Abstract
Initiation of translation on Type II IRESs, such as those of EMCV and FMDV viruses, has been well documented in the recent years. For EMCV, the current model argues for a mechanism in which the key interaction necessary for the pre-initiation complex recruitment is eIF4G binding to the central J-K domains of EMCV-IRES. Here we demonstrate that, in contrast with the current model, the molecular mechanism of EMCV-IRES involves direct recruitment of the 40S subunit. Importantly, we identified a specific structural element that prevents the correct positioning of the initiation codon in the close vicinity of the ribosomal P site. This work clarifies how this interaction could not be anticipated by earlier studies and allows us to propose a new model for initiation complex assembly on EMCV-IRES. The role attributed to eIF4G/4A can thus be refined as stabilizing/promoting the conformational changes that are necessary for IRES function, thus resembling the role conventionally assigned to ITAFs. This raises the interesting possibility that IRESs are primarily ribosome binders, some of which having partly lost the ability to fold into the active structure without the help of proteins.
INTRODUCTION
Most eukaryotic messenger ribonucleic acids (mRNAs) are translated through the classical cap-dependent pathway that relies on the recognition of the 5′-cap structure to recruit the 43S complex (composed of the 40S subunit, tRNA, eIF2 and eIF3). This is mediated by the eIF4F complex that allows cap recognition through eIF4E and unwinding of mRNA secondary structures by the eIF4A helicase. The eIF4G platform coordinates 40S and mRNA association through its connection with eIF4E, eIF3 and Poly(A) Binding Protein (PABP) (1,2). Viruses have developed many ways to manipulate their hosts’ translation machinery, including internal ribosome entry, which allows direct recruitment of the initiation complex without requiring a 5′-cap (3,4). This mechanism relies on RNA sequences and structures called IRESs (Internal Ribosome Entry Sites), generally present in the 5′-UTR (5' Untranslated Region). Numerous IRESs have been identified in both viral and cellular mRNAs. The differences between them lead to the conception that several mechanisms of ribosome internal entry exist (5). For example, IRES-dependent initiation may occur by direct binding of the 40S subunit to the mRNA, or may be mediated by a subset of the canonical initiation factors, and in some cases necessitates specific cellular proteins called IRES trans-acting factors (ITAFs) (6). Initiation of translation on Type II IRESs, such as those present in the encephalomyocarditis virus (EMCV) and in the foot and mouth disease virus (FMDV), has been well documented in the recent years (see (5) for a review). More precisely, translation initiation of Type II IRESs requires the core of the 43S complex, eIF4A and the central domain of eIF4G. Initiation complex formation is only modestly influenced by the presence of eIF4B, eIF1 and eIF1A and does not require the cap-binding protein eIF4E. Moreover, it is enhanced by the cellular proteins ITAF45 or the pyrimidine tract binding protein (PTB) depending on the IRES considered. It is noteworthy that EMCV-IRES dependence toward PTB is only moderate in the wild-type strain (A-loop composed of 6A; see Figure 2A) (7) as compared to variants presenting a different A-loop size (5A or 7A) or a different downstream reporter (8). For EMCV, the current model argues for a mechanism in which the key interaction necessary for the recruitment of the 43S pre-initiation complex (PIC) is eIF4G binding to the central J–K domains of EMCV-IRES (9). Nonetheless, these domains are not sufficient to sustain IRES activity (10). EIF4G binding is strengthened by eIF4A, which in the presence of adenosine triphosphate (ATP) restructures the 3′ border of the IRES (11). These conformational changes are thought to be necessary for the recruitment of 43S PICs via the eIF4G/eIF3 interaction. However, eIF4G mutants lacking the ability to contact eIF3 still promote initiation on EMCV-IRES (12). This prompted us to evaluate the possibility of a direct binding of the 40S subunit to EMCV-IRES.
Figure 2.

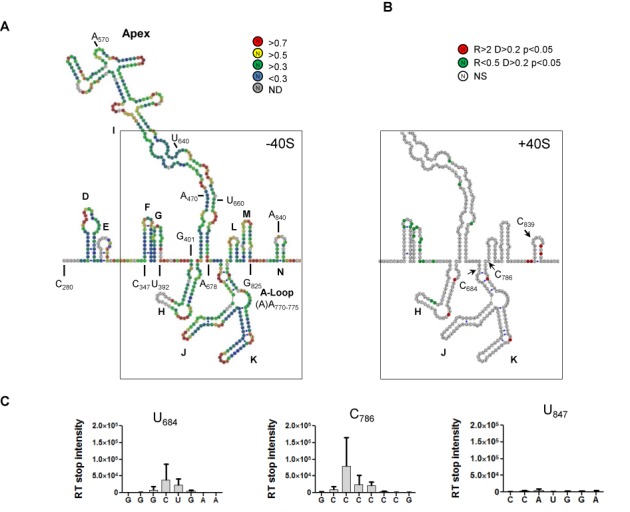
(A) SHAPE probing of EMCV-RLuc in the presence of MgCl2. The secondary structure model of EMCV IRES is a composite of those published by (22,26,27) that is modified according to the data presented here. The color of the circle surrounding each nucleotide corresponds to relative reactivity to 1M7 (see the scale shown on the right). Each sphere corresponds to a nucleotide and side by side spheres indicate a base pair. Domains are indicated in the figure as well as relevant nucleotide positions. SHAPE reactivity corresponds to the mean of at least four independent experiments. The scheme was designed using VaRNA (21). (B) Schematic representation of the differential SHAPE reactivities obtained when probing EMCV-RLuc in the absence or presence of ribosomal 40S. Nucleotides presenting a higher reactivity in the presence of ribosomal 40S are indicated in red while a lower reactivity is indicated in green. Arrows indicate toe-print positions U684, C786 and U847. R: reactivity ratio; D: reactivity difference; P: probability of the Student Test; NS: non-significant. (C) Toe-print analysis of the 40S-EMCV-RLuc binary complexes by measuring the differential intensity of reverse transcriptase stops in the absence or presence of ribosomal 40S. The positions of toe-prints that correspond to 40S-EMCV-Rluc complexes are indicated. Numbering is from the 5′UTR, U847 corresponding to position +14 from the AUG codon. Error bars are the SEM of at least three independent experiments.
In the present study, we focused on 40S ribosomal subunit interaction with the viral RNA and determined that the 40S subunit interacts directly with EMCV-IRES leading to the formation of a stable binary complex. This interaction involves the H and I domains of the IRES that are necessary for 40S subunit binding. A novel role can now be assigned to these domains and elucidate their requirement for EMCV-IRES activity (9,13). Furthermore, we were able to identify a specific structure at the 3′ border of the IRES that prevents the correct positioning of the initiation codon in the close vicinity of the ribosomal P site. Taken together, these results demonstrate the presence of an RNA determinant in EMCV-IRES that allows ribosomal 40S subunit binding, although it is not sufficient to correctly position the AUG in the ribosomal P site. This allows us to propose a new model for initiation complex assembly on EMCV-IRES and to refine the role attributed to eIF4G/4A as stabilizing/promoting the conformational changes that are necessary for IRES function, thus resembling the role typically assigned to ITAFs. Far from contradicting previous studies, our work further elucidates the anticipated ‘lacking determinant’ of 43S complex recruitment by EMCV-IRES (14).
MATERIALS AND METHODS
Plasmids and RNA
EMCV IRES (nt 280–839) was fused with the luciferase open reading frame (ORF) (nt 1–102) in pGEM vector. EMCV-L plasmid was obtained by cloning EMCV IRES (nt 280–905) into pCR2.1 (Invitrogen). Functional variant of the A bulge and EMCV-L Mut2nt were obtained by site-directed mutagenesis (Stratagene) following the manufacturer's instructions. All RNAs were directly transcribed using the T7 RNA polymerase from polymerase chain reaction (PCR) products containing the T7 RNA polymerase promoter sequence (5′-TAATACGACTCACTATAGG-3′) and purified as previously described (15). Radiolabeled RNAs were transcribed as above in the presence of α32-UTP (3000 mCi/mmol) and purified by size exclusion chromatography. Primers for site-directed mutagenesis were: EMCV5A F/R (5′-ATGTGTTTAGTCGAGGTTAAAAAGCGTCTAGGCCCC-3′), EMCV6A F/R (5′-TGGTTTAGTCGAGGTTAAAAAACGTCTAGGCCCC-3′) and Mut2nt F/R (5′-GATAATATGGCCACAACCAACGAACAAGAGACTTGCGCGC-3′). Primers for PCR were as follows: T7EMCVFwd (5′-CCCCTCTCCCTCCCCCTAACGT-3′), T75′D (5′-GTACTGGCCGAAGCCG-3′), T75′E (5′-GCGTTTGTCTATATGTTATTTTCCACC-3′), T75′F (5′-GTTTCCACCATATTGCCGTC-3′), T75′H (5′-GACGAGCATTCCTAGGGGTCTTTCC-3′), T75′I (5′-GTGTTGAATGTCGTGAAGGAAGCAG-3′), T75′J (5′-AAGGGGCTGAAGGATGCCCAGAAGG-3′), M3′ (5′-CCTTTGAAAAACACGATGATAATATGGCCA-3′), K3′(5′-GGCCTAGACGTTTTTTAACCTCGAC-3′), I3′ (5′-TTGTTGAATACGCTTGAGGAGAGCC-3′), EMCV-L DELTA O (5′-CGATGATAATATGGCCACAACCATGGAACAAGAGACTAATGCTCTGCTCTACAATACCGTAATGGATTTTACCTGCTAAAGTATGATGAAGAATGGT-3) and EMCV-RLuc DELTA O (5′-CGAAAGTTTATGATCCAGAACCAGATGTAAACAAATGAATGTT-3′).
Preparation of ribosomal subunits
Ribosomal subunits were prepared following previously established procedures (16). Cytoplasmic extracts from HeLa cells (Ipracell) or rabbit reticulocyte lysate (Green Hectares) was centrifuged in Buffer A (20-mM HEPES-KOH pH 7.5, 50-mM KCl 4-mM MgCl2, 2-mM Dithiothreitol (DTT) and 0.25-M sucrose) at 14000 g for 15 min to remove mitochondria. The supernatant was layered onto a 50% sucrose cushion and centrifuged for 5 h at 44000 rpm in a 45 Ti rotor. The pellet (P1) was further resuspended in Buffer A (OD260 ± 100) and treated with 1-mM puromycin (Sigma) for 10 min on ice followed by 10 min at 37°C. KCl was then added to a final concentration of 500 mM and the solution was incubated on ice for 30 min. The suspension was centrifuged for 1 h 40 at 70000 rpm in a 70 Ti rotor. The obtained pellet (P2) was resuspended in Buffer B (20-mM HEPES-KOH pH 7.5, 0.5-M KCL, 4-mM MgCl2 and 2-mM DTT) and layered onto a 10–30% sucrose gradient prepared in Buffer B. Gradients were run 17 h at 22000 rpm in an SW32 Ti rotor. Fractions were collected, 10 μl of each was loaded onto an agarose gel and the fractions corresponding to 40S ribosomal subunits were pooled, dialyzed and concentrated in Buffer C (20-mM HEPES KOH pH 7.5, 100-mM KCl, 2-mM MgCl2, 2-mM DTT and 0.25-M sucrose). The S1 supernatant (corresponding to P1) was also treated with KCl (0.5-M final concentration) to obtain S1S2 and S1P2 after 5 h at 44000 rpm in a 45 Ti rotor. S1P2 was resuspended in Buffer B and treated with the same procedures as described above for P2. Absence of eIF4G contamination (less than 5%) was ascertained by semi-quantitative western blot (Supplementary Figure S1) using 0.75 to 6 pmol of purified recombinant eIF4G (p100, as described in (17)) run in parallel to 20 pmol of purified 40S subunits. The membrane was probed with anti-eIF4G (Santa Cruz, sc-100730) and anti-S14 (Santa Cruz, sc-68873) antibodies and the appropriate HRP-secondary antibodies (Abcam), revealed with the supersignal West Pico Kit (Pierce), and visualized with a LAS 4000 (Fuji).
Filter-binding assays
Radio-labeled RNA (10 nM) was denatured by heating to 80°C for 2 min and then cooled to room temperature in FB buffer (20-mM Tris-Cl pH 7.5, 100-mM KOAc, 200-mM KCl, 2.5-mM MgCl2, 2-mM DTT). Serial dilutions from a 2-μM solution of 40S were prepared extemporaneously, added to a 10-μl reaction and incubated at 37°C for 10 min. The reactions were then used for filter binding assays. Filter binding was accomplished essentially as previously described using two filters (18): from top to bottom, a nitrocellulose filter and a charged nylon filter. The filters were presoaked in FB buffer, assembled in the dot blot apparatus and the reactions were applied and directly vacuum filtered. The filters were then rinsed with FB buffer, removed and radioactivity was quantified using a storm phosphorImager (GE Healthcare). To determine the apparent Kd, the data were fit to the Langmuir isotherm described by the equation θ = P/[P + Kd], where θ is the fraction of RNA bound and P is 40S subunit concentration.
Mobility of complexes during sucrose density gradient centrifugation
Ribonucleoprotein complexes were assembled as described for the filter binding assay experiment. A 200-μl mixture containing 25–50-nM 32P-labeled RNA and 200–400 nM of purified ribosomal subunits was resolved by centrifugation through 10 to 30% sucrose gradients (25-mM Tris-Cl pH 7.5, 6-mM MgCl2, 75-mM KCl) for 4 h at 39000 rpm at 4°C in a Beckman SW40 Ti rotor. Three-hundred-microliter fractions were collected, one-third of which were vacuum blotted onto Hybond N+ membrane (Amersham), exposed and scanned. The amount of RNA in each fraction was determined and expressed as the percentage of total counts.
RNA probing by ‘SHAPE’ and 40S foot printing experiments
The secondary structures of EMCV-Rluc and EMCV-L were probed using 1-methyl-7-nitroisatoic anhydride (1 M7) in the presence or absence of magnesium chloride (SHAPE experiments with and without MgCl2), and in the presence or absence of the purified 40S subunits (foot-print experiments). Modifications were revealed by selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) process (19,20). Briefly, 6 pmol of RNA were resuspended in 96 μl of water and denatured for 2 min at 80°C. Then, 12 μl of 10X FB buffer containing 25-mM or no MgCl2 were added and the mixture was slowly cooled to 37°C. After a 20 min of incubation at 37°C, a 4-fold excess of the purified 40S ribosomal subunit was added to the mixture, and incubated at 37°C for 20 min (for foot-print experiments). Then, 1M7 (for a final concentration of 2 mM) or the same volume of Dimethyl Sulfoxyde (DMSO) (negative control) was added and the mixture was incubated at 37°C for 3 min. The concentration of the modification agent was established to generate at the most one modification per molecule. The reaction was then immediately precipitated in dry ice with ethanol, 0.5-M ammonium acetate and 20 μg of glycogen. The pellet was washed with 70% ethanol and resuspended in 10 μl of nuclease-free water. Modifications were revealed by elongating fluorescently labeled primers using a reverse transcriptase (RT) as described below.
Primer extension analysis
Unlabeled EMCV RNA (6 pmol) was annealed to primer RLucToe (5′-aacattcatttgtttacatctg-3′, 6 pmol) and placed on ice. EMCV RNA was further diluted to 25–50 nM in RT (20-mM Tris pH 8.3, 75-mM KCl, 3-mM MgCl2 and 10-mM DTT) buffer and incubated in the absence or presence of 200–400 nM of purified ribosomal 40S subunits for 10 min at 37°C. The reaction mix was then placed on ice and incubated for 45 min at 37°C after addition of 500-μM deoxyribonucleoside triphosphate (dNTP[I]: dATP, dCTP, dTTP and dITP) each and 2 μl of RNAse H− M-MLV RT (Promega). Thirty microliters of stop mix (200-μg/ml glycogen, 2-mM ethylenediaminetetraacetic acid, 60-mM NaOAc) were then added and reactions were placed on ice. Sequencing reactions were performed in parallel with 6 pmol of RNA incubated in 10% DMSO for 3 min at 94°C followed by 3 min on ice. RNA was further diluted in 1X RT buffer containing 500-μM dNTP[I], 500-μM ddA or ddT and 1 μl of RNAse H− M-MLV RT (Promega). Elongation was performed for 2 min at 35°C, 30 min at 42°C followed by 5 min at 55°C, 5-μl stop mix was added and the reactions were placed on ice. The products of primer extension reactions were pooled, ethanol precipitated, vacuum-dried and resuspended in 40-μl formamide. Each final sample contained an elongation performed in the presence of ribosomal 40S subunits, an elongation performed in the absence of ribosomal 40S subunits and two sequencing reactions. The complementary deoxyribonucleic acids formed in the primer extension reactions were analyzed by capillary gel electrophoresis (Beckman Coulter CEQ8000). Data were then interpreted and analyzed using the software ‘Shapefinder’ (http:bioinfo.unc.edu). Two reactivity values were considered significantly different when their relative ratio was >2 or <0.5 and their difference was >0.2 and when the bilateral Student test was <0.05. Schemes were designed using VaRNA (21)
RESULTS
EMCV IRES directly recruits the 40S ribosomal subunit
The starting construct used in this work contains EMCV-IRES (C280–A839) followed by the 102 first nucleotides of the Renilla Luciferase (RLuc) ORF. In order to detect any potential direct binding of the 40S subunit to EMCV-IRES, we performed filter binding assays and determined that EMCV IRES binds the 40S subunit with an apparent equilibrium dissociation constant (Kd) of 55 ± 10 nM, in the same order of magnitude as that of HIV-1 gag IRES (15 nM, (15)) and HCV IRES (17 nM, not shown), but contrasts with FMDV-IRES and a capped globin control (Figure 1A). 40S binding holds true for the functional variants containing a 5A or 7A bulge (A)770AAAAAG/A775 (Figure 1B) independently of their different requirement for the PTB to mediate internal ribosome entry (8). Worth noting, 40S binding saturates at 80%, suggesting that not all the molecules are in the active conformation. EMCV-IRES activity relies on five structural domains named H to L (3,22), with the JKL domains constituting the eIF4G/4A binding site (9). To date, the only function attributed to the H and I domains is PTB binding (23,24), although these are essential to sustain IRES activity (9,13). In a preliminary attempt to determine which EMCV domains interact with the 40S subunit, we performed similar binding experiments with RNAs containing up to domain I (I3′, C280–A678) and RNAs starting at the J domain (5′J, starting at A678). In both cases, the truncated RNAs (Figure 1C) were unable to mimic full length IRES but displayed better binding properties than globin RNA (Figure 1A), indicating that EMCV-IRES/40S interaction necessitates both modules and providing the first evidence that the minimum domains required for IRES activity correspond to those required for 40S ribosomal subunit interaction.
Figure 1.

EMCV-RLuc interaction with the ribosomal 40S subunit. (A) Binding curves of 32P-labeled EMCV-RLuc (•), HIV-1 Gag (▪) and FMDV-IRES (▾) and globin control (▴) to purified 40S ribosomal subunit. (B) Binding curves of 32P-labeled EMCV-Rluc presenting a 5A- (•), 6A- (▪) or 7A-bulge (▴) to purified 40S ribosomal subunit. (C) Binding curves of 32P-labeled EMCV-RLuc (•), I3′ (C280–A678, ▪) and 5′J (starting at A678, ▴) truncated mutants to purified 40S subunit. (D) Fractionation by 10–30% sucrose density gradient of EMCV-RLuc in the absence (∘) or presence (•) of excess-purified 40S subunits. OD260 profile in the presence of purified 40S is indicated in gray. The results are the mean of at least three independent experiments ± standard deviation.
To further investigate the ability of 40S subunits to form stable complexes on EMCV-IRES, we analyzed the mobility of the complexes during sucrose density (SDG) gradient centrifugation. As observed in Figure 1D, the profile obtained with EMCV RNA in the presence of 40S subunits shows an additional peak, with a mobility similar to that of the 40S subunit (5 ml from the top of the gradient), when compared to the mobility of EMCV RNA alone (2 ml from the top of the gradient). These results are consistent with the formation of stable binary RNA/40S complexes and prompted us to investigate the accessibility of the viral RNA to chemical probes in the presence of ribosomal 40S.
The secondary structure was probed for the first time by RNA SHAPE analysis (see the Materials and Methods section), using 1-methyl-7-nitroisatoic anhydride (1M7) that reacts with flexible ribose groups of nucleotides, thus revealing that the cognate base is not involved in a stable interaction (25). The SHAPE reactivities obtained (Figure 2A) show high consistency and are essentially compatible with the existing structure models derived from enzymatic probing (22,26). Nonetheless, the high reactivities obtained for U392–U400 (between the G and H domains) are compatible with a single-stranded region and the low reactivity of G401–C403 would be in favor of these nucleotides being part of the H domain as proposed by (8). Considering the 3′ border of the IRES, the reactivities obtained for C786 to A832 are mainly in the low to medium range that does not favor a single-stranded region as proposed by Hoffman et al. (27). Our probing results are thus more compatible with the model proposed by Duke et al. (22) with the presence of the L (C786–G800), M (C802–G825) and N (U833–G848) domains. Consequently, our secondary structure model is a composite of the previously described models with nucleotides 401–450 defining the H domain while the L, M and N domains correspond to those proposed by (22).
We next probed the RNA using 1M7 in the presence of ribosomal 40S subunits and compared the data with those generated in the absence of 40S subunits (Supplementary Figure S2). A position was considered as protected (green) or exposed (red) when the reactivity value was at least 2-fold lower (or higher) than that without 40S and statistically different according to a bilateral Student test. As can be observed in Figure 2B, significant protection is seen upon addition of ribosomal 40S. Specifically, we observe a strong protection of the G stem loop (A372–G373, A381–C384, U392 and U396), of the H domain (A405, U409–A414, U422, U430–C431 and U442), as well as of the central part of domain I (G471, G480, A482–U484 and U488–U490 for the 5′ region and G641–A644, A651, U653 and U659 for the 3′ region). The absence of protection in the JK domains indicates that not only the 40S and the eIF4G binding sites are non-overlapping but also confirms the absence of eIF4G contamination in purified 40S. Although protections can result from direct footprint, from a structural rearrangement or from both, they unambiguously reflect the direct interaction between EMCV IRES and the 40S ribosomal subunit.
To further localized the ribosome, we use capillary toe-printing (28,29) with the reverse transcription primed on the luciferase coding sequence present downstream to the initiation codon 834AUG836. RT premature stops can reflect the position of the leading edge of the ribosome or a structural rearrangement of the IRES induced by the interaction. We observed three premature stops (Figure 2B and Supplementary Figures S3 and S4), two at the bottom of the JK domain (U684, C786) and one 14 nucleotides downstream 834AUG836 (U847), indicating that the initiation codon is positioned close to the P site (30,31). Premature stops at the base of JK are identical to the signals induced by eIF4G/4A (32) and could reflect a structural rearrangement inducing a sharp kink between the I and J domains (33), difficult for the RT to resolve. Similar results were obtained with 5A and 7A-bulge EMCV variants, confirming that the size of the A-bulge does not interfere with 40S binding and positioning (not shown). Taken together, these results clearly indicate that not only can the 40S subunit directly interact with EMCV IRES but also that this interaction positions the ribosomal subunit at the vicinity of AUG834 in this construct.
The nature of the ORF influences EMCV IRES structure and its interaction with the ribosomal 40S
Previous results on EMCV translation indicate that the nature of the downstream reporter gene influences translation efficiency (8). To evaluate whether EMCV IRES followed by its cognate ORF (EMCV-L) behaves like EMCV-RLuc, we performed the same series of experiments as described for EMCV-RLuc (Figure 3). The direct interaction with similar efficiency was confirmed by filter binding assay (Kd = 69.9 ± 9.3 nM; Figure 3A) and sucrose density gradient (Figure 3B). These results confirm that the nature of the ORF does not interfere with the binding properties of the ribosomal 40S subunit. To further define the minimal domains required for ribosomal 40S subunit interaction with EMCV-IRES, we generated RNA constructs truncated at their 5′ or 3′ end that were then tested for 40S ribosomal subunit binding (Figure 3C and Table 1). Deletion of the region upstream of the H domain does not influence 40S subunit binding affinities, as can be observed with the 5′H RNA. Significantly, removal of the H domain (5′I) induces a 2–3-fold increase in the apparent Kd, revealing that there is a binding determinant in this domain. Further truncation of the I domain also impacts on the apparent Kd, confirming its involvement in ribosomal 40S subunit binding. Regarding the 3′-end truncations, removal of the majority of the ORF portion of the RNA (M3′) drastically affects the maximum binding (2-fold reduction). Thus, the main contribution of the N domain and the ORF (71st nucleotides) is to favor effective conformation of the IRES, indicating that nucleotides A834–C905 (belonging to the ORF) are part of EMCV IRES. Finally, further truncation of the L and M domains (K3′) results in a roughly 2-fold increase in the apparent Kd, indicating the presence of a binding determinant in this domain. Taken together, our data are consistent with the requirement of multiple determinants (H, I and L domains) for optimal 40S ribosomal subunit binding, which are part of those previously identified for IRES activity, i.e. the H–L domains (10,13,22,34).
Figure 3.

EMCV-L interaction with ribosomal 40S. (A) Binding curves of 32P-labeled EMCV-RLuc (∘) or EMCV-L (•) to purified 40S ribosomal subunit. (B) Fractionation by 10–30% sucrose density gradient of EMCV-L in the absence (∘) or presence (•) of excess-purified 40S subunits. OD260 profile in the presence of purified 40S is indicated in gray. (C) Binding curves of 32P-labeled 5′- (left panel) or 3′-end (right panel) deletion mutants to purified 40S ribosomal subunit. 5′F: U340−A905; 5′H: G401−C905; 5′I: U451−C905; 5′J: A678−C905; K3′: C280−C785; M3′: C280−A840. The results are the mean of at least three independent experiments ± standard deviation.
Table 1.
40S binding affinities of EMCV IRES deletion mutants
| RNA | Nucleotides | Kd (nM) mean ± SEM | Binding max (%) mean ± SEM |
|---|---|---|---|
| 5′F | U340–C905 | 31.9 ± 5.7 | 81.8 ± 3.4 |
| 5′H | G401–C905 | 24.4 ± 4.8 | 79.6 ± 3.5 |
| 5′I | U451–C905 | 63.0 ± 17.6 | 65.6 ± 4.4 |
| 5′J | A678–C905 | 107.4 ± 29.6 | 60.0 ± 4.9 |
| M3′ | C280–A840 | 24.7 ± 12.0 | 47.8 ± 5.2 |
| K3′ | C280–C785 | 43.8 ± 15.8 | 42.3 ± 4.4 |
| I3′ | C280–A679 | 76.5 ± 21.7 | 58.5 ± 4.7 |
| EMCV-L | C280–C905 | 69.9 ± 9.3 | 87.9 ± 3.0 |
| EMCV-RLuc | C280–U941 | 54.5 ± 10.5 | 84.8 ± 3.8 |
The results are the mean and standard deviation of at least three independent experiments.
We then probed the secondary structure of the IRES in the presence or absence of Mg2+ ions, which are necessary for stabilizing tertiary interactions (Supplementary Figures S5 and S6). Interestingly, the profiles obtained with both EMCV-L and EMCV-RLuc are very similar. However they do exhibit some differences, not only within the coding region but also in the common IRES sequence (Supplementary Figure S7), suggesting structural interactions between these two elements. The differences mostly lie within internal bulges or terminal loops, some of which, like the apex of I, being protected in EMCV-L upon Mg2+ addition. This could be consistent with the presence of tertiary interactions in this construct. Regarding the coding region, important differences are observed in the N and O domains, which is expected considering the divergence of the primary sequences. Further ribosomal footprinting analysis indicates that although similar in both RNAs, the protected regions are not identical (Figure 4A and B and Supplementary Figure S8). More precisely, the strong protection of the F and G stem loops (C376–C389) is maintained but that of the H domain is less pronounced. This can partly be attributed to the poor reactivity of positions A408 to G416 in the context of the cognate ORF (EMCV-L), even in the absence of 40S subunit (Figure 4A). Taking into consideration the filter binding assay experiments, the strong protection observed in the F and G domains rather suggests that ribosomal 40S subunit binding to the IRES has structural/conformational implications outside the minimal binding site and that the H/I domains are involved in 40S recruitment. Importantly, when EMCV-L and EMCV-RLuc reactivity profiles obtained in the presence of ribosomal 40S subunit were compared, only sparse differences could be identified, mainly in the ORF (Supplementary Figure S9), suggesting that the binding of the 40S subunit is similar in both constructs but in the coding region vicinity. Accordingly, when performed on EMCV-L construct, toe-printing procedures detected RT stops at U684, C786 and AUG+6 (C839, Supplementary Figure S10) but none at AUG+14 (Figure 4C), although the ribosome binds with a similar efficiency. This result strongly suggests that the upstream position of the ribosome could be influenced by the nature of the ORF.
Figure 4.
(A) SHAPE probing of EMCV-L in the presence of MgCl2. The secondary structure model of EMCV IRES is derived from (22,26). The color of the circle surrounding each nucleotide corresponds to relative reactivity to 1M7 (see the scale shown on the right). Each sphere corresponds to a nucleotide and side by side spheres indicate a base pair. Domains are indicated in the figure as well as relevant nucleotide positions. SHAPE reactivity corresponds to the mean of at least four independent experiments. The scheme was designed using VaRNA (21). (B) Schematic representation of the differential SHAPE reactivities obtained when probing EMCV-L in the absence or presence of ribosomal 40S subunit. Nucleotides presenting a higher reactivity in the presence of ribosomal 40S subunit are indicated in red while a lower reactivity is indicated in green. Arrows indicate toe-print positions U684, C786 and C839. R: reactivity ratio; D: reactivity difference; P: probability of the Student Test; NS: non-significant. (C) Toe-print analysis of the 40S-EMCV-L binary complexes by measuring the differential intensity of reverse transcriptase stops in the absence or presence of ribosomal 40S subunit. The positions of toe-prints that correspond to 40S-EMCV-L complexes are indicated. Numbering is from the 5′UTR, U847 corresponding to position +14 from the AUG codon. Error bars are the SEM of at least three independent experiments.
Modeling the structure of the 3′ border with the SHAPE reactivities as constraints showed a potential role of the N and O domains (Figure 5A). First, the O domain of EMCV-RLuc is more reactive to 1M7 than that of EMCV-L indicating that it is probably less constrained. Second, in EMCV-L and in contrast to EMCV-Rluc, 834AUG836 is embedded within the N stem, thus possibly precluding the observation of the +14 toe-print. In an attempt to link the differential ribosomal positioning to a difference in the IRES structure, we performed several mutations and evaluated the consequences on the induced toe-prints. Deletion of the O domain in each construct did not result in modification of the +14 toe-print as compared to the reference construct (Figure 5B), indicating that the O domain has no or very little influence on the relative position of the AUG and the 40S ribosome. We next destabilized the N stem (Mut2nt, U847→A and G848→C) to render the initiation codon accessible and evaluated the consequence of these mutations on ribosomal 40S position. As can be observed in Figure 5C, Mut2nt/40S interaction preserves stable binary complex formation. More importantly, upon 40S binding on the mutant a toe-print can be observed at A846. We note that the observed toe-print is 13 nucleotides downstream the initiation codon which could indicate that the mutation is not sufficient to recover a fully correct positioning. However, these results clearly indicate that the IRES structure and the N domain influence 40S positioning at the vicinity of the initiation codon.
Figure 5.

Influence of the 3′ N and O domains on ribosomal 40S position. (A) Proposed model for EMCV-RLuc (left panel) or EMCV-L (right panel) O domain. The color of the circle surrounding each nucleotide reveals their relative reactivity to 1M7 and corresponds to the scale shown on the right. Each sphere corresponds to a nucleotide and adjacent spheres indicate a base pair. Domains are indicated in the figure as well as relevant nucleotide positions. SHAPE reactivity is the mean of at least four independent experiments. The scheme was designed using VaRNA (21). (B) Toe-print analysis of EMCV-Rluc DeltaO (left panel) and EMCV-L DeltaO (right panel) in the presence of purified 40S. Toe-print positions corresponding to positions 844–850 are indicated. Error bars are the SEM of at least three independent experiments. (C) Fractionation by 10–30% sucrose density gradient of EMCV-L Mut2nt in the absence (∘) or presence (•) of excess-purified 40S subunits (left panel) and toe-printing analysis of the 40S-EMCV-L Mut2nt binary complexes by measuring the differential intensity of reverse transcriptase stops in the absence or presence of 40S subunit (right panel). Error bars are the SEM of at least three independent experiments. Mutated residues are indicated by small letters.
DISCUSSION
The currently accepted model of EMCV molecular mechanism stipulates that the initial event is the recruitment of eIF4G/4A. Indeed, the helicase activity of the complex has been shown to restructure the vicinity of the initiation codon (11). More importantly, eIF4G was proposed as indirectly recruiting the 40S subunit of the ribosome through their respective interaction with eIF3. Here, we show through different approaches that the small ribosomal subunit can be directly recruited by EMCV IRES, with no need for initiation factors or for ITAF.
Regarding the ribosomal 40S/EMCV IRES interaction, our deletion and footprinting analyses reveal that important binding determinants for the 40S ribosome are located all along the IRES, more exactly, in the H, I and L domains. Such a large interacting domain is consistent with what is observed with Type III or IV IRESs, for which cryoEM studies clearly show that a large surface of the RNA is in contact with the ribosome (35,36). A role in 40S subunit recruitment can now be assigned to the H and I domains and their significant requirement for EMCV IRES activity is clarified. The revealed contribution of the H and I domains in ribosomal 40S subunit binding clearly illuminates the earlier, puzzling, observation that the J/K domains, which are responsible for eIF4G/4A binding and should have been able to recruit the 43S through eIF3/4G interaction, are not endowed with IRES activity. Taken together, our results favor a model in which the initial binding of EMCV-IRES to the ribosomal 40S subunit occurs through the H and I domains. Ribosomal interaction leads to a general rearrangement of the IRES structure as ascertained by the protection of the FG stem loops (which are not part of the minimal domain) as well as the presence of the U684 and C786 and stops at the bottom of the JK domains. It is important to note that these two premature RT stops correspond to those identified following eIF4G interaction and were suggested to be the result of the eIF4G-induced sharp kink between the J and I domains (33). As the presence of an eIF4G contamination of the ribosomal 40S preparation can be ruled out (see the Materials and Methods section and Supplementary Figure S1), they rather indicate that EMCV-IRES interaction with the 40S ribosomal subunit results in a similar structural rearrangement of the IRES structure.
In addition, when the IRES is followed by its cognate coding sequence (EMCV-L), we observed an anomalous toe-print (C839) that is 6 nucleotides downstream of the initiation codon. This position is also 14 nucleotides downstream of an out-of-frame 826AUG828 triplet. It could originate from a spurious recognition of 826AUG828; or otherwise, the relative position of this toe-print may be fortuitous since there is no reason to assume that the ribosome could recognize an AUG per se in the absence of a correct structural context and in the absence of initiator tRNA. It is of note that aberrant toe-prints have been reported in the context of the scanning mechanism when the initiation codon is in the close proximity of a stable stem loop that cannot be unwound by the 48S complex (37). In the case of EMCV-L, the AUG is concealed in a short stem loop (−2.7 kcal) and we assumed that it could be an obstacle to the correct positioning of the initiation codon in the naked ribosomal subunit when devoid of accessory helicase activity. This possibility is more likely as upon destabilization of this short stem loop, a bona fide toe-print testifies the correct positioning of the initiation codon in the vicinity of the ribosomal P site. We thus propose that the aberrant toe-print (C839) reflects an erroneous accommodation of the RNA in the decoding groove of the ribosome rather than an erroneous interaction of the ribosome with the IRES structure. In such a hypothesis and in agreement with a previous report (11), eIF4G/4A would mediate the conformational rearrangement that frees the initiation codon. How such a mechanism is selected in contrast to the direct positioning observed with the EMCV-RLuc RNA remains obscure. We can only speculate about it: it may ensure the selection of the accurate initiation codon in a sequence where many AUG triplets are in close vicinity to the bona fide translation start site or else set the basis for a physiological regulation yet to be uncovered.
Altogether our data lead us to propose a revised molecular model for the mechanism of internal initiation mediated by EMCV IRES (Figure 6). It is clear that the 40S subunit and eIF4G/4A can bind EMCV-IRES independently and appear to have different binding sites, which render the eIF3/eIF4G interaction clearly dispensable for the recruitment of the ribosomal subunit. This sheds light on a previously confusing result showing that a mutant version of eIF4G unable to interact with eIF3 still efficiently promotes internal initiation (12). Because eIF4G/eIF4A induces the JI kink and affects the accessibility to the initiation codon, we favor a mechanism whereby eIF4G/eIF4A remodels the IRES structure to promote EMCV-IRES/40S interaction and rearranges the coding region to correctly accommodate the AUG in the ribosome mRNA cleft. Then, the 43S complex can undergo a structural rearrangement due to both the AUG/tRNA recognition and close contact between the ribosome and the IRES. This would allow the 60S to join the complex and initiate translation.
Figure 6.
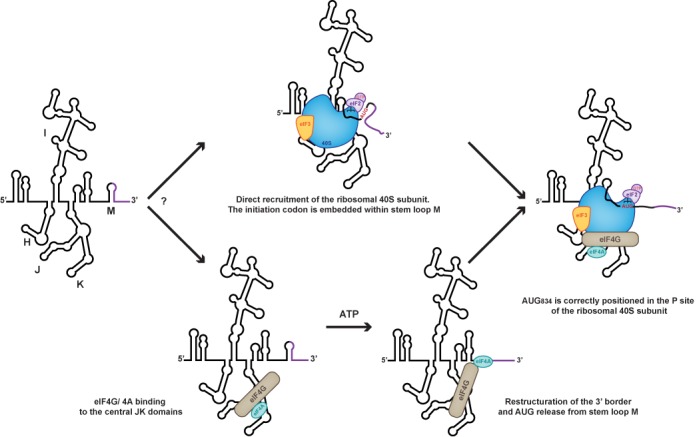
Our current working model hypothesizes that an early event of initiation complex formation onto EMCV IRES is the direct recruitment of the ribosomal 43S PIC. The recruitment of eIF4G/eIF4A, whether ordered or concomitant, remodels the 3′ border of the IRES to release the initiation codon thereby allowing functional 48S assembly.
One might wonder how general this molecular mechanism is, and even if it is restricted to EMCV IRES, or could be extended to all class II IRESs, since the second archetype of these sequences present in the FMDV does not appear to specifically bind the 40S subunit. FMDV and EMCV IRESs have been shown to adopt similar secondary structures and to promote initiation through a similar pathway. However, they do not require the same ITAF, and the dependency on these cellular proteins is more pronounced for FMDV than it is for EMCV (33,38). It is thus possible that the determinants for ribosomal binding also lie within the FMDV sequence and structure. However, to reach the correct conformation they would require the intervention of an ITAF, which could be the previously identified PTB (39) or ITAF45 (40). A parallel could be drawn with what has been observed for most RNA enzymes, from the ‘simple’ self-cleaving introns to the sophisticated ribosome, which require proteins to fold in a conformation where the RNA determinant, holding the catalytic activity, is in the active form. These enzymes are supposed to be ancient RNA ‘only’ enzymes that have evolved to today's situation where they require proteins (41,42). Similarly, we propose that all viral IRESs are primarily ribosome binders that have evolved to cooperate with cellular proteins, but the primordial role of which is to manipulate the ribosome to render it competent for translation. This raises the interesting possibility of a continuum between the different IRESs, whose common property is to directly bind the ribosome although through different RNA structures. Direct interaction already documented for Type III, IV and HIV Gag IRESs may not be observed in some other IRESs, not because the ribosome binding determinant is missing, but because they have lost the ability to structure into the active conformation without the help of cellular proteins. This provocative speculation is not only based on our results but also on the accumulating evidence that IRESs belonging to various types do not only bind to the 40S subunit, but intimately manipulate it to promote translation (36,43).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank T. Ohlmann, T. Poyry and R. Jackson for generous plasmid gift. The authors want to thank P. Romby, T. Ohlmann and S. De Breyne for critical reading of the manuscript.
FUNDING
Centre National pour la Recherche Scientifique (CNRS, Programme ATIP); Agence Nationale de la Recherche contre le Sida [AO12011-11188]; Université Paris-Descartes; Ministère de la Recherche et de l'Enseignement Supérieur [to J.D.]. Funding for open access charge: Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
REFERENCES
- 1.Aitken C.E., Lorsch J.R. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 2.Jackson R.J., Hellen C.U., Pestova T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jang S.K., Krausslich H.G., Nicklin M.J., Duke G.M., Palmenberg A.C., Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 5.Balvay L., Soto Rifo R., Ricci E.P., Decimo D., Ohlmann T. Structural and functional diversity of viral IRESes. Biochim. Biophys. Acta. 2009;1789:542–557. doi: 10.1016/j.bbagrm.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Semler B.L., Waterman M.L. IRES-mediated pathways to polysomes: nuclear versus cytoplasmic routes. Trends Microbiol. 2008;16:1–5. doi: 10.1016/j.tim.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Pestova T.V., Hellen C.U., Shatsky I.N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaminski A., Jackson R.J. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA. 1998;4:626–638. doi: 10.1017/s1355838298971898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolupaeva V.G., Pestova T.V., Hellen C.U., Shatsky I.N. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 1998;273:18599–18604. doi: 10.1074/jbc.273.29.18599. [DOI] [PubMed] [Google Scholar]
- 10.Evstafieva A.G., Ugarova T.Y., Chernov B.K., Shatsky I.N. A complex RNA sequence determines the internal initiation of encephalomyocarditis virus RNA translation. Nucleic Acids Res. 1991;19:665–671. doi: 10.1093/nar/19.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolupaeva V.G., Lomakin I.B., Pestova T.V., Hellen C.U. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol. Cell. Biol. 2003;23:687–698. doi: 10.1128/MCB.23.2.687-698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomakin I.B., Hellen C.U., Pestova T.V. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang S.K., Wimmer E. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 1990;4:1560–1572. doi: 10.1101/gad.4.9.1560. [DOI] [PubMed] [Google Scholar]
- 14.Yu Y., Sweeney T.R., Kafasla P., Jackson R.J., Pestova T.V., Hellen C.U. The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J. 2011;30:4423–4436. doi: 10.1038/emboj.2011.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Locker N., Chamond N., Sargueil B. A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Res. 2011;39:2367–2377. doi: 10.1093/nar/gkq1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pisarev A.V., Unbehaun A., Hellen C.U., Pestova T.V. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
- 17.Ali I.K., McKendrick L., Morley S.J., Jackson R.J. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J. 2001;20:4233–4242. doi: 10.1093/emboj/20.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieft J.S., Zhou K., Jubin R., Doudna J.A. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortimer S.A., Weeks K.M. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 20.Mortimer S.A., Weeks K.M. Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nat. Protoc. 2009;4:1413–1421. doi: 10.1038/nprot.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darty K., Denise A., Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duke G.M., Hoffman M.A., Palmenberg A.C. Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J. Virol. 1992;66:1602–1609. doi: 10.1128/jvi.66.3.1602-1609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kafasla P., Morgner N., Poyry T.A., Curry S., Robinson C.V., Jackson R.J. Polypyrimidine tract binding protein stabilizes the encephalomyocarditis virus IRES structure via binding multiple sites in a unique orientation. Mol. Cell. 2009;34:556–568. doi: 10.1016/j.molcel.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 24.Kolupaeva V.G., Hellen C.U., Shatsky I.N. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA. 1996;2:1199–1212. [PMC free article] [PubMed] [Google Scholar]
- 25.McGinnis J.L., Dunkle J.A., Cate J.H., Weeks K.M. The mechanisms of RNA SHAPE chemistry. J. Am. Chem. Soc. 2012;134:6617–6624. doi: 10.1021/ja2104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pilipenko E.V., Blinov V.M., Chernov B.K., Dmitrieva T.M., Agol V.I. Conservation of the secondary structure elements of the 5′-untranslated region of cardio- and aphthovirus RNAs. Nucleic Acids Res. 1989;17:5701–5711. doi: 10.1093/nar/17.14.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman M.A., Palmenberg A.C. Revertant analysis of J-K mutations in the encephalomyocarditis virus internal ribosomal entry site detects an altered leader protein. J Virol. 1996;70:6425–6430. doi: 10.1128/jvi.70.9.6425-6430.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gould P.S., Bird H., Easton A.J. Translation toeprinting assays using fluorescently labeled primers and capillary electrophoresis. Biotechniques. 2005;38:397–400. doi: 10.2144/05383ST02. [DOI] [PubMed] [Google Scholar]
- 29.Shirokikh N.E., Alkalaeva E.Z., Vassilenko K.S., Afonina Z.A., Alekhina O.M., Kisselev L.L., Spirin A.S. Quantitative analysis of ribosome-mRNA complexes at different translation stages. Nucleic Acids Res. 2009;38:e15. doi: 10.1093/nar/gkp1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pestova T.V., Shatsky I.N., Fletcher S.P., Jackson R.J., Hellen C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson J.E., Pestova T.V., Hellen C.U., Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 32.Pestova T.V., Shatsky I.N., Hellen C.U. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Y., Abaeva I.S., Marintchev A., Pestova T.V., Hellen C.U. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 2011;39:4851–4865. doi: 10.1093/nar/gkr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shih D.S., Park I.W., Evans C.L., Jaynes J.M., Palmenberg A.C. Effects of cDNA hybridization on translation of encephalomyocarditis virus RNA. J. Virol. 1987;61:2033–2037. doi: 10.1128/jvi.61.6.2033-2037.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spahn C.M., Jan E., Mulder A., Grassucci R.A., Sarnow P., Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 36.Filbin M.E., Vollmar B.S., Shi D., Gonen T., Kieft J.S. HCV IRES manipulates the ribosome to promote the switch from translation initiation to elongation. Nat. Struct. Mol. Biol. 2013;20:150–158. doi: 10.1038/nsmb.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abaeva I.S., Marintchev A., Pisareva V.P., Hellen C.U., Pestova T.V. Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning. EMBO J. 2011;30:115–129. doi: 10.1038/emboj.2010.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kafasla P., Lin H., Curry S., Jackson R.J. Activation of picornaviral IRESs by PTB shows differential dependence on each PTB RNA-binding domain. RNA. 2011;17:1120–1131. doi: 10.1261/rna.2549411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hellen C.U., Witherell G.W., Schmid M., Shin S.H., Pestova T.V., Gil A., Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl Acad. Sci. U.S.A. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pilipenko E.V., Pestova T.V., Kolupaeva V.G., Khitrina E.V., Poperechnaya A.N., Agol V.I., Hellen C.U. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 41.Cech T.R. Crawling out of the RNA world. Cell. 2009;136:599–602. doi: 10.1016/j.cell.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 42.Lehman N. RNA in evolution. Wiley Interdiscip. Rev. RNA. 2010;1:202–213. doi: 10.1002/wrna.37. [DOI] [PubMed] [Google Scholar]
- 43.Malygin A.A., Kossinova O.A., Shatsky I.N., Karpova G.G. HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res. 2013;41:8706–8714. doi: 10.1093/nar/gkt632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


