Background: The retroviral preintegration complex (PIC) exhibits full fidelity of integration in vitro.
Results: Application of a protein library to the microplate-based integration assay identified RFPL3 as an enhancer of HIV-1 PIC activity.
Conclusion: This screening platform permits efficient identification of cellular factors modulating PIC.
Significance: This approach provides an advantage in advancing our understanding of virus-host interactions in retrovirus integration.
Keywords: E3 Ubiquitin Ligase, HIV-1 Protease, Host-Pathogen Interaction, Recombinant Protein Expression, Retrovirus, Microtiter Plate-based Integration Assay, Preintegration Complex, Protein Screening, Wheat Germ Cell-free Protein Production System
Abstract
Integration, one of the hallmarks of retrovirus replication, is mediated by a nucleoprotein complex called the preintegration complex (PIC), in which viral DNA is associated with many protein components that are required for completion of the early phase of infection. A striking feature of the PIC is its powerful integration activity in vitro. The PICs from a freshly isolated cytoplasmic extract of infected cells are able to insert viral DNA into exogenously added target DNA in vitro. Therefore, a PIC-based in vitro assay is a reliable system for assessing protein factors influencing retroviral integration. In this study, we applied a microtiter plate-based in vitro assay to a screening study using a protein library that was produced by the wheat germ cell-free protein synthesis system. Using a library of human E3 ubiquitin ligases, we identified RFPL3 as a potential stimulator of human immunodeficiency virus, type 1 (HIV-1) PIC integration activity in vitro. This enhancement of PIC activity by RFPL3 was likely to be attributed to its N-terminal RING domain. To further understand the functional role of RFPL3 in HIV infection, we created a human cell line overexpressing RFPL3. Immunoprecipitation analysis revealed that RFPL3 was associated with the human immunodeficiency virus, type 1 PICs in infected cells. More importantly, single-round HIV-1 infection was enhanced significantly by RFPL3 expression. Our proteomic approach displays an advantage in the identification of new cellular proteins affecting the integration activity of the PIC and, therefore, contributes to the understanding of functional interaction between retroviral integration complexes and host factors.
Introduction
Human immunodeficiency virus, type 1 (HIV-1)6 relies heavily on the interplay between viral proteins and various host cofactors throughout its replication cycle, including the early stage of HIV-1 infection (1). After entry into the host cell, the HIV-1 RNA genome is reverse-transcribed to produce a double-stranded copy of viral DNA in a complex derived from the virion core. Subsequently, the newly synthesized viral DNA remains associated with multiple proteins to form a high-ordered nucleoprotein complex named the preintegration complex (PIC) in the cytoplasm (2). Critical cross-talk between specific cellular factors and viral proteins within the PIC is thought to be important for facilitating the nuclear translocation of the viral DNA genome and its subsequent integration into the host chromosome, which are essential steps for all retrovirus replications (1, 2). Although the integration process, including the DNA breakage and joining reaction, is mainly catalyzed by a critical viral enzyme, integrase (IN), biochemical studies have shown that the simple in vitro reaction with a DNA substrate containing viral DNA ends and catalyzed by purified IN lacks the full fidelity of the integration observed in infected cells (3). On the other hand, PICs isolated from infected cells can efficiently insert both viral DNA ends into a target DNA in a concerted manner in vitro, replicating a hallmark of in vivo integration (4–6). This evidence indicates that the PIC harbors additional host factors required for authentic integration in infected cells. As demonstrated by the recent development of IN strand transfer inhibitors as first-line antiretroviral drugs (7), integration is the critical rate-determining step within the HIV-1 replication cycle. Hence, it remains an attractive target in the treatment of HIV-1-infected individuals. In this context, anti-HIV agents targeting the interaction between viral and host factors within the integration complex will constitute a novel class of antiretroviral drugs that block integration with minimized chances of resistant virus development (8).
Because the PIC-based in vitro assay has been reported to be a more selective system that better replicates in vivo integration conditions, which cannot be reproduced in the reaction assay using the IN protein alone, extensive efforts have been made to uncover the organization and full composition of retroviral PICs (9). However, information on the protein components that affect PIC function is mostly restricted to immunoprecipitation studies and sensitive immunoblotting analysis because of the low abundance of PICs in cell extracts (10–13). An alternative approach taken for the study of a host cofactor of the PIC is the in vitro reconstitution of salt-disrupted PIC integration activity by engaging purified recombinant proteins (14–16). However, these conventional biochemical procedures targeting a particular protein or proteins are often laborious, presenting technical obstacles for high-throughput screening studies using a wide variety of proteins for comprehensive functional analysis of cellular factors involved in PIC integration.
In this study, we developed an efficient screening system allowing for the identification of novel cellular proteins that modulate the integration activity of HIV-1 PICs. To this end, we employed a microtiter plate-based in vitro assay reported previously for the HIV-1 PIC that allows for a rapid measure of PIC integration activity on a 96-well plate, thus enabling the handling of large scale sample sets (17). In addition, as a novel aspect of our study, the microtiter plate-based PIC integration assay was combined with a library of human proteins that had been produced using wheat germ cell-free technology to screen for new cellular modulators of HIV-1 PIC integration activity in vitro (18). The wheat germ cell-free system, which is an in vitro eukaryotic protein translation technology using a wheat embryo, can reportedly produce high-quality proteins that are soluble and folded properly (18, 19). This method offers a variety of potential advantages over other well known protein production approaches, such as Escherichia coli and rabbit reticulocyte systems. In particular, it can be easily adapted to high-throughput operations for the production of functional proteins from cDNA libraries (18, 19).
By exploiting the high-throughput capabilities of both the microtiter plate-based PIC integration assay and the wheat germ cell-free protein production system, we performed a screening study for the identification of cellular proteins that regulate the integration activity of the HIV-1 PIC in vitro. By using a library of human RING finger type E3 ubiquitin ligases, a Ret finger protein-like 3 (RFPL3) was found to enhance the in vitro integration activity of the HIV-1 PIC. The enhancing effect of RFPL3 in PIC activity was attributed to its N-terminal RING finger domain. More importantly, a cell-based overexpression experiment revealed that RFPL3 was able to associate with HIV-1 PICs in infected cells and appeared to augment the infection of single-round HIV-1 infection. Hence, our work offers a proof of concept approach for the identification of novel host modulators of PIC function, which allows us to better understand the molecular aspects of HIV-1 integration.
EXPERIMENTAL PROCEDURES
Isolation of PICs
HEK 293T cells were grown in DMEM supplemented with 10% FCS (Invitrogen), 100 units/ml of penicillin, and 100 μg/ml of streptomycin. To produce vesicular stomatitis virus G protein (VSV-G)-pseudotyped lentiviral vectors (LTR-Tat-IRES-GFP (20)), 2 × 106 of 293T cells were seeded in a 100-mm tissue culture dish 1 day before transfection and transfected with an HIV-1-derived vector plasmid (pEV731), a packaging vector (pMDLg/pRRE), a REV expression vector (pRSV-Rev), and a VSV-G expression vector (pMD.G) using a calcium phosphate method as described previously (21). A supernatant was harvested 48 h after transfection, filtered, and treated with DNase I (New England Biolabs). For infection, 3.3 × 106 293T cells were exposed to 10 ml of the DNase-treated conditioned medium in a 100-mm dish (multiplicity of infection of ∼10). Cells were harvested using trypsin 7 h after infection, washed with cold buffer K (150 mm KCl, 20 mm HEPES-NaOH (pH 7.4), and 10 mm EDTA), and permeabilized in 500 μl of buffer K in the presence of 1 mm DTT, 20 μg/ml of aprotinin (Sigma), and 0.025% digitonin (Sigma) for 5 min on ice. Lysates were centrifuged, and the supernatant containing cytoplasmic PICs was mixed with 100 μl of buffer K containing 40% sucrose for storage at −80 °C. A Moloney murine leukemia virus (MMLV) PIC was prepared as described previously (22).
Preparation of the Target DNA-coated Microtiter Plate
Immobilization of target DNA to a 96-well plate was performed as described previously (17), with slight modifications. 1 μg of pUC19 plasmid (2.7 kb) that had been linearized with EcoRI was suspended in 75 μl of 20 mm 1-methyl-imidazole (pH 7.0) (Sigma) and 200 mm 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (Sigma) and added to a Covalink amine-coated plate (Corning) that had been prewashed three times with 20 mm 1-methyl-imidazole (pH 7.0). The plate was incubated at 50 °C for 3 h, after which the wells were washed five times with buffer A (20 mm HEPES (pH 7.4), 1 m NaCl, 1% SDS, and 10 mm EDTA) at 65 °C (with the third wash performed at 68 °C for 20 min) and five times with buffer K at 50 °C. The wells were then incubated at 50 °C in buffer K with 10 mm citraconic anhydride (Sigma) for 30 min, after which it was replaced with buffer K containing 100 μg/ml of tRNA (Sigma) and 0.2% BSA for storage at 4 °C until use.
Microtiter Plate-based Integration Assay
The standard in vitro HIV-1 PIC integration assay was carried out as follows. 25 μl of HIV-1 PIC was incubated with 5 μl of recombinant protein (or buffer) in a 96-well plate at 37 °C for 1 h. In certain experiments using GST-VRK1, the PIC was treated in the presence of 10 μm ATP (23). A microtiter plate immobilized with target DNA was rinsed with buffer K five times before use, and 30 μl of the PIC mixture was added into each well with 30 μl of 2× integration mix (20 mm HEPES-NaOH (pH 7.4), 10 mm MgCl2, 150 mm KCl, 20 mm DTT, 200 μg/ml BSA, and 30% glycerol). Reactions were incubated at 37 °C for 30 min to allow integration of viral DNA from PICs into immobilized target DNA. 5 mg/ml of proteinase K and 5% SDS were added to stop the reaction, with incubation at 37 °C for another hour. Wells were then washed five times with buffer A at 65 °C (with the third wash done at 68 °C for 20 min) and five times with buffer K. Integrated DNA products were detached from the plate by incubating with 30 μl of 0.04 N NaOH at 50 °C for 10 min before neutralizing with 30 μl of 0.04 N HCl and 50 mm HEPES (pH 7.5) (17). Quantitative PCR (qPCR) for the detection of an eluted HIV-1 DNA product was performed using a primer pair (AA55 and M667) amplifying the R-U5 region of the LTR and a specific fluorogenic probe (HIV-FAM) as described previously (24). A microtiter plate-based integration assay of the MMLV PIC was performed by the same protocol as that for the HIV-1 PIC, except for the qPCR using a primer pair (sense primer, 5′-GCGCCAGTCCTCCGATTGACTG-3′; antisense primer, 5′-CTGACGGGTAGTCAATCACTCAG-3′) and a fluorogenic probe (5′-ATCCGACTTGTGGTCTCGCTGTTC-3′) detecting the MMLV LTR.
Production of a Human E3 Ubiquitin Ligase Library
Generation of a DNA template by split-primer PCR, in vitro transcription (IVT), and cell-free protein synthesis by wheat germ extract were performed as described previously (25–27). ORFs of human E3 ubiquitin ligases were amplified from a set of National Institutes of Health mammalian gene correction clones by PCR using target protein-specific sense primers beginning with the S1 sequence and the antisense primers AODA2306 (for pBluescriptR-based clones (27)) or pDONR221_1stA4080 (5′-ATCTTTTCTACGGGGTCTGA-3′ for pCMV-SPORT6, pOTB7, pCR4-TOPO, and pCR-BluntII-TOPO-based clones). A second split-primer PCR was carried out using an SPu sense primer containing an SP6 promoter sequence, a deSP6E01 sense primer containing a GST tag sequence (28), and the respective AODA2303 (for AODA2306 (27)) or pDONR221_2ndA4035 (5′-ACGTTAAGGGATTTTGGTCA-3′ for pDONR221_1stA4080) antisense primer. Expression of GST-tagged recombinant proteins was performed on 96-well plates using the GenDecoder 1000 protein synthesizer (CellFree Sciences, Japan) according to the protocol of the manufacturer. A transcript of each E3 ligase was made on the final PCR product by IVT using an SP6 RNA polymerase. The mRNA product was precipitated and resuspended in the translation mix containing the wheat germ extract (catalog no. WEPRO 1240G, CellFree Sciences), creatine kinase, and 1× SUB-AMIX® (CellFree Sciences), forming the lower layer of the translation reaction. After placing 1× SUB-AMIX® as the upper layer, the bilayer reaction mixture was incubated at 16 °C for 20 h. To purify the GST-tagged proteins, 300 μl of crude translation reaction was mixed with 350 μl of buffer P (10 mm sodium phosphate (pH 7.4) and 500 mm NaCl) and 50 μl of glutathione-Sepharose 4 Fast Flow beads (GE Healthcare, 50% slurry in buffer P) in a 96-well format and rotated at 4 °C for 1 h. The protein-beads mixture was filtered through a 96-well filter unit (Unifilter, Whatman) via centrifugation at 100 × g for 4 min and washed with 1 ml of buffer P. Proteins were eluted by incubation with 60 μl of buffer E (50 mm Tris-HCl (pH 8.0), 10% glycerol, 1 mm DTT, 5 mm EDTA, 10 mm reduced glutathione (Sigma)) at 4 °C for 30 min before being collected by centrifugation. E3 ubiquitin ligases in elutes were separated with 10% SDS-PAGE gels, followed by Coomassie Brilliant Blue (CBB) staining (Wako Pure Chemical Industries) or immunoblotting analysis as described below. A GST-tagged barrier to autointegration factor (GST-BAF) and vaccinia-related kinase 1 (GST-VRK1) were also produced by the same split-primer PCR method using antisense primers, AODS (for the first PCR), and AODS-3 (for the second PCR) (CellFree Sciences) and a control protein, GST-tagged bacterial dihydrofolate reductase (GST-DHFR), was produced using AODA2306 and AODA2303 from the plasmid DNAs described previously (23, 29, 30).
Production of GST Fusion Proteins in E. coli
ORFs of RFPL3, RNF25, STUB1, and TRIM52 were amplified from a cDNA library and cloned into the SmaI site of pGEX-2T (GE Healthcare). The pGEX-2T constructs were transformed into the E. coli BL21(DE3) strain (Stratagene), and protein expression was induced as described previously (23). Bacterium cells were harvested 5 h after induction, resuspended in buffer R (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 1 mm EDTA), and lysed in buffer L (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 5 mm EDTA, 0.028% β-mercaptoethanol, and 0.4 mg/ml of lysozyme (Sigma)) at 4 °C for 1 h. After centrifugation, GST fusion proteins in the supernatant were bound to a glutathione-Sepharose 4B column (GE Healthcare), washed with buffer G (50 mm Tris-HCl (pH 8.0), 500 mm NaCl, 10% glycerol, and 5 mm DTT), and eluted with buffer G containing 15 mm reduced glutathione. Eluted fractions containing GST fusion proteins were further subjected to gel filtration chromatography using a Superdex 200 column (GE Healthcare) with buffer G. Preparation of BAF (non-tagged) and GST-tagged VRK1 by E. coli were performed as described previously (23, 29). Protein concentration was determined by the Bradford method (Bio-Rad) using BSA as a protein standard.
Production of RFPL3 Mutants Using a Wheat Germ Cell-free System
Amplification of template cDNAs encoding GST-tagged full-length RFPL3 (GST-RFPL3 FL), its N-terminal deletion mutants (GST-RFPL3 Δ36, GST-RFPL3 Δ98, and GST-RFPL3 Δ146), and a control protein (GST-DHFR) was performed by split-primer PCR as described above (25, 31). After IVT, cell-free expression of proteins was performed with 6 ml of translation reaction using wheat germ extracts optimized for GST affinity purification (catalog no. WEPRO 1240G, CellFree Sciences) according to the protocol of the manufacturer. Batch purification was performed using glutathione-Sepharose 4 Fast Flow beads and buffer P, and the GST fusion proteins were finally eluted in buffer E. The proteins were further purified with a 5 ml HiTrap desalting column (GE Healthcare) in buffer G. Protein concentration was determined by the Bradford method.
Immunoblotting Analysis
Protein samples were denatured in an SDS sample buffer, separated by 10% SDS-PAGE gel, and blotted onto an Immobilon P membrane (Millipore). GST fusion proteins and actin were detected by probing with anti-GST mouse monoclonal IgG (Santa Cruz Biotechnology) and anti-actin mouse monoclonal IgG (Sigma), respectively, followed by HRP-conjugated anti-mouse monoclonal IgG (Cell Signaling Technology). FLAG-tagged proteins were detected by HRP-conjugated anti-FLAG mouse monoclonal IgG (M2, Sigma). Proteins were visualized using an ImageQuant LAS 4000 mini chemiluminescent image analyzer (GE Healthcare).
Gel Mobility Shift Assay
A 516-bp DNA fragment (substrate DNA) derived from XcmI-digested pGEX-2T (250 pm) was incubated with GST-RFPL3, GST-DHFR, and BAF (25 or 100 nm) in 10 μl of binding buffer (20 mm HEPES (pH 7.5), 1 mm DTT, and 100 ng/ml BSA) at 30 °C for 1 h. The mixtures were separated by 0.4% agarose gel electrophoresis in Tris acetate-EDTA buffer and blotted to a GeneScreen Plus membrane (PerkinElmer Life Sciences). Southern blotting analysis was performed using a Gene Images AlkPhos Direct labeling and detection system (GE Healthcare) and an alkaline phosphatase-labeled substrate DNA as a probe, followed by visualization via ImageQuant LAS 4000 mini.
Establishment of 293T Cell Lines Stably Expressing FLAG-tagged RFPL3
The ORF of RPL3 and DHFR was first cloned into the EcoRV site of p3xFLAG-CMV14 (Sigma). The inserted genes were then amplified along with the C-terminal FLAG sequences using PCR, followed by insertion of these fragments into an entry plasmid, pDONR221, through a Gateway BP reaction (Invitrogen). We constructed a Gateway-compatible lentiviral vector, pYK005C-Bla, in which the hrGFP gene of pYK005C (21) had been replaced with a blasticidin resistance gene. Individual genes were then transferred to pYK005C-Bla by LR reaction (Invitrogen). A VSV-G-pseudotyped lentiviral vector was produced using 293T cells by cotransfection of pYK005C-Bla encoding FLAG-RFPL3 or FLAG-DHFR with pMDLg/pRRE, pRSV-Rev, and pMD.G (21) and used to transduce 293T cells. Transduced cells were selected in the presence of 10 μg/ml of blasticidin (Invivogen). Expression of FLAG-tagged RFPL3 and DHFR in the stable cell lines was confirmed by immunoblotting using a 12% SDS-PAGE gel.
Immunofluorescence Analysis
To stain for FLAG-tagged RFPL3 in the stable cell lines, 5 × 104 of cells preseeded in Lab-Tek II 8-well chamber slides (Thermo) 1 day before staining were fixed with 4% paraformaldehyde for 30 min, permeabilized with 0.1% Triton X-100 in PBS for 10 min, and blocked with 10% FBS and 5% BSA in PBS for 30 min at room temperature. The cells were stained with a primary antibody, anti-FLAG rabbit IgG (Sigma), followed by a secondary antibody, Alexa Fluor 488-conjugated goat anti-rabbit IgG (Invitrogen). A slide was mounted with a ProLong Gold antifade reagent containing DAPI (Invitrogen) and observed under an Olympus IX81 fluorescence microscope. Images were captured with the CellSens Dimension software (Olympus).
Immunoprecipitation Analysis of HIV-1 PIC
HIV-1 PICs were isolated from 293T cells expressing FLAG-tagged RFPL3 or DHFR by infection of a VSV-G-pseudotyped lentiviral vector, LTR-Tat-IRES-GFP, as described above. A volume of 125 μl of the PIC was mixed with 250 μl of buffer C (20 mm HEPES-NaOH (pH 7.4), 5 mm MgCl2, 150 mm KCl, 6 mm EDTA, 0.04% BSA, 0.1% Nonidet P-40, and protease inhibitors), and 75 μl of the mixture was kept as an input fraction. The rest of the mixture (300 μl) was incubated with 10 μg of anti-FLAG mouse monoclonal IgG (M2, Sigma) at 4 °C for 2 h with rotation, followed by the addition of 30 μl of protein A/G-agarose beads (Santa Cruz Biotechnology) and another 2 h incubation at 4 °C. The immune complex was washed three times with 500 μl of buffer C and then deproteinized by proteinase K and SDS. Viral DNA was recovered from phenol/chloroform extraction and ethanol precipitation, followed by resuspension in 10 μl of a Tris-EDTA buffer containing 20 μg/ml of RNase A (Qiagen). Detection of viral DNA in precipitates was done via a 28-cycle PCR with AA55 and M667 primers using a KOD-Plus DNA polymerase (Toyobo). PCR products were subjected to 1.5% agarose gel electrophoresis using Tris acetate-EDTA buffer and visualized by ethidium bromide staining and a UV transilluminator (Insta BioAnalytik).
Luciferase Assay on RFPL3-expressing 293T Cells
An HIV-1-based vector bearing a luciferase gene (HIV-Luc) was produced using 293T cells by the cotransfection of pMDLg/pRRE, pRSV-Rev, pMD.G, and pYK005-Luc, in which a Renilla luciferase gene had been cloned into the Gateway unit of pYK005C (21), as described above. The supernatant was harvested 48 h after transfection, and the p24CA in a viral supernatant was quantified using an HIV-1 p24 antigen ELISA kit (Zetrometrix). In a single-round infection experiment, FLAG-tagged RFPL3 or DHFR-expressing stable cell lines, which had been seeded in a 12-well plate at 1 × 105/well density 1 day before infection, were exposed to HIV-Luc containing 10 ng of p24CA at 37 °C for 2 h and cultured after washing twice. 48 h post-infection, cells were harvested and lysed with 200 μl of mammalian protein extraction reagent (Thermo). The lysate was centrifuged at 10,000 rpm for 5 min, and the supernatant was subjected to a luciferase activity assay using the Renilla luciferase glow assay kit (Thermo). The luminescence level was detected using a Synergy H1 hybrid multimode microplate reader. The protein concentration of each sample supernatant was quantified using the Bradford kit to normalize the luciferase activity measured.
Assessment of HIV-1 Integration in Infected Cells
HIV-Luc containing 40 ng of p24CA was pretreated with RNase-free DNase I (New England Biolabs) at 37 °C for 30 min to remove contaminating transfected plasmid DNA. FLAG-tagged RFPL3 or DHFR-expressing 293T cells, which had been seeded in a 12-well plate at 2 × 105/well density 1 day before infection, were exposed to DNase-treated HIV-Luc at 37 °C for 2 h and cultured after washing three times. The cells were harvested 24 h after infection, and total DNA was extracted by a QIAamp DNA mini kit (Qiagen). The level of integrated viral DNA was analyzed by an Alu-HIV nested PCR procedure as described previously (32), with minor modifications. Briefly, the first round of PCR was carried out using 100 ng of DNA by primers specific to the Alu element (5′-GCCTCCCAAAGTGCTGGGATTACAG3′ (32)), and the Renilla luciferase gene (5′-CCACTGAGGCCCAGTGATCATGC-3′) that targets the pYK005-Luc genome but not the pYK005C-Bla-based genome used for creation of the FLAG-RFPL3 or FLAG-DHFR cell line. The first (Alu-RLuc) PCR product was diluted 100-fold to reduce the background from the pYK005C-Bla-based genome and unintegrated DNA (24) and subjected to the second round of PCR (qPCR) using HIV-1 LTR-specific primers (AA55 and M667) and a fluorogenic probe (HIV-FAM). A first-round PCR without the Alu element primer (i.e. RLuc primer only), followed by a same second-round qPCR was also performed to confirm that the Alu-RLuc signal was specific to integrated DNA (32).
Statistical Analysis
Student's t test was used to determine statistical significance. p < 0.05, p < 0.01, and p < 0.001 were considered significant.
RESULTS
Application of the Microtiter Plate-based HIV-1 PIC Assay to the Evaluation of Cellular Proteins Involved in Integration
The development of a rapid and efficient HIV-1 PIC integration assay using DNA-coated microtiter plates was first described in a study by Hansen et al. (17) in which a library of chemical compounds with structures related to known IN inhibitors had been screened for the ability to inhibit PIC integration activity in vitro. For the purpose of our study, the microtiter plate-based PIC integration assay was adapted for a screen with recombinant proteins to identify potential cellular modulators of the HIV-1 PIC. In our assay, EcoRI-digested pUC19 (2.7 kb) was covalently immobilized on a Covalink 96-well microtiter plate as the target DNA, and HIV-1 PICs were produced in 293T cells with the infection of a VSV-G-pseudotyped lentiviral vector, LTR-Tat-IRES-GFP (20). The specificity of the assay for detection of in vitro integration was first tested using freshly isolated active PICs and control PICs that had been inactivated in advance with proteinase K and SDS. The copy number of viral DNA detected from the reaction using 25 μl of active HIV-1 PICs was 5957.6 ± 427.1, which was about 34 times higher than that of the background DNA detected with an inactivated PIC (180.8 ± 12.7 copies, Fig. 1A). We next evaluated the microtiter plate-based integration assay using previously identified cellular modulators of PIC activity, BAF (29) and VRK1 (23). BAF has been reported as a component of the MMLV and HIV-1 PICs and has been found to enhance in vitro integration activity of the PICs (15, 29). On the other hand, VRK1 has been shown to abrogate the effect of BAF by phosphorylating the latter, causing its dissociation from the MMLV PIC and, thereby, inhibiting integration activity (23). For evaluation, HIV-1 PIC samples were initially incubated with BAF, GST-tagged VRK1, and control protein (GST) at 37 °C for 1 h and then incubated in the target DNA-immobilized microtiter plate wells to allow in vitro integration. Taking the amount of integrated product yield in the GST control protein-treated PIC to be 100% at the respective protein concentrations, treatment of PICs with 1 and 10 nm of BAF presented an increment in the yield of integrated products to 147.3 ± 11.6% and 256.9 ± 35.9%, respectively (Fig. 1B). Similarly, an inhibitory effect in PIC integration activity by GST-VRK1 treatment was observed, which was dependent on the concentration of the recombinant proteins (55.8 ± 6.0% at 50 nm, 30.6 ± 4.4% at 100 nm, and 5.2 ± 2.7% at 500 nm when compared with the GST control protein at their respective concentrations, Fig. 1C). In addition, it is of note that the dose-dependent and specific inhibition of in vitro PIC activity has also been shown in the treatment with a well documented HIV-1 IN strand transfer inhibitor, elvitegravir (data not shown), with an IC50 of 2.6 nm, which was a similar, subnanomolar range of EC50 obtained by cell-based HIV infection assay (33). Taken together, these data show that the microtiter plate-based assay is an adequate system to quantitatively detect in vitro HIV-1 PIC integration events and their modulation by cellular proteins.
FIGURE 1.
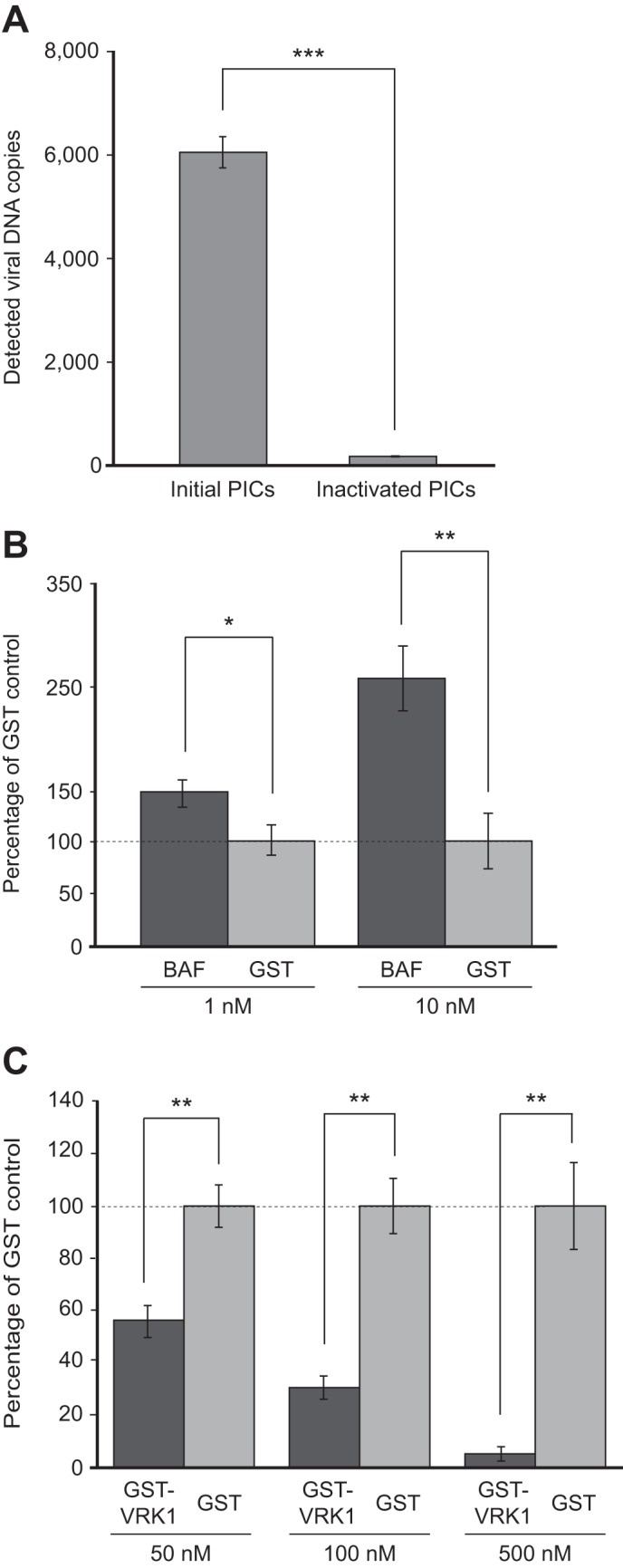
Validation of microtiter plate-based HIV-1 PIC assay. A, quantification of integrated DNA produced by HIV-1 vector-derived PICs. A PIC sample (25 μl) was incubated in the well of a microtiter plate in which 2.7-kb target DNA was covalently immobilized at 37 °C for 30 min. The reaction was then inactivated by treatment with proteinase K and SDS, and, after washing, integrated DNA were released using NaOH to be quantified via qPCR using LTR-specific primers (17). Inactivated PICs were prepared by pretreatment with proteinase K/SDS before being subjected to the microtiter plate-based PIC assay to check the background level of viral DNA. The experiment was done in triplicate, and data are expressed as mean ± S.D. B and C, modulation of PIC integration activity by BAF and VRK1 proteins. HIV-1 PICs were incubated with increasing concentrations of BAF (1 and 10 nm), GST-VRK (50, 100, and 500 nm), and respective concentration of control protein (GST) at 37 °C for 1 h and subjected to a microtiter plate-based integration assay. The level of integrated products quantified was expressed as a relative mean percentage with error bars indicating S.D. over that of GST control protein (100%, dashed line) at the respective concentration for three independent experiments. Statistical significance was determined by Student's t test. *, p < 0.05; **, p < 0.01, ***, p < 0.001.
Screen for Human E3 Ubiquitin Ligases Modulating HIV-1 PIC Activity
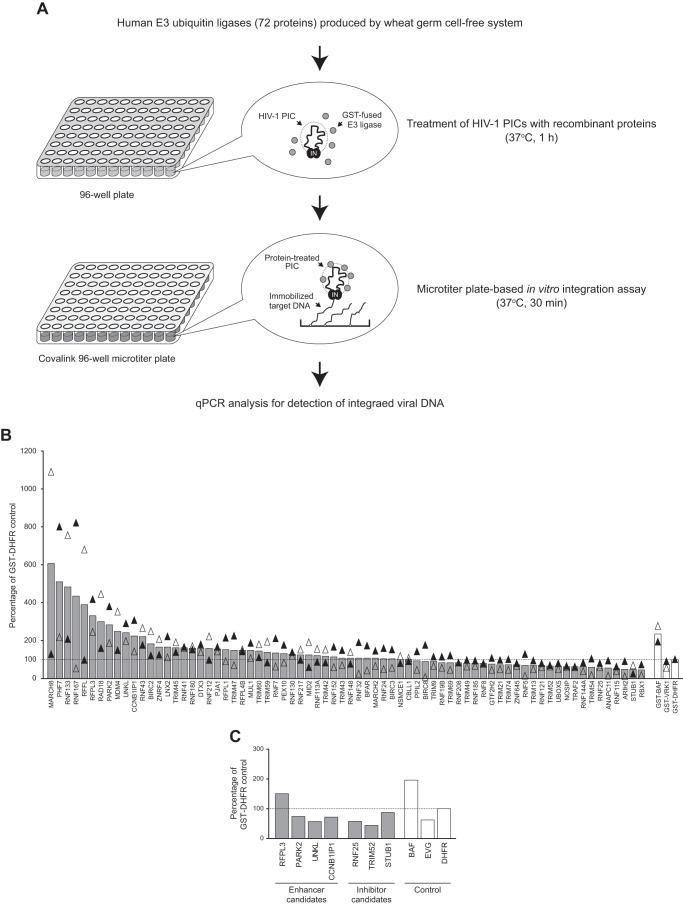
Next, the microtiter plate-based integration assay was adopted as a protein screening platform to identify potential cellular modulators of HIV-1 PIC activity. Previous studies have reported that a host ubiquitin-proteasome system is likely to be implicated in HIV-1 integration or the stability of IN (34–38). This motivated us to investigate E3 ubiquitin ligases influencing the integration activity of HIV-1. To this end, a library of human E3 ligases was generated by the wheat germ cell-free protein production system. This eukaryotic cell-based in vitro translation method offers several advantages over the E. coli system, particularly allowing the high-throughput expression of recombinant proteins from cloned ORF resources in vitro (18, 19). In this study, to generate the protein library, template DNAs containing an ORF were amplified from a collection of National Institutes of Health mammalian gene correction clones encoding 96 RING finger-type E3 ubiquitin ligases and fused with a 5′ terminal SP6 promoter and GST tag sequences by split primer PCR (28, 39). Agarose gel electrophoresis and ethidium bromide staining of the split PCR product confirmed that 92 DNA templates were synthesized successfully (supplemental Fig. 1). Using the DNA fragments as templates, IVT by SP6 RNA polymerase and subsequent in vitro translation using wheat germ extract were performed in a 96-well format (26), and resultant GST-fused proteins were further purified on glutathione-Sepharose beads through a 96-well filter unit. Among the 92 eluted fractions from the affinity purification, 57 proteins were visualized by SDS-PAGE and CBB staining analysis with the expected molecular mass of each E3 ubiquitin ligase, and an additional 15 proteins with low expression levels, which could not be visualized from the CBB staining, were further detected by immunoblot analysis using an anti-GST antibody (supplemental Fig. 2). Altogether, 72 E3 ubiquitin ligases (75%) were produced successfully through the wheat germ cell-free system in combination with a human ORF library.
To search for human E3 ubiquitin ligases that influence the in vitro activity of HIV-1 PICs, each of the 72 proteins was incubated with a PIC for 1 h at 37 °C, and the mixtures were then subjected to the microtiter plate-based integration assay (Fig. 2A). As control proteins for the screen, GST-BAF (enhancer control), GST-VRK1 (inhibitor control), and GST-DHFR (negative control) were included as well. Fig. 2B shows the screening result, in which the average value of the copy number of the integrated viral DNA from two independent screening assays using different batches of proteins and HIV-1 PICs is presented as a percentage compared with copy numbers detected in GST-DHFR-treated reactions. On average, 12 E3 ubiquitin ligases enhanced in vitro integration activity of the HIV-1 PIC more than 200%, whereas 13 E3 ligases exhibited an inhibition of the integration activity by 30% or more (Fig. 2B). However, preliminary experiments using the first batch of protein showed that four proteins (RFPL3, PARK2, UNKL, and CCNB1IP1) still exhibited 2-fold or more enhancement activities, and 3 E3 ligases (RNF25, TRIM52, and STUB1) showed an inhibition effect repeatedly in the preliminary assay (data not shown). Therefore, we carried out an additional round of assays for the four potential enhancers and three potential inhibitors using new batches of recombinant protein and PIC preparations. The results revealed that RFPL3 was still able to enhance the integration activity. Similarly, RNF25, TRIM52, and STUB1 repeatedly retained an inhibiting effect on the HIV-1 PIC activity in vitro (Fig. 2C).
FIGURE 2.
Screening of human E3 ubiquitin ligases using the microtiter plate-based PIC assay. A, workflow of the screening assay. A library of human E3 ubiquitin ligases was generated by the wheat germ cell-free protein production system and further affinity purified using glutathione-Sepharose beads through a 96-well format filter unit. In total, 72 E3 ligases were incubated with PICs that had been isolated from VSV-G-pseudotyped HIV-1 vector-infected cells, and the reaction was subjected to the microtiter plate-based integration assay. The copy number of HIV-1 DNA integrated in immobilized target DNA was measured by qPCR analysis using HIV-1 LTR-specific primers. B, activity of HIV-1 PICs treated with E3 ubiquitin ligases (gray bars) and control proteins (GST-BAF (enhancer control), GST-VRK-1 (inhibitor control), and GST-DHFR (negative control), white bars) were analyzed by microtiter plate-based integration assay. Integrated viral DNA is shown as relative mean percentage over that of GST-DHFR-treated PICs (100%, dashed line) from two independent screening assays using different batches of proteins and HIV-1 PICs. Black and white triangles represent the first and second screening assay results, respectively. C, seven E3 ubiquitin ligases were selected as potential enhancers and inhibitors of HIV-1 PIC integration activity on the basis of the result of the first two rounds of screening assays and subjected to an additional microtiter plate-based integration assay together with control reactions (PICs treated with GST-BAF, 1 μm elvitegravir (EVG) and GST-DHFR). Integrated viral DNA detected is presented as a percentage of that of negative control protein (GST-DHFR, 100%, dashed line).
We next intended to validate the modulating activity of the candidate proteins and also examine whether the modulation of PIC activity exhibited by the wheat germ cell-free system-derived candidates were a universal effect for the recombinant proteins produced by other system. To this end, four candidate proteins were produced as GST fusion proteins using an E. coli-based protein production system (Fig. 3A). A fixed concentration (1 and 10 nm) of each protein was then used in the microtiter plate-based integration assay. Bacterially expressed BAF, GST-VRK1, and GST were also included as control reactions. When the amount of integrated DNA was compared with that detected in a GST-treated control reaction (taken as 100%) at 1 and 10 nm, incubation of HIV-1 PICs with GST-RFPL3 resulted in a significant and dose-dependent increase in the level of viral DNA detected (117.5 ± 12.9% and 170.5 ± 38.3% for 1 and 10 nm GST-RFPL3, respectively, Fig. 3B). Of note, enhancement by a positive control, BAF, was similar to that observed for GST-RFPL3, at 115.0 ± 9.9% and 167.7 ± 25.6% for 1 and 10 nm, respectively (Fig. 3B), indicating that RFPL3 had a reproducible enhancing effect on the in vitro integration activity of the HIV-1 PIC, which was comparable with that of the known cellular enhancer BAF.
FIGURE 3.
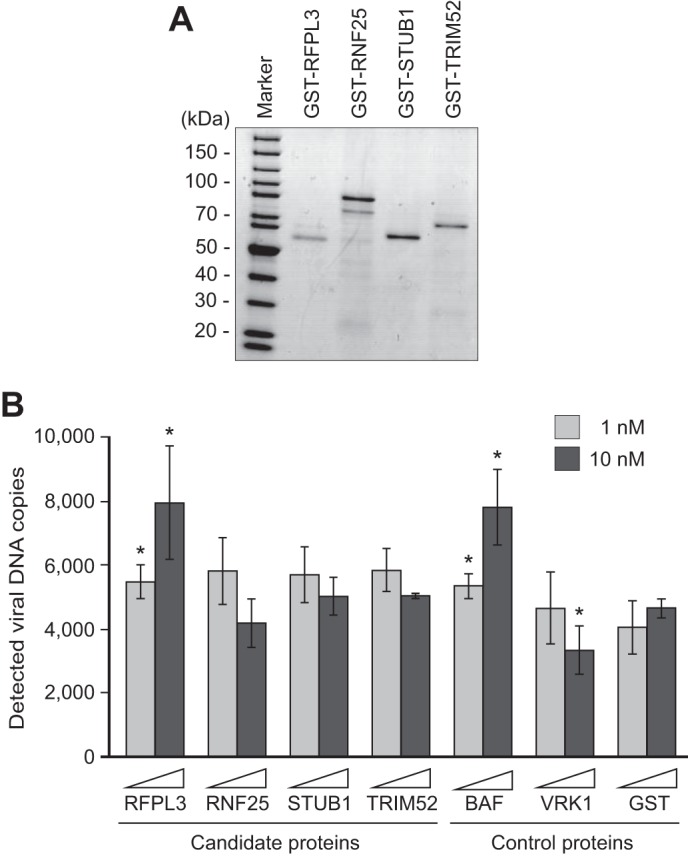
Validation of the modulative effect of candidate proteins in vitro. A, ORF of the four candidate E3 ubiquitin ligases chosen by the screen assay were cloned into the pGEX-2T bacterial expression vector, expressed as GST fusion proteins in E. coli, and purified by glutathione-Sepharose beads, followed by size exclusion chromatography. Purified proteins were checked by SDS-PAGE and CBB staining analysis. The first lane shows a molecular weight marker. B, effect of E. coli-produced candidate proteins on HIV-1 PIC integration activity. Two different concentrations (1 and 10 nm) of GST fusion proteins and a control protein (GST) were incubated with HIV-1 PICs, and the reactions were subjected to the microtiter plate-based integration assay. The amount of integrated products was quantified by qPCR. The level of integration was expressed as the mean value of viral DNA copies of three independent experiments, with error bars indicating S.D. The p value versus GST control protein at the respective concentration was determined by Student's t test (*, p < 0.05).
In contrast, the inhibitor candidate proteins (RNF25, STUB1, and TRIM52) had a minimal effect on the in vitro integration activity of the HIV-1 PIC, whereas incubation with 10 nm GST-VRK1 was able to exhibit its significant inhibiting ability, reducing the PIC integration activity by 28.5% of that of the GST control-treated reaction (Fig. 3B). Although 10 nm RNF25 showed some inhibitory effect on PIC integration activity (Fig. 3B), it was not significant (p = 0.118). Because RFPL3 reproducibly stimulated in vitro integration activity of the PIC with different sources of the recombinant protein (i.e. the wheat germ cell-free system and the E. coli system), we focused on this protein in subsequent experiments.
Domain of RFPL3 Required for Its Enhancement Effect on HIV-1 PIC
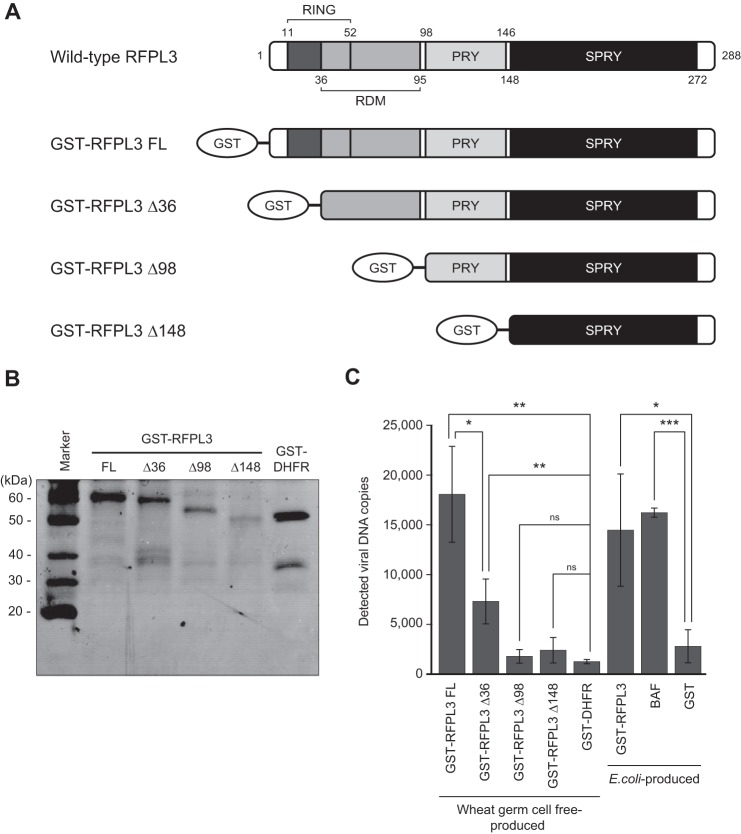
The human RFPL3 is part of the Ret finger protein-like (RFPL) protein family, which is similar in its genetic sequences to the Ret finger protein (also known as TRIM27) (40), and RFPL genes have been identified recently as targets of a transcription factor, Pax6 (41). Although little is known about the cellular activity of RFPL3, protein domain information can be extrapolated on the basis of the conserved amino acid sequences. Human RFPL3 has been reported to be composed of the N-terminal RING finger domain encoding putative E3 ligase activity, whereas the C-terminal B30.2 domain contains the PRY and SPRY motifs that are commonly found in the TRIM (tripartite motif) family proteins. These two domains flanked the ends of the RFPL-defining motif (RDM) that is exclusively restricted to RFPL family proteins (Fig. 4A) (40–42). To determine the RFPL3 domain that is necessary to enhance HIV-1 PIC integration activity, N-terminal-truncated mutants of RFPL3, as well as full-length (288 amino acids) RFPL3 were produced as GST fusion proteins by the wheat germ cell-free system. We chose to produce the mutant proteins using the wheat germ cell-free system because the use of a split PCR allows for efficient generation of the various mutant IVT templates conveniently from the same plasmid DNA (28). The first mutant (GST-RFPL3 Δ36) has a partially truncated RING finger domain, leaving the coiled-coil motif of the domain intact. The second mutant (GST-RFPL3 Δ98) lacks the full extent of the RING finger domain as well as the RDM domain but contains the PRY/SPRY domain. The last mutant (GST-RFPL3 Δ148) is the shortest, containing only the SPRY domain at the C-terminal end (Fig. 4A). All four proteins were expressed in wheat germ cell-free reactions and affinity-purified using glutathione-Sepharose beads, followed by a buffer exchange to remove reduced glutathione from elutes (Fig. 4B). Fig. 4C shows the effect of RFPL3 domain mutants on the HIV-1 PIC activities as measured by microtiter plate-based integration assay. Wheat germ cell-free-produced GST-RFPL3 FL (10 nm) displayed a 14.3-fold enhancement of the PIC integration activity (18,065.3 ± 4818.1 copies) when compared with the GST-DHFR control (1264.3 ± 214.8 copies). This is comparable with the integrated products quantified from PICs treated with E. coli–produced RFPL3 (14,467.9 ± 5636.4 copies) and BAF (16,224.1 ± 456.6 copies) (Fig. 4C). With the partial removal of the RING domain, GST-RFPL3 Δ36 showed a significant reduction in its ability to enhance the integration activity of the HIV-1 PIC but still exhibited a significant enhancement in the integration activity, being able to enhance 5.8 times more than the GST-DHFR control (7302.6 ± 2247.5 copies, Fig. 4C). However, with the entire removal of the RING domain in the remaining two mutants, which contained C-terminal PRY and/or SPRY domains, the enhancement effect by RFPL3 was completely abrogated (1781.3 ± 687.0 and 2402.0 ± 1284.1 copies in GST-RFPL3 Δ98- and GST-RFPL3 Δ148-treated reactions, respectively, Fig. 4C). Therefore, these results indicate that the N-terminal RING finger domain is the essential domain responsible for the functional enhancement of HIV-1 PIC activity.
FIGURE 4.
Domain of RFPL3 responsible for the enhancement of in vitro HIV-1 PIC activity. A, schematic of FL RFPL3 and its N-terminal truncation mutants (Δ36, Δ98, and Δ148) used for the assay. All proteins were expressed as GST fusion proteins by the wheat germ cell-free system and affinity-purified. B, SDS-PAGE and CBB staining analysis of purified proteins. A negative control protein, GST-DHFR, was also produced. The first lane shows a molecular weight marker. C, effect of RFPL3 mutants on the HIV-1 PIC integration activity. A microtiter plate-based PIC integration assay was performed in the presence of 10 nm recombinant proteins. GST-RFPL3, BAF, and a control GST protein produced by E. coli were also included in the assay as control reactions. The experiment was done in triplicates, and data are expressed as mean value of viral DNA copies, with error bars indicating S.D. Statistical significance was determined by Student's t test. *, p < 0.05; **, p < 0.01, ***, p < 0.001; ns, no significance.
Functional Specificity of RFPL3 in Modulation of PIC Activity
Although the full composition of the HIV-1 PIC has yet to be determined, it can be largely separated into viral DNA and both viral and cellular protein components (2). With respect to the cellular cofactors of the PIC function, many previously identified factors, including BAF, are DNA-binding proteins (12, 14, 15, 43, 44). In addition, because the Ret finger protein (TRIM27), which has been reported originally to show a similarity to RFPL family proteins, has been described as a nonspecific DNA-binding protein (40, 45), we initially reasoned that RFPL3 might have DNA-binding activity, thereby associating with HIV-1 PICs through viral DNA. To address this possibility, a gel mobility shift assay was performed using GST-RFPL3. In the assay, BAF, which interacts with double-stranded DNA with no detectable sequence specificity, was included as a positive control (46). A short DNA substrate (0.5 kb) was incubated with the respective proteins before being separated via gel electrophoresis, followed by Southern blotting detection of the substrate DNA to check the formation of a nucleoprotein complex during the incubation. As shown in Fig. 5, left panel, a slower migrating band was observed in a reaction containing 25 nm BAF (Fig. 5, lane 4), indicating the DNA binding activity of this cellular protein (23). In contrast, the nucleoprotein complex migrating at a slower rate during gel electrophoresis was not detected in reactions with GST-RFPL3 (Fig. 5, lane 2) or the control GST-DHFR (Fig. 5, lane 3), which coincided with a reaction containing substrate DNA alone (Fig. 5, lane 1). In the presence of a higher concentration of proteins (100 nm), the binding of BAF to substrate DNA prevented the nucleoprotein complex from entering the agarose gel because of DNA aggregation caused by the intramolecular DNA bridging property of BAF (Fig. 5, lane 8) (29). However, despite a similarly high concentration of proteins, incubation with 100 nm GST-RFPL3 again did not produce any nucleoprotein complex with substrate DNA (Fig. 5, lane 6). Therefore, this in vitro result suggests that RFPL3 does not possess a DNA-binding property.
FIGURE 5.
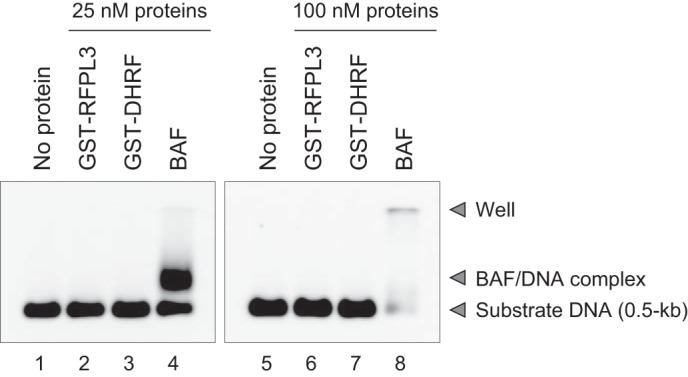
Gel mobility shift assay to examine the DNA-binding activity of RFPL3. Substrate DNA (0.5 kb, 250 pm) was incubated with two different concentrations (25 nm, left panel; 100 nm, right panel) of wheat germ cell-free-produced GST-RFPL3 (lanes 2 and 6), GST-DHFR (control protein, lanes 3 and 7) and E. coli-produced BAF (lanes 4 and 8) for 1 h at 30 °C. The mixtures were separated by agarose gel electrophoresis, and then Southern blotting analysis was carried out to detect substrate DNA using an alkaline phosphatase-labeled DNA probe. Lanes 1 and 5 are reaction of substrate DNA without proteins.
We also examined the influence of RFPL3 on the PIC integration activity of MMLV, which is a gammaretrovirus, to analyze the specificity of RFPL3 on the promotion of retroviral PIC activities because its protein composition has been reported to differ from that of HIV-1 PICs (2, 9). The MMLV PIC was prepared by the coculture of NIH3T3 cells and clone 4 cells, a MMLV-producing cell line, as described previously (6), treated with GST-RFPL3 or BAF (10 nm), and subjected to the microtiter plate-based in vitro integration assay. The result does not show a statistically significant enhancement of PIC integration activity upon treatment with GST-RFPL3 (140.2 ± 59.2% of GST control protein-treated PICs, Fig. 6). On the other hand, treatment with BAF showed a substantial enhancement effect on the MMLV PIC (335.8 ± 139.2% of GST control protein-treated PICs, Fig. 6). This indicates that, unlike BAF, which stimulates both HIV-1 and MMLV PIC activities (15, 29), the enhancing effect of RFPL3 is specific to the HIV-1 PIC. Taken together with the gel mobility shift assay results, these data suggest that RFPL3 is likely to interact with HIV-1 PICs through a protein component unique to HIV-1 but not the viral DNA within the PIC.
FIGURE 6.
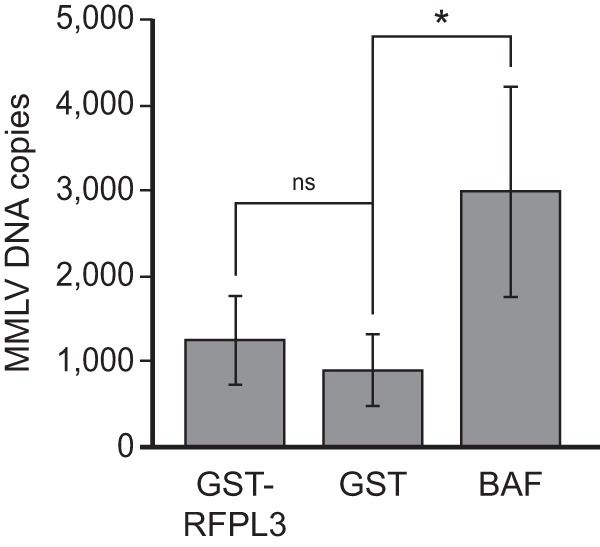
No effect of RFPL3 for the integration activity of MMLV PIC. MMLV PICs were isolated from the coculture of NIH3T3 and MMLV-producing clone 4 cells and treated with 10 nm GST-RFPL3, BAF, and GST control protein, followed by microtiter plate-based integration assay using MMLV LTR-specific primers. The level of integration was expressed as the mean value of viral DNA copies of three independent experiments, with error bars indicating S.D. The p value versus GST control protein at the respective concentration was determined by Student's t test. *, p < 0.05; ns, no significance.
Augmentation of HIV-1 Infection by RFPL3 Expression
To investigate the potential contribution of RFPL3 to HIV-1 infection, 293T-based cell lines stably expressing FLAG-tagged RFPL3 or a control protein (FLAG-DHFR) were established by lentiviral vector transduction (Fig. 7A) and infected with the lentiviral vector LTR-Tat-IRES-GFP. To analyze the association of RFPL3 with the HIV-1 PIC in infected cells, PICs were isolated from FLAG-RFPL3-expressing and control FLAG-DHFR-expressing cells (Fig. 7A), followed by an immunoprecipitation assay using an anti-FLAG antibody. HIV-1 DNA extracted from the coimmunoprecipitated PIC samples were amplified via PCR using primers detecting the LTR R-U5 region and then visualized by gel electrophoresis. As shown in Fig. 7B, specific amplification of viral DNA was detected only in the PICs derived from FLAG-RFPL3-expressing cells, not from FLAG-DHFR-expressing cells, indicating that RFPL3 could be a component of HIV-1 PICs in infected cells.
FIGURE 7.
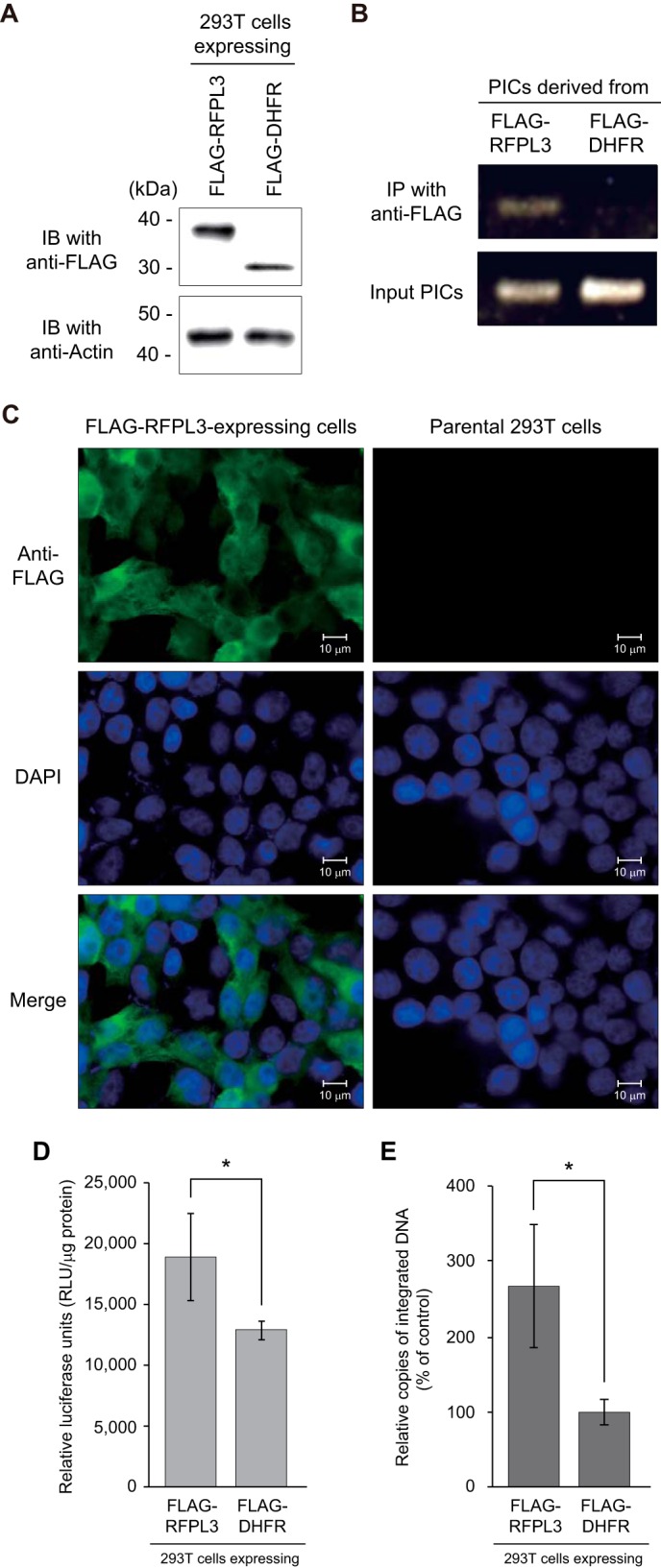
Augmentation of infection efficiency of HIV-Luc by RFPL3. A, establishment of the 293T cell line expressing FLAG-tagged RFPL3. Stable cell lines expressing FLAG-RFPL3 and control protein (FLAG-DHFR) were created by lentiviral vector transduction and blasticidin selection. Expressions of FLAG-tagged proteins were analyzed by immunoblot (IB) analysis with HRP-conjugated anti-FLAG antibody (top panel). Bottom panel, immunoblot analysis to detect actin in each cell lysate. Molecular weight standards are indicated at the left. B, coimmunoprecipitation analysis of HIV-1 PICs derived from RFPL3-expressing cells. 293T cells expressing FLAG-RFPL3 and FLAG-DHFR were infected with a VSV-G-pseudotyped HIV-1 vector, and the cytoplasmic fraction containing PICs was extracted and subjected to immunoprecipitation (IP) using anti-FLAG antibody. Viral DNA in immunoprecipitates was extracted and detected by PCR using LTR R-U5 region-specific primers. The PCR product was visualized on a 1.5% agarose gel through ethidium bromide staining. C, subcellular localization of RFPL3. FLAG-RFPL3-expressing 293T (left column) and parental (right column) cells that had been seeded in a chamber slide were fixed with paraformaldehyde, permeabilized with Triton X-100, and stained with anti-FLAG antibody, followed by detection with Alexa Fluor 488-conjugated secondary antibody (top row). Cell nuclei were stained with DAPI (center row). Merged images are shown in the bottom row. D, single-round HIV-1 luciferase assay on RFPL3-expressing 293T cells. FLAG-RFPL3- and FLAG-DHFR-expressing cells were inoculated with a VSV-G-pseudotyped HIV-1 vector carrying the luciferase reporter gene (HIV-Luc). Cells were lysed 48 h post-infection, and the luciferase activity in the cell lysate was measured, normalized to total protein concentration. The experiment was done in triplicate, and data are expressed as mean relative luciferase units/milligram of protein, with error bars indicating S.D. Statistical significance was determined by Student's t test (*, p < 0.05). E, level of integration in HIV-Luc-infected cells. FLAG-RFPL3- and FLAG-DHFR-expressing 293T cells were exposed with DNase-treated HIV-Luc, and total DNA was isolated 24 h post-infection. The first PCR was performed using specific primers for the Alu element in the human genome and the Renilla luciferase gene in the HIV-Luc genome. A second, nested qPCR was carried out using HIV-1 LTR-specific primers and a fluorogenic probe. Integration in RFPL3 cells relative to DHFR control cells (set to 100%) is shown. Data are expressed as the mean of three independent infections, with error bars indicating S.D., which was confirmed in other independent experiments. Statistical significance was determined by Student's t test (*, p < 0.05).
Cellular localization of RFPL3 was also analyzed through immunofluorescence analysis. FLAG-RFPL3-expressing cells were fixed with paraformaldehyde, followed by permeabilization with Triton X-100 and staining with an anti-FLAG primary antibody and Alexa Fluor 488-conjugated secondary antibody. The immunofluorescence analysis results for 293T cells expressing FLAG-RFPL3 showed that a higher concentration of FLAG-RFPL3 was observed in the cytoplasm, whereas some expression was also found in the nucleus (Fig. 7C, left column). Because the PIC is formed in the cytoplasm of infected cells (2), it is reasonable to assume that cytoplasmic RFPL3 is able to associate with PICs in HIV-1-infected cells.
Finally, we tested whether the increased expression of RFPL3 had an effect on the infectivity of HIV-Luc. FLAG-RFPL3- and FLAG-DHFR-expressing 293T cells were infected with a VSV-G-pseudotyped HIV-1 vector carrying a luciferase gene (HIV-Luc), and luciferase activity was measured 48 h after infection. As shown in Fig. 7D, the mean relative luciferase units detected in RFPL3-expressing cells was 1.46 times higher than that in the DHFR-expressing control cells. Parallel to the level of luciferase activity, increased amounts of integrated DNA, which were detected by a PCR between Alu elements in the human genome and the Renilla luciferase gene in the HIV-Luc genome, were also observed in the FLAG-RFPL3-expressing cells (Fig. 7E).
DISCUSSION
Although the PIC-based in vitro integration assay exhibits full fidelity of in vivo integration (4–6), its technical complication, particularly in the use of radioisotope and Southern blotting analysis, was an impediment to adapting the assay to a high-throughput screening study. However, a development was made by Hansen et al. (17) in which integration activity of HIV-1 PIC could be monitored in target DNA-coated microtiter plates that simplified the in vitro PIC assay. Although the streamlined HIV-1 PIC assay was used initially for the screening of integration inhibitors (17), it holds promise for application in other high-throughput operations. Here, in combination with a library of human E3 ubiquitin ligases generated by a wheat germ cell-free protein synthesis system, we developed an in vitro assay platform to screen for proteins potentially modulating the integration activity of the HIV-1 PIC in a 96-well plate setting.
The wheat germ cell-free system has been shown to permit parallel production of diverse proteins from eukaryotic sources and has been employed to generate libraries of proteins for functional in vitro screening studies (19, 27, 47, 48). In this study, we attempted to produce a set of human E3 ubiquitin ligases using National Institutes of Health mammalian gene correction clones, of which, of 96 cDNAs, 75% of E3 ligases (72 proteins) were confirmed to be synthesized by the wheat germ cell-free system, and they were subsequently purified using glutathione-Sepharose beads. When the protein library was examined to analyze the effect on HIV-1 PIC activity using a microtiter plate-based integration assay, several E3 ubiquitin ligases were shown to enhance or inhibit the in vitro integration activity of PICs (Fig. 2). Because the level of expression varied among proteins (supplemental Fig. 2), we cannot rule out the possibility that other E3 ubiquitin ligases possessing PIC modulation activities might be left out by their poor expression in the wheat germ cell-free reaction, having their effects masked. Although attempts to control for such false negatives by their low yield could be taken into account in future experiments, the strength of the modulation effect exhibited by candidate proteins identified in the screening assay was not likely to be correlated with their level of expression and availability (supplemental Fig. 2).
During the HIV-1 replication cycle, IN has been reported to undergo posttranslational modifications, including ubiquitination, phosphorylation, acetylation, and SUMOylation (34–37, 49–51). Among these major posttranslational modifications, acetylation and phosphorylation have been shown to enhance the DNA affinity or stability of HIV-1 IN, thereby increasing IN enzymatic activity and integration (35, 52, 53). In contrast, it is well known that HIV-1 IN is a metabolically unstable protein and subject to proteasome-mediated degradation via ubiquitination (36). Although IN is likely to escape from the degradation through interaction with other cellular proteins, such as Ku70 (51, 54), the E3 ubiquitin ligase that leads IN into proteasomal degradation remains undetermined (51). This study presented a pilot screening using a limited number of E3 ubiquitin ligases to document the feasibility of proteomic screening for the PIC modulators. However, it has been reported that more than 600 potential RING finger-type ubiquitin ligases are encoded in the human genome (55). Because the wheat germ cell-free system has been used to successfully synthesize nearly 15,000 human proteins in vitro (19), production of a larger collection of proteins by the wheat germ cell-free system would make it feasible for us to perform genome-wide proteomic screening on the microtiter plate-based PIC assay to identify new ubiquitin-proteasome proteolytic factors that drive the degradation of HIV-1 IN in a manner that restricts the integration process. In addition, this screening assay platform would allow us to unveil novel posttranslational modifications that modulate the integration activity of the HIV-1 PIC.
In the initial screening assays using wheat germ cell-free-produced proteins, RFPL3, RNF25, STUB1, and TRIM52 reproducibly enhanced or inhibited in vitro HIV-1 PIC activity (Fig. 2). We next validated the modulating effects of these candidates using E. coli-derived recombinant proteins because the amount of purified protein prepared by a high-throughput expression by the wheat germ cell-free system from cloned ORF resources was limited. Additionally, it was needed to examine whether the effects were specific to wheat germ cell-free-produced proteins. However, a microtiter plate-based integration assay using a fixed concentration of E. coli-derived recombinant proteins revealed that the modulative effect on HIV-1 PIC activity was observed only with RFPL3 (Fig. 3). This inconsistent outcome may be partly explained by the different protein production systems used between the screening (wheat germ cell-free system) and the validation (E. coli-based system) experiments. Eukaryotic cell-free translation systems, including the wheat germ system, have been shown to possess the ability for posttranslational modifications of the target proteins being synthesized, which are deficient in E. coli-based protein production (18, 27, 56, 57). Therefore, when the candidate E3 ubiquitin ligases were synthesized in E. coli cells for the validation assay, RNF25, STUB1, and TRIM52 might not have undergone certain modifications that usually occur in mammalian cells and are required for their functions. In addition, the wheat germ cell-free system has been demonstrated to have a higher capability of generating properly folded eukaryotic proteins than the E. coli-based system (18, 19), which may impact the activity of RNF25, STUB1, and TRIM52 in the inhibition of the HIV-1 PIC integration. Therefore, we cannot entirely exclude RNF25, STUB1, and TRIM52 as potential cellular inhibitors of the PIC. Further analysis will be required for confirmation of the modulative activities of these three E3 ubiquitin ligases in HIV-1 integration.
Nevertheless, through multiple rounds of assays using two different sources of proteins (Figs. 2 and 3), RFPL3 was found to reproducibly enhance the integration activity of the HIV-1 PIC in vitro. RFPL3 is part of the RFPL family (RFPL1, 2, and 3) of proteins that share homology with the Ret finger protein (RFP). RFP, alternatively named TRIM27, is a nuclear protein and belongs to the large B-box RING finger protein family, which has been reported to regulate cell growth and differentiation and is also involved in oncogenicity (58, 59). RFP and RFPL proteins display similarity mainly in the RING finger and the B30.2 domains bridged by a coiled-coil domain, and these domains are believed to be important in mediating protein-protein interactions by promoting homo- or heterodimerization (40). As is well known, the RING domain is the functional domain RING finger E3 ubiquitin ligase, which exhibits binding activity toward E2 ubiquitin-conjugating enzymes to mark proteins for proteasomal degradation (60). The B30.2 domain is comprised of the PRY and SPRY domains that have been identified in 11 different human protein families, but most prominently in TRIM family proteins, including TRIM5α (42). In addition, the RDM is found in RFPL family proteins (Fig. 4A). However, other than high expression of the RFPL1, 2, and 3 genes during neurogenesis in differentiating human embryonic stem cells and in neocortex development (41), the physiological functions and roles of RFPL3 in human cells remain largely unknown.
A cellular localization study performed on 293T cells expressing FLAG-tagged RFPL3 revealed that RFPL3 was likely to be localized mainly in the cytoplasm, where PICs were formed (Fig. 7C). Supporting this evidence, an intracellular association of FLAG-RFPL3 with the HIV-1 PIC was confirmed by coimmunoprecipitation analysis using an anti-FLAG antibody (Fig. 7B). Although the endogenous levels and localization of RFPL3 in human cells remain to be determined because of the absence of a good antibody for RFPL3 detection, similar localization of RFPL family proteins has been shown in a previous study using a pan-hRFPL1,2,3 antibody (41). Therefore, our data suggest that RFPL3 is recruited into the PIC before the translocation to the nucleus in infected cells. More importantly, as measured by luciferase activity, the infection efficiency of single-round HIV-1 was increased significantly by the overexpression of RFPL3 (Fig. 7D), which was likely to coincide with an enhanced level of integrated DNA in infected cells (Fig. 7E). Given the in vitro result of the PIC integration activity, these results suggest that RFPL3 acts to promote the integration process of HIV-1 through association with the PIC in the cytoplasm. Nevertheless, on the basis of the single-round HIV-Luc reporter assay result alone, the functional aspect of RFPL3 in HIV-1 infection is still unclear. In this study, we were unable to use replication-competent HIV-1 to assess the role of RFPL3 in the virus replication cycle because of the lack of biosafety level 3 facilities in our university and restrictions on the use of live HIV in Singapore. However, our future attempts to determine the step in HIV-1 replication responsible for the enhancing effect will point toward a functional role of RFPL3 in virus infection.
One question to ponder would be how RFPL3 modulates the integration activity of HIV-1 PICs. The microtiter plate-based PIC assay using domain mutants of RFPL3 showed that removal of the RING domain resulted in a significant loss of the enhancement of PIC function by WT RFPL3 (Fig. 4C). E3 ligase activity of RFPL3 may be involved in the enhancement of HIV-1 PIC integration activity because this domain is crucial for RING finger-type E3 ubiquitin ligases to interact with E2-conjugating enzymes for subsequent proteasomal degradation of target proteins (61). Interestingly, a mutant with partial truncation of the RING domain (RFPL3 Δ36) was still able to retain a minimal but significant enhancing effect, whereas deletion of the whole extent of the RING domain and the RDM domain almost completely abolished any enhancement (RFPL3 Δ98). This suggests that the C-terminal stretch of the RING domain and/or the RFPL family-specific RDM domain contains the essential region necessary for RFPL3 function on HIV-1 PIC activity.
Our biochemical analysis of using a gel mobility shift assay indicated that RFPL3 is not a DNA-binding protein (Fig. 5). The enhancement effect on in vitro integration by RFPL3 was not detected in the MMLV PIC (Fig. 6). On the basis of these observations, it could be speculated that RFPL3 probably associates with the HIV-1 PIC through a protein component unique to HIV-1 but not present in the MMLV PIC. A preliminary experiment using an AlphaScreen luminescent proximity assay with recombinant proteins (62) showed no in vitro interaction between RFPL3 and HIV-1 IN (data not shown). Although a bona fide target protein (i.e. substrate) of RFPL3 remains unknown and its ubiquitination activity has also yet to be confirmed, RFPL3 may act to remove an unwanted protein component of the HIV-1 PIC that otherwise impedes the subsequent integration reaction. Considering the fact that TRIM5α, which has a domain structure similar to that of RFPL3, blocks HIV-1 replication by associating with the capsid through its PRYSPRY (B30.2) domain and induces disassembly of the capsid lattice (63), it would be intriguing to examine the potential of RFPL3 to recognize the HIV-1 capsid. Alternatively, RFPL3 may stabilize the component of the HIV-1 PIC through ubiquitination for the enhancement of the integration process. In summary, this proteomic approach will facilitate the understanding of molecular interactions between HIV integration complexes and host cells.
Supplementary Material
Acknowledgments
We thank Eric Verdin (Gladstone Institute of Virology and Immunology) for pEV731 and Yoshio Koyanagi (Kyoto University) for pYK005C. We also thank the members of our laboratory for helpful discussions.
This work was supported by National Medical Research Council (Singapore) Grants NMRC/BNIG/1079/2012 (to Y. S.) and NMRC/1273/2010 (to N. Y.) and by National University of Singapore SoM Start-up Grants R-182-000-160-733 and R-182-000-160-133 (to N. Y.).

This article contains supplemental Figs. 1 and 2.
- HIV-1
- human immunodeficiency virus, type 1
- PIC
- preintegration complex
- IN
- integrase
- VSV-G
- vesicular stomatitis virus G protein
- MMLV
- Moloney murine leukemia virus
- qPCR
- quantitative PCR
- IVT
- in vitro transcription
- CBB
- Coomassie Brilliant Blue
- DHFR
- dihydrofolate reductase
- Luc
- luciferase
- RFPL
- Ret finger protein-like
- RDM
- RFPL-defining motif
- FL
- full-length
- RFP
- Ret finger protein.
REFERENCES
- 1. Goff S. P. (2007) Host factors exploited by retroviruses. Nat. Rev. Microbiol. 5, 253–263 [DOI] [PubMed] [Google Scholar]
- 2. Suzuki Y., Craigie R. (2007) The road to chromatin: nuclear entry of retroviruses. Nat. Rev. Microbiol. 5, 187–196 [DOI] [PubMed] [Google Scholar]
- 3. Bushman F. D., Craigie R. (1991) Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc. Natl. Acad. Sci. U.S.A. 88, 1339–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bowerman B., Brown P. O., Bishop J. M., Varmus H. E. (1989) A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 3, 469–478 [DOI] [PubMed] [Google Scholar]
- 5. Farnet C. M., Haseltine W. A. (1990) Integration of human immunodeficiency virus type 1 DNA in vitro. Proc. Natl. Acad. Sci. U.S.A. 87, 4164–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fujiwara T., Mizuuchi K. (1988) Retroviral DNA integration: structure of an integration intermediate. Cell 54, 497–504 [DOI] [PubMed] [Google Scholar]
- 7. Arribas J. R., Eron J. (2013) Advances in antiretroviral therapy. Curr. Opin. HIV AIDS 8, 341–349 [DOI] [PubMed] [Google Scholar]
- 8. Al-Mawsawi L. Q., Neamati N. (2007) Blocking interactions between HIV-1 integrase and cellular cofactors: an emerging anti-retroviral strategy. Trends Pharmacol. Sci. 28, 526–535 [DOI] [PubMed] [Google Scholar]
- 9. Lewinski M. K., Bushman F. D. (2005) Retroviral DNA integration: mechanism and consequences. Adv. Genet. 55, 147–181 [DOI] [PubMed] [Google Scholar]
- 10. Farnet C. M., Haseltine W. A. (1991) Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol. 65, 1910–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bukrinsky M. I., Sharova N., McDonald T. L., Pushkarskaya T., Tarpley W. G., Stevenson M. (1993) Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. U.S.A. 90, 6125–6129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li L., Olvera J. M., Yoder K. E., Mitchell R. S., Butler S. L., Lieber M., Martin S. L., Bushman F. D. (2001) Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20, 3272–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miller M. D., Farnet C. M., Bushman F. D. (1997) Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71, 5382–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Farnet C. M., Bushman F. D. (1997) HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell 88, 483–492 [DOI] [PubMed] [Google Scholar]
- 15. Chen H., Engelman A. (1998) The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc. Natl. Acad. Sci. U.S.A. 95, 15270–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Turlure F., Devroe E., Silver P. A., Engelman A. (2004) Human cell proteins and human immunodeficiency virus DNA integration. Front. Biosci. 9, 3187–3208 [DOI] [PubMed] [Google Scholar]
- 17. Hansen M. S., Smith G. J., 3rd, Kafri T., Molteni V., Siegel J. S., Bushman F. D. (1999) Integration complexes derived from HIV vectors for rapid assays in vitro. Nat. Biotechnol. 17, 578–582 [DOI] [PubMed] [Google Scholar]
- 18. Madono M., Sawasaki T., Morishita R., Endo Y. (2011) Wheat germ cell-free protein production system for post-genomic research. N. Biotechnol. 28, 211–217 [DOI] [PubMed] [Google Scholar]
- 19. Goshima N., Kawamura Y., Fukumoto A., Miura A., Honma R., Satoh R., Wakamatsu A., Yamamoto J., Kimura K., Nishikawa T., Andoh T., Iida Y., Ishikawa K., Ito E., Kagawa N., Kaminaga C., Kanehori K., Kawakami B., Kenmochi K., Kimura R., Kobayashi M., Kuroita T., Kuwayama H., Maruyama Y., Matsuo K., Minami K., Mitsubori M., Mori M., Morishita R., Murase A., Nishikawa A., Nishikawa S., Okamoto T., Sakagami N., Sakamoto Y., Sasaki Y., Seki T., Sono S., Sugiyama A., Sumiya T., Takayama T., Takayama Y., Takeda H., Togashi T., Yahata K., Yamada H., Yanagisawa Y., Endo Y., Imamoto F., Kisu Y., Tanaka S., Isogai T., Imai J., Watanabe S., Nomura N. (2008) Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat. Methods 5, 1011–1017 [DOI] [PubMed] [Google Scholar]
- 20. Jordan A., Defechereux P., Verdin E. (2001) The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20, 1726–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kawano Y., Yoshida T., Hieda K., Aoki J., Miyoshi H., Koyanagi Y. (2004) A lentiviral cDNA library employing λ recombination used to clone an inhibitor of human immunodeficiency virus type 1-induced cell death. J. Virol. 78, 11352–11359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee M. S., Craigie R. (1994) Protection of retroviral DNA from autointegration: involvement of a cellular factor. Proc. Natl. Acad. Sci. U.S.A. 91, 9823–9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Suzuki Y., Ogawa K., Koyanagi Y., Suzuki Y. (2010) Functional disruption of the Moloney murine leukemia virus preintegration complex by vaccinia-related kinases. J. Biol. Chem. 285, 24032–24043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suzuki Y., Misawa N., Sato C., Ebina H., Masuda T., Yamamoto N., Koyanagi Y. (2003) Quantitative analysis of human immunodeficiency virus type 1 DNA dynamics by real-time PCR: integration efficiency in stimulated and unstimulated peripheral blood mononuclear cells. Virus Genes 27, 177–188 [DOI] [PubMed] [Google Scholar]
- 25. Sawasaki T., Hasegawa Y., Tsuchimochi M., Kamura N., Ogasawara T., Kuroita T., Endo Y. (2002) A bilayer cell-free protein synthesis system for high-throughput screening of gene products. FEBS Lett. 514, 102–105 [DOI] [PubMed] [Google Scholar]
- 26. Sawasaki T., Gouda M. D., Kawasaki T., Tsuboi T., Tozawa Y., Takai K., Endo Y. (2005) The wheat germ cell-free expression system: methods for high-throughput materialization of genetic information. Methods Mol. Biol. 310, 131–144 [DOI] [PubMed] [Google Scholar]
- 27. Takahashi H., Nozawa A., Seki M., Shinozaki K., Endo Y., Sawasaki T. (2009) A simple and high-sensitivity method for analysis of ubiquitination and polyubiquitination based on wheat cell-free protein synthesis. BMC Plant Biol. 9, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sawasaki T., Ogasawara T., Morishita R., Endo Y. (2002) A cell-free protein synthesis system for high-throughput proteomics. Proc. Natl. Acad. Sci. U.S.A. 99, 14652–14657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee M. S., Craigie R. (1998) A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. U.S.A. 95, 1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takahashi H., Takahashi C., Moreland N. J., Chang Y. T., Sawasaki T., Ryo A., Vasudevan S. G., Suzuki Y., Yamamoto N. (2012) Establishment of a robust dengue virus NS3-NS5 binding assay for identification of protein-protein interaction inhibitors. Antiviral Res. 96, 305–314 [DOI] [PubMed] [Google Scholar]
- 31. Takahashi H., Ozawa A., Nemoto K., Nozawa A., Seki M., Shinozaki K., Takeda H., Endo Y., Sawasaki T. (2012) Genome-wide biochemical analysis of Arabidopsis protein phosphatase using a wheat cell-free system. FEBS Lett. 586, 3134–3141 [DOI] [PubMed] [Google Scholar]
- 32. Liszewski M. K., Yu J. J., O'Doherty U. (2009) Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods 47, 254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shimura K., Kodama E., Sakagami Y., Matsuzaki Y., Watanabe W., Yamataka K., Watanabe Y., Ohata Y., Doi S., Sato M., Kano M., Ikeda S., Matsuoka M. (2008) Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137). J. Virol. 82, 764–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mulder L. C., Muesing M. A. (2000) Degradation of HIV-1 integrase by the N-end rule pathway. J. Biol. Chem. 275, 29749–29753 [DOI] [PubMed] [Google Scholar]
- 35. Mousnier A., Kubat N., Massias-Simon A., Ségéral E., Rain J. C., Benarous R., Emiliani S., Dargemont C. (2007) von Hippel Lindau binding protein 1-mediated degradation of integrase affects HIV-1 gene expression at a postintegration step. Proc. Natl. Acad. Sci. U.S.A. 104, 13615–13620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Devroe E., Engelman A., Silver P. A. (2003) Intracellular transport of human immunodeficiency virus type 1 integrase. J. Cell Sci. 116, 4401–4408 [DOI] [PubMed] [Google Scholar]
- 37. Manganaro L., Lusic M., Gutierrez M. I., Cereseto A., Del Sal G., Giacca M. (2010) Concerted action of cellular JNK and Pin1 restricts HIV-1 genome integration to activated CD4+ T lymphocytes. Nat. Med. 16, 329–333 [DOI] [PubMed] [Google Scholar]
- 38. Mulder L. C., Chakrabarti L. A., Muesing M. A. (2002) Interaction of HIV-1 integrase with DNA repair protein hRad18. J. Biol. Chem. 277, 27489–27493 [DOI] [PubMed] [Google Scholar]
- 39. Sawasaki T., Hasegawa Y., Morishita R., Seki M., Shinozaki K., Endo Y. (2004) Genome-scale, biochemical annotation method based on the wheat germ cell-free protein synthesis system. Phytochemistry 65, 1549–1555 [DOI] [PubMed] [Google Scholar]
- 40. Seroussi E., Kedra D., Pan H. Q., Peyrard M., Schwartz C., Scambler P., Donnai D., Roe B. A., Dumanski J. P. (1999) Duplications on human chromosome 22 reveal a novel Ret finger protein-like gene family with sense and endogenous antisense transcripts. Genome Res. 9, 803–814 [DOI] [PubMed] [Google Scholar]
- 41. Bonnefont J., Nikolaev S. I., Perrier A. L., Guo S., Cartier L., Sorce S., Laforge T., Aubry L., Khaitovich P., Peschanski M., Antonarakis S. E., Krause K. H. (2008) Evolutionary forces shape the human RFPL1,2,3 genes toward a role in neocortex development. Am. J. Hum. Genet. 83, 208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ozato K., Shin D. M., Chang T. H., Morse H. C., 3rd. (2008) TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8, 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Llano M., Vanegas M., Fregoso O., Saenz D., Chung S., Peretz M., Poeschla E. M. (2004) LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 78, 9524–9537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Raghavendra N. K., Shkriabai N., Graham R. L., Hess S., Kvaratskhelia M., Wu L. (2010) Identification of host proteins associated with HIV-1 preintegration complexes isolated from infected CD4+ cells. Retrovirology 7, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Isomura T., Tamiya-Koizumi K., Suzuki M., Yoshida S., Taniguchi M., Matsuyama M., Ishigaki T., Sakuma S., Takahashi M. (1992) RFP is a DNA binding protein associated with the nuclear matrix. Nucleic Acids Res. 20, 5305–5310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zheng R., Ghirlando R., Lee M. S., Mizuuchi K., Krause M., Craigie R. (2000) Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc. Natl. Acad. Sci. U.S.A. 97, 8997–9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miyakawa K., Sawasaki T., Matsunaga S., Tokarev A., Quinn G., Kimura H., Nomaguchi M., Adachi A., Yamamoto N., Guatelli J., Ryo A. (2012) Interferon-induced SCYL2 limits release of HIV-1 by triggering PP2A-mediated dephosphorylation of the viral protein Vpu. Sci. Signal. 5, ra73. [DOI] [PubMed] [Google Scholar]
- 48. Mizutani Y., Matsuoka K., Takeda H., Shiogama K., Inada K., Hayakawa K., Yamada H., Miyazaki T., Sawasaki T., Endo Y., Tsutsumi Y. (2013) Novel approach to identifying autoantibodies in rheumatoid synovitis with a biotinylated human autoantigen library and the enzyme-labeled antigen method. J. Immunol. Methods 387, 57–70 [DOI] [PubMed] [Google Scholar]
- 49. Allouch A., Di Primio C., Alpi E., Lusic M., Arosio D., Giacca M., Cereseto A. (2011) The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe 9, 484–495 [DOI] [PubMed] [Google Scholar]
- 50. Zamborlini A., Coiffic A., Beauclair G., Delelis O., Paris J., Koh Y., Magne F., Giron M. L., Tobaly-Tapiero J., Deprez E., Emiliani S., Engelman A., de Thé H., Saïb A. (2011) Impairment of human immunodeficiency virus type-1 integrase SUMOylation correlates with an early replication defect. J. Biol. Chem. 286, 21013–21022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zheng Y., Yao X. (2013) Posttranslational modifications of HIV-1 integrase by various cellular proteins during viral replication. Viruses 5, 1787–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cereseto A., Manganaro L., Gutierrez M. I., Terreni M., Fittipaldi A., Lusic M., Marcello A., Giacca M. (2005) Acetylation of HIV-1 integrase by p300 regulates viral integration. EMBO J. 24, 3070–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Terreni M., Valentini P., Liverani V., Gutierrez M. I., Di Primio C., Di Fenza A., Tozzini V., Allouch A., Albanese A., Giacca M., Cereseto A. (2010) GCN5-dependent acetylation of HIV-1 integrase enhances viral integration. Retrovirology 7, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zheng Y., Ao Z., Wang B., Jayappa K. D., Yao X. (2011) Host protein Ku70 binds and protects HIV-1 integrase from proteasomal degradation and is required for HIV replication. J. Biol. Chem. 286, 17722–17735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li W., Bengtson M. H., Ulbrich A., Matsuda A., Reddy V. A., Orth A., Chanda S. K., Batalov S., Joazeiro C. A. (2008) Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PloS ONE 3, e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Katzen F., Chang G., Kudlicki W. (2005) The past, present and future of cell-free protein synthesis. Trends Biotechnol. 23, 150–156 [DOI] [PubMed] [Google Scholar]
- 57. Carraher C., Nazmi A. R., Newcomb R. D., Kralicek A. (2013) Recombinant expression, detergent solubilisation and purification of insect odorant receptor subunits. Protein Expr. Purif. 90, 160–169 [DOI] [PubMed] [Google Scholar]
- 58. Cao T., Duprez E., Borden K. L., Freemont P. S., Etkin L. D. (1998) Ret finger protein is a normal component of PML nuclear bodies and interacts directly with PML. J. Cell Sci. 111, 1319–1329 [DOI] [PubMed] [Google Scholar]
- 59. Shimono Y., Murakami H., Hasegawa Y., Takahashi M. (2000) RET finger protein is a transcriptional repressor and interacts with enhancer of polycomb that has dual transcriptional functions. J. Biol. Chem. 275, 39411–39419 [DOI] [PubMed] [Google Scholar]
- 60. Lorick K. L., Jensen J. P., Fang S., Ong A. M., Hatakeyama S., Weissman A. M. (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 96, 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Metzger M. B., Hristova V. A., Weissman A. M. (2012) HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 125, 531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hou Y., McGuinness D. E., Prongay A. J., Feld B., Ingravallo P., Ogert R. A., Lunn C. A., Howe J. A. (2008) Screening for antiviral inhibitors of the HIV integrase-LEDGF/p75 interaction using the AlphaScreen luminescent proximity assay. J. Biomol. Screen. 13, 406–414 [DOI] [PubMed] [Google Scholar]
- 63. Grütter M. G., Luban J. (2012) TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling. Curr. Opin. Virol. 2, 142–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.