Abstract
Biogenesis of eukaryotic box C/D small nucleolar ribonucleoprotein particles (C/D snoRNPs) involves conserved trans-acting factors, which are proposed to facilitate the assembly of the core proteins Snu13p/15.5K, Nop58p/NOP58, Nop56p/NOP56 and Nop1p/Fibrillarin on box C/D small nucleolar RNAs (C/D snoRNAs). In yeast, protein Rsa1 acts as a platform, interacting with both the RNA-binding core protein Snu13 and protein Pih1 of the Hsp82–R2TP chaperone complex. In this work, a proteomic approach coupled with functional and structural studies identifies protein Hit1 as a novel Rsa1p-interacting partner involved in C/D snoRNP assembly. Hit1p contributes to in vivo C/D snoRNA stability and pre-RNA maturation kinetics. It associates with U3 snoRNA precursors and influences its 3′-end processing. Remarkably, Hit1p is required to maintain steady-state levels of Rsa1p. This stabilizing activity is likely to be general across eukaryotic species, as the human protein ZNHIT3(TRIP3) showing sequence homology with Hit1p regulates the abundance of NUFIP1, the Rsa1p functional homolog. The nuclear magnetic resonance solution structure of the Rsa1p317–352–Hit1p70–164 complex reveals a novel mode of protein–protein association explaining the strong stability of the Rsa1p–Hit1p complex. Our biochemical data show that C/D snoRNAs and the core protein Nop58 can interact with the purified Snu13p–Rsa1p–Hit1p heterotrimer.
INTRODUCTION
In eukaryotes, snoRNPs (small nucleolar RNPs) and scaRNPs (small Cajal body RNPs) are essential to biogenesis of ribosomes and spliceosomes, respectively (for review (1–3)). They mediate site-specific modification of ribosomal RNAs (rRNAs) and U-rich small nuclear RNAs (UsnRNAs); box C/D snoRNPs and scaRNPs (C/D sno/scaRNPs) catalyze 2′-O-methylation of riboses, while box H/ACA snoRNPs and scaRNPs (H/ACA sno/scaRNPs) catalyze conversion of uridine to pseudouridine. Furthermore, some snoRNPs, C/D snoRNPs U3 and U8 (4,5), and mammalian U17 and yeast snR30 H/ACA snoRNPs (6,7), are not involved in nucleotide modifications but are implicated in pre-RNA endonucleolytic cleavages.
C/D snoRNPs each contain a box C/D small nucleolar RNA (C/D snoRNA) and a set of four core proteins, Snu13p, Nop1p, Nop56p and Nop58p in the yeast Saccharomyces cerevisiae, and 15.5K, Fibrillarin, NOP56 and NOP58 in human cells (8–11). Snu13p and 15.5K belong to the L7Ae family of RNA binding proteins (12) which members are components of essential RNPs: snRNP U4 (13), H/ACA snoRNPs (14), telomerase (15) and SECIS mRNPs (16,17). The L7Ae-like proteins specifically recognizes kink-turn (K-turn) or kink-loop (K-loop) RNA motifs (10,18).
Biogenesis of snoRNPs is a complex process including transcription and maturation of snoRNAs, synthesis and nuclear import of core proteins, and their assembly on snoRNAs. Mature RNPs are then delivered to their functional sites. Several studies have highlighted the involvement of trans-acting factors during snoRNP assembly. Two specific factors Shq1p/SHQ1 and Naf1p/NAF1, which both bind to Cbf5 core protein, play a key role in H/ACA sno/scaRNP biogenesis. Shq1p functions as an RNA mimic to prevent unspecific RNA binding (19), while Naf1p facilitates the H/ACA snoRNP co-transcriptional assembly by interacting with the RNA polymerase II CTD (20). In addition, assembly of C/D, H/ACA snoRNPs and telomerase require the Hsp82/Hsp90 co-chaperone R2TP complex (21–24), which is composed of the two AAA+ hexameric helicases Rvb1p/TIP49 and Rvb2p/TIP48, Tah1p/hSPAGH (RPAP3), and Pih1p/hPIH1 (PIH1D1, NOP17). The Hsp82/Hsp90 chaperone interacts with the R2TP complex by binding to the TPR domain of protein Tah1 (23,25). Data obtained in human cells indicate that in association with the R2TP complex, Hsp90 controls the stability of RNP core proteins (15.5K, NOP58, NHP2, SBP2) and participates in the assembly process (21,22,25,26). The ATPase activities of the TIP48 and TIP49 helicases are also involved in this process, since the A and B Walker motifs of TIP48 are required for yeast C/D snoRNA accumulation (21,22,27,28).
Another protein Rsa1/NUFIP1 plays a central role in R2TP recruitment on pre-snoRNPs. It interacts with Pih1p/hPIH1D1, Rvb1p/hTIP49 and Rvb2p/hTIP48, and with the RNP core proteins (21,22). In particular, a conserved stretch of 32 amino acids (yPEP in Rsa1p and hPEP in NUFIP1) is dedicated to the interaction with the C/D snoRNP protein Snu13p/15.5K (21). We identified residues required for the Snu13p–Rsa1p interaction and showed that the RNA and Rsa1p interact with opposite faces of Snu13p (29). Rsa1p/NUFIP1 and in a nucleotide-dependent manner, the Rvb1p/TIP49 and Rvb2p/TIP48 components of the R2TP complex, cooperate together to scaffold C/D snoRNP assembly (28).
By interacting both with proteins Tah1 and Nop58, Pih1p plays an important role in C/D snoRNP assembly (25,26,30,31). This is illustrated by reduced amounts of C/D core proteins (26) and their mislocation in the cytosol and the nucleoplasm (31) in a ΔPIH1 strain. Structural information on the Pih1p–Tah1p interaction was recently gained by nuclear magnetic resonance (NMR) and X-ray studies (32,33).
Despite these recent advances in identification of cellular proteins involved in C/D snoRNPs assembly, their precise mechanism of action remains poorly understood, and functional maturation complexes have not been purified yet. There is even no evidence that the full set of C/D snoRNP assembly factors has been identified. In the present work, we identify a new participant in C/D snoRNP assembly, protein Hit1 (High Temperature growth, YJR055W). This 164 amino acid protein was originally identified in a screen for temperature sensitive mutations in the yeast Saccharomyces cerevisiae (34), but due to the absence of further analysis, the Hit1p function remained obscure. A potential zinc finger-like domain is present in its N-terminal end. Here, we identify a functional role of Hit1p. The starting point was our co-purification of Hit1p with TAP-tagged Rsa1p in a cellular extract. Then, by in vitro assays and co-expression in Escherichia coli, we demonstrated a direct and stable interaction between the two proteins and explained this stability by NMR structural analysis of the interacting domains. In parallel, we showed that Hit1p and Rsa1p are functionally linked and that the Rsa1p steady-state level depends upon Hit1p expression. We showed that the purified recombinant Snu13p–Rsa1p–Hit1p heterotrimer binds C/D snoRNAs and Nop58p. Altogether, the data reveal a role of Hit1p in C/D snoRNP assembly and bring new insights into the assembly complexes involved in this process. In addition, according to our study of ZNHIT3(TRIP3), a human homologue of Hit1p, some of our conclusions can be extended to vertebrates.
MATERIALS AND METHODS
Plasmids, yeast and bacterial strains used
For protein co-expression in E. coli, PCR-amplified fragments were cloned between the NdeI and BamHI sites of plasmids pnEA-3CH (His6-tagged protein) and pnCS, as described (29). Snu13p was expressed using plasmid pGEX-6P-1::SNU13 (35). Derivatives of plasmid pACTII expressing Gal4-AD-Rsa1p and Gal4-AD-Rsa1230–381 (21) were used for Y2H assays and we built a pAS2 plasmid expressing Gal4-BD-Hit1p. Plasmid pASZ11 was used to produce U3Δ2,3,4 in yeast as described (35). The yeast strains (MATa, ade2, arg4, leu2–3,112, trp1–289, ura3–52) were used for TAP-Tagged experiments (SC4186 (RSA1-TAP), SC3760 (HIT1-TAP) and SC3165 (PIH1-TAP)) according to (36), and knocked out strains derivatives of the S. cerevisiae BY4741 strain (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) were provided by Euroscarf. The S. cerevisiae strains Y190 and Y187 were used for Y2H assays (Clontech). The E. coli strains CodonPlus and pRare2 (Novagen) were used for co-expression assays and protein production, respectively.
Complex purification by protein A selection affinity and their analysis by mass spectrometry
Saccharomyces cerevisiae strains expressing TAP-tagged proteins, as well as the untagged strain BY4741, were grown at 30°C in YPD to A600 ∼0.8–1. Cells were lysed by bead-beating in breaking buffer (100 mM NaCl, 8% glycerol, 0.1 mM DTT, 100 mM Tris-HCl, pH 8). Protein A-based purifications were performed using cell amounts corresponding to 2500 A600 U of culture (37). Total cellular extracts were incubated for 12 h at 4°C with IgG-Sepharose beads (Sigma-Aldrich). Beads were washed in washing buffer (100 mM NaCl, 0.1% IGEPAL, 0.1 mM DTT, 20 mM Tris-HCl, pH 7.4), using Mobicol columns (MoBiTec). After TEV cleavage, eluted proteins were fractionated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Colloidal Coomassie Blue. Gel slices were picked systematically each 2 mm along the gel. In gel digestion of SDS-PAGE gel slices and mass spectrometry (MS) analysis of peptide extracts were performed as previously (38), and details are available in Supplemental Materials and Methods.
RNA co-IP and protein co-IP assays
Yeast cells expressing the TAP-tagged proteins were grown as above. Cells were lysed by bead-beating in breaking buffer; lysates were added to IgG-Sepharose beads (Sigma-Aldrich) and incubated 2 h at 4°C. Beads were washed four times in washing buffer. RNAs were extracted with phenol-chloroform and analyzed by RT-PCR using the oligonucleotides given in Supplementary Table S2 in Supplementary material. cDNAs were generated using primer OG-Rev containing sequence exogenous to the yeast genome, and used as templates for PCR amplification. PCR was performed with the 5′ oligonucleotide OG-5′PCRa or the OG-5′PCRb and the 3′ oligonucleotide OG-3′PCR.
Northern blot analyses
Total RNAs were extracted from exponentially growing cells (A600 ∼0.8). Northern blot was carried on 5 μg of total RNA fractionated on polyacrylamide denaturing gels (for small RNAs) or 1.2% glyoxal/DMSO agarose gels (for rRNAs). After transfer to Zeta-Probe membrane (Biorad), RNAs were detected using specific 5′ 32P-radiolabeled oligonucleotides probes (Supplementary Table S2 and Supplementary materials).
Protein detection by western blot
Crude cell extracts were prepared from pellets of 7 × 108 yeast cells resuspended in 300 μl of buffer (150 mM KCl, 5 mM MgCl2, 0,05% TRITON-X100, 20 mM Tris-HCl, pH 7,5), and lysed by bead beating. The lysates were centrifuged twice at 10 000 g for 5 min. Equivalent amounts of proteins were fractionated on 12.5% SDS-PAGE and western blots were performed according to standard procedures using rabbit commercial Peroxidase Anti-Peroxidase (PAP), anti-GFP, anti-HA, or anti-PMA1 (provided by B. André, LPMC, IBMM-ULB, Bruxelles, Belgium) and anti-AspRS (provided by C. Allmang-Cura and G. Eriani, IBMC, Strasbourg, France).
Immunoprecipitations using HeLa cells and analysis by SDS-PAGE and western blotting were performed as previously described (39).
Yeast two hybrid assays
For Y2H assays, appropriate pACTII and pAS2 plasmids were used to transformed haploid yeast cells (strains Y190 and Y187, respectively), which were then crossed. Diploids were selected on Leu−, Trp− medium and plated on Leu−, Trp−, His− medium to test for the interaction. Various concentrations of 3-amino-1,2,4-triazol (3AT) (Sigma) were used to evaluate the strength of the interactions. Growth was assessed following three days of incubation at 30°C.
Tests for synthetic lethality
Diploid strains were obtained by cross-breeding on YPD plates of simple knock-out haploid strains BY4741 (MATa) and BY4742 (MATα) from Euroscarf. Diploid cells were resuspended in 2 ml of sporulation medium (1% potassium acetate; 0.05% zinc acetate). Sporulation cultures were incubated on a roller wheel for five days at 25°C, then for three days at 30°C. Cells were dissected with a micromanipulator and separated on YPD plates. Strain genotypes were confirmed by genomic DNA extraction and PCR analysis.
RNAi experiments and analysis
HeLa cells were transfected using the Calphos Mammalian Transfection Kit (Clontech) with ON-TARGET plus SMART pool siRNAs targeting human ZNHIT3 (TRIP3) (ThermoScientific) or firefly luciferase siRNAs as a negative control. After 48 h of incubation, total extracts were prepared and analysed by western blotting. The following antibodies were used: anti-SRP68 (Proteintech), anti-β-tubulin (Sigma), anti-TRIP3 (Abcam) and anti-NUFIP1 (Proteintech). Quantification was performed using Fusion Solo bio-ID software (Vilber Lourmat).
Production of recombinant proteins
The recombinant GST-Snu13 protein was purified from cellular extract under native conditions as described (29).
Complex production by co-expressed in E. coli
The recombinant plasmids pnEA-3CH encoding His6Snu13p, His6Rsa1p, His6Rsa1p230–375, or His6Tah1p, and the recombinant plasmid pnCS encoding Hit1p, Rsa1p230–375, or Pih1p, were simultaneously transformed into pRare2 E. coli host cells (Novagen). Co-expression tests were carried out using the described procedure (29,40).
His-tagged protein pull-down
Complex formation was tested in 20 μl of buffer D (20 mM HEPES, pH 7.9, 150 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 10% glycerol) in the presence of recombinant proteins at a 2.5 μM concentration and 4 μl of 35S radiolabeled Nop58p, obtained by using the E. coli T7 S30 Extract System for Circular DNA (Promega). After 20 min at 30°C, 180 μl of binding buffer (10 mM Tris-HCl, pH 8, 150 mM NaCl, 0.1% Igepal, 5 mM imidazole) and 20 μl of cobalt-sepharose beads (GE Healthcare) were added. The mixture was incubated on a spinning wheel at 4°C for 90 min. Beads were washed three times with binding buffer, and retention of radiolabeled protein was analyzed by 7.5% SDS-PAGE.
Native MS
MS experiments were performed on a hybrid quadrupole/ion mobility separator/time-of-flight instrument (Synapt HDMS G2, Waters, Manchester, UK) equipped with an automated chip-based nanoelectrospray source (Triversa Nanomate, Advion Biosciences, Ithaca, NY, USA) operating in the positive ion mode. External calibration was performed with the multiply charged ions produced by 2 μM horse heart myoglobin diluted in 1:1 (v/v) water/acetonitrile acidified with 1% (v/v) formic acid. Prior to noncovalent analysis, protein buffer was exchanged against 500 mM or 1 M ammonium acetate buffer, pH 7.5, using microcentrifuge gel-filtration columns (Zeba 0.5 ml, Thermo Scientific, Rockford, IL, USA). Mass measurements under denaturing conditions were carried out by diluting samples to 2 μM in water/acetonitrile/formic acid (50:50:1). The Accelerating voltage (Vc) was set to 20 or 40 V, while pressure in the interface region (Pi) was 1.1 mbar. Either 500 mM (for Rsa1p230–375–Hit1p) or 1 M (for Snu13p−Rsa1p230−375−Hit1p) ammonium acetate buffer at pH 7.5 was used. Optimal interface parameters (Vc, Pi) were used in order to allow for detection of intact noncovalent complexes under conditions of efficient ion desolvation and ion transmission in the mass spectrometer (41). Data analysis was performed with MassLynx 3.5 (Waters, Manchester, UK).
Electrophoresis mobility shift assays
Yeast U14 snoRNA, U3Δ2,3,4 snoRNA (35) and tRNAHis were in vitro transcribed and 5’-end labeled as described (21). Five hundred nanomolar of various proteins or complexes was mixed and incubated for 20 min at 4°C with 5 fmol of 5′-end 32P-radiolabeled RNA in buffer D. The RNA–protein complexes formed were visualized by native gel electrophoresis.
NMR spectroscopy and structure calculation
One millimolar 15N and 13C-15N labeled samples of Rsa1p317−352−Hit1p70−164 in buffer (10 mM sodium phosphate, pH 6.4, 150 mM NaCl, 10% D2O) were used for NMR experiments. Essentially complete resonance assignment was achieved using a classical approach based on 3D NMR spectra recorded at 293 K on 600 and 950 MHz Bruker spectrometers, both equipped with cryoprobes. Dynamics and folding of the backbone were evaluated through measurement of the 1H-15N heteronuclear nOe and HetSOFAST ratios (42) recorded in the same conditions as the ones used for assignment. For structure determination, distance and dihedral angle restraints were derived respectively from NOESY-HSQC 1H-15N and 1H-13C NMR experiments and from TALOS+ (43). The NMR structures were calculated using the automated procedure of CYANA 3.0 (44). The NOE assignments were carefully checked after the final iteration. Finally, RECOORD scripts were used to generate 200 water-refined structures and the 20 structures with the lowest overall energies were selected as the most representative (45). In the final structures, 99.0% of residues were in favored to generously allowed regions of Ramachandran space and 1.0% were outside.
RESULTS
Association of Rsa1p with Hit1p in yeast cells
To better characterize complexes formed during C/D snoRNP biogenesis, we tried to identify partners of proteins Rsa1 and Pih1 in yeast cells. We used a single-step immunoglobulin G (IgG) affinity purification procedure based on TAP-tagged Rsa1p and Pih1p, followed by a proteomics-based analysis. After cleavage with the TEV protease, the Rsa1p and Pih1p co-purified proteins were fractionated by 1D SDS-PAGE. Gel slices were cut systematically along the gel, and protein contents were identified by MS.
As illustrated by the spectral counts of unique peptides detected for the identified proteins, the Rvb1p, Rvb2p and Tah1p components of the R2TP complex were uniquely found in significant amounts in the Pih1p-TAP purification (Table 1) and an unexpected partner of Rsa1p, protein Hit1, was detected in the Rsa1p-TAP purification (Table 1). In both TAP-tag experiments, only three of the C/D core proteins were detected: Snu13p, Nop1p in both experiments, together with either Nop56p (Rsa1p-TAP) or Nop58p (Pih1p-TAP), and these proteins were represented by a limited number of peptides. Absence of detectable Rsa1p in the Pih1p-TAP purification and of Pih1p in the Rsa1p-TAP purification was surprising. It might reflect short life span of the interactions in vivo, or their destabilization during extraction. The TAP sequences might have reinforced the instability.
Table 1.
Identification of Hit1p as an interacting partner of Rsa1p
| No TAP | PIH1-TAP | RSA1-TAP | HIT1-TAP | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein name | Accession number | MW (Da) | Unique pep (% cover) | Spectral count | Unique pep (% cover) | Spectral count | Unique pep (% cover) | Spectral count | Unique pep (% cover) | Spectral count | |
| Rsa1p | Q08932 | 43981 | - | - | - | - | 10 (35,4) | 81 | 10 (32,0) | 77 | |
| Hit1p | P46973 | 18358 | - | - | 2 (20,7) | 2 | 7 (54,9) | 31 | 7 (54,9) | 49 | |
| Box C/D core proteins | Snu13p | P39990 | 13551 | - | - | 2 (36,5) | 6 | 2 (36,5) | 9 | 2 (36,5) | 8 |
| Nop1p | P15646 | 34195 | 2 (10,4) | 4 | 4 (21,4) | 5 | 5 (24,5) | 9 | 6 (27,8) | 9 | |
| Nop56p | Q12460 | 56849 | - | - | - | - | 5 (20,4) | 7 | - | - | |
| Nop58p | A6ZPE5 | 57176 | - | - | 2 (6,8) | 3 | - | - | - | - | |
| R2TP proteins | Pih1p | P38768 | 39503 | - | - | 13 (51,7) | 58 | - | - | - | - |
| Tah1p | P25638 | 12493 | - | - | 4 (69,4) | 34 | - | - | - | _ | |
| Rvb1p | Q03940 | 50436 | - | - | 16 (47,7) | 26 | - | - | - | - | |
| Rvb2p | Q12464 | 51595 | - | - | 17 (46,9) | 34 | - | - | - | - | |
List of the C/D snoRNP biogenesis related proteins identified by proteomic analysis after tandem affinity experiments using Rsa1p-TAP, Hit1p-TAP, Pih1p-TAP and an untagged negative control (no TAP).
Nevertheless, detection of a Rsa1p–Hit1p association was interesting and to verify this association, we performed an IgG affinity purification with cells expressing Hit1p-TAP. Rsa1p was found to be associated with Hit1p, as 10 unique Rsa1p peptides were detected at high levels in the eluted Hit1p-TAP samples. Here again, no peptide from the R2TP complex was observed (Table 1). Taken together, these data suggested the existence in yeast cells of a complex containing Rsa1p and Hit1p. By screening the literature, we noticed that a possible interaction between Rsa1p and Hit1p had also been found in a large yeast two-hybrid (Y2H) screen for protein–protein interactions in yeast (46). This prompted us to test for possible direct interaction between these two proteins.
The Rsa1p–Hit1p interaction is direct and compatible with the Snu13p–Rsa1p interaction
First, we showed by Y2H assays that the Rsa1p C-terminal fragment extending from Pro230 to Lys381 (Rsa1p230−381), which ectopic expression restores normal growth of a slow growing ΔRSA1 strain (21), is sufficient for association with Hit1p (Figure 1A). As this fragment carries the yPEP domain (residues 230−263), which constitutes the Snu13p-binding site of Rsa1p (21,29), we tested by co-expression in various combinations of His6-tagged Snu13p, Hit1p and Rsa1p in E. coli cells, whether the Rsa1p−Hit1p interaction is direct and whether Rsa1p can simultaneously interact with Snu13p and Hit1p. Co-expressed proteins were co-purified from bacterial extracts by immobilized metal ion affinity chromatography, fractionated by SDS-PAGE and analyzed by MS. As MS analysis revealed spontaneous proteolysis of the six C-terminal amino acids of Rsa1p230−381 (data not shown), we expressed the Rsa1p230−375 fragment lacking these residues. Hit1p co-purified with both His6Rsa1p and His6Rsa1p230−375 (Figure 1B). After cleavage of the His6 tag, the release Rsa1p230−375–Hit1p heterodimer could be purified at homogeneity by gel filtration, demonstrating the direct interaction of the two proteins (Figure 1D). In addition, co-expressed Rsa1p230−375 and Hit1p formed a heterotrimer with His6Snu13p (Figure 1C) that was purified at homogeneity (Figure 1E). Native MS confirmed the respective 1:1 and 1:1:1 stoichiometry of the heterodimer and heterotrimer (Figure 1F). These data demonstrated the capability of Rsa1p to interact simultaneously with Hit1p and Snu13p, which was a strong hint for its possible involvement in snoRNP assembly. Therefore, we tested the effect of HIT1 gene disruption on this process.
Figure 1.
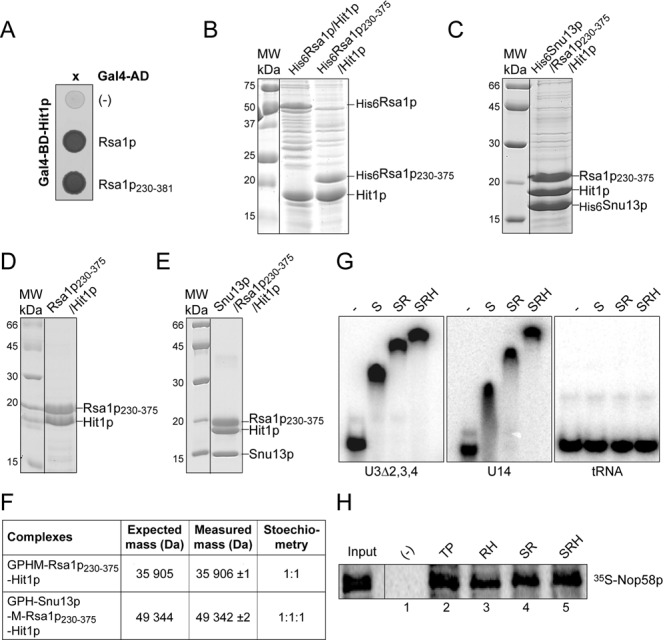
Rsa1p and Hit1p form a stable heterodimer able to bind Snu13p and Nop58p. (A) Y2H assay. Rsa1p and Rsa1p230−381 fused to the Gal4 activation domain (AD) interact with Hit1p fused to the Gal4 DNA binding domain (BD) as evidenced by growth on a His deprived medium. (B, C,D and E) Co-expression assays in E. coli and complex purifications. In (B) His6-taged Rsa1p and Rsa1p230−375 were co-expressed with Hit1p, in (C) His6-taged Snu13p was co-expressed with Rsa1p230−375 and Hit1p. Complexes were selected from crude extracts by Immobilized Metal Ion Affinity Chromatography (IMAC), followed by SDS-PAGE and Coomassie Blue staining. Molecular weight markers (MW) (in kDa) were loaded on the left. (D and E) complexes released by cleavage of the His6 tag with the PreScission protease were purified by size exclusion chromatography (Superdex 200 prep grade column, GE Healthcare) using GF buffer (10 mM HEPES pH 7.5, 150 mM NaCl, and 5 mM β-mercaptoethanol). (F) Native MS characterization of the complexes purified by gel filtration in (D) and (E). NanoESI mass spectra performed under nondenaturing conditions as described in Materials and Methods confirmed the presence of a 1:1 Rsa1p230−375−Hit1p and a 1:1:1 Snu13p−Rsa1p230−375−Hit1p complex. Removal of the His6 tag generates the N-terminal sequences GPHM for Rsa1p and GPH for Snu13p. (G) The trimeric complex His6Snu13p–Rsa1p230−375–Hit1p interacts in vitro with RNAs containing a K-turn RNA motif. Formation of RNA–protein complexes was assessed by EMSA. Radiolabeled U3Δ2,3,4 and U14 RNAs and a control tRNA were incubated with His6Snu13p (lane S), His6Snu13p–Rsa1p230−375 (lane SR), or His6Snu13p–Rsa1p230−375–Hit1p (lane SRH) complexes obtained by IMAC followed by elution in presence of 200 mM imidazole and purified as in (D) and (E). The RNP complexes formed were resolved on a 6% non-denaturing polyacrylamide gels. (H) His-pull down assays with Nop58p. Complexes formed between 35S Nop58p translated in vitro in bacterial S30 lysate and purified recombinant His6Tah1p–Pih1p (lane TP), His6Rsa1p230−375–Hit1p (lane RH), and SR and SRH complexes of panel (G) were bound to cobalt-Sepharose beads and analyzed by SDS-PAGE. 10% of the 35S Nop58p used per assay was loaded in the Input lane.
Synthetic lethality tests reveal functional links between Hit1p, Rsa1p and Pih1p
We previously observed decreased levels of snoRNA and growth defects upon RSA1 gene disruption (21,29), therefore, for further analysis of Hit1p function, we compared the growth phenotypes of S. cerevisiae cells carrying individual deletions of the HIT1 and RSA1 genes, and of the PIH1 gene encoding another snoRNP assembly factor. Very similar slow growth phenotypes were observed after HIT1 or RSA1 gene inactivation, while a twice less marked effect was found for PIH1 gene disruption (Figure 2A). In addition, to test for possible functional links between these three genes, we crossed single knock-out haploid strains to obtain heterozygous diploid strains. Tetrads of haploid cells were produced by meiosis during sporulation and dissected, and growth of the haploid cells was tested on YPD media plates (Figure 2B). The genotypes of the haploids were identified by PCR amplification. Interestingly, the ΔRSA1-ΔHIT1 double knock-out haploid strain did not reveal any cumulative growth retardation relative to the phenotypes of the single knock-out strains. Altogether, the data strongly suggested that Hit1p and Rsa1p are involved in the same cellular process(es). Moreover, the absence of growth found for cells combining null mutations of RSA1 and PIH1 genes, or null mutations of HIT1 and PIH1 genes (Figure 2B), was a strong hint for a functional connection between these three genes and snoRNP assembly.
Figure 2.
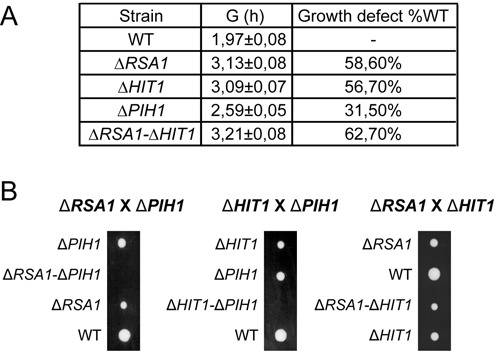
Growth of ΔRSA1, ΔHIT1, ΔPIH and ΔRSA1-ΔHIT1 BY4741 mutant cells. (A) Growth of WT and mutant strains in YPD medium was monitored over time by A600 measuring. Doubling times (G) were calculated and growth defects are expressed as a percentage of the doubling time of the WT strain. (B) Strains lacking Rsa1p and Pih1p, or Hit1p and Pih1p are not viable. Diploid strains generated from crossbreeding between ΔRSA1, ΔPIH1 or ΔHIT1 haploid BY4741 (MATa) strains with BY4742 (MATα) strain were placed under starvation conditions for meiosis and spore formation. Tetrads were dissected and haploid spores were deposited on YPD plates and incubated for one week at 30°C. A representative dissection of tetrads is shown for each crossbreeding.
Hit1p is required to maintain the steady-state level of C/D snoRNAs
For complete demonstration of the involvement of Hit1p in snoRNA biogenesis, we compared the levels of C/D and H/ACA snoRNAs, as well as UsnRNAs, in the ΔHIT1, ΔRSA1 and ΔPIH1 mutants and the isogenic wild-type (WT) strains. As previously observed in the RSA1 knock-out strain (29), lower levels of the tested C/D snoRNAs, except U3, and unchanged levels of H/ACA snoRNAs and UsnRNAs were observed in the three mutant strains (Figure 3A), which strongly reinforced the idea of an implication of Hit1p in snoRNP biogenesis. Noticeably, in accord with the stronger growth phenotypes found for the ΔRSA1 and ΔHIT1 mutants compared to the ΔPIH1 mutant, a stronger snoRNA destabilization effect was observed for the ΔRSA1 and ΔHIT1 mutants compared to the ΔPIH1 mutant.
Figure 3.
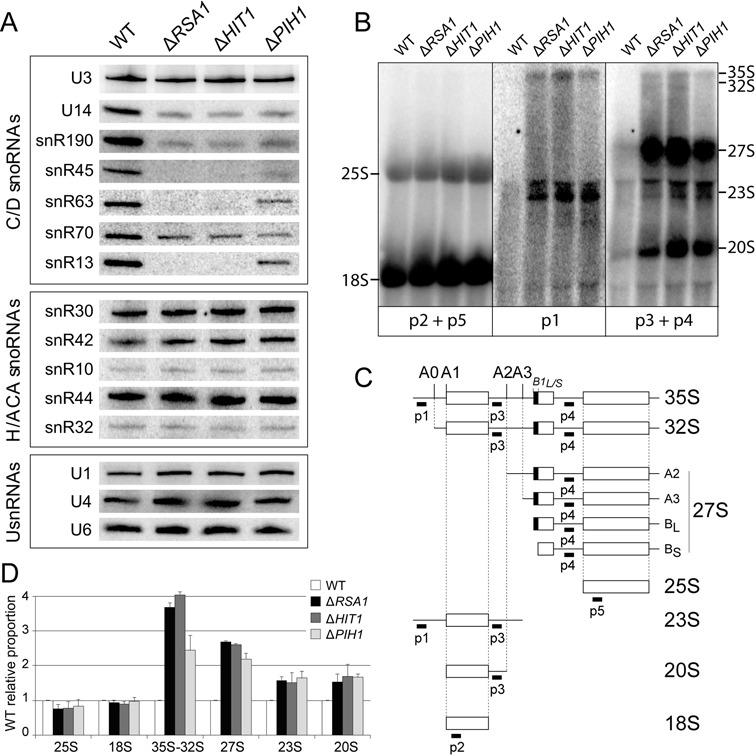
Rsa1p, Hit1p and Pih1p expressions are required for accumulation of C/D snoRNAs and pre-rRNA processing. (A) Northern blot analysis of steady-state levels in exponential phase of various RNAs (C/D snoRNAs, H/ACA snoRNAs and UsnRNAs) in the WT and the null mutant strains. Hybridizations were performed with specific radiolabeled probes (see Supplementary Table S2). (B, C, D) Analysis of pre-rRNA processing. In (B), total RNA from WT and mutant BY4741 strains were extracted in log phase and fractionated on 1.2% glyoxal/DMSO agarose gels. Northern blotting was performed by hybridization with 32P labeled probes p1, p2, p3, p4 and p5 (see Supplementary Table S2), allowing detection of mature rRNAs (p2 + p5), or the pre-rRNA processing intermediates schematically represented in (C) (p1, or p3 + p4). In (C), sequences targeted by the probes are indicated under the schemes ofS. cerevisiae pre-rRNA processing intermediates. In (D) quantification of data from northern blot experiments (B). Radioactivity in the bands of gel was quantified with the ImageQuant software after exposure of a phosphorimager screen. The signal for each RNA species was divided by the accrued signal of all RNA species in the same lane. The ratios corresponding to the mature forms (25S and 18S), the precursors (35S and 32S) and the maturation intermediates (27S A2 + A3 + BS + BL, 23S and 20S) in WT strain were arbitrary considered as having a value of 1, and the ratios of the corresponding RNAs in the mutants were expressed relative to this value. Error bars represent the standard deviation obtained from the mean value of two independent experiments.
Given that snoRNPs participate in ribosome biogenesis, we further analyzed the effects of the ΔRSA1, ΔHIT1 and ΔPIH1 disruptions on 35S pre-ribosomal RNA (35S pre-rRNA) processing, using previously described protocols (47). Total RNAs fractionated on agarose gels were transferred on membrane and probed with a series of 5’-end labeled probes to test for accumulation of pre-18S and pre-25S intermediates (probes p1 to p4, Figure 3B and C), and to evaluate relative levels of mature 18S and 25S rRNAs (p2 and p5) (Figure 3B and C). A marked accumulation of 27S precursors of the 5.8S and 25S rRNAs (27SA2, A3, BS and/or BL) and increased steady-state levels of the 35S pre-rRNA, 32S, 23S and 20S intermediates were observed for the three strains (Figure 3B and C). Here again, the similar features observed for the ΔRSA1 and ΔHIT1 mutants reinforced the idea of a functional link between Rsa1p and Hit1p. Accumulation of the same pre-rRNA intermediates in the ΔPIH1 mutant, demonstrated its role in pre-rRNA maturation and its possible functional link with Rsa1p and Hit1p.
Hit1p influences U3 snoRNA 3′-terminal processing
U3 snoRNA level did not seem to be markedly modified in the absence of Rsa1p, Hit1p or Pih1p (Figure 3A). However, U3 snoRNA production in S. cerevisiae is a complex process including the production by RNA polymerase II of a precursor RNA (pre-U3) containing one intron, which is spliced by the spliceosome (48), and an extended 3′-end (49). Unspliced pre-U3 and spliced pre-U3 could be detected by RT-PCR using a reverse-primer hybridizing in the 3′-extension (Figure 4A). Due to efficient splicing, unspliced pre-U3 was detected in low amounts. Nevertheless, we used such detection assay to test for association of U3 precursors with the TAP–tagged snoRNP core proteins, the already described assembly factors, and Hit1p. The results strongly suggested the in vivo association of both the spliced and unspliced pre-U3 containing a 3′ extension with the Nop1p, Nop56p and Nop58p core proteins (Figure 4B), and the Rsa1p, Hit1p, Pih1p, Rvb1p and Rvb2p proteins (Figure 4C). This prompted us to test for a possible influence of Hit1p expression on maturation of the U3 snoRNA 3′-extension. Truncations within the sequence coding for the 3′ structural domains of S. cerevisiae U3A snoRNA gene (U3Δ2,3,4) generate the accumulation of 3′-end maturation intermediates I’ and I (49) and the amounts of these maturation intermediates increase in the absence of Rsa1p expression (21). We show here that the absence of Hit1p and Rsa1p expressions has similar effects on accumulation of U3Δ2,3,4 intermediates (Figure 4D), with a parallel decrease of mature U3Δ2,3,4 RNA, indicating that both Hit1p and Rsa1p likely influence the kinetics of U3 snoRNA 3′-end processing.
Figure 4.
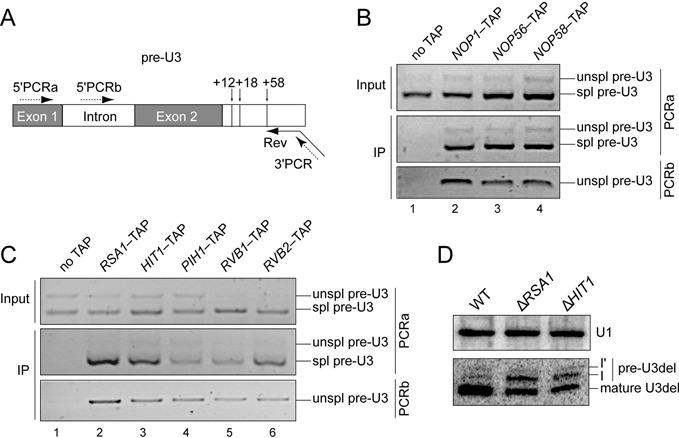
Core proteins and assembly factors are associated with U3 precursors, and Rsa1p and Hit1p are involved in its processing. (A) Scheme of the U3 transcript showing its intron and extended 3′-end. Positions of cleavages in the 3’-extension are indicated by vertical arrows. Synthesis of cDNAs was primed by oligonucleotide Rev (broken arrow) interacting with the U3 3′-extension. Primers 5′PCRa, 5′PCRb, and 3′PCR used for the PCR amplifications are indicated by dotted arrows. (B, C) Log phase extracts prepared from various TAP-tagged strains (core proteins (B) and assembly factors (C)) were purified on IgG-Sepharose beads and analyzed by RT-PCR using primer 5′PCRb specific for unspliced (unspl) or 5’PCRa amplifying both spliced (spl) and unspliced U3 precursors carrying the full length 3′-extension. An untagged strain was used as negative control. Inputs correspond to 10% of cellular extracts used in IPs. (D) Northern blot analysis of processing of the U3Δ2,3,4 variant RNA in the WT, ΔRSA1 and ΔHIT1 strains reveals the accumulation of pre-U3Δ2,3,4 precursors in the two mutant strains, especially precursors cleaved at positions +12 (band I) or +18 (band I’). U1 snRNA was used as a loading control.
Hit1p regulates the steady-state level of Rsa1p and this feature is conserved in human cells
The above data all revealed identical defects upon the absence of expression of Rsa1p or Hit1p. Therefore, we tried to determine the reason for this interdependency and found that Hit1p controls the cellular concentration of Rsa1p. Indeed, we observed a lower level of Rsa1p-TAP in the ΔHIT1 knock-out strain (Figure 5A, compare lanes 1 and 2), while ectopic expression of HA-Hit1p was associated with high level of Rsa1p-TAP protein (lane 3). The influence of Hit1p on the abundance of Rsa1p was confirmed by comparing the amounts of plasmid-expressed GFP-Rsa1p fusion protein in a ΔRSA1 strain and in the double ΔRSA1–ΔHIT1 knock-out strain (Figure 5B, compare lanes 2 and 3). In contrast, the absence of Rsa1p did not show a strong effect on the level of Hit1p fused to the HA epitope from influenza hemagglutinin (lanes 5 and 6).
Figure 5.
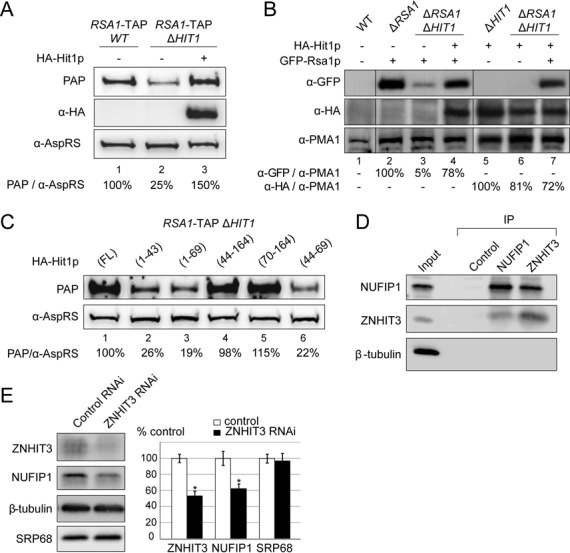
Hit1p/ZNHIT3 is required for accumulation of Rsa1p/NUFIP1. (A, B) Hit1p stabilizes Rsa1p. Cells were grown to A600 ∼0.8–1. Equal amounts of cells (30 A600 U) were used for crude cell extract preparation. Western blot analysis of equal amounts of total cell lysates from the RSA1-TAP strain (A, lane 1), the isogenic ΔHIT1 mutant (A, lanes 2–3), WT BY4741 (B, lane 1), the isogenic ΔRSA1 (B, lane 2), ΔHIT1 (B, lane 5), and ΔRSA1ΔHIT1 (B, lanes 3–4, 6–7) mutants with or without ectopic expression of HA-Hit1p (A, B) and/or GFP-Rsa1p (B). Rsa1p tagged proteins were detected with PAP (A) or anti-GFP antibodies (B). Anti-HA (α-HA) were used for Hit1p (A, B). The PMA1 and AspRS proteins used as loading controls were detected using specific anti-PMA1 (α-PMA1) and anti-AspRS (α-AspRS) antibodies. Relative amounts of Rsa1p (PAP/α-AspRS, or α-GFP/α-PMA1) or of Hit1p (α-HA/α-PMA1) were determined from the ratio of band intensities in each lane. They are expressed as a percentage of the value obtained for lane 1 (A) or lanes 2 and 5 (B) (100%). (C) Western blot analysis of equal amounts of total cell lysates from the HIT1 null mutant strain expressing the Rsa1-TAP protein (strain RSA1-TAP/ΔHIT1) transformed with plasmids expressing various fragments of HA-Hit1p. Tagged proteins were detected as in panel (A). The relative amounts of Rsa1p indicated below each lane were estimated as in panel (A). (D) ZNHIT3 co-precipitates with NUFIP1. Immunoprecipitation (IP) experiments were carried out on HeLa total extracts using anti-NUFIP1 or anti-ZNHIT3 antibodies bound to Protein A Sepharose. Protein A Sepharose alone was used as control. Co-immunoprecipitated proteins were analyzed by SDS-PAGE and western blotting with the antibodies indicated on the left; 5% of total proteins used per assay were loaded in lane Input. (E) ZNHIT3 concentration influences NUFIP1 steady-state level. HeLa cells were transfected with negative control Gl2 firefly luciferase (Control RNAi) or ZNHIT3 siRNAs (ZNHIT3 RNAi). Total extracts were analyzed by western blotting. β-tubulin and SRP68 served as loading controls. Experiments were performed in triplicate and relative amounts of proteins in HeLa cells transfected with ZNHIT3 siRNAs were plotted as a% of proteins in HeLa cells transfected with the control siRNAs. Error bars indicate the standard error of the mean values (s.e.m); (*P < 0.05).
Then, in order to identify Hit1p sequences required to maintain the cellular concentration of Rsa1p, we complemented the ΔHIT1 strain with several constructs expressing various Hit1p fragments (Supplementary Figure S1). In particular, we tested fragments carrying the first 43 amino acids proposed to correspond to a Zn-finger domain. In fact, residues 70−164 were found to be sufficient to restore the WT growth phenotype. In accord with the proposed role of Hit1p, there was a good correlation between the ability of the fragments to complement the growth defect and the measured intracellular levels of Rsa1p (Figure 5C). Therefore, we concluded that the Zn-finger domain of Hit1p is not essential for this function.
By co-immunoprecipitation (co-IP) assays carried out with HeLa cell total extracts and anti-NUFIP1 or anti-ZNHIT3 antibodies bound to Protein A-Sepharose beads, we showed that NUFIP1 and ZNHIT3, the human homologues of Rsa1p and Hit1p, also associate in human cell extract (Figure 5D). The co-IP specificity was verified by the absence of selection of β-tubulin used as a control. We next tested whether ZNHIT3 is also important to control the cellular concentration of NUFIP1. RNA interference knockdown experiments were performed using ZNHIT3-specific siRNAs, and the firefly luciferase siRNA as a negative control (Figure 5E). The ZNHIT3 protein level was reduced by ∼50%, leading to a significant decrease in the level of NUFIP1 (∼40%), whereas the levels of the β-tubulin and SRP68 proteins used as negative controls remained stable. Our results therefore support the conclusion that ZNHIT3 is also required to maintain the steady-state level of NUFIP1 in HeLa cells.
Purified Snu13p−Rsa1p230−375−Hit1p heterotrimer can bind C/D RNAs and Nop58p
Having shown the functional link between Rsa1p and Hit1p and developed conditions for purification to homogeneity of the Snu13p−Rsa1p230−375−Hit1p complex, we used this complex to get information on complexes that may be formed during C/D snoRNP assembly. First, as Snu13p is known to interact with C/D snoRNAs and with Rsa1p (29), we tested by electrophoresis mobility shift assays (EMSA) whether the Rsa1p−Hit1p interaction is compatible with binding of Rsa1p to a [C/D snoRNA–Snu13p] complex. As illustrated in Figure 1G, when incubated with a Snu13p–Rsa1p–Hit1p purified ternary complex, a radiolabeled C/D snoRNA is found in a complex displaying a slower mobility compared to the complex formed with the purified Snu13p–Rsa1p heterodimer. Therefore, binding of Hit1p does not impair the capacity of Snu13p–Rsa1p to interact with RNA, and the Rsa1p–Hit1p interaction is stable in the presence of the RNA.
Furthermore, as association of Nop58p with Rsa1p (21) and Pih1p (31) had been detected by Y2H assay, it was interesting to test whether 35S radiolabeled Nop58p produced in E. coli S30 extract can interact with purified recombinant His6Tah1p–Pih1p, His6Rsa1p230−375–Hit1p and His6Snu13p–Rsa1p230−375–Hit1p complexes immobilized on Ni-Sepharose beads. Nop58p was retained on the His6Tah1p–Pih1p (Figure 1H, lane 2), His6Rsa1p230−375–Hit1p (lane 3), His6Snu13p–Rsa1p230−375 (lane 4) and His6Snu13p–Rsa1p230−375–Hit1p complexes (lane 5), but not in the absence of His6-tagged complex (lane 1). Therefore, Nop58p can associate directly with Pih1p and with Rsa1p in the absence of RNA, and the binding of Tah1p to Pih1p and of Hit1p to Rsa1p does not impair formation of these interactions. Furthermore, in agreement with the idea that Hit1p can participate to large pre-snoRNP assembly complexes (Figure 4C), our data indicate that the Rsa1p–Hit1p complex can interact simultaneously with Snu13p and Nop58p.
3D. NMR structure of the Rsa1p–Hit1p interaction explains its high stability
For complete understanding of the Rsa1p–Hit1p interaction, we solved the 3D structure of their interacting domains by multi-dimensional NMR spectroscopy, following co-expression of fragments of both proteins in 13C, 15N-isotopically-enriched minimal medium. First, by using various conditions for limited proteolysis and native MS analyses of the Snu13p–Rsa1p230−375–Hit1p, we identify the Rsa1p230−375–Hit1p70–164 sub-complex suitable for structural analysis. Secondly, NMR 1H-15N HSQC analysis showed that segments P230−E316 and S353−A375 of Rsa1p are not required for the Rsa1p–Hit1p interaction (Supplementary Figure S2). With a remarkable homogeneity and very few resonance overlaps, the 1H-15N HSQC spectrum of a complex formed between Rsa1p317−352 and Hit1p70–164 displayed the features of a well-folded entity suitable for 3D solution structure determination (Figure 6).
Figure 6.
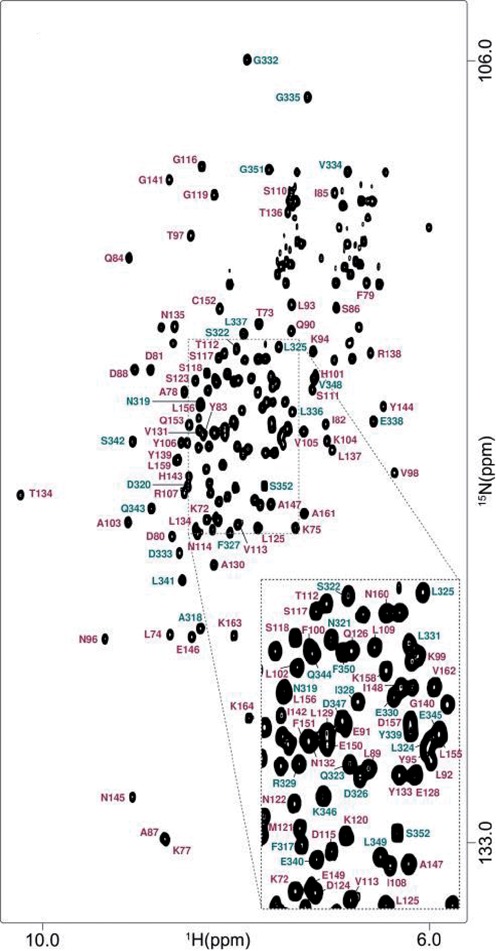
1H-15N HSQC spectrum of the complex formed between Rsa1p317−352 and Hit1p70−164. The spectrum was recorded on a 600 MHz NMR spectrometer in NaPi 10 mM, pH 6.4, NaCl 150 mM and at 293K. Assignment of the main-chain amide groups is figured in teal blue and in magenta for Rsa1p317−352 and Hit1p70−164, respectively.
The water-refined NMR structure of the Rsa1p317−352–Hit1p70–164 complex yielded secondary structure RMSD values of 0.53 ± 0.08 Å and 1.03 ± 0.28 Å, measured on backbone and heavy atoms respectively (Figure 7; for structural statistics, see Supplementary Table S1). Hit1p70−164 comprises five α-helices and one long internal disordered loop (between α3 and α4), while Rsa1p317−352 contains two α-helices α′1 and α′2 linked by a well-defined linear fragment. This secondary structure pattern composed of helices is in good agreement with the 1H-15N heteronuclear and HetSOFAST ratios (Supplementary Figure S3). In the complex, Hit1p70−164 acts as a jaw which almost completely encloses Rsa1p317−352 (Figure 7B). Among the 130 amino acids of the two Hit1p and Rsa1p fragments, 58 residues including 31 mainly hydrophobic, 14 charged and 11 polar residues along with 2 glycines form the protein interface (i.e. distance of separation less than 3.5 Å) (Figure 8). These residues (33 from Hit1p70−164 and 25 from Rsa1p317−352) bury a mean surface area of 3267 ± 128 Å2, which represents almost 40% of the solvent-exposed surface of the Rsa1p317−352−Hit1p70−164 complex. More specifically, the five α-helices of Hit1p70−164 form a ‘claw’ which locks helix α1′ of Rsa1p317−352, while the Rsa1p317−352 helix α2′ packs against the exposed surface of the Hit1p70−164 helix α3. Thus helix α1′ of the Rsa1p320−334 forms the architectural base for the heterodimeric complex. Some of the stabilizing non-polar or polar interactions are highlighted in panels B and C of Figure 8. Interestingly, interfacing residues of the Rsa1p317−352−Hit1p70−164 complex are conserved among species such as L93, L102, L109 in α2, α3 helices or L156, L/V159 in α5 helix of Hit1p70−164, and E330, L331 and G332 in the keystone α1’ helix of Rsa1p317−352 (Figure 8A). Added to co-IP and siRNA assays, we may assume that the structural interleaved mode of interaction between Rsa1p/NUFIP1 and Hit1p/ZNHIT3 is similar in yeast and mammals. Nonetheless, multiple sequence alignment reveals insertions in NUFIP1 and deletions in ZNHIT3, indicating that the mammal complex could exhibit significant changes compared to what is observed in yeast. By site-directed mutagenesis of both Rsa1p and Hit1p, we tried to abolish the capability of interaction of the two proteins in in vivo and in vitro assays. However, the amino acids involved in the interaction turned to be required for both stability and solubility of each of the two partners and we could not get soluble mutant proteins having lost their interaction capacities.
Figure 7.
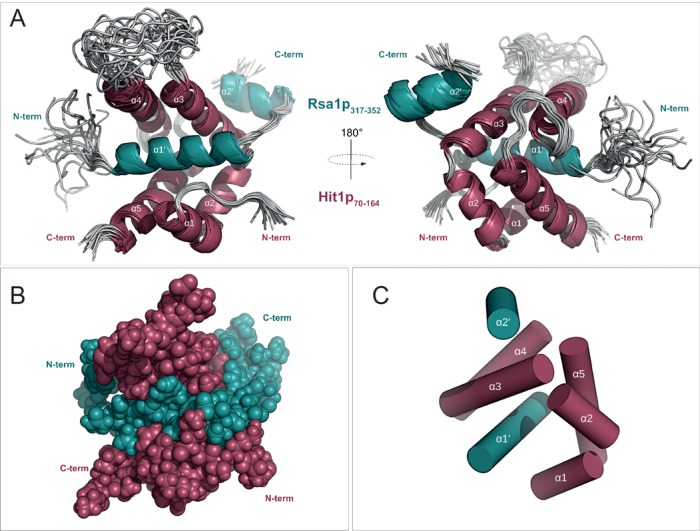
NMR solution structure of the complex formed between Rsa1p317−352 and Hit1p70−164. Two-side view of the cartoon representations of the 20 lowest energy NMR structures (A) and sphere representation of the best NMR structure (B) calculated with RECOORD scripts. (C) Spatial organization of the seven α-helices. Proteins Rsa1p317−352 and Hit1p70−164 are represented in teal blue and in magenta, respectively.
Figure 8.
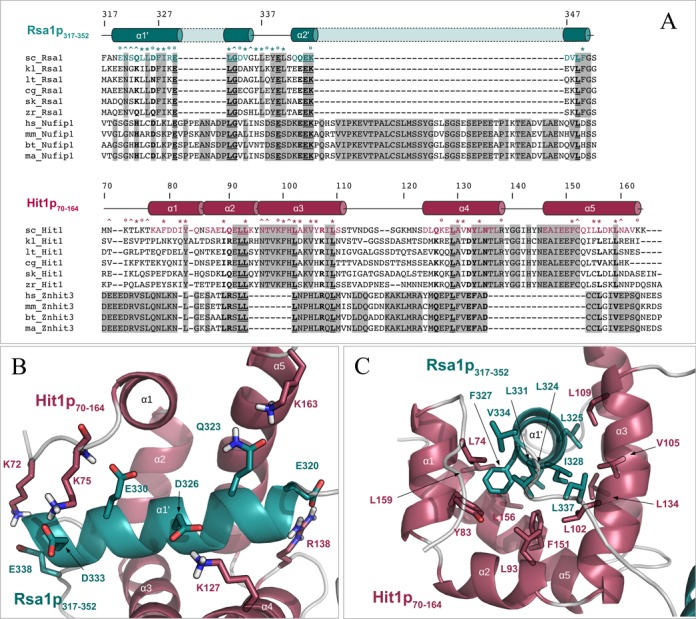
The protein–protein interface between Rsa1p317−352 and Hit1p70−164 is maintained by hydrophobic and polar contacts involving conserved residues. (A) Multiple amino acid sequence alignments of proteins Rsa1/NUFIP1 and Hit1/ZNHIT3 present in Saccharomyces cerevisiae (sc_), Kluyveromyces lactis (kl_), Lachancea thermotolerans (lt_), Candida glabrata (cg_), Saccharomyces (Lachancea) kluyveri (sk_), Zygosaccharomyces rouxii (zr_), Homo sapiens (hs_), Mus musculus (mm_), Bos taurus (bt_) and Macaca mulatta (ma_), and constructed using CLUSTALW. Strictly matching residues in yeast or in mammals are highlighted using gray squares. Strictly conserved inter-species positions are underlined and highlighted in bold. Inter-species residues with groups of strongly similar properties are indicated in bold. Secondary structures extracted from the Rsa1p317−352−Hit1p70−164 3D structure are represented above the protein sequences, using cylinders for α-helices and lines for loops. Residues located at the interface between the two proteins (according to an intermolecular distance cut-off of 3.5 Å) are highlighted using circles, stars and carets for charged, mainly hydrophobic and neutral to polar amino acids respectively. (B) and (C) Representation of some hydrophobic (B) and polar (C) contacts located at the protein–protein interface of the Rsa1p317−352−Hit1p70−164 complex. Non-polar hydrogens are not represented.
Remarkably, DALI (50) and VAST (51) servers failed to identify similar global structures of protein–protein complexes in the Protein Data Bank, highlighting the unique and novel feature of this protein association. Due to its particular structural features, we propose to name the fold of Hit1p70−164, the Pac-Hit fold that mimics the figure of Pac-Man, the iconic video game character. Overall, these data reveal an intricate interaction of Rsa1p with Hit1p explaining the participation of Hit1p together with Rsa1p in common steps of box C/D snoRNP biogenesis.
DISCUSSION
Rsa1p−Hit1p is a specific sub-complex of C/D snoRNP assembly in yeast
Although yeast Hit1p was discovered many years ago, its function has not been evident, aside from the fact that disruption of its encoding gene leads to a thermo-sensitive phenotype (34). Here, we demonstrate that Hit1p is a component of the C/D snoRNP assembly machinery, which is required to control the abundance of Rsa1p, one of the key factors involved in C/D snoRNP biogenesis (21), and which is also involved in 60S ribosomal subunit production (52). Deletions of either the HIT1 or the RSA1 genes results in similar growth defects in yeast cells. According to our data, these results are explained by the decreased levels of several C/D snoRNAs and reduced rates of some pre-rRNA maturation steps. Taken together, these data reinforce the idea that Rsa1p plays an important role in C/D snoRNP assembly and ribosome biogenesis.
As previously observed for yeast RSA1 gene deletion, the HIT1 gene deletion did not alter H/ACA snoRNA levels, showing that activity of the Rsa1p−Hit1p complex is dedicated to C/D snoRNP assembly. Therefore, biogenesis of the two classes of yeast snoRNPs depends both upon a common factor, the R2TP complex (21,22,24,26,28), and specific factors: Shq1p and Naf1p for H/ACA snoRNPs (for review (20)), and Rsa1p and Hit1p for C/D snoRNPs. Previous observations (26) suggested that R2TP components are not each required to the same extent for C/D and H/ACA RNP assembly. Here, we confirm that Pih1p is necessary for maintaining C/D snoRNA levels, while its absence has no marked effect on H/ACA snoRNA levels (26), an observation which contrasts with the strong requirement for the Rvb1 and Rvb2 AAA+ helicases for H/ACA compared to C/D snoRNP (26,28).
Rsa1p−Hit1p constitutes a functional module for the early stages of C/D snoRNP assembly
In the current model of C/D snoRNP assembly, an initial interaction between proteins Snu13p/15.5K in eukaryotes or L7Ae in archaea and the K-turn motif present in C/D snoRNAs is required for subsequent assembly of the other RNP proteins (53–57). However, how these RNPs are assembled in vivo and the precise functions of the assembly factors are not well understood. The K-turn may be initially bound by the free form of Snu13p and subsequently by the Rsa1p–Hit1p sub-complex or directly by a Snu13p–Rsa1p–Hit1p complex (Figure 9, Pathway A, Step 1). This scenario is plausible because such stable heterotrimeric complex can be purified (Figure 1E), and because association of Rsa1p and Hit1p with Snu13p does not impair the RNA binding capacity of Snu13p (Figure 1G). The C-terminal Rsa1p230−381 segment, which represents the minimal fragment able to restore normal growth in yeast (21), contains several binding sites for proteins involved in C/D snoRNP assembly. In this manner, it constitutes a platform for several non-mutually exclusive interactions: the Rsa1p230−261 segment binds Snu13p (29), the Rsa1p317−352 segment binds Hit1p, and according to the present data, another segment, which has yet to be identified, interacts directly with Nop58p. The formation of these interactions is probably favored by the low level of internal structuration of Rsa1p, as evidenced by secondary structure predictions. This flexibility may explain its high sensitivity to proteolysis and the need for protection by Hit1p. Nevertheless, we cannot exclude the possibility of specific sites for ubiquitinylation in Rsa1p, which may contribute to its rapid degradation in yeast cells.
Figure 9.
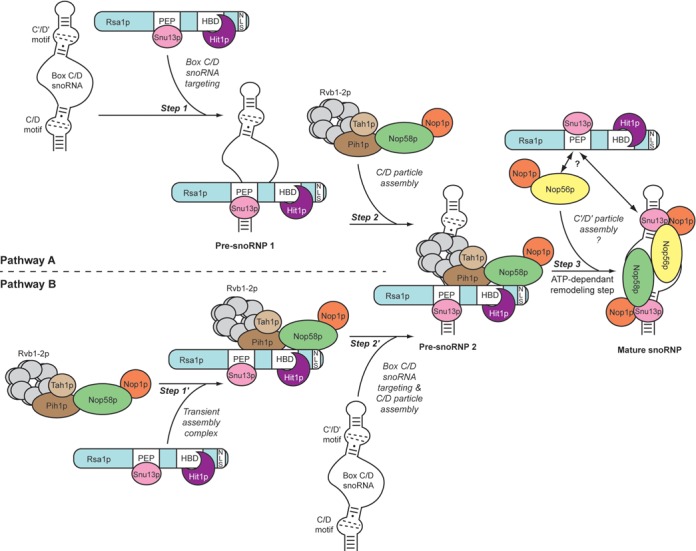
Model for the box C/D snoRNP assembly pathway. The particles [RNA−Snu13p−Nop58p−Nop1p] and [RNA−Snu13p−Nop56p−Nop1p], respectively nucleated on the C/D and C′/D′ motifs, are linked by a coiled-coil dimer in the mature snoRNP (59). Two alternative assembly pathways are proposed: the primary targeting of the snoRNA by the complex Snu13p−Rsa1p−Hit1p (Pathway A) or the initial formation of a large transient RNA-free complex (Pathway B). The existence of the pre-snoRNP 2, common to the two scenarios, is based on the detection of the various assembly factors on the same U3 snoRNA precursors (Figure 4). The Snu13p-binding site (PEP), the Hit1p-binding site (HBS) and the nuclear localization signal (NLS) are displayed on the functional C-terminal part of Rsa1p. Bcd1p specifically controls stability of the C/D snoRNAs and is essential for cell viability. It was not identified in our proteomic analysis but should play an important role in C/D snoRNP biogenesis.
Given that Snu13p does not possess a nuclear localization signal (NLS), one possibility would be that Rsa1p or Hit1p serves as protein cargo for the import of Snu13p into the nucleus. Interestingly, an NLS signal is present in the functional C-terminal region of Rsa1p. Hence, Snu13p–Rsa1p–Hit1p may represent a pre-formed module for nuclear import and RNA binding (Figure 9, Step 1).
Transient functional association between Rsa1p–Hit1p and R2TP
Given that Snu13p–Rsa1p–Hit1p can be associated with Nop58p (Figure 1H), it is also possible that a larger multiprotein complex containing the full set of proteins, but that was not detected by our proteomic analysis, can be recruited to the snoRNA (Figure 9, pathway B, Steps 1′ and 2′). GST pull down and Y2H experiments previously revealed a possible interaction between Rsa1p and Pih1p (21), and accordingly, we found that Rsa1p is required to tether Pih1p−Tah1p to a [C/D RNA−Snu13p] complex (29). Nop58p was initially shown to interact with Pih1p (31), and previous Y2H assays (21) and the present in vitro binding assays (Figure 1H) have revealed that Nop58p also associates with the purified Rsa1p230−375–Hit1p and Snu13p–Rsa1p230−375–Hit1p complexes. Thus, at some point during C/D snoRNP biogenesis, Nop58p could associate with or be exchanged between Pih1p and the Snu13p–Rsa1p–Hit1p complex (Figure 9, Step 2). The intricate network of interactions between the [RNA−Snu13p−Rsa1p−Hit1p] complex (pre-snoRNP 1 in Figure 9), R2TP components and Nop58p can explain the fact that we detected the association of pre-U3 snoRNA with proteins of these complexes using the sensitive RT-PCR approach, and interestingly, that Rsa1p and Hit1p influence U3 3′-terminal maturation. However, with the exception of Rsa1p−Hit1p, such a complete protein complex was not detected by proteomic analysis of yeast cellular extracts. This suggests that (i) in cellulo interactions may be transient and rapidly disrupted by ATPase activity, so that free Rsa1p−Hit1p and free R2TP are predominant in the extract; (ii) the TAP sequence may decrease complex stability; (iii) accessibility of the TAP sequence may be low within large complexes and/or (iv) complexes might be tightly associated with chromatin or components of nucleolar bodies.
If a transient cooperation between the two sub-complexes exists, Rsa1p could facilitate the recruitment of the Nop proteins to the [RNA–Snu13p] pre-RNP (21,22,28), while R2TP could participate in a conformational remodeling required for the progression of the RNP assembly by the helicase activity of the Rvb1p–Rvb2p proteins (21,28,58) (Figure 9, Step 3).
Finally, many lines of evidence suggest that the assembly of Nop56p on the box C′/D′ couple is a secondary event. Indeed, the C′/D′ motif is a sub-optimal K-turn, exhibiting a lower affinity for Snu13p, and the nucleation of a particle [C′/D′ motif-Snu13p−Nop56p−Nop1p] seems highly dependent on the establishment of a coiled-coil dimer between Nop56p and Nop58 pre-positioned on the box C/D particle (59,60). In contrast to Nop58p which is mainly associated with the R2TP complex components, we found Nop56p associated with Rsa1p in our TAP-tag assay (Table 1). This observation raises the question of the role played by Rsa1p−Hit1p in the assembly of the C′/D′ particle (Figure 9, Step 3). A future issue to address will be to determine the stoichiometry of the assembly process and if a second Snu13p−Rsa1p−Hit1p module is involved in this final assembly step.
Possible additional functions of the Rsa1p−Hit1p complex
ZNHIT3 and Hit1p both carry a Zn finger domain. However, this domain might have another functional role, as the Hit1p Zn finger domain is not required to maintain cellular concentrations of Rsa1p. In this context, it is worth noting that Rsa1p was originally identified as being required for production of ribosomal 60S subunit (52). Interestingly, deletion of RSA1, HIT1 or PIH1 induces marked defects in the pre-rRNA maturation steps occurring after cleavages at sites A1 and A2 (Figure 3). Indeed, the released 20S and 27S intermediates accumulate (Figure 3). One attractive possibility is that some 2′-O modifications catalyzed by C/D RNPs increase the efficiency of 20S maturation into 18S and processing of 27S into 5.8S and 25S precursors, which may contribute to the effect of RSA1 deletion on 60S production (52). A direct role for Rsa1p−Hit1p in these steps is also possible.
ZNHIT3, also known as TRIP3 (Thyroid Hormone Receptor Interacting protein 3), is a co-activator of several transcription factors (thyroid hormone receptor RXR (61), nuclear hepatocyte factor 4α (62), peroxisome proliferator activated receptor γ (63)). Furthermore, the cellular level of ZNHIT3 influences proliferation of cancer cells (64). It remains to be defined whether this property relies on its capacity to control cell growth through its involvement in nuclear-receptor dependant transcription, or by promoting C/D snoRNP biogenesis and ribosome synthesis.
ACCESSION NUMBERS
Chemical shifts data referenced to DSS were deposited in the Biological Magnetic Resonance Data Bank under entry 19626. The corresponding coordinates and restraints for structure calculations have been deposited in the Protein Data Bank with accession code 2MJF.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We are grateful to E. Bertrand (IGM Montpellier) who provided the GFP-Rsa1p expression vector and shared unpublished data. We thank the CNRS TGIR-RMN-THC Fr3050 facilities, the Proteomic French Infrastructure (ANR-10-INSB-08–03), the SCBIM platform of UL and FR BMCT CNRS 3209 for access to the 600 MHz NMR spectrometer.
Footnotes
Present addresses:
Benjamin Rothé, Ecole polytechnique fédérale de Lausanne (EPFL) SV ISREC, Station 19, CH-1015 Lausanne, Switzerland.
Jean-Michel Saliou, Plateforme de Protéomique et Peptides Modifiés (P3M), Institut Pasteur de Lille, CNRS, Université Lille Nord, 1 rue du Professeur Calmette, BP 245, 59019 Lille Cedex, France.
Régis Back, Laboratoire de Biochimie, CNRS UMR7654, Ecole Polytechnique, Route de Saclay, 91128 Palaiseau, France.
FUNDING
The Centre National de la Recherche Scientifique, University of Lorraine, University of Strasbourg; the Agence Nationale de la Recherche [ANR-06-BLAN-0208; ANR-11-BSV8–01503]; the Pôle de Recherche Scientifique et Technologique IMTS of Région Lorraine; the European Associated Laboratory on pre-mRNA splicing (CNRS, Lorraine University, Montpellier Universities and the Max Planck Institut); the Fondation pour la Recherche Médicale, the Région Alsace. B.R. and R.B. were fellows of the French Ministère de l'Enseignement Supérieur et de la Recherche.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 2.Watkins N.J., Bohnsack M.T. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA. 2012;3:397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 3.Matera A.G., Terns R.M., Terns M.P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 4.Kass S., Tyc K., Steitz J.A., Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 5.Peculis B.A. The sequence of the 5’ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol. Cell. Biol. 1997;17:3702–3713. doi: 10.1128/mcb.17.7.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atzorn V., Fragapane P., Kiss T. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol. Cell. Biol. 2004;24:1769–1778. doi: 10.1128/MCB.24.4.1769-1778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrissey J.P., Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol. Cell. Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lafontaine D.L., Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA. 1999;5:455–467. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lafontaine D.L., Tollervey D. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol. 2000;20:2650–2659. doi: 10.1128/mcb.20.8.2650-2659.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watkins N.J., Segault V., Charpentier B., Nottrott S., Fabrizio P., Bachi A., Wilm M., Rosbash M., Branlant C., Luhrmann R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 11.Watkins N.J., Newman D.R., Kuhn J.F., Maxwell E.S. In vitro assembly of the mouse U14 snoRNP core complex and identification of a 65-kDa box C/D-binding protein. RNA. 1998;4:582–593. doi: 10.1017/s1355838298980128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koonin E.V., Bork P., Sander C. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme. Nucleic Acids Res. 1994;22:2166–2167. doi: 10.1093/nar/22.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nottrott S., Hartmuth K., Fabrizio P., Urlaub H., Vidovic I., Ficner R., Luhrmann R. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5’ stem-loop of U4 snRNA. EMBO J. 1999;18:6119–6133. doi: 10.1093/emboj/18.21.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henras A., Henry Y., Bousquet-Antonelli C., Noaillac-Depeyre J., Gelugne J.P., Caizergues-Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998;17:7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell J.R., Wood E., Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 16.Allmang C., Carbon P., Krol A. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA. 2002;8:1308–1318. doi: 10.1017/s1355838202020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Copeland P.R., Fletcher J.E., Carlson B.A., Hatfield D.L., Driscoll D.M. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vidovic I., Nottrott S., Hartmuth K., Luhrmann R., Ficner R. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 19.Walbott H., Machado-Pinilla R., Liger D., Blaud M., Rety S., Grozdanov P.N., Godin K., van Tilbeurgh H., Varani G., Meier U.T., et al. The H/ACA RNP assembly factor SHQ1 functions as an RNA mimic. Genes Dev. 2011;25:2398–2408. doi: 10.1101/gad.176834.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiss T., Fayet-Lebaron E., Jady B.E. Box H/ACA small ribonucleoproteins. Mol. Cell. 2010;37:597–606. doi: 10.1016/j.molcel.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 21.Boulon S., Marmier-Gourrier N., Pradet-Balade B., Wurth L., Verheggen C., Jady B.E., Rothe B., Pescia C., Robert M.C., Kiss T., et al. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 2008;180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKeegan K.S., Debieux C.M., Boulon S., Bertrand E., Watkins N.J. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol. Cell. Biol. 2007;27:6782–6793. doi: 10.1128/MCB.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao R., Houry W.A. Hsp90: a chaperone for protein folding and gene regulation. Biochem. Cell Biol. 2005;83:703–710. doi: 10.1139/o05-158. [DOI] [PubMed] [Google Scholar]
- 24.Machado-Pinilla R., Liger D., Leulliot N., Meier U.T. Mechanism of the AAA+ ATPases pontin and reptin in the biogenesis of H/ACA RNPs. RNA. 2012;18:1833–S1845. doi: 10.1261/rna.034942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Back R., Dominguez C., Rothe B., Bobo C., Beaufils C., Morera S., Meyer P., Charpentier B., Branlant C., Allain F.H., et al. High-resolution structural analysis shows how Tah1 tethers Hsp90 to the R2TP complex. Structure. 2013;21:1834–1847. doi: 10.1016/j.str.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 26.Zhao R., Kakihara Y., Gribun A., Huen J., Yang G., Khanna M., Costanzo M., Brost R.L., Boone C., Hughes T.R., et al. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J. Cell Biol. 2008;180:563–578. doi: 10.1083/jcb.200709061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King T.H., Decatur W.A., Bertrand E., Maxwell E.S., Fournier M.J. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol. Cell. Biol. 2001;21:7731–7746. doi: 10.1128/MCB.21.22.7731-7746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKeegan K.S., Debieux C.M., Watkins N.J. Evidence that the AAA+ proteins TIP48 and TIP49 bridge interactions between 15.5K and the related NOP56 and NOP58 proteins during box C/D snoRNP biogenesis. Mol. Cell. Biol. 2009;29:4971–4981. doi: 10.1128/MCB.00752-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothe B., Back R., Quinternet M., Bizarro J., Robert M.C., Blaud M., Romier C., Manival X., Charpentier B., Bertrand E., et al. Characterization of the interaction between protein Snu13p/15.5K and the Rsa1p/NUFIP factor and demonstration of its functional importance for snoRNP assembly. Nucleic Acids Res. 2014;42:2015–2036. doi: 10.1093/nar/gkt1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckert K., Saliou J.M., Monlezun L., Vigouroux A., Atmane N., Caillat C., Quevillon-Cheruel S., Madiona K., Nicaise M., Lazereg S., et al. The Pih1-Tah1 cochaperone complex inhibits Hsp90 molecular chaperone ATPase activity. J. Biol. Chem. 2010;285:31304–31312. doi: 10.1074/jbc.M110.138263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzales F.A., Zanchin N.I., Luz J.S., Oliveira C.C. Characterization of Saccharomyces cerevisiae Nop17p, a novel Nop58p-interacting protein that is involved in Pre-rRNA processing. J. Mol. Biol. 2005;346:437–455. doi: 10.1016/j.jmb.2004.11.071. [DOI] [PubMed] [Google Scholar]
- 32.Manival X., Jacquemin C., Charpentier B., Quinternet M. 1H, 15N and 13C resonance assignments of the yeast Pih1 and Tah1 C-terminal domains complex. Biomol. NMR Assignments. 2014. [Epub ahead of print] [DOI] [PubMed]
- 33.Pal M., Morgan M., Phelps S.E., Roe S.M., Parry-Morris S., Downs J.A., Polier S., Pearl L.H., Prodromou C. Structural basis for phosphorylation-dependent recruitment of tel2 to hsp90 by pih1. Structure. 2014;22:805–818. doi: 10.1016/j.str.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawakami K., Shafer B.K., Garfinkel D.J., Strathern J.N., Nakamura Y. Ty element-induced temperature-sensitive mutations of Saccharomyces cerevisiae. Genetics. 1992;131:821–832. doi: 10.1093/genetics/131.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marmier-Gourrier N., Clery A., Senty-Segault V., Charpentier B., Schlotter F., Leclerc F., Fournier R., Branlant C. A structural, phylogenetic, and functional study of 15.5-kD/Snu13 protein binding on U3 small nucleolar RNA. RNA. 2003;9:821–838. doi: 10.1261/rna.2130503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 37.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 38.Miguet L., Bechade G., Fornecker L., Zink E., Felden C., Gervais C., Herbrecht R., Van Dorsselaer A., Mauvieux L., Sanglier-Cianferani S. Proteomic analysis of malignant B-cell derived microparticles reveals CD148 as a potentially useful antigenic biomarker for mantle cell lymphoma diagnosis. J. Proteome Res. 2009;8:3346–3354. doi: 10.1021/pr801102c. [DOI] [PubMed] [Google Scholar]
- 39.Piazzon N., Rage F., Schlotter F., Moine H., Branlant C., Massenet S. In vitro and in cellulo evidences for association of the survival of motor neuron complex with the fragile X mental retardation protein. J. Biol. Chem. 2008;283:5598–5610. doi: 10.1074/jbc.M707304200. [DOI] [PubMed] [Google Scholar]
- 40.Romier C., Ben Jelloul M., Albeck S., Buchwald G., Busso D., Celie P.H., Christodoulou E., De Marco V., van Gerwen S., Knipscheer P., et al. Co-expression of protein complexes in prokaryotic and eukaryotic hosts: experimental procedures, database tracking and case studies. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1232–1242. doi: 10.1107/S0907444906031003. [DOI] [PubMed] [Google Scholar]
- 41.Sanglier S., Atmanene C., Chevreux G., Dorsselaer A.V. Nondenaturing mass spectrometry to study noncovalent protein/protein and protein/ligand complexes: technical aspects and application to the determination of binding stoichiometries. Methods Mol. Biol. 2008;484:217–243. doi: 10.1007/978-1-59745-398-1_15. [DOI] [PubMed] [Google Scholar]
- 42.Schanda P., Forge V., Brutscher B. HET-SOFAST NMR for fast detection of structural compactness and heterogeneity along polypeptide chains. Magn. Reson. Chem. 2006. pp. S177–S184. 44 Spec No. [DOI] [PubMed]
- 43.Shen Y., Delaglio F., Cornilescu G., Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez-Mendez B., Guntert P. Automated protein structure determination from NMR spectra. JACS. 2006;128:13112–13122. doi: 10.1021/ja061136l. [DOI] [PubMed] [Google Scholar]
- 45.Nederveen A.J., Doreleijers J.F., Vranken W., Miller Z., Spronk C.A., Nabuurs S.B., Guntert P., Livny M., Markley J.L., Nilges M., et al. RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins. 2005;59:662–672. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- 46.Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. U.S.A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marmier-Gourrier N., Clery A., Schlotter F., Senty-Segault V., Branlant C. A second base pair interaction between U3 small nucleolar RNA and the 5’-ETS region is required for early cleavage of the yeast pre-ribosomal RNA. Nucleic Acids Res. 2011;39:9731–9745. doi: 10.1093/nar/gkr675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myslinski E., Segault V., Branlant C. An intron in the genes for U3 small nucleolar RNAs of the yeast Saccharomyces cerevisiae. Science. 1990;247:1213–1216. doi: 10.1126/science.1690452. [DOI] [PubMed] [Google Scholar]
- 49.Kufel J., Allmang C., Chanfreau G., Petfalski E., Lafontaine D.L., Tollervey D. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell. Biol. 2000;20:5415–5424. doi: 10.1128/mcb.20.15.5415-5424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holm L., Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gibrat J.F., Madej T., Bryant S.H. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 52.Kressler D., Doere M., Rojo M., Linder P. Synthetic lethality with conditional dbp6 alleles identifies rsa1p, a nucleoplasmic protein involved in the assembly of 60S ribosomal subunits. Mol. Cell. Biol. 1999;19:8633–8645. doi: 10.1128/mcb.19.12.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cahill N.M., Friend K., Speckmann W., Li Z.H., Terns R.M., Terns M.P., Steitz J.A. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 2002;21:3816–3828. doi: 10.1093/emboj/cdf376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Omer A.D., Ziesche S., Ebhardt H., Dennis P.P. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl Acad. Sci. U.S.A. 2002;99:5289–5294. doi: 10.1073/pnas.082101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rashid R., Aittaleb M., Chen Q., Spiegel K., Demeler B., Li H. Functional requirement for symmetric assembly of archaeal box C/D small ribonucleoprotein particles. J. Mol. Biol. 2003;333:295–306. doi: 10.1016/j.jmb.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 56.Tran E.J., Zhang X., Maxwell E.S. Efficient RNA 2’-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C’/D’ RNPs. EMBO J. 2003;22:3930–3940. doi: 10.1093/emboj/cdg368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watkins N.J., Dickmanns A., Luhrmann R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 2002;22:8342–8352. doi: 10.1128/MCB.22.23.8342-8352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watkins N.J., Lemm I., Ingelfinger D., Schneider C., Hossbach M., Urlaub H., Luhrmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 59.Lin J., Lai S., Jia R., Xu A., Zhang L., Lu J., Ye K. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature. 2011;469:559–563. doi: 10.1038/nature09688. [DOI] [PubMed] [Google Scholar]
- 60.Qu G., van Nues R.W., Watkins N.J., Maxwell E.S. The spatial-functional coupling of box C/D and C′/D′ RNPs is an evolutionarily conserved feature of the eukaryotic box C/D snoRNP nucleotide modification complex. Mol. Cell. Biol. 2011;31:365–374. doi: 10.1128/MCB.00918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee J.W., Choi H.S., Gyuris J., Brent R., Moore D.D. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol. Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 62.Iwahashi H., Yamagata K., Yoshiuchi I., Terasaki J., Yang Q., Fukui K., Ihara A., Zhu Q., Asakura T., Cao Y., et al. Thyroid hormone receptor interacting protein 3 (trip3) is a novel coactivator of hepatocyte nuclear factor-4alpha. Diabetes. 2002;51:910–914. doi: 10.2337/diabetes.51.4.910. [DOI] [PubMed] [Google Scholar]
- 63.Koppen A., Houtman R., Pijnenburg D., Jeninga E.H., Ruijtenbeek R., Kalkhoven E. Nuclear receptor-coregulator interaction profiling identifies TRIP3 as a novel peroxisome proliferator-activated receptor gamma cofactor. Mol. Cell. Proteomics. 2009;8:2212–2226. doi: 10.1074/mcp.M900209-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brito G.C., Fachel A.A., Vettore A.L., Vignal G.M., Gimba E.R., Campos F.S., Barcinski M.A., Verjovski-Almeida S., Reis E.M. Identification of protein-coding and intronic noncoding RNAs down-regulated in clear cell renal carcinoma. Mol. Carcinog. 2008;47:757–767. doi: 10.1002/mc.20433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


