Background: JH antagonizes the 20E pathway.
Results: JH induces Broad protein BrZ7 phosphorylation in the lepidopteran insect Helicoverpa armigera.
Conclusion: JH induces the phosphorylation of BrZ7 to inhibit 20E-mediated metamorphosis.
Significance: Our study reveals a mechanism of JH antagonizing 20E-activating metamorphosis.
Keywords: Development, Endocrinology, Gene Regulation, Insect, Phosphorylation, Transcription Factor, Broad, Juvenile Hormone, Metamorphosis, Non-genomic Pathway
Abstract
The steroid hormone 20-hydroxyecdysone (20E) initiates insect molting and metamorphosis. By contrast, juvenile hormone (JH) prevents metamorphosis. However, the mechanism by which JH inhibits metamorphosis remains unclear. In this study, we propose that JH induces the phosphorylation of Broad isoform Z7 (BrZ7), a newly identified protein, to inhibit 20E-mediated metamorphosis in the lepidopteran insect Helicoverpa armigera. The knockdown of BrZ7 in larvae inhibited metamorphosis by repressing the expression of the 20E response gene. BrZ7 was weakly expressed and phosphorylated during larval growth but highly expressed and non-phosphorylated during metamorphosis. JH regulated the rapid phosphorylation of BrZ7 via a G-protein-coupled receptor-, phospholipase C-, and protein kinase C-triggered pathway. The phosphorylated BrZ7 bound to the 5′-regulatory region of calponin to regulate its expression in the JH pathway. Exogenous JH induced BrZ7 phosphorylation to prevent metamorphosis by suppressing 20E-related gene transcription. JH promoted non-phosphorylated calponin interacting with ultraspiracle protein to activate the JH pathway and antagonize the 20E pathway. This study reveals one of the possible mechanisms by which JH counteracts 20E-regulated metamorphosis by inducing the phosphorylation of BrZ7.
Introduction
The development of holometabolous insects, including growth and metamorphosis, is mainly modulated by two hormones, 20-hydroxyecdysone (20E)2 and juvenile hormone (JH) (1). The homeostasis of these two hormones determines the nature of each developmental transition (2). 20E mainly initiates and coordinates molting and metamorphosis (3); JH prevents 20E-induced metamorphosis by modulating the activity of 20E (4).
The molecular basis of 20E activity is well studied. 20E binds to the ecdysone nuclear receptor (EcR), and EcR then interacts with ultraspiracle protein (USP) to direct metamorphosis by mediating the expression of 20E-response genes (5), including several transcription factors, such as hormone receptor 3 (HR3) (6), earlier response gene (E74A) (7), and Broad-Complex (Br-C, also known as Br) (8). The expressions of late genes, including prodeath serine/threonine protein kinase (prodeath-S/TK) (9), caspase-1 (10), and protein phosphatase 6 (PP6) (11), are sequentially regulated. In the JH pathway, JH modulates downstream gene transcription through the intracellular receptor methoprene-tolerant (Met) (12). Downstream genes include the anti-metamorphosis transcription factor Kruppel homolog 1 (Kr-h1) (13), RNA-binding protein (RBP) (14), and an actin-binding protein, calponin (Cal) (15). USP (16), Br-C (17), E75 (18), Kr-h1 (13), or Met (19) may be implicated in 20E and JH cross-talk. In Tribolium castaneum, JH suppresses the expression of Br via Met/Kr-h1 to block metamorphosis (13). In Drosophila melanogaster, JH counteracts Met or GCE (germ cell-expressed) to block 20E-induced metamorphosis (20). In D. melanogaster S2 cells, JH-induced ectopic E75A can repress the ecdysone activation of early genes (21). In Apis mellifera, the JH analog methoprene suppresses 20E-mediated midgut remodeling by inhibiting the expressions of EcRB, USP, and Br-C (22). These data reveal that JH antagonizes 20E signaling by suppressing 20E response gene expression (23).
The transcription factor Br-C/Br is a “pupal specifier” that initiates insect metamorphosis (24) in Manduca sexta (25) and D. melanogaster (26). Br-C encodes a family of alternately spliced isoforms (27), which contain a common BTB (Broad-Tramtrack-Bric-a-brac) protein-protein interaction domain at the N terminus and different zinc finger domains at the C terminus (28). Four isoforms (BrZ1–Z4) have been identified in D. melanogaster and M. sexta (25); five isoforms (BrZ1–Z5) with various C-terminal zinc finger domains have been detected in T. castaneum (29, 30). However, no similar proteins have been found in mammals by a basic local alignment search tool in the NCBI database. Br-C significantly influences early metamorphic events in response to 20E (31). In D. melanogaster, Br-C null mutants fail to pupariate (32). In T. castaneum, Br knockdown results in larval-pupal-adult intermediate characteristics (33). These studies have revealed the important functions of Br in 20E-mediated metamorphosis. However, the exposure of pupae to JH at the onset of adult development induces Br re-expression and results in a second pupal cuticle in M. sexta and in an abdominal pupal cuticle in D. melanogaster (25). In T. castaneum, exogenous JH or its mimic methoprene at the onset of adult development also induces Br-C expression and inhibits the pupal-adult transition (34). These results suggest that Br is also involved in the JH pathway. However, the mechanism of Br action in the JH pathway remains unclear.
In addition to genomic induction by the intracellular receptor Met (35), JH may induce rapid non-genomic events via unknown membrane receptors (36) and the protein kinase C (PKC) signaling pathway (37). In Locusta migratoria, JH III binds to the membrane preparations of the ovaries, suggesting the presence of a membrane receptor for JH III (38). Protein phosphorylation and dephosphorylation are typical characteristics of the non-genomic pathway; however, limited information on JH-modulated protein phosphorylation is available. Transcription factors function via post-translational modifications, such as phosphorylation and ubiquitylation (39). In D. melanogaster, 20E-regulated EcR phosphorylation functions in the 20E pathway (40). However, whether or not JH regulates the post-translational modification of the transcription factor Br remains unknown.
In this study, the Broad isoform Z7 (BrZ7) was identified in Helicoverpa armigera, a severe agricultural pest. BrZ7 contains an N-terminal BTB domain and two C-terminal zinc finger domains; however, BrZ7 is different from the Br isoforms (Z1–Z4) found in Bombyx mori. The knockdown of BrZ7 blocks 20E-activated metamorphosis. BrZ7 is weakly expressed and phosphorylated during the larval growth stage but highly expressed and non-phosphorylated during metamorphosis. JH regulates the rapid phosphorylation of BrZ7 via a G-protein-coupled receptor-, phospholipase C-, and protein kinase C-triggered pathway. The phosphorylated BrZ7 participates in JH-regulated gene transcription. The exogenous JH III induces BrZ7 phosphorylation and suppresses 20E response gene expression. These studies have revealed a mechanism by which JH antagonizes 20E-mediated metamorphosis by inducing BrZ7 phosphorylation.
EXPERIMENTAL PROCEDURES
Experimental Animals and Reagents
H. armigera larvae were raised according to methods described previously (41). The larvae were fed with an artificial diet in our laboratory at 26 ± 1 °C under a 14-h/10-h light/dark cycle.
20E, JH III, G-protein-coupled receptor (GPCR) inhibitor suramin sodium salt, phospholipase C (PLC) inhibitor U73122, and PKC-specific inhibitor chelerythrine chloride were purchased from Sigma. The reagents were dissolved in DMSO to obtain the desired concentrations and stored at −20 °C. Different inhibitors were used for 1 h before treatment was administered.
Recombinant Expression of BrZ7 and Antiserum Preparation
The open reading frame of BrZ7 was inserted into the pET30a(+) vector. Recombinant pET30a-BrZ7 was transformed into Escherichia coli BL21 (DE3). pET30a-BrZ7 plasmid-transfected E. coli was cultured in a Luria-Bertani medium (1.0% tryptone, 0.5% yeast extract, and 1.0% NaCl). At A600 = 0.5, E. coli was exposed to 0.4 mm isopropyl-β-d-thiogalactopyranoside. After 4 h, the cells were harvested and sonicated. Recombinant BrZ7 protein was purified with His-Bind resin (Ni2+-resin; Novagen, Darmstadt, Germany). The purified recombinant BrZ7 protein (200 μg) was mixed with the same volume of Freund's complete adjuvant (Sigma) and then injected into a rabbit. After 3 weeks, 500 μg of protein was mixed with the same volume of Freund's incomplete adjuvant. The serum was collected after 2 weeks, and the specificity of the antiserum was analyzed by Western blot. The secondary antibody used in this study was alkaline phosphatase goat anti-rabbit IgG (H+L; Zhongshan, Beijing, China).
RNA Interference in Larvae
A fragment containing a 684-bp gene-specific region with T7 promoter sequences on both ends was amplified with primers to synthesize BrZ7 double-stranded RNA (dsRNA) by using the MEGAscriptTM RNA interference (RNAi) kit (Ambion, Austin TX) according to the manufacturer's instructions. The PCR primers are listed in Table 1. Approximately 1 μg of dsBrZ7 was injected into the sixth instar 72 h larvae (6th-72 h). Controls were treated with the same volume of the green fluorescent protein (GFP) dsRNA (dsGFP). The knockdown phenotypes were examined after 4 days. In the experiments, the number of animals per treatment was 60 larvae. The experiments were conducted in triplicate with independent experimental samples.
TABLE 1.
Primer sequences for RNAi and qRT-PCR
| Primers | Nucleotide sequence (5′ → 3′) |
|---|---|
| BrZ7T7F1 | gcgtaatacgactcactataggcagtcaaatcatgcagccat |
| BrZ7T7R1 | gcgtaatacgactcactatagggaaaacctaatgtaacaaa |
| HHR3QRTF | aagggtttcttcaggcgatc |
| HHR3QRTR | gttggtatttgcgtgtgcttc |
| E74AQRTF | tgcaggaccgcgagtact |
| E74AQRTR | gctggtagtagtagcgca |
| ProdeathQRTF | atgccgggacctgatgta |
| ProdeathQRTR | tcatgatatcttattcaa |
| Caspase-1QRTF | cattcacagcctaaagtctcgtaccgg |
| Caspase-1QRTR | cattcccaactcgccgtgagttaacac |
| Kr-h1QRTF | ggctaacgctgtccactc |
| Kr-h1QRTR | atcgctcgccgtaatg |
| CalQRTF | atgggcgactatcgtgcg |
| CalQRTR | ttacatctgtcgcctggtg |
| RBPQRTF | aggaagcacaggaagagga |
| RBPQRTR | aagcaccagtccacggaaa |
| BrZ7QRTF | ggtgactgtccttactgcggcat |
| BrZ7QRTR | ttaattcctttgaccatgact |
| USP1QRTF | ggtcctgacagcaatgtt |
| USP1QRTR | ttccagctccagctgactgaag |
| Met1QRTF | tatccaagccactcacagcg |
| Met1QRTR | tccgccgttgtttttctgc |
| BrZ7OVF | tactcagagctcatggctgatcaattctgt |
| BrZ7OVR | tactcactgcagattcctttgaccatgact |
| GFPOVF | tactcaggtaccatgagcaagggcgaggaactg |
| GFPOVR | tactcagcggccgccttgtacagctcgtccatgcc |
| β-ActinF | cctggtattctgaccgtatgc |
| β-ActinR | ctgttggaaggtggagagggaa |
| CalPF | acatcatcaaaccttaattat |
| CalPR | aactagccttgttagaggccc |
Protein Preparation
An epidermal cell line of H. armigera (HaEpi) was incubated at 27 °C in a 6-well plate on Grace's insect medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen). At a density of 2 × 106, the cells were exposed to 1 μm 20E or JH III. Proteins were isolated after 0, 5, 15, 30, 60, and 180 min of induction by hormones for Western blot analysis.
20E (storage concentration = 20 mm) or JH III (38 mm) was dissolved in DMSO and diluted to 100 ng/μl in phosphate-buffered saline (PBS; 140.0 mm NaCl, 2.7 mm KCl, 10.0 mm Na2HPO4, and 1.8 mm KH2PO4). Each larva at 6th-72 h was injected with 500 ng of 20E or JH III. The control group received the same volume of diluted DMSO. For Western blot analysis, the proteins were extracted from the fat body at 0, 6, 12, and 24 h after each hormone was injected. A total of 60 larvae/treatment were used, and the experiments were conducted in triplicate with independent experimental samples.
RNAi in HaEpi Cells
Transient transfection was performed using RNAfectin reagent (Tiangen, Beijing, China) according to the manufacturer's instructions. HaEpi cells were seeded into a 6-well plate for 2 days before transfection. These cells were cultured in 1 ml of Grace's medium with dsRNA and RNAfectin reagent but without FBS. The final concentrations of the dsRNA and RNAfectin transfection reagent were 2 and 4 μg/ml, respectively. After 12 h, the cells were refed in a fresh medium with FBS containing JH III with a final concentration of 1 μm. The control group was treated with equivalent amounts of DMSO. After 6 h of growth, RNA was isolated, and the concentrations were determined using a spectrophotometer. The first-strand cDNA was synthesized using a Moloney murine leukemia virus reverse transcriptase (BioTeke Corp., Beijing, China). Quantitative real-time reverse-transcriptase PCR (qRT-PCR) was performed using 2× SYBR RT-PCR premixture (BioTeke Corp., Beijing, China) with a CFX96 TM real-time system (Bio-Rad). The relative expression levels of the genes of interest were quantified, with H. armigera β-actin expression levels used as internal controls according to 2−ΔΔCT [ΔΔCt = (target gene Ct − β-actin Ct) in experiment − (target gene Ct − β-actin Ct) in control]. The qRT-PCR primers used in this study are listed in Table 1.
Vector Construction and Overexpression in HaEpi Cells
Appropriate DNA sequences encoding BrZ7 and different truncated mutants (BrZ7Δ1–100, BrZ7Δ101–200, BrZ7Δ201–300, BrZ7Δ301–400, and BrZ7Δ401–477) were inserted into the pIEx-4-RFP-His plasmid (with a C-terminal red fluorescent protein tag) or the pIEx-4-His plasmid (without an RFP tag). The inserts were verified by DNA sequencing, and the correct expression was confirmed by Western blot with anti-His antibody. HaEpi cells were incubated in a 24-well tissue culture plate containing 500 μl of Grace's medium with 10% FBS at a density of 70–90%. Before transfection was performed, cells were preincubated in Grace's medium for 1 h. Afterward, 2 μg of DNA with 2 μg of DNAfectin transfection reagent (Tiangen, Beijing, China) was suspended in 50 μl of Grace's medium, incubated for 20 min, and added to the medium. After 12 h, the cells were refed in Grace's medium containing 10% FBS. After 48 h, the cells were fixed in freshly prepared 4% paraformaldehyde solution at room temperature for 20 min and then washed with PBS. Nuclei were stained with 1 μg/ml DAPI at room temperature. The negative control group was transfected with the same amount of the pIEx-4-RFP-His plasmids. Fluorescence signal was visualized using an Olympus BX51 fluorescence microscope (Tokyo, Japan).
Protein Phosphorylation Assay
Proteins were extracted and analyzed by SDS-PAGE using 7.5% gel. The λ-phosphatase (Millipore, Temecula, CA) was used according to the manufacturer's specifications. The levels of protein phosphorylation were detected by the Phosphoprotein Phosphate Estimation Assay Kit purchased from Sangon Co. (Shanghai, China) according to the manufacturer's instructions.
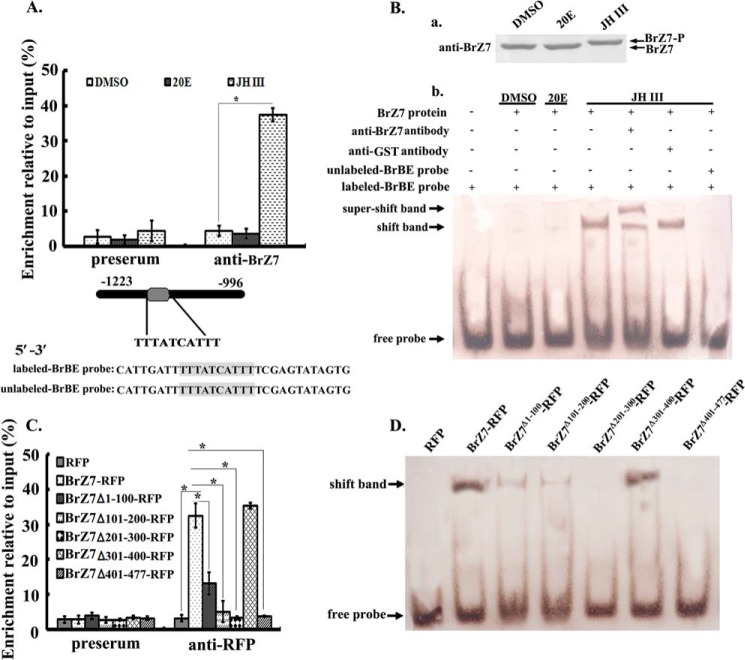
Chromatin Immunoprecipitation (ChIP)
The HaEpi cells with a density of 2 × 106 were treated with 1 μm JH III or 20E, and the control group received the same volume of DMSO. After 3 h, the cells were cross-linked using 0.5% formaldehyde at 37 °C for 10 min and then incubated at 0.125 m glycine at room temperature for 10 min to stop the reaction. The cells were washed twice with ice-cold 1× PBS and harvested. Afterward, the cells were resuspended in SDS-lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.1) and sonicated to yield average DNA fragments with a length of 200–1000 bp. The effect of chromatin sonication was determined using 0.8% agarose gel. After centrifugation was performed, the lysates were precleared with protein A resin at 4 °C for 1 h and then incubated with anti-BrZ7 antibody at 4 °C overnight. The input is the amount of chromatin DNA used for immunoprecipitation. Preserum was used as negative control treatment. Immunoprecipitated protein and DNA complexes were incubated with protein A resin at 4 °C. After 2 h, the complexes were washed once with low-salt buffer (0.1% SDS, 1.0% Triton X-100, 2 mm EDTA, 200 mm Tris-HCl, pH 8.0, 150 mm NaCl), high-salt wash buffer (0.1% SDS, 1.0% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.0, 500 mm NaCl), and LiCl wash buffer (10 mm Tris-HCl, pH 8.1, 0.25 m LiCl, 1 mm EDTA, 1% Nonidet P-40, 1% deoxycholate); these complexes were then washed twice with TE buffer (10 mm Tris-HCl, pH 8.1, 1 mm EDTA). The bound proteins were eluted using elution buffer (1% SDS, 0.1 m NaHCO3). DNA-protein cross-links were reversed at 65 °C overnight and then treated with RNase and proteinase K. DNA was purified by ethanol precipitation and then with phenol/chloroform. The DNA purified by ChIP was analyzed by qRT-PCR (CalPF/CalPR primers are listed in Table 1). Enrichment relative to input was calculated using the equation, enrichment (%) = (antibody − preserum)/input × 100%.
Electrophoretic Mobility Shift Assay (EMSA)
The 5′-regulatory region of Cal was cloned using the genome walker method. The genomic DNA was isolated with the MagExtractor genomic DNA purification kit (TOYOBO, Osaka, Japan). The cells were incubated with 1 μm 20E or JH III, and the control was treated with the same volume of DMSO. After 6 h, the cells were lysed with radioimmune precipitation assay lysis buffer (50 mm Tris-HCl, 150 mm NaCl, 1.0% Triton X-100, and 0.1% SDS). BrZ7 protein was purified using anti-BrZ7 antibody- CNBr-activated Sepharose 4B (60 mg; Amersham Biosciences); the purified protein levels were analyzed by Western blot. The probes used in the experiment were synthesized by Sangon Co. Approximately 5 μg (5 μl) of purified proteins in a binding buffer (Beyotime Institute of Biotechnology, Shanghai, China) was incubated with 100 fmol of digoxigenin-labeled probe. In the supershift experiment, 1 μl of rabbit BrZ7 antibody was added to the proteins and incubated in an ice bath for 10 min. The nonspecific anti-GST antibody was used as negative control treatment. In competition experiments, a 100-fold excess of unlabeled probe was preincubated with the purified proteins for 10 min before digoxigenin-labeled probe was added for another 20 min at room temperature. The reaction ran on a 6.5% polyacrylamide, 0.5× TBE gel at 80 V; afterward, the sample was transferred onto a nylon membrane (IMMOBILON-NY+, Millipore, Milford, MA). The membrane was blocked for 30 min and then incubated with anti-digoxigenin phosphatase antibody (1:10,000 in blocking solution; Roche Applied Science). After 1 h, the signal was visualized with 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium chloride.
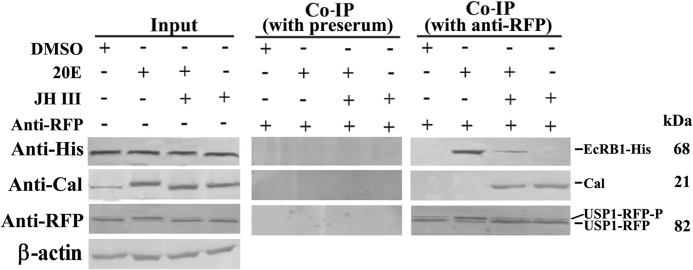
Co-immunoprecipitation (Co-IP)
The plasmids pIEx-4-EcRB1-His and pIEx-4-USP1-RFP-His were transiently co-transfected in HaEpi cells. After 48 h, the cells were incubated with 1 μm 20E, JH III, or 20E+JH III. After 6 h, the cells were collected and lysed in 400 μl of radioimmune precipitation assay lysis buffer. The lysate was incubated in an ice bath with constant rotation for 30 min and centrifuged at 12,000 × g at 4 °C for 15 min. The supernatant was pretreated with protein A resin for 30 min to remove nonspecific binding. The lysate was incubated with anti-RFP antibody at 4 °C with constant rotation overnight. The same amount of preserum was used as negative control treatment. The mixture was cultured with protein A at 4 °C for another 2 h. Afterward, the supernatant was discarded; protein A resin was washed with radioimmune precipitation assay buffer three times. The samples containing loading buffer and SDS were heated for 5 min in a boiling water bath. The proteins were detected by Western blot with rabbit anti-RFP, anti-His, or anti-Cal antibodies on immunoblots.
RESULTS
BrZ7 Is a Newly Identified Broad Protein
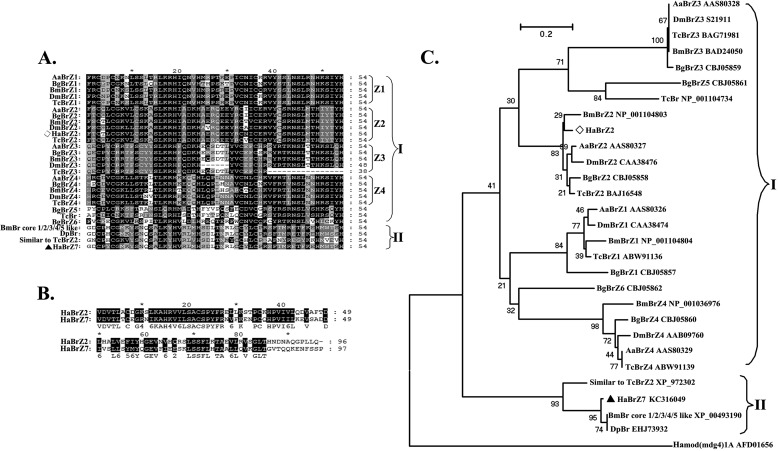
A 3,291-bp full-length cDNA was obtained from the transcriptome of HaEpi and contains a 120-bp 5′-untranslated region (UTR), a 1,434-bp open reading frame (ORF), and a 1,737-bp 3′-UTR. The ORF encodes 477 amino acids, including an N-terminal BTB domain and the two C-terminal zinc finger domains (Fig. 1). BrZ7 shares identities with the “broad-complex core protein isoforms 1/2/3/4/5-like” from B. mori, “putative broad” from Danaus plexippus, and “similar to broad-complex Z2” from T. castaneum at 78, 70, and 66%, respectively. However, BrZ7 shares less identity with BrZ2 from H. armigera, D. melanogaster, B. mori, T. castaneum, and Aedes aegypti at 23, 22.63, 26.57, 23.51, and 22.69%, respectively. The zinc finger domains of BrZ7 are different from the isoforms (Z1–Z4) from B. mori and other insects (Fig. 2A), and the BTB domain of BrZ7 exhibits low identity (48%) with the BTB domain of HaBrZ2 (Fig. 2B). Phylogenetic tree analysis indicated that all of the Br proteins can be classified into two groups (Fig. 2C). Group I consists of different BrZ1–Z4 isoforms from different insects; group II comprises BrZ7 and other Br proteins.
FIGURE 1.
Full-length cDNA sequence and predicted amino acid sequence of BrZ7. The ORF encodes a 477-amino acid protein with a predicted molecular mass of 52.6 kDa. The underlined sequence corresponds to the BTB domain (aa 30–125); the dotted line shows the two zinc finger domains (aa 420–443 and 450–473) by SMART EMBL analysis. The sequence for RNAi is shaded.
FIGURE 2.
The alignments and phylogenetic analysis of Br. A, alignment of zinc finger regions between BrZ7 from H. armigera and Br isoforms from other insects. B, alignment of BTB domain between BrZ2 and BrZ7 from H. armigera. Aa, Aedes aegypti; Bg, Blattella germanica; Bm, B. mori; Dm, D. melanogaster; Tc, T. castaneum; Dp, D. plexippus; Ha, H. armigera. BmBr core 1/2/3/4/5 like, broad-complex core protein isoforms 1/2/3/4/5-like from B. mori. C, phylogenetic tree analysis of BrZ7 and BrZ2 from H. armigera with different Br isoforms from other insects. Hamod(mdg4)1A, a modifier of Mod protein from H. armigera containing a BTB domain and a zinc finger domain (FLYWCH), is an outgroup member.
BrZ7 Is Critical for Metamorphosis
The 6th-72 h larvae were injected with BrZ7 dsRNA that was designed at the C terminus (Fig. 1) to investigate the function of BrZ7 in H. armigera development. After 4 days of RNAi with dsGFP injection, 98% of the control larvae (58 of 60, n = 60) molted into normal pupae, except a few larvae (2 of 60) that died because of injection injury. However, only 8% of the larvae (5 of 60, n = 60) injected with dsBrZ7 molted into normal pupae, whereas 65% of the larvae (39 of 60) developed into abnormal pupae and died soon after ecdysis. In addition, ∼27% of the larvae (16 of 60) were blocked at the prepupal stage and died 2 days later (Fig. 3, A and B). The expression levels of the 20E response genes prodeath-S/TK, caspase-1, and PP6 were decreased (Fig. 3C) when BrZ7 was knocked down. These results confirm that BrZ7 regulates the expression of 20E response gene and is required for metamorphosis.
FIGURE 3.
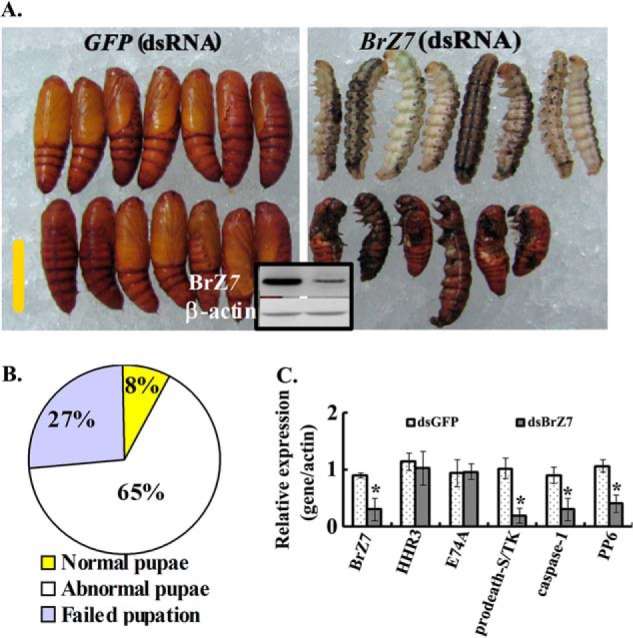
BrZ7 silencing inhibited the metamorphosis and 20E response gene expression. The 6th-72 h larvae were injected with 1 μg of dsBrZ7. The control larvae received the equivalent volume of dsGFP. Sixty larvae were used in every treatment, and the experiments were conducted in triplicate with independent experimental samples. A, phenotypes after dsGFP or dsBrZ7 injection. The black square shows that BrZ7 was knocked down in the fat body of 6th-96 h, as analyzed by Western blot with 7.5% SDS-PAGE. β-Actin was used as an internal control treatment. Scale bar, 1 cm. B, percentage of different phenotypes after dsBrZ7 was injected. C, relative expression of the 20E-induced genes analyzed by qRT-PCR. RNA was isolated from the fat body of the 6th-96 h larvae. The results were based on the 2−ΔΔCT method in the presence of normalized β-actin gene. Asterisks, significant differences (Student's t test; *, p < 0.05).
BrZ7 Is Regulated on Its Expression Level and Phosphorylation Status during Development
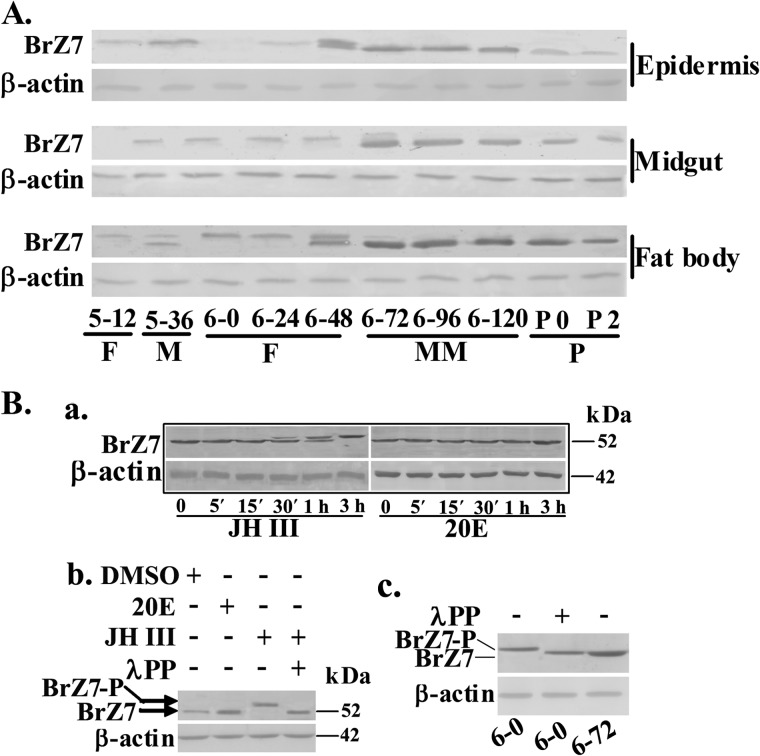
To determine the hormonal regulatory mechanism of BrZ7, we prepared the antibody as described under “Experimental Procedures” and analyzed the expression profiles of BrZ7 during development by Western blot. BrZ7 expression was low during the larval growth stage from fifth instar 12 h to 6th-48 h. However, the expression was significantly increased during the metamorphic stage from 6th-72 h to pupae in the epidermis, midgut, and fat body (Fig. 4A). Unexpectedly, BrZ7 yielded a high molecular mass during the larval growth stage and a low molecular mass at the metamorphic stage. These results show that BrZ7 is highly expressed during metamorphosis and is probably modified post-translationally during development.
FIGURE 4.
JH-induced BrZ7 phosphorylation analyzed by Western blot. The SDS-PAGE gel was 7.5%. A, the expression profiles of BrZ7 during development were detected using anti-BrZ7 antibody. 5–12 to 6–120 indicate the larval developmental stages from fifth instar 12 h to 6th-120 h, respectively. P0–P2, days of pupal development. F, feeding; M, molting; MM, metamorphic molting; P, pupae. B, a, cells were incubated with 1 μm 20E or JH III for 5, 15, 30, 60, and 180 min, respectively. Proteins were then extracted for Western blot analysis with anti-BrZ7 antibody. b, cells were treated with 1 μm 20E or JH III for 3 h; proteins were isolated and incubated with λ-phosphatase at 5 μm for 30 min. In c, 6–0 indicates the proteins from the fat body of 6th instar 0 h larvae, and 6–72 indicates the proteins from 6th instar 72 h larvae, analyzed by Western blot. BrZ7-P, phosphorylated BrZ7.
To investigate the possible post-translational modification, we incubated HaEpi cells with 20E or JH III and analyzed the BrZ7 protein. Although lepidopteran insect JH II is a native hormone (42), H. armigera (43) and B. mori (44) respond to JH III. Thus, JH III was used in the study. Western blot results showed that JH III treatment caused the loss of the lower band and increased the intensity of the upper band from 30 min to 3 h; by comparison, 20E treatment only enhanced the intensity of the lower band after 3 h (Fig. 4B, a). After the cells were treated with λ-phosphatase, the upper protein band disappeared (Fig. 4B, b). The fat body protein was extracted from 6th-0 h. After being treated with λ-phosphatase, the molecular mass was decreased to the same level as the protein from 6th-72 h fat body (Fig. 4B, c). These results suggest that the change in molecular weight is due to the rapid phosphorylation induced by JH III.
JH III Regulates BrZ7 Phosphorylation through a GPCR-, PLC-, and PKC-triggered Pathway
BrZ7-His was overexpressed in HaEpi cells by using pIEx-4-BrZ7-His plasmid to reveal the pathway through which JH induces BrZ7 phosphorylation. The overexpressed BrZ7-His appeared both non-phosphorylated and phosphorylated at ∼58 kDa in the DMSO-treated control cells by anti-His antibody detection. JH III treatment converted the non-phosphorylated BrZ7 to phosphorylated BrZ7. The GPCR inhibitor suramin, PLC inhibitor U73122, and PKC inhibitor chelerythrine chloride repressed JH III-induced BrZ7 phosphorylation (Fig. 5A). These results indicate that JH III regulates BrZ7 phosphorylation via a GPCR-, PLC-, and PKC-triggered pathway.
FIGURE 5.
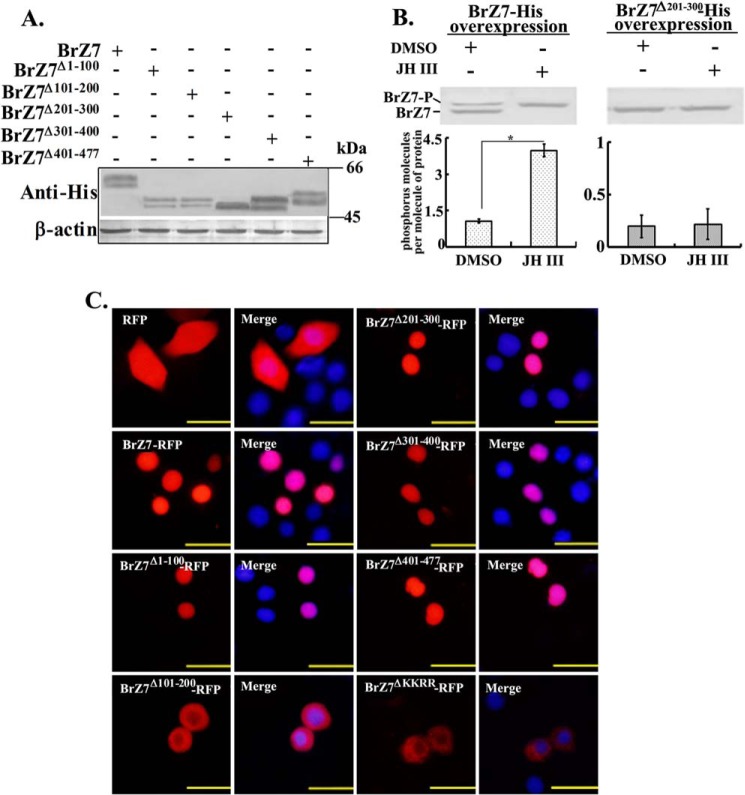
JH III regulated BrZ7 phosphorylation through a GPCR-, PLC-, and PKC-triggered pathway. A, the inhibitors of GPCR, PLC, and PKC suppressed JH III-induced BrZ7 phosphorylation. Cells were transfected with pIEx-4-BrZ7-His for 48 h, pretreated with different inhibitors for 1 h, and incubated with 1 μm JH III for 3 h. Control cells were transfected with pIEx-4-RFP-His. Western blot was performed on 7.5% SDS-PAGE gel with anti-His antibody. B, predicted putative phosphorylation sites of BrZ7 are underlined. C, mutant plasmids were constructed. Boxed amino acids correspond to the mutated amino acids. D, the phosphorylation of BrZ7 was not affected by the overexpression of different mutant plasmids using pIEx-4-His in HaEpi cells as revealed by Western blot.
The phosphorylation sites on BrZ7 were investigated by constructing various mutations fused with His tag. Approximately 10 putative and high-percentage (>90%) phosphorylation sites (Ser-161, Ser-162, Ser-164, Ser-265, Ser-270, Tyr-274, Ser-352, Ser-355, Thr-460, and Thr-464) on BrZ7 were predicted by NetphosK version 2.0 (Fig. 5B). Unfortunately, individual mutation of these sites did not affect BrZ7 phosphorylation (Fig. 5, C and D). Thus, five truncated mutants (BrZ7Δ1–100, BrZ7Δ101–200, BrZ7Δ201–300, BrZ7Δ301–400, and BrZ7Δ401–477) were constructed by deleting the related numbered 100 amino acids (aa) once and were overexpressed in HaEpi cells by using pIEx-4-His plasmid. JH III-induced phosphorylation levels were obviously decreased in the mutant BrZ7Δ201–300 when aa 201–300 were deleted (Fig. 6A). The level of JH III-induced BrZ7 phosphorylation was determined as 3 phosphates/molecule of BrZ7 protein by a phosphoprotein phosphate estimation assay kit. After deletion of aa 201–300, the phosphorylation of BrZ7 by JH III induction was not detected (Fig. 6B). These findings confirm that the putative phosphorylation sites are distributed in the aa 201–300 region.
FIGURE 6.
Analysis of the phosphorylation sites and subcellular location of BrZ7. A, cells were transfected with various plasmids expressing different BrZ7 mutants fused with His tag. Proteins were extracted, and Western blot was performed with anti-His antibody using 7.5% SDS-polyacrylamide gel. B, number of molecules of phosphorus per molecule of BrZ7-His or BrZ7Δ201–300-His by DMSO or JH III treatments analyzed by a phosphoprotein phosphate estimation assay kit. The cells were transfected with pIEx-4-BrZ7-His or pIEx-4-BrZ7Δ201–300-His plasmids in HaEpi cells. After 48 h of overexpression, the cells were incubated with DMSO or 1 μm JH III for 3 h, and the proteins were purified by His-Bind resin for the measurement. About 50 μg of protein was used per assay. Statistical significance (*, p < 0.05) was based on three biologically independent repeats and analyzed by the Student's t test. C, BrZ7 and various truncated BrZ7-RFP mutants were overexpressed in HaEpi cells, and pIEx-4-RFP-transfected plasmid was used as a control sample. The nuclei were stained with DAPI. Fluorescence signal was visualized using an Olympus BX51 fluorescence microscope. Scale bar, 25 μm. Error bars, S.E.
To determine the subcellular location where BrZ7 phosphorylation occurs, we observed the location of BrZ7 and various truncated mutants overexpressed in HaEpi cells by using pIEx-4-RFP-His plasmid. RFP was located in the whole cell. The BrZ7-RFP and BrZ7 mutants BrZ7Δ1–100-RFP, BrZ7Δ201–300-RFP, BrZ7Δ301–400-RFP, and BrZ7Δ401–477-RFP were located in the nucleus regardless of the occurrence of phosphorylation. However, mutant BrZ7Δ101–200-RFP (aa 101–200 truncated mutant), which lost the KKRR motif (aa 157–160), was localized in the cytoplasm (Fig. 6C), and this mutant was phosphorylated (Fig. 6A), suggesting that the phosphorylation of BrZ7 occurs in the cytoplasm. The KKRR motif (aa 157–160) deletion mutant BrZ7ΔKKRR-RFP was located in the cytoplasm (Fig. 6C), suggesting that the KKRR motif determines the nuclear location.
Phosphorylated BrZ7 Is Required for Cal Transcription in JH Pathway
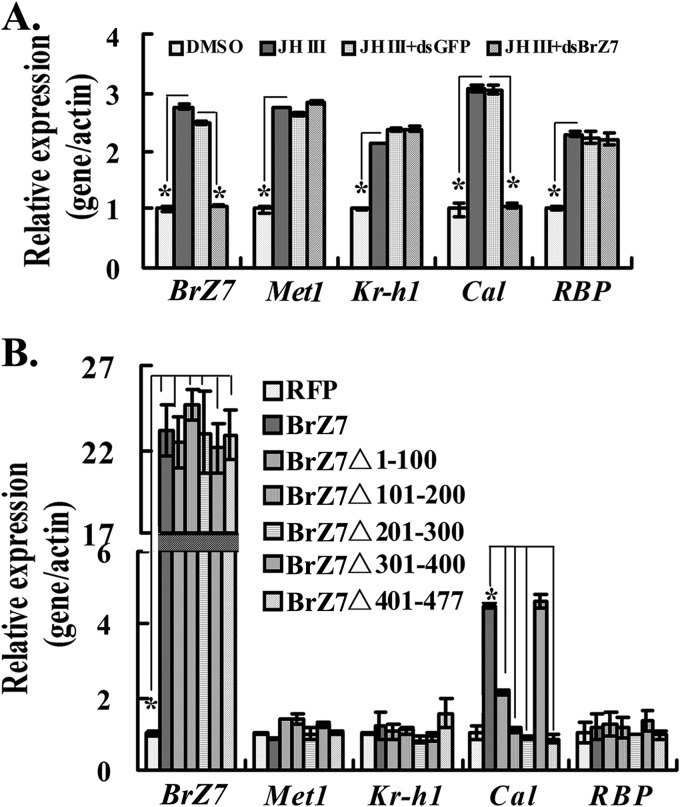
To reveal the function of BrZ7 in the JH pathway, we knocked down BrZ7 in HaEpi cells. The knockdown of BrZ7 did not affect the JH III-induced expression of Met1, Kr-h1, and RBP. However, the silencing of BrZ7 decreased the mRNA level of Cal by ∼2-fold compared with the dsGFP-treated control (Fig. 7A). These results reveal that BrZ7 is involved in JH III-induced Cal expression. This conclusion was confirmed by the overexpression of the truncated mutants of BrZ7 in the HaEpi cell line. The Cal transcript was up-regulated by BrZ7 overexpression but not up-regulated by overexpression of the following mutants: BrZ7Δ1–100 (BTB domain for protein interaction), BrZ7Δ101–200, (BTB domain for protein interaction), BrZ7Δ201–300 (the putative phosphorylational region), and BrZ7Δ401–477 (zinc finger motif for DNA binding) (Fig. 7B). These results suggest that these domains are required for JH-induced Cal expression.
FIGURE 7.
qRT-PCR analysis of the function of BrZ7 in JH pathway in HaEpi cells. A, knockdown of BrZ7. Cells were transfected with dsBrZ7 and dsGFP. After 24 h, cells were subjected to JH III for 6 h. RNA was isolated for qRT-PCR. B, overexpression of BrZ7. Cells were transfected with different plasmids, including BrZ7 and its mutants, for 48 h and induced with 1 μm JH III for 6 h. RNA was isolated for qRT-PCR. Results were based on the 2−ΔΔCT method in the presence of the normalized β-actin gene. Asterisks indicate significant differences (Student's t test; *, p < 0.05). Error bars, S.E.
We performed ChIP with anti-BrZ7 antibody to determine the mechanism by which the transcription factor BrZ7 regulates Cal expression in the JH pathway. We cloned the Cal 5′-regulatory region and found one putative BrZ2-binding element (BrBE, TTTATCATTT), which changed CT in D. melanogaster salivary gland secretion protein gene Sgs4 (TTTACTATTT) (8) to TC (Fig. 8). A small amount of the DNA product was amplified by qRT-PCR from the precipitate obtained from the preserum negative control sample. By contrast, the DNA fragment containing BrBE was amplified from the precipitate by anti-BrZ7 antibody from the JH III-induced cells but not from DMSO- or 20E-treated cells (Fig. 9A). These results show that BrZ7 binds to the Cal 5′-regulatory region under JH-induced conditions.
FIGURE 8.
5′-regulatory region of Cal gene. Double-underlined sequence corresponds to the BrZ7 binding sequence BrBE. The sequence used to detect the BrBE fragment in the ChIP assay is shaded.
FIGURE 9.
ChIP and EMSA assays to determine BrZ7 binding to BrBE. A, ChIP assays by qRT-PCR detecting the BrBE (TTTATCATTT) fragment from the precipitates by anti-BrZ7 antibody. Cells were immunoprecipitated with anti-BrZ7 antibody after the cells were treated with 1 μm 20E or JH III for 3 h. Preserum was used as a negative control of nonspecific binding. Chromatin sample before immunoprecipitate was used as an input and positive control. Enrichment relative to input was calculated as the percentage of chromatin input according to various treatments. B, a, Western blots showing the expression level and the phosphorylation status of the purified BrZ7 from DMSO-, 20E-, or JH III-treated cells (1 μm 20E or JH III for 3 h). b, EMSA detected the shift band of BrZ7 binding to digoxigenin-labeled BrBE and the supershift band produced by anti-BrZ7 antibody. Anti-GST antibody was used as a nonspecific antibody control. Unlabeled BrBE probe competed in the shift band. BrZ7 protein was purified with CNBr-activated Sepharose 4B resin bound with anti-BrZ7 antibody after 1 μm 20E or JH III incubation for 3 h. C, ChIP assays by qRT-PCR revealed the effect of BrZ7 mutants on binding to the BrBE fragment. Cells were transfected with various plasmids expressing different BrZ7 proteins for 48 h and incubated with 1 μm JH III for 3 h. pIEx-4-RFP was used as a negative control sample. Preserum was used as a negative control sample of nonspecific binding. D, EMSA analysis of the effect of BrZ7 mutants on binding to BrBE. The different BrZ7 proteins were purified with CNBr-activated Sepharose 4B resin bound with anti-RFP antibody after the treatments as in C. Error bars, S.E.
EMSA was performed using a digoxigenin-labeled BrBE probe and purified endogenous BrZ7 protein to confirm whether or not BrZ7 directly binds to BrBE. In DMSO- or 20E-induced conditions, BrZ7 was not phosphorylated, and a few shift bands were detected. In JH III-treated cells, BrZ7 was phosphorylated, and a distinct shift band was detected. The added anti-BrZ7 antibody produced a supershift band; by comparison, anti-GST antibody (nonspecific antibody control) did not affect the complex. Binding was competed by an unlabeled BrBE probe (Fig. 9B). The results suggest that BrZ7 directly binds to BrBE under JH-induced conditions.
To confirm that BrZ7 phosphorylation was essential for BrZ7 binding to BrBE, we examined the binding activities of BrZ7 and the mutants with 1 μm JH III incubation. A small amount of the DNA product was detected in the precipitates produced by preserum by ChIP analysis. By contrast, the DNA containing BrBE was amplified from the precipitate by using anti-RFP antibody in BrZ7-RFP-transfected cells, compared with RFP-transfected cells. However, a small amount of the DNA was detected from the mutants, including BrZ7Δ1–100-RFP, BrZ7Δ101–200-RFP, BrZ7Δ201–300-RFP, and BrZ7Δ401–477-RFP (Fig. 9C). To confirm the results, we performed EMSA with the purified BrZ7-RFP and the mutated proteins. RFP itself could not produce a shift band, but BrZ7-RFP produced an obvious shift band. However, the mutants BrZ7Δ1–100-RFP, BrZ7Δ101–200-RFP, BrZ7Δ201–300-RFP, and BrZ7Δ401–477-RFP produced much weaker shift bands (Fig. 9D). These results reveal that the BTB domain (aa 1–200), phosphorylated sites (aa 201–300), and zinc finger motifs (aa 401–477) of BrZ7 are necessary to induce BrZ7 to bind to BrBE.
JH Inhibits Metamorphosis by Inducing BrZ7 Phosphorylation
To determine the mechanism by which JH inhibits metamorphosis, we injected JH III into the 6th-72 h larvae, at the onset of metamorphosis. Approximately 43% of the larvae (26 of 60, n = 60) showed larval-pupal intermediate characteristics and soon died, and 45% (27 of 60) molted into deficient pupae 4 days after JH III was injected (Fig. 10, A and B). In 20E-injected larvae, the non-phosphorylated BrZ7 was up-regulated; however, BrZ7 protein was increased and phosphorylated after JH III injection (Fig. 10C). 20E injection up-regulated the expressions of 20E-response genes, including HHR3, E74A, prodeath-S/TK, caspase-1, and PP6 (Fig. 10D). However, JH III injection repressed these 20E-response gene expressions but enhanced BrZ7 and Cal expression (Fig. 10E). These results suggest that exogenous JH III regulates the high expression and phosphorylation of BrZ7 and expression of Cal but suppresses 20E response gene expression.
FIGURE 10.
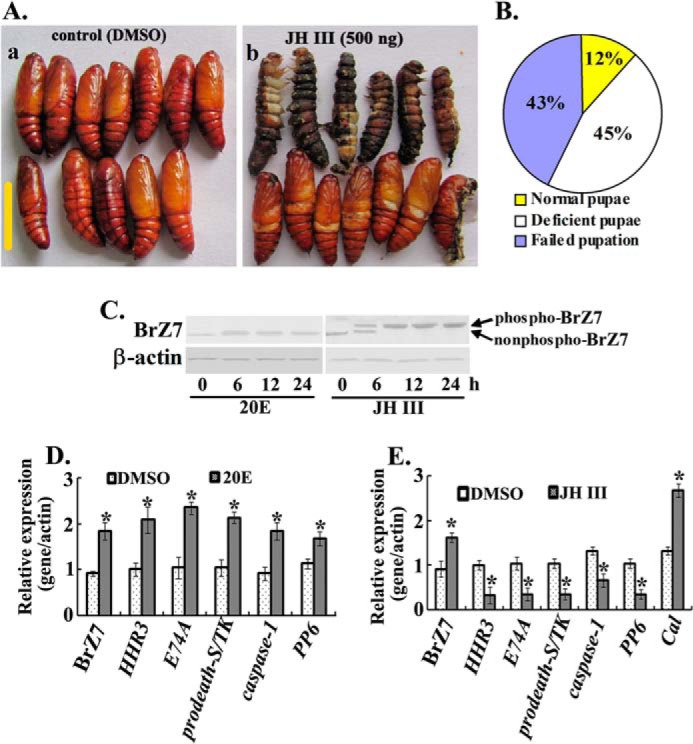
JH III injection induced BrZ7 expression and phosphorylation and inhibited metamorphosis by suppressing 20E response gene expression. A, phenotypes of DMSO or JH III injection. The 6th-72 h larvae were injected with JH III or 20E (500 ng/larva, 3 × 60 larvae). Scale bar, 1 cm. B, percentage of different phenotypes after JH III injection. C, Western blot showing the BrZ7 expression and phosphorylation by 20E or JH III injection with 7.5% SDS-polyacrylamide gel. Proteins from fat body were obtained at 0, 6, 12, and 24 h after each hormone was injected and analyzed using anti-BrZ7 antibody. D and E, the mRNA levels of various genes were analyzed by qRT-PCR. The RNA of the fat body of 6th-96 h was isolated from 20E- or JH III-injected larvae, respectively. β-actin was used as an internal control sample. Asterisks indicate significant differences (Student's t test; *, p < 0.05). Error bars, S.E.
In the 20E pathway, 20E binds to its nuclear receptor, EcR, and regulates the formation of a transcription complex to direct gene expression (5, 45). The JH analog methoprene regulates non-phosphorylated Cal interacting with USP1 to induce gene expression in the JH pathway of H. armigera (15). To determine the mechanism by which JH suppresses 20E-induced gene expression, we performed co-IP and examined the interaction of EcRB1, USP1, and Cal under hormone induction. EcRB1-His and USP1-RFP were overexpressed by co-transfecting HaEpi cells with pIEx-4-EcRB1-His and pIEx-4-USP1-RFP-His plasmids. In the input positive control cells, Cal, EcRB1-His, and USP1-RFP were detected evenly; Cal and USP1-RFP were phosphorylated induced by 20E. In the co-immunoprecipitates produced by preserum negative control, no Cal, EcRB1-His, or USP1-RFP was detected. In the co-immunoprecipitates produced by anti-RFP, EcRB1-His and USP1-RFP were detected from the 20E-treated cells, and no Cal was detected. In 20E plus JH III induction, USP1-RFP and non-phosphorylated Cal were detected; the amount of detected EcRB1-His was decreased. In JH-incubated cells, non-phosphorylated Cal and USP1-RFP were detected (Fig. 11). These results reveal that JH III regulates the interaction between non-phosphorylated Cal and USP1 to activate the JH pathway and inhibit the formation of 20E transcription complex, therefore repressing 20E response gene expression.
FIGURE 11.
Co-IP showing the interactions of different proteins. HaEpi cells were transiently co-transfected with pIEx-4-EcRB1-His and pIEx-4-USP1-RFP-His for 48 h and incubated with DMSO, 20E, 20E plus JH III, or JH III for 6 h. Input showed the protein levels of Cal, the overexpressed EcRB1-His, and USP1-RFP-His in the cells before immunoprecipitation. Preserum was used as the negative control for co-IP with anti-RFP antibody. Co-IP performed with anti-RFP antibody showed the interactions of USP1, Cal, and EcRB1. Anti-His, anti-H. armigera Cal, and anti-RFP antibodies were used to detect the corresponding proteins, respectively. USP1-RFP-P, phosphorylated USP1.
DISCUSSION
Numerous examples have been presented to demonstrate the crosstalk between 20E and JH (3). However, the molecular basis by which JH prevents 20E-induced metamorphosis remains unclear. In the research, JH induces the phosphorylation of a newly identified Broad protein, BrZ7, to block 20E-induced metamorphosis. JH III regulates BrZ7 phosphorylation via a GPCR-, PLC-, and PKC-triggered pathway. The phosphorylated BrZ7 directly regulates Cal expression in the JH pathway. JH-induced Cal interacts with USP1 to suppress 20E response gene transcription and metamorphosis. These findings elucidated one of the mechanisms by which JH inhibits metamorphosis.
Broad Protein BrZ7 Is Involved in 20E-mediated Metamorphosis
In this research, a Broad protein, BrZ7, was identified in the presence of an N-terminal BTB domain and C-terminal zinc finger domains from H. armigera. Different BrZ1–Z4 isoforms share a common BTB domain but different zinc finger domain in a particular species (28); BTB domains are highly conserved in different insects (90–97% identity) (34). The zinc finger domain in an isoform of BrZ1–Z4 is highly conserved, respectively, in different insects (90–97%) (46). However, the zinc finger domains of BrZ7 are different from the isoforms (Z1–Z4) from B. mori, and the BTB domain of BrZ7 exhibits low identity (48%) with the BTB domain of H. armigera BrZ2. By contrast, BrZ7 exhibits higher similarities to other Br proteins, including “broad-complex core protein isoforms 1/2/3/4/5-like” from B. mori (78%), “putative broad” from D. plexippus (70%), and “similar to broad-complex Z2” from T. castaneum (66%). The phylogenetic tree shows that BrZ7 belongs to the group II Broad protein. Therefore, we named it as BrZ7, which was referred to as BrZ2 or Br in our previous studies (15, 43, 45, 47). We also found that BrZ7 is necessary to induce 20E response gene expression and 20E-mediated metamorphosis in H. armigera. Although the amino acid sequence of BrZ7 is different from the Br isoforms of other insects (group I Broad protein), BrZ7 performs a similar function in regulating metamorphosis.
JH Regulates Cal Transcription via Phosphorylated BrZ7
We found that JH induced the rapid phosphorylation of BrZ7 via a GPCR-, PLC-, and PKC-triggered pathway. The GPCR-, PLC-, and PKC-triggered axis is a classical pathway involved in membrane receptors (48). These findings suggested the presence of a JH non-genomic pathway and presented a good readout for further studies on the non-genomic JH pathway. We also revealed that the aa 201–300 region of BrZ7 is the putatively phosphorylated region, and phosphorylation occurs in the cytoplasm. Approximately 9 possible phosphorylation sites (>80%) were predicted in the region of aa 201–300, and there might be three phosphorylation sites in aa 201–300. However, the single site mutations of the high-percentage phosphorylation sites (Ser-265, 98%; Ser-270, 99%) had no effect on the phosphorylation of BrZ7. Therefore, further investigations are needed to identify the exact phosphorylation sites.
In D. melanogaster, the BrZ2-binding element is identified as TTTACTATTT in the promoter of the Sgs4 glue gene in the salivary gland (8). We obtained a BrZ2-binding element (BrBE) (TTTATCATTT) in the upstream region of Cal and found that phosphorylated BrZ7 directly binds to BrBE to regulate Cal expression under JH induction conditions. Cal is an actin-binding domain (Chd)-containing protein; this protein is implicated in the 20E and JH pathways (15). In D. melanogaster, a 21-kDa calponin-like protein (Chd64) mediates the cross-talk between 20E and JH pathways by interacting with DmEcR, DmUSP, or DmMet (49). Our findings revealed the direct target gene Cal of BrZ7 in the JH pathway.
JH Inhibits 20E-induced Metamorphosis by Inducing BrZ7 Phosphorylation
Br, USP1, Met, and heat shock protein 90 (43) are involved in the 20E-JH regulatory cross-talk. In T. castaneum (13), endogenous JH inhibits metamorphosis by repressing Br expression via Met/Kr-h1; however, the application of exogenous JH at the pupal-adult transition induces Br re-expression via Met/Kr-h1 to repress metamorphosis. Br plays an important role in JH antagonizing the 20E pathway (12). In H. armigera, 20E up-regulates the expressions of non-phosphorylated BrZ7 and 20E response genes. JH III induces BrZ7 phosphorylation and blocks 20E response gene expression. These results suggest that the functions of BrZ7 differ in the 20E and JH pathways because of the altered phosphorylation status of BrZ7. Non-phosphorylated BrZ7 is involved in 20E-induced metamorphosis; phosphorylated BrZ7 functions in the JH pathway. At the onset of metamorphosis, exogenous JH III induces BrZ7 expression and phosphorylation to activate the JH pathway and to block the 20E pathway. These findings suggest a mechanism by which JH inhibits metamorphosis. However, the mechanism of the lower expression of BrZ7 during larval growth and high expression during metamorphosis is unclear.
20E regulates gene transcription by forming a transcription complex (2). In our research, exogenous JH inhibits metamorphosis; this result is consistent with that in previous studies involving other insects (25, 34). 20E regulates EcRB1 interacting with phosphorylated USP1 induced by 20E (45). In the presence of JH, the non-phosphorylated Cal interacts with non-phosphorylated USP1, and the formation of an EcR·USP1 complex is blocked. These results indicate a possible mechanism by which JH suppresses 20E response gene expression.
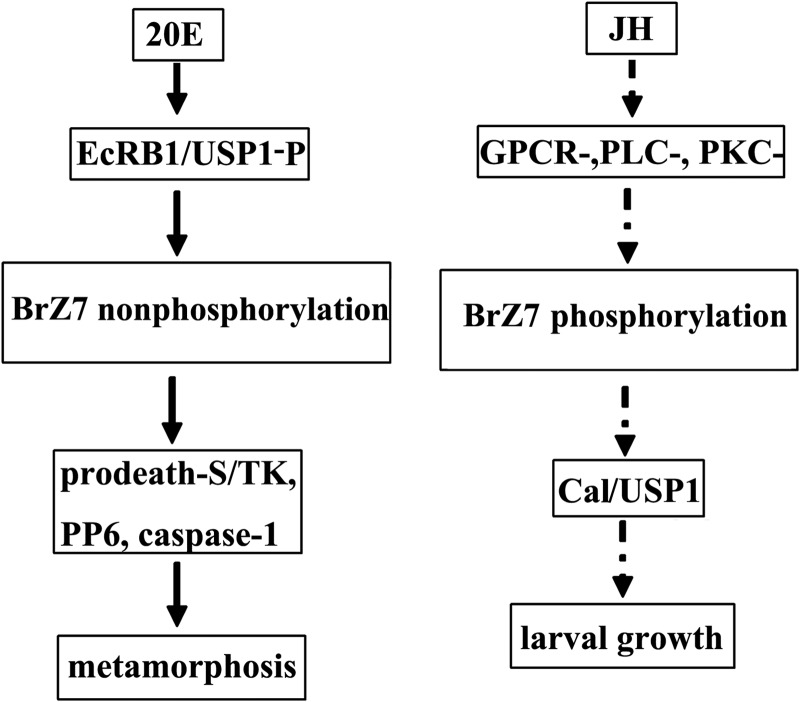
In summary, we reveal a new mechanism through which that JH inhibits the 20E pathway by regulating BrZ7 phosphorylation. During the larval growth stage, natural JH regulates the weak expression and phosphorylation of BrZ7 for JH response gene expression to direct larval growth. At the metamorphic stage, 20E regulates the high expression of BrZ7 and maintains the non-phosphorylation of BrZ7 to direct 20E-induced gene expression and metamorphosis. Exogenous JH III induces the high expression and phosphorylation of BrZ7 to promote Cal expression. JH mediates non-phosphorylated Cal to interact with non-phosphorylated USP1 and repress the formation of the 20E transcription complex EcRB1·USP1-P and block the 20E response gene expression and metamorphosis (Fig. 12).
FIGURE 12.
A summary of the mechanism by which BrZ7 participates in 20E and JH pathways. 20E induces the interaction of EcRB1 with phosphorylated-USP1, regulates BrZ7 high expression, and maintains BrZ7 non-phosphorylation to modulate 20E response gene expression for metamorphosis. JH induces BrZ7 phosphorylation via a GPCR-, PLC-, and PKC-signaling pathway. The phosphorylated BrZ7 directs Cal transcription, and the non-phosphorylated Cal interacts with non-phosphorylated USP1 for larval growth.
Acknowledgments
We thank Dr. Marek Jindra and Masako Asahina (Czech Academy of Sciences and University of South Bohemia, Czech Republic) for providing the plasmid pIEx-4.
This work was supported by National Natural Science Foundation of China Grant 31230067, National Basic Research Program of China (973 Program) Grant 2012CB114101, and Ph.D. Programs Foundation of the Ministry of Education of China Grant 20120131110025.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) KC316049.
- 20E
- 20-hydroxyecdysone
- BrZ7
- Broad isoform Z7
- JH
- juvenile hormone
- GPCR
- G-protein-coupled receptor
- PLC
- phospholipase C
- EcR
- ecdysone nuclear receptor
- USP
- ultraspiracle protein
- Met
- methoprene-tolerant
- Kr-h1
- Kruppel homolog 1
- RBP
- RNA-binding protein
- Cal
- calponin
- HaEpi
- an epidermal cell line from H. armigera
- BrBE
- Br-binding element
- co-IP
- co-immunoprecipitation
- qRT-PCR
- quantitative RT-PCR
- aa
- amino acid(s)
- 6th
- sixth instar.
REFERENCES
- 1. Riddiford L. M. (1993) Hormone receptors and the regulation of insect metamorphosis. Receptor 3, 203–209 [PubMed] [Google Scholar]
- 2. Riddiford L. M., Hiruma K., Zhou X., Nelson C. A. (2003) Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem. Mol. Biol. 33, 1327–1338 [DOI] [PubMed] [Google Scholar]
- 3. Dubrovsky E. B. (2005) Hormonal cross talk in insect development. Trends Endocrinol. Metab. 16, 6–11 [DOI] [PubMed] [Google Scholar]
- 4. Riddiford L. M. (1996) Juvenile hormone: the status of its “status quo” action. Arch. Insect Biochem. Physiol. 32, 271–286 [DOI] [PubMed] [Google Scholar]
- 5. Baehrecke E. H. (2000) Steroid regulation of programmed cell death during Drosophila development. Cell Death Differ. 7, 1057–1062 [DOI] [PubMed] [Google Scholar]
- 6. Lan Q., Hiruma K., Hu X., Jindra M., Riddiford L. M. (1999) Activation of a delayed-early gene encoding MHR3 by the ecdysone receptor heterodimer EcR-B1-USP-1 but not by EcR-B1-USP-2. Mol. Cell Biol. 19, 4897–4906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fletcher J. C., Burtis K. C., Hogness D. S., Thummel C. S. (1995) The Drosophila E74 gene is required for metamorphosis and plays a role in the polytene chromosome puffing response to ecdysone. Development 121, 1455–1465 [DOI] [PubMed] [Google Scholar]
- 8. von Kalm L., Crossgrove K., Von Seggern D., Guild G. M., Beckendorf S. K. (1994) The Broad-Complex directly controls a tissue-specific response to the steroid hormone ecdysone at the onset of Drosophila metamorphosis. EMBO J. 13, 3505–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu C. Y., Liu W., Zhao W. L., Wang J. X., Zhao X. F. (2013) Upregulation of the expression of prodeath serine/threonine protein kinase for programmed cell death by steroid hormone 20-hydroxyecdysone. Apoptosis 18, 171–187 [DOI] [PubMed] [Google Scholar]
- 10. Yang D., Chai L., Wang J., Zhao X. (2008) Molecular cloning and characterization of Hearm caspase-1 from Helicoverpa armigera. Mol. Biol. Rep. 35, 405–412 [DOI] [PubMed] [Google Scholar]
- 11. Wang C. X., Zheng W. W., Liu P. C., Wang J. X., Zhao X. F. (2012) The steroid hormone 20-hydroxyecdysone upregulated the protein phosphatase 6 for the programmed cell death in the insect midgut. Amino Acids 43, 963–971 [DOI] [PubMed] [Google Scholar]
- 12. Jindra M., Palli S. R., Riddiford L. M. (2013) The juvenile hormone signaling pathway in insect development. Annu. Rev. Entomol. 58, 181–204 [DOI] [PubMed] [Google Scholar]
- 13. Minakuchi C., Namiki T., Shinoda T. (2009) Kruppel homolog 1, an early juvenile hormone-response gene downstream of Methoprene-tolerant, mediates its anti-metamorphic action in the red flour beetle Tribolium castaneum. Dev. Biol. 325, 341–350 [DOI] [PubMed] [Google Scholar]
- 14. Yang X. H., Liu P. C., Zheng W. W., Wang J. X., Zhao X. F. (2011) The juvenile hormone analogue methoprene up-regulates the Ha-RNA-binding protein. Mol. Cell Endocrinol. 333, 172–180 [DOI] [PubMed] [Google Scholar]
- 15. Liu P. C., Wang J. X., Song Q. S., Zhao X. F. (2011) The participation of calponin in the cross-talk between 20-hydroxyecdysone and juvenile hormone signaling pathways by phosphorylation variation. PLoS One 6, e19776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fang F., Xu Y., Jones D., Jones G. (2005) Interactions of ultraspiracle with ecdysone receptor in the transduction of ecdysone- and juvenile hormone-signaling. FEBS J. 272, 1577–1589 [DOI] [PubMed] [Google Scholar]
- 17. Zhou B., Riddiford L. M. (2001) Hormonal regulation and patterning of the broad-complex in the epidermis and wing discs of the tobacco hornworm, Manduca sexta. Dev. Biol. 231, 125–137 [DOI] [PubMed] [Google Scholar]
- 18. Zhou B., Hiruma K., Jindra M., Shinoda T., Segraves W. A., Malone F., Riddiford L. M. (1998) Regulation of the transcription factor E75 by 20-hydroxyecdysone and juvenile hormone in the epidermis of the tobacco hornworm, Manduca sexta, during larval molting and metamorphosis. Dev. Biol. 193, 127–138 [DOI] [PubMed] [Google Scholar]
- 19. Zou Z., Saha T. T., Roy S., Shin S. W., Backman T. W., Girke T., White K. P., Raikhel A. S. (2013) Juvenile hormone and its receptor, methoprene-tolerant, control the dynamics of mosquito gene expression. Proc. Natl. Acad. Sci. U.S.A. 110, E2173–E2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y., Sheng Z., Liu H., Wen D., He Q., Wang S., Shao W., Jiang R. J., An S., Sun Y., Bendena W. G., Wang J., Gilbert L. I., Wilson T. G., Song Q., Li S. (2009) Juvenile hormone counteracts the bHLH-PAS transcription factors MET and GCE to prevent caspase-dependent programmed cell death in Drosophila. Development 136, 2015–2025 [DOI] [PubMed] [Google Scholar]
- 21. Dubrovsky E. B., Dubrovskaya V. A., Berger E. M. (2004) Hormonal regulation and functional role of Drosophila E75A orphan nuclear receptor in the juvenile hormone signaling pathway. Dev. Biol. 268, 258–270 [DOI] [PubMed] [Google Scholar]
- 22. Wu Y., Parthasarathy R., Bai H., Palli S. R. (2006) Mechanisms of midgut remodeling: juvenile hormone analog methoprene blocks midgut metamorphosis by modulating ecdysone action. Mech. Dev. 123, 530–547 [DOI] [PubMed] [Google Scholar]
- 23. Riddiford L. M. (2012) How does juvenile hormone control insect metamorphosis and reproduction? Gen. Comp. Endocr. 179, 477–484 [DOI] [PubMed] [Google Scholar]
- 24. Emery I. F., Bedian V., Guild G. M. (1994) Differential expression of Broad-Complex transcription factors may forecast tissue-specific developmental fates during Drosophila metamorphosis. Development 120, 3275–3287 [DOI] [PubMed] [Google Scholar]
- 25. Zhou X., Riddiford L. M. (2002) Broad specifies pupal development and mediates the “status quo” action of juvenile hormone on the pupal-adult transformation in Drosophila and Manduca. Development 129, 2259–2269 [DOI] [PubMed] [Google Scholar]
- 26. Erezyilmaz D. F., Riddiford L. M., Truman J. W. (2006) The pupal specifier broad directs progressive morphogenesis in a direct-developing insect. Proc. Natl. Acad. Sci. U.S.A. 103, 6925–6930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. DiBello P. R., Withers D. A., Bayer C. A., Fristrom J. W., Guild G. M. (1991) The Drosophila Broad-Complex encodes a family of related proteins containing zinc fingers. Genetics 129, 385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bayer C. A., Holley B., Fristrom J. W. (1996) A switch in broad-complex zinc-finger isoform expression is regulated posttranscriptionally during the metamorphosis of Drosophila imaginal discs. Dev. Biol. 177, 1–14 [DOI] [PubMed] [Google Scholar]
- 29. Suzuki Y., Truman J. W., Riddiford L. M. (2008) The role of Broad in the development of Tribolium castaneum: implications for the evolution of the holometabolous insect pupa. Development 135, 569–577 [DOI] [PubMed] [Google Scholar]
- 30. Uhlirova M., Foy B. D., Beaty B. J., Olson K. E., Riddiford L. M., Jindra M. (2003) Use of Sindbis virus-mediated RNA interference to demonstrate a conserved role of Broad-Complex in insect metamorphosis. Proc. Natl. Acad. Sci. U.S.A. 100, 15607–15612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karim F. D., Guild G. M., Thummel C. S. (1993) The Drosophila Broad-Complex plays a key role in controlling ecdysone-regulated gene expression at the onset of metamorphosis. Development 118, 977–988 [DOI] [PubMed] [Google Scholar]
- 32. Kiss I., Beaton A. H., Tardiff J., Fristrom D., Fristrom J. W. (1988) Interactions and developmental effects of mutations in the Broad-Complex of Drosophila melanogaster. Genetics 118, 247–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parthasarathy R., Tan A., Bai H., Palli S. R. (2008) Transcription factor broad suppresses precocious development of adult structures during larval-pupal metamorphosis in the red flour beetle, Tribolium castaneum. Mech. Dev. 125, 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Konopova B., Jindra M. (2008) Broad-Complex acts downstream of Met in juvenile hormone signaling to coordinate primitive holometabolan metamorphosis. Development 135, 559–568 [DOI] [PubMed] [Google Scholar]
- 35. Zhao W. L., Liu C. Y., Liu W., Wang D., Wang J. X., Zhao X. F. (2014) Methoprene-tolerant1 regulates gene transcription to maintain insect larval status. J. Mol. Endocrinol. 53, 93–104 [DOI] [PubMed] [Google Scholar]
- 36. Gorelick-Feldman J., Cohick W., Raskin I. (2010) Ecdysteroids elicit a rapid Ca2+ flux leading to Akt activation and increased protein synthesis in skeletal muscle cells. Steroids 75, 632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wheeler D. E., Nijhout H. F. (2003) A perspective for understanding the modes of juvenile hormone action as a lipid signaling system. BioEssays 25, 994–1001 [DOI] [PubMed] [Google Scholar]
- 38. Kim Y., Davari E. D., Sevala V., Davey K. G. (1999) Functional binding of a vertebrate hormone, L-3,5,3′-triiodothyronine (T3), on insect follicle cell membranes. Insect Biochem. Mol. Biol. 29, 943–950 [DOI] [PubMed] [Google Scholar]
- 39. Whitmarsh A. J., Davis R. J. (2000) Regulation of transcription factor function by phosphorylation. Cell. Mol. Life Sci. 57, 1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rauch P., Grebe M., Elke C., Spindler K. D., Spindler-Barth M. (1998) Ecdysteroid receptor and ultraspiracle from Chironomus tentans (Insecta) are phosphoproteins and are regulated differently by molting hormone. Insect Biochem. Mol. Biol. 28, 265–275 [DOI] [PubMed] [Google Scholar]
- 41. Zhao X. F., Wang J. X., Wang Y. C. (1998) Purification and characterization of a cysteine proteinase from eggs of the cotton boll worm, Helicoverpa armigera. Insect Biochem. Mol. Biol. 28, 259–264 [Google Scholar]
- 42. Schooley D., Baker F., Tsai L., Miller C., Jamieson G. (1984) in Biosynthesis, Metabolism and Mode of Action of Invertebrate Hormones, pp. 373–383, Springer, New York [Google Scholar]
- 43. Liu W., Zhang F. X., Cai M. J., Zhao W. L., Li X. R., Wang J. X., Zhao X. F. (2013) The hormone-dependent function of Hsp90 in the crosstalk between 20-hydroxyecdysone and juvenile hormone signaling pathways in insects is determined by differential phosphorylation and protein interactions. Biochim. Biophys. Acta 1830, 5184–5192 [DOI] [PubMed] [Google Scholar]
- 44. Deng H., Zheng S., Yang X., Liu L., Feng Q. (2011) Transcription factors BmPOUM2 and BmbetaFTZ-F1 are involved in regulation of the expression of the wing cuticle protein gene BmWCP4 in the silkworm, Bombyx mori. Insect Mol. Biol. 20, 45–60 [DOI] [PubMed] [Google Scholar]
- 45. Liu W., Cai M. J., Zheng C. C., Wang J. X., Zhao X. F. (2014) Phospholipase C γ1 connects the cell membrane pathway to the nuclear receptor pathway in insect steroid hormone signaling. J. Biol. Chem. 289, 13026–13041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ijiro T., Urakawa H., Yasukochi Y., Takeda M., Fujiwara Y. (2004) cDNA cloning, gene structure, and expression of Broad-Complex (BR-C) genes in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 34, 963–969 [DOI] [PubMed] [Google Scholar]
- 47. Cai M. J., Liu W., He H. J., Wang J. X., Zhao X. F. (2012) Mod(mdg4) participates in hormonally regulated midgut programmed cell death during metamorphosis. Apoptosis 17, 1327–1339 [DOI] [PubMed] [Google Scholar]
- 48. Rane S. G., Walsh M. P., McDonald J. R., Dunlap K. (1989) Specific inhibitors of protein kinase C block transmitter-induced modulation of sensory neuron calcium current. Neuron 3, 239–245 [DOI] [PubMed] [Google Scholar]
- 49. Li Y., Zhang Z., Robinson G. E., Palli S. R. (2007) Identification and characterization of a juvenile hormone response element and its binding proteins. J. Biol. Chem. 282, 37605–37617 [DOI] [PMC free article] [PubMed] [Google Scholar]