovery. In the absence of actinomycin D, the expression of Alu RNA or 4.5S RNA had no significant effect on the time course of disassembly (Figure 7E, -ActD). In contrast, the presence of the Alu RNA, but not of 4.5S RNA partially, but significantly accelerated SG disassembly in actinomycin D-treated cells (Alu + ActD, 4.5S + ActD). The rescue was only partial, because, not surprisingly, actinomycin D also impaired Alu RNA expression from the plasmid. However, Alu RNA levels remained higher in transfected cells during recovery after actinomycin D treatment as compared to non transfected cells (compare Figure 7D to Supplementary Figure S7D, ActD) and can therefore explain the observed rescue. These results therefore confirmed a role of the dimeric Alu RNA in SG disassembly.
To correlate Alu RNA expression to SRP9/14 binding, we expressed the RNA binding-defective 14-9VNABD3 protein, which localized with 60% efficiency to SGs (Figure 4E), in HeLa cells. We quantified SG-positive cells over 120 min of recovery of the transfected (Figure 7F, 14-9VN, ABD3) as well as of the untransfected cells (ctrl). Overexpression of 14-9VN did not significantly affect the time course of SG disassembly as compared to untransfected cells indicating that the level of the protein was not limiting for SG disassembly (Figure 7F). In contrast, at 90 and 120 min after arsenite removal, significantly more cells expressing 14-9VNABD3 contained SGs as compared to cells expressing WT 14-9VN and untransfected cells. The protein 14-9VNABD3 behaved like a dominant-negative mutant. It significantly delayed the overall disassembly of SGs validating a role of Alu RNA and the Alu RNA binding function of SRP9/14 in stress recovery.
DISCUSSION
This study provides evidence for a role of SRP9/14 and Alu RNA in the cellular stress response. The ability of SRP9/14 to bind directly to 40S ribosomal subunits and the capacity of Alu RNAs to compete with 40S subunits for SRP9/14 binding are at the heart of SRP9/14 and Alu RNA functions in SG assembly and disassembly. These results are consistent with the model shown in Supplementary Figure S7E. In this model, stress-induced polysome disassembly leads to the binding of SRP9/14 to the 40S subunits, thereby resulting in the maintenance of 40S subunits in an inactive state. In contrast, the physiological upregulation of Alu RNA following stress will facilitate SG disassembly by disengaging SRP9/14 from 40S during stress recovery.
Alu elements are also present in RNA polymerase II-transcribed genes and can even be found in mature mRNAs. However, SRP9/14 is not bound to Alu sequences in SG-sequestered mRNAs, since several of the mutated proteins with a functional Alu RNA binding domain are absent from SGs (Figure 4E, H, 14-9VNA5, A6–12 and A12). Only the mutations in SRP9/14 that abolish 40S binding strongly reduce SG localization. Our results therefore support the interpretation that SRP9/14 is bound to 40S subunits in SGs. The elements in SRP9/14 required for 40S binding are a C-terminal region of SRP14 (residues 96–107) and, since SRP9/14 cannot bind both Alu RNA and 40S subunits at the same time, most likely also the Alu RNA binding domain. Both have a strongly basic character and are likely to make contacts with 18S rRNA.
Based on these results, we propose two possible explanations for the less efficient SG localization of the 14-9VN proteins with mutations in the Alu RNA binding domain (ABD1–3 proteins): (i) the mutations in the Alu RNA binding domain might reduce the stability of the SRP9/14–40S complex and (ii) scAlu and scB1 RNPs, and not the free protein, might serve as reservoirs for SRP9/14 recruitment to 40S subunits. In this model, the Alu RNA-binding deficient 14-9VN proteins would assemble inefficiently with scAlu RNA and therefore accumulate with lower efficiency in SGs. Such Alu RNP-mediated delivery of SRP9/14 to 40S subunits has been identified as the molecular mechanism, which explains why only the Alu RNP, but not the protein alone (11) inhibits translation initiation (E.I., A.B., A.S., E. Alkalaeva and K.S., manuscript in preparation).
SRP9/14 promotes SG formation as illustrated by a decrease in SG size and number of SG-positive cells after its depletion. SRP9/14 is not a direct target of stress signaling, since it functions after shutdown of translation by stress kinases (Figure 5E). Moreover, it is not likely that it participates in SG formation via the oligomerization of its human-specific alanine-rich tail, since the murine protein lacks this sequence. The role of SRP9/14 in SGs appears to be linked to its ability to bind 40S subunits, since the expression of 40S binding-deficient proteins could not rescue the defect in SG assembly of SRP9/14 knockdown cells. Taking into account that the Alu RNP can interfere with polysome assembly (11), the most plausible function of SRP9/14 in SGs is to stabilize an inactive state of 40S subunits. Thus, it would contribute to create and maintain a high concentration of free mRNAs, which is required for SG formation (50).
Following stress, maximal levels of Alu RNA coincided with the beginning of SG disassembly (Figure 7B and C) and SG disassembly was delayed if Alu RNA accumulation was impaired, or if an RNA binding-defective SRP9/14 protein was expressed. As mentioned before, these findings suggest that the newly synthesized Alu RNA competes with 40S subunits for SRP9/14 binding and thereby promotes the dissociation of SRP9/14 from 40S subunits (Supplementary Figure S7E). The same function can be postulated for B1 RNA, which also increases in response to stress (19,21). Interestingly, it is primarily the cellular level of the dimeric Alu RNA, and less so of the scAlu RNA, which increases following stress (Figure 7B) (19). Since the dimeric Alu RNA has a shorter half-live than scAlu RNA, its expression level can be rapidly reduced following stress, which might represent an advantage for cells. Indeed, Alu RNA expression is tightly regulated in absence of stress most likely to keep retrotransposition events, which can be detrimental for cells, at a low frequency (15). Therefore, the accumulation of Alu RNA during the first hour after stress most likely results from an increase in transcription due to better chromatin accessibility (51). Interestingly, dicer can process dimeric Alu RNA (52,53) and it might be involved in the rapid degradation of Alu RNA. Dicer has specifically been shown to play a role in protecting cells against the degeneration of the retinal pigmented epithelium by preventing Alu RNA accumulation (52).
Exogenous expression of Alu RNA before stress caused SG formation to be transiently delayed, presumably by preventing SRP9/14 binding to 40S subunits. The already known effects on transcription (54) and on translation (20,55) assigned exclusively to Alu RNA, but not to scAlu RNA, were excluded as a cause for the delay, since it was also observed in scAlu RNA-expressing cells.
Interestingly, infection of cells with different viruses such as HIV (56), herpes simplex virus (57) and adenovirus 5 (15,23) increases Alu RNA levels. Hence, the sequestration of SRP9/14 by the virally induced expression of Alu RNA might delay SG formation in infected cells. This response might contribute to the complex interplay between host cells and viruses, which results in oscillations of translation and SG formation (58,59) thereby allowing the virus to replicate while keeping the host cell functional.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
K.S. acknowledges financial support from the Swiss National Science Foundation [31003A-143844] and the Canton of Geneva. R.M. acknowledges a grant from the Natural Sciences and Engineering Council of Canada [MOP-702406] and a New Investigator Scholarship Award from the Canadien Institute of Health Research. A.B. was a recipient of a one-year Roche Research Foundation fellowship.
Supplementary Material
Acknowledgments
We gratefully acknowledge Drs. C. Mary and L. Terzi for the preparation of recombinant proteins, Dr. A. Lakkaraju for technical help and plasmid construction, Dr. H. Tiedge for the plasmid pSP6-U6 and Dr. J. Pelletier for hippuristanol. We also thank Drs. F. Stutz and E. Radosta for helpful discussions, Drs. D. Picard and K. Maundrell for proofreading the manuscript.
Conflict of interest statement. None declared.
REFERENCES
- 1.Holcik M., Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 2.Buchan J.R., Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tourriere H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Zhang J., Okabe K., Tani T., Funatsu T. Dynamic association-dissociation and harboring of endogenous mRNAs in stress granules. J. Cell Sci. 2011;124:4087–4095. doi: 10.1242/jcs.090951. [DOI] [PubMed] [Google Scholar]
- 6.Kedersha N., Cho M.R., Li W., Yacono P.W., Chen S., Gilks N., Golan D.E., Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bovia F., Fornallaz M., Leffers H., Strub K. The SRP9/14 subunit of the signal recognition particle (SRP) is present in more than 20-fold excess over SRP in primate cells and exists primarily free but also in complex with small cytoplasmic Alu RNAs. Mol. Biol. Cell. 1995;6:471–484. doi: 10.1091/mbc.6.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang D.Y., Hsu K., Maraia R.J. Monomeric scAlu and nascent dimeric Alu RNAs induced by adenovirus are assembled into SRP9/14-containing RNPs in HeLa cells. Nucleic Acids Res. 1996;24:4165–4170. doi: 10.1093/nar/24.21.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lakkaraju A.K., Mary C., Scherrer A., Johnson A.E., Strub K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell. 2008;133:440–451. doi: 10.1016/j.cell.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mary C., Scherrer A., Huck L., Lakkaraju A.K., Thomas Y., Johnson A.E., Strub K. Residues in SRP9/14 essential for elongation arrest activity of the signal recognition particle define a positively charged functional domain on one side of the protein. RNA. 2010;16:969–979. doi: 10.1261/rna.2040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Häsler J., Strub K. Alu RNP and Alu RNA regulate translation initiation in vitro. Nucleic Acids Res. 2006;34:2374–2385. doi: 10.1093/nar/gkl246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984;312:171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- 13.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 14.Deininger P.L., Jolly D.J., Rubin C.M., Friedmann T., Schmid C.W. Base sequence studies of 300 nucleotide renatured repeated human DNA clones. J. Mol. Biol. 1981;151:17–33. doi: 10.1016/0022-2836(81)90219-9. [DOI] [PubMed] [Google Scholar]
- 15.Berger A., Strub K. Multiple Roles of Alu-Related Noncoding RNAs. Prog. Mol. Subcell. Biol. 2011;51:119–146. doi: 10.1007/978-3-642-16502-3_6. [DOI] [PubMed] [Google Scholar]
- 16.Matera A.G., Hellmann U., Schmid C.W. A transpositionally and transcriptionally competent Alu subfamily. Mol. Cell. Biol. 1990;10:5424–5432. doi: 10.1128/mcb.10.10.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang D.Y., Maraia R.J. A cellular protein binds B1 and Alu small cytoplasmic RNAs in vitro. J. Biol. Chem. 1993;268:6423–6428. [PubMed] [Google Scholar]
- 18.Bovia F., Wolff N., Ryser S., Strub K. The SRP9/14 subunit of the human signal recognition particle binds to a variety of Alu-like RNAs and with higher affinity than its mouse homolog. Nucleic Acids Res. 1997;25:318–326. doi: 10.1093/nar/25.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W.M., Chu W.M., Choudary P.V., Schmid C.W. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu W.M., Ballard R., Carpick B.W., Williams B.R., Schmid C.W. Potential Alu function: regulation of the activity of double-stranded RNA-activated kinase PKR. Mol. Cell. Biol. 1998;18:58–68. doi: 10.1128/mcb.18.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T., Spearow J., Rubin C.M., Schmid C.W. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene. 1999;239:367–372. doi: 10.1016/s0378-1119(99)00384-4. [DOI] [PubMed] [Google Scholar]
- 22.Jang K.L., Latchman D.S. HSV infection induces increased transcription of Alu repeated sequences by RNA polymerase III. FEBS Lett. 1989;258:255–258. doi: 10.1016/0014-5793(89)81667-9. [DOI] [PubMed] [Google Scholar]
- 23.Panning B., Smiley J.R. Activation of RNA polymerase III transcription of human Alu repetitive elements by adenovirus type 5: requirement for the E1b 58-kilodalton protein and the products of E4 open reading frames 3 and 6. Mol. Cell. Biol. 1993;13:3231–3244. doi: 10.1128/mcb.13.6.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panning B., Smiley J.R. Activation of RNA polymerase III transcription of human Alu elements by herpes simplex virus. Virology. 1994;202:408–417. doi: 10.1006/viro.1994.1357. [DOI] [PubMed] [Google Scholar]
- 25.Russanova V.R., Driscoll C.T., Howard B.H. Adenovirus type 2 preferentially stimulates polymerase III transcription of Alu elements by relieving repression: a potential role for chromatin. Mol. Cell. Biol. 1995;15:4282–4290. doi: 10.1128/mcb.15.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hausner T.P., Giglio L.M., Weiner A.M. Evidence for base-pairing between mammalian U2 and U6 small nuclear ribonucleoprotein particles. Genes Dev. 1990;4:2146–2156. doi: 10.1101/gad.4.12a.2146. [DOI] [PubMed] [Google Scholar]
- 27.Shyu Y.J., Liu H., Deng X., Hu C.D. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques. 2006;40:61–66. doi: 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- 28.Lakkaraju A.K., Luyet P.P., Parone P., Falguieres T., Strub K. Inefficient targeting to the endoplasmic reticulum by the signal recognition particle elicits selective defects in post-ER membrane trafficking. Exp. Cell Res. 2007;313:834–847. doi: 10.1016/j.yexcr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 30.Devys D., Lutz Y., Rouyer N., Bellocq J.P., Mandel J.L. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat. Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- 31.Terzi L., Pool M.R., Dobberstein B., Strub K. Signal recognition particle Alu domain occupies a defined site at the ribosomal subunit interface upon signal sequence recognition. Biochemistry. 2004;43:107–117. doi: 10.1021/bi0353777. [DOI] [PubMed] [Google Scholar]
- 32.Weichenrieder O., Wild K., Strub K., Cusack S. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature. 2000;408:167–173. doi: 10.1038/35041507. [DOI] [PubMed] [Google Scholar]
- 33.Pisarev A.V., Unbehaun A., Hellen C.U., Pestova T.V. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
- 34.Strub K., Walter P. Assembly of the Alu domain of the signal recognition particle (SRP): dimerization of the two protein components is required for efficient binding to SRP RNA. Mol. Cell. Biol. 1990;10:777–784. doi: 10.1128/mcb.10.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Politz J.C., Yarovoi S., Kilroy S.M., Gowda K., Zwieb C., Pederson T. Signal recognition particle components in the nucleolus. Proc. Natl. Acad. Sci. U.S.A. 2000;97:55–60. doi: 10.1073/pnas.97.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazroui R., Huot M.E., Tremblay S., Filion C., Labelle Y., Khandjian E.W. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 2002;11:3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- 37.Kedersha N.L., Gupta M., Li W., Miller I., Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kedersha N., Chen S., Gilks N., Li W., Miller I.J., Stahl J., Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Souquere S., Mollet S., Kress M., Dautry F., Pierron G., Weil D. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J. Cell Sci. 2009;122:3619–3626. doi: 10.1242/jcs.054437. [DOI] [PubMed] [Google Scholar]
- 40.Mazroui R., Sukarieh R., Bordeleau M.E., Kaufman R.J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bordeleau M.E., Mori A., Oberer M., Lindqvist L., Chard L.S., Higa T., Belsham G.J., Wagner G., Tanaka J., Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- 42.Bui N., Wolff N., Cusack S., Strub K. Mutational analysis of the protein subunits of the signal recognition particle Alu-domain. RNA. 1997;3:748–763. [PMC free article] [PubMed] [Google Scholar]
- 43.Bovia F., Strub K. The signal recognition particle and related small cytoplasmic ribonucleoprotein particles. J. Cell Sci. 1996;109(Pt 11):2601–2608. doi: 10.1242/jcs.109.11.2601. [DOI] [PubMed] [Google Scholar]
- 44.Bovia F., Bui N., Strub K. The heterodimeric subunit SRP9/14 of the signal recognition particle functions as permuted single polypeptide chain. Nucleic Acids Res. 1994;22:2028–2035. doi: 10.1093/nar/22.11.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uversky V.N. What does it mean to be natively unfolded. Eur. J. Biochem. 2002;269:2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 46.Weichenrieder O., Stehlin C., Kapp U., Birse D.E., Timmins P.A., Strub K., Cusack S. Hierarchical assembly of the Alu domain of the mammalian signal recognition particle. RNA. 2001;7:731–740. doi: 10.1017/s1355838201010160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bensaude O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription. 2011;2:103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu W.M., Liu W.M., Schmid C.W. RNA polymerase III promoter and terminator elements affect Alu RNA expression. Nucleic Acids Res. 1995;23:1750–1757. doi: 10.1093/nar/23.10.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li T.H., Schmid C.W. Alu's dimeric consensus sequence destabilizes its transcripts. Gene. 2004;324:191–200. doi: 10.1016/j.gene.2003.09.036. [DOI] [PubMed] [Google Scholar]
- 50.Bounedjah O., Desforges B., Wu T.D., Pioche-Durieu C., Marco S., Hamon L., Curmi P.A., Guerquin-Kern J.L., Pietrement O., Pastre D. Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res. 2014;42:8678–8691. doi: 10.1093/nar/gku582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li T.H., Kim C., Rubin C.M., Schmid C.W. K562 cells implicate increased chromatin accessibility in Alu transcriptional activation. Nucleic Acids Res. 2000;28:3031–3039. doi: 10.1093/nar/28.16.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaneko H., Dridi S., Tarallo V., Gelfand B.D., Fowler B.J., Cho W.G., Kleinman M.E., Ponicsan S.L., Hauswirth W.W., Chiodo V.A., et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu Q., Tanasa B., Trabucchi M., Li W., Zhang J., Ohgi K.A., Rose D.W., Glass C.K., Rosenfeld M.G. DICER- and AGO3-dependent generation of retinoic acid-induced DR2 Alu RNAs regulates human stem cell proliferation. Nat. Struct. Mol. Biol. 2012;19:1168–1175. doi: 10.1038/nsmb.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mariner P.D., Walters R.D., Espinoza C.A., Drullinger L.F., Wagner S.D., Kugel J.F., Goodrich J.A. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 55.Rubin C.M., Kimura R.H., Schmid C.W. Selective stimulation of translational expression by Alu RNA. Nucleic Acids Res. 2002;30:3253–3261. doi: 10.1093/nar/gkf419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jang K.L., Collins M.K., Latchman D.S. The human immunodeficiency virus tat protein increases the transcription of human Alu repeated sequences by increasing the activity of the cellular transcription factor TFIIIC. J. Acquir. Immune Defic. Syndr. 1992;5:1142–1147. [PubMed] [Google Scholar]
- 57.Jang K.L., Latchman D.S. The herpes simplex virus immediate-early protein ICP27 stimulates the transcription of cellular Alu repeated sequences by increasing the activity of transcription factor TFIIIC. Biochem. J. 1992;284(Pt 3):667–673. doi: 10.1042/bj2840667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lloyd R.E. Regulation of stress granules and P-bodies during RNA virus infection. Wiley Interdiscip. Rev. RNA. 2013;4:317–331. doi: 10.1002/wrna.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruggieri A., Dazert E., Metz P., Hofmann S., Bergeest J.P., Mazur J., Bankhead P., Hiet M.S., Kallis S., Alvisi G., et al. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe. 2012;12:71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


