Background: How nuclease activity of Mre11 is controlled by Rad50 is poorly understood.
Results: Removal of the C-terminal domain of T4 Mre11 enhances its nuclease activity.
Conclusion: The C terminus of Mre11 is autoinhibitory, and complex formation with Rad50 relieves this inhibition.
Significance: This autoinhibition provides a mechanism for restricting the nuclease activity of Mre11 in the absence of Rad50.
Keywords: Deoxyribonuclease (DNase), DNA Recombination, DNA Repair, DNA-Protein Interaction, Protein Domain, Autoinhibition, DNA Exonuclease, Double Strand Break Repair
Abstract
Mre11 and Rad50 form a stable complex (MR) and work cooperatively in repairing DNA double strand breaks. In the bacteriophage T4, Rad50 (gene product 46) enhances the nuclease activity of Mre11 (gene product 47), and Mre11 and DNA in combination stimulate the ATPase activity of Rad50. The structural basis for the cross-activation of the MR complex has been elusive. Various crystal structures of the MR complex display limited protein-protein interfaces that mainly exist between the C terminus of Mre11 and the coiled-coil domain of Rad50. To test the role of the C-terminal Rad50 binding domain (RBD) in Mre11 activation, we constructed a series of C-terminal deletions and mutations in bacteriophage T4 Mre11. Deletion of the RBD in Mre11 eliminates Rad50 binding but only has moderate effect on its intrinsic nuclease activity; however, the additional deletion of the highly acidic flexible linker that lies between RBD and the main body of Mre11 increases the nuclease activity of Mre11 by 20-fold. Replacement of the acidic residues in the flexible linker with alanine elevates the Mre11 activity to the level of the MR complex when combined with deletion of RBD. Nuclease activity kinetics indicate that Rad50 association and deletion of the C terminus of Mre11 both enhance DNA substrate binding. Additionally, a short peptide that contains the flexible linker and RBD of Mre11 acts as an inhibitor of Mre11 nuclease activity. These results support a model where the Mre11 RBD and linker domain act as an autoinhibitory domain when not in complex with Rad50. Complex formation with Rad50 alleviates this inhibition due to the tight association of the RBD and the Rad50 coiled-coil.
Introduction
DNA double strand breaks (DSB)3 are considered to be one of the most lethal forms of DNA damage and could be induced by external agents, such as ionizing radiation, genotoxic chemicals, and internal agents, such as reactive oxygen species and proteins that stall the replication fork (1). If not repaired properly, DSBs will lead to chromatin rearrangement and ultimately result in tumorigenesis or cell death. Three pathways exist for DSB repair, homologous recombination (HR), nonhomologous end-joining, and microhomology-mediated end-joining (2, 3). The Mre11/Rad50 (MR) complex is involved in the initial steps of the HR and microhomology-mediated end-joining pathways, but its role is best elucidated in HR (4, 5). The MR complex is among the first proteins that respond to DSBs in yeast (6). Upon arriving at the DSB, the MR complex processes the dsDNA ends to produce a short 3′ ssDNA overhang at the break site. Following further resection of 5′ end of the DNA by SgsI/Top3/RmiI, Dna2, and RPA or ExoI and RPA, the recombinase (Rad51) promotes homolog pairing and 3′ ssDNA strand invasion (7). The D-loop created from strand invasion is then used as primer for strand extension. The extended strand can be dissociated from its template by a DNA helicase, or the Holliday junction can be processed with a DNA nuclease and ligase (8). In eukaryotes, the MR complex also acts as a DSB sensor by recruiting and stimulating ataxia telangiectasia-mutated protein kinase to initialize checkpoint signal cascade (9, 10).
The MR complex is evolutionarily conserved, with orthologs of Mre11 and Rad50 existing in all kingdoms of life (4). Eukaryotic MR complex requires a third protein component, Nbs1 in human (11) and Xrs2 in yeast (12). MR complexes in eukaryotes, archaea, and bacteriophages all catalyze DNA resection at DSBs; in contrast, the bacterial MR complex is involved in processing of cruciform structure during DNA replication (13) and DSB resection is carried out by the RecBCD helicase-nuclease complex (14). The MR complex from bacteriophage T4 functions in a similar fashion as its eukaryotic homologs and is required for both DSB repair and origin-independent DNA replication (15–18).
Rad50 belongs to the ATP-binding cassette (ABC) protein superfamily (19). All ABC proteins contain a conserved nucleotide binding domain (NBD) that dimerizes upon ATP binding at the dimer interface. ABC proteins act as chemo-mechanical engines, utilizing the energy of ATP binding and hydrolysis to fuel various biological functions, including membrane transportation, DNA repair, chromosome condensation, RNA-protein remodeling, and protein translation (19, 20). The ATPase activity of Rad50 may facilitate translocation of the MR complex along its DNA substrate and is required for repetitive resection of DNA (21). Structures of the NBD domain of Rad50 from Pyrococcus furiosus, Thermatoga maritima, and Methanocaldococcus jannaschii have been determined in the presence and absence of ATP (22–24). The Rad50 NBD domains dimerize in a head-to-tail fashion. The WalkerA, WalkerB, Q-loop, and H-loop motifs from one monomer and the Signature and the D-loop motifs from the adjacent monomer together create an intact active site. A long coiled-coil domain embeds between the N- and C-terminal halves of the NBD domain in Rad50. A highly conserved CXXC motif in the middle of the coiled-coil domain forms a zinc finger-like structure with the CXXC motif from adjacent Rad50 (25). The CXXC motif is believed to mediate Rad50-dependent DNA tethering (4, 18, 26).
As a member of Ser/Thr protein phosphatase superfamily (27), Mre11 is responsible for the nuclease activity of the MR complex (21). The nuclease activity of Mre11 is metal-dependent, and the highest levels of nuclease activity are observed with Mn2+. Mg2+ supports activity to a lesser extent and an altered product profile is produced (21). The conserved Mre11 core contains a nuclease domain, a capping domain, and a C-terminal Rad50 binding domain (RBD), with a flexible linker bridging the capping domain and RBD (28, 29). The nuclease domain of Mre11 forms a dimer with an interface that appears highly flexible and may adopt multiple functional states during catalysis of the MR complex (30–34).
MR forms a stable heterotetrameric complex with two subunits of Mre11 and two subunits of Rad50 (21, 23, 24). Mre11 and DNA activate the ATPase activity of T4 Rad50 by ∼20-fold (21). Extensive, repetitive nuclease activity is completely reliant on the presence of Rad50 and ATP hydrolysis. Several structures of the MR complex have been solved recently with Rad50 constructs lacking most of the coiled-coil domain (23, 24, 35). MR stays in an extended conformation in the absence of ATP and a compacted conformation in the presence of ATP (36). Mre11 embraces Rad50 on the surface of Rad50 opposing the coiled-coil domain. There are two protein-protein interfaces between Mre11 and Rad50. The first is the capping domain of Mre11 that interacts with the distal face of Rad50 (the side distal to the coiled-coil) with a surface area of 686 Å2 (24). The second interface is the RBD of Mre11 and the root of the coiled-coil domain in Rad50, which includes a surface area of 1334 Å2 (24). Both of these interfaces are far away from the active site of Mre11 (Fig. 1). The structural mechanism of how the binding of Rad50 stimulates the nuclease activity of Mre11 has been elusive. In this report, we explored the role of Mre11 RBD and the linker domain in the allosteric communication of MR complex. We find that Mre11 is autoinhibited by its RBD and the flexible linker. Deletion of the RBD of Mre11 eliminates Rad50 binding but only has a moderate effect on basal nuclease activity of Mre11; however, the additional deletion of the flexible linker between RBD and main body of Mre11 increases the Mre11 nuclease activity by 20-fold. Mutations of the negatively charged residues in the flexible linker to alanine elevate the Mre11 activity to the level of MR complex when combined with deletion of RBD. Kinetics experiments indicate that Rad50 association and deletion of the C terminus of Mre11 both enhance DNA binding to a similar degree. The results here suggest a simple model for the role of Rad50 in ATP-independent nuclease activity. The RBD and flexible linker of Mre11 block a portion of its DNA-binding site, thus reducing its affinity for the DNA substrate and inhibiting its nuclease activity. This inhibition is relieved upon complex formation with Rad50 as its coiled-coil domain sequesters the RBD and linker of Mre11.
FIGURE 1.
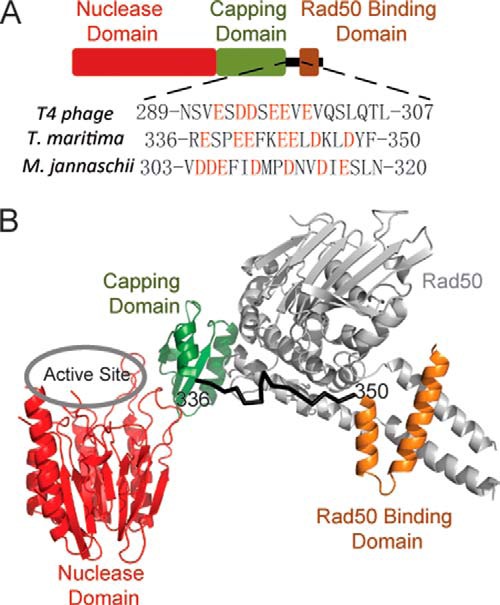
Domain structure of Mre11. A, nuclease domain, capping domain, and Rad50 binding domain are colored in red, green, and brown, respectively. The sequences of the flexible linker between capping domain and Rad50 binding domain are shown, and the negatively charged residues are highlighted in red. B, structure of T. maritima open MR complex in the absence of ATP is exhibited (Protein Data Bank code 3QG5, chains A and C). The Rad50 is colored in gray, and nuclease domain, capping domain, and Rad50 binding domain are colored in red, green, and brown, respectively.
EXPERIMENT PROCEDURES
Materials
Coupling enzymes (pyruvate kinase and lactate dehydrogenase), dNTPs, NADH, and nickel-agarose came from Sigma. Phosphoenolpyruvate was from Alfa Aesar. Adenosine 5′-triphosphate was purchased from United States Biochemical Corp. Chitin beads were from New England Biolabs. Chemicals, buffers, and media components came from Fisher. The M13 phage single-stranded DNA was obtained using PEG precipitation and phenol/chloroform extraction (37).
Mutagenesis, Protein Expression, and Purification
The open reading frames for the truncated forms of Mre11 were amplified from previous T4Mre11-PTYB1 construct (21) using PCR primers that contained NdeI cutting site on the forward primer and SapI cutting site on the reverse primer. The PCR products were then subcloned into pTYB1 vector. Site-directed mutations of Mre11 were generated following Stratagene QuikChange mutagenesis protocol. The sequences of all DNA primers used are available upon request. The sequences and integrity of all the constructs were verified by DNA sequencing provided by Iowa State University DNA facility. Bacterial expression and purification of the wild type (WT) and mutated Mre11 and Rad50 proteins were carried out as described previously (21, 38).
Steady-state ATPase Kinetics
ATPase activity of Rad50 or MR complex was measured spectrofluorometrically with a standard coupling assay (39). ADP formation was coupled to NADH consumption through the enzymatic action of pyruvate kinase and lactate dehydrogenase. All assays were carried out at 30 °C using excitation and emission wavelengths of 340 and 460 nm, respectively, on a Cary Eclipse spectrofluorometer (Varian). The reaction buffer contained 50 mm Tris-HCl, pH 7.6, 50 mm KCl, 5 mm MgCl2, 50 μm NADH, 150 μm phosphoenolpyruvate, 6.67 units/ml pyruvate kinase, 10 units/ml lactate dehydrogenase, and 0.1 mg/ml BSA. All ATPase assays were initialized with the addition of Rad50 or preassembled MR complex. Initial velocity (v) was measured at different ATP concentrations ([S]), and the maximum velocity (Vmax), Michaelis constant (Km), and Hill coefficient (n) were estimated according to Equation 1 with GraFit (Erithacus Software Ltd., UK).
 |
The sequence of the oligonucleotides used to create the DNA substrate is identical to those used previously (21).
Determination of Nuclease Activity
Nuclease activity was determined fluorometrically using DNA substrates with a fluorescent 2-aminopurine deoxyribonucleotide at the 1st position relative to the 3′ end of the substrate. The fluorescence level increases as the 2-aminopurine deoxyribonucleotide is excised from DNA through the nuclease activity of Mre11 (40). The assay was carried out at 30 °C using excitation and emission wavelengths of 310 and 375 nm, respectively, on a Cary Eclipse (Varian) spectrofluorometer. The reaction buffer contained 50 mm Tris-HCl, pH 7.6, 50 mm KCl, 5 mm MgCl2, 0.3 mm MnCl2 (standard reaction buffer), and 1.3 μm DNA substrate. Various DNA substrate concentrations were used (0.5 to 20 μm) in determining the kcat and Michaelis constant (Km) of DNA using Equation 1.
The steady-state product profile of the WT-Mre11, ΔC289-Mre11, and their complexes with Rad50 was examined using denaturing urea-PAGE. Assays were carried out at 30 °C in the standard reaction buffer with 1.3 μm hexachlorofluorescein-labeled DNA substrate. The hexachlorofluorescein label was attached to the 5′ end of the ds50-F primer. A phosphorothioate linkage between the 1st and 2nd nucleotides of the ds50-R substrate (relative to the 3′ end) prevented Mre11 from excising nucleotides from the unlabeled DNA strand. Reactions were quenched with an equal volume of quench buffer containing 50% formamide and 100 mm EDTA at time points 0, 10, 20, 40, and 120 min. Reaction products were resolved with 16% denaturing PAGE containing 7.5 m urea in TBE buffer. Gels were run for 3 h at a constant power of 60 watts. The gel was visualized on a Typhoon PhosphorImager using a 532-nm laser and a 555-nm bandpass filter and analyzed with the ImageJ software (National Institutes of Health).
The ssDNA endonuclease activity was carried out at 30 °C in the standard reaction buffer with 1 μg of M13 phage single-stranded DNA per time point. The reaction products were monitored using a 0.8% TAE-agarose gel. The reaction time points were quenched in 5 mm Tris-HCl, pH 7.5, 200 mm EDTA, 0.2% orange G, 0.015% bromphenol blue, 0.015% xylene cyanol FF, and 7.5% Ficoll 400. The agarose gels were stained with 10 μg/ml ethidium bromide for 30 min prior to visualization under UV light.
Gel Filtration Analysis
A Superdex 200 10/300 GL column (GE Healthcare) was used to determine the oligomerization states of WT and mutant forms of Mre11 and MR complex. The column was equilibrated with buffer containing 20 mm Tris, pH 8.0, and 400 mm NaCl. For each run, 100 μl of sample with Mre11, Rad50, or MR complex was injected. The protein concentrations are 100 μm for Mre11 and 50 μm for Rad50.
RESULTS
Nuclease Activity of C-terminal Deletions of Mre11
The fluorescence 2-AP assay was used to examine the 3′ to 5′exonuclease activity of WT and mutant Mre11 proteins. A 50-base pair DNA substrate with 2-AP at the first position was used to assess the ATP-independent nuclease activity.
Previous experiments on the P. furiosus MR complex indicate the RBD is strictly required for MR complex formation (30). To test the role of the C-terminal element in Mre11 activation, we designed a series of Mre11 deletion constructs. Deletion of the RBD (ΔC307, deletion of residues 308–339) completely eliminates Rad50 activation, verifying the indispensable role of RBD in the MR interaction. However, the deletion causes only a minimal effect in nuclease activity of Mre11 (Fig. 2A). Further deletions of residues 301–339 (ΔC300), 299–339 (ΔC298), 297–339 (ΔC296), and 292–339 (ΔC292) gradually increase the nuclease activity of Mre11. Removal of the entire flexible linker (ΔC289, deletion of residues 290–339) elevates the nuclease activity of Mre11 by 20-fold. Addition of Rad50 slightly reduces the nuclease activity of ΔC307, ΔC298, ΔC296, and A6ΔC300, but this effect is absent or not easily detectable for ΔC292 and ΔC289 due to the noise associated with these mutants. Rad50 alone binds DNA tightly and may compete for DNA substrate in the nuclease assay (34).
FIGURE 2.
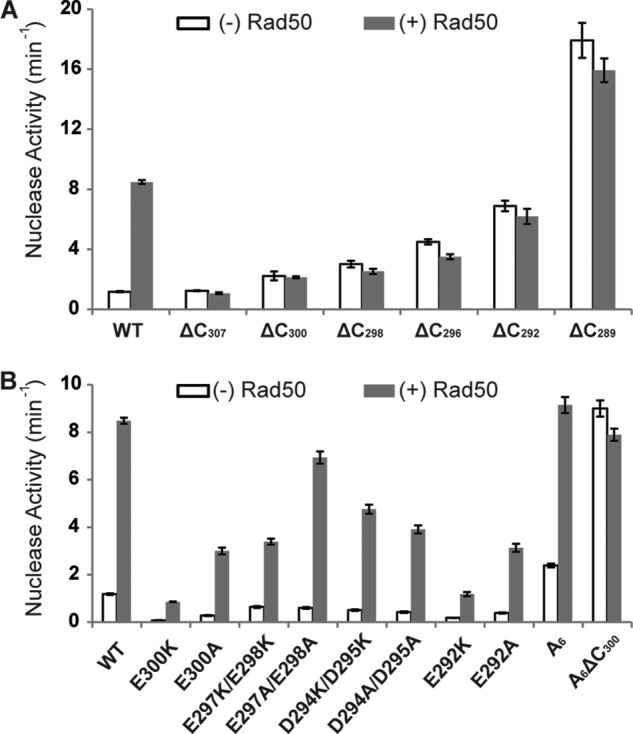
Effects of C-terminal deletions (A) or point mutations (B) on the nuclease activity of Mre11. The activity was measured in the absence (white bar) or presence (gray bar) of Rad50. The substrate used is a blunt-ended 50-bp dsDNA with a fluorescent nucleotide analog (2-aminopurine) located its the 3′ ends. Nuclease activity is monitored using the excision of 2-aminopurine as a reporter. The assays contained 0.3 mm Mn2+ and 1.3 μm DNA. Error bars represent the standard deviation from three replicates.
Nuclease Activity of Mutations on Flexible Linker of Mre11
Results from C-terminal deletions of Mre11 highlight residues 290–300; ΔC300 shows only a 2-fold increase in nuclease activity, whereas ΔC289 exhibits a 20-fold increase in nuclease activity (Fig. 2A). Residues 290–300 are highly negatively charged, with aspartic acid at positions 294 and 295 and glutamic acid at positions 292, 297, 298, and 300 (Fig. 1A). Mutations of one or two of these negatively charged residues to alanine or lysine do not disturb the Rad50 binding, as indicated by at least a 10-fold activation of Rad50 (Fig. 2B). However, the nuclease activities decrease for all single and double mutants tested. Most striking are the E300K-Mre11 and E292K mutants, which lower the nuclease activity of both Mre11 alone and the MR complex by at least 10-fold. The cause of this reduced nuclease activity is unclear, but it appears that these residues must have additional role(s) in promoting the fully active state of Mre11. Considering the flexibility of the loop and the enrichment of the negatively charged residues, the role of one or two negatively charged residues may be compensated by their neighbors. To control for this possibility, we mutated all negatively charged residues to alanine (A6-Mre11). In contrast to single or double mutants whose nuclease activities (without Rad50) drop significantly, A6-Mre11 exhibits a 2-fold increase in nuclease activity. Rad50 activates A6-Mre11 by ∼5-fold, significantly lower than the activation of WT or the single and double Mre11 mutants. However, the final activity of A6-MR and WT-MR is comparable. The results imply that the Rad50 binding compensates for defects in the flexible linker.
Both ΔC300 and the mutation of all negatively charged residues (A6-Mre11) increase the nuclease activity by around 2-fold. However, the changes are minor compared with the 10-fold Rad50 activation for WT-Mre11 or a 20-fold increase that occurs with the ΔC289-Mre11 mutant. There are two possible causes for this, either other residues in flexible linker hinder Mre11 activity or the negatively charged residues work cooperatively with the RBD to inhibit Mre11 activity. Combination of ΔC300 and A6 mutations (A6ΔC300-Mre11) shows a similar level of activity as the WT-MR complex, supporting the latter possibility.
Gel Filtration Analysis
Gel filtration analysis was utilized to examine the oligomeric state and Rad50 interaction of WT and selected Mre11 mutants (Fig. 3). According to their elution time, WT-Mre11 and Rad50 remain as monomer and dimer, respectively. The MR complex sample included an excess amount of Mre11. If a tight MR complex forms, then the Rad50 peak should disappear and a new peak will emerge at an earlier elution volume when compared with the Rad50 peak, as it does in the case of the WT-MR complex. A6-Mre11 exhibits similar peak shift behavior as a WT-MR complex. Consistent with the nuclease assay, Rad50 does not interact with any C-terminal deletions of Mre11 (ΔC289, ΔC300, and A6-ΔC300).
FIGURE 3.
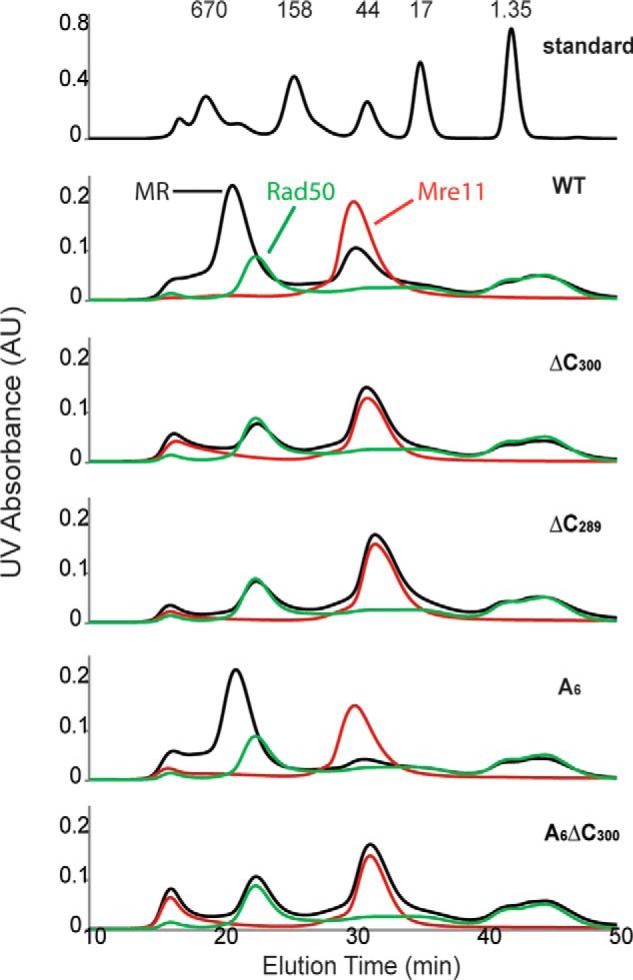
Gel filtration analysis of WT and mutant Mre11 (red), Rad50 (green), and MR (black) complex. For each run, 100 μl of sample with Mre11, Rad50, or MR complex was injected. The protein concentrations are 100 μm for Mre11 and 50 μm for Rad50. The WT·MR and A6·MR complex peaks shift to earlier elution times relative to Rad50 alone, indicating the formation of MR complex, whereas none of the C-terminal deletions of Mre11 interact with Rad50. AU, absorbance units.
Steady-state Kinetics of the Nuclease Activity for WT and Mutant Mre11
The same fluorescence 2-AP assay was used for the steady-state kinetics characterization. For WT-, A6-, and ΔC300-Mre11, no saturation can be achieved in the range of 0.5 to 20 μm DNA substrate, suggesting that their Km-DNA values are significantly higher than 20 μm. In contrast, the WT-MR complex is able to be saturated with the DNA substrate, and the kinetic data can be fitted with the Michaelis-Menten equation (Km-DNA of 3.8 μm, Table 1). The results here are consistent with previous results from anisotropic fluorescence DNA binding assay, where MR complex binds DNA tightly but the DNA binding for Mre11 alone is barely detectable (34). The A6-MR complex and ΔC289-Mre11 exhibit ∼30% decrease in Km-DNA as compared with the WT-MR complex, and the ΔC-A6-Mre11 has a Km-DNA that is essentially identical to the WT-MR complex. All the mutants and WT-MR have about the same kcat values with the exception of ΔC289-Mre11, which is ∼1.5-fold higher.
TABLE 1.
Nuclease activity of WT and mutant proteins
Fluorometric assays were performed as described “Experimental Procedures.” The DNA substrate was varied from 0.5 to 20 μm with 2-AP at first position.
| Km-DNA | kcat | |
|---|---|---|
| μm | min−1 | |
| WT-Mre11 | NDa | ND |
| WT-MR | 3.8 ± 0.7 | 41 ± 3 |
| Α6-Mre11 | ND | ND |
| A6-MR | 2.6 ± 0.4 | 36 ± 2 |
| ΔC300-Mre11 | ND | ND |
| ΔC289-Mre11 | 2.3 ± 0.2 | 64 ± 2 |
| ΔC289-Mre11/Cpepb | ND | ND |
| A6ΔC300-Mre11 | 4.7 ± 0.4 | 45 ± 2 |
a ND means not determined. The nuclease activity increased linearly with respect to substrate, indicating the Km is beyond the concentration range of DNA substrate.
b The concentration of Cpep was held at 160 μm.
Both the C-terminal deletion of Mre11 (ΔC289) and WT-MR complex display a high level of nuclease activity for the excision of the first nucleotide. To monitor the processivity of ΔC289-Mre11, the nuclease activity for WT-MR and ΔC289-Mre11 were assayed using denaturing PAGE. A hexachlorofluorescein label is attached to the 5′ end of the ds50-F primer. A phosphorothioate linkage between the 1st and 2nd nucleotides of the ds50-R substrate (relative to the 3′ end) prevents Mre11 from excising nucleotides from the unlabeled DNA strand. As shown in Fig. 4, ΔC289-Mre11 exhibits much greater nuclease activity compared with WT-Mre11. The WT-MR complex can processively remove nucleotides in an ATP-dependent manner, whereas ΔC289-Mre11 is not processive, as indicated by the absence of short DNA products. This is consistent with our previous experiments that demonstrate both Rad50 and ATP hydrolysis are required for repetitive resection of DNA (21).
FIGURE 4.
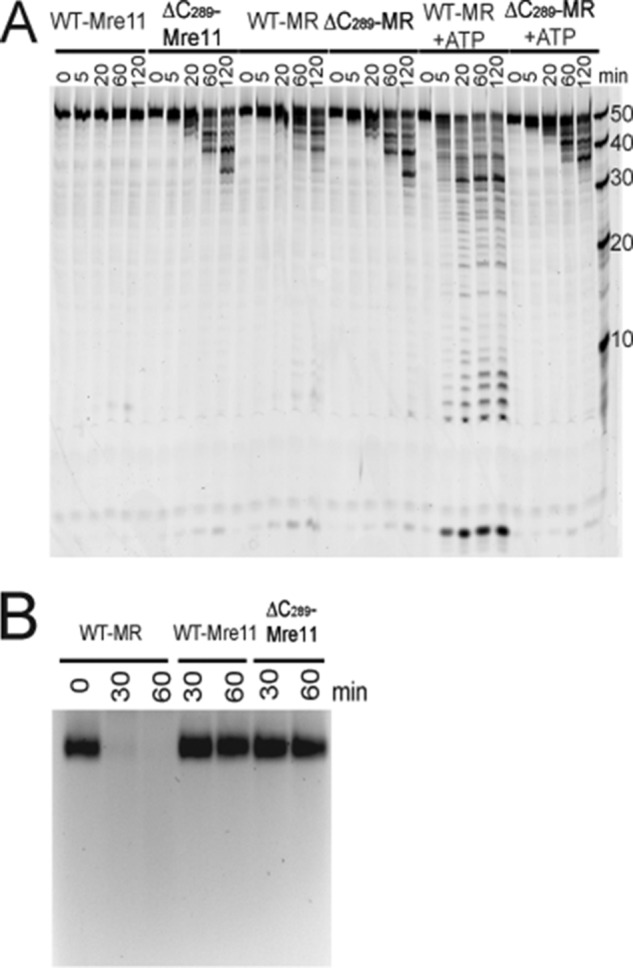
Nuclease activity of WT and ΔC289 Mre11. A, nuclease activity with a 50-bp blunt-ended dsDNA substrate. The reaction products are separated using 16% urea-PAGE and are visualized using a hexachlorofluorescein dye located at the 5′ end of the DNA. The reactions were carried out at 30 °C, and the time points for each protein are 0, 5, 20, 60, and 120 min. Each assay consisted of 220 nm Rad50, 200 nm Mre11, 1.3 μm DNA substrate, 5 mm MgCl2, and 0.3 mm MnCl2. The ATP concentration was 1 mm when included. B, nuclease activity with a 7250-base circular ssDNA substrate. The reaction products are separated using 0.8% agarose gel electrophoresis, and the DNA is visualized using ethidium bromide. The reactions were carried out at 30 °C, and the time points for each protein are 30 and 60 min. Each assay consisted of 330 nm Rad50, 300 nm Mre11, 1 μg of DNA substrate, 5 mm MgCl2, and 0.3 mm MnCl2.
Construction and Characterization of Cpep-Mre11
The C-terminal residues 290–339 (Cpep-Mre11) were cloned and constructed into pTYB1 vector. Following the standard purification procedure for Mre11, the Cpep-Mre11 was purified to homogeneity as indicated by SDS-PAGE (data not shown). The fluorescence 2-AP assay was used to test the inhibition of Cpep-Mre11 on Mre11. As shown in Fig. 5A, Cpep-Mre11 inhibits ΔC289-Mre11 effectively, with an IC50 of 47 ± 6 μm. As expected, the inhibition of Cpep-Mre11 on WT-Mre11 is significantly weaker, with an IC50 estimate of 250 ± 60 μm. The Cpep-Mre11 inhibition appears to be competitive with respect to DNA, as the IC50 increases 2-fold for ΔC289-Mre11 when the concentration of the DNA is doubled (IC50 of 90 ± 16 μm) and the Km-DNA (apparent) is elevated at least 10-fold in the presence of 160 μm Cpep (Table 1).
FIGURE 5.
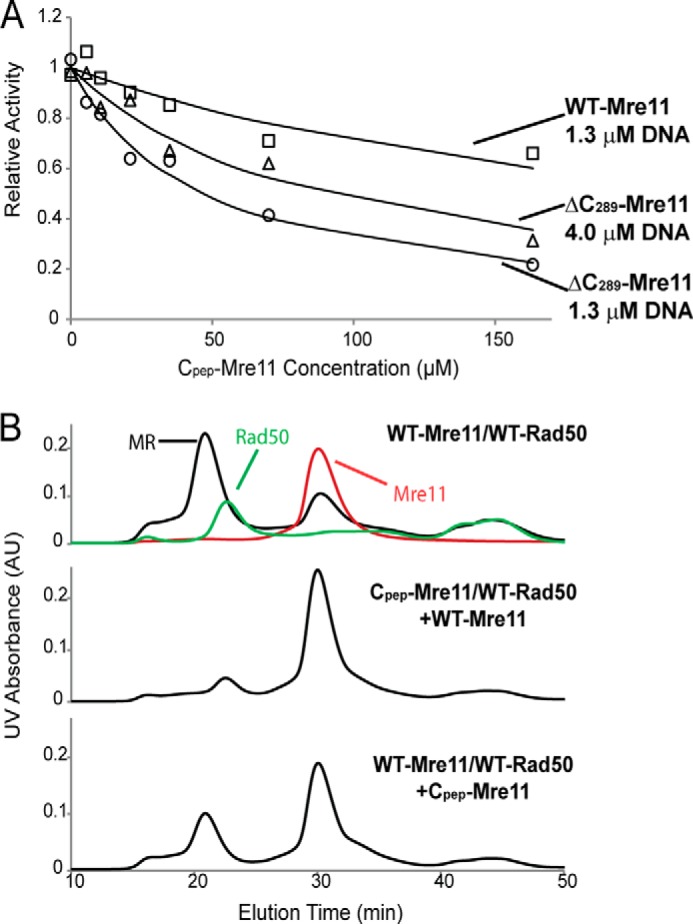
Characterization of C-terminal Mre11. A, plot of relative activity of WT-Mre11 (□) and ΔC289Mre11 (Δ and ○) versus Cpep-Mre11 concentration. Relative activity (Vi/Vo) is defined as the activity in the presence of Cpep-Mre11 (Vi) divided by the activity in the absence of Cpep-Mre11 (Vo). The 2-AP-labeled DNA concentration was 1.3 μm for(□ and ○ and 4 μm for ▵. B, Cpep-Mre11 affects MR complex assembly. The top panel displayed the gel filtration profiles for Mre11 (red), Rad50 (green), and MR complex (black). If Cpep-Mre11 and Rad50 were mixed before the addition of WT-Mre11 (middle), then the MR complex does not form. However, if Mre11 and Rad50 were mixed prior to addition of Cpep-Mre11, MR complex forms normally.
Complexes of Cpep-Mre11 and Rad50 from P. furiosus and T. maritima have been co-crystallized (23, 30). As seen in the crystal structures, Cpep-Mre11 binds to Rad50 at the same site in the presence or absence of main body of Mre11. To test whether the same scenario applies in the T4 system, we employed the gel filtration assay to explore the interaction between Rad50 and Cpep-Mre11. Because Cpep-Mre11 is too small to be directly detectable via gel filtration analysis, we performed a simple competition assay. Previous pulldown experiments indicate tight binding and slow dissociation of MR complex (41). If Cpep-Mre11 binds strongly to the same site on Rad50 as does WT-Mre11, then the MR complex should not form in the presence of Cpep-Mre11. We incubated Cpep-Mre11 with Rad50 for 1 h and then added Mre11 prior to injection. The elution profile indicates the absence of MR complex (Fig. 5B). If Rad50 and WT-Mre11 were incubated prior to the addition of Cpep-Mre11, then the MR complex peak is still evident, although to a somewhat lesser extent.
Steady-state ATPase Activity
The addition of WT-Mre11 and dsDNA to Rad50 results in around 30-fold increase in kcat-ATP, a 3-fold increase in Km-ATP, and a Hill coefficient increase from 1.3 to 2.5 (Table 2). The kcat-ATP for A6-Mre11/Rad50·DNA complex is 2-fold lower than that of WT·MR complex, yet it is 15-fold higher compared with Rad50 alone. As indicated by the nuclease assays and gel filtration analysis, ΔC289-Mre11 does not interact with Rad50, thus having a negligible effect on ATPase activity of Rad50.
TABLE 2.
ATP hydrolysis activity of WT and mutant proteins
Fluorometric assays were performed as described under “Experimental Procedures.” Errors represent the standard errors of the fit.
| Km-ATP | kcat | Hill | |
|---|---|---|---|
| μm | s−1 | ||
| Rad50 | 19 ± 1 | 0.088 ± 0.003 | 1.28 ± 0.07 |
| Rad50-DNA | 37 ± 2 | 0.065 ± 0.001 | 1.11 ± 0.06 |
| WT-MR | 53 ± 5 | 0.25 ± 0.01 | 1.3 ± 0.1 |
| WT-MR-DNA | 57 ± 1 | 2.89 ± 0.05 | 2.5 ± 0.1 |
| A6-MR | 24 ± 1 | 0.222 ± 0.004 | 1.37 ± 0.08 |
| A6-MR-DNA | 33 ± 1 | 1.34 ± 0.02 | 1.7 ± 0.1 |
| ΔC289-MR | 20 ± 1 | 0.091 ± 0.003 | 1.28 ± 0.08 |
| ΔC289-MR-DNA | 41 ± 4 | 0.087 ± 0.003 | 1.11 ± 0.08 |
DISCUSSION
The MR complex is involved in various aspects of DSB repair, including sensing the DSB, triggering signal pathways, and facilitating DSB repair through different pathways (42). The nuclease activity of Mre11 is essential for meiotic HR and ataxia telangiectasia-mutated signaling yet dispensable for other functions of MR. Because of the multiple functions of MR in diverse cellular contexts, the nuclease activity of Mre11 must be under tight control.
In this report, we described a simple model for the regulation of T4 Mre11 exonuclease activity by its own C terminus. Mre11 exonuclease activity is auto-inhibited by its C-terminal RBD and flexible linker. Upon complex formation with Rad50, the RBD of Mre11 is sequestered by the coiled-coil domain of Rad50, thereby relieving inhibition. The data suggest that inhibition of Mre11 by its C terminus is competitive with respect to DNA. Removal of the RBD and flexible linker of Mre11 greatly decreases the Km-DNA and stimulates its exonuclease activity. Two structural elements, the RBD and the clustered negatively charged residues in the flexible linker, are involved in the auto-inhibition. Individual deletion of the RBD or mutation of all negatively charged residues within the linker has only a moderate effect on nuclease activity, whereas the combination elevates the exonuclease activity to the level of the MR complex. Although the location of the C-terminal domain binding site on Mre11 is uncertain because all structures of Mre11 alone have this domain removed, the net charge of the T4 Mre11 capping domain is highly positive with a pI = 9.16, whereas the flexible linker domain of T4 is highly negative with a pI of 3.2. As these two domains are adjacent to each other, it is quite possible that they interact. Although charge reversal mutations of one or two of the negatively charged residues on the flexible linker results in a reduction of activity, mutation of all the negatively charged residues causes the activity to increase. This suggests that the interaction between the flexible linker and the Mre11 main body may be variable and nonspecific. Thus, we propose that the flexible linker loosely interacts with positively charged residues on the surface of Mre11, likely within the capping domain, and this interaction sterically disrupts DNA binding (Fig. 6). The x-ray crystal structure of Pfu-Mre11 in complex with branched DNA reveals ionic interactions between the capping domain and the DNA, consistent with the proposal that a putative interaction between the linker domain and the capping domain would compete with DNA binding (43). Similar negatively charged residue clusters can be found in the flexible linker in both M. jannaschii and T. maritima Mre11 (Fig. 1), suggesting a conserved regulatory mechanism during evolution. However, these negatively charged residue clusters cannot be detected in either human or yeast Mre11. Instead, eukaryotic Mre11 could be regulated by post-translational modifications of their C-terminal regions. Human and yeast Mre11 have an extended the C-terminal domain that contains a glycine-arginine-rich domain and DNA binding domains. Methylation of arginine residues within the glycine-arginine-rich domain by PRMT1 is essential in regulating DNA binding and nuclease activity of human Mre11 (44). Moreover, phosphorylation of Mre11 at position Ser-676 inhibits Mre11 (45). These post-translational modifications of Mre11 highlight the essential regulatory role of C-terminal Mre11.
FIGURE 6.
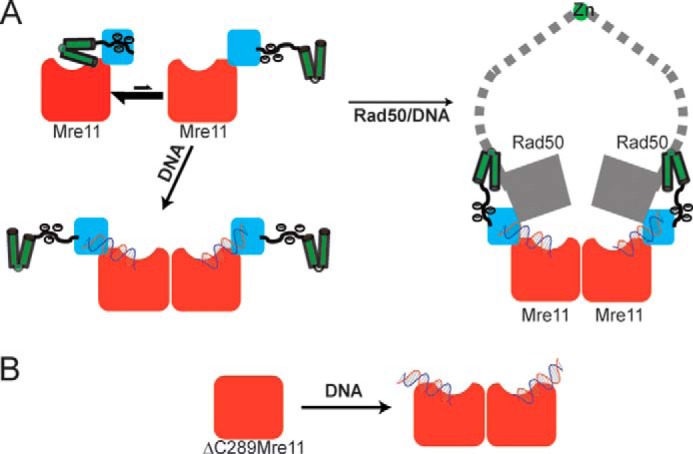
Proposed model for Mre11 activation. A, nuclease domain, capping domain, the negatively charged flexible linker, RBD, and Rad50 are colored in red, blue, black, green, and gray, respectively. The C-terminal flexible linker and RBD may exist in two distinct conformations. In the absence of Rad50, the Mre11 is autoinhibited with the negatively charged linker bound to capping domain, which partially occludes the DNA-binding site. In the presence of Rad50, the coiled-coil domain binds to the RBD of Mre11 and relieves the autoinhibition by the flexible linker allowing DNA to bind. B, removal of the negatively charged linker and RBD from Mre11 exposes the capping domain allowing ΔC289-Mre11 to bind DNA with high affinity in the absence of Rad50.
In contrast, deletion of the C-terminal element of Mre11 does not affect its endonuclease activity. DNA substrates for endo- and exonuclease activity may bind distinctly in Mre11. Specific inhibitors for either endo- or exonuclease activity have been identified (46). Mutation of His-52 in Pfu-Mre11 eliminates exonuclease activity but only has a minor effect on endonuclease activity (43). In Pfu-Mre11, two blunt-ended DNA substrates can bind to the Mre11 dimer simultaneously, whereas only a single branched DNA substrate occupies the DNA-binding site in the Mre11 dimer (43). Moreover, mutations that disrupt dimerization in Pfu-Mre11 reduce ssDNA binding by 30–50-fold while only causing a 3–10-fold decrease for dsDNA (47). The DNA substrate for endonuclease activity should bind to Mre11 in a capping domain independent mode and thus is not affected by C-terminal deletion. Conversely, Rad50 enhances both endo- and exonuclease activity of Mre11. Besides chelating the C-terminal domain of Mre11, Rad50 could also strengthen the dimerization of Mre11 and promote DNA binding. T4 Mre11 exists primarily as a monomer at low concentrations, whereas the MR complex is a stable heterotetramer (38). Meanwhile, both ssDNA and dsDNA bind strongly to Rad50 with sub-micromolar affinity (34). Rad50 may help to align the ssDNA substrate in the active site of Mre11.
The endo- and exonuclease activities of Mre11 act at different stages during HR (46). Endonuclease activity initiates DSB resection and may affect pathway choice in DSB repair, whereas its exonuclease activity functions downstream. The differential regulation of Mre11 by Rad50 may correlate to different roles of endo- and exonuclease activity in vivo. Moreover, ssDNA is a common intermediate produced during DNA replication, transcription, recombination, and DNA repair, making the control of ssDNA endonuclease activity a strong requirement. Indeed, NbsI, a third component in eukaryotic MRN complex, stimulates the endonuclease activity of the MR complex on hairpin substrates without enhancing exonuclease activity (47). However, the exact regulatory mode of different activity of Mre11 needs to be further explored.
Autoinhibition is a relatively widespread regulatory strategy, and the presence of an autoinhibitory domain suggests that the protein or enzyme possesses an activity that must be tightly controlled. Autoinhibition is particularly common in proteins involved in cellular signaling and regulation of transcription. The autoinhibitory domain of protein kinases is often considered to be a “pseudosubstrate” that competes with the binding of the true substrate (48). Similarly, the autoinhibitory domains of most transcription factors behave as DNA mimics whose binding site overlaps with DNA (49). Autoinhibition is less common among nucleases, but there are several well established examples. The Dicer protein, which uses its endonuclease activity to process dsRNA into short fragments as part of the RNAi pathway, is autoinhibited by its N-terminal helicase domain (50). In this case, the inhibition appears to be allosteric in nature as the removal of the N-terminal region increases the kcat of the reaction rather than the Km-RNA. The restriction endonuclease EcoRII is also autoinhibited by its N-terminal domain, which sterically occludes the active site of the enzyme to prevent DNA binding (51). Perhaps the most similar example of the mechanism we have proposed for T4 Mre11 is the regulatory mechanism of human exonuclease I (hExoI) (52). Orans et al. (52) have proposed that the C terminus of hExoI acts as an autoinhibitory domain that down-regulates its nuclease activity when not in complex with other DNA repair pathway-specific proteins. In the case of mismatch repair, the authors (52) propose that MutSα activates the exonuclease activity of hExoI by interacting with its C-terminal autoinhibitory domain.
The benefits of controlling spurious nuclease activity of Mre11 seem clear, and it appears that Mre11 does not have specific functions outside of the MR complex (15, 53). Very little is known regarding the relative expression levels of Mre11 and Rad50 in T4 phage, but the intrinsic autoregulatory mechanism described here for T4 Mre11 ensures that it will not localize to DNA until MR complex formation. Once the MR complex is formed, stable binding to DNA becomes ATP-dependent and additional factors such as gp32 or UvsY may play a role in recruiting the MR complex to the ends of dsDNA (21, 34). Testing the in vivo consequences of autoinhibition removal will require an Mre11 mutant that lacks autoinhibition but retains the ability to interact with Rad50. This may be possible if the Mre11- and Rad50- binding sites on the RBD of Mre11 are nonoverlapping.
This work was supported by Carver Trust Young Investigator Grant 10-3603 (to S. W. N.), Iowa State University institutional support (to S. W. N), and National Science Foundation Grant MCB-1121693 (to S. W. N).
- DSB
- double strand break
- T4 phage
- bacteriophage T4
- HR
- homologous recombination
- MR
- Mre11/Rad50
- NBD
- nucleotide binding domain
- RBD
- Rad50 binding domain
- ssDNA
- single-stranded DNA.
REFERENCES
- 1. Povirk L. F. (2006) Biochemical mechanisms of chromosomal translocations resulting from DNA double strand breaks. DNA Repair 5, 1199–1212 [DOI] [PubMed] [Google Scholar]
- 2. Bassing C. H., Alt F. W. (2004) The cellular response to general and programmed DNA double strand breaks. DNA Repair 3, 781–796 [DOI] [PubMed] [Google Scholar]
- 3. McVey M., Lee S. E. (2008) MMEJ repair of double strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 24, 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Connelly J. C., Leach D. R. (2002) Tethering on the brink: the evolutionarily conserved Mre11-Rad50 complex. Trends Biochem. Sci. 27, 410–418 [DOI] [PubMed] [Google Scholar]
- 5. San Filippo J., Sung P., Klein H. (2008) Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 6. Lisby M., Barlow J. H., Burgess R. C., Rothstein R. (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699–713 [DOI] [PubMed] [Google Scholar]
- 7. Mimitou E. P., Symington L. S. (2009) DNA end resection: many nucleases make light work. DNA Repair 8, 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krogh B. O., Symington L. S. (2004) Recombination proteins in yeast. Annu. Rev. Genet. 38, 233–271 [DOI] [PubMed] [Google Scholar]
- 9. Lee J.-H., Paull T. T. (2004) Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304, 93–96 [DOI] [PubMed] [Google Scholar]
- 10. Lee J.-H., Paull T. T. (2005) ATM activation by DNA double strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308, 551–554 [DOI] [PubMed] [Google Scholar]
- 11. Carney J. P., Maser R. S., Olivares H., Davis E. M., Le Beau M., Yates J. R., 3rd, Hays L., Morgan W. F., Petrini J. H. (1998) The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double strand break repair to the cellular DNA damage response. Cell 93, 477–486 [DOI] [PubMed] [Google Scholar]
- 12. Usui T., Ohta T., Oshiumi H., Tomizawa J., Ogawa H., Ogawa T. (1998) Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95, 705–716 [DOI] [PubMed] [Google Scholar]
- 13. Connelly J. C., de Leau E. S., Leach D. R. (1999) DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 27, 1039–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dillingham M. S., Kowalczykowski S. C. (2008) RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72, 642–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mickelson C., Wiberg J. S. (1981) Membrane-associated DNase activity controlled by genes 46 and 47 of bacteriophage T4D and elevated DNase activity associated with the T4 das mutation. J. Virol. 40, 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Woodworth D. L., Kreuzer K. N. (1996) Bacteriophage T4 mutants hypersensitive to an antitumor agent that induces topoisomerase-DNA cleavage complexes. Genetics 143, 1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mosig G. (1998) Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu. Rev. Genet. 32, 379–413 [DOI] [PubMed] [Google Scholar]
- 18. Almond J. R., Stohr B. A., Panigrahi A. K., Albrecht D. W., Nelson S. W., Kreuzer K. N. (2013) Coordination and processing of DNA ends during double strand break repair: the role of the bacteriophage T4 Mre11/Rad50 (MR) complex. Genetics 195, 739–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hopfner K. (2003) Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr. Opin. Struct. Biol. 13, 249–255 [DOI] [PubMed] [Google Scholar]
- 20. Oldham M. L., Davidson A. L., Chen J. (2008) Structural insights into ABC transporter mechanism. Curr. Opin. Struct. Biol. 18, 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Herdendorf T. J., Albrecht D. W., Benkovic S. J., Nelson S. W. (2011) Biochemical characterization of bacteriophage T4 Mre11/Rad50 complex. J. Biol. Chem. 286, 2382–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hopfner K. P., Karcher A., Shin D. S., Craig L., Arthur L. M., Carney J. P., Tainer J. A. (2000) Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double strand break repair and the ABC-ATPase superfamily. Cell 101, 789–800 [DOI] [PubMed] [Google Scholar]
- 23. Lim H. S., Kim J. S., Park Y. B., Gwon G. H., Cho Y. (2011) Crystal structure of the Mre11-Rad50-ATP S complex: understanding the interplay between Mre11 and Rad50. Genes Dev. 25, 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lammens K., Bemeleit D. J., Möckel C., Clausing E., Schele A., Hartung S., Schiller C. B., Lucas M., Angermüller C., Söding J., Strässer K., Hopfner K.-P. (2011) The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double strand break repair. Cell 145, 54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hopfner K.-P., Craig L., Moncalian G., Zinkel R. A., Usui T., Owen B. A., Karcher A., Henderson B., Bodmer J.-L., McMurray C. T., Carney J. P., Petrini J. H., Tainer J. A. (2002) The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418, 562–566 [DOI] [PubMed] [Google Scholar]
- 26. van Noort J., van Der Heijden T., de Jager M., Wyman C., Kanaar R., Dekker C. (2003) The coiled-coil of the human Rad50 DNA repair protein contains specific segments of increased flexibility. Proc. Natl. Acad. Sci. U.S.A. 100, 7581–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barford D., Das A. K., Egloff M. P. (1998) The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 27, 133–164 [DOI] [PubMed] [Google Scholar]
- 28. Taylor A. M., Groom A., Byrd P. J. (2004) Ataxia-telangiectasia-like disorder (ATLD)–its clinical presentation and molecular basis. DNA Repair 3, 1219–1225 [DOI] [PubMed] [Google Scholar]
- 29. Hopfner K. P., Karcher A., Craig L., Woo T. T., Carney J. P., Tainer J. A. (2001) Structural biochemistry and interaction architecture of the DNA double strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105, 473–485 [DOI] [PubMed] [Google Scholar]
- 30. Williams G. J., Williams R. S., Williams J. S., Moncalian G., Arvai A. S., Limbo O., Guenther G., SilDas S., Hammel M., Russell P., Tainer J. A. (2011) ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat. Struct. Mol. Biol. 18, 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Das D., Moiani D., Axelrod H. L., Miller M. D., McMullan D., Jin K. K., Abdubek P., Astakhova T., Burra P., Carlton D., Chiu H.-J., Clayton T., Deller M. C., Duan L., Ernst D., Feuerhelm J., Grant J. C., Grzechnik A., Grzechnik S. K., Han G. W., Jaroszewski L., Klock H. E., Knuth M. W., Kozbial P., Krishna S. S., Kumar A., Marciano D., Morse A. T., Nigoghossian E., Okach L., Paulsen J., Reyes R., Rife C. L., Sefcovic N., Tien H. J., Trame C. B., van den Bedem H., Weekes D., Xu Q., Hodgson K. O., Wooley J., Elsliger M.-A., Deacon A. M., Godzik A., Lesley S. A., Tainer J. A., Wilson I. A. (2010) Crystal structure of the first eubacterial Mre11 nuclease reveals novel features that may discriminate substrates during DNA repair. J. Mol. Biol. 397, 647–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park Y. B., Chae J., Kim Y. C., Cho Y. (2011) Crystal structure of human Mre11: understanding tumorigenic mutations. Structure 19, 1591–1602 [DOI] [PubMed] [Google Scholar]
- 33. Schiller C. B., Lammens K., Guerini I., Coordes B., Feldmann H., Schlauderer F., Möckel C., Schele A., Strässer K., Jackson S. P., Hopfner K.-P. (2012) Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat. Struct. Mol. Biol. 19, 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Albrecht D. W., Herdendorf T. J., Nelson S. W. (2012) Disruption of the bacteriophage T4 Mre11 dimer interface reveals a two-state mechanism for exonuclease activity. J. Biol. Chem. 287, 31371–31381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Möckel C., Lammens K., Schele A., Hopfner K.-P. (2012) ATP-driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res. 40, 914–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wyman C., Lebbink J., Kanaar R. (2011) Mre11-Rad50 complex crystals suggest molecular calisthenics. DNA Repair 10, 1066–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sambrook J. F., Maniatis T., Sambrook J. F. (1989) Molecular Cloning: A Laboratory Manual, pp. 3.30–3.32, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 38. Herdendorf T. J., Nelson S. W. (2011) Functional evaluation of bacteriophage T4 Rad50 signature motif residues. Biochemistry 50, 6030–6040 [DOI] [PubMed] [Google Scholar]
- 39. Gilbert S. P., Mackey A. T. (2000) Kinetics: a tool to study molecular motors. Methods 22, 337–354 [DOI] [PubMed] [Google Scholar]
- 40. Bloom L. B., Otto M. R., Eritja R., Reha-Krantz L. J., Goodman M. F., Beechem J. M. (1994) Pre-steady-state kinetic analysis of sequence-dependent nucleotide excision by the 3′-exonuclease activity of bacteriophage T4 DNA polymerase. Biochemistry 33, 7576–7586 [DOI] [PubMed] [Google Scholar]
- 41. De la Rosa M. B., Nelson S. W. (2011) An interaction between the Walker A and D-loop motifs is critical to ATP hydrolysis and cooperativity in bacteriophage T4 Rad50. J. Biol. Chem. 286, 26258–26266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Williams G. J., Lees-Miller S. P., Tainer J. A. (2010) Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double strand breaks. DNA Repair 9, 1299–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Williams R. S., Moncalian G., Williams J. S., Yamada Y., Limbo O., Shin D. S., Groocock L. M., Cahill D., Hitomi C., Guenther G., Moiani D., Carney J. P., Russell P., Tainer J. A. (2008) Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Déry U., Coulombe Y., Rodrigue A., Stasiak A., Richard S., Masson J.-Y. (2008) A glycine-arginine domain in control of the human MRE11 DNA repair protein. Mol. Cell. Biol. 28, 3058–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen C., Zhang L., Huang N.-J., Huang B., Kornbluth S. (2013) Suppression of DNA-damage checkpoint signaling by Rsk-mediated phosphorylation of Mre11. Proc. Natl. Acad. Sci. U.S.A. 110, 20605–20610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shibata A., Moiani D., Arvai A. S., Perry J., Harding S. M., Genois M.-M., Maity R., van Rossum-Fikkert S., Kertokalio A., Romoli F., Ismail A., Ismalaj E., Petricci E., Neale M. J., Bristow R. G., Masson J.-Y., Wyman C., Jeggo P. A., Tainer J. A. (2014) DNA double strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 53, 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Paull T. T., Gellert M. (1999) Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13, 1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pufall M. A., Graves B. J. (2002) Autoinhibitory domains: modular effectors of cellular regulation. Annu. Rev. Cell Dev. Biol. 18, 421–462 [DOI] [PubMed] [Google Scholar]
- 49. Graves B. J., Cowley D. O., Goetz T. L., Petersen J. M., Jonsen M. D., Gillespie M. E. (1998) Autoinhibition as a transcriptional regulatory mechanism. Cold Spring Harbor Symp. Quant. Biol. 63, 621–629 [DOI] [PubMed] [Google Scholar]
- 50. Ma E., MacRae I. J., Kirsch J. F., Doudna J. A. (2008) Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 380, 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Szczepek M., Mackeldanz P., Möncke-Buchner E., Alves J., Krüger D. H., Reuter M. (2009) Molecular analysis of restriction endonuclease EcoRII from Escherichia coli reveals precise regulation of its enzymatic activity by autoinhibition. Mol. Microbiol. 72, 1011–1021 [DOI] [PubMed] [Google Scholar]
- 52. Orans J., McSweeney E. A., Iyer R. R., Hast M. A., Hellinga H. W., Modrich P., Beese L. S. (2011) Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell 145, 212–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Miller E. S., Kutter E., Mosig G., Arisaka F., Kunisawa T., Rüger W. (2003) Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67, 86–156 [DOI] [PMC free article] [PubMed] [Google Scholar]


