Background: The individual role of members of the poly(ADP-ribose) polymerase family is unclear.
Results: PARP12 displays a dual subcellular localization and effector function, controlling both protein translation and NF-κB signaling.
Conclusion: PARP12 mediates two important effector mechanisms linked to the establishment of an anti-viral state.
Significance: ADP-ribosylation may play an important role in the innate control of microbial infections.
Keywords: ADP-ribosylation, Interferon, NF-κB (NF-κB), Stress Granule, Translation Regulation, Interferon-stimulated Gene (ISG), Poly(ADP-ribose) Polymerase (PARP)
Abstract
Transcriptome analyses have recently identified PARP12, a member of a large family of ADP-ribosyl transferases, as an interferon-induced gene (ISG), whose function remains incompletely characterized. We demonstrate herein that PARP12 is a genuine ISG, whose expressed protein displays at least two distinct subcellular locations and related functions. Upon ectopic expression or exposure to oxidative stress, PARP12 is recruited to stress-granules (SGs), known sites of mRNA translational arrest. Accordingly, PARP12 was found to block mRNA translation, possibly upon association to the translational machinery. Both the N-terminal domain (containing an RNA-binding domain characterized by the presence of five CCCH-type Zn-fingers) and integrity of the catalytic domain are required for this suppressive function. In contrast, stimulation with LPS leads to the localization of PARP12 to p62/SQSTM1 (an adaptor protein involved in innate signaling and autophagy) containing structures, unrelated to SGs. Deletion of the N-terminal domain promotes the association of the protein to p62/SQSTM1, suggesting that the RNA-binding domain is responsible for the subcellular localization of PARP12. Association to p62/SQSTM1 was found to correlate with increased NF-κB signaling, suggesting a role for PARP12 in inflammation. Collectively, these observations suggest that PARP12 can alternate between two distinct subcellular compartments associated to two distinct cellular functions. The present work therefore identifies PARP12 as an ISG with a potential role in cellular defenses against viral infections.
Introduction
Poly(ADP-ribose) polymerases (PARPs)3 represent a family of enzymes present in almost all nucleated cells of mammals, plants, lower eukaryotes (with the notable exception of yeasts), as well as eubacteria, archaebacteria, and double-stranded DNA viruses (1, 2). PARPs catalyze the transfer of negatively charged ADP-ribose subunits to target proteins using NAD+ as a substrate. PARP1, the founding member of this family, is an abundant nuclear protein catalyzing the polymerization of multiple ADP-ribose units on target proteins, resulting in the synthesis of large, linear or branched, polymers that can profoundly affect protein function and/or location (3). In addition to its widespread expression among living organisms, the reversible nature of this post-translational modification suggests an important regulatory role for this family of proteins.
Originally thought to be involved solely in cellular responses to DNA injury (recruiting DNA repair and checkpoint proteins to sites of damage), PARP1 has been recently demonstrated to play a role in a diverse set of biological processes including chromatin modification, transcription regulation, and cell death (2, 4, 5). Although PARP1 is an abundant and catalytically very active protein possibly accounting for over 90% of the overall cellular PARP activity, several proteins sharing a similar catalytic domain (referred to as the “PARP” signature) have been identified, in particular in the human and murine genomes (6). Despite the presence of a common PARP motif, altered catalytic sites have been identified in several members of the family, suggesting a wider range of enzymatic activities than previously anticipated (7). Recent experimental evidence using recombinant and/or purified forms of several members of this family have confirmed this functional heterogeneity. These studies have experimentally confirmed that while PARP-1 and PARP-2 represent bona fide poly(ADP-ribose) polymerases, other members, such as PARP-10 and PARP14 catalyze the transfer of a single ADP-ribose moiety to an acceptor protein, and should therefore be considered as mono-ADP-ribose transferases (8). Sequence comparison analyses and mutagenesis studies have led to the identification of a key glutamate residue in the PARP1 catalytic domain (Glu988) required for the enzymatic polymerase activity of PARP1 (9). Notably, this residue is missing in PARP10, an observation that recently led several authors to propose an alternative classification of PARP family members based on sequence homology and enzymatic (poly versus mono ADP-ribosyl transferase) activity (7). Accordingly, several PARP members (PARP-3, -6, -7, -8, -10, -11, -12, -14, -15, and -16) have been tentatively re-classified as mono-ADP ribosyltransferases, while others (PARP-1, -2, and -4) are presently considered as poly-ADP-ribosyl transferases. Of note, two members of this family (PARP-9 and -13), appear to lack any enzymatic activity. Despite the presence of a glutamate in the catalytic domain (Glu514), PARP-3 only expresses mono-ADP-ribosylating capacities (10), indicating thus that the active-site glutamate is not the sole feature that determines poly-ADP-ribosyl transferase activity. It is worth stressing however that the classification of PARP members in either mono or poly-ADP-ribosyl transferases is hampered by the lack of knowledge of their natural substrate(s) and often relies of the analysis of the transferred ADP-ribosyl moiety during an in vitro automodification reaction.
In any event, the identification of a new set of intracellular proteins possibly endowed with mono-ADP-ribosyl transfer capacities is of particular interest. Indeed, until recently, only extracellular enzymes and bacterial toxins were thought to-mono-ADP-ribosylate cellular proteins, with important functional consequences. Despite evidence of endogenous mono-ADP-ribosylation of several of these substrates, the cellular proteins responsible for these post-translational modifications have not been clearly identified. It is therefore tempting to speculate that some of the previously described PARP members may in fact be the endogenous proteins that trans-ADP-ribosylate cellular substrates previously identified as bacterial toxin targets.
In addition to these diversified catalytic properties, members of the PARP superfamily are multi-modular proteins possessing a wide variety of domains including zinc fingers, breast cancer-1 protein (BRCA1) C terminus-like (BRCT) motifs, ankyrin repeats, macro domains, and WWE domains, compatible with their role in a wide range of biological responses (2, 4, 5).
The aim of the present study was to further investigate the potential biological role of PARP12, a member of the PARP family expressing a typical “mono-ADP-ribosyl-transferase” active site signature. PARP12 has been recently identified as a putative anti-viral gene, belonging to a large family of interferon-stimulated genes (ISGs) whose expression is often induced during viral infections (11, 12). More recently, expression of PARP12 was also found elevated in tissues from mice subjected to bacterial superantigen-(SEB) mediated toxic shock (13), suggesting a potential role of this protein during immune activation. PARP12 belongs to a subgroup of PARP family members characterized by the presence of typical CCCH zinc fingers known to bind to viral (14, 15), but also cytoplasmic RNAs (16, 17). Accordingly, PARP12 has been recently found to accumulate in cytoplasmic stress granules known to regulate mRNA translation and stability in response to stress (18), and to inhibit viral translation by directly binding to polysomes of Venezuelan equine encephalitis infected cells (19). The present study was undertaken to better understand the potential biological role of the functional protein domains of PARP12. We confirm herein that PARP12 is a mono-ADP-ribosyl-transferase with auto-modification activity, whose expression appears to be tightly controlled by the type I IFN pathway. Unlike many PARP members, PARP12 is largely excluded from the nucleus, and appears to localize into distinct cytoplasmic structures in a protein domain-dependent fashion, revealing a possible complex role for this protein during viral infection.
EXPERIMENTAL PROCEDURES
Reagents, Antibodies, and Mice
3-Aminobenzamide (Sigma-Aldrich), NAD (Sigma), [32P]NAD (Amersham Biosciences), Lipopolysaccharide from Escherichia coli 0111:B4 (Enzo Life Sciences), CpG ODN 1826 (Invivogen), poly(I:C) (Sigma-Aldrich), recombinant interferon-β (R&D Systems) were purchased from their respective manufacturers. The rabbit polyclonal anti-actin, anti-p62/SQSTM1, and mouse monoclonal anti-Flag M2 were purchased from Sigma-Aldrich. The rabbit polyclonal anti-PARP1 and anti-eIF3η (sc-16377), mouse monoclonal anti-HSP90, and anti-ubiquitin (clone P4D1) and goat polyclonal anti-vimentin (C-20) were purchased from Santa Cruz Biotechnology. The anti-poly(ADP-ribose) 10H antibody has been described elsewhere (20). The IFNAR−/− mice and wild-type controls were from (B&K Universal). Sodium arsenite was from Sigma-Aldrich. Alexa Fluor secondary antibodies were from Invitrogen.
Constructs
The murine PARP12 cDNA was cloned into the pCMV-SPORT6 vector using the EcoRI/XhoI sites. The Flag-tag was added by PCR. Point mutations in the PARP12 cDNA (C104H, C205H-H208R, H208R, G575W, G575P, G575R, and H574E) were generated by site-directed PCR-mutagenesis. PARP12ΔZnF was generated by two PCR reactions with primers amplifying the 5′ and 3′ fragments of PARP12. The YFP-PARP12 fusion proteins were generated by cloning different PARP12 constructs into the pEYFP-C1 vector using the EcoRI/SalI sites. Myc-PARP12, PARP12-G575W, and PARP12ΔZnF were generated by PCR from their respective templates and re-cloned into the pCMV-SPORT6 inbetween the EcoRI/XhoI sites. The PARP12-MS2 fusion constructs were generated by cloning PARP12 into the pcNMS2 vector (21) between the XhoI/XbaI sites. Murine Flag-tagged TRIF and RIPK1 were amplified by PCR and respectively cloned into the pCMV-SPORT6 vector between the EcoRI/XhoI and AgeI/XhoI sites. CFP-TIAR, RFP-Dcp1a, YFP-CIRP, the bidirectional Renilla/Firefly luciferase constructs containing 0 or 8 MS2-loops and the NFKB-Luciferase reporter construct have been described elsewhere (22–25). The pGL4.74 plasmid was from Clontech.
Generation of the MP12 Anti-PARP12 Antibody
His6-tagged full-length mPARP12 was expressed in E. coli (SE1) using the pSCodon1 vector from Delphi Genetics. The bacteria were lysed using a French press in 6 m guanidine, and the recombinant protein was purified by affinity chromatography using HisPur Cobalt Resin (Pierce) and eluted with imidazole. Recombinant PARP12 was dialyzed overnight against 20 mm acetic acid, and BALB/C mice were immunized three times with 20 μg of protein at a 2-week interval, successively in CFA, then IFA and finally in PBS. Mice were given a booster dose (20 μg protein in PBS) 3 days before the fusion of spleen cells with SP2/O myeloma cells in presence of PEG 1500 (Roche). Hybridomas producing specific antibodies recognizing PARP12 were screened by ELISA on plates coated with the recombinant protein and cloned by limiting dilutions. The selected antibody (clone MP12) recognizes murine PARP12 by ELISA, Western blot, immunofluorescence, and FACS. The antibody was purified with Melon Gel (Pierce) and coupled to Alexa Fluor 488 (Invitrogen) according to manufacturer's indications.
Cell Culture and Transfection
HEK293T (ATCC, CRL-11268), HeLa, and RAW264.7 (ATCC, TIB-71) were grown at 37 °C and 5% CO2 in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with fetal bovine serum to a final concentration of 10%, 1 mm sodium pyruvate and non-essential amino acids (MEM NEAA, Invitrogen). HEK293T cells were transfected with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions.
Purification of Dendritic Cells
Splenic dendritic cells were purified from mice as previously described (26). Briefly, spleen cells were separated into low and high density fractions on a Nycodenz gradient (Nycomed). The low density fraction was separated by MACS (Miltenyi Biotec) according to CD11c expression by incubation with anti-CD11c-coupled microbeads. Immature dendritic cells were immediately lysed in the appropriate buffer, while the mature cells were cultured overnight in RPMI supplemented with 10% FCS and different TLR ligands.
PARP Activity Assay
PARP12 was purified from transfected HEK293T cells by immunoprecipitation with the anti-Flag M2 antibody. The Sepharose beads were washed twice with PARP reaction buffer (50 mm Tris-HCL, pH 8.0, 4 mm MgCl2, and 200 μm DTT, dithiothreitol) with or without 3-aminobenzamide. The beads were incubated in the PARP reaction buffer for 30 min at 30 °C in presence of 5 μCi of [32P]NAD and various concentrations of cold NAD. The beads were then washed to eliminate the unbound radioactivity, the proteins eluted with Tris-glycine SDS Sample Buffer (Invitrogen) and loaded onto a Tris-glycine 8% gel (Invitrogen). After SDS-PAGE, the gels were fixed, dehydrated, and the radioactive signals were detected by autoradiography.
SDS-PAGE and Western Blot
Cells were lysed in RIPA buffer (PBS with 1% Nonidet P-40, 0.5% Na deoxycholate and 0.1% SDS) containing protease inhibitors (complete Mini Protease Inhibitor Mixture Tablet, Roche). Protein concentration was measured with the Micro BCA Protein Assay kit (Pierce) and 20–40 μg of protein was loaded onto a 12% Bis-Tris polyacrylamide gel. Gels were run in NuPAGE MOPS SDS Running Buffer (Invitrogen) at 150 V for 1 h. Proteins were transferred onto a PVDF membrane (Amersham Biosciences) for 1 h 45 min at 125 V, blocked in 5% BSA and detected with appropriate antibodies. The bands were quantified with GeneTools software (Syngene).
RNA Extraction and qPCR
RNA was extracted using Trizol Reagent (Invitrogen) following manufacturer's instructions. Plasmid DNA was removed from RNA extracted from transfected cells by washing the RNA pellet with a solution containing one volume of DNAzol (Invitrogen) and 0.3 volumes of ethanol after isopropanol precipitation. The RNA was then washed twice in 100% ethanol and resuspended in 20 μl of ultrapure water. Genomic DNA was removed by treating extracted RNA with DNase I (Roche) for 30 min at 37 °C. The RNA was retrotranscribed into cDNA using the SuperScript II RT (Invitrogen), and the DNA was quantified by SYBR Green qPCR (Fermentas) on a StepOneplus real type PCR system (Applied Biosystems). The quantities of endogenous DNA were normalized to the corresponding quantities of ribosomal protein L32 (RPL32). The quantities of Renilla luciferase (R.Luc) mRNA were normalized to the corresponding quantities of Firefly luciferase (F.Luc) mRNA. The primers were designed on separate exons when possible using the primer3 software.
Immunofluorescence
Cells were grown on sterile cover slips. Following stress granule induction with sodium arsenite (1 mm for 30 min), cells were washed twice in PBS, fixed in 4% paraformaldehyde, then permeabilized in −20 °C methanol for 10 min at −20 °C. The cells were slowly rehydrated in PBS, blocked in PBS containing 10% BSA (1 h) and incubated with the primary antibodies (1 h). The cells were then washed three times in PBS containing 0.1% Tween 20 and incubated with the secondary antibodies. Finally, the cells were washed again in PBS-Tween, rinsed rapidly in distilled water and mounted on slides in fluorescence mounting medium (Dako) containing 100 ng/ml of DAPI.
Luciferase and ELISA Assays
Cell lysis and luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega) following manufacturer's instructions. The luminescence was measured using a TD-20/20 luminometer (Turner Designs) with at least three independent experiments. Statistical analyses were performed with GraphPad Prism software. Upon transfection with WT and mutant PARP12 constructs, HEK293T cells were stimulated with recombinant human TNF (10 ng/ml) and, after 16 h of culture, IL-8 accumulation in the supernatant was assayed by ELISA (R&D Human CXCL8/IL-8 Quantikine ELISA Kit) following manufacturer's instructions.
Sucrose Gradient Polysome Fractionation
4,8 × 106 HEK293T cells or 12 × 106 RAW264.7 were cultured in 175 cm2 flasks. 5 min after the addition of cycloheximide, the cells were recovered and centrifuged at 400 × g for 7 min, then resuspended in 180 μl of lysis buffer (25 mm HEPES pH 6.9, 100 mm KCl, 5 mm MgCl2, 100 units/ml RNaseOUT (Invitrogen), 10 μg/ml cycloheximide, 0.5% Nonidet P-40, and 1 μg/ml heparin). The lysates were cleared by centrifugation at 13,000 rpm (10 min) and loaded onto linear 15–50% sucrose gradients prepared in sucrose buffer (25 mm HEPES pH 6.9, 100 mm KCl, 5 mm MgCl2, and 10 μg/ml cycloheximide). Gradients were separated by ultracentrifugation at 39,000 × g for 2 h using a Beckman SW41 rotor. The fractions were recovered using a Brandel BR-188 Density Gradient Fractionation System. Proteins were extracted from each fraction as previously described (27). For extraction of RNA, 250 μl of Trizol (Invitrogen) was added to 200 μl of each fraction, and the manufacturer's instructions were followed. The RNA was then loaded onto a 1% agarose gel and visualized after ethidium bromide staining.
Immunoprecipitation
Proteins were immunoprecipitated from cell lysates with μMACS anti-c-myc MicroBeads (Miltenyi Biotech) following manufacturers indications. Briefly, cells were lysed in low-salt lysis buffer (1% Nonidet P-40 and 50 mm Tris-HCl pH 8.0) containing protease inhibitors (cOmplete Mini Protease Inhibitor Mixture Tablet, Roche). The lysate was cleared by centrifugation (14,000 × g, 15min at 4 °C) and incubated with 40 μl of anti-c-myc beads for 1 h on ice. After the incubation, the cell lysate was applied to a μ-column, washed four times, and eluted with 50 μl pre-heated (95 °C) Elution Buffer (50 mm TrisHCl, pH 6.8, 50 mm DTT, 1% SDS, 1 mm EDTA, 0.005% bromphenol blue, 10% glycerol). The eluate was analyzed by SDS-PAGE.
RESULTS
PARP12 Is An Interferon-stimulated Gene (ISG) Coding for a Mono-ADP-ribosyl-transferase
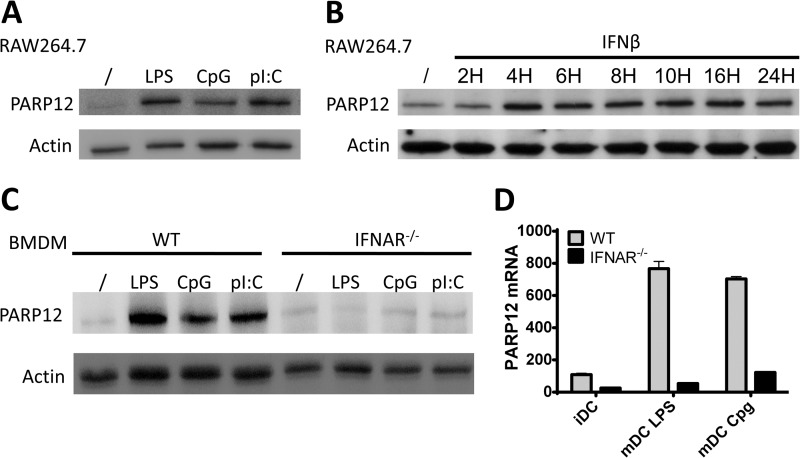
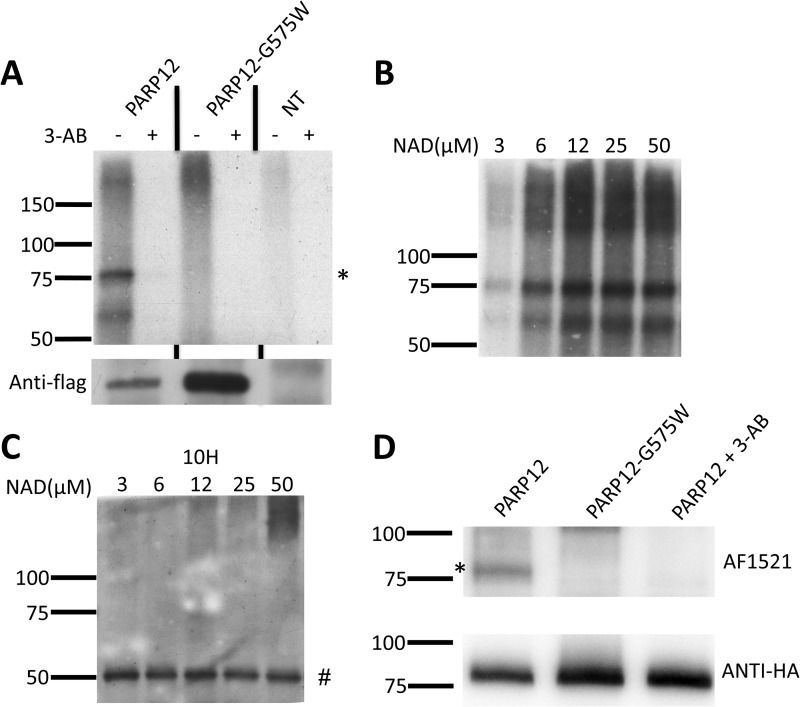
A monoclonal antibody specific to murine PARP12 was generated to monitor expression of this PARP member under different experimental conditions (see “Experimental Procedures” for details). In keeping with previously published observations and microarray data (28), a murine macrophage-like cell line (RAW264.7 cells) was found to constitutively express detectable, albeit weak levels of PARP12, that could be further up-regulated in response to toll-like receptor (TLR) ligands such as LPS (TLR4) poly(I:C) (TLR3) and CpG DNA (TLR9, Fig. 1A). Analyses of the 1000-base pair region upstream of the transcription initiation site of mPARP12 revealed several putative IRF-binding and ISRE (interferon-stimulated response element) sites (data not shown), suggesting a potential major role for type I IFNs in regulating PARP12 expression. Accordingly, a kinetic analysis revealed a rapid (4 h) and sustained (up to 24 h post-stimulation) up-regulation of PARP12 expression in response to IFNβ (Fig. 1B). To evaluate the role of type I IFNs in regulating PARP12 expression in response to TLR ligands, bone marrow-derived macrophages were generated and splenic dendritic cells were purified from wild-type or interferon-α/β receptor1 (IFNAR1) knock-out mice and stimulated overnight with LPS, CpG or polyIC. Cells lacking type I IFN receptors failed to upregulate PARP12 protein and mRNA expression in response to all TLR ligand tested (Fig. 1, C and D), indicating that, at least for murine innate cells, PARP12 is a genuine interferon stimulated gene or ISG. Finally, the capacity of PARP12 to express ADP-ribose transferase catalytic activity was confirmed using a series of in vitro assays using [32P]NAD and a recombinant form of the Af1521 protein from the archaebacteria Archaeoglobus fulgidus, possessing an ADP-ribose recognizing module (termed macro domain) know to bind to both free and protein-bound mono and poly-ADP-ribose moieties (29, 30) (Fig. 2). This assay also showed that the putative PARP12 variant, generated by mutating the glycine of the R/H-G-T/S motif into a tryptophan (G575W, see Fig. 4A), lacked enzymatic activity. Note also that the lack of a mobility shift usually detected by the formation of high molecular weight poly(ADP-ribose) polymers on genuine poly(ADP-ribose) polymerases (PARP1, PARP2, and PARP5a/b) (31–33), even in the presence of increasing doses of NAD (Fig. 2B) confirmed that as previously shown for PARP10 (8), PARP12 is a mono-ADP-ribosyl transferase.
FIGURE 1.
Expression of PARP12 is type-I interferon dependent. A, RAW264.7 macrophages were left untreated or activated for 8 h with LPS (10 μg/ml), CpG (100 ng/ml), or polyI:C (1 μg/ml), and PARP12 protein expression was detected by Western blot with the MP12 antibody. Actin was blotted as loading control. B, RAW264.7 cells were stimulated with IFNβ for the indicated time and expression of PARP12 was detected as in A. C, expression of the PARP12 protein in WT and IFNAR−/− BMDM untreated or activated for 8 with LPS (10 μg/ml) CpG (100 ng/ml) or polyI:C (1 μg/ml). D, relative accumulation of PARP12 mRNA in WT and IFNAR−/− splenic dendritic cells activated overnight with TLR ligands. Error bars represent the standard deviation of experimental duplicates. The data are representative of three independent experiments.
FIGURE 2.
PARP12 is a bona fide ADP-ribosyl transferase and with automodification activity in vitro. A, Flag-tagged PARP12, and the catalytic mutant PARP12-G575W were immunoprecipitated (IP) and submitted to ADP-ribosylation reaction in vitro in the presence of [32P]NAD and in the presence or absence of the PARP inhibitor, 3-aminobenzamide (3-AB). The products were analyzed by SDS-PAGE followed by autoradiography (left) or by anti-Flag Western blot as IP control (bottom). A band corresponding to automodified PARP12 (*) was visible at the expected molecular weight. This band was absent when a pan-PARP-inhibitor (3-AB) was added during the in vitro PARP-reaction, or when cells were left untransfected or transfected with a PARP12 catalytic mutant (G575W). B, immunopurified wild-type PARP12 was incubated in the presence of [32P]NAD and increasing concentrations of unlabeled NAD, and the reaction product was analyzed by autoradiography. C, automodified PARP12 is not detectable with an anti-poly(ADP-ribose) antibody. The reaction was performed as in B but without radiolabeled NAD. The reaction product was detected using an anti-poly(ADP-ribose) antibody (clone 10H). The 50-kDa band (#) corresponds to the Ig heavy chain. D, HA-tagged PARP12, and the catalytic mutant PARP12-G575W were immunoprecipitated (IP) and submitted to ADP-ribosylation reaction in vitro in the presence of NAD and the PARP inhibitor, 3-aminobenzamide (3-AB). The products were analyzed by AF1521 Western blot (top) or by anti-HA Western blot as an IP control (bottom). A band corresponding to automodified PARP12 (*) was only detected following incubation of WT, but not catalytically inactive PARP12. As previously shown, addition of 3-AB inhibited this reaction.
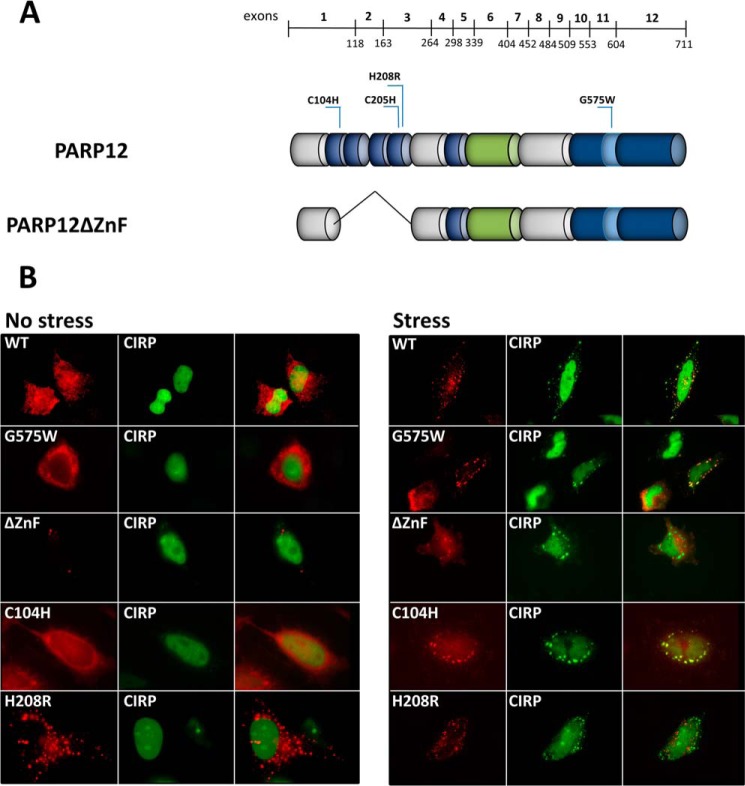
FIGURE 4.
The integrity of fourth zinc finger domain is critical for the localization of PARP12 to stress granules. A, domain organization and point mutants of PARP12. The mutations of different amino acids generated by site-directed PCR mutagenesis are indicated (C104H, C205H, H208R, and 575W) B, HeLa cells were transfected with DNA constructs encoding WT or mutated Flag-tagged PARP12 in combination with YFP-CIRP. Stress granules were induced with 1 mm sodium arsenite for 30 min, and PARP12 was detected with M2 anti-Flag followed by anti-mouse Alexa594.
PARP12 Localization to Stress Granules Requires a Functional CCCH Zn Finger Domain
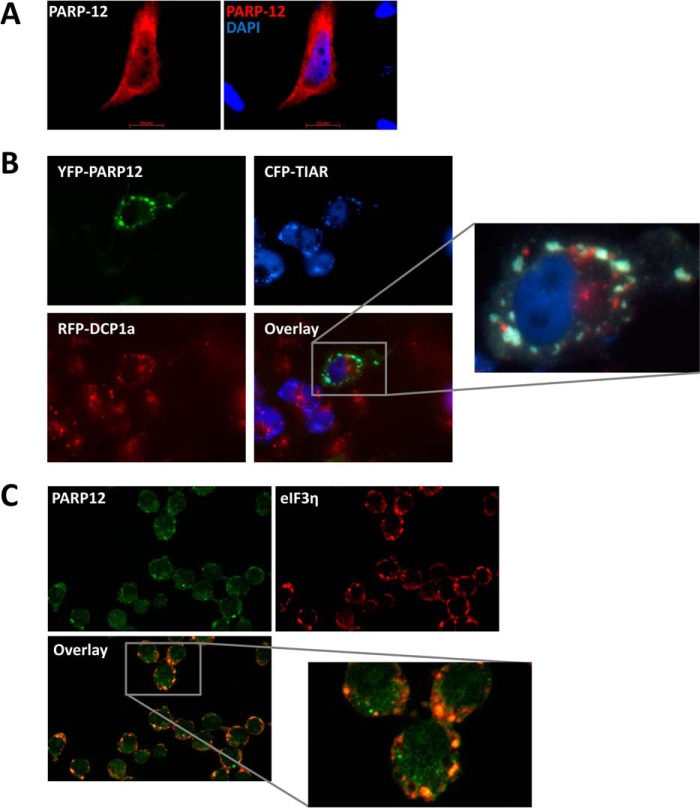
Although several PARPs described to date display a nuclear localization (34) human PARP12 has been previously found in the cytoplasm, often associated to subcellular structures known as stress granules. To confirm these observations, HeLa cells were transfected with PARP12, followed by immunofluorescent detection. Although most of the protein was found in the cytoplasm, an occasional faint signal was detectable in the nucleus (Fig. 3A). Association of PARP12 to stress granules was confirmed following co-transfection of HeLa cells with vectors coding for yellow fluorescent protein-tagged PARP12 (YFP-PARP12), cyan fluorescent protein-tagged TIAR (CFP-TIAR) and red fluorescent protein-tagged DCP1a (RFP-DCP1a). TIAR is a well-defined component of stress granules (sites of untranslated mRNA storage), while the decapping complex subunit DCP1a is predominantly found in processing bodies, cytoplasmic structures involved in mRNA decay (35). As shown in Fig. 3B, YFP-PARP12 selectively accumulated in stress granules (SG) in response to arsenite-induced oxidative stress, as shown by its preferential colocalization with CFP-TIAR but not RFP-DCP1a. The localization of the endogenous protein confirms these data obtained upon ectopic, and probably supra-physiological expression of PARP12: RAW264.7 macrophages were incubated with IFNβ for 4 h to induce the expression of PARP12, and cells were exposed to oxidative stress. As shown in Fig. 3C, PARP12 and eIF3η, a known component of stress granules (36), were found to colocalize in cytoplasmic puncta. To analyze the domains that contribute to SG localization, several mutant forms of PARP12 were constructed (see Fig. 4A) and transfected into HeLa cells together with a plasmid encoding for eYFP-tagged CIRP, a recently described SG component (24). The transfected cells were incubated in the presence of arsenite and SG visualized by fluorescent microscopy. The catalytic activity of PARP12 did not appear to play an important role in the capacity of PARP12 to be recruited to stress granules, since the point mutant G575W and PARP12 forms lacking the catalytic domain could both be found in these structures upon arsenite exposure (Fig. 4B and data not shown). Mutation of cysteine 104 had little or no effect on the localization of PARP12, while mutation of the histidine 208 residue, and the deletion of the Zn fingers-containing domains, led to a profound alteration of PARP12 subcellular localization. Indeed, these mutant forms were found in punctate foci distinct from SGs even in the absence of stress, and were not recruited to stress granules in response to arsenite (Fig. 4B). Collectively, these observations indicate an important role for the zinc finger domains in the capacity of PARP12 to be recruited to SGs in response to cellular stress.
FIGURE 3.
PARP12 is a component of cytoplasmic stress granules but not of processing bodies. A, HeLa cells were transfected with a PARP12-flag DNA construct and the protein was detected by immunostaining and fluorescence microscopy using anti-Flag M2 and anti-mouse Alexa594 antibodies. DAPI was used to stain the nuclei. B, HeLa cells were transfected with YFP-tagged PARP12 in combination with CFP-TIAR as a stress granule marker or with RFP-dcp1 as a processing body marker. Stress granules were induced by the addition of 1 mm sodium arsenite for 30 min. The overlay image shows colocalization between YFP-PARP12 (green) and cytoplasmic CFP-TIAR (blue) positive stress granules. C, RAW264.7 cells were stimulated with 1000 units/ml of recombinant IFNβ for 4 h to induce PARP12 expression and stress granules were induced with 1 mm sodium arsenite for 20 min. Endogenous proteins were detected using the MP12-Alexa488 and anti-eIF3η followed by staining with anti-rabbit Alexa594 antibodies. The overlay image shows the colocalization between PARP12 (green) and eIF3η (red) in stress granules.
PARP12 Inhibits the Translation of Target mRNAs
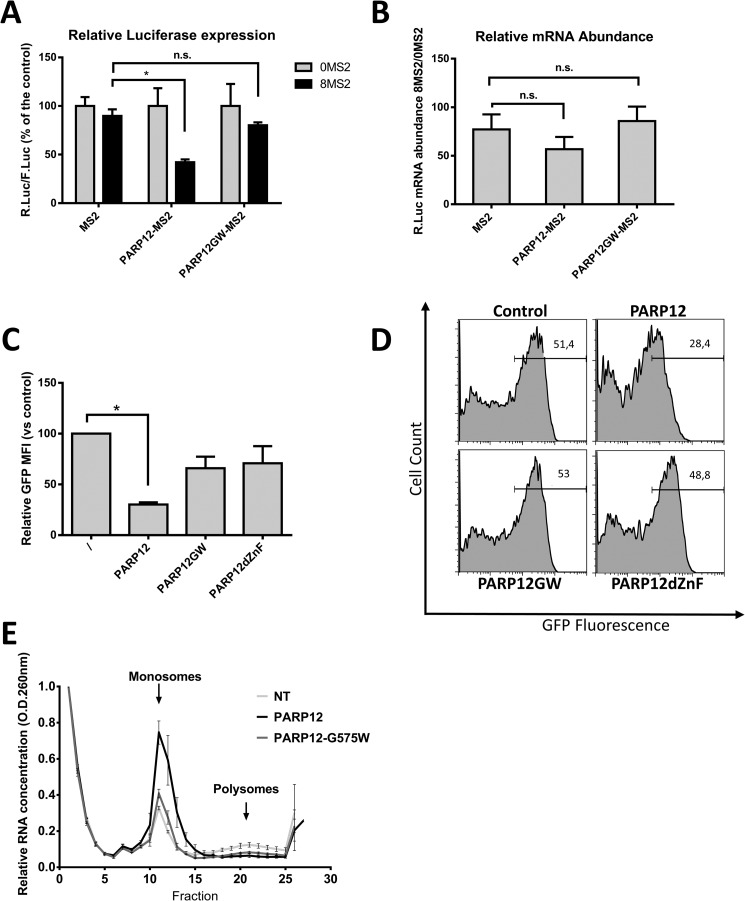
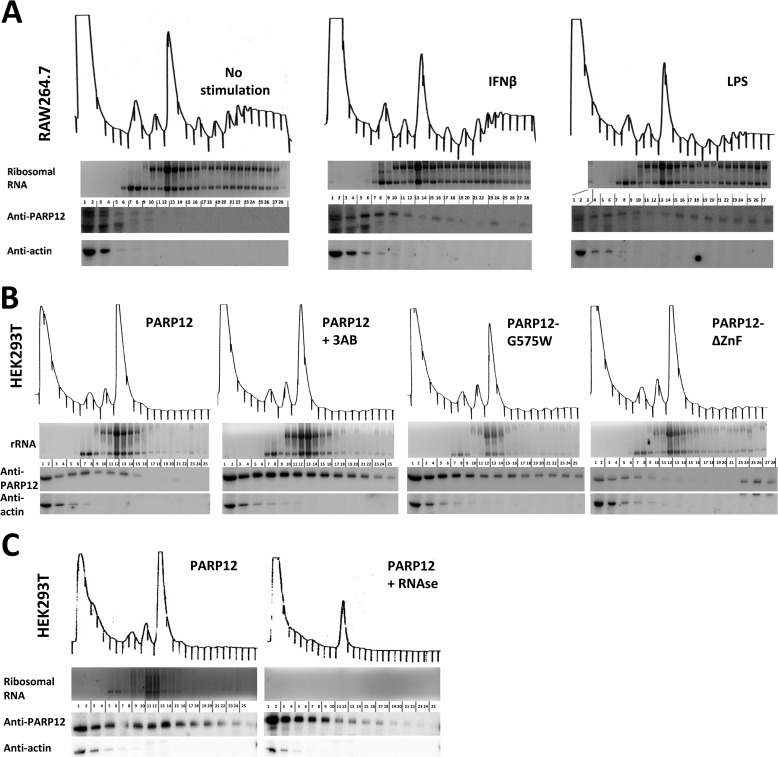
The localization of PARP12 to stress granules, the presence of a RNA-binding domain and previous observations (19) all concur to indicate a role for this member of the PARP family in the regulation of mRNA translation. To further validate this hypothesis, a tethering approach, allowing the binding of PARP12 to a reporter gene mRNA was used, as previously described (37). PARP12 was fused to the MS2 coat protein from the bacteriophage λ known to bind with high affinity a specific 21 nucleotide stem-loop structure in mRNAs (38). HEK293T cells were transfected with a vector encoding both the Firefly and the Renilla luciferase constructs under the control of a bidirectional CMV promoter. The 3′UTR of the Renilla luciferase mRNA contained either 0 or 8 MS2 stem-loops while the Firefly luciferase served as an internal control. The cells were also transfected with the MS2 protein alone or with PARP12 fused to the MS2 protein. Forced binding of PARP12 to the Renilla luciferase mRNA via the MS2 protein led to a significant inhibition of luciferase protein expression (Fig. 5A). Of note, binding of the point-mutant catalytically inactive PARP12-G575W to the same reporter mRNA had no significant effect on protein expression, strongly suggesting an important role for the catalytic activity of PARP12 in the negative control of mRNA translation. Expression of the fused PARP12-MS2 construct had no effect on the accumulation of target R.Luc mRNAs (Fig. 5B), indicating that, in agreement with its subcellular localization, PARP12 appears to selectively inhibit mRNA translation with no detectable effect on mRNA stability. To further confirm the putative role for PARP12 in regulating mRNA translation, PARP12 and mutant forms of the protein were co-transfected into HEK293T cells with a GFP-encoding plasmid. Flow cytometry analysis of GFP-fluorescence (see a typical fluorescence histogram in Fig. 5D) revealed the capacity of PARP12 to down-regulate expression of a co-transfected plasmid, in a catalytic domain- and zinc finger-dependent fashion (Fig. 5C). We next evaluated the translational status of HEK293T cells overexpressing PARP12 by examining the distribution of ribosomes on cellular messenger RNAs. HEK293T cells were transfected with PARP12 or PARP12-G575W and the distribution of mono- and polysomes analyzed following sucrose gradient fractionation. As shown in Fig. 5E, PARP12 expression led to a profound shift in the polysome profile compatible with a translational blockade, as suggested by the accumulation of monosomes and the loss of actively translated mRNAs (polysomes). Overexpression of the PARP12 catalytic mutant did not cause monosome accumulation, stressing the importance of the ADP-ribosyl transferase activity in the regulation of mRNA translation. To test whether PARP12 is associated with polysomes, we evaluated the distribution of PARP12 following sucrose gradient fractionation, in HEK293T cells and in RAW264.7 cells. PARP12 was faintly detectable in the unbound fractions in unstimulated RAW cells (due to low expression levels), but was associated with the mono- and polysomal fractions in cells stimulated with IFNβ or LPS (Fig. 6A). In HEK293T cells, PARP12 was associated with monosomal fractions and polysomal fractions, independently of its catalytic activity (shown with the catalytic mutant PARP12-G575W, and with the WT protein in cells treated with 3-AB, Fig. 6B). Mutation of the zinc-finger (PARP12ΔZnF), however decreases its association with the monosomes and polysomes (Fig. 6B). Of note, PARP12ΔZnF is detectable in the final fractions of the sucrose gradient, possibly due to its aggregation to previously described cytoplasmic structures. RNase treatment led to a significant loss of PARP12 from the high density samples, confirming that its association to the polysome-containing fractions required the presence of RNA (Fig. 6C). Collectively, these findings suggest that PARP12 is recruited to the translational machinery in a ZnF-domain-dependent fashion, where it exerts a negative influence of mRNA translation (in a catalytic activity-dependent fashion).
FIGURE 5.
PARP12 induces translational repression. A, tethering of PARP12 to the 3′-UTR of a reporter gene mRNA blocks its expression. The luciferase activities measured in the cell extracts are reported as the normalized (0MS2 = 100%) ratio of the R.Luc to F.Luc activities. The data represent the mean and S.E. of three independent experiments (*: p value < 0.05; n.s.: not statistically significant: Wilcoxon matched-pairs signed rank test). B, quantitative PCR analysis of R.Luc mRNA expression. The data are shown as the ratio of R.Luc-8MS2 divided by R.Luc-0MS2 mRNA, F.Luc serving as endogenous control. The data represent the mean and S.E. of three independent experiments. C, WT PARP12 decreases the expression of a co-transfected plasmid. HEK293T cells were transfected with PARP12- and GFP-encoding plasmids, and GFP fluorescence was measured by flow cytometry. Data represent mean GFP fluorescence intensity normalized to cells transfected with GFP and a control plasmid. D, representative histograms of GFP expression measured by flow cytometry. E, analysis of polysomes by sucrose gradient fractionation. HEK293T cells were either not transfected (NT) or transfected with wild-type (PARP12) or catalytic mutant (G575W) PARP12. The data represent mean and S.D. of the optical density (A260) measured for each fraction and normalized to the first fraction (= 1). E, PARP12 associates with the translation machinery.
FIGURE 6.
PARP12 is associated with the cellular translation machinery. A, RAW264.7 cells were left untreated or stimulated with interferon β (1000u/ml) for 4 h or LPS (10 μg/ml) for 8 h. The polysomes were analyzed by sucrose gradient fractionation and proteins associated with each fraction were extracted and analyzed by Western blotting using MP12 and anti-actin antibodies. The RNA was extracted from each fraction and analyzed by agarose gel electrophoresis followed by ethidium bromide staining. The visible bands correspond to 28 S and 18 S ribosomal RNA. B, HEK293T cells were transfected with PARP12, the catalytic mutant PARP12-G575W or the zinc-finger mutant (PARP12ΔZnF) and the proteins and RNA were analyzed as in A. C, association of PARP12 with the translation machinery is RNA-dependent. HEK293T cells were transfected with PARP12, and the polysomes were analyzed by sucrose gradient fractionation following treatment with RNase. The proteins and RNA were analyzed as in A.
PARP12 Is Recruited to Aggresome-like Induced Structures (ALIS)
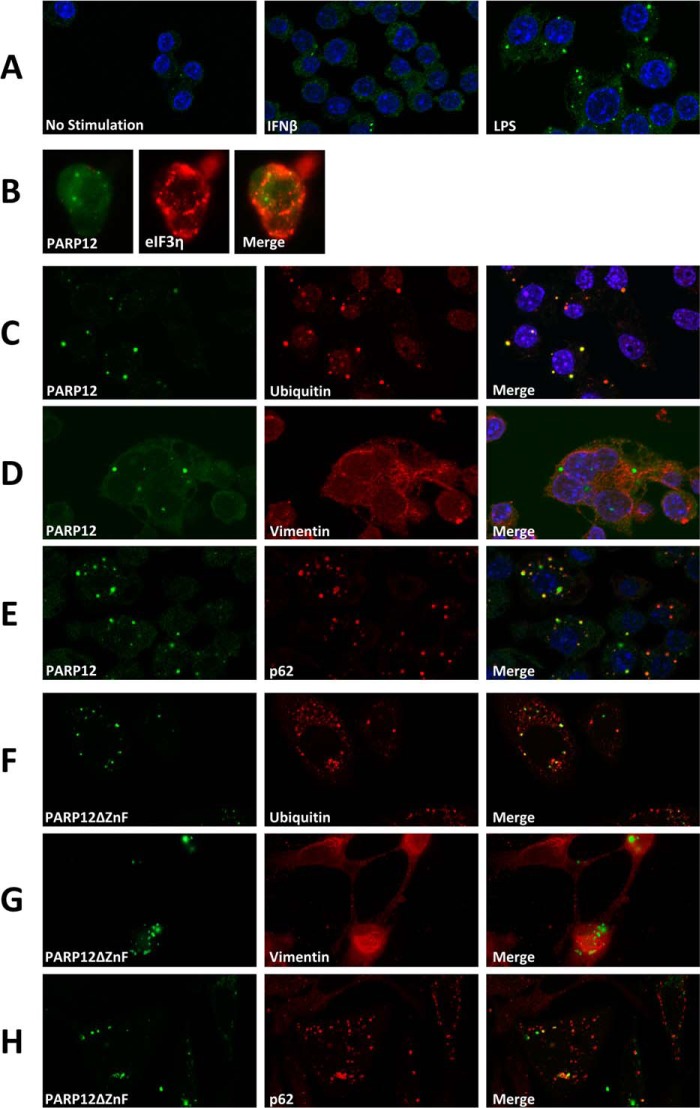
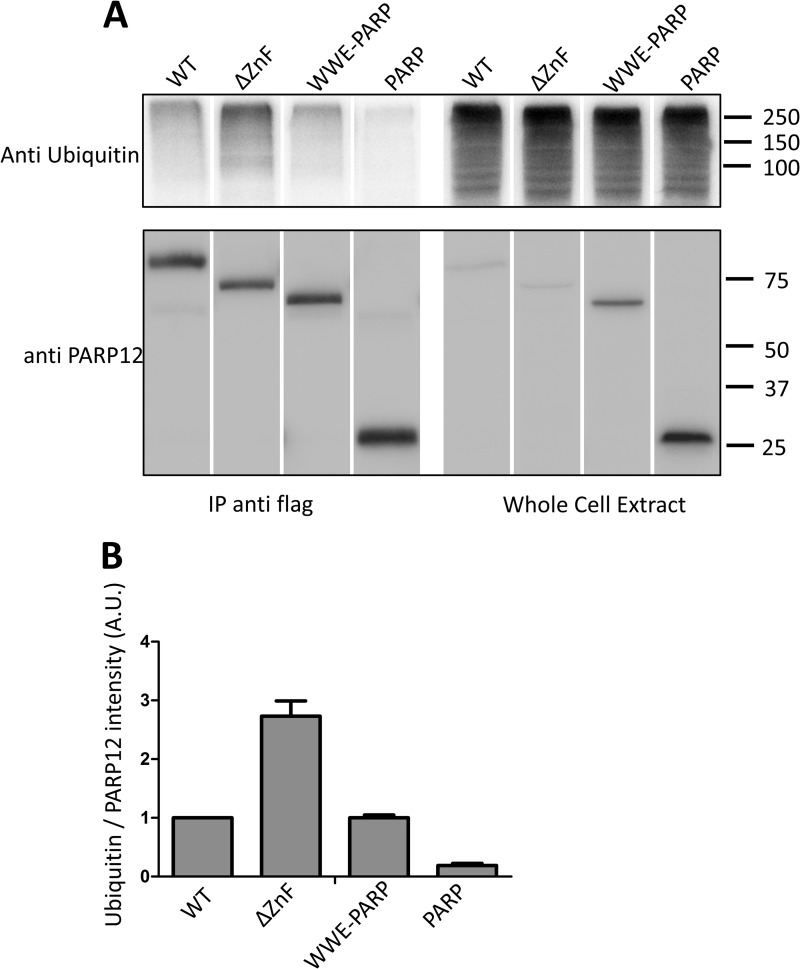
Taking advantage of a monoclonal antibody enabling us to detect expression of the endogenous PARP12 protein in untransfected cells, we undertook a detailed analysis of the intracellular localization of this protein in the RAW264.7 cell line. In control cells, the expression of PARP12 was barely detectable by immunofluorescence, and appeared as a faint, mostly cytoplasmic, diffuse signal (Fig. 7A). In keeping with observations depicted in Fig. 1, stimulation of cells with rIFNβ induced PARP12 expression (as revealed by an increase in cytoplasmic fluorescence) with the distinctive appearance of punctate structures in a sizable proportion of cells (Fig. 7A). These PARP12-containing cytoplasmic puncta were more abundant and clearly visible in cells stimulated by LPS in comparison to cells exposed to rIFNβ (Fig. 7A). These structures were distinct from the previously described stress granules, as determined by co-localization studies using antibodies to eIF3η (Fig. 7B). Further co-localization studies led us to identify these PARP12-containing granules as aggresome-like structures or ALIS. These structures, originally identified in dendritic cells and later found in macrophages and other cell types (39, 40), are thought to represent storage sites for ubiquitinated proteins possibly awaiting degradation by the proteasome or the autophagic machinery, and differ from classical aggresomes by the absence of a vimentin-cage (39). Accordingly, PARP12 colocalized with ubiquitin or ubiquitinated proteins in LPS-stimulated RAW264.7 cells, in non-vimentin-containing structures (Fig. 7, C and D). To confirm these findings, we also investigated the possible co-localization of PARP12 with p62 (also known as sequestosome-1 or SQSTM1), a previously identified ALIS component required for the degradation of the ubiquitinated proteins by autophagy (41). The observation depicted in Fig. 7E confirms that PARP12 and p62 do belong to the same complexes in LPS activated macrophages. Finally, to confirm that these observations could be reproduced in another setting, we analyzed the cytoplasmic localization of a mutant form of PARP12 lacking the Zn finger domain (PARP12ΔZnF) and previously shown to be largely excluded from SGs. Following transfection in HeLa cells, this mutant form of PARP12 was found to associate with ubiquitin- and p62-containing granules (Fig. 7, F–H) lacking a vimentin-cage (Fig. 7G), further confirming that under adequate experimental conditions, PARP12 is recruited to aggresome-like structures in both immune and non-immune cells. Immunoprecipitation and Western blot analysis in transfected HEK293 cells confirmed that PARP12 and mutant forms expressing a WWE domain were associated with high molecular weight, ubiquitin-containing complexes, confirming the previous co-localization studies (Fig. 8).
FIGURE 7.
PARP12 is recruited to ALIS following TLR stimulation. A, RAW264.7 macrophages were grown on coverslips and stimulated with IFNβ or LPS for 8 h. Cells were fixed, permeabilized, incubated with 24G2 for 30 min to block the Fc-receptors, and stained for endogenous PARP12 (green) using the monoclonal MP12.1 antibody coupled to Alexa488. The nuclei (DNA) were labeled with DAPI (blue). (B) PARP12-containing puncta are not stress granules. RAW264.7 cells were grown on coverslips and stimulated with IFNβ and stress granules were induced by the addition of 1 mm sodium arsenite for 30′. PARP12 was detected as in A, and eiF3η was detected with a goat anti-eiF3B and an Alexa594-coupled donkey anti-goat (C–E) PARP12 is recruited to ALIS following TLR stimulation. RAW264.7 cells grown on coverslips were stimulated with 1 μg/ml LPS for 8 h and stained with mouse monoclonal anti-ubiquitin, goat anti-vimentin, or rabbit anti-p62/SQSTM1 antibody, followed by a goat anti-mouse Ig Alexa594 antibody, a donkey anti-goat Alexa594 antibody or an anti-rabbit Alexa594 antibody. The coverslips were treated with normal mouse serum and incubated with a mouse monoclonal anti-PARP12 (MP12.1 Alexa488). Nuclei were stained with DAPI (blue). The colocalization of PARP12 and ubiquitin (D) and p62 (F) are visible in the merged images (D). These structures are not surrounded by a vimentin cage (E). G–I, overexpressed PARP12ΔZnF is a component of ALIS in HeLa cells. HeLa cells were transfected with YFP-tagged PARP12ΔZnF (green). Fixed and permeabilized cells were stained for endogenous ubiquitin, vimentin, and p62 as in D–F. The images were acquired with a Zeiss LSM710 confocal microscope with a 63× objective.
FIGURE 8.
PARP12 interacts with ubiquitin. HEK293T cells were transfected with different constructs coding for Flag-tagged PARP12 (WT, zinc finger mutant, ΔZnF, deletion mutant containing only the WWE and PARP domains - WWE-PARP, and the PARP domain alone, PARP). The cells were lysed and Flag-PARP12 was immunoprecipitated using anti-Flag (M2) and protein G microbeads (Miltenyi). A, ubiquitin was detected with the FK2 anti-ubiquitin antibody and PARP12 was detected with the MP12.1 antibody. The spliced lanes were merged from a single immunoblot. B, mean + S.D. of the densitometric quantification of ubiquitin normalized to PARP12 band intensities (arbitrary units) (n = 2).
Co-localization and Physical Interaction of PARP12 with Members of the NF-κB Signaling Pathway
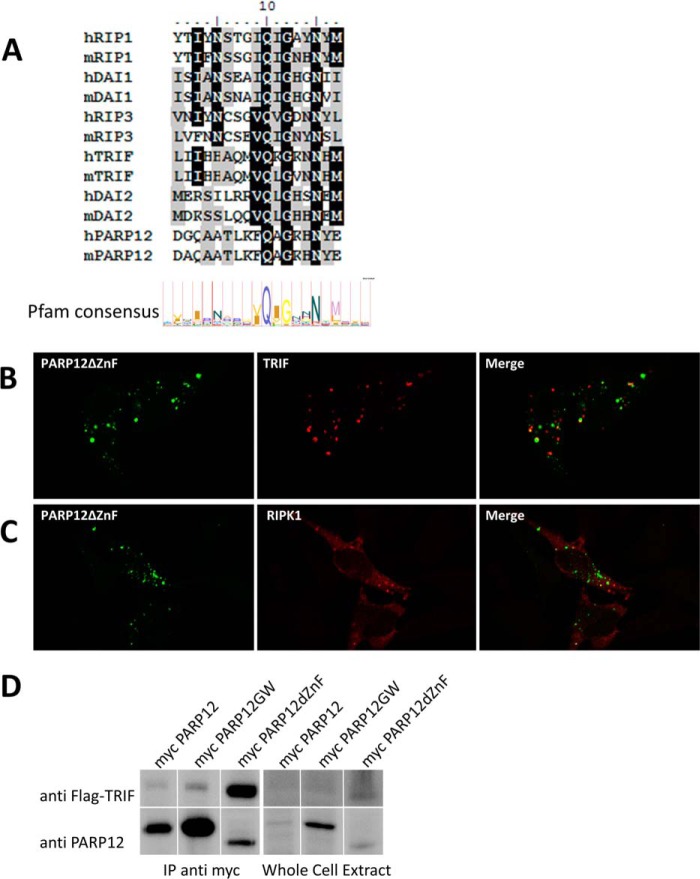
The analysis of the sequence of PARP12 revealed the presence of a putative RHIM (RIP homotypic interaction motif), as illustrated in Fig. 9A. The RHIM motif is known to mediate homotypic associations between signaling proteins involved in programmed necrosis and TLR signaling (42–44). Based on these observations, HeLa cells were co-transfected with plasmids encoding YFP-tagged PARP12ΔZnF and tagged version of RHIM-containing proteins such as TRIF/TICAM1 (a Toll/IL1R domain containing signaling adaptor known to play a role in TLR signaling) and RIPK1 (an important kinase in several signaling pathways including NF-κB activation). As previously published (45), we confirmed the capacity of overexpressed TRIF to spontaneously accumulate into punctate structures in the cytoplasm. As shown in Fig. 9B, PARP12ΔZnF partially co-localized with TRIF-containing structures, although single fluorescent structures (containing PARP12 or TRIF) could be observed. A similar, partial co-localization was observed in cells over-expressing PARP12ΔZnF and RIPK1, further suggesting a dynamic interaction between PARP12, TRIF, and RIPK1 under these experimental conditions (Fig. 9C). A direct interaction between PARP12ΔZnF and TRIF could be confirmed by co-immunoprecipitation, as shown in Fig. 9D. Note that in agreement with the co-localization data, only the mutant form of PARP12 lacking the CCCH Zn finger domain displayed significant binding to TRIF, while the native form of the protein failed to efficiently interact with TRIF.
FIGURE 9.
Co-localization and physical interaction of PARP12 with members of the NF-κB signaling pathway. A, PARP12 possesses a putative RHIM motif. The sequence of PARP12 was aligned to the RHIM motifs of known RHIM-containing proteins using the ClustalW algorithm, and compared with the Pfam RHIM consensus motif. B, HeLa cells were transfected with YFP-tagged PARP12ΔZnF (green) and Flag-tagged TRIF, grown at 37 °C for 24 h, fixed, and permeabilized, and TRIF (red) was detected with a monoclonal anti-Flag antibody and secondary goat anti-mouse Alexa594. The merged images (right panel) show colocalization between PARP12ΔZnF and TRIF. B, HeLa cells were transfected with YFP-PARP12ΔZnF (green) and Flag-tagged RIP1, and RIP1 (red) was detected with M2 anti-Flag and anti-mouse Alexa594 antibodies. The images were acquired with a Zeiss LSM710 confocal microscope (63×). C, HEK293T cells were transfected with different constructs coding for myc-tagged PARP12 (WT, catalytic mutant, GW, zinc finger mutant, dZnF) and Flag-tagged TRIF. The cells were lysed and myc-PARP12 was immunoprecipitated using anti-myc microbeads (Miltenyi). TRIF was detected with the M2 anti-Flag antibody, and PARP12 was detected with the MP12.1 antibody. The spliced lanes were merged from a single immunoblot.
PARP12 Is a Component of the Inflammatory Signaling Cascade
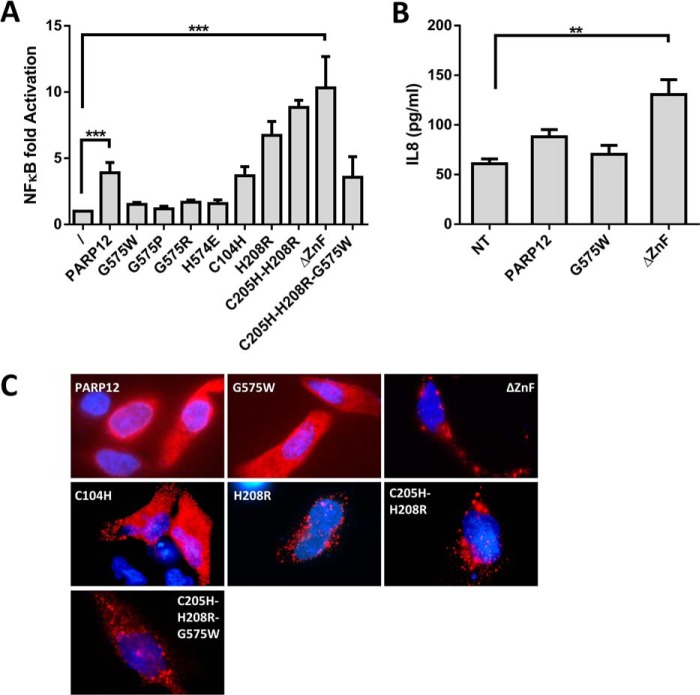
The previously documented colocalization of PARP12 with TRIF suggested that in addition to mRNA translation regulation, PARP12 could also affect the signaling cascade leading to NF-κB activation. To reveal a possible role for PARP12 in this signaling pathway, HEK293T cells were transfected with a NF-κB reporter plasmid together with constructs coding for wild-type or mutated forms of PARP12. Expression of PARP12 led to a significant increase (3–5-fold) in NF-κB-dependent reporter activity that was further enhanced upon deletion of the ZnF domain (10–15-fold) (Fig. 10A). Notably, the capacity of PARP12 to promote NF-κB-dependent gene transcription required the presence of an active catalytic domain, as evidenced by the lack of gene reporter activation in cells expressing the previously characterized G575W mutant. Assay of multiple mutant forms of PARP12 confirmed the lack of transactivating activity in a series of catalytically inefficient mutants (G575P, G575R, and H574H) and increased NF-κB activation in cells transfected with PARP12 proteins displaying altered localization (Fig. 10C) caused by mutations in their Zn-finger containing domains (ΔZnF, H208R, and C205H-H208R). To confirm the capacity of the PARP12 isoform lacking the Zn-finger containing domain to facilitate NF-κB activation, HEK293T cells were transfected with WT or mutated PARP12 constructs and stimulated overnight with rhTNF, a well-characterized NF-κB activating stimulus (46). As shown in Fig. 10B, overexpression ΔZnF PARP12 led to a significant and reproducible increase in the secretion of the NF-κB target IL-8, thus confirming the pro-inflammatory activity of PARP12 on an endogenous gene target.
FIGURE 10.
Activation of the NF-κB pathway by PARP12. HEK293T cells were transfected with the indicated constructs of PARP12 and assayed for A, luciferase expression when co-transfected with an NFκB-sensitive promoter (pNFκB-Luc, Firefly) and a control plasmid encoding for a Renilla Luciferase (pGL4.74). The data represent the mean ± S.E. of the ratio of Firefly/Renilla activity normalized to the sample not transfected with PARP12 (***: p value < 0.001, Friedman test with Dunn's multiple comparisons) and B, IL-8 secretion upon stimulation with rhTNF (10 ng/ml). The data represent mean ± S.E. of four independent experiments (**: p value < 0.01 - Kruskal-Wallis test with Dunn's multiple comparisons. C, subcellular localization of the different mutants of PARP12 in HeLa cells. The NFκB-activating mutants of PARP12 spontaneously form cytoplasmic puncta.
DISCUSSION
Type I interferons exert their immune regulatory and protective role through the coordinate expression of hundreds of genes (known as interferon stimulated genes or ISGs) of which only a fraction has been thoroughly characterized to date (47). The present work was undertaken in order to identify the functional properties of PARP12, a NAD-dependent ADP-ribosyltransferase whose expression had been previously shown to be induced by IFNs. Taking advantage of a PARP12-specific monoclonal antibody generated during the course of this study, we formally demonstrate herein that PARP12 is a strict ISG, whose expression (confirmed at protein level) appears to require IFNAR signaling even in response to TLR ligands. We next wished to establish the subcellular localization of PARP12 and evaluate the role of its distinct protein domains in mediating both subcellular localization and functional properties of the protein.
In keeping with its multimodular structure, PARP12 can be found in at least two distinct subcellular localizations, possibly associated to distinct functional properties with relevance to the anti-microbial activity of IFNs.
Upon ectopic expression of the complete protein, PARP12 is found in cytoplasmic structures known as stress granules (SGs), in which mRNA translation is halted until stress diminishes and protein synthesis can resume (48). Accordingly, overexpression of PARP12 led to a general translation arrest in BHK-21 (19) and HEK293T cells (this study). The ZnF-containing domain played an important role in addressing PARP12 to SGs, as shown by altered localization of PARP12 mutant lacking the ZnF domain and of the fourth ZnF point mutated forms (see Fig. 4). Experiments conducted in untransfected cell lines strongly suggest that localization of PARP12 to SGs has physiological relevance. Indeed, oxidative stress led to the recruitment of endogenous PARP12 (whose expression had previously been induced by exposing cells to IFNβ) to SGs. This observation suggests that physiological levels of PARP12 do not cause SG appearance in the absence of oxidative stress. The role of several members of the PARP family in SG assembly has been previously addressed (18). In this previous study, six members of the human PARP family (PARP-5a, -12, -13.1, -13.2, -14, and -15) were found to localize to SGs (note that PARP15 is absent from the murine genome (49)). Consistent with the present work, overexpression of most of these proteins led to the de novo assembly of SGs, even in the absence of exogenous stress. In our own study, overexpression of catalytically inefficient forms of PARP12 failed to induce SGs4 suggesting an important role for ADP-ribosylation in SG assembly. Exposure of cells to oxidative stress led however to a recruitment of catalytically inactive PARP12 to SGs (Fig. 4), suggesting that the capacity of PARP12 to initiate SGs formation requires its catalytic activity, while its recruitment to these granules probably relies on its putative RNA-binding domain. These observations are best explained by assuming that nucleation of SGs can be induced by an ADP-ribosylation event initiated by one or several members of the SG-associated PARPs (i.e. PARP5a, -12, -13, and -15) with the notable exception of PARP13, which lacks enzymatic activity). The reason for this high functional redundancy between PARP isoforms and the precise individual role of each PARP members in response to stress awaits further investigations.
While the catalytic activity of PARP12 was dispensable for recruitment to SGs, it was strictly required for down-regulation of mRNA translation. Although these results, obtained in experimental setting in which PARP12 was ectopically expressed at supra-physiological levels, need to be considered with caution, they suggest a possible role for PARP12 in regulating protein synthesis in cells exposed to an inflammatory environment. Experiments performed with mutant forms of PARP12 affected in their N-terminal domain suggest an important role for mRNA recognition in PARP12 effector function, although the putative RNA associated structure and/or motif recognized by PARP12 remains elusive. Note that the tethering approach used in this study concurs with the idea that direct binding of PARP12 to a target RNA is a prerequisite for down-regulation of translational efficiency. The evidence accumulated to date favors a model in which PARP12 binds to a highly conserved mRNA structure, thus explaining its capacity to inhibit translation of virtually all cellular mRNAs (Ref. 19 and Fig. 5). Further studies, including electrophoretic mobility shift assays, will be required to confirm the capacity of PARP12 to directly interact with RNA molecules in a Zn-finger domain-dependent fashion. The precise role of the catalytic activity of PARP12 in mediating translational arrest also remains elusive. Despite numerous attempts, we have been unable to identify a cellular substrate for PARP12. PARP12, like many members of this family, is capable of automodification. However, the link between this posttranslational modification and translational arrest is difficult to establish. In a previous study, a mutant form of PARP12, lacking a putative autoribosylation site has been generated based on sequence homology with an experimental validated site found on PARP10 (19). Although the catalytic activity of this mutant protein was not evaluated, the authors concluded from their studies that mutants lacking a catalytic activity and/or displaying a modified automodification site were equally inefficient in mediating translational arrest. These observations are compatible with a model in which the catalytic activity of PARP12 serves as a molecular switch enabling, possibly through a conformational change, the efficient translational blockade of numerous cellular mRNAs. This conclusion is also supported by the high abundance of PARP12 mRNA in oocytes (a site of intense translational control (50).
Of particular relevance to these observations, formation of SGs has been previously identified as a host response to virus infection (51) while disruption of these cytoplasmic structures may represent a mechanism adopted by selected viruses (52, 53) to escape cell innate control. Expression of PARP12 in response to IFN may therefore contribute to the cellular antiviral response by increasing the efficiency SG-mediated silencing of viral and cellular RNAs.
Ectopic expression of PARP12 mutant forms lacking RNA binding activity suggested an additional functional role for this protein, with relevance to innate immune responses. These altered forms of the protein were found to localize to non-SG structures, characterized by the presence of ubiquitin and the p62/sequestosome-1 protein (SQSTM1), and the absence of a vimentin cage. SQSTM1 is a scaffold protein containing several protein interaction domains and with multiple functions in signal transduction, cell death and inflammation (54, 55). A growing body of evidence suggests in particular a role for p62 in directing ubiquitinated proteins for autophagic destruction, and as a platform for NF-κB signaling. Our own observations support a role for PARP12 in NF-κB activation. Indeed, overexpression of PARP12, and especially of the zinc-finger mutant form that interacts and colocalizes with TRIF and ubiquitin-modified proteins, activates a NF-κB reporter plasmid and promotes IL-8 secretion in response to an extracellular ligand. The importance of ubiquitin, and ubiquitin ligases, in NF-κB signal transduction has been amply recognized. Notably, ubiquitination not only regulates IkB (a cellular NF-κB inhibitor) degradation, but also regulates the signaling cascade leading to NF-κB-mediated gene transcription (56). A possible link between ADP-ribosylation and ubiquitination has been suggested by the shared presence of WWE domains among several enzymes catalyzing these two post-translational modifications (57). The best characterized WWE domain (from the E3 ubiquitin ligase RNF146) has been shown to bind to poly(ADP-ribose) through recognition of the iso-ADP-ribose moiety only found in poly-, but not mono-ADP-ribosyl modified proteins (58). However, other WWE domains display weak to no affinity for iso-ADP-ribose, suggesting that other members of the ubiquitin ligase family may interact with mono-ADP-ribose and possibly mediate proteasome-dependent degradation of mono-ribosylated substrates.
In addition to its role in signaling, p62/SQSTM1 also represents a central player in the induction of autophagy (54). p62 possesses an LC3-interacting region mediating binding to LC3, a protein involved in phagophore elongation for autophagosome formation. p62 has been shown to act as an autophagy receptor for the selective autophagic clearance of misfolded and/or aggregated proteins, damaged organelles or pathogens, illustrating a close link between autophagy and innate resistance to infection. Of relevance to this study, autophagy has also been recently identified as a mechanism for SGs clearance (59). PARP12 recruitment to p62-containing cytoplasmic puncta was found in the present study to be enhanced by TLR4 signaling, an observation that suggests a possible role for this PARP member in receptor signaling leading to NF-κB and/or the autophagic clearance of intracellular pathogens. Moreover, PARP10 has also recently been found in association with p62-containing granules (60), a finding that further suggests a possible functional redundancy among members of this complex family. Further work is therefore required to identify the specific and unique role of PARP12 during an IFN-mediated response.
As a conclusion, the available evidence suggests a complex functional role for PARP12 during an innate immune response. Exposure of cells to IFNs induces PARP12 expression in virtually all cells tested to date. The original observations performed in the present study using a novel reagent to detect endogenous forms of murine PARP12 reveal a relatively homogenous, cytoplasmic distribution of the protein in a fibroblast (data not shown) and a macrophage-like cell line. Upon a specific signal (oxidative versus TLR-initiated), PARP12 appears to relocate to SGs or p62-containing structures related to ALIS. Experiments in which wild type and mutant forms of the protein were ectopically expressed suggest an important role for the Zn-finger containing domain in determining PARP12 subcellular location. Indeed, lack of this functional domain appears to preclude its recruitment to SGs, while favoring its colocalization with p62. Overexpression studies suggested that recruitment to SGs was linked to the capacity of PARP12 to halt mRNA translation, while interaction of PARP12 with p62-containing structures correlated with enhanced NF-κB signaling. Since both cytoplasmic localizations could be observed in untransfected cells lines expressing the wild type form of the protein, it is tempting to speculate that the subcellular distribution of PARP12 may be regulated by a signaling event leading to the post-translational modification of the N-terminal domain of the protein. Accordingly, several studies have identified PARP12-derived N-terminal peptides displaying phosphorylated residues (61, 62). PARP12 may therefore represent an ISG with dual functional properties both of which may contribute to the establishment of an antimicrobial state in cells exposed to IFNs.
This work was supported by the Belgian Program in Interuniversity Poles of Attraction initiated by Science Policy Programming, Belgium, the Research Concerted Action and the “Fonds pour la formation à la Recherche dans l'Industrie et l'Agronomie” (F.R.I.A.) of the “Fédération Wallonie Bruxelles,” the “Fonds David et Alice Van Buuren,” and the Walloon Region.
I. Welsby and D. Hutin, unpublished observations.
- PARP
- poly(ADP-ribose) polymerase
- ISG
- interferon-stimulated gene
- ALIS
- aggresome-like induced structure
- SG
- stress granule.
REFERENCES
- 1. Citarelli M., Teotia S., Lamb R. S. (2010) Evolutionary history of the poly(ADP-ribose) polymerase gene family in eukaryotes. BMC Evol. Biol. 10, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hassa P. O., Hottiger M. O. (2008) The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 13, 3046–3082 [DOI] [PubMed] [Google Scholar]
- 3. Krishnakumar R., Kraus W. L. (2010) The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell 39, 8–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schreiber V., Dantzer F., Ame J.-C., de Murcia G. (2006) Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7, 517–528 [DOI] [PubMed] [Google Scholar]
- 5. Gibson B. A., Kraus W. L. (2012) New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13, 411–424 [DOI] [PubMed] [Google Scholar]
- 6. Amé J.-C., Spenlehauer C., de Murcia G. (2004) The PARP superfamily. Bioessays 26, 882–893 [DOI] [PubMed] [Google Scholar]
- 7. Hottiger M. O., Hassa P. O., Lüscher B., Schüler H., Koch-Nolte F. (2010) Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 35, 208–219 [DOI] [PubMed] [Google Scholar]
- 8. Kleine H., Poreba E., Lesniewicz K., Hassa P. O., Hottiger M. O., Litchfield D. W., Shilton B. H., Lüscher B. (2008) Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell 32, 57–69 [DOI] [PubMed] [Google Scholar]
- 9. Rolli V., O'Farrell M., Ménissier-de Murcia J., de Murcia G. (1997) Random mutagenesis of the poly(ADP-ribose) polymerase catalytic domain reveals amino acids involved in polymer branching. Biochemistry 36, 12147–12154 [DOI] [PubMed] [Google Scholar]
- 10. Loseva O., Jemth A.-S., Bryant H. E., Schüler H., Lehtiö L., Karlberg T., Helleday T. (2010) PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J. Biol. Chem. 285, 8054–8060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Veer M. J., Holko M., Frevel M., Walker E., Der S., Paranjape J. M., Silverman R. H., Williams B. R. (2001) Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69, 912–920 [PubMed] [Google Scholar]
- 12. Liu S.-Y., Sanchez D. J., Aliyari R., Lu S., Cheng G. (2012) Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl. Acad. Sci. U.S.A. 109, 4239–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferreyra G. a, Elinoff J. M., Demirkale C. Y., Starost M. F., Buckley M., Munson P. J., Krakauer T., Danner R. L. (2014) Late multiple organ surge in interferon-regulated target genes characterizes staphylococcal enterotoxin B lethality. PLoS One 9, e88756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo X., Carroll J. W., Macdonald M. R., Goff S. P., Gao G. (2004) The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J. Virol. 78, 12781–12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo X., Ma J., Sun J., Gao G. (2007) The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl. Acad. Sci. U.S.A. 104, 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liang J., Song W., Tromp G., Kolattukudy P. E., Fu M. (2008) Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS One 3, e2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hall T. M. T. (2005) Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Biol. 15, 367–373 [DOI] [PubMed] [Google Scholar]
- 18. Leung A. K. L., Vyas S., Rood J. E., Bhutkar A., Sharp P. A., Chang P. (2011) Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 42, 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Atasheva S., Frolova E. I., Frolov I. (2014) Interferon-stimulated poly(ADP-Ribose) polymerases are potent inhibitors of cellular translation and virus replication. J. Virol. 88, 2116–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kawamitsu H., Hoshino H., Okada H., Miwa M., Momoi H., Sugimura T. (1984) Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures. Biochemistry 23, 3771–3777 [DOI] [PubMed] [Google Scholar]
- 21. Lykke-Andersen J., Shu M. D., Steitz J. A. (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103, 1121–1131 [DOI] [PubMed] [Google Scholar]
- 22. Barreau C., Watrin T., Beverley Osborne H., Paillard L. (2006) Protein expression is increased by a class III AU-rich element and tethered CUG-BP1. Biochem. Biophys. Res. Commun. 347, 723–730 [DOI] [PubMed] [Google Scholar]
- 23. Mollet S., Cougot N., Wilczynska A., Dautry F., Kress M., Bertrand E., Weil D. (2008) Translationally repressed mRNA transiently cycles through stress granules during stress. Mol. Biol. Cell 19, 4469–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Leeuw F., Zhang T., Wauquier C., Huez G., Kruys V., Gueydan C. (2007) The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp. Cell Res. 313, 4130–4144 [DOI] [PubMed] [Google Scholar]
- 25. Plaisance S., Vanden Berghe W., Boone E., Fiers W., Haegeman G. (1997) Recombination signal sequence binding protein Jkappa is constitutively bound to the NF-kappaB site of the interleukin-6 promoter and acts as a negative regulatory factor. Mol. Cell Biol. 17, 3733–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oldenhove G., de Heusch M., Urbain-Vansanten G., Urbain J., Maliszewski C., Leo O., Moser M. (2003) CD4+ CD25+ regulatory T cells control T helper cell type 1 responses to foreign antigens induced by mature dendritic cells in vivo. J. Exp. Med. 198, 259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morello L. G., Hesling C., Coltri P. P., Castilho B. A., Rimokh R., Zanchin N. I. T. (2011) The NIP7 protein is required for accurate pre-rRNA processing in human cells. Nucleic Acids Res. 39, 648–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C. L., Haase J., Janes J., Huss J. W., 3rd, Su A. I. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karras G. I., Kustatscher G., Buhecha H. R., Allen M. D., Pugieux C., Sait F., Bycroft M., Ladurner A. G. (2005) The macro domain is an ADP-ribose binding module. EMBO J. 24, 1911–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rosenthal F., Feijs K. L. H., Frugier E., Bonalli M., Forst A. H., Imhof R., Winkler H. C., Fischer D., Caflisch A., Hassa P. O., Lüscher B., Hottiger M. O. (2013) Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 20, 502–507 [DOI] [PubMed] [Google Scholar]
- 31. Nikiforov A., Dölle C., Niere M., Ziegler M. (2011) Pathways and subcellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J. Biol. Chem. 286, 21767–21778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith S., Giriat I., Schmitt A., de Lange T. (1998) Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282, 1484–1487 [DOI] [PubMed] [Google Scholar]
- 33. Amé J. C., Rolli V., Schreiber V., Niedergang C., Apiou F., Decker P., Muller S., Höger T., Ménissier-de Murcia J., de Murcia G. (1999) PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 274, 17860–17868 [DOI] [PubMed] [Google Scholar]
- 34. Leung A. K. L. (2014) Poly(ADP-ribose): An organizer of cellular architecture. J. Cell Biol. 205, 613–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anderson P., Kedersha N. (2009) RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10, 430–436 [DOI] [PubMed] [Google Scholar]
- 36. Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. (2005) Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Coller J., Wickens M. (2002) Tethered function assays using 3′ untranslated regions. Methods 26, 142–150 [DOI] [PubMed] [Google Scholar]
- 38. Johansson H. E., Liljas L., Uhlenbeck O. C. (1997) RNA Recognition by the MS2 Phage Coat Protein. Semin. Virol. 8, 176–185 [Google Scholar]
- 39. Lelouard H., Gatti E., Cappello F., Gresser O., Camosseto V., Pierre P. (2002) Transient aggregation of ubiquitinated proteins during dendritic cell maturation. Nature 417, 177–182 [DOI] [PubMed] [Google Scholar]
- 40. Szeto J., Kaniuk N. A, Canadien V., Nisman R., Mizushima N., Yoshimori T., Bazett-Jones D. P., Brumell J. H. (2006) ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy 2, 189–199 [DOI] [PubMed] [Google Scholar]
- 41. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J.-A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 42. Meylan E., Burns K., Hofmann K., Blancheteau V., Martinon F., Kelliher M., Tschopp J. (2004) RIP1 is an essential mediator of Toll-like receptor 3-induced NF-κB activation. Nat. Immunol. 5, 503–507 [DOI] [PubMed] [Google Scholar]
- 43. Rebsamen M., Heinz L. X., Meylan E., Michallet M.-C., Schroder K., Hofmann K., Vazquez J., Benedict C. A., Tschopp J. (2009) DAI/ZBP1 recruits RIP1 and RIP3 through RIP homotypic interaction motifs to activate NF-κB. EMBO Rep. 10, 916–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sun X., Yin J., Starovasnik M. A., Fairbrother W. J., Dixit V. M. (2002) Identification of a novel homotypic interaction motif required for the phosphorylation of receptor-interacting protein (RIP) by RIP3. J. Biol. Chem. 277, 9505–9511 [DOI] [PubMed] [Google Scholar]
- 45. Funami K., Sasai M., Ohba Y., Oshiumi H., Seya T., Matsumoto M. (2007) Spatiotemporal mobilization of Toll/IL-1 receptor domain-containing adaptor molecule-1 in response to dsRNA. J. Immunol. 179, 6867–6872 [DOI] [PubMed] [Google Scholar]
- 46. Imamura R., Konaka K., Matsumoto N., Hasegawa M., Fukui M., Mukaida N., Kinoshita T., Suda T. (2004) Fas ligand induces cell-autonomous NF-κB activation and interleukin-8 production by a mechanism distinct from that of tumor necrosis factor-α. J. Biol. Chem. 279, 46415–46423 [DOI] [PubMed] [Google Scholar]
- 47. Schoggins J. W., Wilson S. J., Panis M., Murphy M. Y., Jones C. T., Bieniasz P., Rice C. M. (2011) A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kedersha N., Ivanov P., Anderson P. (2013) Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38, 494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li H., Coghlan A., Ruan J., Coin L. J., Hériché J.-K., Osmotherly L., Li R., Liu T., Zhang Z., Bolund L., Wong G. K.-S., Zheng W., Dehal P., Wang J., Durbin R. (2006) TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 34, D572–D80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Richter J. D., Lasko P. (2011) Translational control in oocyte development. Cold Spring Harb. Perspect. Biol. 3, a002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Okonski K. M., Samuel C. E. (2013) Stress granule formation induced by measles virus is protein kinase PKR dependent and impaired by RNA adenosine deaminase ADAR1. J. Virol. 87, 756–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ng C. S., Jogi M., Yoo J.-S., Onomoto K., Koike S., Iwasaki T., Yoneyama M., Kato H., Fujita T. (2013) Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J. Virol. 87, 9511–9522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Khaperskyy D. A., Hatchette T. F., McCormick C. (2012) Influenza A virus inhibits cytoplasmic stress granule formation. FASEB J. 26, 1629–1639 [DOI] [PubMed] [Google Scholar]
- 54. Moscat J., Diaz-Meco M. T. (2009) P62 At the Crossroads of Autophagy, Apoptosis, and Cancer. Cell 137, 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Manley S., Williams J. a, Ding W.-X. (2013) Role of p62/SQSTM1 in liver physiology and pathogenesis. Exp. Biol. Med. 238, 525–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Clark K., Nanda S., Cohen P. (2013) Molecular control of the NEMO family of ubiquitin-binding proteins. Nat. Rev. Mol. Cell Biol. 14, 673–685 [DOI] [PubMed] [Google Scholar]
- 57. Aravind L. (2001) The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 26, 273–275 [DOI] [PubMed] [Google Scholar]
- 58. Wang Z., Michaud G. A., Cheng Z., Zhang Y., Hinds T. R., Fan E., Cong F., Xu W. (2012) Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 26, 235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Buchan J. R., Kolaitis R.-M., Taylor J. P., Parker R. (2013) Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kleine H., Herrmann A., Lamark T., Forst A. H., Verheugd P., Lüscher-Firzlaff J., Lippok B., Feijs K. L., Herzog N., Kremmer E., Johansen T., Müller-Newen G., Lüscher B. (2012) Dynamic subcellular localization of the mono-ADP-ribosyltransferase ARTD10 and interaction with the ubiquitin receptor p62. Cell Commun. Signal. 10, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gu T.-L., Cherry J., Tucker M., Wu J., Reeves C., Polakiewicz R. D. (2010) Identification of activated Tnk1 kinase in Hodgkin's lymphoma. Leukemia 24, 861–865 [DOI] [PubMed] [Google Scholar]