Background: The Beclin 1-Vps34 protein-protein interaction network is critical for autophagy regulation.
Results: Nrbf2 is a component of the Atg14L-containing Beclin 1-Vps34 protein complex. Nrbf2 deficiency disrupts Atg14L-Vps34/Vps15 interactions and increases intracellular PI3P levels and autophagic flux.
Conclusion: Nrbf2 is important for the Beclin 1-Vps34 interaction network to achieve tight autophagy regulation.
Significance: Our work identifies a novel aspect of autophagy regulation.
Keywords: Autophagy, Cellular Regulation, Protein Degradation, Protein-Protein Interaction, Subcellular Organelle, Beclin 1, Nrbf2, Vps34, Autophagosome Biogenesis
Abstract
Autophagy is a tightly regulated lysosomal degradation pathway for maintaining cellular homeostasis and responding to stresses. Beclin 1 and its interacting proteins, including the class III phosphatidylinositol-3 kinase Vps34, play crucial roles in autophagy regulation in mammals. We identified nuclear receptor binding factor 2 (Nrbf2) as a Beclin 1-interacting protein from Becn1−/−;Becn1-EGFP/+ mouse liver and brain. We also found that Nrbf2-Beclin 1 interaction required the N terminus of Nrbf2. We next used the human retinal pigment epithelial cell line RPE-1 as a model system and showed that transiently knocking down Nrbf2 by siRNA increased autophagic flux under both nutrient-rich and starvation conditions. To investigate the mechanism by which Nrbf2 regulates autophagy, we demonstrated that Nrbf2 interacted and colocalized with Atg14L, suggesting that Nrbf2 is a component of the Atg14L-containing Beclin 1-Vps34 complex. Moreover, ectopically expressed Nrbf2 formed cytosolic puncta that were positive for isolation membrane markers. These results suggest that Nrbf2 is involved in autophagosome biogenesis. Furthermore, we showed that Nrbf2 deficiency led to increased intracellular phosphatidylinositol-3 phosphate levels and diminished Atg14L-Vps34/Vps15 interactions, suggesting that Nrbf2-mediated Atg14L-Vps34/Vps15 interactions likely inhibit Vps34 activity. Therefore, we propose that Nrbf2 may interact with the Atg14L-containing Beclin 1-Vps34 protein complex to modulate protein-protein interactions within the complex, leading to suppression of Vps34 activity, autophagosome biogenesis, and autophagic flux. This work reveals a novel aspect of the intricate mechanism for the Beclin 1-Vps34 protein-protein interaction network to achieve precise control of autophagy.
Introduction
Autophagy is an essential lysosomal degradation pathway that eukaryotic cells employ to recycle cytoplasmic contents for providing building blocks and energy. Macroautophagy, the primary form of autophagy, referred to as autophagy hereafter, is characterized by sequestration of the bulk of the protein- and organelle-containing cytoplasm into de novo-generated, double-membraned structures called autophagosomes (i.e. autophagosome biogenesis) and subsequent fusion of autophagosomes with late endosomes or lysosomes for degradation of the enwrapped cytoplasmic contents (i.e. autophagosome maturation) (1). Although autophagy deficiency has been implicated in a broad spectrum of human diseases, including liver diseases, cardiovascular diseases, cancer, and neurodegeneration (2), hyperactive autophagy may lead to cell death (e.g. autosis (3)). Therefore, autophagy must be tightly regulated for fitness and survival.
Becn1, the ortholog of the yeast Vps30/Atg6 gene, is one of the first discovered mammalian autophagy genes (4). Beclin 1, encoded by Becn1, was originally reported as a Bcl-2-interacting protein through a yeast two-hybrid screen of an adult mouse brain cDNA library (5, 6). Beclin 1/Vps30 plays important roles in autophagy regulation through its numerous interacting proteins. In yeast, Vps30 forms two distinct protein complexes with Vps34, the only known class III phosphatidylinositol (PI)2 3-kinase that catalyzes the conversion of PI to phosphatidylinositol 3-phosphate (PI3P) (7, 8). One Vps30-Vps34 complex is recruited to the preautophagosomal structure by Atg14 (9) and functions to produce PI3P during autophagosome biogenesis. The other Vps30-Vps34 complex contains Vps38 and is involved in the vacuolar protein sorting (Vps) pathway (10). Likewise, in mammals, Beclin 1 and its interactors form multiple protein complexes for diverse functions (reviewed comprehensively in Refs. 11–13). Some of the Beclin 1-interacting proteins, e.g. Vps34 (14), Atg14L (15, 16) (also named Atg14 (17) and Barkor (18)), p110β (19), Ambra1 (20), VMP1 (21, 22), and HGMB1 (23), positively regulate autophagy. Others, e.g. Rubicon (15, 16) and Bcl-2/XL (24), are negative regulators of autophagy. In particular, Beclin 1 also forms two major protein complexes with Vps34. One is the Atg14L-containing Beclin 1-Vps34 protein complex that is involved in autophagosome biogenesis (15–18, 25, 26), and the other is the ultraviolet irradiation resistance-associated gene (UVRAG)-containing Beclin 1-Vps34 protein complex that is involved in endocytic trafficking (17, 27) and, with some controversy, in autophagy regulation (17, 28). In short, on the basis of the extensive investigation reviewed above, a theme has emerged for the presence of a central autophagy regulation hub composed of a dynamic Beclin 1-Vps34 protein-protein interaction network.
Despite the importance of Beclin 1 in regulating autophagy and in tumor suppression (4, 28–32), development (20, 30, 33), aging (34, 35), and neurodegeneration (36, 37), the molecular details of how the Beclin 1-Vps34 protein-protein interaction network regulates autophagy are not fully understood. In this work, we set out to determine additional important players in the Beclin 1-Vps34 protein-protein interaction network and identified nuclear receptor binding factor 2 (Nrbf2) as a Beclin 1-interacting protein. Nrbf2 was originally reported to interact with nuclear receptors as a coregulator (38, 39). Here we determined a novel role for Nrbf2 in autophagy regulation through modulating the Beclin 1-Vps34 protein-protein interaction network.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
NuPAGE® BisTris gels, Western blot transfer buffer (catalog no. NP0006), MOP (catalog no. NP0001) and MES (catalog no. NP0002) SDS running buffers, NuPAGE® antioxidant (catalog no. NP0005), glutamine (catalog no. 25030), FBS (catalog no. 26140), SlowFade® Gold antifade reagent (catalog no. S36937), Opti-MEM® I reduced serum medium (catalog no. 31985), the Ambion® Micropoly (A)PuristTM kit (catalog no. AM1919), the SuperScriptTM III first-strand synthesize system for RT-PCR (catalog no. 18080-051), SYBR® safe DNA gel stain (catalog no. S33102), the pENTRTM directional TOPO® cloning kit (K2400-20), the TOPO® TA cloning kit (catalog no. K450001), Lipofectamine 2000 (catalog no. 11668), Lipofectamine RNAi MAX (catalog no. 13778), and ProLong® Gold antifade reagent (catalog no. P36934) were purchased from Invitrogen. Restriction enzymes and the quick ligation kit (catalog no. M2200S) were purchased from New England BioLabs (Ipswich, MA). The TaKaRa LA PCR kit 2.1 (catalog no. RR013A) and the pmCherry-C1 vector (catalog no. 632524) were purchased from Clontech (Mountain View, CA). The Wizard® Plus SV Minipreps DNA purification system (catalog no. A1330) was purchased from Promega (Madison, WI). The Plasmid Plus Maxi kit (catalog no. 12963) and the RNeasy mini kit (catalog no. 74014) were purchased from Qiagen (Valencia, CA). The iScriptTM cDNA synthesis kit (catalog no. 170-8890) and SsoAdvancedTM Universal SYBR® Green Supermix (catalog no. 172-5271) were purchased from Bio-Rad. Bafilomycin A1 (Baf) from Streptomyces griseus (catalog no. B1793), TCA (catalog no. T6399), BSA (catalog no. A7906), and trypsin-EDTA solution (catalog no. T4049) were purchased from Sigma. EDTA-free protease inhibitor mixture tablets (catalog no. 11836170001) and PhosSTOP phosphatase inhibitor mixture tablets (catalog no. 04906837001) were purchased from Roche Diagnostics. Pierce Halt protease and phosphatase inhibitor mixture (catalog no. 78443), the subcellular protein fractionation kit (catalog no. 78840), SuperSignal West Pico chemiluminescent substrate (catalog no. 34080), SuperSignal West Femto Maximum substrate (catalog no. 34096), the micro BCA protein assay reagent kit (catalog no. 23235), and Dharmacon siRNAs were purchased from Thermo Scientific (Rockford, IL). The Immobilon-P 0.2-μm PVDF membrane (catalog no. ISEQ00010) and 10× radioimmunoprecipitation assay lysis buffer (catalog no. 20-188) were purchased from Millipore (Billerica, MA). The PI3P Mass StripTM kit (catalog no. K-3600) and PI3P mass ELISA kit (catalog no. K-3300) were purchased from Echelon Biosciences (Salt Lake City, UT). DMEM (Cellgro 10-013-CV) was purchased from Corning (Manassas, VA). l-leucine (catalog no. L2020-05) and DMEM high glucose with pyridoxal·HCl without l-glutamine and leucine (catalog no. D9816) were purchased from USBiological (Marblehead, MA). Econo-Safe economical biodegradable counting mixture (catalog no. 111175) was purchased from Research Products International Corp. (Mount Prospect, IL). Four-well slides coated with type I collagen (catalog no. 354557) were purchased from BD Biosciences. Recombinant protein A beads (catalog no. IPA-300) was purchased from Repligen (Waltham, MA).
For Western blotting, Beclin 1 (H300) antibody (catalog no. sc-11427, 1:500), Ulk1 (H-240) antibody (catalog no. sc-33182, 1:500), anti-rabbit IgG-HRP (catalog no. sc-2004) and anti-mouse IgG-HRP (catalog no. sc-2005) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Nrbf2 antibody (catalog no. A301-852A, 1:5000) and Vps15/p150 (catalog no. A302-571A, 1:4000) were purchased from Bethyl Laboratories (Montgomery, TX). Guinea pig anti-p62 antibody (catalog no. 03-GP62-C, 1:1700) was purchased from American Research Products (Waltham, MA). Anti-LC3 (clone 2G6) antibody (catalog no. 0260-100, 1:1000) was purchased from nanoTools Antikörpertechnik GmbH & Co. KG (Teningen, Germany). Phospho-p70 S6 kinase (Thr-389) antibody (catalog no. 9205, 1:1000), p70 S6 kinase antibody (clone 49D7) (catalog no. 2708, 1:1000), Vps34 antibody (clone D9A5) (catalog no. 4263, 1:1000), and Atg14L antibody (catalog no. 5504, 1:1000) were purchased from Cell Signaling Technology (Danvers, MA). Peroxidase-rabbit anti-guinea pig IgG (H+L) was purchased from Invitrogen. Anti-β-actin (clone AC-15) mouse monoclonal antibody (catalog no. A5441, 1:10,000) was purchased from Sigma.
For immunoprecipitation (IP), Atg14L (catalog no. PD026) and UVRAG (catalog no. M160-3) antibodies were purchased from MBL International (Woburn, MA). Vps34 antibody (catalog no. Z-R015) was purchased from Echelon Biosciences. Beclin 1 antibody (catalog no. A302-567A) was purchased from Bethyl Laboratories).
For immunofluorescent imaging, c-myc antibody (9E10, catalog no. sc-40, 1:1000) was purchased from Santa Cruz Biotechnology, and FIP200 antibody (10043-2-AP, 1:400) was pursed from ProteinTech Group, Inc. (Chicago, IL). Rhodamine Red-XTM goat anti-mouse IgG (H+L, catalog no. R-6393) and Alexa Fluor 488 goat anti-rabbit IgG (H+L) antibody (catalog no. A11034) were purchased from Invitrogen.
Plasmid Construction
Total mRNA was isolated from HepG2 cells and purified using the Micropoly (A)PuristTM kit. Human Nrbf2 (Ensembl transcript ID ENST00000277746) cDNA was PCR-amplified using the SuperScriptTM III first-strand synthesize system. Full-length Nrbf2 inserts intended for N- and C-terminal tagging were generated by N- and C-terminal-specific primers, respectively. For N-terminal tagging, the forward primer was 5′ CAC CCA AAT GGA AGT AAT GGA AGG AC 3′, and the reverse primer was 5′ CTA ATT ATT CAT AAA TCC TTT CAG AAT 3′. For C-terminal tagging, the forward primer (#5) was 5′ CAC CAT GGA AGT AAT GGA AGG ACC CC 3′, and the reverse primer (#8) was 5′ ATT ATT CAT AAA TCC TTT CAG AAT AT 3′. To generate the C-terminal tagged Nrbf2 truncation mutants M1-M6, the reverse primers for M1 was 5′ CTG CTC TGA CTG TGT CAG CTT CAT GGC TT 3′, the forward primer for M2 was 5′ CAC CAT GGC CAT GAA GCT GAC ACA GTC AGA 3′, the reverse primer for M3 was 5′ GCT GTA CTT CTG AGA AAG GGG ACT CTG 3′, the forward primer for M4 was 5′ CAC CAT GGC TCC TTC CAC AGA GAA ATG CCT G 3′, the reverse primer for M5 was 5′ ATC AGC ATC TAC ATC CAG CTC CTT TTC 3′, and the reverse primer for M6 was 5′ GGC TTT GCT TCC AAT ACA TGG CTC TGC T 3′. These primers were used in combination with primers #5 and #8. Inserts containing the cDNAs of full-length Nrbf2 and the truncation mutants were originally inserted into the Gateway pENTRTM/D-TOPO entry vector, and the correctness of the insertions was confirmed by sequencing. C-terminal cycle 3 GFP-tagged full-length Nrbf2 and the truncation mutants were generated using the Gateway destination vector pcDNATM-DEST47. Full-length Nrbf2 cDNA in the Gateway entry vector was amplified using TaKaRa LA PCR kit 2.1 as well as forward primer 5′ CCG GCT AGA TCT ATG GAA GTA ATG GAA GGA CCC 3′ and reverse primer 5′ CCG GCT GTC GAC TTA ATT ATT CAT AAA TCC TT 3′. The Nrbf2 amplicon was later subcloned into the pCR2.1-TOPO vector, excised using the restriction enzymes BglII and SalI, and cloned into the mCherry-C1 vector using the New England BioLabs quick ligation kit. All positive clones were verified by restriction digestion and sequencing. All oligonucleotides were synthesized by Integrated DNA Technologies. For cloning, all plasmid DNAs were prepared using Escherichia coli DH5α or TOP10 cells and the Promega Wizard® Plus SV Minipreps DNA purification system. For transfection into mammalian cells, all plasmids were prepared using the Qiagen Plasmid Plus Maxi kit.
Cell Culture and Biochemical Analyses
Unless noted otherwise, RPE-1 cells were cultured in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin in a cell culture incubator (set at 37 °C and 5% CO2). For overexpression, cells at 70–80% confluency in 6-cm dishes were transfected with 4 μg of mCherry C1 vector or human mCherry-Nrbf2 plasmid using Opti-MEM® I reduced serum medium and Lipofectamine 2000 following the instructions of the manufacturer. Twenty-four hours after transfection, cells were washed with PBS and cultured in either nutrient-rich or starvation (i.e. 18 h serum starvation or 1 h Hanks' buffer starvation) conditions in the presence or absence of 200 nm Baf for 18 h. Different cells showed different sensitivities in response to Baf. We chose to treat RPE-1 cells with 200 nm Baf for 18 h in this study because, in our hands, this treatment resulted in prominent changes in LC3-phosphatidylethanolamine (i.e. LC3II) levels in RPE-1 cells, whereas 2 h of 100 nm Baf treatment had a minimal effect.
For siRNA treatments, cells were reverse-transfected with 20 nm of each siRNA using Opti-MEM® I reduced serum medium and Lipofectamine RNAiMAX following the instructions of the manufacturer. In brief, 6–8 × 105 cells were suspended in 3 ml of DMEM supplemented with 10% FBS without antibiotics. siRNAs were added to the cell suspensions to a final concentration of 20 nm each. These siRNAs were Dharmacon ON-TARGETplus non-targeting siRNA pool (catalog no. D-001810-10-20) and SMARTpool siRNA targeting human Nrbf2 (catalog no. L-014648-00-0005), Atg14L (catalog no. L-020438-01-0005), Beclin 1 (catalog no. L-010552-00-0005), and Vps34 (catalog no. L-005250-00-0005). The sequence of the human UVRAG siRNA was 5′ UCA CUU GUG UAG UAC UGA AUU 3′, with the target sequence (28) underlined. The sequence of the human Nrbf2 3′UTR siRNA was 5′ UGU GAA AUG CGC UGC GUA UUU 3′, with the target sequence underlined. The cell-siRNA mixtures were seeded onto 6-cm dishes. Cells were cultured for 48–72 h before being subjected to either nutrient-rich or starvation conditions in the presence or absence of 200 nm Baf for 18 h.
To harvest cells for biochemical analysis, cell monolayers were washed with ice-cold PBS and collected on ice using scrapers. Cell pellets were lysed on ice with 1× radioimmunoprecipitation assay lysis buffer supplemented with protease and phosphatase inhibitor mixture using 1-ml syringes and 27G1/2 needles. Cell lysates were centrifuged at 14,000 × g and 4 °C for 5 min, and the supernatant was used for Western blot analyses on an equal protein basis (15–40 μg). For resolving LC3II and its un-lipidated precursor LC3I, cell lysates were run on 10 or 12% BisTris gels using MOP or MES buffer. For probing all other proteins, cell lysates were run on 4–12% BisTris gels using MOP buffer. Proteins were transferred to PVDF membranes using a Mini Trans-Blot apparatus (Bio-Rad) at 30 V overnight in Western blot transfer buffer supplemented with NuPAGE® antioxidant, 20% methanol (MeOH), and 0.025% SDS. Membranes were then incubated with primary (at 4 °C overnight) and secondary (at 25 °C for 1 h) antibodies, and signals were detected by SuperSignal West Pico/Femto chemiluminescent substrate. Films were exposed for various time periods and scanned using positive mode to ensure that data were obtained within a linear range of exposure. The p62 (or LC3II) levels were quantified using ImageJ and normalized to corresponding actin levels. The p62 (or LC3II)/actin ratios were further normalized to the samples with mCherry empty vector or non-targeting siRNA treatments. Comparisons were made in Microsoft Excel using one-sample, two-tailed Student's t test. For subcellular fractionation, harvested cell pellets were processed using the Pierce subcellular protein fractionation kit following the instructions of the manufacturer.
IP was carried out as reported (40). In brief, RPE-1 cells grown in 10-cm dishes were transfected with different siRNA as described above. After 48 h, cells were lysed with 1 ml of mild lysis buffer (MLB) (10 mm Tris (pH 7.5), 2 mm EDTA, 100 mm NaCl, 1% Nonidet P-40, 50 mm NaF, 1 mm Na3VO4, EDTA-free protease inhibitor mixture (Roche), and Pierce Halt protease and phosphatase inhibitor mixture). Cell lysates, except for 100-μl aliquots saved for probing the input, were used for IP. Anti-Beclin 1, anti-Nrbf2, anti-Atg14L, and anti-Vps34 antibodies were coupled with Repligen immobilized protein A-agarose beads overnight at 4 °C in TBST (20 mm Tris (pH 8.0), 170 mm NaCl, and 0.05% Tween 20) supplemented with 1% BSA. The immune complex was then added to cell lysates and incubated at 4 °C for 3 h. The resulting beads were washed with MLB five times before the antigen and their interacting proteins were eluted by SDS-PAGE loading buffer.
Long-lived Protein Degradation
Long-lived protein degradation was measured following a published protocol (41), with minor modifications. In brief, RPE-1 cells were reverse-transfected with 20 nm of non-targeting siRNA and human Nrbf2 SMARTpool siRNA using Opti-MEM® I reduced serum medium and Lipofectamine RNAiMAX in either hot or cold medium. The hot medium was composed of l-glutamine- and leucine-free DMEM supplemented with 10% FBS, 4 mm l-glutamine, 65 μm of cold l-leucine, and 1 μCi/ml 3H-l-leucine. The cold medium had all compositions of the hot medium except for hot l-leucine. After being pulse-labeled for 24 h, cells were washed twice with sterile PBS (at 25 °C) and cultured in cold medium for 20 h to chase out short-lived proteins. Cells were then washed twice with sterile PBS (at 25 °C) and cultured in 2 ml of cold medium either without or with 10% FBS for 8 h to degrade long-lived proteins. Long-lived protein degradation was calculated from the TCA-soluble radioactivity of the medium and the TCA-precipitated radioactivity of the cell monolayers. A volume of 860 μl of medium was taken from each plate and mixed thoroughly with 140 μl of 71.4% TCA (final TCA concentration, 10%). The mixtures were incubated on ice for 1 h before being centrifuged at 17,720 × g for 10 min at 4 °C. An aliquot of 0.4 ml of the supernatant was mixed with 4 ml of scintillation counting mixture for measuring the content of tritium in the medium using a scintillation counter. The total counts in the supernatant were labeled as A. The cell monolayers were washed with ice-cold PBS once. Cells were fixed to the plate by 2 ml of ice-cold 10% TCA for 40 min at 4 °C. The cell monolayers were washed with 2 ml of ice-cold 10% TCA once, lysed in 2 ml of 0.2 m NaOH (prewarmed to 37 °C), and incubated at 37 °C for 1 h. An aliquot of 0.4 ml of the lysate was mixed with 4 ml of scintillation counting mixture to measure the tritium content in the cell monolayer. The total counts in the pellet were labeled as B. The average rate of long-lived protein degradation during the 8 h for degrading long-lived proteins was calculated as A / (A + B). Data from three independent experiments were averaged, with each experiment having triplicated samples. Comparisons were made using analysis of variance.
Fluorescence Microscopy
RPE-1 cells were seeded in type I collagen-coated 4-well slides at a density of 5 × 104 cells/well. For transient expression, cells that had been seeded for 24 h were transfected with a total of 1 μg of plasmids in each well using Lipofectamine 2000 and Opti-MEM® I reduced serum medium. Twenty-four hours after transfection of plasmids, cells were washed once with Hanks' buffer and cultured in Hanks' buffer at 37 °C in the presence of 5% CO2 for 1 h. Cells were then fixed in 4% paraformaldehyde in PBS for 15 min, permeabilized with 50 μg/ml digitonin for 10 min, and blocked with 2% goat serum and 2% BSA in PBS for 30 min at 25 °C. Cells were incubated in primary antibodies against c-myc and FIP200, respectively, overnight at 4 °C, followed by dye-conjugated secondary antibodies for 1 h at 25 °C. After four washes, the liquid in the wells was aspirated before the slides were mounted with ProLong® Gold antifade reagent and 1.5 mm-thick coverglass. Confocal fluorescent images were collected on a Nikon A1 confocal microscope equipped with an Apo TIRF 60× oil objective lens (numerical aperture 1.49) and quantified manually using Nikon NIS-Elements AR 3.2 software. Cells with low to medium expression levels and with good morphology were imaged. Image intensity levels were processed linearly to enhance contrast. Comparisons were made in Microsoft Excel using two-sample, two-tailed Student's t test.
Protein-Lipid Overlay and ELISA Assays
Extraction and detection of total intracellular PI3P were carried out using a PI3P mass strip kit and a PI3P mass ELISA kit following the instructions of the manufacturer. To extract the acidic lipids including PI3P, cells were first fixed with 0.5 m ice-cold TCA on ice for 5 min. Fixed cells were harvested on ice using scrapers. The cell suspension was centrifuged at 1500 rpm for 5 min at 4 °C, and cell pellets were washed twice with 5% TCA in the presence of 1 mm EDTA. Neutral lipids were extracted with MeOH:chloroform (CHCl3) (2:1) for 10 min at 25 °C, vortexing three times. After centrifugation at 1500 rpm for 5 min at 4 °C, the supernatant containing neutral lipids was discarded. Extraction and removal of neutral lipids were repeated once. Acidic lipids, including PI3P, were then extracted by CHCl3:MeOH:12 m HCl (40:80:1) for 15 min at 25 °C, vortexing four times. After centrifugation at 1500 rpm for 5 min at 4 °C, the supernatant containing acidic lipids was transferred to new tubes. CHCl3 and 0.1 m HCl were added sequentially to the supernatant. The tubes were vortexed and then centrifuged at 1500 rpm for 5 min at 4 °C. The organic (lower) phase containing acidic lipids was transferred to new tubes and dried in a vacuum dryer. Cell pellets were lysed by incubation in 0.1 m NaOH overnight in a 37 °C shaker. The protein concentrations of lysates were measured by a Micro BCA protein assay reagent kit following the instructions of the manufacturer.
To carry out the protein-lipid overlay assay, dried acidic lipids were reconstituted with 10 μl of CHCl3:MeOH:H2O (1:2:0.8). The samples were vortexed and sonicated in an ice water bath. Lipid solutions were spotted onto PI3P strips and dried at 25 °C. The strips were blocked with 3% BSA and then incubated with 5 μg of PI3P-binding protein. The PI3P spots were detected with SuperSignal West Femto Maximum substrate. Intracellular PI3P levels were quantified using ImageJ and normalized to total protein levels. Comparisons were made in Microsoft Excel using two-sample, one-tailed Student's t test.
To measure total intracellular PI3P by quantitative and competitive ELISA assay, dried acidic lipids were reconstituted in PBST (PBS and 0.05% Tween 20) with 3% protein stabilizer. Reconstituted samples were incubated with PI3P detector protein before being transferred to a microplate precoated with PI3P for competitive binding. A peroxidase-conjugated antibody against PI3P detector protein was added, and colorimetric detection was carried out to determine PI3P detector protein binding on the microplate. Total intracellular PI3P quantities were calculated using the standard curve from logarithmic fitting of manufacturer-supplied PI3P standards and normalized to total protein levels. Comparisons were made in Microsoft Excel using one-tailed, paired Student's t test.
Sequence Alignment
Sequence alignment was carried out in the Biology Workbench using ClustalW and ClustalWPROF with default settings. The results were displayed using TEXSHADE.
Quantitative RT-PCR (qRT-PCR)
RPE-1 cells that were reverse-transfected with non-targeting, SMARTpool, and 3′UTR Nrbf2 siRNA were seeded into 60-mm dishes and cultured for 48 h before total mRNA was extracted using the Qiagen RNeasy mini kit following the instructions of the manufacturer. Purified mRNA (1 μg) from each sample was converted to cDNA in 20-μl reaction mixtures using the iScriptTM cDNA synthesis kit following the protocols of the manufacturer. Subsequent qRT-PCR of target transcripts was performed in 20-μl reaction mixtures, each composed of 2 μl (20 ng) of cDNA, 10 μl of SsoAdvancedTM Universal SYBR® Green Supermix, 1–2 μl of qRT-PCR primer probe set, and 6–7 μl of nuclease-free water. The qRT-PCR primers used for human p62/SQSTM1 transcript (Hs.PT.56a.40413286.g, 5′ GGA GGA GAT GAT GAC TGG AC 3′ (forward) and 5′ CAG AGA GCT TGG CCC TTC 3′ (reverse)) and the control human ACTB transcript (Hs.PT.56a.19461448.g, 5′ GCG AGA AGA TGA CCC AGA T 3′ (forward) and 5′ CCA GTG GTA CGG CCA GA 3′ (reverse)) were purchased from Integrated DNA Technologies. Forty cycles of amplification were performed in a Stratagene Mx3005P QPCR system (Agilent Technologies) according to the specifications of the manufacturer. Three replicates were performed for each probe-cDNA combination. The fold change of the human p62/SQSTM1 transcript in response to the SMARTpool or 3′UTR Nrbf2 siRNA treatment was calculated from the cycle number at the threshold SYBR® Green signal (Ct) as follows:
 |
Note that, in this equation, ACTB was used as the control to account for the variations in the total mRNA quantity.
RESULTS
Identification of Nrbf2 as a Beclin 1-interacting Protein
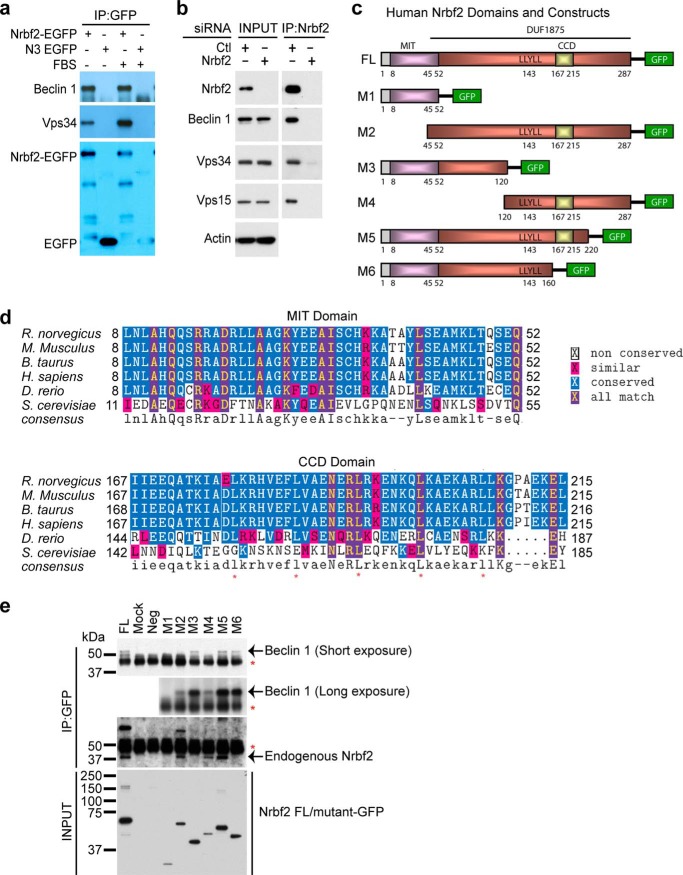
Utilizing the bacterial artificial chromosome technique, we previously generated compound mouse Becn1-EGFP/+;Becn1−/−, which expressed Beclin 1-EGFP under the endogenous Becn1 promoter and in the genetic background of the homozygous Beclin 1 knockout mouse Becn1−/− (15). Remarkably, Beclin 1-EGFP rescued embryonic lethality of Becn1−/−, suggesting that the transgene is functional in the compound mouse (15). We previously also affinity-purified and mass spectrometrically identified a number of Beclin 1-interacting proteins from the brain and liver of the rescued mice, including Vps34, Vps15, UVRAG, Atg14L, and Rubicon (Fig. 1b in Ref. 15). One additional Beclin 1-interacting protein with an apparent mass of 38 kDa was identified as Nrbf2 (mass spectra not shown). Here, using an anti-GFP antibody to perform reciprocal IP in HEK 293 cells stably expressing Nrbf2-EGFP, we showed that Nrbf2-EGFP was capable of pulling down endogenous Beclin 1 and Vps34 under both nutrient-rich and serum starvation conditions (Fig. 1a), confirming the Nrbf2-Beclin 1 interaction. Moreover, to assess endogenous interactions between Nrbf2 and the core of the Beclin 1-Vps34 protein-protein interaction network, we used an anti-Nrbf2 antibody to perform IP from the retinal pigment epithelium cell line RPE-1 cultured under nutrient-rich conditions. Our result shows that anti-Nrbf2 antibody was capable of pulling down endogenous Beclin 1, Vps34, and Vps15 from RPE-1 cells transfected with non-targeting siRNA but not from RPE-1 cells transfected with Nrbf2 siRNA (Fig. 1b). This result suggests that, at endogenous levels, Nrbf2 interacts with the core of the Beclin 1-Vps34 protein-protein interaction network. The Nrbf2-Beclin 1 interaction was also discovered independently by Harper and co-workers in a large scale proteomic study of the human autophagy network (42). Taken together, our results demonstrate that Nrbf2 is a component of the Beclin 1-Vps34 protein-protein interaction network.
FIGURE 1.
Identification of Nrbf2 as a novel Beclin 1-interacting protein. a, anti-EGFP antibody pulled down endogenous Beclin 1 and Vps34 in HEK293 cells stably expressing mouse Nrbf2-EGFP under both nutrient-rich and serum starvation conditions. Immunoprecipitated Nrbf2-EGFP and EGFP were labeled in the anti-GFP blot. b, anti-Nrbf2 antibody pulled down endogenous Beclin 1, Vps34, and Vps15 from RPE-1 cells transfected with non-targeting siRNA but not from those transfected with the SMARTpool Nrbf2 siRNA. Of note, both cell extracts before IP (INPUT) contain similar levels of Beclin 1, Vps34, Vps15, and actin. c, human Nrbf2 domain structures and diagrams of the C-terminal Cycle3 GFP-tagged full-length (FL) human Nrbf2 and truncation mutants (M1-M6) of Nrbf2. d, sequence alignment of the Nrbf2 MIT and CCD domains from different species. The five conserved leucines in the CCD are marked by red asterisks. Note that Atg38, the yeast ortholog of Nrbf2, has low sequence homology to the mammalian Nrbf2, particularly lacking the abovementioned five leucines in the CCD. e, anti-GFP IP of Cycle3 GFP-tagged full-length and mutant human Nrbf2 constructs from HepG2 cells revealed that the N-terminal 120 residues of Nrbf2 were required and sufficient for the Nrbf2-Beclin 1 interaction. Red asterisks mark the IgG bands. GFP-tagged full-length and mutant Nrbf2 constructs in the INPUT were probed with anti-GFP antibody.
The human Nrbf2 protein is 287 amino acids long. It has an N-terminal microtubule-interacting and trafficking (MIT) domain (amino acids 8–52) that has been reported in vacuolar sorting proteins and a C-terminal domain of unknown function (DUF1875, amino acids 45–287) that contains a coiled-coil domain (CCD, amino acids 167–215) for protein-protein interactions (Fig. 1, c and d). To determine which domain of Nrbf2 interacts with Beclin 1, we generated C-terminal GFP-tagged full-length and truncation mutants of Nrbf2 (Fig. 1c). We transiently transfected HepG2 cells with these constructs and performed anti-GFP IP from cell extracts. Our result shows that full-length Nrbf2 and truncation mutants M3 (amino acids 1–120), M5 (amino acids 1–220), and M6 (amino acids 1–160) pulled down endogenous Beclin 1. In contrast, truncation mutants M1 (amino acids 1–52), M2 (amino acids 45–287), and M4 (amino acids 120–287) were largely unable to pull down endogenous Beclin 1 (Fig. 1e, Beclin 1 short exposure), suggesting that the N terminus of Nrbf2 (i.e. amino acids 1–120) is important for Nrbf2-Beclin 1 interaction. To detect any trace of M1/M2/M4 mutant Nrbf2-Beclin 1 interaction, we exposed the film for a longer time and found that a small amount of endogenous Beclin 1 was pulled down by the truncation mutants M2 and M4 but not M1 (Fig. 1e, Beclin 1 long exposure). Interestingly, full-length Nrbf2 and truncation mutants M2, M4, and M5, all of which contain the CCD, pulled down endogenous Nrbf2 (Fig. 1e), suggesting that Nrbf2 is likely capable of self-association through its CCD. Therefore, our observation that a small amount of endogenous Beclin 1 was pulled down by M2 and M4 likely resulted from the CCD-mediated interactions between the truncation mutant M2/M4 and the full-length Nrbf2. Taken together, our results suggest that the N terminus of Nrbf2 (i.e. amino acids 1–120, including both the MIT domain and amino acids 53–120) is necessary and sufficient for the Nrbf2-Beclin 1 interaction.
Nrbf2 Negatively Regulates Autophagic Flux
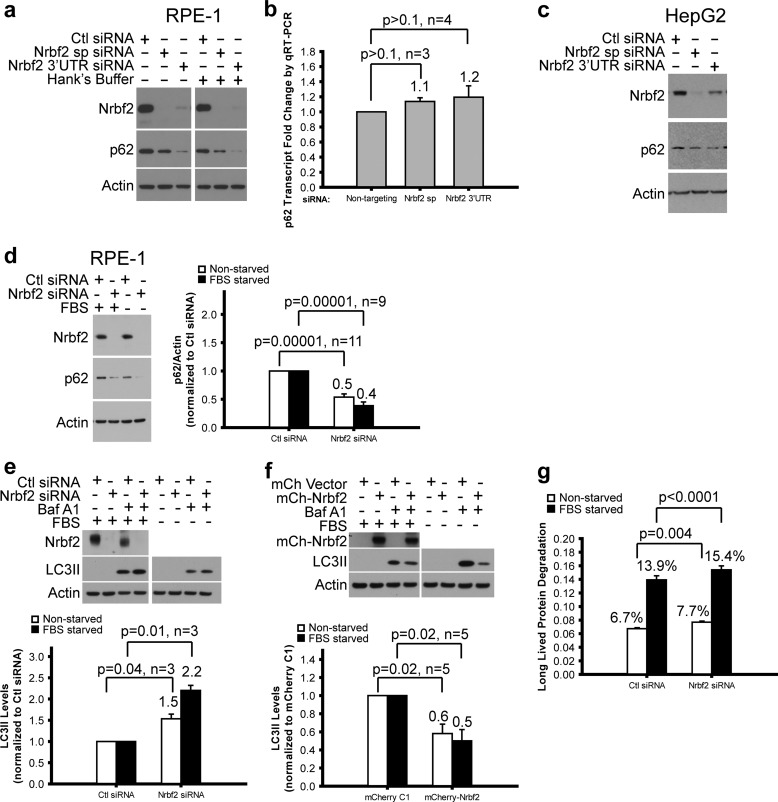
Because the Beclin 1-Vps34 protein-protein interaction network is a key regulatory hub for autophagy, we next assessed the role of Nrbf2 on autophagic activity. To do that, we carried out three widely used autophagic flux assays. The first involved measuring the steady-state levels of p62/SQSTM1 protein, an adaptor for delivering autophagy substrates to autophagosomes while the adaptor itself is degraded along with the substrates in autolysosomes (43). We treated RPE-1 cells with non-targeting siRNA, a pool of four siRNAs against the ORFs of human Nrbf2 (SMARTpool Nrbf2 siRNA), or a single siRNA against the 3′ untranslated region (3′UTR) of human Nrbf2. Our data show that treating cells with either the SMARTpool or the 3′UTR Nrbf2 siRNA, compared with non-targeting siRNA, decreased p62 protein levels under both nutrient-rich and Hanks' buffer starvation conditions (Fig. 2a). We further examined whether decreased p62 protein levels upon Nrbf2 siRNA treatments resulted from reduced p62 transcript levels. To do that, we performed RNA sequencing (RNA-Seq) analysis of RPE-1 cells treated with non-targeting, SMARTpool, and 3′UTR Nrbf2 siRNAs. We found that p62 transcripts were highly abundant in RPE-1 cells and not significantly changed upon Nrbf2 siRNA treatment (data not shown). We also performed qRT-PCR analysis of RPE-1 cells treated with non-targeting, SMARTpool, and 3′UTR Nrbf2 siRNAs. Our results confirm that p62 transcripts in RPE-1 cells were not significant changed upon Nrbf2 siRNA treatment (Fig. 2b). Therefore, the reduction in the p62 protein levels in the Nrbf2-deficient RPE-1 cells was likely the result of an increased autophagic flux. Moreover, we observed a similar decrease of p62 protein levels in the HepG2 cells treated with either SMARTpool or 3′UTR Nrbf2 siRNA compared with non-targeting siRNA (Fig. 2c), suggesting that the increase of autophagic flux in response to Nrbf2 deficiency is likely a common phenomenon for human cells. Therefore, in the rest of the study, we focused on RPE-1 cells. Of note, although the SMARTpool Nrbf2 siRNA depleted Nrbf2 protein more effectively than the 3′UTR Nrbf2 siRNA in both RPE-1 and HepG2 cells, the 3′UTR Nrbf2 siRNA reduced p62 protein levels more effectively than the SMARTpool Nrbf2 siRNA (Fig. 2, a and c). It is possible that either 3′UTR Nrbf2 siRNA has an off-target effect that increases p62 degradation by the ubiquitin proteasome system or that the SMARTpool Nrbf2 siRNA has an off-target effect that decreases p62 degradation by the ubiquitin proteasome system. We have no data to favor one over the other. Therefore, we chose to use the SMARTpool Nrbf2 siRNAs hereafter, solely on the basis of knockdown efficiency. In addition, Nrbf2 siRNA (compared with non-targeting siRNA) decreased p62 protein levels under serum starvation conditions (Fig. 2d), similar to the observation under either nutrient-rich or Hanks' buffer starvation conditions, suggesting that the increase of autophagic flux in response to Nrbf2 deficiency is likely a common phenomenon for different nutrient conditions. Therefore, in the rest of the study, we used either Hanks' buffer starvation or serum starvation.
FIGURE 2.
Characterization of Nrbf2 as a negative regulator of autophagic flux under both nutrient-rich and starvation conditions in human cell lines. Western blot analysis of p62 in RPE-1 (a) and HepG2 (c) cells transfected with non-targeting control (Ctl) siRNA, human SMARTpool (sp) and 3′UTR Nrbf2 siRNAs showed decreased p62 levels upon Nrbf2 siRNA treatment. a, cells were grown under nutrient-rich or 1-h Hanks' buffer starvation conditions. c, cells were grown under nutrient-rich conditions. b, qRT-PCR quantification of the relative p62 transcript levels in the RPE-1 cells transfected with Ctl, Nrbf2 sp, and Nrbf2 3′UTR siRNAs. d, Western blot analysis of p62 levels in RPE-1 cells transfected with Ctl or Nrbf2 sp siRNA showed decreased p62 levels upon Nrbf2 siRNA treatment under both nutrient-rich and 18-h serum starvation conditions. e, Western blot analysis of LC3II levels in RPE-1 cells transfected with Ctl siRNA or Nrbf2 sp siRNA and treated either without or with 200 nm Baf showed a greater Baf-induced increase of LC3II levels upon Nrbf2 siRNA treatment under both nutrient-rich and 18-h serum starvation conditions. f, Western blot analysis of LC3II levels in RPE-1 cells transfected with mCherry empty vector or mCherry-tagged human Nrbf2 and treated either without or with 200 nm Baf showed a lesser Baf-induced increase of LC3II levels upon Nrbf2 overexpression under both nutrient-rich and 18-h serum starvation conditions. The statistical significance for the LC3II assays in e and f is lower than that for the p62 assay in d, likely because of fewer numbers of independent experiments. g, the long-lived protein degradation rate increased upon Nrbf2 siRNA treatment under both nutrient-rich and serum starvation conditions. For all quantifications, means ± S.E. were plotted with means labeled.
The second autophagic flux assay involves monitoring the changes in the levels of LC3II in response to lysosomal inhibitors (e.g. Baf, an inhibitor of the vacuolar H+-ATPase and, therefore, lysosomal acidification and autophagosome-lysosome fusion (44–46)). Using this assay, we observed that, under both nutrient-rich and serum starvation conditions, transfecting Nrbf2 siRNA (compared with non-targeting siRNA) led to a greater increase in the LC3II levels in response to Baf, suggesting that Nrbf2 deficiency increases autophagic flux (Fig. 2e). Conversely, overexpressing mCherry-tagged Nrbf2 (compared with mCherry empty vector) led to a smaller increase in the LC3II levels upon Baf treatment, suggesting that Nrbf2 overexpression decreases autophagic flux (Fig. 2f).
The third autophagic flux assay directly measured the rate of long-lived protein degradation by pulse-chasing with a radioactively labeled amino acid (e.g. 14C-Leu). Our data show that, compared with non-targeting siRNA, Nrbf2 siRNA treatment increased long-lived protein degradation under both nutrient-rich and serum starvation conditions (Fig. 2g). Collectively, these autophagic flux data suggest that Nrbf2 negatively regulates basal and starvation-induced autophagy.
Nrbf2 Is a Component of the Atg14L-containing Beclin 1-Vps34 Protein Complex
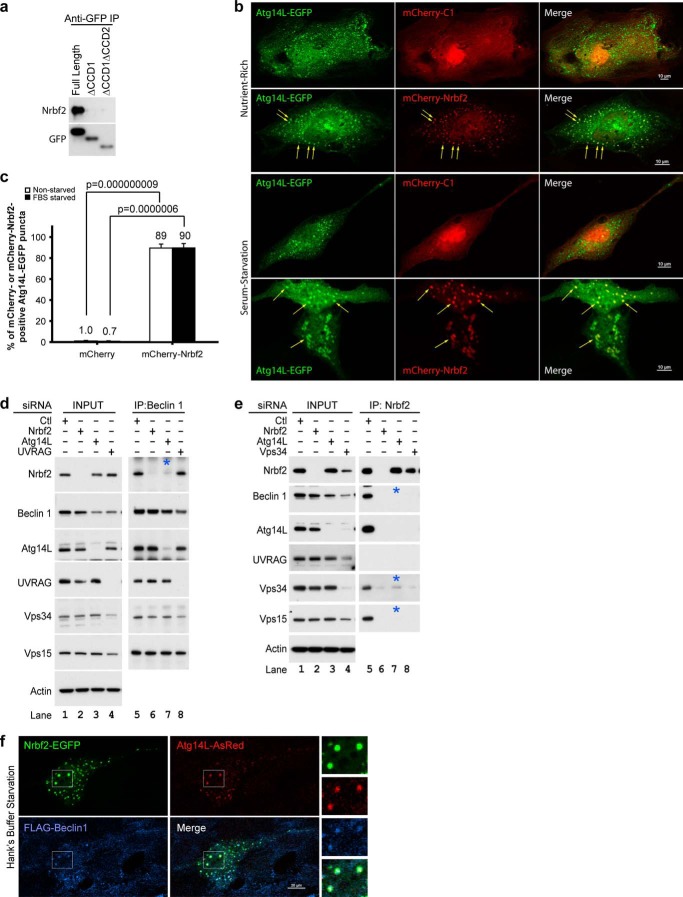
Because we identified Nrbf2 as a Beclin 1-interacting protein and because it is well known that there are at least two Beclin 1-Vps34 protein complexes, we asked with which Beclin 1-Vps34 protein complex Nrbf2 was associated. The two main Beclin 1-Vps34 protein complexes contain exclusively Atg14L or UVRAG, with the Atg14L-containing complex known to be involved in autophagosome biogenesis (15–18, 25, 26). We first tested whether Nrbf2 was associated with the Atg14L-containing Beclin 1-Vps34 protein complex. To do that, we transiently transfected cells with EGFP-tagged full-length Atg14L or mutant Atg14L with either the first or both of the CCDs deleted (labeled ΔCCD1 and ΔCCD1ΔCCD2, respectively) (15) and performed anti-GFP IP. Our data show that EGFP-tagged full-length Atg14L, but not the ΔCCD1 or ΔCCD1ΔCCD2 mutant of Atg14L, pulled down endogenous Nrbf2 (Fig. 3a), suggesting that Nrbf2 interacts with Atg14L in an Atg14L CCD-dependent manner.
FIGURE 3.
Nrbf2 is a component of the Atg14L-containing Beclin 1-Vps34 protein complex. a, transiently expressed, EGFP-tagged, full-length Atg14L, but not the CCD-deletion mutants of Atg14L, pulled down endogenous Nrbf2 in HeLa cells. Immunoprecipitated EGFP-tagged full-length and mutant Atg14L were probed with anti-GFP antibody. b, ectopically expressed mCherry-tagged human Nrbf2 colocalized with Atg14L-EGFP on punctate structures (arrows) in RPE-1 cells under nutrient-rich and 18-h serum-starvation conditions. In contrast, mCherry alone showed diffused cytoplasmic and nuclear localization. Scale bars = 10 μm. c, quantification of colocalization of mCherry or mCherry-Nrbf2 with Atg14L-EGFP as measured by the percentage of Atg14L-EGFP puncta that were either mCherry- or mCherry-Nrbf2-positive. Cells with more than five Atg14L-EGFP puncta were used for quantification. The numbers of cells used for quantification were five (for mCherry, nutrient-rich), seven (for mCherry, serum starvation), nine (for mCherry-Nrbf2, nutrient-rich), and seven (for mCherry-Nrbf2, serum starvation). These cell numbers are sufficient to reveal a statistical significance of colocalization between mCherry-Nrbf2 and Atg14L-EGFP using two-tailed Student's t test. d, anti-Beclin 1 IP from RPE-1 cell treated with non-targeting, Nrbf2, Atg14L, or UVRAG siRNA. e, anti-Nrbf2 IP from RPE-1 cells treated with non-targeting, Nrbf2, Atg14L, or Vps34 siRNA. Note that Vps34 siRNA treatment led to unstable Nrbf2, Beclin 1, Atg14L, UVRAG, and Vps15, whereas Atg14L siRNA treatment only slightly destabilized Beclin 1 and did not destabilize Nrbf2, UVRAG, Vps34, or Vps15. d and e, interactions that required Atg14L are marked by blue asterisks. f, ectopically expressed Nrbf2-EGFP, Atg14L-AsRed, and FLAG-Beclin 1 colocalized on punctate structures in RPE-1 cells under 1-h Hanks' buffer starvation conditions. Scale bars = 20 μm.
We then tested whether Nrbf2 and Atg14L colocalized. Our data show that ectopically expressed mCherry-Nrbf2 and Atg14L-EGFP largely colocalized on punctate structures in RPE-1 cells under both nutrient-rich and serum starvation conditions (Fig. 3b, arrows; quantified in Fig. 3c). This result is consistent with the presence of the Nrbf2-Atg14L interaction.
To dissect the role of the Nrbf2-Atg14L interaction, we performed anti-Beclin 1 IP from RPE-1 cells treated with non-targeting, Nrbf2, Atg14L, or UVRAG siRNA. Our data show that Atg14L, but not UVRAG, was required for the Beclin 1-Nrbf2 interaction (Fig. 3d, lanes 7 and 8). In contrast, Nrbf2 was not required for the interaction of Beclin 1 with Atg14L, UVRAG, Vps34, or Vps15 (Fig. 3d, lane 6). We further performed anti-Nrbf2 IP from RPE-1 cells treated with non-targeting, Nrbf2, Atg14L, or Vps34 siRNA. Our data show that Nrbf2 pulled down not only Beclin 1, Vps34, and Vps15 but also Atg14L. However, Nrbf2 was unable to pull down UVRAG (Fig. 3e, lane 5). Our data also show that anti-Nrbf2 IP from RPE-1 cells treated with Atg14L siRNA was unable to pull down Beclin 1 (Fig. 3e, lane 7) even though Beclin 1 was reasonably stable in the absence of Atg14L (Fig. 3e, lane 3), confirming the requirement of Atg14L for the Beclin 1-Nrbf2 interaction. In addition, anti-Nrbf2 IP from RPE-1 cells treated with Atg14L siRNA was also unable to pull down Vps15 and only capable of pulling down a small fraction of Vps34 (Fig. 3e, lane 7), even though both Vps34 and Vps15 were stable in the absence of Atg14L (Fig. 3e, lane 3). These results suggest that the association of Nrbf2 with the Beclin 1-Vps34 protein complex is primarily mediated by Atg14L and that Nrbf2 is largely associated with the Atg14L-containing, rather than the UVRAG-containing, Beclin 1-Vps34 protein complex.
We reported previously that ectopically coexpressed Beclin 1 and Atg14L colocalized on cytosolic punctate structures that were enwrapped with double membranes and contained electron-dense materials, consistent with a role for the Atg14L-containing Beclin 1-Vps34 protein complex in autophagosome biogenesis (15). Here we further showed that, in RPE-1 cells transiently cotransfecting with Nrbf2-EGFP, FLAG-Beclin 1, and Atg14L-AsRed, Nrbf2-EGFP localized on the FLAG-Beclin 1- and Atg14L-AsRed-positive cytosolic puncta known to be wrapped by double membranes (Fig. 3f). Therefore, our data collectively suggest that Nrbf2 is primarily associated with the Atg14L-containing Beclin 1-Vps34 protein complex that is known to participate in autophagosome biogenesis.
Nrbf2 Is Involved in Autophagosome Biogenesis
We directly examined whether Nrbf2 was involved in autophagosome biogenesis. Using subcellular fractionation, we first showed that, in HEK293 cells stably expressing Nrbf2-EGFP, Nrbf2-EGFP was primarily present in the cytosol fraction (Fig. 4a) and so was endogenous Nrbf2 in untransfected HEK293 cells (Fig. 4b).
FIGURE 4.
Nrbf2 forms cytoplasmic puncta that are positive for isolation membrane markers in RPE-1 cells, particularly under starvation conditions. Subcellular fractionation revealed that both stably expressed Nrbf2-EGFP (a) and endogenous Nrbf2 (b) were primarily localized in the cytosol in HEK293 cells under nutrient-rich conditions. A1, cytosolic fraction as marked by SOD1; A2, membrane fraction as marked by EGFR; A3, soluble nuclear fraction as marked by SP1; A4, chromatin-bound nuclear fraction as marked by histone H3; A5, cytoskeletal fraction; A6, pellet fraction. c, ectopically expressed Nrbf2-EGFP formed cytoplasmic punctate structures under both nutrient-rich and Hanks' buffer starvation conditions. The numbers of cells used for quantification were 18 (for EGFP, nutrient-rich), 24 (for EGFP, Hanks' buffer starvation), 22 (for Nrbf2-EGFP, nutrient-rich), and 22 (for Nrbf2-EGFP, Hanks' buffer starvation). d, ectopically expressed mCherry-Nrbf2 and EGFP-Atg5 colocalized on punctate structures under Hanks' buffer starvation conditions. The numbers of cells used for quantification were 14 (for mCherry) and 12 (for mCherry-Nrbf2). e, ectopically expressed Nrbf2-EGFP and myc-tagged Ulk1 WT colocalized on punctate structures under Hanks' buffer starvation conditions. The numbers of cells used for quantification were nine (for EGFP) and four (for Nrbf2-EGFP). f, ectopically expressed mCherry-Nrbf2 and endogenous FIP200 colocalized on punctate structures under Hanks' buffer starvation conditions. The numbers of cells used for quantification were three (for mCherry) and eight (for mCherry-Nrbf2). g, ectopically expressed Nrbf2-EGFP and myc-tagged Ulk1 K46I mutant colocalized on punctate structures under Hanks' buffer starvation conditions. The numbers of cells used for quantification were six (for EGFP) and nine (for Nrbf2-EGFP). d–g, note that mCherry or EGFP was primarily diffused, whereas mCherry-Nrbf2 or Nrbf2-EGFP formed puncta under Hanks' buffer starvation conditions. h, ectopically expressed Nrbf2-EGFP formed cytoplasmic puncta in both FIP200+/+ and FIP200−/− MEFs under Hanks' buffer starvation conditions. The numbers of cells used for quantification were six (for EGFP, FIP200+/+), 11 (for Nrbf2-EGFP, FIP200+/+), eight (for EGFP, FIP200−/−), and ten (for Nrbf2-EGFP, FIP200−/−). NS, not significant. Scale bars = 10 μm (c–f) and 20 μm (g and h).
Moreover, using confocal microscopy, we found that, under nutrient-rich conditions, ectopically expressed Nrbf2-EGFP is primarily diffused in the cytosol, forming only a small number of cytoplasmic puncta. In contrast, upon 1 h starvation in Hanks' buffer, the number of cytoplasmic Nrbf2-EGFP puncta increased significantly (Fig. 4c). During autophagosome biogenesis, Atg14L localizes to the rough endoplasmic reticulum (25) and, specifically, the endoplasmic reticulum-mitochondrion contact site (47), leading to the recruitment of Beclin 1 and Vps34 and subsequent production of PI3P in a Ulk1-FIP200 protein complex-dependent manner (25, 26) at the autophagosome initiation site (often referred to as isolation membrane, also termed omegasome because of its Ω shape (48)). Ulk1 and Atg5, which synchronously form puncta in a PI3P-independent manner (49), accumulate in a PI3P-dependent manner at the PI3P-enriched autophagosome initiation sites (50). Subsequently, the Atg12-Atg5 and LC3-phosphatidylethanolamine ubiquitin-like conjugation systems were recruited and assembled, leading to the expansion of isolation membranes. Therefore, to further characterize the starvation-induced Nrbf2 punctate structures, we examined the presence of isolation membrane markers on these structures. Of note, because starvation resulted in considerably more Nrbf2 puncta than nutrient-rich conditions, we monitored the colocalization of fluorescently tagged Nrbf2 and isolation membrane markers in cells only under starvation conditions. We showed that ectopically expressed mCherry-Nrbf2, but not mCherry alone, colocalized with EGFP-Atg5 (Fig. 4d) and that ectopically expressed Nrbf2-EGFP, but not EGFP alone, colocalized with myc-Ulk1 (Fig. 4e). We further showed that ectopically expressed mCherry-Nrbf2, but not mCherry alone, colocalized with endogenous FIP200 (Fig. 4f). Taken together, our results demonstrate colocalization of Nrbf2 with the isolation membrane markers Atg5, Ulk1 and FIP200, suggesting the involvement of Nrbf2 in autophagosome biogenesis.
Because the Ulk1-FIP200 protein complex is critical for initiating autophagosome biogenesis, we next investigated whether the Ulk1 kinase activity or FIP200 was required for forming the Nrbf2 puncta. Ulk1 K46I has been reported to be a dominant negative mutation, impairing Ulk1 kinase activity (51). Similarly, loss of FIP200 has been reported to compromise Ulk1 stability, localization, and activity (26, 52, 53). We showed that Nrbf2-EGFP puncta formed in both RPE-1 cells overexpressing Ulk1 K46I mutant (Fig. 4g) and FIP200−/− mouse embryonic fibroblasts (MEFs) (Fig. 4h). These results suggest that, in contrast to the Atg14L puncta, which cannot form in either HEK293 cells overexpressing dominant negative Ulk1 K46N mutant (25) or FIP200−/− MEFs (26), the Nrbf2 puncta were able to form independently of the Ulk1 kinase activity or FIP200.
Nrbf2 Suppresses Intracellular PI3P Levels
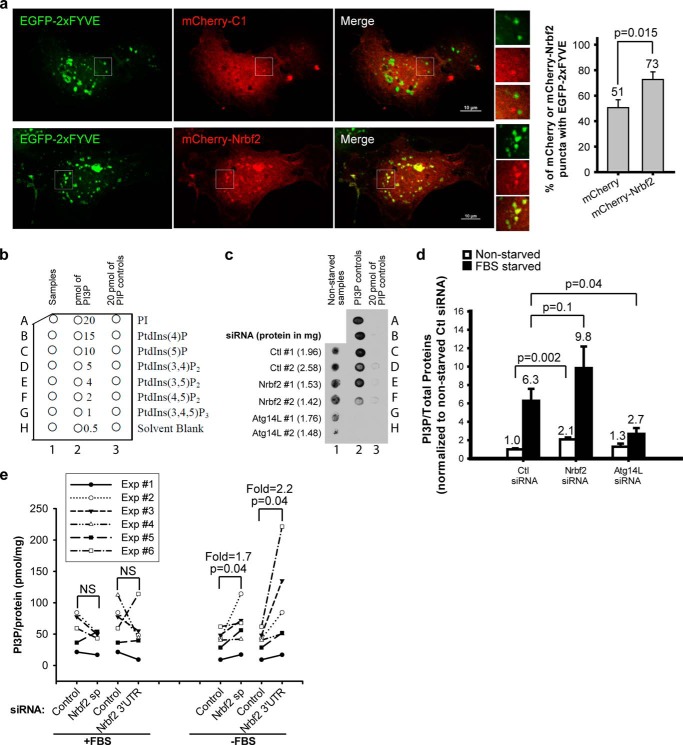
The conversion of PI to PI3P by Vps34 is important for autophagosome formation in both yeast and mammals (7, 8, 14). Because our data presented above suggest that Nrbf2 is a component of the Atg14L-containing Beclin 1-Vps34 protein complex, we hypothesized that Nrbf2 may negatively regulate autophagic flux through suppressing the lipid kinase activity of Vps34. Utilizing an EGFP-2×FYVE plasmid that contains two consecutive specific PI3P-binding Fab1/YOTB/Vac1/EEA1 (FYVE) motifs (54, 55), we showed that ectopically expressed mCherry-Nrbf2, but, to a significantly lesser degree, mCherry alone, colocalized with EGFP-2×FYVE on punctate structures under serum starvation conditions, suggesting localization of Nrbf2 on PI3P-enriched lipid domains (Fig. 5a).
FIGURE 5.
Nrbf2 reduces intracellular PI3P levels in RPE-1 cells. a, ectopically expressed mCherry-Nrbf2 and EGFP-2×FYVE colocalized on punctate structures under serum starvation conditions. Scale bars = 10 μm. The number of cells used for quantification was 18 for either mCherry or mCherry-Nrbf2. Note that because EGFP-2xFYVE was capable of sequestering cytoplasmic constituents, even mCherry alone formed some punctate structures that were positive for EGFP-2xFYVE. However, mCherry-Nrbf2 clearly formed more distinctive puncta and colocalized with EGFP-2xFYVE to a higher degree. b–d, monitoring intracellular PI3P levels in RPE-1 cells treated with non-targeting (Ctl), Nrbf2, or Atg14L siRNA by protein-lipid overlay assay. b, layout of the PI3P mass strip. c, a representative dot blot showing extracted acidic lipids as detected by SuperSignal West Femto chemiluminescent substrate. Corresponding amounts of total proteins are labeled in parentheses. Note that PI, PIP3, PIP2s, and other phosphatidylinositol phosphate controls in lane 3 and 0.5–1 pmol of PI3P in lane 2 showed barely detectable signals. d, comparison of extracted intracellular PI3P levels (normalized to total protein levels) upon non-targeting, Nrbf2, or Atg14L siRNA treatment in RPE-1 cells for data collected in dot blots like c. Four independent experiments were performed. Mean ± S.E. was plotted with means labeled. e, comparison of extracted intracellular PI3P levels (normalized to total protein levels) upon non-targeting, Nrbf2 sp, or Nrbf2 3′UTR siRNA treatment in RPE-1 cells using the PI3P mass ELISA kit. Six independent experiments were performed, with each experiment plotted individually to best show the effect of Nrbf2 siRNA treatment.
To investigate whether Nrbf2 modulates intracellular PI3P levels, we initially used a semiquantitative protein-lipid overlay assay to monitor intracellular PI3P levels in RPE-1 cells treated with non-targeting or Nrbf2 siRNA. The dot blot (layout shown in Fig. 5b) clearly shows that the protein-lipid overlay assay specifically recognized PI3P but not PI, PIP3, PIP2s, or other phosphatidylinositol phosphates (Fig. 5c, lane 3). Because we and others reported previously that Atg14L facilitates Vps34 lipid kinase activity (15, 18, 25), here we used Atg14L siRNA as a control. Our data demonstrate that Atg14L deficiency resulted in significantly decreased intracellular PI3P levels under serum starvation conditions (Fig. 5d), consistent with previous reports (15, 18, 25). We further showed that Nrbf2 deficiency, contrary to Atg14L deficiency, led to increased intracellular PI3P levels (p = 0.002 for nutrient-rich conditions and p = 0.1 for serum starvation conditions) (Fig. 5d). Because the PI3P mass strip kit for the protein-lipid overlay assay was discontinued by the company during our study, we later monitored intracellular PI3P levels using a replacement PI3P ELISA assay. Our ELISA data show that knocking down Nrbf2 by either the SMARTpool or 3′UTR Nrbf2 siRNA resulted in significantly increased intracellular PI3P levels under serum starvation conditions, whereas, under nutrient-rich conditions, the effect of Nrbf2 deficiency on intracellular PI3P levels was undetectable (Fig. 5e). Because ELISA assays are generally thought to be more quantitative than dot blot assays (e.g. the protein-lipid overlay assay), collectively, our results suggest that Nrbf2 suppresses intracellular PI3P levels under starvation conditions, likely through suppressing the Vps34-catalyzed conversion of PI to PI3P, leading to negative regulation of autophagic flux.
Nrbf2 Is Required for the Atg14L-Vps34/Vps15 Interactions
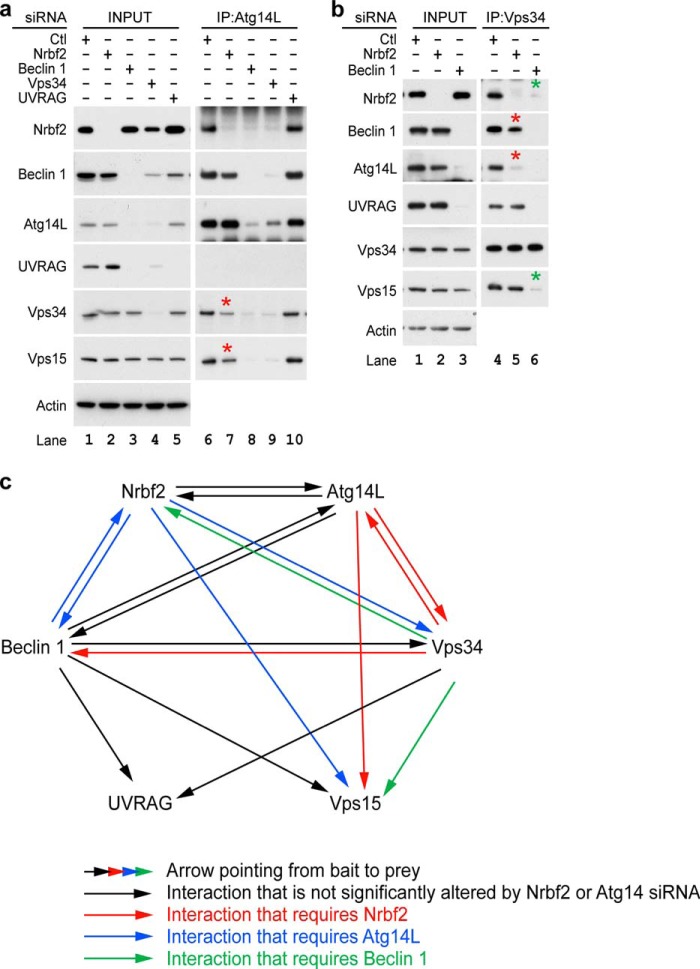
To further investigate the mechanism by which Nrbf2 regulates Vps34 activity, we performed IP to determine whether Nrbf2 was required for protein-protein interactions within the Atg14L-containing Beclin 1-Vps34 protein complex. We first performed IP of endogenous Atg14L from RPE-1 cells treated with non-targeting, Nrbf2, Beclin 1, Vps34, or UVRAG siRNA. Our data show that, consistent with the literature (15, 17), both Atg14L and UVRAG were unstable upon Beclin 1 or Vps34 siRNA treatment (Figs. 6a, INPUT, lanes 3 and 4). We also observed that Atg14L was unable to pull down Nrbf2 upon Beclin 1 or Vps34 siRNA treatment (Fig. 6a, IP: Atg14L, lanes 8 and 9), which primarily reflected the loss of Atg14L as a result of Beclin 1 or Vps34 deficiency rather than the requirement of Beclin 1 or Vps34 for the Atg14L-Nrbf2 interaction. In addition, our data show that Atg14L was unable to pull down UVRAG (Fig. 6a, IP: Atg14L, lane 6), consistent with the notion that UVRAG and Atg14L are mutually exclusive (17, 56). Most importantly, despite the fact that Atg14L, Vps34 and Vps15 proteins were stable upon Nrbf2 siRNA treatment (Fig. 6a, INPUT, lanes 1 and 2), endogenous Atg14L pulled down much less Vps34 or Vps15 upon Nrbf2 siRNA treatment compared with non-targeting siRNA treatment (Fig. 6a, IP: Atg14L, lanes 6 and 7), suggesting that Nrbf2 is required for the Atg14L-Vps34/Vps15 interactions. Of note, our data also show that Nrbf2 was minimally needed for the Beclin 1-Atg14L interaction (Fig. 3d, IP: Beclin 1, lanes 5 and 6, and Fig. 6a, IP: Atg14L, lanes 6 and 7).
FIGURE 6.
Nrbf2 is required for the Atg14L-Vps34/Vps15 interactions. a, IP of endogenous Atg14L in RPE-1 cell treated with non-targeting, Nrbf2, Beclin 1, Vps34, or UVRAG siRNA under nutrient-rich conditions. b, IP of endogenous Vps34 in RPE-1 cell treated with non-targeting, Nrbf2, or Beclin 1 siRNA under nutrient-rich conditions. a and b, interactions that required Nrbf2 and Beclin 1 are marked by red and green asterisks, respectively. c, diagram summarizing the interactions identified in this work through endogenous IPs shown in Fig. 3, d and e, and this figure, a and b. Each arrow is pointed from bait to prey.
To further examine the requirement of Nrbf2 for the Atg14L-Vps34 interaction, we performed IP of endogenous Vps34 from RPE-1 cell treated with non-targeting, Nrbf2, or Beclin 1 siRNA. Our data show that the Vps34-UVRAG interaction was not changed upon Nrbf2 siRNA treatment (Fig. 6b, IP: Vps34, lanes 4 and 5). In contrast, despite the fact that both Atg14L and Vps34 proteins were stable upon Nrbf2 siRNA treatment (Fig. 6b, INPUT, lanes 1 and 2), endogenous Vps34 pulled down much less Atg14L upon Nrbf2 siRNA treatment compared with non-targeting siRNA treatment (Fig. 6b, IP: Vps34, lanes 4 and 5). These results further support the hypothesis that Nrbf2 is required for the Atg14L-Vps34 interaction. In addition, Nrbf2 appears to be required for the Beclin 1-Vps34 interaction to a lesser degree (Fig. 6b, IP: Vps34, lanes 4 and 5). Furthermore, despite the fact that Nrbf2, Vps34, and Vps15 protein levels were unchanged upon Beclin 1 deficiency (Fig. 6b, INPUT, lanes 1 and 3), the Nrbf2-Vps34 interaction or the Vps34-Vps15 interaction was abolished as a result of Beclin 1 siRNA treatment (Fig. 6b, IP: Vps34, lanes 4 and 6), suggesting that Beclin 1 is required for Vps34 to interact with other components of the Beclin 1-Vps34 protein-protein interaction network.
DISCUSSION
To elucidate the molecular details of autophagy regulation by the Beclin 1-Vps34 protein-protein interaction network, we set out to identify novel components in the network and characterized their roles in regulating autophagy (15). To this end, we identified Nrbf2 as a Beclin 1-interacting protein with its N terminus required for binding to Beclin 1 and determined a novel role for Nrbf2 in negatively regulating autophagic flux. Using endogenous IP, we showed that Nrbf2 interacted with Beclin1, Vps34, and Vps15, the components of the core complex in the Beclin 1-Vps34 protein-protein interaction network. Interestingly, Nrbf2 also interacted with Atg14L, the adaptor to recruit the core complex for autophagosome biogenesis, but not UVRAG, the adaptor to recruit the core complex for endocytic trafficking. Moreover, Atg14L was required for the Beclin 1-Nrbf2 interaction, suggesting that Nrbf2 may interact with Beclin 1 through Atg14L. In addition, ectopically expressed Nrbf2 was colocalized with either ectopically expressed Atg14L alone or coexpressed Beclin 1 and Atg14L on cytoplasmic punctate structures. Taken together, we hypothesize that Nrbf2, by being a component of the Atg14L-containing Beclin 1-Vps34 protein complex, was involved in autophagosome biogenesis. Indeed, our data show that ectopically expressed Nrbf2 formed cytoplasmic puncta positive for the isolation membrane markers Atg5, Ulk1, and FIP200 in a process that was independent of the Ulk1 kinase activity or FIP200. Therefore, our current working model for the role of Nrbf2 in autophagy is that Nrbf2 likely suppresses autophagy through negatively regulating Atg14L-containing Beclin 1-Vps34 protein complex-mediated autophagosome biogenesis, although our results do not exclude additional roles for Nrbf2 in autophagosome maturation.
To unravel the molecular mechanism by which Nrbf2 negatively regulates autophagosome biogenesis and autophagy, we showed that Nrbf2 localized on PI3P-enriched lipid domains and suppressed intracellular PI3P levels, consistent with a potential role of Nrbf2 in inhibiting Vps34 activity. We further showed that Nrbf2 was critical for the Atg14L-Vps34/Vps15 interactions, whereas the Beclin 1-Atg14L or Beclin1/Vps34-UVRAG interactions were unaffected by Nrbf2 deficiency. We summarize the protein-protein interactions identified in this work in Fig. 6c. It is possible that the Nrbf2-mediated Atg14L-Vps34/Vps15 interactions may contribute to the role of Nrbf2 in suppressing Vps34 activity through a conformational regulation-based mechanism. Therefore, in our working model, we propose that Nrbf2 modulates protein-protein interactions in the Atg14L-containing Beclin 1-Vps34 protein complex, leading to the suppression of Vps34 activity, autophagosome biogenesis, and autophagic flux.
The roles of the Beclin 1-Vps34 protein-protein interaction network are largely context-dependent because components of the Beclin 1-Vps34 protein-protein interaction network have different, even opposing effects on autophagy. For example, Vps34 is essential for autophagy (7, 8, 14). Paradoxically, during amino acid stimulation, Vps34 positively regulates mammalian target of rapamycin complex 1, leading to autophagy inhibition (57–59). As another example, Beclin 1, when interacting with Atg14L, promotes autophagosome biogenesis (15–18). In contrast, Beclin 1 is involved in inhibition of autophagic activity when interacting with either 14-3-3 (60) (as a result of Beclin 1 phosphorylation by Akt) or Bcl-2 (24) (interaction weakened by death-associated protein kinase-mediated Beclin 1 phosphorylation (61, 62)). In addition, UVRAG has been suggested to either direct Beclin 1 and Vps34 to endocytic trafficking (17) or promote autophagosome maturation (27) and, with some controversy, autophagosome biogenesis (17, 28). Therefore, the Beclin 1-Vps34 protein-protein interaction network appears to be dynamic, serving as a key regulatory hub to integrate extracellular and intracellular cues for tight control of autophagic activity. On the basis of our data presented here, Nrbf2 is an important component of this context-dependent Beclin 1-Vps34 protein-protein interaction network. Under certain conditions (such as those reported in this work), Nrbf2 negatively regulates autophagy and likely Vps34 activity through modulating protein-protein interactions in the Beclin 1-Vps34 network.
As a final note, while our manuscript was under review, three studies reported the identification of human (63) and mouse (64) Nrbf2 as well as the yeast ortholog Atg38 (65) as a component of the Beclin 1/Vps30-Vps34 protein-protein interaction network. Our biochemical data are largely consistent with the above reports that Nrbf2 is primarily a component of the Atg14L/Atg14-containing Beclin 1/Vps30-Vps34 protein complex and is critical for the assembly of the Atg14L/Atg14-Beclin1/Vps30 and Vps34-Vps15 subcomplexes. Our results are also consistent with the above reports that Nrbf2 is involved in autophagosome biogenesis. However, contrary to the above reports showing a positive role of Nrbf2 in regulating autophagy, using a variety of independent autophagic flux assays, we showed that human Nrbf2 suppressed autophagic flux in RPE-1 and HepG2 cells. Although we do not understand what causes the difference in our study and those of the other groups, we postulate that the discrepancy is likely rooted in the context dependence of autophagy regulation. For example, the MIT and CCD domains of yeast Atg38 are evolutionarily more divergent than those of the mammalian Nrbf2 (Fig. 1d). In particular, the CCD of yeast Atg38 lacks the five leucines that are consecutive at every seven residues in the mammalian Nrbf2 CCD (Fig. 1d). These sequence deviations of yeast Atg38 from mammalian Nrbf2 may result in an alteration of function. Moreover, we also participated in one of the reported studies where Nrbf2 was observed to enhance autophagy and Vps34 activity in NIH 3T3 cells and MEFs (64). Even in our own hands, Nrbf2 siRNA-treated NIH 3T3 cells showed increased p62 levels (data not shown). Therefore, it is possible that Nrbf2 may function differently in human versus mouse cells. Supporting this notion, it has been reported that expression levels of Beclin 1 and Atg14L, as well as the stoichiometry of the Beclin 1-Vps34 protein complexes, were drastically different in mouse (NIH 3T3 and MEFs) and human (HeLa) cell lines (40). Furthermore, our study utilized siRNA to transiently (2∼3 days) knock down Nrbf2, whereas all abovementioned reports utilized either genetic knockout (64, 65) or stable knockdown (63). It is possible that lacking Nrbf2 for an extended period of time may alter the cellular context. In short, Nrbf2 is an important player for tight autophagy regulation, maybe in a species-, cell type-, or stress-specific manner. Further investigation is necessary to reveal the context dependence of autophagy regulation by Nrbf2.
Acknowledgments
We thank J. Guan (University of Cincinnati) for FIP200+/+ and FIP200−/− MEF cells; H. Stenmark (University of Oslo) for the EGFP-2×FYVE plasmid; S. Tooze (London Research Institute) for the myc-Ulk1 and myc-Ulk1 K46I constructs; Z. Yue (Mount Sinai School of Medicine) for the mouse Nrbf2-EGFP, Atg14L-AsRed, and FLAG-Beclin 1 plasmids; and X. Jiang (Memorial Sloan Kettering Cancer Center) for the EGFP-Atg5 plasmid. We also thank the Imaging Core of the National Institutes of Health Center of Biomedical Research Excellence (COBRE) at the University of Kentucky for providing access to microscopes. We further thank J. Simpson for technical support and Drs. K. L. Guan and J. Kim for advice regarding Beclin 1-Vps34 complex immunoprecipitation.
This work was supported, in whole or in part, by National Institutes of Health Grants P20GM103486 (to Q. J. W.) and RR00862 and RR022220 (to B. T. C.). This work was also supported by the Ellison Medical Foundation (to Q. J. W.).
- PI
- phosphatidylinositol
- PI3P
- phosphatidylinositol 3-phosphate
- Vps
- vacuolar protein sorting
- Baf
- bafilomycin A1
- IP
- immunoprecipitation
- LC3II
- LC3-phosphatidylethanolamine
- qRT-PCR
- quantitative RT-PCR
- EGFP
- enhanced GFP
- sp
- SMARTpool
- MEF
- mouse embryonic fibroblast
- FYVE
- Fab1/YOTB/Vac1/EEA1
- UVRAG
- ultraviolet irradiation resistance-associated gene
- CCD
- coiled-coil domain
- MIT
- microtubule-interacting and trafficking
- PIP
- phosphatidylinositol 3-phosphate, phosphatidylinositol 4-phosphate, or phosphatidylinositol 5-phosphate
- PIP2
- phosphatidylinositol 3,4-bisphosphate, phosphatidylinositol 3,5-bisphosphate, or phosphatidylinositol 4,5-bisphosphate
- PIP3
- phosphatidylinositol 3,4,5-triphosphate.
REFERENCES
- 1. Suzuki K., Ohsumi Y. (2007) Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 581, 2156–2161 [DOI] [PubMed] [Google Scholar]
- 2. Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu Y., Shoji-Kawata S., Sumpter R. M., Jr., Wei Y., Ginet V., Zhang L., Posner B., Tran K. A., Green D. R., Xavier R. J., Shaw S. Y., Clarke P. G., Puyal J., Levine B. (2013) Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc. Natl. Acad. Sci. U.S.A. 110, 20364–20371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 5. Liang X. H., Kleeman L. K., Jiang H. H., Gordon G., Goldman J. E., Berry G., Herman B., Levine B. (1998) Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 72, 8586–8596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maiuri M. C., Le Toumelin G., Criollo A., Rain J. C., Gautier F., Juin P., Tasdemir E., Pierron G., Troulinaki K., Tavernarakis N., Hickman J. A., Geneste O., Kroemer G. (2007) Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 26, 2527–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stack J. H., Emr S. D. (1994) Vps34p required for yeast vacuolar protein sorting is a multiple specificity kinase that exhibits both protein kinase and phosphatidylinositol-specific PI 3-kinase activities. J. Biol. Chem. 269, 31552–31562 [PubMed] [Google Scholar]
- 8. Volinia S., Dhand R., Vanhaesebroeck B., MacDougall L. K., Stein R., Zvelebil M. J., Domin J., Panaretou C., Waterfield M. D. (1995) A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 14, 3339–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Obara K., Sekito T., Ohsumi Y. (2006) Assortment of phosphatidylinositol 3-kinase complexes: Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol. Biol. Cell 17, 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kihara A., Noda T., Ishihara N., Ohsumi Y. (2001) Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Funderburk S. F., Wang Q. J., Yue Z. (2010) The Beclin 1-VPS34 complex: at the crossroads of autophagy and beyond. Trends Cell Biol. 20, 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. He C., Levine B. (2010) The Beclin 1 interactome. Curr. Opin. Cell Biol. 22, 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kang R., Zeh H. J., Lotze M. T., Tang D. (2011) The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 18, 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jaber N., Dou Z., Chen J. S., Catanzaro J., Jiang Y. P., Ballou L. M., Selinger E., Ouyang X., Lin R. Z., Zhang J., Zong W. X. (2012) Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc. Natl. Acad. Sci. U.S.A. 109, 2003–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhong Y., Wang Q. J., Li X., Yan Y., Backer J. M., Chait B. T., Heintz N., Yue Z. (2009) Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11, 468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T., Akira S., Noda T., Yoshimori T. (2009) Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11, 385–396 [DOI] [PubMed] [Google Scholar]
- 17. Itakura E., Kishi C., Inoue K., Mizushima N. (2008) Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 19, 5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun Q., Fan W., Chen K., Ding X., Chen S., Zhong Q. (2008) Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. U.S.A. 105, 19211–19216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dou Z., Chattopadhyay M., Pan J. A., Guerriero J. L., Jiang Y. P., Ballou L. M., Yue Z., Lin R. Z., Zong W. X. (2010) The class IA phosphatidylinositol 3-kinase p110-β subunit is a positive regulator of autophagy. J. Cell Biol. 191, 827–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fimia G. M., Stoykova A., Romagnoli A., Giunta L., Di Bartolomeo S., Nardacci R., Corazzari M., Fuoco C., Ucar A., Schwartz P., Gruss P., Piacentini M., Chowdhury K., Cecconi F. (2007) Ambra1 regulates autophagy and development of the nervous system. Nature 447, 1121–1125 [DOI] [PubMed] [Google Scholar]
- 21. Ropolo A., Grasso D., Pardo R., Sacchetti M. L., Archange C., Lo Re A., Seux M., Nowak J., Gonzalez C. D., Iovanna J. L., Vaccaro M. I. (2007) The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J. Biol. Chem. 282, 37124–37133 [DOI] [PubMed] [Google Scholar]
- 22. Molejon M. I., Ropolo A., Re A. L., Boggio V., Vaccaro M. I. (2013) The VMP1-Beclin 1 interaction regulates autophagy induction. Sci. Rep. 3, 1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang D., Kang R., Livesey K. M., Cheh C. W., Farkas A., Loughran P., Hoppe G., Bianchi M. E., Tracey K. J., Zeh H. J., 3rd, Lotze M. T. (2010) Endogenous HMGB1 regulates autophagy. J. Cell Biol. 190, 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., Levine B. (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939 [DOI] [PubMed] [Google Scholar]
- 25. Matsunaga K., Morita E., Saitoh T., Akira S., Ktistakis N. T., Izumi T., Noda T., Yoshimori T. (2010) Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J. Cell Biol. 190, 511–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Itakura E., Mizushima N. (2010) Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6, 764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liang C., Lee J. S., Inn K. S., Gack M. U., Li Q., Roberts E. A., Vergne I., Deretic V., Feng P., Akazawa C., Jung J. U. (2008) Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10, 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liang C., Feng P., Ku B., Dotan I., Canaani D., Oh B. H., Jung J. U. (2006) Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 8, 688–699 [DOI] [PubMed] [Google Scholar]
- 29. Aita V. M., Liang X. H., Murty V. V., Pincus D. L., Yu W., Cayanis E., Kalachikov S., Gilliam T. C., Levine B. (1999) Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 59, 59–65 [DOI] [PubMed] [Google Scholar]
- 30. Yue Z., Jin S., Yang C., Levine A. J., Heintz N. (2003) Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. U.S.A. 100, 15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E. L., Mizushima N., Ohsumi Y., Cattoretti G., Levine B. (2003) Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112, 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Takahashi Y., Coppola D., Matsushita N., Cualing H. D., Sun M., Sato Y., Liang C., Jung J. U., Cheng J. Q., Mulé J. J., Pledger W. J., Wang H. G. (2007) Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 9, 1142–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qu X., Zou Z., Sun Q., Luby-Phelps K., Cheng P., Hogan R. N., Gilpin C., Levine B. (2007) Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 128, 931–946 [DOI] [PubMed] [Google Scholar]
- 34. Meléndez A., Tallóczy Z., Seaman M., Eskelinen E. L., Hall D. H., Levine B. (2003) Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391 [DOI] [PubMed] [Google Scholar]
- 35. Shibata M., Lu T., Furuya T., Degterev A., Mizushima N., Yoshimori T., MacDonald M., Yankner B., Yuan J. (2006) Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J. Biol. Chem. 281, 14474–14485 [DOI] [PubMed] [Google Scholar]
- 36. Yue Z., Horton A., Bravin M., DeJager P. L., Selimi F., Heintz N. (2002) A novel protein complex linking the Δ 2 glutamate receptor and autophagy: implications for neurodegeneration in lurcher mice. Neuron 35, 921–933 [DOI] [PubMed] [Google Scholar]
- 37. Diskin T., Tal-Or P., Erlich S., Mizrachy L., Alexandrovich A., Shohami E., Pinkas-Kramarski R. (2005) Closed head injury induces upregulation of Beclin 1 at the cortical site of injury. J. Neurotrauma 22, 750–762 [DOI] [PubMed] [Google Scholar]
- 38. Yasumo H., Masuda N., Furusawa T., Tsukamoto T., Sadano H., Osumi T. (2000) Nuclear receptor binding factor-2 (NRBF-2): a possible gene activator protein interacting with nuclear hormone receptors. Biochim. Biophys. Acta 1490, 189–197 [DOI] [PubMed] [Google Scholar]
- 39. Flores A. M., Li L., Aneskievich B. J. (2004) Isolation and functional analysis of a keratinocyte-derived, ligand-regulated nuclear receptor comodulator. J. Invest. Dermatol. 123, 1092–1101 [DOI] [PubMed] [Google Scholar]
- 40. Kim J., Kim Y. C., Fang C., Russell R. C., Kim J. H., Fan W., Liu R., Zhong Q., Guan K. L. (2013) Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gao W., Shen Z., Shang L., Wang X. (2011) Upregulation of human autophagy-initiation kinase ULK1 by tumor suppressor p53 contributes to DNA-damage-induced cell death. Cell Death Differ. 18, 1598–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Behrends C., Sowa M. E., Gygi S. P., Harper J. W. (2010) Network organization of the human autophagy system. Nature 466, 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klionsky D. J., Elazar Z., Seglen P. O., Rubinsztein D. C. (2008) Does bafilomycin A(1) block the fusion of autophagosomes with lysosomes? Autophagy 4, 849–850 [DOI] [PubMed] [Google Scholar]
- 45. Menzies F. M., Moreau K., Puri C., Renna M., Rubinsztein D. C. (2012) Measurement of autophagic activity in mammalian cells. Curr. Protoc. Cell Biol. Chapter 15:Unit 15.16 [DOI] [PubMed] [Google Scholar]
- 46. Mizushima N., Yoshimori T., Levine B. (2010) Methods in mammalian autophagy research. Cell 140, 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., Amano A., Yoshimori T. (2013) Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393 [DOI] [PubMed] [Google Scholar]
- 48. Axe E. L., Walker S. A., Manifava M., Chandra P., Roderick H. L., Habermann A., Griffiths G., Ktistakis N. T. (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Koyama-Honda I., Itakura E., Fujiwara T. K., Mizushima N. (2013) Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy 9, 1491–1499 [DOI] [PubMed] [Google Scholar]
- 50. Karanasios E., Stapleton E., Manifava M., Kaizuka T., Mizushima N., Walker S. A., Ktistakis N. T. (2013) Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J. Cell Sci. 126, 5224–5238 [DOI] [PubMed] [Google Scholar]
- 51. Chan E. Y., Longatti A., McKnight N. C., Tooze S. A. (2009) Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell Biol. 29, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ganley I. G., Lam du H., Wang J., Ding X., Chen S., Jiang X. (2009) ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J. L., Mizushima N. (2008) FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stenmark H., Aasland R., Toh B. H., D'Arrigo A. (1996) Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J. Biol. Chem. 271, 24048–24054 [DOI] [PubMed] [Google Scholar]
- 55. Gillooly D. J., Morrow I. C., Lindsay M., Gould R., Bryant N. J., Gaullier J. M., Parton R. G., Stenmark H. (2000) Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19, 4577–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li X., He L., Che K. H., Funderburk S. F., Pan L., Pan N., Zhang M., Yue Z., Zhao Y. (2012) Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat. Commun. 3, 662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Backer J. M. (2008) The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem. J. 410, 1–17 [DOI] [PubMed] [Google Scholar]
- 58. Byfield M. P., Murray J. T., Backer J. M. (2005) hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J. Biol. Chem. 280, 33076–33082 [DOI] [PubMed] [Google Scholar]
- 59. Nobukuni T., Joaquin M., Roccio M., Dann S. G., Kim S. Y., Gulati P., Byfield M. P., Backer J. M., Natt F., Bos J. L., Zwartkruis F. J., Thomas G. (2005) Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. U.S.A. 102, 14238–14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang R. C., Wei Y., An Z., Zou Z., Xiao G., Bhagat G., White M., Reichelt J., Levine B. (2012) Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338, 956–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zalckvar E., Berissi H., Eisenstein M., Kimchi A. (2009) Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy 5, 720–722 [DOI] [PubMed] [Google Scholar]
- 62. Zalckvar E., Berissi H., Mizrachy L., Idelchuk Y., Koren I., Eisenstein M., Sabanay H., Pinkas-Kramarski R., Kimchi A. (2009) DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 10, 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cao Y., Wang Y., Abi Saab W. F., Yang F., Pessin J. E., Backer J. M. (2014) NRBF2 regulates macroautophagy as a component of Vps34 complex I. Biochem. J. 461, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lu J., He L., Behrends C., Araki M., Araki K., Wang Q. J., Catanzaro J. M., Friedman S. L., Zong W. X., Fiel M. I., Li M., Yue Z. (2014) NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L-linked phosphatidylinositol-3 kinase III activity. Nat. Commun. 5, 3920–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Araki Y., Ku W. C., Akioka M., May A. I., Hayashi Y., Arisaka F., Ishihama Y., Ohsumi Y. (2013) Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J. Cell Biol. 203, 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]