Background: Hsp90 binding immunophilins may be regulators of the NF-κB mechanism of action.
Results: FKBP51 and FKBP52 show antagonistic properties on the nuclear accumulation and transcriptional activity of NF-κB.
Conclusion: Both immunophilins modulate NF-κB trafficking and NF-κB transcription when they are recruited to the promoter regions of target genes.
Significance: The competitive effect between both immunophilins in different cell types may explain the pleiotropic actions of NF-κB.
Keywords: 70-Kilodalton Heat Shock Protein (Hsp70), Chromatin Immunoprecipitation (ChIP), Heat Shock Protein 90 (Hsp90), NF-kappaB (NF-κB), Protein Translocation, FKBP51, FKBP52, PPIase, Protein Trafficking, RelA
Abstract
Hsp90 binding immunophilins FKBP51 and FKBP52 modulate steroid receptor trafficking and hormone-dependent biological responses. With the purpose to expand this model to other nuclear factors that are also subject to nuclear-cytoplasmic shuttling, we analyzed whether these immunophilins modulate NF-κB signaling. It is demonstrated that FKBP51 impairs both the nuclear translocation rate of NF-κB and its transcriptional activity. The inhibitory action of FKBP51 requires neither the peptidylprolyl-isomerase activity of the immunophilin nor its association with Hsp90. The TPR domain of FKBP51 is essential. On the other hand, FKBP52 favors the nuclear retention time of RelA, its association to a DNA consensus binding sequence, and NF-κB transcriptional activity, the latter effect being strongly dependent on the peptidylprolyl-isomerase activity and also on the TPR domain of FKBP52, but its interaction with Hsp90 is not required. In unstimulated cells, FKBP51 forms endogenous complexes with cytoplasmic RelA. Upon cell stimulation with phorbol ester, the NF-κB soluble complex exchanges FKBP51 for FKBP52, and the NF-κB biological effect is triggered. Importantly, FKBP52 is functionally recruited to the promoter region of NF-κB target genes, whereas FKBP51 is released. Competition assays demonstrated that both immunophilins antagonize one another, and binding assays with purified proteins suggest that the association of RelA and immunophilins could be direct. These observations suggest that the biological action of NF-κB in different cell types could be positively regulated by a high FKBP52/FKBP51 expression ratio by favoring NF-κB nuclear retention, recruitment to the promoter regions of target genes, and transcriptional activity.
Introduction
Because NF-κB2 was discovered in 1986 as a transcription factor able to bind to the enhancer element of the immunoglobulin κ light chain of activated B cells (1), it became clear that in addition to having a crucial role in innate immunity, it is also able to regulate many other basic functions of the cell such as inflammatory responses, cell development, programmed cell death, proliferation control, and tumorigenesis (see Refs. 2–4) for recent comprehensive updates). NF-κB is actually regarded as a family of structurally related homologues that in mammals comprises p50 (NF-κB1), p52 (NF-κB2), p65 (RelA), RelB, and c-Rel. All of these proteins share a highly conserved DNA binding and dimerization domain known as RHD (Rel homology domain) and could potentially form up to 15 types of dimers. Nonetheless, the physiological existence of all of these dimers has not been demonstrated to date, the p50·RelA heterodimer (the p65/RelA subunit will be named RelA onward) being unquestionably the most abundant in all cell types (5). On the other hand, the NF-κB family may be divided from the transcriptional perspective into two groups based on the occurrence of the CT-transactivation domains, which are only present in RelA, RelB, and c-Rel (5).
All Rel factors form homodimers or heterodimers with the sole exception of RelB, which forms only heterodimers. The relative abundance of different NF-κB proteins may vary in different tissues and cell types, whereas the p50·RelA heterodimer is highly ubiquitous and the most frequently detected complex in most cell types of all tissues (6).
In unstimulated cells p50·RelA heterodimers are normally retained in the cytosol in an inactive form due to their association with the inhibitory factor IκB through an ankyrin repeat domain (7). Upon cell stimulation, IκB is phosphorylated by the kinase IKK, dissociated from the complex with NF-κB proteins, and targeted to proteasomal degradation. This allows the nuclear translocation of the NF-κB heterodimer (8). Nonetheless, a dynamic nuclear-cytoplasmic shuttling of NF-κB complexes always takes place (9–11) allowing a low basal transcriptional activity of NF-κB given the fact that the IκB·NF-κB complex is also subject to dynamic dissociation/reassociation events. The nuclear-cytoplasmic shuttling of NF-κB resembles that observed of steroid receptors where the inactive cytoplasmic form of these ligand-dependent transcription factors must translocate to the nucleus upon stimulation with steroid hormones (12–14).
In previous studies our laboratory and others have reported that the 51- and 52-kDa FK506-binding proteins FKBP51 and FKBP52 are responsible in a mutually exclusive fashion for the retrotransport mechanism of GR (15, 16) and mineralocorticoid receptor (17, 18). Both FKBPs are also regulators of the ligand-dependent transcriptional activity for those receptors (18–20) and other members of the family such as PR (21, 22), AR (23, 24), and to a minor degree, estrogen receptor α (22, 25). These Hsp90 binding immunophilins are highly homologous and share 60% homology and 75% similarity (26). They are structurally characterized by the presence of two key sequences: the TPR domain, through which they bind to Hsp90, and the peptidyl-prolyl isomerase (PPIase) domain (27), where the macrolide FK506 and also the dynein·dynactin motor complex bind. Both domains are essential for the retrotransport mechanism of steroid receptors (14, 28), although the enzymatic activity of the PPIase domain does not appear to be essential. Upon steroid binding, FKBP51 is released from the receptor·Hsp90 heterocomplex and is replaced by FKBP52, which recruits dynein·dynactin motor proteins favoring the transport of the receptor to the nucleus on microtubule tracks (29).
Inasmuch as both types of transcription factors, steroid receptors and NF-κB, show similar requirements for subcellular redistribution upon the onset of a given activating stimulus, we asked whether NF-κB proteins could also be regulated by immunophilins in a similar fashion as GR and mineralocorticoid receptor. Therefore, we investigated the potential association of FKBP51 and FKBP52 with RelA, whether such complex affects the nuclear translocation of RelA, and how these immunophilins influence the transcriptional activity of NF-κB.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments
Human embryonic kidney HEK 293-T cells were grown at 37 °C in a humidified atmosphere of 5% CO2, 95% O2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum and antibiotics (50 units/ml penicillin and 50 mg/ml streptomycin). Human placenta choriocarcinoma BeWo cells were grown in DMEM/F-12 (1:1) medium supplemented with 10% fetal bovine serum and antibiotics. Transcriptional activity was measured in cells transfected with pNF-κB cis-reporter plasmid (Stratagene, La Jolla, CA, catalog no. 219078-51) as described previously (18). After 24 h post-transfection, cells were washed, and the culture medium was replaced by serum-free DMEM. Cells were stimulated with 100 ng/ml phorbol 12-myristate 13-acetate (PMA) (Sigma) for 7 h. Results were expressed as the percentage of NF-κB activation after discounting the basal activity and normalized to the β-galactosidase expression. Overexpression of FKBP proteins (wild type or mutants) was achieved by transfection of 1 μg of pCI-neo-hFKBPs or empty vector per well of 35 mm containing 1 ml of medium following the calcium phosphate precipitation standard method (18). The HEK-FKBP51 cell line stably transfected with hFKBP51 (termed 51+) was obtained by transfection of HEK 293 cells with pCI-neo-hFKBP51 followed by selection in 400 μg/ml Geneticin (G418; Invitrogen)-containing medium for a period of 30 days. The stably transfected cells were usually cultured in DMEM supplemented with 10% fetal bovine serum and 200 μg/ml G418.
Zymographies
Gelatin zymography assays for PMA-stimulated BeWo cells were achieved following a standard method described in the literature (30). Briefly, the assays were performed running a 10% SDS-PAGE with 1 mg/ml gelatin polymerized in the gel (0.75-mm thickness). Gels were loaded with samples diluted (1:1) in non-reducing buffer (12.5% 0.5 m Tris-HCl, pH 6.8, 10% glycerol, 4% SDS, and 0.05% bromphenol blue). After electrophoresis, gels were washed twice with 2.5% Triton X-100 in PBS for 15 min, incubated with the reaction buffer (50 mm Tris-HCl, 30 mm CaCl2, 0,15 mm NaCl, pH 7.6) for 18 h at 37 °C, stained with 0.5% Coomassie Brilliant Blue R-250 solution (containing 30% methanol and 10% acetic acid) for 30 min under gentle shaking, and destained twice for 20 min with a solution containing 5% ethanol and 8% acetic acid. Gels were scanned with a charge-coupled device (Fuji Film) and analyzed by densitometry with National Institutes of Health Scion-Image Software.
IL-6 Expression
BeWo cells were plated in 24-well plates at a concentration of 3 × 103 cells per well. The next day cells were transfected with the proper pCI-neo plasmid encoding for the proper immunophilin (or empty vector), and 24 h after the culture, medium was replaced by serum-free DMEM-F-12 and incubated with or without 100 ng/ml PMA for 24 or 48 h or with IL-1β (0.5 ng/ml). The concentration of IL-6 en the culture supernatant was measured using commercial ELISA kit from Roche Applied Science according to the manufacturer's guidelines. Results were normalized by the total amount of protein of each well.
Indirect Immunofluorescence Assays
Cells grown on polylysine-coated coverslips were fixed with 3% p-formaldehyde for 30 min and permeabilized with 0.1% Triton X-100 as described (31). Indirect immunofluorescence was performed using 1/100 dilutions of the following primary antibodies: rabbit monoclonal IgG anti-FKBP51 (Affinity BioReagents, Golden, CO), MG19 mouse monoclonal IgG anti-FKBP51 (32), UP30 rabbit antiserum against FKBP52 (33), and anti-HA or anti-RelA mouse monoclonal antibodies (Santa Cruz Biotechnology, Dallas, TX) to detect transfected RelA protein or endogenous RelA protein, respectively. These incubations were followed by a subsequent incubation with 1/200 dilutions of the proper Alexa-labeled secondary antibody (Molecular Probes, Eugene, OR). The subcellular distribution of these proteins was monitored by confocal microscopy using an Olympus Fluoview 1100 microscope. Image analyses for co-immunolocalization of FKBPs and RelA were carried out using the colocalization threshold plug-in of the Image-J program (v.1.45) from the National Institutes of Health as described (31).
Immunoprecipitation Assays
After the indicated treatment, cells were washed 3 times with phosphate-buffered saline at pH 7.3, scrapped, and homogenized in HEM buffer (10 mm Hepes buffer at pH 7.4, 1 mm EDTA, and 20 mm Na2MoO4). After a centrifugation for 30 min at 4 °C at 120,000 × g, RelA was immunoprecipitated from 300 μl of cytosol (∼4 mg/ml protein) with 2 μl of anti-RelA (or anti-FKBP51) antibody and 30 μl of 50% g/v protein A-Sepharose (Sigma) under rotation for 2 h at 4 °C. Pellets were washed 5 times with 1 ml of HEM buffer, and proteins were resolved by Western blotting (0.05%v/v dilution for primary antibodies and 0.02% v/v dilution for horseradish peroxidase-labeled secondary antibodies) and visualized by enhanced chemiluminescence.
Electrophoretic Mobility Shift Assays (EMSA)
Nuclear fractions of HEK 293-T cells were isolated as previously described (34). An oligonucleotide containing the consensus sequence of NF-κB (AGTTGAGGGGACTTTCCCAGG; catalog no. E3292; Promega, San Luis Obispo, CA) was end-labeled with [γ-32P]ATP (PerkinElmer Life Sciences) using T4 polynucleotide kinase (Promega, Madison, WI) and purified using ChromaSpin-10 columns (Sigma). Samples were incubated for 20 min at room temperature in binding buffer (10 mm Tris-HCl buffer, pH 7.5, 20% (v/v) glycerol, 1 mm MgCl2, 0.5 mm EDTA, 0.5 mm dithiothreitol, 50 mm NaCl, and 0.05 mg/ml poly(dI-dC)) with labeled oligonucleotide (2–3 × 105 cpm). For supershift assays, samples were incubated in the presence of mouse antibodies anti-RelA, anti-FLAG (clone M2, Sigma), anti-HA, or mouse non immune IgG (Santa Cruz Biotechnology., Dallas, TX) before the addition of labeled oligonucleotide. The products were separated by electrophoresis in a 5% (w/v) non-denaturing polyacrylamide gel using 45 mm Tris borate, 1 mm EDTA as the running buffer. Gels were dried, the radioactivity was revealed with Kodak x-ray film, and images were scanned in a Fuji Film Intelligent Dark Box II with a Fuji film LAS-1000 digital camera (Stanford, CA). The specificity of the shifted complexes was assessed after a pretreatment of 30 min with a 1–100-fold molar excess of cold competitor (κB) and by using non-immune antibodies.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed following a modification of a previously described method (35). Briefly, proteins and DNA were cross-linked after HEK 293-T cell treatment with 1% formaldehyde for 10 min at 37 °C and neutralized with glycine added at a final concentration of 0.125 m. Cells were rinsed twice with cold PBS, scraped, and centrifuged at 800 × g for 5 min at 4 °C. Cells were washed with PBS and homogenized in SDS lysis buffer (1% (w/v) SDS, 10 mm EDTA, 50 mm Tris-HCl buffer, pH 8.1) containing protease inhibitors (protease inhibitor mixture, Roche Applied Science). Chromatin was fragmented in 500–800-bp fragments by sonication, lysates were centrifuged at 12,000 rpm for 10 min at 4 °C, and the supernatants were diluted 10-fold in ChIP dilution buffer (1% (v/v) Triton X-100, 2 mm EDTA, 150 mm NaCl, 20 mm Tris-HCl buffer, pH 8.1). Extracts were precleared with protein A-agarose 50% slurry (antibody binding beads, Diagenode, Denville, NJ 07834) for 30 min at 4 °C. Immunoprecipitations were performed overnight at 4 °C using the UP30 rabbit antiserum against FKBP52 or the MG19 mouse monoclonal IgG against FKBP51. Samples were incubated with protein A-agarose for 1 h, and the immune complexes were collected by centrifugation (1000 rpm at 4 °C), washed, and extracted with 1% SDS, 0.1 m NaHCO3. The cross-linking was reversed by heating with 5 m NaCl at 65 °C for 4 h. Chromatin-associated proteins were digested with proteinase K, and the samples were extracted with phenol/chloroform followed by precipitation with ethanol. The pellets were lysed in nuclease-free water and subjected to real-time PCR. The MMP9 primers used for PCR were synthesized by GenBiotech (Buenos Aires, Argentina), and their sequences are CCCTCCCTCCCTTTCATACAGTTC (forward) and GCTTACACCACCTCCTCCTCTC (reverse).
Statistics
Data were analyzed by one-way analysis of variance using Statview 5.0 (SAS Institute Inc., Cary, NC). Fisher's least significance difference test was used to examine differences between group means. A p < 0.05 was considered statistically significant. Values are shown as the means ± S.E.
RESULTS
FKBP51 and FKBP52 Affect NF-κB Nuclear Translocation
In previous studies we showed that FKBP52 favors the nuclear translocation of steroid receptors (15, 17, 36), p53 mutant isoforms (37), and RAC3 (38). This is achieved by linking the corresponding client protein·Hsp90 complex to the dynein·dynactin motor complex. On the other hand, it is regarded that FKBP51 impairs this mechanism (18, 19, 39). Recently, it has been reported that the RelA subunit of NF-κB is also associated to components of the dynein·dynactin complex (11, 40). Inasmuch as all of these transcription factors (steroid receptors, p53, RAC3, and NF-κB) are primarily localized in the cytoplasm and must be transported to the nucleus upon proper stimulation, we hypothesized that NF-κB nuclear translocation could also be modulated by Hsp90 binding immunophilins in a similar fashion.
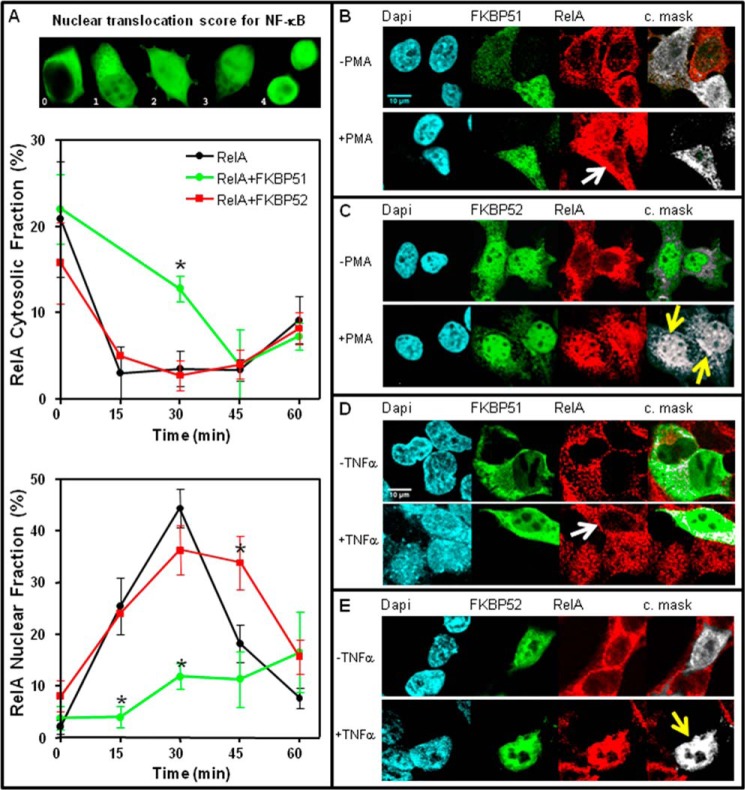
To test that premise we used HEK 293-T cells as they not only show high efficiency for transfections but also a relatively low level of expression of endogenous FKBP51 and FKBP52 compared with other cell types, making these cells a highly sensitive system to evidence changes in the biological responses due to variations in the expression level of these immunophilins. Cells were cotransfected with pcDNA3-HA-RelA and pCI-neo empty vector, pCI-neo-FLAG-hFKBP51, or pCI-neo-hFKBP52. After 24 h the cells were stimulated with a time-course of PMA (0–60 min), and the nuclear localization of RelA was analyzed by confocal microscopy and scored (Fig. 1A). Although RelA reaches a maximal nuclear accumulation after 30 min of stimulation with PMA and cycles back to cytoplasm very rapidly (Fig. 1A, lower panel), the overexpression of FKBP51 strongly delays nuclear accumulation such that RelA remains for longer periods of time in the cytoplasm (Fig. 1A, middle panel). On the other hand, the overexpression of FKBP52 does not affect the nuclear import rate of RelA but significantly increases the nuclear retention time of the transcription factor. The rapid cycling to the cytoplasm of NF-κB observed in control cells after 30 min agrees with previously reported data (9, 34, 41, 42), whereas both effects of the immunophilins, i.e. the nuclear translocation rate and nuclear retention time of the associated cargo, are quite similar to those previously reported for steroid receptors (14). This observation suggests a similar regulatory mechanism by both TPR-domain immunophilins for both unrelated transcription factors.
FIGURE 1.
FKBP51 and FKBP52 influence NF-κB nuclear translocation. A, RelA nuclear translocation rate. HEK 293-T cells were transfected with pcDNA3-HA-RelA and pCI-neo (black circles), pcDNA3-HA-RelA and pCI-neo-FLAG-hFKBP51 (green circles), or pcDNA3-HA-RelA and pCI-neo-hFKBP52 (red squares). After 24 h, cells were incubated with PMA (100 ng/ml), fixed at the indicated times, and scored for the nuclear and cytoplasmic localization of RelA after performing an indirect immunofluorescence by confocal microscopy. Data are the mean ± S.E. after scoring 100 cells in each experiment (n = 4). The upper panel shows cell images that are an example of the score assigned according to the distribution of HA-RelA. The cytosolic fraction depicted in the middle panel represents the percentage of transfected cell showing a score equal to 0 (i.e. cytosolic distribution only), and the RelA nuclear fraction depicted in the lower panel represents the percentage of transfected cell showing a score equal to 4 (nuclear distribution only). *, Significantly different compared with the group of cells not transected with the immunophilin. B, endogenous NF-κB colocalizes with FKBPs. HEK 293-T cells transfected with pCI-neo-FLAG-hFKBP51 (panels B and D) or pCI-neo-hFKBP52 (panels C and E) were incubated for 30 min with (+) or without (−) 100 ng/ml PMA (panels B and C) or 10 ng/ml TNFα (panels D and E). An indirect immunofluorescence was performed using anti-FKBP51 or anti-FKBP52 antibodies (green) and anti-RelA antibody (red). DAPI was used for nuclear staining (cyan). The extreme right images correspond to a colocalization mask (c.mask) analysis between the immunophilins and RelA fluorescent signals by using the colocalization threshold plug-in of the ImageJ program (v.1.45) from the National Institutes of Health. Bar scale, 10 μm. Arrows are pointing to those cells described in the text.
Fig. 1, B–E, show indirect immunofluorescence images by confocal microscopy of the RelA subunit of NF-κB (red) and either immunophilin FKBP51 or FKBP52 (green). The far right images of all panels show a colocalization analysis performed with the ImageJ program to analyze the actual colocalization mask between both proteins (shown in white). Note the poor accumulation of RelA in the nucleus of the cell that overexpresses FKBP51 (Fig. 1, B and D, white arrows) compared with the other untransfected cells of the field. Interestingly, both immunophilins increased the respective nuclear pool colocalizing with NF-κB upon cell stimulation with either stimulus, PMA, or TNFα (Fig. 1, C and E, yellow arrows), suggesting the potential existence of nuclear interactions.
FKBP51 and FKBP52 Form Complexes with NF-κB
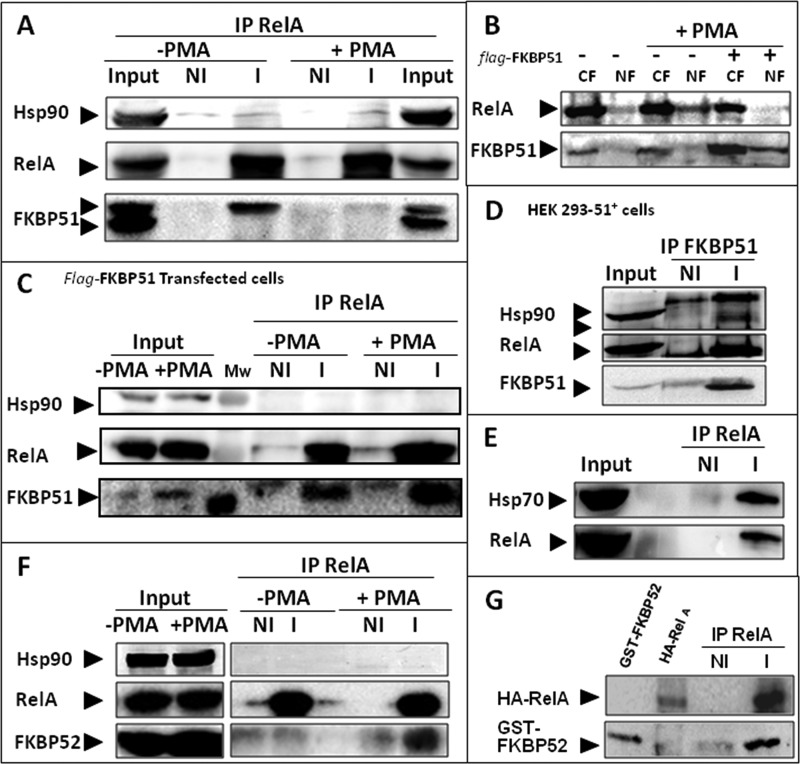
In view of the previous observations we asked whether both immunophilins form complexes with NF-κB. In this sense, we noted some differences in the subcellular localization of endogenously and exogenously expressed FKBP51. Fig. 2A shows a co-immunoprecipitation assay of endogenous FKBP51 with endogenous RelA in unstimulated cells (−PMA). The lack of significant concentration of FKBP51 in the nucleus observed by confocal microscopy in Fig. 1 was confirmed by cell fractionation (Fig. 2B), demonstrating that RelA·FKBP51 complexes are mostly cytoplasmic unless FKBP51 is highly expressed, a situation where a significant fraction of FKBP51 is also recovered in the nuclear fractions of stimulated cells. This observation resembles the well known action of this immunophilin on the subcellular distribution of steroid receptors (17, 18, 43). Consistent with those images observed in Fig. 1 and the cell fractionation shown in Fig. 2B, Fig. 2C also shows that higher levels of expression of FKBP51 make the immunophilin stay more associated to NF-κB in PMA-stimulated cells than in unstimulated cells (compare with Fig. 2A for endogenous FKBP51). Fig. 2D confirms the interaction by reverse co-immunoprecipitation, i.e. RelA was co-immunoadsorbed with FKBP51. As expected, Hsp90 is also co-immunoadsorbed as the immunophilin forms several complexes with Hsp90. However, it is noteworthy that the interaction of FKBP51 with the transcription factor is Hsp90-independent (Fig. 2A). This is a structural difference regarding the steroid receptors complexes, where the association of TPR-domain immunophilins with the transcription factor is entirely dependent on Hsp90. Fig. 2E also demonstrates that endogenous Hsp70 is part of an endogenous complex with RelA, as was recently suggested in similar assays with overexpressed proteins (44).
FIGURE 2.
FKBP51 and FKBP52 form complexes with NF-κB. A, HEK 293-T cells were incubated with (+) or without (−) 100 ng/ml PMA for 30 min, and endogenous RelA was immunoadsorbed with a specific IgG (I) or a non-immune IgG (NI). IP, immunoprecipitate. B, HEK 293-T cell transfected with pCI-neo empty vector (−) or pCI-neo-FLAG-hFKBP51 (+) were incubated with 100 ng/ml PMA for 30 min (+PMA) and then subjected to subcellular fractionation into cytosolic (CF) and nuclear (NF) fractions as described in a previous work (34). C, RelA was immunoprecipitated with a specific IgG (I) or a non-immune IgG (NI) from whole extracts of cells co-transfected with pcDNA3-HA-RelA and pCI-neo-FLAG-hFKBP51. D, FKBP51 was immunoprecipitated from whole extracts of the HEK 293-FKBP51 cell line that constitutively overexpresses FKBP51. E, endogenous RelA was immunoprecipitated from whole cell extracts of HEK 293-T and hsp70 was revealed by Western blot. F, HEK 293-T cells were incubated with (+) or without (−) 100 ng/ml PMA for 30 min, and RelA was immunoprecipitated with a specific IgG (I) or a non-immune IgG (NI). F, HEK 293-T cells were incubated with (+) or without (−) 100 ng/ml PMA for 30 min. Endogenous RelA was immunoadsorbed, and proteins were resolved by Western blot. G, direct interaction of FKBP52 with RelA. HA-tagged RelA was immunopurified, stripped of associated proteins with RIPA buffer/1 M NaCl, and incubated with GST-FKBP52 recombinant protein purified as described in a previous work (17).
Inasmuch as FKBP51 is associated with steroid receptors in a ligand-free medium and is exchanged by FKBP52 upon steroid binding (16–18), whether or not this is the case for NF-κB activation was investigated. Fig. 2F shows that FKBP52 is recruited to RelA in an Hsp90-independent manner when cells were incubated with PMA. Because FKBP51 is no longer recovered with RelA in the presence of PMA (Fig. 2A), it is likely that FKBP51 is swapped with FKBP52 as was previously described for steroid receptors. In contrast to the case for steroid receptors, the binding of immunophilins to NF-κB does not depend on the presence of Hsp90. Therefore, it was speculated that FKBP52 could bind directly to RelA. This is supported by the direct association of purified RelA and recombinant FKBP52 shown in Fig. 2G. This direct association strengthens the hierarchical role of these immunophilins in signal transduction cascades as it implies that these proteins exert regulatory roles by themselves.
FKBP51 and FKBP52 Regulate the Transcriptional Activity of NF-κB by Recruitment to Promoter Regions of Target Genes
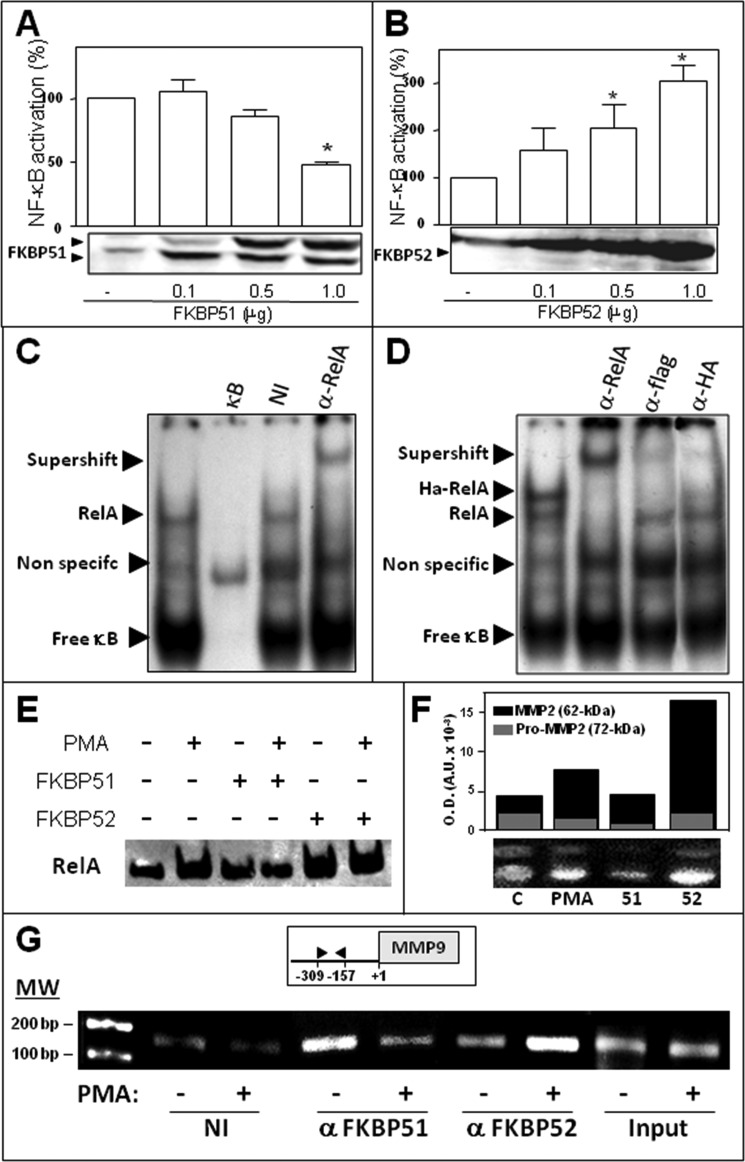
To determine if the expression level of immunophilins affects NF-κB transcriptional activity, FKBP51 or FKBP52 were expressed at different levels, and NF-κB-dependent transcription was triggered with PMA. Fig. 3A shows that FKBP51 inhibits NF-κB transcriptional activity, whereas Fig. 3B shows that FKBP52 strongly favored such activity.
FIGURE 3.
FKBP51 and FKBP52 modulate NF-κB transcriptional activity by recruitment to promoter regions of target genes. HEK 293-T cells were transfected with increasing amounts of pCI-neo-FLAG-hFKBP51 (A) or pCI-neo-hFKBP52 plasmid (B); the cells were stimulated with 100 ng/ml PMA for 7 h, and NF-κB transcriptional activity was measured with a luciferase reporter-gene and normalized by β-galactosidase activity. The results are expressed as the percentage of NF-κB activation after subtracting the basal activity (mean ± S.E. of five independent experiments). *, Significantly different compared with the control group transected with empty vector (p < 0.01). C, RelA binding to an NF-κB consensus sequence of DNA was analyzed by EMSA using the nuclear fractions of HEK 293-T cells stimulated with 100 ng/ml PMA for 60 min. The specificity of the shifted complexes detected by EMSA was evaluated after a pretreatment of 30 min with an excess of cold competitors (κB), nonspecific immunoglobulin (NI), and a specific antibody anti-RelA (α-RelA). D, HEK 293-T cells were co-transfected with pcDNA3-HA-RelA and pCI-neo-FLAG-hFKBP51 and incubated with 100 ng/ml PMA for 60 min, and the extracts were pretreated with a specific antibody anti-RelA (α-RelA), anti-FLAG tag antibody (α-FLAG), and anti-hemagglutinin tag antibody (α-HA). Arrowheads show specific DNA-protein complexes, antibody-DNA-protein complexes (supershift), or free κB consensus sequences. E, profiles of RelA binding to DNA in cells overexpressing FKBP51 or FKBP52 that were stimulated (or not) with PMA. F, zimography for MMP2 in BeWo cells. C, control cells; PMA, cells treated with 100 ng/ml PMA; 51 and 52, cells transfected with 4 μg of pCI-neo-hFKBP51 or pCI-neo-hFKBP52 and then treated with PMA. The bar graph represents the optical density for both bands MMP2 full-length (black) and pro-MMP2 (gray). G, ChIP using the NF-κB promoter site of MMP-9 in HEK 293-T cells treated (+) or not (−) with 100 ng/ml PMA for 30 min. ChIPs were performed using antibodies against both immunophilins or a nonspecific immunoglobulin (NI), followed by quantitative real-time PCR. Input chromatin (1%) was removed from samples before immunoprecipitation and also subjected to quantitative real-time PCR to control any potential variation of the starting material.
We next performed an EMSA assay to evaluate the potential association of NF-κB with a consensus sequence that is recognized by NF-κB. The supershift observed in Fig. 3C with an anti-RelA antibody demonstrates the specificity of signal for endogenous RelA. Fig. 3D shows loss of signal for HA-RelA with both antibodies, anti-FLAG and anti-HA, in samples derived from FLAG-FKBP51 and HA-RelA co-transfected cells. Fig. 3E shows the increased recruitment of RelA to the DNA in nuclear cell extracts treated with PMA, a property that is impaired by FKBP51 and favored by FKBP52.
To assay the biological response of an endogenous gene, BeWo cells were used. This placenta choriocarcinoma cell line produces high amounts of matrix metallopeptidase 2 (MMP2) (30) and responds to PMA stimulation via NF-κB activation (45). In agreement with the results described above, the zymographies shown in Fig. 3F demonstrate that a high level of expression of FKBP51 reduced the release of MMP2 to the medium, whereas the opposite effect was observed for FKBP52. Importantly, all these observations related to the role of immunophilins on the biological response of NF-κB target genes were confirmed by ChIP assays for endogenous immunophilins (Fig. 3G). Consistent with the endogenous immunophilin swapping observed in Fig. 2, A and F, this ChIP assay demonstrates that endogenous immunophilins exchange on the promoter sequence of the matrix metallopeptidase 9 (MMP9) gene, a known classic target of NF-κB (46). In contrast to other MMPs that are cell-specific, MMP9 is widely expressed in various cell types (47, 48). Therefore, the regulatory mechanisms for this MMP should be more complex than for others. These results suggest that the expression balance of FKBP51 and FKBP52 may be part of such still poorly understood mechanism of regulation.
FKBP51 and FKBP52 Regulate Endogenous NF-κB-dependent Biological Responses Triggered by Various Stimuli
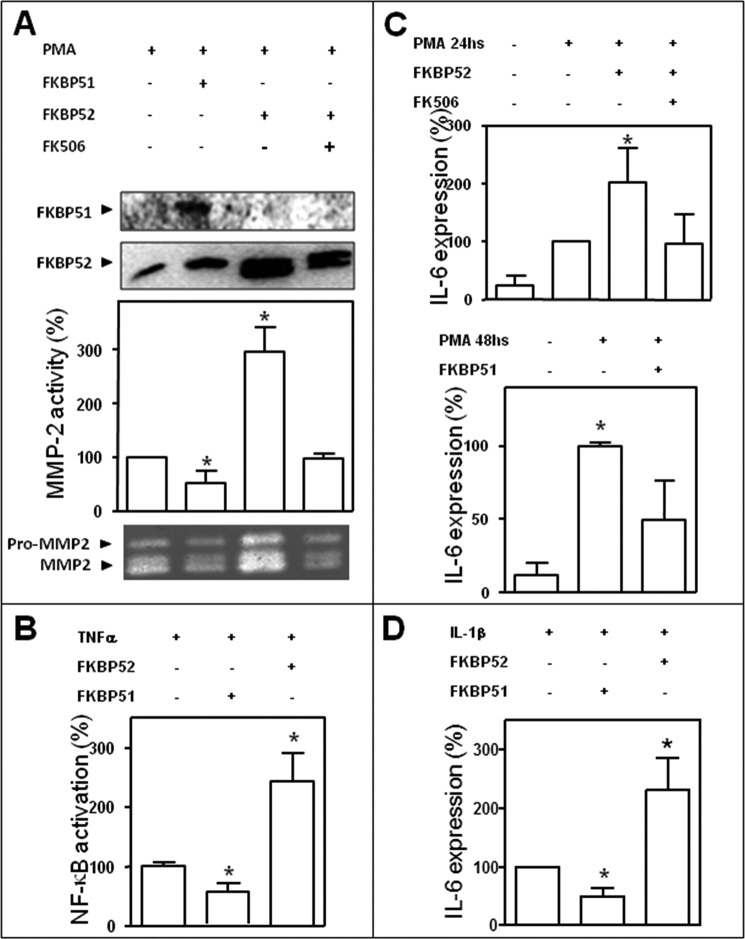
Because the experiments shown in Fig. 3, F and G, demonstrate that FKBP52 is a strong activator of the NF-κB-dependent biological response, we asked whether the ligand macrolide FK506 could affect that response. The zymography shown in Fig. 4A demonstrates that the endogenous biological response is fully abrogated by preincubation of the cells with the inhibitory macrolide FK506, suggesting an apparent dependence of the biological action of the immunophilin with its PPIase enzymatic activity. FKBP51 was included as an internal control. In a separate zymography assay, FK506 was also used to prevent the inhibitory action of FKBP51, but no differences with the control were observed (data not shown), suggesting that at variance of FKBP52, the PPIase activity appears not to be required for FKBP51.
FIGURE 4.
FKBP51 and FKBP52 regulate the expression and activity of endogenous NF-κB target genes in a stimulus-independent manner. A, a gelatin zymography assay performed in BeWo cells incubated with PMA as in Fig. 3F shows that the stimulatory action of FKBP52 on NF-κB action is impaired by cell preincubation of cells with 1 μm FK506 for 30 min. Bar graphs are the optical densities scanned for four independent gels (mean ± S.E.). Results are the percentage of induced MMP activity after subtracting the basal activity. *, Significantly different compared with the control group (p < 0.003). B, HEK 293T cells were stimulated with 10 ng/ml TNFα for 6 h, and NF-κB activation was assessed by the induction of a luciferase reporter genes. IL-6 expression was measured after treatment of BeWo cells with 100 ng/ml PMA for 24 or 48 h (C) or with 0.5 ng/ml IL-1β for 24 h (D). All the transfections with immunophilins or empty vector used 1 μg of DNA. Results of panels B, C, and D represent the mean ± S.E. of three independent experiments. *, Significantly different from controls at p < 0.05.
To determine if immunophilins affect NF-κB transcriptional activity triggered by other stimuli than PMA, TNFα also was assayed. HEK 293T cells were treated with 10 ng/ml TNFα for 6 h. Luciferase activity induction confirmed that FKBP51 has inhibitory action, and FKBP52 is a strong enhancer of NF-κB activity (Fig. 4B).
It has been previously reported that BeWo cells produce IL-6 via NF-κB upon stimulation with phorbol esters or IL-1β (49, 50). Therefore, these cells were stimulated with PMA for 24 h and 48 h (Fig. 4C) or IL-1β (Fig. 4D), and IL-6 expression was measured confirming the roles of both immunophilins on different types of NF-κB target genes.
Role of the FKBP51 and FKBP52 TPR Domains
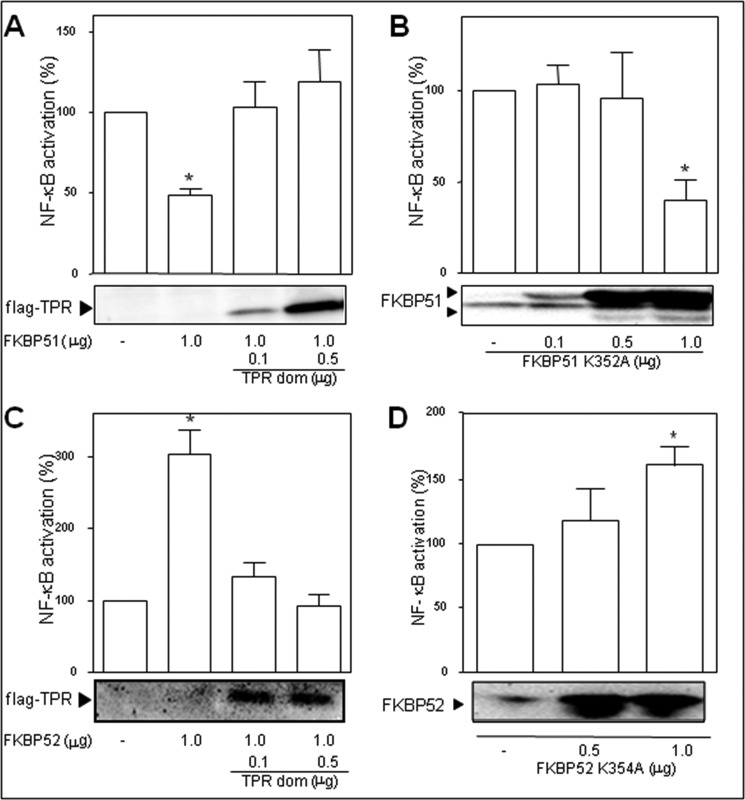
Usually, FKBP51 and FKBP52 form heterocomplexes with Hsp90 via their TPR domains, such that both immunophilins form a functional complex with this essential chaperone. Nonetheless, the co-immunoprecipitation assays shown in Fig. 2 demonstrate that the complexes of these FKBPs with NF-κB are Hsp90-independent. We next analyzed such unexpected property from the functional perspective. The potential involvement of the TPR domain of FKBP51 and FKBP52 on the regulation of the transcriptional activity of NF-κB was assessed by competition with the TPR domain. Both the inhibitory action of FKBP51 (Fig. 5A) and the stimulatory effect of FKBP52 (Fig. 5C) were reversed by the increased expression of the TPR peptide. To analyze whether this effect is due to the ability of these immunophilins to interact with Hsp90 in some other point of the signaling cascade, point mutants in the TPR domain that are unable to interact with Hsp90 were assayed. The inhibitory action of FKBP51 was not modified (Fig. 5B) compared with controls transfected with wild type FKBP51, whereas the stimulatory action of FKBP52 was still observed (Fig. 5D). This observation agrees with the functional recruitment of FKBP52 shown in the co-immunoprecipitation assays of Fig. 2. Taken together, these results suggest that the regulatory action of both immunophilins may involve their TPR-domains but not due to association with Hsp90.
FIGURE 5.
Contribution of the TPR domain to NF-κB activation. Cells were co-transfected with 1 μg of pCI-neo-hFKBP51 (A) or 1 μg of pCI-neo-hFKBP52 (C) and increasing amounts of pCMV6 FLAG-TPR (A and C) or were transfected with increasing amounts of point mutants in the TPR domain that are unable to interact with Hsp90, i.e. pCI-neo-FLAG-hFKBP51 K352A (B) or pCI-neo-hFKBP52K354A (D). After 7 h of stimulation with 100 ng/ml PMA, transcriptional activity was measured as in the previous figures. Results are the mean ± S.E. of three independent experiments performed in quadruplicate. *, Significantly different compared with the control group that was not transfected with FKBPs (p < 0.05).
FKBP52 Regulatory Action Is PPIase Activity-dependent
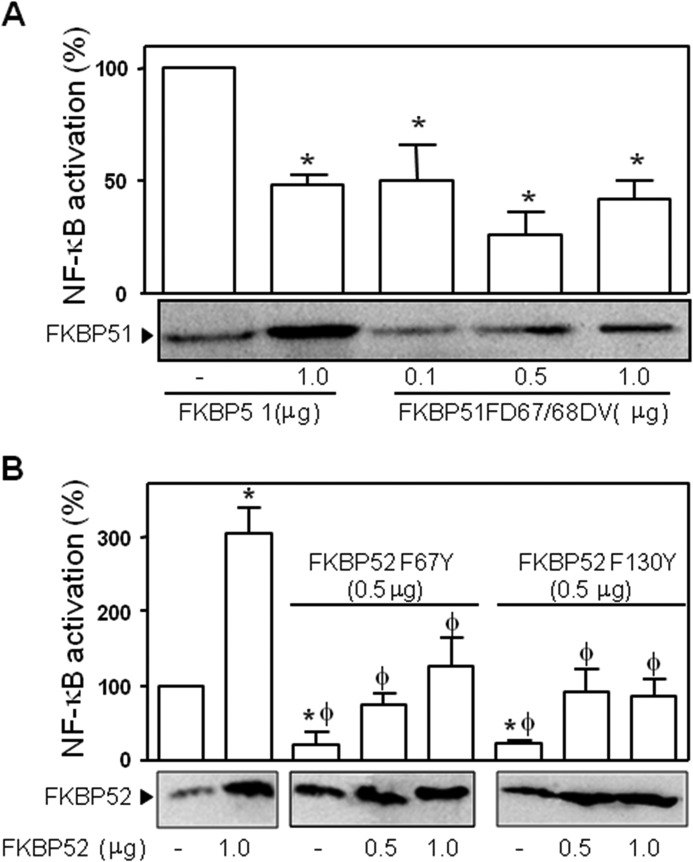
Because the PPIase activity can affect the conformation and orientation of small peptides and the helixes of larger proteins, we analyzed the potential role of such enzymatic activity in the regulation of the biological response of NF-κB. The first approach was to analyze the transcriptional activity of NF-κB in cells stimulated with PMA in the presence of FK506, a pharmacological inhibitor of the PPIase activity of FKBP proteins. Although the overexpression of FKBP51 reduced the transcriptional activity of NF-κB to 48.7 ± 4% compared with controls transfected with empty vector, the presence of 1 μm FK506 did not modify that effect of the immunophilin (47.2 ± 7.5%), suggesting that the inhibitory action of FKBP51 does not require its PPIase activity. This result agrees with the lack of effect of this drug on MMP2 release upon stimulation of BeWo cells with PMA as discussed before. Nonetheless, this is a pharmacological approach. A conclusive confirmatory experiment was the transfection of an FKBP51 mutant devoid of PPIase activity (FD67/68DV) (Fig. 6A). Therefore, it is concluded that the effect of FKBP51 on NF-κB transcriptional activity depends on the presence of the immunophilin itself and does not lay on its PPIase activity.
FIGURE 6.
Contribution of the respective PPIase domains to NF-κB activation. Cells were transfected with pCI-neo-hFKBP51 or pCI-neo-hFKBP51 FD67/68DV (A) or with pCI-neo-hFKBP52 or either plasmid pCI-neo-hFKBP52 F67Y or pCI-neo-hFKBP52F130Y encoding for PPIase inactive FKBP52 (B). After 7 h of stimulation with 100 ng/ml PMA, transcriptional activity was measured as in the previous figures. Results are expressed as the percentage of NF-κB activation after subtracting the basal activity (mean ± S.E. of four independent experiments performed in quadruplicate). *, Significantly different with regard to the group that was not transfected with FKBPs (p < 0.05). φ, Significantly different versus the FKBP52 transfected group (p < 0.05). #, Significantly different versus the FKBP51 transfected group (p < 0.01).
On the other hand the highly stimulatory action of FKBP52 (303.7 ± 34.7%) was almost fully abolished by FK506 (31.5 ± 15.0%). Similarly, the overexpression of two PPIase inactive mutants of FKBP52 (F67Y and F1300Y) also decreased the transcriptional activity of NF-κB to a similar extent (Fig. 6B). This demonstrates a relevant role for the PPIase activity of FKBP52 on the stimulatory action of NF-κB. As expected, the co-transfection of increasing amounts of wild type FKBP52 partially reversed the inhibitory action of the mutants to the levels measured for controls.
The Stimulatory Effect of FKBP52 Is Antagonized by FKBP51 and the GR
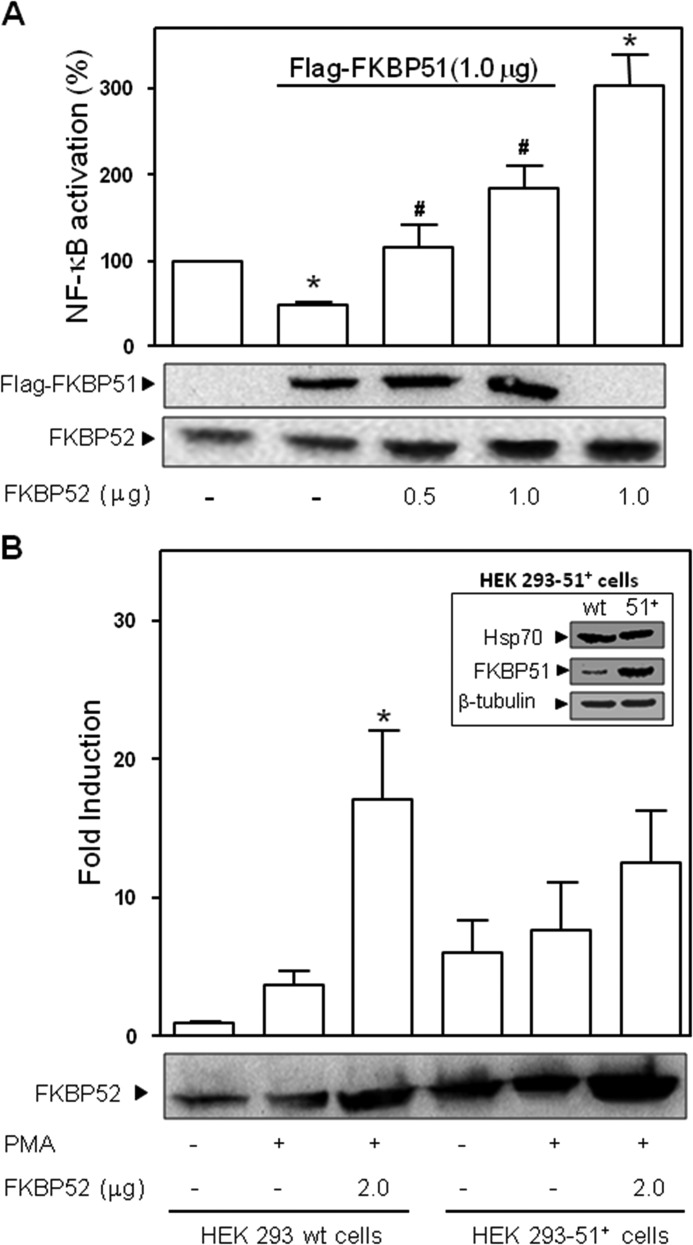
Given the fact that FKBP51 and FKBP52 share high homology and because they usually exhibit opposite effects in other systems, we hypothesize that the opposite biological action evidenced in this work could be due to competition between both proteins. Cells were transfected with a constant amount of FKBP51 and increasing concentrations of FKBP52 (Fig. 7A). As expected, the inhibitory action of FKBP51 was reversed by FKBP52. Note that the high stimulatory effect of 1 μg FKBP52 alone is reduced when a similar amount of FKBP51 is also co-transfected. In line with these observations, a cell line that overexpresses FKBP51 in a stable manner shows lower activation by FKBP52 (Fig. 7B).
FIGURE 7.
Competition of FKBP51 and FKBP52 regulates NF-κB activation. A, transcriptional activity measured in cells treated with PMA that were previously co-transfected with 1 μg of pCI-neo-FLAG-hFKBP51 and increased amounts of pCI-neo-hFKBP52 (mean ± S.E. of four independent experiments). *, Significantly different versus the control group not transfected with FKBP51 or FKBP52 (p < 0.05). #, Significantly different versus the FKBP51 transfected group (p < 0.01). B, inhibition of the stimulatory action of FKBP52 in HEK 293 (no T) fibroblasts that constitutively overexpress FKBP51. The FKBP52-dependent potentiation of NF-κB transcriptional activity measured in HEK no T wild type cells (HEK 293 wt cells) stimulated with PMA is significantly attenuated in the same cell type overexpressing FKBP51 (HEK 293–51+ cells). *, Significantly different from untreated control groups (p < 0.01).
Because GR is able to form heterocomplexes with NF-κB and also recruits immunophilins to its own chaperone complex, it was speculated that the biological action of NF-κB could also be antagonized by GR. The transfection of pSV2Wrec-GR encoding for rat GR did not affect either basal (0.8 ± 0.3%) or PMA-induced activity of endogenous NF-κB (91.5 ± 9.6%). Co-transfection of FKBP51 showed equivalent inhibitory action on NF-κB transcriptional activity (48.2 ± 3.2%), but the stimulatory effect of co-transfecting FKBP52 could not be evidenced in GR expressing cells (63.0 ± 2.5%). In agreement with the expected results, the lack of stimulatory effect was assigned to the known fact that FKBP52 greatly potentiates GR activation (19, 51) and consequently increases the number of GR·NF-κB heterocomplexes able to transrepress the biological response of NF-κB (52).
DISCUSSION
In previous works we demonstrated that steroid receptor action is regulated in an antagonistic manner by the high molecular weight immunophilins FKBP51 and FKBP52. In both cases, the association of the immunophilins with Hsp90 was essential. In this study we clearly demonstrate that the biological activity of NF-κB is affected in a similar manner, such that FKBP51 shows inhibitory effects and FKBP52 shows stimulatory effects. In contrast to steroid receptors, these biological actions are Hsp90-independent for NF-κB. This is confirmed by the fact that point mutants in the TPR domains of the FKBPs unable to bind Hsp90 show similar effects as the wild type immunophilins. Nevertheless, the overexpression of a TPR peptide inhibits the biological action of both proteins, suggesting that the domain itself is required at some point in the mechanism of action, perhaps for interacting with other regulatory factors such as cofactors (53). On the other hand, the PPIase activity of FKBP52 is very important for its stimulatory action, although this enzymatic activity is not required for the FKBP51 inhibitory action. A similar independent effect on the PPIase activity was described for the regulation of the GR by FKBP51 (19, 54), whereas a PPIase-dependent mechanism is implicated for FKBP52 (19, 51).
One interesting extrapolation of these effects is that the biological action of NF-κB may be regulated in different tissues and cell types by the overall expression balance of both immunophilins, which could contribute in part to the pleiotropic actions of NF-κB. Moreover, our assays showed that Hsp70 is also a RelA-interactor, which is in agreement with a very recent report in neurons (44), where the nuclear translocation of both RelA and Hsp70 was postulated to occur as a protein-protein complex. Interestingly, the up-regulation of Hsp70 was also reported to induce nuclear translocation of RelA in rat liver cells (55).
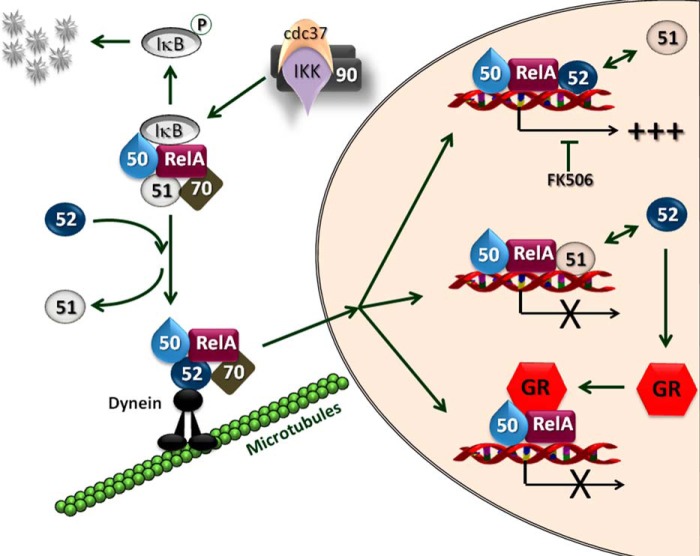
Upon phosphorylation of the NF-κB inhibitory protein IκB by a kinase located upstream, the IKK complex, NF-κB dimers are transported to the nucleus. Even so, a dynamic nuclear-cytoplasmic shuttling of NF-κB complexes always takes place (9–11) such that a basal transcriptional activity of NF-κB is always present because the IκB·NF-κB complex is also subject to dynamic dissociation/reassociation events. Experiments with IKKα knock-out mice (56) demonstrated defective cell proliferation and differentiation and have also shown that IKKα is dispensable for IκB degradation. Moreover IKKα has been reported to be required for the termination of NF-κB activation (57). A physical interaction between FKBP51 and the IKK complex has been demonstrated, most likely via the IKKα subunit bound to Hsp90 (58, 59), but the biological function of FKBP51 on IKK signaling is still unclear (59). Although down-modulation of Hsp90α and Hsp90β likewise resulted in reduced kinase activity, it has been shown that FKBP51 is not a constitutively associated component of the IKK complex (59), and its down-modulation interfered with neither TNFα-induced IKK activity nor IκBα degradation and RelA translocation. Actually, the prevailing complex for the IKK·Hsp90 complex to generate an activated state is the one that recruits Cdc37 rather than FKBP51 (59), both factors being transiently associated with NF-κB. Interestingly, it was reported that TNFα is unable to modify the association of IKK with those interacting factors (59). In short, the role of FKBP51 on NF-κB signaling cascade remains still elusive. Our study demonstrates an inhibitory action for this immunophilin on NF-κB signaling and, therefore, suggests the possibility that NF-κB biological activity could be mitigated in those tissues where the expression of FKBP51 is constitutively high or induced by stimuli. The opposite reasoning is equally valid for tissues that express different amounts of FKBP52, a clear stimulating factor of NF-κB activity. In line with these concepts, it should be emphasized that both the subcellular distribution and transcriptional activity of steroid receptors is modulated by the expression level of these two immunophilins (17, 18, 43). Consequently, based on the experimental data shown in this work, we would like to propose a novel mechanism of regulation of NF-κB by FKBPs that affects NF-κB activation at different levels, i.e. nuclear transport, nuclear retention, and transcriptional activity (Fig. 8).
FIGURE 8.
Model for the regulation of the biological action of NF-κB by FKBPs. The p50·RelA dimer is associated to FKBP51 in its inactive cytoplasmic state (Fig. 2A). Hsp70 is also part of the heterocomplex (Fig. 2E). Upon cell stimulation, the kinase activity of IKK is activated by phosphorylation via the cdc37·(Hsp90)2 interacting complex (59). This results in IκB phosphorylation, its dissociation from the NF-κB complex, and the subsequent degradation of IκB via proteasome. Active NF-κB replaces FKBP51 by FKBP52 (Fig. 2F), which is able to interact with dynein·dynactin motors proteins (15), favoring both the retrotransport of NF-κB (11, 40) and its interaction with the nuclear sites of action. FKBP52 greatly favors (+++) NF-κB biological action (Fig. 3B) in a PPIase-dependent manner (inhibited by FK506) when the immunophilin is recruited to the promoter sites of NF-κB target genes (Fig. 3G). On the other hand, the recruitment of FKBP51 to those promoters inhibits the NF-κB biological response (Fig. 3A). Both immunophilins compete one another (↔) and can hamper the original effect of the other (Fig. 7). The steroid-dependent activation of the GR, which is also improved by FKBP52 (19, 51), also prevents NF-κB effects via its known mechanism of tans-repression (52).
To the best of our knowledge this work is the first that shows such antagonistic activity of high molecular weight immunophilins on NF-κB biological action and also shows the first evidence that both endogenous immunophilins are dynamically recruited to the promoter sites of target genes. In line with our findings where endogenous proteins were analyzed, a recent report by Romano et al. (60) has also shown that overexpressed FLAG-tagged FKBP51 forms part of the transcriptional complexes of ABCG2 (ATP binding cassette transporters) in melanoma cells.
A crucial nuclear mechanism for gene expression is the modification of the chromatin environment of the respective genes. It has been shown that when NF-κB is activated, histone phosphorylation can be mediated by nuclear IKKα that is recruited to the promoter sites of NF-κB-regulated genes (61, 62). Among a number of chromatin remodelers is the PPIase protein Pin1 (63), which is an immunophilin that targets RelA (64). Because PPIase-induced conformational changes have functional effects on target proteins, the action of Pin1 on RelA is reflected in a more efficient nuclear accumulation of RelA and also a greater stability by preventing its ubiquitin-mediated proteolysis (64). In certain types of cancer cells, Pin1 is usually up-regulated (65–67), whereas the E3-ubiquitin-ligase of RelA, SOCS1, is down-regulated (68–70) or mutated (71), all of which may contribute to the constitutive activation of NF-κB in those cancers. A similar mechanism can be proposed here for the expression balance of FKBP51 and FKBP52, in particular for the latter immunophilin, which shows an important stimulatory action dependent on its PPIase activity.
Given the role played in the initiation and progression of cancer, the NF-κB signaling pathway is a potent node of pharmacological interference in the clinic. Because NF-κB is also an essential protein in the immunological response against cancer, there has been a reluctance to use NF-κB-targeting inhibitors for the treatment of such malignancies. Nevertheless, combining classical chemotherapeutics with inhibitors of NF-κB activation seems to result in a promising synergistic strategy (72, 73). Elevated NF-κB activity and/or higher half-life persistence in the nucleus of cancer cells (like that evidenced with greater amounts of FKBP52 in Fig. 1A) provide a survival mechanism by up-regulating anti-apoptotic genes, thereby representing a major causative factor for drug resistance (74). In line with this concept, it has been shown that 0.1 μm rapamycin abolishes doxorubicin-induced activation of NF-κB and thereby enhances drug-induced apoptosis, an effect that was first assigned to FKBP51 inhibition (75). However, that drug concentration cannot guarantee specificity of action as rapamycin can equally interact with FKBP51 (Ki < 5 nm) (76, 77) and FKBP52 (Ki ≈ 8 nm) (78, 79). Even though FKBP51 can interact with IKK (58), an up-stream regulator of the NF-κB signaling cascade, it has been demonstrated that FKBP51 is not a constitutive member of the IKK complex (59), and the biological consequence of such association (when it takes place) is unknown at this point. Nonetheless, we cannot rule out the possibility that differences among cell types may exist such that NF-κB is regulated in different manner.
Here, we demonstrate that in the biological systems we assayed, FKBP51 shows inhibitory rather stimulatory action on the overall activity of NF-κB and that this effect is not dependent on the PPIase activity (which is inhibited by rapamycin or FK506). Because FKBP52 also shows a stimulatory action on NF-κB action in a PPIase-dependent manner, we propose that a likely explanation for all these experimental facts, ours and those reported by others, is the PPIase-dependent (drug-inhibited) stimulatory role of FKBP52 on NF-κB activation. Accordingly, zymographies like those shown in Fig. 3F performed in the presence of FK506 fully abolished the production of MMP2 in BeWo cells stimulated with PMA (Fig. 4A).
The development of immunophilin ligands appears to have promising perspectives in the coming years (80). Thus, the ability to regulate the functions of a specific protein using cell-permeable small molecules like those that bind FKBPs is an unquestionably powerful method, not only to study biological systems, but also a desired alternative to be used in therapeutic treatments. Ideally, the biological function of certain nuclear factors could be regulated if we can influence the mechanisms by which they reach their sites of action. In this sense, because NF-κB is active in many cancer cells and its persistent localization in the nucleus strengthens or directly leads to tumor development, we propose that targeting specifically the PPIase activity of FKBP52 could be a novel regulatory approach to inhibit NF-κB activity.
Acknowledgments
We are indebted to Dr. Paola De Luca and Dr. Adriana De Siervi for the helpful advance on ChIP assay and to Dr. William B. Pratt from the University of Michigan for the kind gift of the UP30 antibody against FKBP52.
This work was supported, in whole or in part, by National Institutes of Health Grants 5 G12 RR008124 (Border Biomedical Research Center from the National Center for Research Resources) and 8 G12 MD007592 (National Institute on Minority Health and Health Disparities (to M. B. C.). This work was also supported by Agencia Nacional de Promoción Científica y Tecnológica de Argentina Grants PICT-2010-1170 and PICT-2011-1715 (to M. D. G.), PICT-2010-2215 (to A. G. E.), and PICT-2012-2612 (to G. P.-P.) and University of Buenos Aires Grants UBACYT-2011/14-GC (to M. D. G.), UBACYT 2011/13-IJ, and UBACYT 2013/15-IF (to A. G. E.).
- NF-κB
- nuclear factor-κB
- IκB
- inhibitor of κB
- IKK
- IκB kinase
- Hsp90
- heat-shock protein of 90 kDa
- FKBP51
- FK506-binding protein of 51-kDa
- FKBP52
- FK506-binding protein of 52-kDa
- TPR
- tetratricopeptide repeats
- PPIase
- peptidylprolyl-isomerase
- PMA
- phorbol 12-myristate 13-acetate
- MMP
- matrix metallopeptidase
- GR
- glucocorticoid receptor; mineralocorticoid receptor
- PR
- progesterone receptor.
REFERENCES
- 1. Sen R., Baltimore D. (1986) Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell 46, 705–716 [DOI] [PubMed] [Google Scholar]
- 2. Hoesel B., Schmid J. A. (2013) The complexity of NF-κB signaling in inflammation and cancer. Mol. Cancer 12, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Diamant G., Dikstein R. (2013) Transcriptional control by NF-κB: elongation in focus. Biochim. Biophys. Acta 1829, 937–945 [DOI] [PubMed] [Google Scholar]
- 4. Tornatore L., Thotakura A. K., Bennett J., Moretti M., Franzoso G. (2012) The nuclear factor κB signaling pathway: integrating metabolism with inflammation. Trends Cell Biol. 22, 557–566 [DOI] [PubMed] [Google Scholar]
- 5. Oeckinghaus A., Ghosh S. (2009) The NF-κB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 1, a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoffmann A., Natoli G., Ghosh G. (2006) Transcriptional regulation via the NF-κB signaling module. Oncogene 25, 6706–6716 [DOI] [PubMed] [Google Scholar]
- 7. Ciechanover A., Gonen H., Bercovich B., Cohen S., Fajerman I., Israël A., Mercurio F., Kahana C., Schwartz A. L., Iwai K., Orian A. (2001) Mechanisms of ubiquitin-mediated, limited processing of the NF-κB1 precursor protein p105. Biochimie 83, 341–349 [DOI] [PubMed] [Google Scholar]
- 8. Gilmore T. D. (1999) The Rel/NF-κB signal transduction pathway: introduction. Oncogene 18, 6842–6844 [DOI] [PubMed] [Google Scholar]
- 9. Birbach A., Gold P., Binder B. R., Hofer E., de Martin R., Schmid J. A. (2002) Signaling molecules of the NF-κ B pathway shuttle constitutively between cytoplasm and nucleus. J. Biol. Chem. 277, 10842–10851 [DOI] [PubMed] [Google Scholar]
- 10. Huang T. T., Kudo N., Yoshida M., Miyamoto S. (2000) A nuclear export signal in the N-terminal regulatory domain of IκBα controls cytoplasmic localization of inactive NF-κB/IκBα complexes. Proc. Natl. Acad. Sci. U.S.A. 97, 1014–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mikenberg I., Widera D., Kaus A., Kaltschmidt B., Kaltschmidt C. (2007) Transcription factor NF-κB is transported to the nucleus via cytoplasmic dynein/dynactin motor complex in hippocampal neurons. PLoS ONE 2, e589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elbi C., Walker D. A., Romero G., Sullivan W. P., Toft D. O., Hager G. L., DeFranco D. B. (2004) Molecular chaperones function as steroid receptor nuclear mobility factors. Proc. Natl. Acad. Sci. U.S.A. 101, 2876–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Madan A. P., DeFranco D. B. (1993) Bidirectional transport of glucocorticoid receptors across the nuclear envelope. Proc. Natl. Acad. Sci. U.S.A. 90, 3588–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Galigniana M. D., Echeverría P. C., Erlejman A. G., Piwien-Pilipuk G. (2010) Role of molecular chaperones and TPR-domain proteins in the cytoplasmic transport of steroid receptors and their passage through the nuclear pore. Nucleus 1, 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galigniana M. D., Radanyi C., Renoir J. M., Housley P. R., Pratt W. B. (2001) Evidence that the peptidylprolyl isomerase domain of the hsp90-binding immunophilin FKBP52 is involved in both dynein interaction and glucocorticoid receptor movement to the nucleus. J. Biol. Chem. 276, 14884–14889 [DOI] [PubMed] [Google Scholar]
- 16. Davies T. H., Ning Y. M., Sánchez E. R. (2002) A new first step in activation of steroid receptors: hormone-induced switching of FKBP51 and FKBP52 immunophilins. J. Biol. Chem. 277, 4597–4600 [DOI] [PubMed] [Google Scholar]
- 17. Galigniana M. D., Erlejman A. G., Monte M., Gomez-Sanchez C., Piwien-Pilipuk G. (2010) The hsp90-FKBP52 complex links the mineralocorticoid receptor to motor proteins and persists bound to the receptor in early nuclear events. Mol. Cell. Biol. 30, 1285–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gallo L. I., Ghini A. A., Piwien Pilipuk G., Galigniana M. D. (2007) Differential recruitment of tetratricorpeptide repeat domain immunophilins to the mineralocorticoid receptor influences both heat-shock protein 90-dependent retrotransport and hormone-dependent transcriptional activity. Biochemistry 46, 14044–14057 [DOI] [PubMed] [Google Scholar]
- 19. Wochnik G. M., Rüegg J., Abel G. A., Schmidt U., Holsboer F., Rein T. (2005) FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J. Biol. Chem. 280, 4609–4616 [DOI] [PubMed] [Google Scholar]
- 20. Riggs D. L., Roberts P. J., Chirillo S. C., Cheung-Flynn J., Prapapanich V., Ratajczak T., Gaber R., Picard D., Smith D. F. (2003) The Hsp90 binding peptidylprolyl isomerase FKBP52 potentiates glucocorticoid signaling in vivo. EMBO J. 22, 1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tranguch S., Cheung-Flynn J., Daikoku T., Prapapanich V., Cox M. B., Xie H., Wang H., Das S. K., Smith D. F., Dey S. K. (2005) Cochaperone immunophilin FKBP52 is critical to uterine receptivity for embryo implantation. Proc. Natl. Acad. Sci. U.S.A. 102, 14326–14331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang Z., Wolf I. M., Chen H., Periyasamy S., Chen Z., Yong W., Shi S., Zhao W., Xu J., Srivastava A., Sánchez E. R., Shou W. (2006) FK506-binding protein 52 is essential to uterine reproductive physiology controlled by the progesterone receptor A isoform. Mol. Endocrinol. 20, 2682–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheung-Flynn J., Prapapanich V., Cox M. B., Riggs D. L., Suarez-Quian C., Smith D. F. (2005) Physiological role for the cochaperone FKBP52 in androgen receptor signaling. Mol. Endocrinol. 19, 1654–1666 [DOI] [PubMed] [Google Scholar]
- 24. Periyasamy S., Hinds T., Jr., Shemshedini L., Shou W., Sanchez E. R. (2010) FKBP51 and Cyp40 are positive regulators of androgen-dependent prostate cancer cell growth and the targets of FK506 and cyclosporin A. Oncogene 29, 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schülke J. P., Wochnik G. M., Lang-Rollin I., Gassen N. C., Knapp R. T., Berning B., Yassouridis A., Rein T. (2010) Differential impact of tetratricopeptide repeat proteins on the steroid hormone receptors. PLoS ONE 5, e11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cioffi D. L., Hubler T. R., Scammell J. G. (2011) Organization and function of the FKBP52 and FKBP51 genes. Curr. Opin. Pharmacol. 11, 308–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sivils J. C., Storer C. L., Galigniana M. D., Cox M. B. (2011) Regulation of steroid hormone receptor function by the 52-kDa FK506-binding protein (FKBP52). Curr. Opin. Pharmacol. 11, 314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Storer C. L., Dickey C. A., Galigniana M. D., Rein T., Cox M. B. (2011) FKBP51 and FKBP52 in signaling and disease. Trends Endocrinol. Metab. 22, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Galigniana M. D. (2012) Steroid receptor coupling becomes nuclear. Chem. Biol. 19, 662–663 [DOI] [PubMed] [Google Scholar]
- 30. Fontana V., Coll T. A., Sobarzo C. M., Tito L. P., Calvo J. C., Cebral E. (2012) Matrix metalloproteinase expression and activity in trophoblast-decidual tissues at organogenesis in CF-1 mouse. J. Mol. Histol. 43, 487–496 [DOI] [PubMed] [Google Scholar]
- 31. Quintá H. R., Maschi D., Gomez-Sanchez C., Piwien-Pilipuk G., Galigniana M. D. (2010) Subcellular rearrangement of hsp90-binding immunophilins accompanies neuronal differentiation and neurite outgrowth. J. Neurochem. 115, 716–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gallo L. I., Lagadari M., Piwien-Pilipuk G., Galigniana M. D. (2011) The 90-kDa heat-shock protein (Hsp90)-binding immunophilin FKBP51 is a mitochondrial protein that translocates to the nucleus to protect cells against oxidative stress. J. Biol. Chem. 286, 30152–30160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quintá H. R., Galigniana M. D. (2012) The neuroregenerative mechanism mediated by the Hsp90 binding immunophilin FKBP52 resembles the early steps of neuronal differentiation. Br. J. Pharmacol. 166, 637–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Erlejman A. G., Jaggers G., Fraga C. G., Oteiza P. I. (2008) TNFα-induced NF-κB activation and cell oxidant production are modulated by hexameric procyanidins in Caco-2 cells. Arch. Biochem. Biophys. 476, 186–195 [DOI] [PubMed] [Google Scholar]
- 35. Susperreguy S., Prendes L. P., Desbats M. A., Charó N. L., Brown K., MacDougald O. A., Kerppola T., Schwartz J., Piwien-Pilipuk G. (2011) Visualization by BiFC of different C/EBPβ dimers and their interaction with HP1α reveals a differential subnuclear distribution of complexes in living cells. Exp. Cell Res. 317, 706–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Piwien Pilipuk G., Vinson G. P., Sanchez C. G., Galigniana M. D. (2007) Evidence for NL1-independent nuclear translocation of the mineralocorticoid receptor. Biochemistry 46, 1389–1397 [DOI] [PubMed] [Google Scholar]
- 37. Galigniana M. D., Harrell J. M., O'Hagen H. M., Ljungman M., Pratt W. B. (2004) Hsp90 binding immunophilins link p53 to dynein during p53 transport to the nucleus. J. Biol. Chem. 279, 22483–22489 [DOI] [PubMed] [Google Scholar]
- 38. Colo G. P., Rubio M. F., Nojek I. M., Werbajh S. E., Echeverría P. C., Alvarado C. V., Nahmod V. E., Galigniana M. D., Costas M. A. (2008) The p160 nuclear receptor co-activator RAC3 exerts an anti-apoptotic role through a cytoplasmatic action. Oncogene 27, 2430–2444 [DOI] [PubMed] [Google Scholar]
- 39. Sanchez E. R. (2012) Chaperoning steroidal physiology: lessons from mouse genetic models of Hsp90 and its cochaperones. Biochim. Biophys. Acta 1823, 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mackenzie G. G., Keen C. L., Oteiza P. I. (2006) Microtubules are required for NF-κB nuclear translocation in neuroblastoma IMR-32 cells: modulation by zinc. J. Neurochem. 99, 402–415 [DOI] [PubMed] [Google Scholar]
- 41. Nelson G., Paraoan L., Spiller D. G., Wilde G. J., Browne M. A., Djali P. K., Unitt J. F., Sullivan E., Floettmann E., White M. R. (2002) Multi-parameter analysis of the kinetics of NF-κB signalling and transcription in single living cells. J. Cell Sci. 115, 1137–1148 [DOI] [PubMed] [Google Scholar]
- 42. Hohmann H. P., Remy R., Scheidereit C., van Loon A. P. (1991) Maintenance of NF-κ B activity is dependent on protein synthesis and the continuous presence of external stimuli. Mol. Cell. Biol. 11, 259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Banerjee A., Periyasamy S., Wolf I. M., Hinds T. D., Jr., Yong W., Shou W., Sanchez E. R. (2008) Control of glucocorticoid and progesterone receptor subcellular localization by the ligand-binding domain is mediated by distinct interactions with tetratricopeptide repeat proteins. Biochemistry 47, 10471–10480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klenke C., Widera D., Engelen T., Müller J., Noll T., Niehaus K., Schmitz M. L., Kaltschmidt B., Kaltschmidt C. (2013) Hsc70 is a novel interactor of NF-κB p65 in living hippocampal neurons. PLoS ONE 8, e65280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yura S., Sagawa N., Ogawa Y., Masuzaki H., Mise H., Matsumoto T., Ebihara K., Fujii S., Nakao K. (1998) Augmentation of leptin synthesis and secretion through activation of protein kinases A and C in cultured human trophoblastic cells. J. Clin. Endocrinol. Metab. 83, 3609–3614 [DOI] [PubMed] [Google Scholar]
- 46. Suh J., Rabson A. B. (2004) NF-κB activation in human prostate cancer: important mediator or epiphenomenon? J. Cell. Biochem. 91, 100–117 [DOI] [PubMed] [Google Scholar]
- 47. Nissinen L., Kähäri V. M. (2014) Matrix metalloproteinases in inflammation. Biochim. Biophys. Acta 1840, 2571–2580 [DOI] [PubMed] [Google Scholar]
- 48. Li Y. F., Xu X. B., Chen X. H., Wei G., He B., Wang J. D. (2012) The nuclear factor-κB pathway is involved in matrix metalloproteinase-9 expression in RU486-induced endometrium breakdown in mice. Hum. Reprod. 27, 2096–2106 [DOI] [PubMed] [Google Scholar]
- 49. Tsukihara S., Harada T., Deura I., Mitsunari M., Yoshida S., Iwabe T., Terakawa N. (2004) Interleukin-1β-induced expression of IL-6 and production of human chorionic gonadotropin in human trophoblast cells via nuclear factor-κB activation. Am. J. Reprod. Immunol. 52, 218–223 [DOI] [PubMed] [Google Scholar]
- 50. Fujisawa K., Nasu K., Arima K., Sugano T., Narahara H., Miyakawa I. (2000) Production of interleukin (IL)-6 and IL-8 by a choriocarcinoma cell line, BeWo. Placenta 21, 354–360 [DOI] [PubMed] [Google Scholar]
- 51. Riggs D. L., Cox M. B., Cheung-Flynn J., Prapapanich V., Carrigan P. E., Smith D. F. (2004) Functional specificity of co-chaperone interactions with Hsp90 client proteins. Crit. Rev. Biochem. Mol. Biol. 39, 279–295 [DOI] [PubMed] [Google Scholar]
- 52. Scheinman R. I., Gualberto A., Jewell C. M., Cidlowski J. A., Baldwin A. S., Jr. (1995) Characterization of mechanisms involved in transrepression of NF-κB by activated glucocorticoid receptors. Mol. Cell. Biol. 15, 943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Erlejman A. G., Lagadari M., Toneatto J., Piwien-Pilipuk G., Galigniana M. D. (2014) Regulatory role of the 90-kDa heat-shock protein (Hsp90) and associated factors on gene expression. Biochim. Biophys. Acta 1839, 71–87 [DOI] [PubMed] [Google Scholar]
- 54. Cluning C., Ward B. K., Rea S. L., Arulpragasam A., Fuller P. J., Ratajczak T. (2013) The helix 1–3 loop in the glucocorticoid receptor LBD is a regulatory element for FKBP cochaperones. Mol. Endocrinol. 27, 1020–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dokladny K., Lobb R., Wharton W., Ma T. Y., Moseley P. L. (2010) LPS-induced cytokine levels are repressed by elevated expression of HSP70 in rats: possible role of NF-κB. Cell Stress Chaperones 15, 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sil A. K., Maeda S., Sano Y., Roop D. R., Karin M. (2004) IκB kinase-α acts in the epidermis to control skeletal and craniofacial morphogenesis. Nature 428, 660–664 [DOI] [PubMed] [Google Scholar]
- 57. Lawrence T., Bebien M., Liu G. Y., Nizet V., Karin M. (2005) IKKα limits macrophage NF-κB activation and contributes to the resolution of inflammation. Nature 434, 1138–1143 [DOI] [PubMed] [Google Scholar]
- 58. Bouwmeester T., Bauch A., Ruffner H., Angrand P. O., Bergamini G., Croughton K., Cruciat C., Eberhard D., Gagneur J., Ghidelli S., Hopf C., Huhse B., Mangano R., Michon A. M., Schirle M., Schlegl J., Schwab M., Stein M. A., Bauer A., Casari G., Drewes G., Gavin A. C., Jackson D. B., Joberty G., Neubauer G., Rick J., Kuster B., Superti-Furga G. (2004) A physical and functional map of the human TNF-α/NF-κ B signal transduction pathway. Nat. Cell Biol. 6, 97–105 [DOI] [PubMed] [Google Scholar]
- 59. Hinz M., Broemer M., Arslan S. C., Otto A., Mueller E. C., Dettmer R., Scheidereit C. (2007) Signal responsiveness of IκB kinases is determined by Cdc37-assisted transient interaction with Hsp90. J. Biol. Chem. 282, 32311–32319 [DOI] [PubMed] [Google Scholar]
- 60. Romano S., Staibano S., Greco A., Brunetti A., Nappo G., Ilardi G., Martinelli R., Sorrentino A., Di Pace A., Mascolo M., Bisogni R., Scalvenzi M., Alfano B., Romano M. F. (2013) FK506 binding protein 51 positively regulates melanoma stemness and metastatic potential. Cell Death Dis. 4, e578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Park K. J., Krishnan V., O'Malley B. W., Yamamoto Y., Gaynor R. B. (2005) Formation of an IKKα-dependent transcription complex is required for estrogen receptor-mediated gene activation. Mol. Cell 18, 71–82 [DOI] [PubMed] [Google Scholar]
- 62. Park G. Y., Wang X., Hu N., Pedchenko T. V., Blackwell T. S., Christman J. W. (2006) NIK is involved in nucleosomal regulation by enhancing histone H3 phosphorylation by IKKα. J. Biol. Chem. 281, 18684–18690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Raghuram N., Strickfaden H., McDonald D., Williams K., Fang H., Mizzen C., Hayes J. J., Th'ng J., Hendzel M. J. (2013) Pin1 promotes histone H1 dephosphorylation and stabilizes its binding to chromatin. J. Cell Biol. 203, 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ryo A., Suizu F., Yoshida Y., Perrem K., Liou Y. C., Wulf G., Rottapel R., Yamaoka S., Lu K. P. (2003) Regulation of NF-κB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell 12, 1413–1426 [DOI] [PubMed] [Google Scholar]
- 65. Pang R. W., Lee T. K., Man K., Poon R. T., Fan S. T., Kwong Y. L., Tse E. (2006) PIN1 expression contributes to hepatic carcinogenesis. J. Pathol. 210, 19–25 [DOI] [PubMed] [Google Scholar]
- 66. Lu K. P., Liou Y. C., Zhou X. Z. (2002) Pinning down proline-directed phosphorylation signaling. Trends Cell Biol. 12, 164–172 [DOI] [PubMed] [Google Scholar]
- 67. Wulf G. M., Ryo A., Wulf G. G., Lee S. W., Niu T., Petkova V., Lu K. P. (2001) Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 20, 3459–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Oshimo Y., Kuraoka K., Nakayama H., Kitadai Y., Yoshida K., Chayama K., Yasui W. (2004) Epigenetic inactivation of SOCS-1 by CpG island hypermethylation in human gastric carcinoma. Int. J. Cancer 112, 1003–1009 [DOI] [PubMed] [Google Scholar]
- 69. Oh J., Kim S. H., Ahn S., Lee C. E. (2012) Suppressors of cytokine signaling promote Fas-induced apoptosis through down-regulation of NF-κB and mitochondrial Bfl-1 in leukemic T cells. J. Immunol. 189, 5561–5571 [DOI] [PubMed] [Google Scholar]
- 70. Zhao X. D., Zhang W., Liang H. J., Ji W. Y. (2013) Overexpression of miR −155 promotes proliferation and invasion of human laryngeal squamous cell carcinoma via targeting SOCS1 and STAT3. PLoS ONE 8, e56395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schif B., Lennerz J. K., Kohler C. W., Bentink S., Kreuz M., Melzner I., Ritz O., Trümper L., Loeffler M., Spang R., Möller P. (2013) SOCS1 mutation subtypes predict divergent outcomes in diffuse large B-cell lymphoma (DLBCL) patients. Oncotarget 4, 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fuchs O. (2010) Transcription factor NF-κB inhibitors as single therapeutic agents or in combination with classical chemotherapeutic agents for the treatment of hematologic malignancies. Curr. Mol. Pharmacol. 3, 98–122 [DOI] [PubMed] [Google Scholar]
- 73. Fabre C., Mimura N., Bobb K., Kong S. Y., Gorgun G., Cirstea D., Hu Y., Minami J., Ohguchi H., Zhang J., Meshulam J., Carrasco R. D., Tai Y. T., Richardson P. G., Hideshima T., Anderson K. C. (2012) Dual inhibition of canonical and noncanonical NF-κB pathways demonstrates significant antitumor activities in multiple myeloma. Clin. Cancer Res. 18, 4669–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Salem K., Brown C. O., Schibler J., Goel A. (2013) Combination chemotherapy increases cytotoxicity of multiple myeloma cells by modification of nuclear factor (NF)-κB activity. Exp. Hematol. 41, 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Romano M. F., Avellino R., Petrella A., Bisogni R., Romano S., Venuta S. (2004) Rapamycin inhibits doxorubicin-induced NF-κB/Rel nuclear activity and enhances the apoptosis of melanoma cells. Eur. J. Cancer 40, 2829–2836 [DOI] [PubMed] [Google Scholar]
- 76. Yeh W. C., Bierer B. E., McKnight S. L. (1995) Rapamycin inhibits clonal expansion and adipogenic differentiation of 3T3-L1 cells. Proc. Natl. Acad. Sci. U.S.A. 92, 11086–11090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sinars C. R., Cheung-Flynn J., Rimerman R. A., Scammell J. G., Smith D. F., Clardy J. (2003) Structure of the large FK506-binding protein FKBP51, an Hsp90 binding protein, and a component of steroid receptor complexes. Proc. Natl. Acad. Sci. U.S.A. 100, 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Galat A. (1993) Peptidylproline cis-trans-isomerases: immunophilins. Eur. J. Biochem. 216, 689–707 [DOI] [PubMed] [Google Scholar]
- 79. Peattie D. A., Harding M. W., Fleming M. A., DeCenzo M. T., Lippke J. A., Livingston D. J., Benasutti M. (1992) Expression and characterization of human FKBP52, an immunophilin that associates with the 90-kDa heat shock protein and is a component of steroid receptor complexes. Proc. Natl. Acad. Sci. U.S.A. 89, 10974–10978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gaali S., Gopalakrishnan R., Wang Y., Kozany C., Hausch F. (2011) The chemical biology of immunophilin ligands. Curr. Med. Chem. 18, 5355–5379 [DOI] [PubMed] [Google Scholar]