Background: The nematode Caenorhabditis elegans consumes bacteria as its sole food source.
Results: The bacterium Vibrio cholerae produces a quorum-sensing signal molecule called CAI-1, which C. elegans detects through the AWCON chemosensory neuron.
Conclusion: C. elegans uses bacterial-produced molecules as cues, and these molecules are also physiologically significant to bacteria.
Significance: The V. cholerae molecule CAI-1 enables cross-kingdom chemical interaction.
Keywords: Bacterial Signal Transduction, Caenorhabditis elegans (C. elegans), Chemotaxis, Gene Regulation, Quorum Sensing, V. cholerae
Abstract
In a process known as quorum sensing, bacteria use chemicals called autoinducers for cell-cell communication. Population-wide detection of autoinducers enables bacteria to orchestrate collective behaviors. In the animal kingdom detection of chemicals is vital for success in locating food, finding hosts, and avoiding predators. This behavior, termed chemotaxis, is especially well studied in the nematode Caenorhabditis elegans. Here we demonstrate that the Vibrio cholerae autoinducer (S)-3-hydroxytridecan-4-one, termed CAI-1, influences chemotaxis in C. elegans. C. elegans prefers V. cholerae that produces CAI-1 over a V. cholerae mutant defective for CAI-1 production. The position of the CAI-1 ketone moiety is the key feature driving CAI-1-directed nematode behavior. CAI-1 is detected by the C. elegans amphid sensory neuron AWCON. Laser ablation of the AWCON cell, but not other amphid sensory neurons, abolished chemoattraction to CAI-1. These analyses define the structural features of a bacterial-produced signal and the nematode chemosensory neuron that permit cross-kingdom interaction.
Introduction
Bacterial group behaviors are governed by quorum sensing (QS),2 in which bacteria produce, release, and detect extracellular signal molecules called autoinducers (AIs). Vibrios, which are the model organisms for QS analyses, produce multiple AIs, some of which enable intraspecies communication, some are for intragenera communication, and others promote inter-species communication (1–7). Important for the present work is that Vibrio cholerae, the pathogen that causes the endemic diarrheal disease cholera, produces two AIs: an intragenera AI (S)-3-hydroxytridecan-4-one, called CAI-1 (7–10), and the interspecies AI called AI-2 ((2S,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran borate) (3, 4, 11). The information contained in the AIs is funneled into a phosphorelay signaling cascade that controls virulence, biofilm formation, and other traits (3, 12, 13). CAI-1 is the dominant signal in V. cholerae (7–10).
Chemicals produced by bacteria, the Caenorhabditis elegans food source, stimulate egg-laying, chemotaxis, feeding, and defecation (14–19). A few of the molecules the nematode detects have been defined. For example, C. elegans detects Pseudomonas aeruginosa acyl homoserine lactone AIs (20), the Serratia marcescens cyclic lipodepsipentapeptide, serrawettin W2 (21), and 17 volatile compounds produced by the nematode pathogen Bacillus nematocida (22). However, little is known about the neurons that integrate and process the information encoded in bacterial-produced cues. In the present study we show that C. elegans detects the CAI-1 signal, the same chemical that is used by bacteria to control QS, via the amphid sensory neuron AWCON. We identify specific CAI-1 structural motifs that are required for C. elegans to detect CAI-1.
EXPERIMENTAL PROCEDURES
Chemotaxis Assays
Nematodes were grown at 20 °C on Escherichia coli strain HB101 under well-fed and uncrowded conditions (23). Chemoattraction assays were performed on square plates containing 10 ml of 1.6% agar, 5 mm potassium phosphate, 1 mm calcium chloride, and 1 mm magnesium sulfate (24). Plates were divided into six equal sectors labeled A-F. For population assays, 1 μl each of stimulus and 1 μl of 1 m sodium azide were added in two spots in sector A, and 1 μl each of control diluent (DMSO, LB, or ethanol) and 1 μl of 1 m sodium azide were added in two spots in sector F. Adult animals were washed twice with S-Basal buffer (23) and once with chemotaxis buffer, placed in the center of the assay plate (between sectors C and D) and counted after 1 h. 100–200 worms were assayed per plate, with the exception of the laser ablation studies, in which 15–30 worms were assayed per plate. Each assay was performed in triplicate, with at least three independent experiments. The chemotaxis index was calculated using the formula,
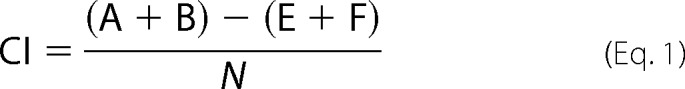 |
where A, B, E, and F are the number of animals in plate sectors A, B, E, and F, respectively, N is the total number of animals in all six sectors of the plate. Positive control odorant dilutions were 1:100 (isoamyl alcohol) and 1:200 (benzaldehyde). All chemotactic responses were verified in multiple experiments. All assays were quantified by two independent investigators. Data for chemoattraction assays were compared using unequal two-tailed t tests.
Laser Ablations
Laser ablations were performed on anesthetized early L4 animals (N2 strain) expressing str-2::GFP using a 2-photon microscope. Only one cell in each animal expressed cytoplasmic GFP. Using GFP as a marker for AWCON neurons enabled us to target the str-2::GFP-expressing cell for ablation. Nematodes were anesthetized using 1.5 mm sodium azide. Worms were mounted on an agar mold with ridges to align animals for serial ablations. Ridges were achieved using a vinyl record as a caste. Worms were imaged on a custom-built two-photon scanning microscope (25) built around an up-right Olympus BX51. Fluorescence photons were collected through a LUMPlanFl/IR 40×, 0.8NA water immersion objective (Olympus) and detected with high quantum efficiency GaAsP photomultipliers (Hamamatsu). The microscope used ScanImage software (26). The laser was administered for 20 pulses at 0.5-ms intervals and 40% power. Images were taken with an excitation wavelength of 920 nm. Successful ablations were marked by diffusion of GFP out of the cell. Animals were allowed to recover on standard NGM plates with E. coli HB101 overnight before assaying for chemotaxis. All worms were verified for target cell gfp expression before behavioral assays.
Bacterial Strains
All V. cholerae strains are derivatives of wild-type C6706str (27). V. cholerae and E. coli mutant and recombinant strains were generated in the Bassler group (Table 1).
TABLE 1.
Bacterial strains and genotypes
C. elegans Strains
Strains were acquired from the Caenorhabditis Genetics Center (CGC) (Table 2) and include CGC strain numbers and genotypes. The wild-type strain, Bristol N2, was acquired from Professor Jean Schwarzbauer. To obtain the tax-2; tax-4 double mutant, tax-2(ks10) and tax-4(p678) animals were mated. The presence of each mutation was confirmed by PCR in the F1 and F2 generations.
TABLE 2.
C. elegans strains and genotypes
| Strain | Relevant mutation | Genotype |
|---|---|---|
| N2 | Wild type | |
| tax-2; tax-4 | tax-2; tax-4 | tax-2(ks10); tax-4(p678) |
| FG125 | ocr-2, osm-9 | ocr-2(ak47) osm-9(ky10) IV |
| FK311 | ceh-36 | ceh-36(ks86) X |
| PR674 | che-1 | che-1(p674) I |
| CX6161 | nsy-5 | inx-19(ky634) I |
| CX4998 | nsy-1 | kyIs140 I; nsy-1(ky397) II |
| CX3695 | str-2::GFP | kyIs140[str-2::GFP + lin-15(+)] I |
| OH3679 | che-1::GFP | otIs114[lim-6p::GFP + rol-6(su1006)] |
| OH3192 | gcy-5::GFP | ntIs1 adEx1262[lin-15(+); gcy-5::GFP] |
| OH3351 | gcy-6::GFP | otIs162[gcy-6::GFP + lin-15(+)] |
Chemical Synthesis
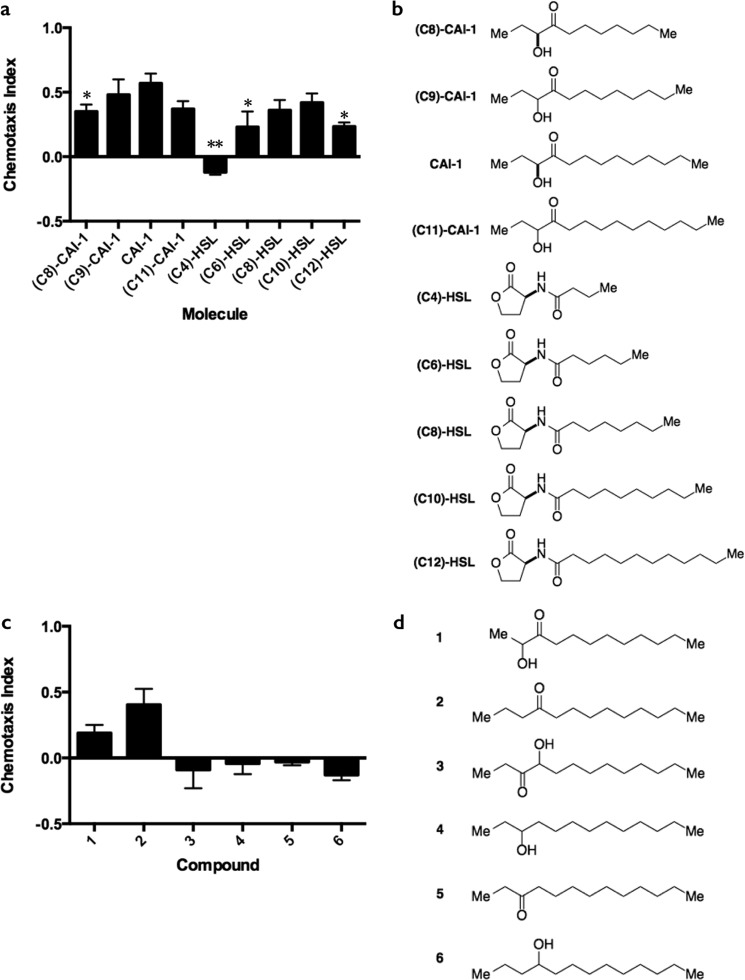
All compounds were synthesized using previously reported procedures and were consistent with the published characterization data for these molecules. (C8)-CAI-1, (C9)-CAI-1, CAI-1, (C11)-CAI-1, and 1–6 were prepared according to procedures described in Bolitho et al. (28). (C4)-HSL, (C6)-HSL, (C8)-HSL, and (C10)-HSL were prepared using established procedures from l-homoserine lactone hydrochloride and the appropriate carboxylic acid (29). (C8)-CAI-1, CAI-1, (C4)-HSL, (C6)-HSL, (C8)-HSL, (C10)-HSL, and (C12)-HSL were analyzed as enantiopure (S) isomers. (C9)-CAI-1 and (C11)-CAI-1 were tested as racemic mixtures. The chemotaxis response to Compound 2 (Fig. 3, c and d) demonstrates that the hydroxyl group does not influence chemotaxis; therefore, the racemic compounds did not require further purification for individual analysis.
FIGURE 3.
The 4-ketone moiety is the key structural feature for C. elegans detection of CAI-1. a, chemotaxis indices for carbon chain length variants of CAI-1 and homoserine lactone AIs at 100 μm. Values are the mean ± S.D. of triplicate assays from three independent experiments. Unequal two-tailed t tests were performed comparing data for each CAI-1 variants and each homoserine lactone molecule to the CAI-1 bar. *, p < 0.05; **, p < 0.01. b, structures corresponding to a. c, chemotaxis indices for compounds with varied positioning of the hydroxyl and/or ketone group of CAI-1. d, structures corresponding to compounds in c. In panels b and d, the stereochemistry of each molecule is depicted. See “Experimental Procedures” for details.
RESULTS
C. elegans Chemotact toward V. cholerae
C. elegans chemotaxis occurs to a variety of organic compounds as well as to different species of bacteria (30). The notion is that chemotaxis toward certain bacterial-derived compounds enables C. elegans to locate food sources, whereas chemotaxis away from noxious bacteria allows C. elegans to avoid danger. Two pathogens, P. aeruginosa and B. nematocida, are more attractive to C. elegans than are harmless laboratory E. coli strains (22, 31). In the case of P. aeruginosa, C. elegans is initially attracted to the bacteria but through associative learning subsequently avoids them (31). It is not understood how C. elegans distinguishes attractive and aversive bacterial-produced cues and translates detection of these chemicals into meaningful behavioral responses. We chose to investigate whether QS AIs are involved in C. elegans chemotactic behavior using the nematode pathogen V. cholerae.
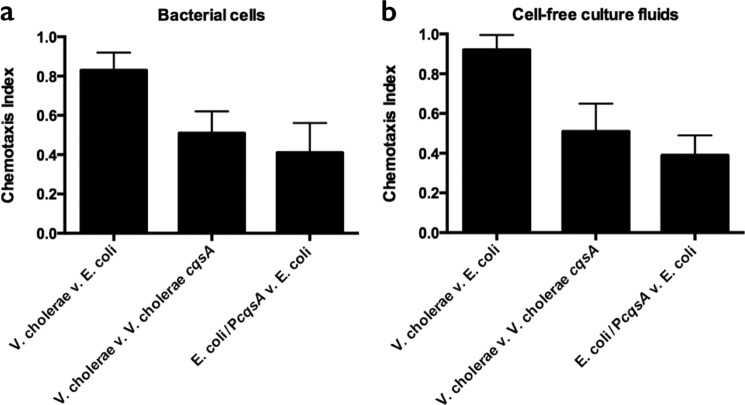
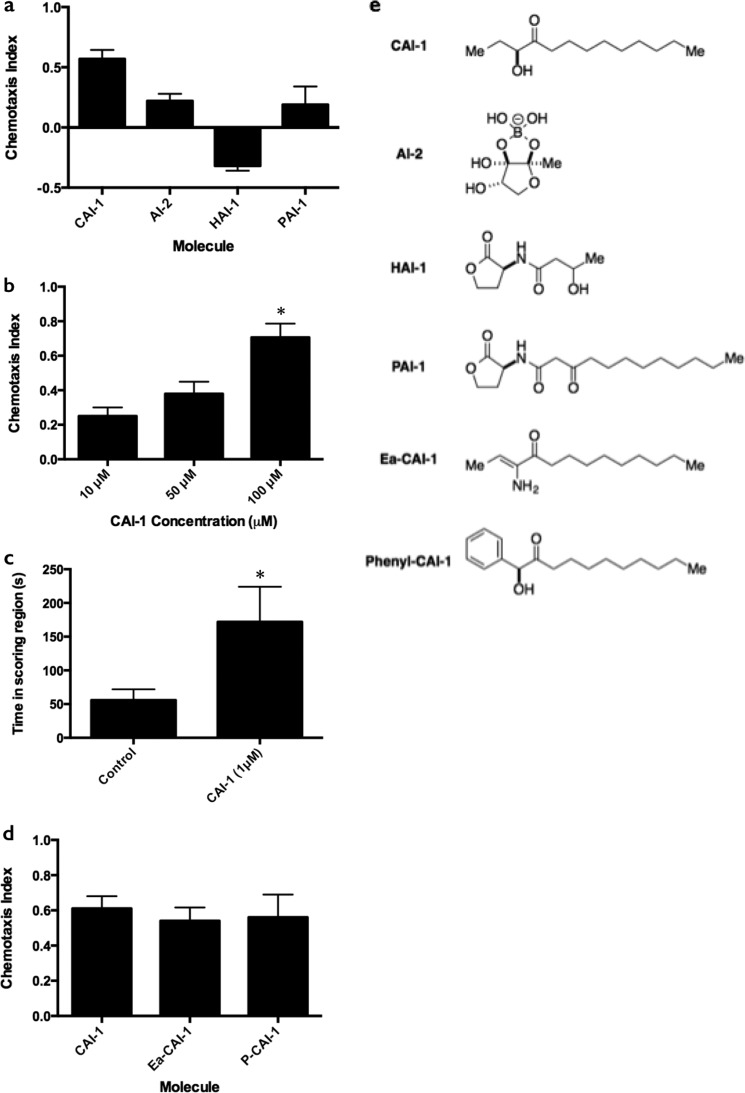
We first tested whether C. elegans can distinguish between its standard laboratory food source, E. coli HB101, and V. cholerae using volatile chemotaxis assays (24). C. elegans displayed a preference for V. cholerae over E. coli (C.I. = 0.83, leftmost bar in Fig. 1a) that is comparable to its preference for P. aeruginosa over E. coli (31). Higher level chemoattraction also occurred to V. cholerae cell-free culture fluids compared with those prepared from E. coli HB101 (C.I. = 0.87, leftmost bar in Fig. 1b). We sought to identify the compounds that mediate this potent attraction. Obvious candidates are bacterial AIs. We examined the C. elegans chemotactic response to purified, synthetic AIs with varying chemical structures, including the intragenera Vibrio signal CAI-1, the interspecies AI, AI-2, the intraspecies AI from the related bacterium Vibrio harveyi, N-(3-hydroxybutanoyl)-l-homoserine lactone (HAI-1), and the P. aeruginosa intraspecies AI, N-(3-oxododecanoyl)-l-homoserine lactone (PAI-1) (Fig. 2a, and structures are shown in Fig. 2e). Among these molecules, C. elegans was most potently attracted to CAI-1, had a far weaker response to AI-2, and the response to PAI-1 was insignificant (C.I. = 0.19, S.D. = 0.15). The nematodes were weakly repulsed by HAI-1. With respect to CAI-1, we observed a striking dose-dependent response to the V. cholerae CAI-1 autoinducer: C.I. = 0.39 at 10 μm CAI-1 and C.I. = 0.56 at 100 μm CAI-1 (Fig. 2b). In V. cholerae bacterial cultures, however, 1 μm CAI-1 saturates the QS response (7), so it was unclear whether chemoattraction to these concentrations of CAI-1 represented a biologically relevant behavior. To address this we adapted the chemotaxis protocol described in Choe et al. (32) (termed the two-spot assay) such that CAI-1 is applied to a lawn of E. coli HB101 and the amount of time each worm spends in the CAI-1 region is compared with the control region. This assay showed that 1 μm CAI-1 is indeed sufficient for C. elegans attraction, as the worms spent 3 times longer in the CAI-1 region than in the control region (Fig. 2c).
FIGURE 1.
C. elegans chemotact toward CAI-1. Chemotaxis indices are shown for the C. elegans response to A600 = 1.0 bacterial cultures (a) and cell-free culture fluids (b). The chemotaxis index was calculated as previously defined (19, 24). Values are the mean ± S.D. of triplicate assays from three independent experiments.
FIGURE 2.
C. elegans detects CAI-1 and CAI-1 variants. a, chemotaxis indices for the V. cholerae AI, CAI-1, the interspecies AI, AI-2, the V. harveyi AI, HAI-1, and the P. aeruginosa AI, PAI-1, all at 100 μm. b, chemotaxis indices at 10, 50, and 100 μm CAI-1. c, 1 μm CAI-1 was applied to one scoring region in an adapted two spot assay (32). The time period (seconds) the nematodes spent in each scoring region was scored from 10 min of video recorded at 1 frame/s. d, chemotaxis indices for enamino-CAI-1 (Ea-CAI-1) and phenyl-CAI-1 (P-CAI-1) are compared with CAI-1. Values are the mean ± S.D. of triplicate assays from three independent experiments. For b–d, unequal two-tailed t tests were performed comparing data to the leftmost bar of each graph. *, p < 0.05. e, structures of molecules.
Although C. elegans chemotacts toward synthetic, purified CAI-1, it was possible that CAI-1 was not a major contributor to C. elegans attraction to V. cholerae bacterial cultures. To examine whether CAI-1 is the attractant, we assayed chemotaxis to the wild-type V. cholerae strain that produces CAI-1 and compared that to a mutant strain that is incapable of CAI-1 production as well as to the cell-free fluids from the same two strains (V. cholerae cqsA, Fig. 1, a and b, middle bars). In addition, we assayed cultures of and cell-free fluids from E. coli carrying the cqsA gene encoding the CAI-1 synthase (E. coli/PcqsA) and compared those preparations to E. coli carrying the empty vector (Fig. 1, a and b, rightmost bars). We showed previously that introduction of cqsA into recombinant E. coli is sufficient for high level production of CAI-1 (7, 33). In all cases C. elegans preferred bacteria that produce CAI-1 to those that do not, and C. elegans preferred fluids collected from bacterial cultures containing CAI-1 compared with those lacking CAI-1. We note, however, that the absence of CAI-1 does not completely eliminate C. elegans attraction to V. cholerae bacterial cultures or cell-free fluids. Therefore, V. cholerae must produce chemoattractants in addition to CAI-1.
C. elegans Recognizes Key Structural Features of CAI-1
Previous structure-activity relationship analyses demonstrate that the V. cholerae CAI-1 receptor, CqsS, can be activated by a set of CAI-1 variants (9, 33). We wondered whether C. elegans also recognizes structural variants of CAI-1. To explore this, we tested enamino-CAI-1, a naturally-occurring CAI-1 variant that potently activates CqsS, and we tested phenyl-CAI-1, which is a CqsS antagonist (9, 33) (see the structures in Fig. 2e). C. elegans displayed statistically similar recognition of each molecule (Fig. 2d), suggesting that the head group of the molecule is not playing a role in specifying the structural requirements for chemoattraction.
To further examine the specificity of CAI-1 detection by C. elegans, we tested molecules with variations in the carbon tail length. For example, V. cholerae CAI-1 has a 10-carbon tail, whereas V. harveyi produces a CAI-1-type molecule with an 8-carbon side chain, (C8)-CAI-1. In addition to assaying these natural molecules, we synthesized and tested (C9)-CAI-1 and (C11)-CAI-1. Analyses of this series of CAI-1 molecules demonstrate that 9- and 10-carbon chain CAI-1 variants produce the most robust C. elegans chemotactic response (Fig. 3a, molecules shown in Fig. 3b). Specifically, we found the order of chemotactic preference to be: CAI-1 = (C9)-CAI-1 > (C8)-CAI-1 = (C11)-CAI-1. Interestingly, this order of preference for tail length exactly parallels the CqsS receptor preference (9, 28, 33). To assess the generality of this response to other bacterial signaling molecules containing fatty acid tails, we examined the responses of C. elegans to a series of acylated homoserine lactone AIs. We observed a similar trend within this series of molecules (Fig. 3, a and b). Specifically, a homoserine lactone with a C10 tail induced high-level chemotaxis, whereas molecules with shorter carbon chains resulted in reduced attraction (i.e. (C6)-HSL) or repulsion (i.e. (C4)-HSL). A molecule harboring a longer chain, (C12)-HSL, also elicited weak chemoattraction (Fig. 3, a and b). Consistent with this result, Fig. 2a shows that C. elegans exhibits similarly low attraction to PAI-1, which is a homoserine lactone harboring a 12-carbon tail and a 4-ketone moiety (Fig. 2, a and e). Together, these data suggest that, similar to the CAI-1 series, a range of carbon chain lengths enable chemotaxis to homoserine lactone AIs with C10 being the optimal length.
In our effort to define the structural features of the CAI-1 molecule that are important for chemotaxis in C. elegans, we prepared a series of related compounds that vary in the position of the hydroxyl or ketone moieties and tested them in the chemotaxis assay. The compounds analyzed can be broadly categorized into molecules that maintain chemoattractant properties (Fig. 3, c and d, compounds 1 and 2) and neutral molecules (Fig. 3, c and d, compounds 3–6). Shifting the positions of the hydroxyl and ketone groups from C3 and C4 to C2 and C3, respectively, reduced the chemoattractive potency of CAI-1 (Fig. 3c, compound 1), whereas removal of the hydroxyl group from CAI-1 while maintaining the 4-ketone functionality had minimal effect on activity (Fig. 3c, compound 2). Compound 2 was previously assayed for agonist activity in V. cholerae QS and had no activity at concentrations up to 50 μm (28). Exchanging the ketone position with the hydroxyl group (compound 3), moving the ketone by one carbon atom to make a 3-ketone molecule (compound 2 versus compound 5), or removal of the ketone (compounds 4 and 6) eliminated chemoattraction (Fig. 3c). In summary, although moderate alterations in the length of the carbon chain and dramatic changes in the head group are tolerated, the presence of a 4-ketone functionality is necessary for robust CAI-1 chemoattractive activity.
C. elegans Detects CAI-1 as an Attractant through the AWCON Sensory Neuron
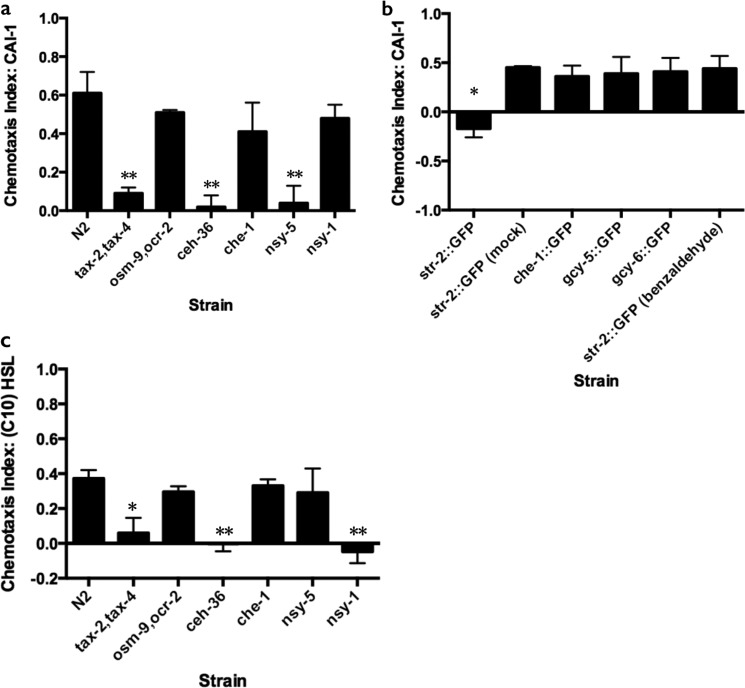
To determine how C. elegans detects CAI-1, we investigated the role of the amphid sensory neurons, which detect many chemical stimuli (30). Most chemosensory neurons in C. elegans use either cGMP-gated channels or TRPV channels for sensory transduction (30). We found that animals with mutations in the cGMP-gated channel encoded by tax-2 and tax-4 are defective in attraction to CAI-1, as they fail to respond to 100 μm synthetic CAI-1 (Fig. 4a). By contrast, animals mutant for the TRPV channels encoded by osm-9 and ocr-2 chemotact to CAI-1-like wild type (Fig. 4a). This result suggests that sensory neurons expressing tax-4 and tax-2 are required to detect CAI-1, whereas neurons expressing osm-9 and ocr-2 are dispensable. tax-2 and tax-4 function in eight classes of amphid chemosensory neurons, including AWB, AWC, and ASE neurons. To pinpoint the cell or cells that mediate the C. elegans response to V. cholerae CAI-1, we next assayed ceh-36 mutant animals, which lack functional AWC and ASEL neurons (34, 35). ceh-36 animals do not chemotact to CAI-1 (Fig. 4a). This result narrows candidate neurons to ASEL and the two AWC cells. che-1 mutants are defective in the development of both ASEL and ASER sensory neurons (34, 36, 37). These mutant animals exhibited wild-type attraction to CAI-1, eliminating the ASE neuron as having a role in the CAI-1 response (Fig. 4a). Finally, to differentiate between the two AWC neurons, we examined nsy-5 mutants. These mutants fail to develop the AWCON neuron, as defined by the expression of str-2::GFP (38), resulting in the formation of two AWCOFF neurons (39, 40). nsy-5 mutant animals do not chemotact toward CAI-1 (Fig. 4a). Conversely, nsy-1 mutants, which fail to develop the AWCOFF neuron, maintain the ability to detect CAI-1 (Fig. 4a). Together, these results suggest that the AWCON neuron is essential for attraction to CAI-1, but the AWCOFF neuron is not.
FIGURE 4.
AWCON mediates detection of CAI-1 and AWCOFF mediates detection of (C10)-HSL. a, chemotaxis indices for mutant C. elegans strains to CAI-1 (100 μm). b, chemotaxis indices for animals with individually ablated or mock-ablated amphid sensory neurons. Target neurons were identified by cell-specific gfp expression. Mock-ablated animals were prepared identically to ablated animals but were not exposed to the laser. c, chemotaxis indices for mutant C. elegans strains to (C10)-HSL (100 μm). Values are the mean ± S.D. of triplicate assays from at least three independent experiments. Unequal two-tailed t tests were performed comparing data to the N2 (wild type) response in (a and c) and compared with the str-2::GFP mock-ablated animals in b. *, p < 0.05; **, p < 0.01
To verify the role of AWCON in CAI-1 detection, we tested transgenic animals in which AWCON neurons expressing GFP (str-2::GFP) were laser-ablated. AWCON-ablated animals were not attracted to CAI-1 (Fig. 4b), whereas animals that were mock ablated or defective in development of neurons other than AWCON, i.e. either defective for the ASE neuron (che-1::GFP), ASER (gcy-5::GFP), or ASEL (gcy-6::GFP), demonstrated wild-type attraction to CAI-1 (Fig. 4b). In addition, AWCON-ablated animals were able to chemotact toward benzaldehyde (Fig. 4b and Ref. 41), demonstrating that ablation of AWCON did not generally impair detection of stimuli or chemotaxis behavior. Thus, AWCON is the neuron that mediates C. elegans attraction to V. cholerae CAI-1. In an analogous series of experiments we show that, surprisingly, animals lacking the AWCON cell (nsy-5 mutants) remained capable of chemotaxis toward (C10)-HSL, whereas animals lacking the AWCOFF cell (nsy-1 mutants) were impaired (Fig. 4c). Therefore, AWCOFF, rather than AWCON, mediates chemoattraction to (C10)-HSL. There are precedents for opposite behaviors for the two AWC neurons. Indeed, our results are consistent with prior studies in which the AWCON neuron plays the primary role in sensation of particular chemicals, such as butanone, whereas the AWCOFF neuron is critical for detection of a different set of stimuli (41). By contrast, in the case of high salt chemotaxis, both the AWCON and AWCOFF neurons are essential (42).
DISCUSSION
Nematodes have behavioral as well as developmental strategies for coping with environmental stresses. Success depends on correctly interpreting the information encoded in the chemical environments in which they reside. One crucial environmental factor for nematode survival is the availability of non-pathogenic bacteria as food. In this study we have demonstrated that C. elegans can intercept bacterial-produced signals; specifically, C. elegans detects the QS AI, CAI-1. We additionally observed chemotaxis to another class of QS signaling molecules, the homoserine lactone AIs. The maximal response to both classes of bacterial signals was to molecules containing 10-carbon fatty acid tails; however, other chain length tails are tolerated. Assessment of the other structural features of CAI-1 reveals that the 4-ketone moiety is an important feature for recognition. CAI-1 appears to mediate only a portion of the C. elegans response to V. cholerae bacterial cells. C. elegans remains attracted to a V. cholerae mutant strain that is incapable of CAI-1 production as well as to cell-free culture fluids prepared from this mutant strain. Thus, other factors must be released by V. cholerae that contribute to attraction.
C. elegans distinguishes small molecules in its environment based on subtle differences in chemical structures. This appears to be the case for both self-produced and bacterial-produced molecules. Nematode-produced ascarosides with fatty acid chains are potent cues that induce an alternative second larval stage, called dauer, which is characterized by morphological changes and metabolic arrest (43–46). The C9 ascaroside is more potent than the C7 ascaroside, and ascarosides with C20 or longer side chains do not induce dauer formation (47, 48). Although the outcome of detection of ascarosides differs from detection of AIs, CAI-1 represents a new bacterial signal that C. elegans can detect as well as distinguish structural variations. C. elegans possess similar capabilities for homoserine lactone AIs (20). Recently, Artyukhin et al. (49) suggested that molecules with short carbon chains act as dispersal signals for C. elegans, whereas molecules with longer carbon chains may signal that there is sufficient nutrition in the environment. This suggestion is consistent with our observation that bacterial-produced molecules containing short carbon chains were either neutral or repulsive in chemotaxis assays, whereas longer chain molecules were attractants (Fig. 3a). Small molecules control dauer formation, chemoavoidance, and chemoattraction. In each case the molecules are detected by distinct neural cells (30); nonetheless, each behavior appears to be differentially influenced by the length of the carbon chain on the small molecule (47–49). Possibly, sensory neuron subtypes distinguish molecules, at least in part, based on carbon chain length.
Our findings in the present study suggest that bacteria and C. elegans can detect similar chemical cues, and we demonstrate a mechanism underpinning how bacterial QS can influence the behavior of potential host animals. The intimate relationship between C. elegans and bacteria, as their food source, could require that C. elegans rely on the detection and interpretation of bacterial-produced cues. For example, while many of the nematode genes involved in dauer formation have been characterized, C. elegans uses an unknown bacterial cue or cues to exit the metabolically arrested dauer phase and continue through development (43). Similarly, in a recent characterization of L1 nematode metabolites, it was found that the C. elegans avoidance response to L1-produced small molecules is overridden by the addition of bacteria to the assay (49). In these cases of bacterial-regulated C. elegans development and behavior, the specific bacterial-produced stimuli remain mysterious. We have begun to examine the relationship between C. elegans behavior and specific bacterial exoproducts. We determined the key chemical moieties (4-ketone and a long carbon tail) along with the nematode sensory neuron (AWCON) required for cross-kingdom interaction between C. elegans and V. cholerae. These relationships provide a promising platform to study prokaryote-eukaryote interactions at the molecular level, and they may reveal how eukaryotes detect and interpret information about nutrition and the microbes that shape their life cycles.
Acknowledgments
We thank the Caenorhabditis Genetics Center and Professor Jean Schwarzbauer for providing C. elegans strains; Kevin T. O'Brien, Samantha Sustek, and Kevin C. Lyman for assistance in the synthesis of Compounds 1–6; Dr. Stephan Thiberge for assistance in the two-photon facility at Princeton University; Professor Cornelia Bargmann and her group for expertise and training in performing chemotaxis assays and analyses; the Bassler group for insightful comments and discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant 5R01GM065859 (to B. L. B.). This work was also supported by the Howard Hughes Medical Institute and National Science Foundation Grant MCB-0343821 (to B. L. B.)
- QS
- quorum sensing
- AI
- autoinducer
- C.I.
- chemotaxis index.
REFERENCES
- 1. Surette M. G., Bassler B. L. (1998) Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. U.S.A. 95, 7046–7050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Surette M. G., Miller M. B., Bassler B. L. (1999) Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. U.S.A. 96, 1639–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miller M. B., Skorupski K., Lenz D. H., Taylor R. K., Bassler B. L. (2002) Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110, 303–314 [DOI] [PubMed] [Google Scholar]
- 4. Chen X., Schauder S., Potier N., Van Dorsselaer A., Pelczer I., Bassler B. L., Hughson F. M. (2002) Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415, 545–549 [DOI] [PubMed] [Google Scholar]
- 5. Mok K. C., Wingreen N. S., Bassler B. L. (2003) Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 22, 870–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henke J. M., Bassler B. L. (2004) Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 186, 6902–6914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Higgins D. A., Pomianek M. E., Kraml C. M., Taylor R. K., Semmelhack M. F., Bassler B. L. (2007) The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450, 883–886 [DOI] [PubMed] [Google Scholar]
- 8. Kelly R. C., Bolitho M. E., Higgins D. A., Lu W., Ng W.-L., Jeffrey P. D., Rabinowitz J. D., Semmelhack M. F., Hughson F. M., Bassler B. L. (2009) The Vibrio cholerae quorum-sensing autoinducer CAI-1: analysis of the biosynthetic enzyme CqsA. Nat. Chem. Biol. 5, 891–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ng W.-L., Perez L. J., Wei Y., Kraml C., Semmelhack M. F., Bassler B. L. (2011) Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol. Microbiol. 79, 1407–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wei Y., Perez L. J., Ng W.-L., Semmelhack M. F., Bassler B. L. (2011) Mechanism of Vibrio cholerae autoinducer-1 biosynthesis. ACS Chem. Biol. 6, 356–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schauder S., Shokat K., Surette M. G., Bassler B. L. (2001) The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41, 463–476 [DOI] [PubMed] [Google Scholar]
- 12. Waters C. M., Lu W., Rabinowitz J. D., Bassler B. L. (2008) Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190, 2527–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hammer B. K., Bassler B. L. (2009) Distinct sensory pathways in Vibrio cholerae El Tor and classical biotypes modulate cyclic dimeric GMP levels to control biofilm formation. J. Bacteriol. 191, 169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ward S. (1973) Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc. Natl. Acad. Sci. U.S.A. 70, 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dusenbery D. B. (1974) Analysis of chemotaxis in the nematode Caenorhabditis elegans by countercurrent separation. J. Exp. Zool. 188, 41–47 [DOI] [PubMed] [Google Scholar]
- 16. Horvitz H. R., Chalfie M., Trent C., Sulston J. E., Evans P. D. (1982) Serotonin and octopamine in the nematode Caenorhabditis elegans. Science 216, 1012–1014 [DOI] [PubMed] [Google Scholar]
- 17. Avery L., Horvitz H. R. (1990) Effects of starvation and neuroactive drugs on feeding in Caenorhabditis elegans. J. Exp. Zool. 253, 263–270 [DOI] [PubMed] [Google Scholar]
- 18. Thomas J. H. (1990) Genetic analysis of defecation in Caenorhabditis elegans. Genetics 124, 855–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bargmann C. I., Hartwieg E., Horvitz H. R. (1993) Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74, 515–527 [DOI] [PubMed] [Google Scholar]
- 20. Beale E., Li G., Tan M.-W., Rumbaugh K. P. (2006) Caenorhabditis elegans senses bacterial autoinducers. Appl. Environ. Microbiol. 72, 5135–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pradel E., Zhang Y., Pujol N., Matsuyama T., Bargmann C. I., Ewbank J. J. (2007) Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 104, 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Niu Q., Huang X., Zhang L., Xu J., Yang D., Wei K., Niu X., An Z., Bennett J. W., Zou C., Yang J., Zhang K.-Q. (2010) A Trojan horse mechanism of bacterial pathogenesis against nematodes. Proc. Natl. Acad. Sci. U.S.A. 107, 16631–16636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Troemel E. R., Kimmel B. E., Bargmann C. I. (1997) Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell 91, 161–169 [DOI] [PubMed] [Google Scholar]
- 25. Denk W., Strickler J. H., Webb W. W. (1990) Two-photon laser scanning fluorescence microscopy. Science 248, 73–76 [DOI] [PubMed] [Google Scholar]
- 26. Pologruto T. A., Sabatini B. L., Svoboda K. (2003) ScanImage: flexible software for operating laser scanning microscopes. Biomed. Eng. Online 2, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thelin K. H., Taylor R. K. (1996) Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 64, 2853–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bolitho M. E., Perez L. J., Koch M. J., Ng W.-L., Bassler B. L., Semmelhack M. F. (2011) Small molecule probes of the receptor binding site in the Vibrio cholerae CAI-1 quorum sensing circuit. Bioorg. Med. Chem. 19, 6906–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Swem L. R., Swem D. L., O'Loughlin C. T., Gatmaitan R., Zhao B., Ulrich S. M., Bassler B. L. (2009) A quorum-sensing antagonist targets both membrane-bound and cytoplasmic receptors and controls bacterial pathogenicity. Mol. Cell 35, 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bargmann C. I. (2006) Chemosensation in C. elegans. WormBook 1–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y., Lu H., Bargmann C. I. (2005) Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438, 179–184 [DOI] [PubMed] [Google Scholar]
- 32. Choe A., Chuman T., von Reuss S. H., Dossey A. T., Yim J. J., Ajredini R., Kolawa A. A., Kaplan F., Alborn H. T., Teal P. E., Schroeder F. C., Sternberg P. W., Edison A. S. (2012) Sex-specific mating pheromones in the nematode Panagrellus redivivus. Proc. Natl. Acad. Sci. U.S.A. 109, 20949–20954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ng W.-L., Wei Y., Perez L. J., Cong J., Long T., Koch M., Semmelhack M. F., Wingreen N. S., Bassler B. L. (2010) Probing bacterial transmembrane histidine kinase receptor-ligand interactions with natural and synthetic molecules. Proc. Natl. Acad. Sci. U.S.A. 107, 5575–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chang S., Johnston R. J., Jr., Hobert O. (2003) A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 17, 2123–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lanjuin A., VanHoven M. K., Bargmann C. I., Thompson J. K., Sengupta P. (2003) Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Dev. Cell 5, 621–633 [DOI] [PubMed] [Google Scholar]
- 36. Lewis J. A., Hodgkin J. A. (1977) Specific neuroanatomical changes in chemosensory mutants of the nematode Caenorhabditis elegans. J. Comp. Neurol. 172, 489–510 [DOI] [PubMed] [Google Scholar]
- 37. Uchida O., Nakano H., Koga M., Ohshima Y. (2003) The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development 130, 1215–1224 [DOI] [PubMed] [Google Scholar]
- 38. Chuang C.-F., Vanhoven M. K., Fetter R. D., Verselis V. K., Bargmann C. I. (2007) An innexin-dependent cell network establishes left-right neuronal asymmetry in C. elegans. Cell 129, 787–799 [DOI] [PubMed] [Google Scholar]
- 39. Troemel E. R., Sagasti A., Bargmann C. I. (1999) Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99, 387–398 [DOI] [PubMed] [Google Scholar]
- 40. Sagasti A., Hisamoto N., Hyodo J., Tanaka-Hino M., Matsumoto K., Bargmann C. I. (2001) The CaMKII UNC-43 activates the MAPKKK NSY-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell 105, 221–232 [DOI] [PubMed] [Google Scholar]
- 41. Wes P. D., Bargmann C. I. (2001) C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature 410, 698–701 [DOI] [PubMed] [Google Scholar]
- 42. Leinwand S. G., Chalasani S. H. (2013) Neuropeptide signaling remodels chemosensory circuit composition in Caenorhabditis elegans. Nat. Neurosci. 16, 1461–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Golden J. W., Riddle D. L. (1984) The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev. Biol. 102, 368–378 [DOI] [PubMed] [Google Scholar]
- 44. Jeong P.-Y., Jung M., Yim Y.-H., Kim H., Park M., Hong E., Lee W., Kim Y. H., Kim K., Paik Y.-K. (2005) Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature 433, 541–545 [DOI] [PubMed] [Google Scholar]
- 45. Butcher R. A., Fujita M., Schroeder F. C., Clardy J. (2007) Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat. Chem. Biol. 3, 420–422 [DOI] [PubMed] [Google Scholar]
- 46. Srinivasan J., Kaplan F., Ajredini R., Zachariah C., Alborn H. T., Teal P. E., Malik R. U., Edison A. S., Sternberg P. W., Schroeder F. C. (2008) A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature 454, 1115–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Butcher R. A., Ragains J. R., Li W., Ruvkun G., Clardy J., Mak H. Y. (2009) Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proc. Natl. Acad. Sci. U.S.A. 106, 1875–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zagoriy V., Matyash V., Kurzchalia T. (2010) Long-chain O-ascarosyl-alkanediols are constitutive components of Caenorhabditis elegans but do not induce dauer larva formation. Chem. Biodivers. 7, 2016–2022 [DOI] [PubMed] [Google Scholar]
- 49. Artyukhin A. B., Yim J. J., Srinivasan J., Izrayelit Y., Bose N., von Reuss S. H., Jo Y., Jordan J. M., Baugh L. R., Cheong M., Sternberg P. W., Avery L., Schroeder F. C. (2013) Succinylated octopamine ascarosides and a new pathway of biogenic amine metabolism in C. elegans. J. Biol. Chem. 288, 18778–18783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hanahan D. (1985) DNA Cloning: A practical approach, pp. 109–135, IRL Press, Oxford, UK [Google Scholar]
- 51. Boyer H.W., Roulland-Dussoix D. (1969) A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41, 459–472 [DOI] [PubMed] [Google Scholar]