Background: The double-stranded RNA-dependent protein kinase PKR plays a critical role in the regulation of protein synthesis, apoptosis, cell proliferation, and stress signaling.
Results: SUMO covalently modifies PKR and induces its activation.
Conclusion: SUMO is a new PKR kinase coactivator.
Significance: SUMO may be implicated in the abnormal activation of PKR found in several diseases.
Keywords: Double-stranded RNA (dsRNA), Protein Kinase RNA-activated (PKR), Sumoylation, Translation Control, Virus
Abstract
The dsRNA-dependent kinase PKR is an interferon-inducible protein with ability to phosphorylate the α subunit of the eukaryotic initiation factor (eIF)-2 complex, resulting in a shut-off of general translation, induction of apoptosis, and inhibition of virus replication. Here we analyzed the modification of PKR by the small ubiquitin-like modifiers SUMO1 and SUMO2 and evaluated the consequences of PKR SUMOylation. Our results indicate that PKR is modified by both SUMO1 and SUMO2, in vitro and in vivo. We identified lysine residues Lys-60, Lys-150, and Lys-440 as SUMOylation sites in PKR. We show that SUMO is required for efficient PKR-dsRNA binding, PKR dimerization, and eIF2α phosphorylation. Furthermore, we demonstrate that SUMO potentiates the inhibition of protein synthesis induced by PKR in response to dsRNA, whereas a PKR SUMOylation mutant is impaired in its ability to inhibit protein synthesis and shows reduced capability to control vesicular stomatitis virus replication and to induce apoptosis in response to vesicular stomatitis virus infection. In summary, our data demonstrate the important role of SUMO in processes mediated by the activation of PKR.
Introduction
Mammalian PKR is a dsRNA-dependent protein kinase that is transcriptionally induced by interferon and becomes activated and undergoes autophosphorylation upon binding to dsRNA (1). Activated PKR causes an inhibition of protein synthesis by phosphorylating the α subunit of translation initiation factor 2 (eIF2α) or nuclear factors NFAR1/2 and interferes with virus propagation (1–3). PKR contains an N-terminal dsRNA binding domain and a C-terminal kinase domain, separated by a flexible linker region. The dsRNA binding domain corresponds to the regulatory domain, which consists of two tandem copies of the dsRNA binding motif, dsRBM1 and dsRBM2. The C-terminal kinase domain (residues 258–551) is the catalytic center. It is not well understood how RNA binding results in PKR activation, and several models have been proposed (4, 5). In addition to dsRNA, PKR can be activated by heparin, PKR-activating protein (PACT), or ISG15 (6–8). Although PKR is a multifunctional host defense enzyme, there are mechanisms of activation that remain to be defined to explain the plethora of cell functions (9). In this study, we evaluated the possible regulation of PKR by SUMO.4
SUMOylation is a reversible post-translational modification that consists of the attachment of the SUMO proteins to a lysine residue of a target protein via an enzymatic cascade analogous to, but distinct from, the ubiquitylation pathway (10). Usually, the target lysine is located in the consensus sequence ψKXE (where ψ is a hydrophobic residue, and X any residue) (11, 12). However, SUMO can be also conjugated to lysine residues located in non-consensus sequences. There are four different human genes coding for SUMO proteins: SUMO1, SUMO2, SUMO3, and SUMO4. SUMO1 is the most similar to the yeast Smt3. SUMO2 and SUMO3, nearly identical in sequence and therefore collectively referred to as SUMO2/3, are characterized by an internal SUMOylation site that allows the formation of SUMO chains, and SUMO4 has been correlated to diabetes (13–15). SUMOylation regulates a wide range of processes, but its main function is to regulate protein-protein interactions (16).
In this study, we show that PKR is modified by SUMO1 and SUMO2 in vitro and in vivo. We identified the lysine residues in PKR that work as main SUMO acceptors. We demonstrate that SUMO increases both PKR-dsRNA binding and PKR dimerization, and therefore, it is required for an efficient activation of PKR. In this sense, we show that SUMO increases the efficiency of PKR to phosphorylate eIF2α in vitro and favors the shut-off of the protein synthesis induced by PKR when expressed from a recombinant vaccinia virus or upon dsRNA treatment. In contrast, Ubc9 down-modulation reduces the phosphorylation of eIF2α in response to VSV infection. Finally, we demonstrate that a PKR SUMOylation mutant is unable to inhibit protein synthesis upon dsRNA treatment, and it is partially impaired in its ability to control VSV replication. In summary, here we identify SUMO as a novel regulator of PKR.
EXPERIMENTAL PROCEDURES
Cells, Transfections, and Virus
3T3-like cells derived from homozygous PKR−/− and wild type animals with the same genetic background (PKR+/+) (both a generous gift of C. Weissmann), African green monkey kidney cells BSC-40, and HEK-293 cells were grown in DMEM supplemented with 10% FBS (Life Technologies), 5 mmol/liter l-glutamine (Life Technologies), and penicillin-streptomycin (Life Technologies). The cells were transfected using Xtreme (Roche Diagnostics) or Lipofectamine 2000 (Life Technologies), as suggested by the manufacturer. The recombinant vaccinia virus expressing isopropyl-1-thio-β-d-galactopyranoside-inducible PKR (VV-PKR) was described previously (17). VSV virus was titered by the standard plaque assay method. For infections, cells were infected with VV-PKR or VSV at a multiplicity of infection of 10 pfu/cell.
Plasmids, siRNAs, and Reagents
Plasmids pcDNA3-PKR/HA (18) was a generous gift of Dr. B. Y. Ahn. pcDNA-PKR-N terminus (residues 1–265) and pcDNA-PKR-C terminus (residues 265–550) (19) were a generous gift from Dr. E. Meurs. Plasmids pcDNA-His6-SUMO1, pcDNA-His6-SUMO2, and pcDNA-Ubc9 were described previously (20, 21). Lysine to arginine mutations were carried out using the QuikChange PCR-based site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions, using pcDNA3-PKR/HA plasmid as template and the oligonucleotides listed in Table 1. SMARTpool siRNAs against Ubc9 (siUbc9) and scramble siRNA (siIRR) were purchased from Dharmacon. GST-SENP1 was purchased from Biomol. PKR recombinant protein was purchased from Merck Millipore. Recombinant eIF2α was purchased from ProSpec. Antibodies to PKR and SUMO1 were purchased from Santa Cruz Biotechnology. Antibodies to phospho-PKR (Thr-451), phospho-eIF2α (Ser-51), and anti-SUMO2 were purchased from Life Technologies. Anti-VSV-M antibody was from KeraFAST. Anti-HA monoclonal antibody was purchased from Covance. Anti-actin antibody was from MP Biomedicals. Anti-VSV-G antibody was a generous gift of Dr. I Ventoso.
TABLE 1.
Oligonucleotides used in site-directed mutagenesis
aa, amino acid.
| Position (aa) | Sequence |
|---|---|
| 60 | Forward, 5′-ggtgaaggtagatcaaggaaggaagcaaaaaatgccg-3′ |
| Reverse, 5′-cggcattttttgcttccttccttgatctaccttcacc-3′ | |
| 150 | Forward, 5′-ggtacaggttctactagacaggaagcaaaacaattggcc-3′ |
| Reverse, 5′-ggccaattgttttgcttcctgtctagtagaacctgtacc-3′ | |
| 304 | Forward, 5′-gttaaatataataacgagagggcggagcgtgaag-3′ |
| Reverse, 5′-cttcacgctccgccctctcgttattatatttaac-3′ | |
| 440 | Forward, 5′-ggacttgtaacatctctgagaaatgatggaaagcg-3′ |
| Reverse, 5′-cgctttccatcatttctcagagatgttacaagtcc-3′ |
In Vitro SUMO Conjugation Assay
In vitro SUMO conjugation assays were performed on [35S]methionine-labeled in vitro-transcribed/translated proteins as described previously (22) using recombinant E1 SUMO-activating enzyme (SAE1/2) (Biomol, Enzo Life Sciences), E2 SUMO-conjugating enzyme (Ubc9), and SUMO1 or SUMO2. The in vitro transcription/translation of proteins was performed by using 1 μg of plasmid DNA and a rabbit reticulocyte-coupled transcription/translation system according to the instructions provided by the manufacturer (Promega).
In Vitro DeSUMOylation Assay
In vitro deSUMOylation assay with GST-SENP1 was performed on PKR-SUMO1 as described previously (23).
λ-Phosphatase Treatment
SUMOylated PKR protein was incubated with 1 μl of λ-phosphatase (New England Biolabs) in buffer for λ-phosphatase treatment (50 mm Tris/HCl, pH 7.5, 100 mm NaCl, 0.1 mm EGTA, 2 mm DTT, 0.01% Brij 35) (New England Biolabs) supplemented with 2 mm MnCl2. The reaction was incubated for 30 min at 30 °C and stopped with SDS-PAGE loading buffer.
Western Blot Analysis
Cells were washed in PBS, scraped into SDS-PAGE loading buffer, and boiled for 5 min. Proteins of total extracts were separated by SDS-PAGE and transferred onto a nitrocellulose membrane. The membranes were incubated with the indicated antibodies, and signals were detected by using chemiluminescence.
PKR Protein Kinase Assay
eIF2α phosphorylation catalyzed by PKR or SUMOylated PKR protein was carried out in the presence of dsRNA, as indicated. The reaction was stopped by the addition of SDS-PAGE loading buffer, and proteins were separated by SDS-PAGE and transferred to nitrocellulose membrane. Phosphorylation of eIF2α was evaluated using the antibody anti-phospho-eIF2α. The blots were probed with anti-PKR and anti-eIF2α to quantify protein levels.
dsRNA Binding Assay
[35S]Methionine-labeled in vitro-translated unmodified or SUMOylated PKR protein was mixed with 50 μl of poly(I:C)-agarose beads in binding buffer (20 mm Tris-HCl, pH 7.5, 0.3 m NaCl, 5 mm MgCl2, 1 mm DTT, 0.1 mm phenylmethylsulfonyl fluoride, 0.5% Nonidet P-40, and 10% glycerol) and incubated at 30 °C for 30 min. Beads were then washed with 500 μl of binding buffer four times. The proteins bound to beads after washing were analyzed by SDS-PAGE followed by fluorography.
Purification of His-tagged Conjugates
The purification of His-tagged conjugates, using Ni2+-nitrilotriacetic acid-agarose beads allowing the purification of proteins that are covalently conjugated to SUMO, was performed as described previously (24).
GST Pulldown
GST pulldown experiments were performed using [35S]methionine-labeled in vitro-transcribed/translated PKR-WT or PKR-SUMOmut protein and the recombinant GST-PKR protein as described previously (24).
In Vitro Translation Inhibition Assay
Cells were co-transfected with the reporter PGL3-control and the indicated plasmids, and 36 h after transfection, cells were incubated or not with poly(I:C) (5 μg/ml) for 7 h. Then, cell extracts were harvested and assayed for luciferase activity after normalizing for the transfection efficiency by measuring the total protein.
Apoptosis Quantification
Apoptosis was quantified by flow cytometry using the caspase-3, active form, mAb apoptosis kit from BD Pharmingen, according to the manufacturer's protocol.
Statistical Analysis
For statistical analysis between control and different groups, the Student's t test was applied. The significance level chosen for the statistical analysis was p < 0.05.
RESULTS
PKR Conjugates to SUMO in Vitro and in Vivo
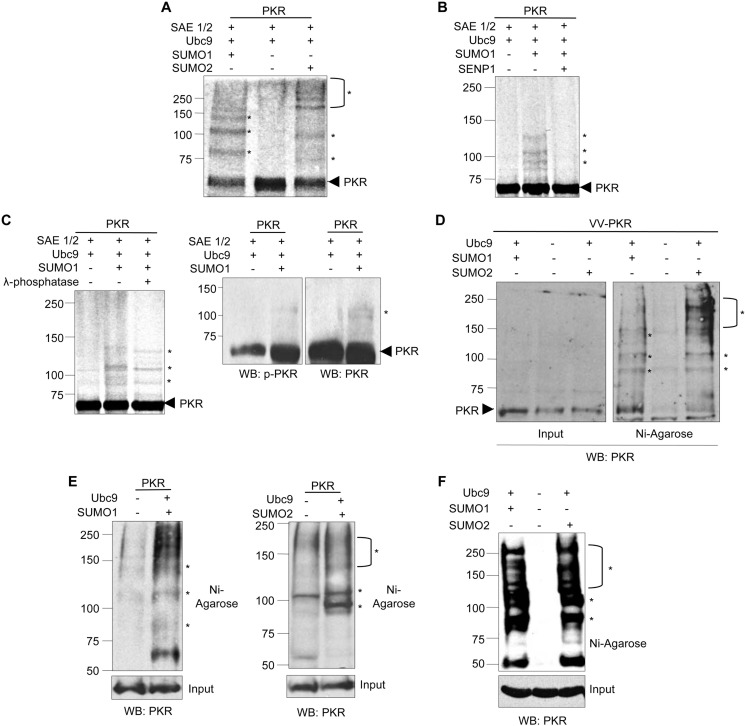
PKR can be modulated by its interaction with ubiquitin and the ubiquitin-like protein ISG15 (7, 25). Thus, we decided to analyze whether PKR can also be modulated by SUMO. First, we carried out a SUMOylation assay using [35S]methionine-labeled in vitro-translated PKR protein as a substrate. As expected, PKR was detected as a band of ∼68 kDa (Fig. 1A, lane 2). The addition of SUMO1 to the SUMOylation reaction led to the appearance of at least three higher molecular mass bands of ∼85, 105, and 125 kDa (Fig. 1A, lane 1) that correspond to PKR-SUMO1. Similarly, when SUMO2 was added to the SUMOylation reaction, we observed two very faint bands of ∼85 and 105 kDa and a smear of additional higher molecular mass bands that correspond to PKR-SUMO2 (Fig. 1A, lane 3). To further demonstrate that the bands observed corresponded to SUMOylated PKR, PKR-SUMO1 protein obtained as described above was incubated with the catalytic domain of the SUMO-specific protease SENP1 fused to GST. The incubation of PKR-SUMO1 with SENP1 led to the disappearance of the higher molecular mass bands corresponding to PKR-SUMO1 (Fig. 1B). These results indicated that PKR can be SUMOylated in vitro. The detection of the PKR-SUMO1 and PKR-SUMO2 proteins as broad bands in these assays, as well as the multiple migration bands that could be detected in some SUMOylation assay experiments, suggested that PKR-SUMO protein might be phosphorylated. To evaluate this hypothesis, PKR-SUMO1 protein obtained as described above was treated or not with λ-phosphatase. As shown in Fig. 1C, left panel, we observed an increase in the electrophoretic mobility of each of the PKR-SUMO1 bands after treatment with λ-phosphatase, suggesting that the SUMOylated PKR protein is phosphorylated. In addition, we analyzed by Western blot the phosphorylation status of in vitro-translated PKR protein subjected to in vitro SUMOylation with SUMO1, using an anti-phospho-PKR antibody. Immunoblot analysis with anti-PKR antibody recognized two bands of ∼105 kDa when SUMO1 was added to the reaction. One of these PKR-SUMO1 bands was also detected by anti-phospho-PKR antibody (Fig. 1C, right panel), confirming that PKR-SUMO1 protein is phosphorylated. Then, we decided to analyze whether PKR is also modified by SUMO in vivo. First, we analyzed the SUMOylation of PKR protein expressed by VV-PKR. HEK-293 cells were transfected with pcDNA, Ubc9, and His6-SUMO1 or Ubc9 and His6-SUMO2, and 36 h after transfection, cells were infected with VV-PKR. At 16 h after infection, whole protein extracts or histidine-tagged purified proteins were analyzed by Western blotting using an anti-PKR antibody. Analysis of the histidine-purified extracts revealed the appearance of bands of the expected molecular masses for PKR-SUMO1 and PKR-SUMO2 only in those cells transfected with His6-SUMO1 or His6-SUMO2, respectively (Fig. 1D), indicating that PKR is modified by SUMO when expressed by a recombinant vaccinia virus. We then evaluated whether PKR can be modified by SUMO when expressed from a plasmid. HEK-293 cells were co-transfected with PKR and pcDNA, His6-SUMO1 and Ubc9, or His6-SUMO2 and Ubc9, and 48 h after transfection, we analyzed the whole cell extracts and histidine-tagged purified proteins by Western blotting with anti-PKR antibody. Analysis of the histidine-purified extracts revealed the appearance of the expected higher molecular mass bands only in those cells co-transfected with His6-SUMO1 or His6-SUMO2 (Fig. 1E), confirming that PKR is SUMOylated in transfected cells. Then, we decided to evaluate the SUMOylation of endogenous PKR protein. HEK-293 cells were transfected with pcDNA, His6-SUMO1 and Ubc9, or His6-SUMO2 and Ubc9, and 48 h after transfection, Western blot analysis of the histidine-tagged purified proteins using anti-PKR antibody was carried out. As shown in Fig. 1F, bands of the molecular mass expected for PKR-SUMO1 and PKR-SUMO2 were exclusively detected in those cells transfected with His6-SUMO1 or His6-SUMO2, respectively, indicating that endogenous PKR can be SUMOylated. Altogether these results demonstrated that PKR is modified by SUMO1 and SUMO2 in vitro and in vivo.
FIGURE 1.
Covalent modification of PKR by SUMO1 and SUMO2 in vitro and in vivo. A, modification of [S35]methionine-labeled in vitro-translated PKR protein by SUMO1 or SUMO2 in vitro. SAE1/2, E1 SUMO-activating enzyme. B, deconjugation of SUMO1 from PKR by SENP1. C, [35S]methionine-labeled SUMOylated PKR protein was treated with λ-phosphatase for 30 min (left panel). In vitro-translated PKR protein incubated in a SUMOylation assay with SUMO1 was analyzed by Western blot (WB) using anti-PKR or anti-phospho-PKR antibody (right panel). D, modification of PKR expressed from VV-PKR with SUMO1 or SUMO2. E, modification of transfected PKR by SUMO1 or SUMO2. F, modification of endogenous PKR by SUMO1 or SUMO2. The position of PKR is indicated by an arrowhead; stars indicate the position of PKR-SUMO1 or PKR-SUMO2 bands.
Identification of the Main SUMO Acceptor Sites in PKR
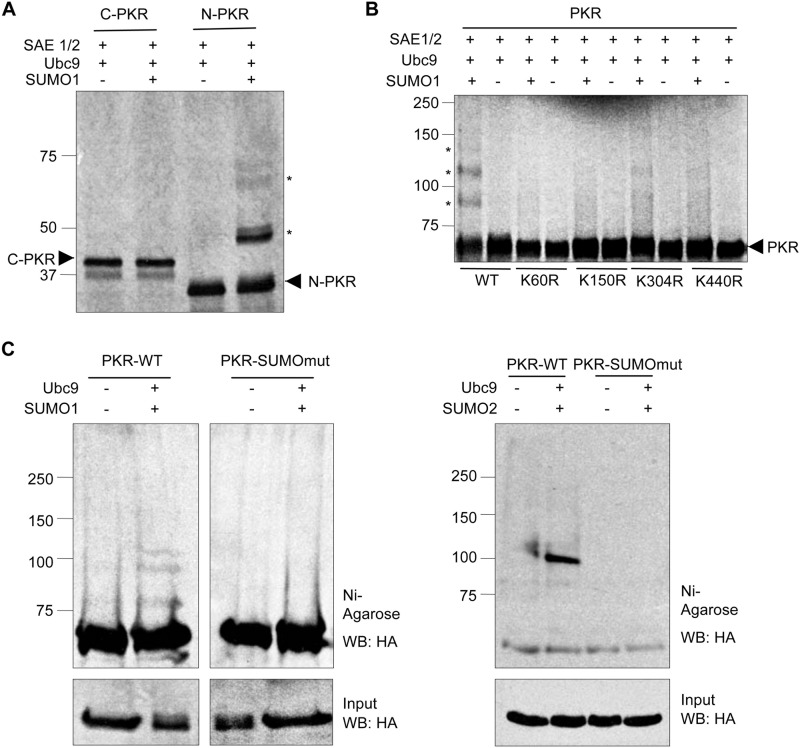
We then decided to identify the main lysine residues that conjugate to SUMO in PKR. First, we carried out a SUMOylation assay using SUMO1 and [35S]methionine-labeled in vitro-translated N-terminal and C-terminal PKR fragments as substrates. We did not detect any additional band in the lane corresponding to the C-terminal PKR fragment after incubation with SUMO1 (Fig. 2A). However, the addition of SUMO1 to the SUMOylation reaction led to the appearance of two additional bands of ∼50 and 65 kDa in the lane corresponding with the N-terminal PKR protein (Fig. 2A), indicating the presence of at least two SUMO acceptors in the N-terminal domain of PKR. To note, at least one shifted band migrating slightly slower than each N-terminal PKR-SUMO1 band was detected, suggesting that SUMO conjugation to the N-terminal PKR domain induces its phosphorylation.
FIGURE 2.
Identification of the main lysine residues in PKR that conjugate SUMO. A, modification of [S35]methionine-labeled in vitro-translated C-terminal or N-terminal PKR fragments (C-PKR and N-PKR, respectively) by SUMO1 in vitro. SAE1/2, E1 SUMO-activating enzyme. B, modification of [S35]methionine-labeled in vitro-translated PKR-WT or the indicated PKR mutant proteins by SUMO1 in vitro. C, modification of transfected PKR-WT or PKR-SUMOmut by SUMO1 or SUMO2. The position of PKR is indicated by an arrowhead; stars indicate the position of PKR-SUMO1 or PKR-SUMO2 bands. WB, Western blot.
In silico analysis of the amino acid sequence of PKR using SUMOsp program revealed at least 5 lysine residues as putative SUMO conjugation sites, including Lys-60 and Lys-150, previously identified as required for PKR-dsRNA interaction (26–28). In addition, the SUMOplot software program revealed the lysine residues 304 and 440, previously implicated in conforming the ring of the cleft important for heparin binding (29), as potential SUMOylation sites. Therefore, we generated a series of mutants in these residues and evaluated their SUMOylation in vitro. As shown in Fig. 2B, we observed a clear reduction in PKR SUMOylation after mutating lysine residue 60, 150, or 440 in PKR. We then constructed a triple mutant, PKR-K60R/K150R/K440R (PKR-SUMOmut), and evaluated its SUMOylation in vivo. HEK-293 cells were co-transfected with PKR-SUMOmut or the control PKR-WT, together with pcDNA, Ubc9 and His6-SUMO1, or Ubc9 and His6-SUMO2, and 48 h after transfection, evaluation of PKR SUMOylation was carried out. Western blotting analysis of the histidine-tagged proteins purified from Ubc9 and His6-SUMO1 or His6-SUMO2 co-transfected cells with anti-HA antibody revealed the appearance of the expected PKR-SUMO1 and PKR-SUMO2 bands, respectively, exclusively in the PKR-WT-expressing cells (Fig. 2C), indicating that PKR-SUMOmut is defective in SUMO1 and SUMO2 conjugation. Altogether these results pointed to lysine residues 60, 150, and 440 as SUMO acceptor sites in PKR.
SUMO Promotes PKR-dsRNA Binding, PKR Dimerization, and the Activity of PKR in Vitro
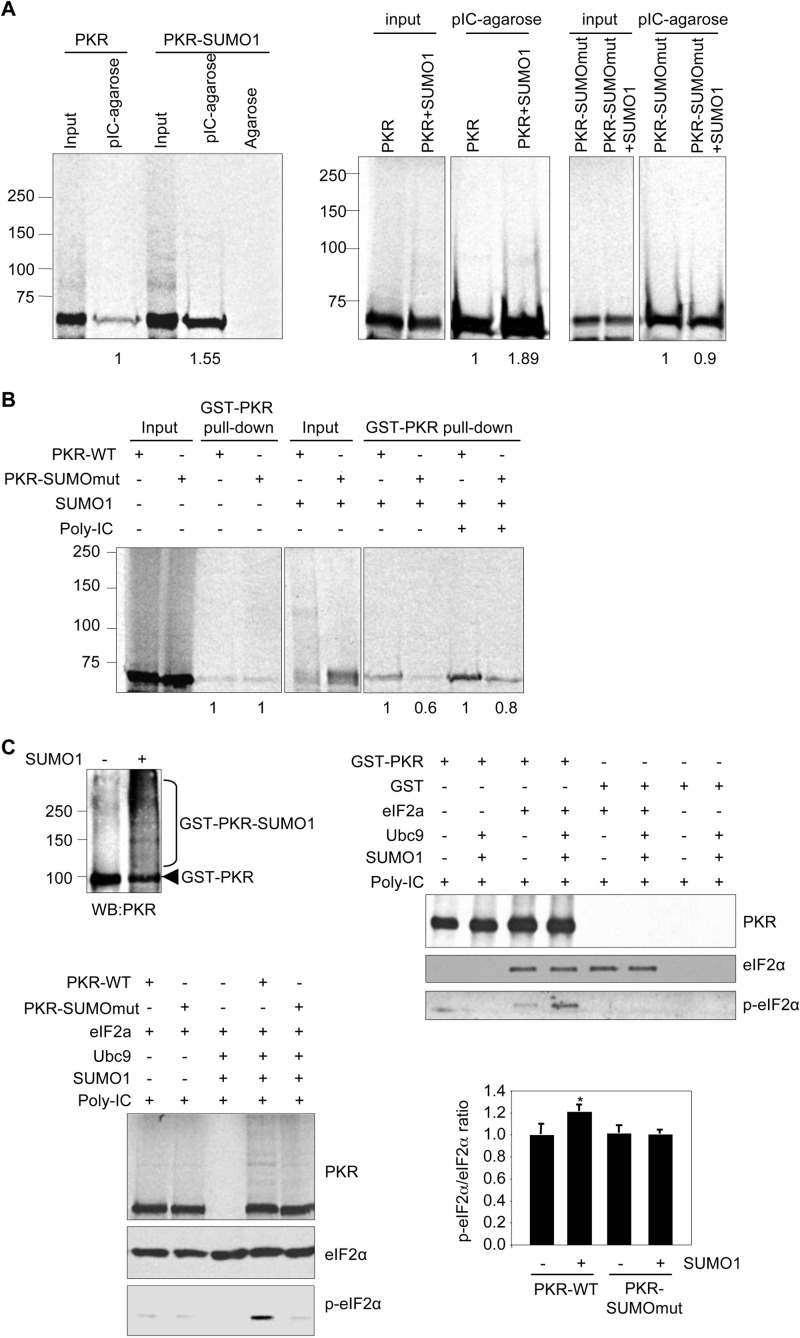
SUMO modification can regulate either protein-protein or protein-nucleic acid interactions (30). Therefore, we decided to evaluate whether SUMO has an effect on the interaction of PKR with dsRNA or the dimerization of the PKR protein. First, we carried out a poly(I:C)-agarose pulldown assay with unmodified or SUMOylated [35S]methionine-labeled in vitro-translated PKR protein, as indicated. PKR interacted with poly(I:C)-agarose, as expected (Fig. 3A, left panel). Interestingly, the interaction between PKR and poly(I:C)-agarose increased in the presence of SUMO (Fig. 3A, left panel). To confirm this result, we carried out a poly(I:C)-agarose pulldown assay with [35S]methionine-labeled in vitro-translated PKR-WT or PKR-SUMOmut protein, previously incubated or not with SUMO1 in a SUMOylation assay. We detected both PKR-SUMO1 protein and an increase in unSUMOylated PKR-WT protein interacting with poly(I:C)-agarose in the presence of SUMO1 (Fig. 3A, right panel). In contrast, the interaction between PKR-SUMOmut and poly(I:C)-agarose was not affected by SUMO1 (Fig. 3A, right panel). Altogether these results indicated that SUMO1 favored PKR-dsRNA binding.
FIGURE 3.
SUMO potentiates PKR-dsRNA binding, PKR dimerization, and PKR activity. A, poly(I:C)-agarose binding activity (pIC-agarose) of the in vitro-translated [S35]methionine-labeled PKR protein previously subjected to in vitro SUMOylation assay in the presence or absence of SUMO1 (left panel). Right panel, poly(I:C)-agarose binding activity of the in vitro-translated [S35]methionine-labeled PKR-WT or PKR-SUMOmut protein previously subjected to in vitro SUMOylation assay in the presence or absence of SUMO1. The ratio of bound protein was calculated as the proportion between the bound protein band and the input band. Relative ratio of bound PKR-WT or PKR-SUMOmut protein in the absence of SUMO1 was normalized to 1. B, in vitro-translated [S35]methionine-labeled PKR-WT or PKR-SUMOmut proteins previously subjected to in vitro SUMOylation assay in the presence or absence of SUMO1 were tested for interaction with GST-PKR protein. The amount of PKR-SUMOmut protein bound to GST-PKR relative to the levels of WT protein bound to GST-PKR in each condition is shown. C, recombinant PKR protein was subjected to in vitro SUMOylation assay in the presence or absence of SUMO1 (upper left panel). Upper right panel, in vitro kinase assay using 25 ng of recombinant PKR protein subjected to in vitro SUMOylation assay in the presence or absence of SUMO1 and recombinant eIF2α protein as a substrate is shown. Lower left panel, in vitro kinase assay with in vitro-translated PKR-WT or PKR-SUMOmut proteins previously subjected to in vitro SUMOylation assay in the presence or absence of SUMO1. Phosphorylation of eIF2α was detected using anti-phospho-eIF2α antibody. Equal loading of eIF2α and PKR per reaction was confirmed by Western blotting with the antibody directed against eIF2α or PKR protein. The lower right panel represents the ratio of phospho-eIF2α (p-eIF2α)/total eIF2α from three independent experiments. Relative eIF2α phosphorylation in the presence of PKR-WT was normalized to 1. Bars, S.E. *, p < 0.05, Student's t test, relative to PKR-WT.
Then, a GST-PKR-WT pulldown assay with unmodified or SUMOylated [35S]methionine-labeled in vitro-translated PKR-WT or PKR-SUMOmut proteins, in the presence or absence of dsRNA, as indicated, was performed. As shown in Fig. 3B, both PKR-WT and PKR-SUMOmut bound with similar efficiency to GST-PKR, in the absence of SUMO. However, after incubation with the SUMOylation components, PKR-WT exhibited stronger interaction with GST-PKR than PKR-SUMOmut, either in the presence or in the absence of dsRNA (Fig. 3B). These results indicated that SUMO promoted PKR dimerization. Therefore, we decided to evaluate whether SUMO can also improve PKR activity in vitro. In vitro kinase assay using unmodified or SUMOylated recombinant GST-PKR protein (Fig. 3C, upper left panel) and recombinant eIF2α as PKR substrate was carried out. GST-PKR induced phosphorylation of eIF2α, as expected (Fig. 3C, upper right panel). A clear increase in the levels of phosphorylated eIF2α was detected when we used SUMOylated GST-PKR protein as the kinase (Fig. 3C, upper right panel). Similarly, we observed an increase in the levels of phosphorylated eIF2α when in vitro-translated PKR-WT protein was previously incubated in an in vitro SUMOylation assay with SUMO1 (Fig. 3C, lower right panel). In contrast, we did not observe significant differences in the amount of eIF2α phosphorylated by PKR-SUMOmut independently of the presence or not of SUMO1 (Fig. 3C, lower right panel). Altogether these results indicated that SUMO favored the kinase activity of PKR.
SUMO Potentiates the Control of Protein Synthesis by PKR
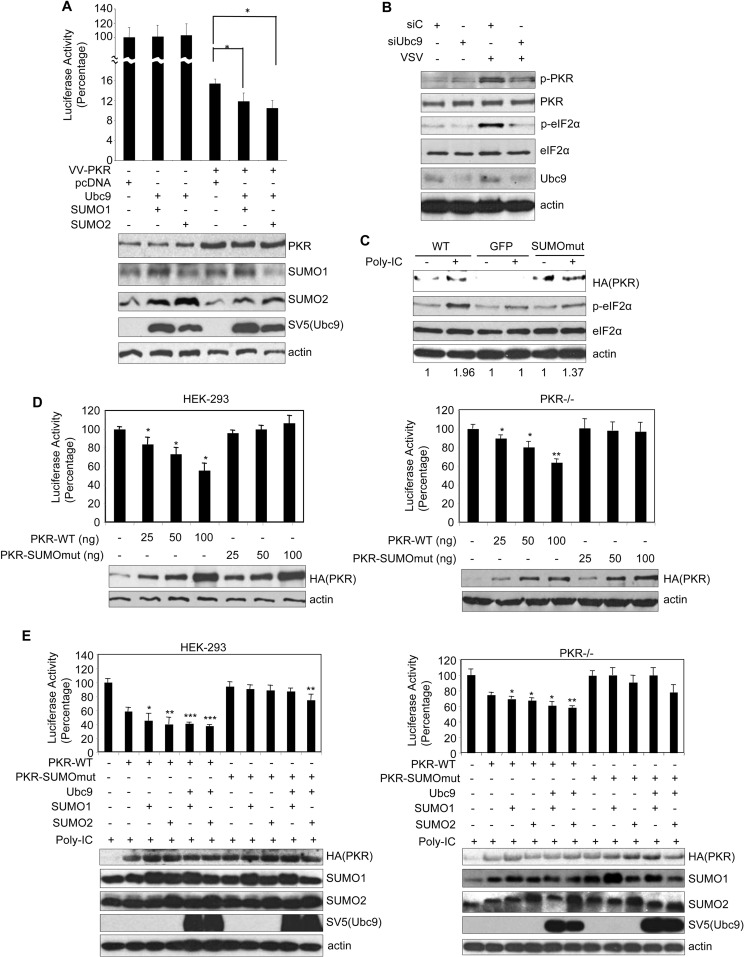
Covalent SUMO conjugation may regulate the subcellular localization or stability of the target proteins. We did not detect any difference between the subcellular localization or stability of PKR-SUMOmut when compared with PKR-WT. Then, we decided to analyze the effect of SUMO on the activity of PKR in vivo. First, we evaluated whether overexpression of Ubc9 and SUMO1 or SUMO2 altered the inhibition of protein synthesis induced by VV-PKR (17), by using a luciferase reporter system. HEK-293 cells were co-transfected with PGL3-control vector and pcDNA, Ubc9 and His6-SUMO1, or Ubc9 and His6-SUMO2, and 36 h after transfection, cells were infected with VV-PKR and treated or not with isopropyl-1-thio-β-d-galactopyranoside to induce PKR expression, as indicated. At 8 h after infection, luciferase expression levels were measured. Expression of PKR reduced luciferase synthesis, in accordance with previous studies (28, 31–33) (Fig. 4A). Interestingly, the inhibition of luciferase synthesis observed after PKR expression was significantly stronger in those cells transfected with Ubc9 and SUMO1 or Ubc9 and SUMO2 than in the pcDNA-transfected cells, suggesting that SUMO favored the shut-off of the protein synthesis induced by PKR. A positive effect of SUMOylation machinery components on endogenous PKR activity was indeed confirmed down-modulating the levels of the E2 SUMO-conjugating enzyme Ubc9. HEK-293 cells were transfected with siRNAs against Ubc9 (siUbc9); at 48 h after transfection, cells were infected with VSV, and 8 h after infection, protein extracts were analyzed by Western blotting with the indicated antibodies. As shown in Fig. 4B, VSV induced the phosphorylation of both PKR and eIF2α in those cells transfected with siRNA control (siC), as expected. Phosphorylation of PKR or eIF2α was clearly reduced in those cells transfected with siUbc9 (Fig. 4B). To further prove a positive role of SUMO on the activity of PKR, we decided to evaluate the ability of PKR-SUMOmut to induce eIF2α phosphorylation in response to dsRNA. PKR-deficient immortalized fibroblasts were transfected with PKR-WT, PKR-SUMOmut, or GFP, and 36 h after transfection, cells were incubated or not with dsRNA for 8 h. Western blot analysis of the protein extracts revealed that incubation with dsRNA induced a strong phosphorylation of eIF2α in those cells expressing PKR-WT protein, as expected (Fig. 4C). The level of phosphorylated eIF2α protein detected in cells expressing PKR-SUMOmut was slightly higher than that detected in cells transfected with GFP and clearly lower than that detected in cells expressing the WT protein (Fig. 4C), suggesting that SUMOylation of PKR contributes to the inhibition of protein synthesis resulting from dsRNA treatment. To evaluate this hypothesis, HEK-293 cells or PKR-deficient cells were co-transfected with PGL3-control and increasing doses of PKR-WT or PKR-SUMOmut, as indicated, and 48 h after transfection, luciferase expression was measured. As shown in Fig. 4D, we detected a decrease in the luciferase reporter synthesis in a PKR-WT dose-dependent manner in both HEK-293 and PKR-deficient cells. However, transfection of PKR-SUMOmut did not induce a significant decrease in the luciferase expression in any of the cells analyzed (Fig. 4D), indicating that a PKR SUMOylation mutant was unable to inhibit protein synthesis in the absence of stimulus. We then evaluated the consequences of increasing the SUMOylation machinery on the activity of PKR-WT or PKR-SUMOmut using this assay. HEK-293 or PKR-deficient cells were co-transfected with PGL3-control and PKR-WT or PKR-SUMOmut together with pcDNA, SUMO1, SUMO2, Ubc9 and SUMO1, or Ubc9 and SUMO2, and then treated or not with dsRNA, as indicated. The luciferase expression levels detected after PKR-WT transfection were not significantly affected by co-transfection of SUMOylation machinery components, either in HEK-293 or in PKR-deficient cells, in the absence of stimulus. Co-transfection of PKR-deficient cells with SUMOylation machinery components induced a significant reduction in the luciferase levels detected after transfection of PKR-WT but did not affect the luciferase expression detected in PKR-SUMOmut-transfected cells, upon stimulation with dsRNA (Fig. 4E). Similarly, overexpression of SUMOylation machinery components also potentiated the inhibition of protein synthesis induced by PKR-WT transfection in dsRNA-treated HEK-293 cells (Fig. 4E). In contrast, we only observed a significant reduction in the luciferase expression levels of dsRNA-treated PKR-SUMOmut-transfected HEK-293 cells after overexpression of Ubc9 and SUMO2 (Fig. 4E), likely due to activation of endogenous PKR. Altogether these results indicated that SUMO potentiated the PKR-mediated dsRNA-induced protein synthesis inhibition.
FIGURE 4.
SUMO potentiates the control of protein synthesis by PKR. A, HEK-293 cells were co-transfected with the reporter plasmid PGL3-control together with the indicated plasmid DNAs and then infected with VV-PKR. Cells were harvested 8 h after infection and assayed for luciferase activity after normalizing for total protein. The relative luciferase activity obtained is represented on the y axis. Each experiment was done in triplicate and repeated three times. Bars, S.E. *, p < 0.05, Student's t test. B, Western blotting of HEK-293 cells transfected with siUbc9 or siC and infected or not with VSV, as indicated. p-PKR, phospho-PKR; p-eIF2α, phospho-eIF2α. C, Western blotting of PKR-deficient cells transfected with HA-PKR, GFP, or HA-PKR SUMOmut and treated or not with poly(I:C), as indicated. The values under the Western blot panels represent the intensities of phospho-eIF2α (p-eIF2α) in each lane normalized with respect to the corresponding total eIF2α. For comparison, the value obtained in the absence of poly(I:C) was set as 1. D, inhibition of protein synthesis induced by transfection of PKR-WT but not PKR-SUMOmut in HEK-293 cells (left panel) or in PKR-deficient cells (right panel). Cells were co-transfected with the reporter plasmid PGL3-control together with the indicated plasmid DNAs, and 48 h after transfection, cells were assayed for luciferase activity after normalizing for total protein. The relative luciferase activity obtained is represented on the y axis. Each experiment was done in triplicate and repeated three times. Bars, S.E. E, overexpression of SUMOylation machinery components potentiates the shut-off of protein synthesis induced by transfection of PKR-WT in HEK-293 (left panel) or in PKR-deficient cells (right panel). Cells were co-transfected with the reporter plasmid PGL3-control together with the indicated plasmid DNAs, and then cells treated with poly(I:C) and 7 h after treatment were assayed for luciferase activity. The relative luciferase activity obtained after normalization to total protein amount is represented on the y axis. Each experiment was done in triplicate and repeated three times. Bars, S.E. *, p < 0.05; **, p < 0.005; ***, p < 0.0005, Student's t test, relative to PKR-WT- or PKR-SUMOmut-transfected cells.
SUMO Contributes to the Antiviral Activity of PKR
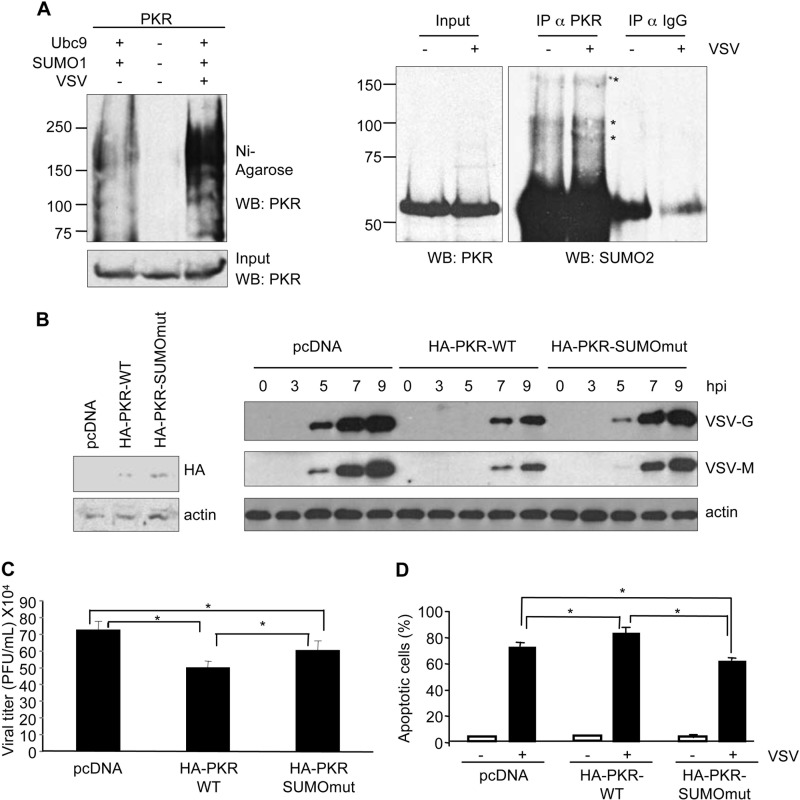
It has been previously reported that VSV infection promotes the SUMOylation of different cellular proteins (23, 34, 35). Therefore, we decided to investigate whether VSV infection also induced SUMOylation of PKR. HEK-293 cells were co-transfected with PKR, Ubc9, and His6-SUMO1, and 36 h after transfection, cells were infected or not with VSV. At 6 h after infection, whole cell extracts and histidine-purified proteins were analyzed by Western blotting using anti-PKR antibody. As shown in Fig. 5A, left panel, VSV infection induced an increase in the levels of PKR-SUMO1 protein. We also evaluated the putative effect of VSV infection on endogenous PKR-SUMO2 modification. Western blot analysis of immunoprecipitated PKR protein using anti-SUMO2 antibody revealed the presence of the expected PKR-SUMO2 bands whose intensity clearly increased in those cells infected with VSV (Fig. 5A, right panel). Therefore, we decided to evaluate the role of SUMO in the antiviral activity of PKR. PKR-deficient cells stably transfected with pcDNA, PKR-WT, or PKR-SUMOmut were infected with VSV, and Western blotting analysis of VSV protein synthesis at different times after infection was carried out. As shown in Fig. 5B, synthesis of VSV proteins was reduced in PKR-WT-expressing cells, in comparison with that detected in PKR-deficient cells, as reported previously (36–38). The level of VSV proteins detected in those cells transfected with PKR-SUMOmut was lower than that detected in the pcDNA-transfected cells but higher than that detected in PKR-WT-expressing cells (Fig. 5B). These results suggested that the PKR-SUMOylation mutant has reduced antiviral activity in comparison with the WT protein. To prove this hypothesis, virus titers 24 h after infection were determined. As shown in Fig. 5C, we could observe only a small but significant reduction in the viral titer when PKR was reintroduced in the PKR-deficient cells. In addition, the titer observed in cells transfected with PKR-SUMOmut was significantly lower than that detected in pcDNA-transfected cells, but higher than the detected in PKR-WT cells. Altogether these results indicated that SUMO contributes to the antiviral effect mediated by PKR. The antiviral activity of PKR is attributed to its ability to block protein synthesis and mediate an apoptotic response (39). Therefore, pcDNA-, PKR-WT-, or PKR-SUMOmut-transfected PKR-deficient cells were analyzed for apoptosis induction in response to VSV infection. As shown in Fig. 5D, the apoptosis detected in PKR-WT cells was significantly higher than that detected in pcDNA or PKR-SUMOmut stably transfected cells. Moreover, the apoptosis observed in those cells expressing PKR-SUMOmut were significantly lower than that detected in the cells transfected with an empty vector (Fig. 5D). These results indicated that SUMO contributes to the induction of apoptosis mediated by PKR in response to VSV infection.
FIGURE 5.
SUMO contributes to the PKR-mediated antiviral activity. A, modification of transfected PKR by SUMO1 in cells infected or not with VSV, as indicated (left panel). SUMO2 modification of endogenous PKR in cells infected or not with VSV, as indicated (right panel). WB, Western blot; IP, immunoprecipitation. B, PKR-deficient cells were co-transfected with a plasmid DNA containing a puromycin resistance gene and the indicated plasmids (1:10 ratio) and selected with puromycin (3 mg/ml) for 3 days. Puromycin-resistant cells were evaluated for PKR expression (left panel), and then cells were infected with VSV at a multiplicity of infection of 10, and at different times after infection, cells were recovered and analyzed by Western blotting using anti-VSV-M and anti-VSV-G antibodies (right panel). hpi, hours after infection. C, PKR-deficient cells transfected with the indicated plasmids, as indicated in Fig. 5B legend, were infected with VSV at a multiplicity of infection of 10, and 24 h after infection, virus titers in supernatants were measured. *, p < 0.05, Student's t test. D, PKR-deficient cells transfected with pcDNA, PKR-WT, or PKR-SUMOmut and infected with VSV at a multiplicity of infection of 10 for 24 h, as indicated in panel C, were subjected to caspase 3 staining according to the manufacturer's specifications (BD Biosciences). Cells were then subjected to flow cytometry analysis by using FACScan. Each experiment was done in triplicate and repeated three times. Bars, S.E. *, p < 0.05, Student's t test.
DISCUSSION
Modification of many signaling proteins by ubiquitin or ubiquitin-like proteins is emerging as an important mechanism to regulate innate immune responses. Here we demonstrate that PKR can be modified by both SUMO1 and SUMO2 in vitro and in vivo. We identified lysine residues Lys-60 and Lys-150, located at the dsRNA binding domain of PKR, and Lys-440, located at the C-terminal domain of the protein, as SUMO acceptors in PKR. The proximity of the SUMO acceptor sites in PKR to the lysine residues implicated in ISG15 conjugation (lysine residues Lys-69 and Lys-159 in PKR) (7), together with those results showing that a mutant of PKR in the lysine residues Lys-60, Lys-150, and Lys-61 was more strongly ISGylated than the WT protein (7), open up the possibility that SUMO and ISG15 may compete for modification of PKR.
Based on the location of the SUMO acceptors in PKR, we hypothesized that SUMO conjugation might regulate dsRNA binding or PKR dimerization, and consequently, kinase activity. Consistent with this notion, our results demonstrated that SUMO promoted PKR-dsRNA binding, dimerization of PKR, and kinase activation in vitro and in vivo, whereas cells with reduced Ubc9 expression exhibit defective responses to VSV infection, as evidenced by failure to fully phosphorylate eIF2α. Moreover, mutation of the SUMOylation sites in PKR abolished its ability to inhibit protein synthesis in response to dsRNA and significantly reduced its proapoptotic and antiviral activities, providing evidences for a critical role of SUMO in the PKR activity.
PKR is an important component of the antiviral machinery of the cell as evidenced by the fact that many viruses have developed mechanisms to inhibit PKR (40). Our results demonstrate that VSV infection promotes PKR SUMOylation and that SUMO contributes to the antiviral activity of PKR, indicating that SUMOylation of PKR is an important antiviral response of the cell. Therefore, it is possible to speculate that recruitment of SUMO to viral factories (41, 42) or targeting of the SUMOylation machinery by virus (43) may work as mechanisms to counteract PKR antiviral activity.
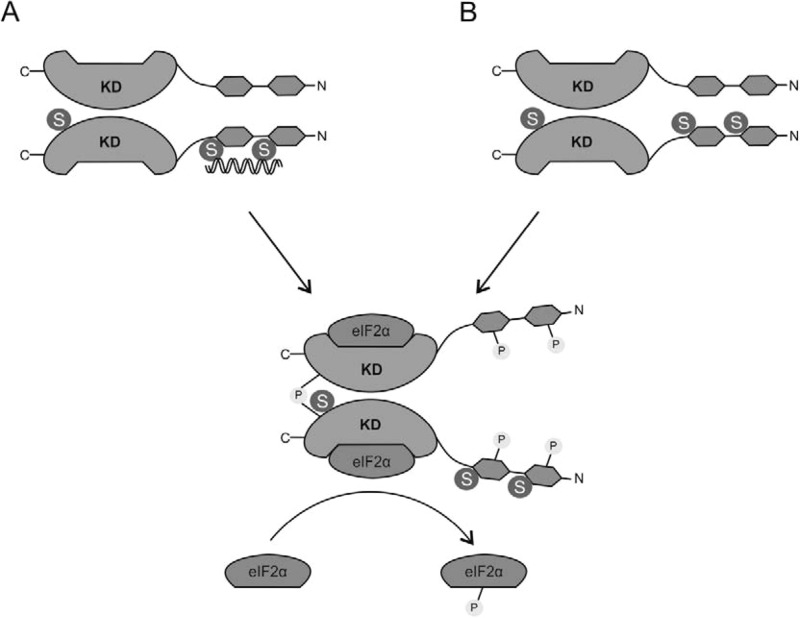
Different models have been proposed to explain the dsRNA activation of PKR (4). Some models suggest that RNA binding promotes dimerization of PKR by inducing conformational changes, and others propose that RNA bring multiple PKR monomers in close proximity, promoting its dimerization. As a result of this interaction, PKR is autophosphorylated at Thr-446, a critical event for binding the substrate (5). However, phosphorylation of PKR in other sites is also required for its correct functioning. In this sense, phosphorylation of the protein at tyrosine residues located at both dsRNA binding and catalytic domains of PKR has been proved needed for optimal Thr-446 phosphorylation, and consequently, it is required for efficient dsRNA binding, dimerization, activation and antiviral activity of PKR (44). According to these models, SUMO may contribute to the full activity of PKR by directly favoring the dimerization of the protein or its interaction with dsRNA, favoring in this way its autophosphorylation (Fig. 6).
FIGURE 6.
Model of PKR activation upon binding to SUMO. SUMO may directly promote PKR-dsRNA binding (A) or support PKR dimerization (B), favoring in this way its autophosphorylation.
Acknowledgment
We are grateful to Dr. I. Ventoso for providing the anti-VSV-G antibody.
This work was supported by Grant BFU-2011-27064 from the Ministry of Economy and Competitiveness of Spain.
- SUMO
- small ubiquitin-like modifier
- VSV
- vesicular stomatitis virus
- VV
- vaccinia virus
- ISG
- interferon-stimulated gene
- mut
- mutant.
REFERENCES
- 1. García M. A., Gil J., Ventoso I., Guerra S., Domingo E., Rivas C., Esteban M. (2006) Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70, 1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Farrell P. J., Balkow K., Hunt T., Jackson R. J., Trachsel H. (1977) Phosphorylation of initiation factor elF-2 and the control of reticulocyte protein synthesis. Cell 11, 187–200 [DOI] [PubMed] [Google Scholar]
- 3. Harashima A., Guettouche T., Barber G. N. (2010) Phosphorylation of the NFAR proteins by the dsRNA-dependent protein kinase PKR constitutes a novel mechanism of translational regulation and cellular defense. Genes Dev. 24, 2640–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cole J. L. (2007) Activation of PKR: an open and shut case? Trends Biochem. Sci. 32, 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dey M., Cao C., Dar A. C., Tamura T., Ozato K., Sicheri F., Dever T. E. (2005) Mechanistic link between PKR dimerization, autophosphorylation, and eIF2α substrate recognition. Cell 122, 901–913 [DOI] [PubMed] [Google Scholar]
- 6. Hovanessian A. G., Galabru J. (1987) The double-stranded RNA-dependent protein kinase is also activated by heparin. Eur. J. Biochem. 167, 467–473 [DOI] [PubMed] [Google Scholar]
- 7. Okumura F., Okumura A. J., Uematsu K., Hatakeyama S., Zhang D. E., Kamura T. (2013) Activation of double-stranded RNA-activated protein kinase (PKR) by interferon-stimulated gene 15 (ISG15) modification down-regulates protein translation. J. Biol. Chem. 288, 2839–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Patel R. C., Sen G. C. (1998) PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 17, 4379–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marchal J. A., Lopez G. J., Peran M., Comino A., Delgado J. R., García-García J. A., Conde V., Aranda F. M., Rivas C., Esteban M., Garcia M. A. (2014) The impact of PKR activation: from neurodegeneration to cancer. FASEB J. 28, 1965–1974 [DOI] [PubMed] [Google Scholar]
- 10. Hay R. T. (2005) SUMO: a history of modification. Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 11. Bernier-Villamor V., Sampson D. A., Matunis M. J., Lima C. D. (2002) Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108, 345–356 [DOI] [PubMed] [Google Scholar]
- 12. Sampson D. A., Wang M., Matunis M. J. (2001) The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276, 21664–21669 [DOI] [PubMed] [Google Scholar]
- 13. Guo D., Li M., Zhang Y., Yang P., Eckenrode S., Hopkins D., Zheng W., Purohit S., Podolsky R. H., Muir A., Wang J., Dong Z., Brusko T., Atkinson M., Pozzilli P., Zeidler A., Raffel L. J., Jacob C. O., Park Y., Serrano-Rios M., Larrad M. T., Zhang Z., Garchon H. J., Bach J. F., Rotter J. I., She J. X., Wang C. Y. (2004) A functional variant of SUMO4, a new IκBα modifier, is associated with type 1 diabetes. Nat. Genet. 36, 837–841 [DOI] [PubMed] [Google Scholar]
- 14. Matic I., van Hagen M., Schimmel J., Macek B., Ogg S. C., Tatham M. H., Hay R. T., Lamond A. I., Mann M., Vertegaal A. C. (2008) In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell. Proteomics 7, 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tatham M. H., Jaffray E., Vaughan O. A., Desterro J. M., Botting C. H., Naismith J. H., Hay R. T. (2001) Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368–35374 [DOI] [PubMed] [Google Scholar]
- 16. Zhao J. (2007) Sumoylation regulates diverse biological processes. Cell. Mol. Life Sci. 64, 3017–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee S. B., Esteban M. (1994) The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology 199, 491–496 [DOI] [PubMed] [Google Scholar]
- 18. Kang J. I., Kwon S. N., Park S. H., Kim Y. K., Choi S. Y., Kim J. P., Ahn B. Y. (2009) PKR protein kinase is activated by hepatitis C virus and inhibits viral replication through translational control. Virus Res. 142, 51–56 [DOI] [PubMed] [Google Scholar]
- 19. Bonnet M. C., Daurat C., Ottone C., Meurs E. F. (2006) The N-terminus of PKR is responsible for the activation of the NF-κB signaling pathway by interacting with the IKK complex. Cell. Signal. 18, 1865–1875 [DOI] [PubMed] [Google Scholar]
- 20. Desterro J. M., Rodriguez M. S., Hay R. T. (1998) SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2, 233–239 [DOI] [PubMed] [Google Scholar]
- 21. Vertegaal A. C., Andersen J. S., Ogg S. C., Hay R. T., Mann M., Lamond A. I. (2006) Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell. Proteomics 5, 2298–2310 [DOI] [PubMed] [Google Scholar]
- 22. Campagna M., Herranz D., Garcia M. A., Marcos-Villar L., González-Santamaría J., Gallego P., Gutierrez S., Collado M., Serrano M., Esteban M., Rivas C. (2011) SIRT1 stabilizes PML promoting its sumoylation. Cell Death Differ. 18, 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. González-Santamaría J., Campagna M., Ortega-Molina A., Marcos-Villar L., de la Cruz-Herrera C. F., González D., Gallego P., Lopitz-Otsoa F., Esteban M., Rodríguez M. S., Serrano M., Rivas C. (2012) Regulation of the tumor suppressor PTEN by SUMO. Cell Death Dis. 3, e393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marcos-Villar L., Lopitz-Otsoa F., Gallego P., Muñoz-Fontela C., González-Santamaría J., Campagna M., Shou-Jiang G., Rodriguez M. S., Rivas C. (2009) Kaposi's sarcoma-associated herpesvirus protein LANA2 disrupts PML oncogenic domains and inhibits PML-mediated transcriptional repression of the survivin gene. J. Virol. 83, 8849–8858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perkins D. J., Qureshi N., Vogel S. N. (2010) A Toll-like receptor-responsive kinase, protein kinase R, is inactivated in endotoxin tolerance through differential K63/K48 ubiquitination. MBio 1, e00239–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McMillan N. A., Carpick B. W., Hollis B., Toone W. M., Zamanian-Daryoush M., Williams B. R. (1995) Mutational analysis of the double-stranded RNA (dsRNA) binding domain of the dsRNA-activated protein kinase, PKR. J. Biol. Chem. 270, 2601–2606 [DOI] [PubMed] [Google Scholar]
- 27. Patel R. C., Stanton P., McMillan N. M., Williams B. R., Sen G. C. (1995) The interferon-inducible double-stranded RNA-activated protein kinase self-associates in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 92, 8283–8287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patel R. C., Stanton P., Sen G. C. (1996) Specific mutations near the amino terminus of double-stranded RNA-dependent protein kinase (PKR) differentially affect its double-stranded RNA binding and dimerization properties. J. Biol. Chem. 271, 25657–25663 [DOI] [PubMed] [Google Scholar]
- 29. Anderson E., Pierre-Louis W. S., Wong C. J., Lary J. W., Cole J. L. (2011) Heparin activates PKR by inducing dimerization. J. Mol. Biol. 413, 973–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson E. S. (2004) Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 31. Cai R., Williams B. R. (1998) Mutations in the double-stranded RNA-activated protein kinase insert region that uncouple catalysis from eIF2α binding. J. Biol. Chem. 273, 11274–11280 [DOI] [PubMed] [Google Scholar]
- 32. Xu Z., Williams B. R. (2000) The B56α regulatory subunit of protein phosphatase 2A is a target for regulation by double-stranded RNA-dependent protein kinase PKR. Mol. Cell. Biol. 20, 5285–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yue Z., Shatkin A. J. (1997) Double-stranded RNA-dependent protein kinase (PKR) is regulated by reovirus structural proteins. Virology 234, 364–371 [DOI] [PubMed] [Google Scholar]
- 34. Kubota T., Matsuoka M., Chang T. H., Tailor P., Sasaki T., Tashiro M., Kato A., Ozato K. (2008) Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J. Biol. Chem. 283, 25660–25670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marcos-Villar L., Pérez-Girón J. V., Vilas J. M., Soto A., de la Cruz-Hererra C. F., Lang V., Collado M., Vidal A., Rodríguez M. S., Muñoz-Fontela C., Rivas C. (2013) SUMOylation of p53 mediates interferon activities. Cell Cycle 12, 2809–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Balachandran S., Roberts P. C., Brown L. E., Truong H., Pattnaik A. K., Archer D. R., Barber G. N. (2000) Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13, 129–141 [DOI] [PubMed] [Google Scholar]
- 37. Durbin R. K., Mertz S. E., Koromilas A. E., Durbin J. E. (2002) PKR protection against intranasal vesicular stomatitis virus infection is mouse strain dependent. Viral Immunol 15, 41–51 [DOI] [PubMed] [Google Scholar]
- 38. Stojdl D. F., Abraham N., Knowles S., Marius R., Brasey A., Lichty B. D., Brown E. G., Sonenberg N., Bell J. C. (2000) The murine double-stranded RNA-dependent protein kinase PKR is required for resistance to vesicular stomatitis virus. J. Virol. 74, 9580–9585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Samuel C. E. (2001) Antiviral actions of interferons. Clin. Microbiol. Rev. 14, 778–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Langland J. O., Cameron J. M., Heck M. C., Jancovich J. K., Jacobs B. L. (2006) Inhibition of PKR by RNA and DNA viruses. Virus Res. 119, 100–110 [DOI] [PubMed] [Google Scholar]
- 41. González-Santamaría J., Campagna M., García M. A., Marcos-Villar L., González D., Gallego P., Lopitz-Otsoa F., Guerra S., Rodríguez M. S., Esteban M., Rivas C. (2011) Regulation of vaccinia virus E3 protein by small ubiquitin-like modifier proteins. J. Virol. 85, 12890–12900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Palacios S., Perez L. H., Welsch S., Schleich S., Chmielarska K., Melchior F., Locker J. K. (2005) Quantitative SUMO-1 modification of a vaccinia virus protein is required for its specific localization and prevents its self-association. Mol. Biol. Cell 16, 2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wilson V. G. (2012) Sumoylation at the host-pathogen interface. Biomolecules 2, 203–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Su Q., Wang S., Baltzis D., Qu L. K., Wong A. H., Koromilas A. E. (2006) Tyrosine phosphorylation acts as a molecular switch to full-scale activation of the eIF2α RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 103, 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]