Abstract
The cell cycle genes homology region (CHR) has been identified as a DNA element with an important role in transcriptional regulation of late cell cycle genes. It has been shown that such genes are controlled by DREAM, MMB and FOXM1-MuvB and that these protein complexes can contact DNA via CHR sites. However, it has not been elucidated which sequence variations of the canonical CHR are functional and how frequent CHR-based regulation is utilized in mammalian genomes. Here, we define the spectrum of functional CHR elements. As the basis for a computational meta-analysis, we identify new CHR sequences and compile phylogenetic motif conservation as well as genome-wide protein-DNA binding and gene expression data. We identify CHR elements in most late cell cycle genes binding DREAM, MMB, or FOXM1-MuvB. In contrast, Myb- and forkhead-binding sites are underrepresented in both early and late cell cycle genes. Our findings support a general mechanism: sequential binding of DREAM, MMB and FOXM1-MuvB complexes to late cell cycle genes requires CHR elements. Taken together, we define the group of CHR-regulated genes in mammalian genomes and provide evidence that the CHR is the central promoter element in transcriptional regulation of late cell cycle genes by DREAM, MMB and FOXM1-MuvB.
INTRODUCTION
Genes expressed periodically during the cell cycle are often regulated on the level of transcription. The cell cycle genes homology region CHR is a central DNA element in many promoters of late cell cycle genes with a maximal expression in G2 and M phases (1). The CHR primarily controls transcriptional repression of these genes in G0 and early G1. Moreover, we recently provided evidence that the CHR is also required for full activation in late cell cycle phases (2). The CHR is bound by LIN9, LIN37, LIN52, LIN54 and RBBP4 proteins, which form the MuvB core complex. It has been suggested that LIN54 is the component which mediates binding to the CHR (3). Depending on the cell cycle phase, several other proteins interact with the MuvB core. In G0 and early G1, the MuvB proteins bind E2F4, DP1 and p130 forming the DREAM complex (4,5). This protein complex represses gene activity when bound to promoters of cell cycle genes (4). In some promoters, binding of DREAM to the CHR is supported by an adjacent cell cycle-dependent element (CDE), a motif rich in guanines and cytosines and located upstream of the CHR and separated from it by a spacer of four nucleotides (2). When a cell progresses through G1 to S phase, E2F4, DP1 and p130 proteins dissociate from MuvB and are replaced by B-MYB forming the MMB (Myb-MuvB) complex. In late S phase, MMB recruits FOXM1 to the promoters of late cell cycle genes. Finally, proteasome-mediated degradation of B-MYB results in maximal expression of these genes through the FOXM1–MuvB complex in G2 and M phases (6–8).
In addition to CHR motifs, three other elements have been implicated in recruiting MuvB-based protein complexes, namely, E2F elements (binding E2F/DP dimers), MBS motifs (MYB-binding sites) and FBS (forkhead sites binding FOXM1) (4,7–9). However, it appears that not all interactions of these transcription factors with their canonical recognition sites are required for the function of MuvB-containing complexes. For instance, FOXM1 binding to promoters of cell cycle genes does not require forkhead-binding sites, but rather depends on CHR elements (6).
The diversity of known CHR sequences is small. To date, four variants of the CHR motif have been described: the most common, TTTGAA, was shown to be a central promoter element in genes such as Ccnb2 (10), Cdk1 (11), Aurkb (12) and Cdca3 (Tome1) (13). The motifs TTTAAA and CTTGAA have been identified to be functional CHRs in the Ccnb1 (14) and CCNA2 (11) promoters, respectively. A CHR-like sequence was identified in the mouse Mybl2 (B-Myb) promoter and named downstream repression site (DRS) (15–17). This motif (TAGGAA) is functional in combination with an adjacent E2F site although it differs in two nucleotides from the common TTTGAA CHR sequence. Another example was established with the TTCAAA sites of the Ube2c and Plk4 promoters, showing that inverse CHR elements are also functional (2,18). Interestingly, in Caenorhabditis elegans and Drosophila, the orthologous DREAM complexes also bind to CHR-like sequences (19,20).
In this study, we search for DREAM, MMB and FOXM1-binding sites in human promoter regions in a genome-wide screen. In our computational meta-analysis based on genome-wide protein-DNA binding, gene expression data and analysis of phylogenetic motif conservation, we identify CHR elements in most late cell cycle genes bound by DREAM, MMB or FOXM1. In contrast, a large number of the observed E2F motifs in DREAM-bound promoters is found in early cell cycle genes. Furthermore, Myb and forkhead-binding sites are underrepresented compared to CHRs in cell cycle regulated genes bound by MMB or FOXM1. Taken together, our findings suggest the CHR as the central promoter element in transcriptional regulation of late cell cycle genes. We propose that regulation of these genes by a sequential binding of DREAM, MMB and FOXM1-MuvB always requires a CHR promoter element.
MATERIALS AND METHODS
Cell culture
NIH3T3, HFF and F9 cells from DSMZ (Braunschweig, Germany) were grown in DMEM supplemented with 10% FCS and penicillin/streptomycin. Cells were synchronized in G0 either by serum starvation (DMEM with 0% FCS) for 60 h or by density-arrest. For cell cycle analyses, NIH3T3 and HFF cells were stimulated to re-enter the cell cycle with 20% FCS in DMEM after the serum-deprivation phase.
Plasmids and DNA probes
Promoters were amplified from genomic DNA extracted from NIH3T3 cells and human foreskin fibroblasts, respectively, by standard PCR. DNA fragments were cloned into the pGL4.10 luciferase reporter vector (Promega). Site-directed mutagenesis was performed following the QuikChange protocol (Stratagene). Sequences of cloning primers as well as of mutagenic primers are provided in Supplementary Table S1. Plasmids used for the amplification of biotinylated DNA probes with CHR elements that deviate from the common TTTGAA sequence in one nucleotide (Supplementary Table S1) were prepared by ligation of double-stranded oligonucleotides into the pGL4.10 vector.
DNA affinity purification
DNA affinity purifications were performed as described earlier (2). Biotinylated DNA probes were amplified by PCR using a biotinylated standard reverse primer and gene-specific forward primers. In order to investigate binding of DREAM components to isolated CDE/CHR elements in an irrelevant DNA context, we amplified DNA probes with a length of 193 nucleotides comprising the pGL4.10 vector backbone and the inserted CDE/CHR elements. Oligonucleotide sequences are available in Supplementary Table S1. The biotinylated DNA probes were subjected to DNA affinity purification with nuclear extracts of density-arrested NIH3T3 cells or proliferating F9 cells, respectively, followed by western blot assaying for binding of the DREAM or MMB components. As an input control, 1% of the nuclear extracts used for the DNA affinity purification was analyzed.
Chromatin immunoprecipitation (ChIP)
ChIPs were performed as described in (2). Bound promoter regions were quantified with the QuantiTect SYBR Green PCR Kit (Qiagen, Hilden, Germany) and the ABI 7300 real-time PCR system (Life Technologies, Carlsbad, CA, USA). Oligonucleotide sequences are available in Supplementary Table S1.
SDS-PAGE and western blot
SDS-PAGE and western blot were performed following standard protocols (21). The following antibodies were applied for detection of DREAM and MMB components, respectively: E2F4 (C-20) and p130 (C-20) (Santa Cruz Biotechnology, Santa Cruz, CA, USA, 1:1000 dilution). The Lin37, Lin9 and Lin54 antibodies (1:1000 dilution) and the monoclonal B-Myb LX015.1 antibody (hybridoma media 1:5) were kind gifts from James DeCaprio (4) and Roger Watson (22).
Transfections and luciferase assays
Analyses of cell cycle-dependent promoter activities with luciferase reporter assays in serum-starved and re-stimulated NIH3T3 cells were performed as described earlier (2).
FACS analysis
Cells were fixed overnight at 4°C in one volume PBS/1 mM EDTA and three volumes of absolute ethanol. DNA was stained with Hoechst 33343 (Invitrogen) at a final concentration of 10 μg/ml for 15 min at 37°C. DNA content per cell was determined by flow cytometry on an LSR II instrument (Becton Dickinson, Franklin Lakes, NJ, USA). Data analysis was performed with BD FACS Diva 6.1 and WinMDI 2.9 software.
Definition of promoter regions
Promoter regions of protein-coding transcripts were compiled as follows: Starting with the set of all coding transcripts given in the UCSC Genes (knownGene) track for the human genome (version GRCh37/hg19) (23), only transcripts with a functional annotation in the GREAT tool version 2.0.2 (24) were selected resulting in a set of verified protein-coding transcripts. Promoter regions of these selected transcripts were defined by the region including the annotated TSS and the 200 bases upstream and downstream ([−200, +200] interval). Due to the high phylogenetic conservation of protein-coding regions that would interfere with the conservation-based motif detection, all parts of a promoter sequence that overlap with coding exons of other transcripts were removed. Promoter regions in which the TSS overlaps with a coding exon were completely excluded from the analyses. The protein-coding genes corresponding to this set of promoter regions are referred to as All Genes. Sequences of promoter regions (fasta format) and genomic coordinates of promoter regions (BED format) are provided in Dataset 1 and Dataset 2, respectively. For visualization of promoter regions in Supplementary Figure S5, the UCSC Genome Browser was utilized (25).
Protein-binding data
Protein-binding data were collected from the literature, namely, ChIP data for DREAM (E2F4, LIN9, LIN54, p130) (4), ChIP data for LIN9 and B-MYB of MMB complex (8) and FOXM1 data (6). When necessary, genomic coordinates were lifted to the latest version of human genome assembly (GRCh37/hg19) using UCSC liftover (26). Intersections of binding data and promoter regions were calculated using BEDTools (27). For MMB, bound regions were defined by the regions bound by both LIN9 and B-MYB proteins. A promoter sequence was defined to be bound by a certain protein if it overlaps with a region bound by that protein. DREAM binding was defined analogously to the original publication (4) by requiring binding of at least E2F4, LIN9 and p130.
Cell cycle expression data
Cell cycle-dependent expression data were compiled from three publications referred to as WF (28), BJ (29) and SV (8). The data extracted from WF (CellCycleGeneList_1134.txt) contains annotations of peak expression for 1134 loci, which were assigned to a total of 600 protein-coding genes using the IDConverter (30) to map Genbank accession numbers to UCSC ID's. The 480 loci with annotated peak expression from BJ (Supplementary SI Table 5) were mapped via the official gene symbol to 367 protein-coding genes. The raw data published by SV were processed following the instructions of the original publication (normalization using RMA from the R Affy package (31) and identification of genes with cell cycle-dependent expression using the R timecourse package (32). Conversion of the Affy IDs to UCSC IDs was performed with the R package biomaRt (33). Using the 1300 top ranked loci and their time point of maximal expression, a total of 767 protein-coding genes were assigned to a cell cycle phase of peak expression by mapping 0h->G1/S, 2h->S-phase, 4h->G2, 6h->G2/M and 8h->M/G1.
For genes having different annotations in different data sets, a single cell cycle phase of peak expression was chosen. Similar to the method used in SV and BJ, annotations from WF were used as reference to classify genes, i.e. the WF annotation was used whenever available. For the 29 genes annotated only in SV and BJ, the cell cycle phase with maximum expression was chosen from SV in cases of differing annotations. In addition, a very recently published data set (34) was incorporated in the analysis of verified CHR elements and referred to as GT. Annotations of peak expression for 1871 loci were mapped to 1543 protein-coding genes, complementing the annotations from the other studies. The new data set was also used to redefine peak expression of genes previously annotated as ‘M/G1’. Thus, a total of 2486 protein-coding genes were assigned to a unique cell cycle phase of maximum expression.
Detection of binding sites in promoter regions
All sequences deviating in at most two positions from the canonical TTTGAA were considered as putative CHR elements. Other transcription factor binding sites were modeled based on published binding sites or JASPAR PWMs (35): CDE [CGT][CG][CG][CG][CG][CG] (1); E2F TT[CG][CG][CG][CG][CG] Jaspar ID: MA0470.1; MBS [CT]AAC[GT]G (36); FBS AAA[CT]A (6). Single binding sites were searched on the plus and minus strand of promoter regions. PhastCons data for the placental mammals with human as a reference (37,38) was used to calculate the average evolutionary conservation over all bases in a binding site. Detected sites were rejected if their conservation was below an average of 0.9.
De novo motif prediction and motif comparison
For de novo motif prediction in the 92 promoter regions that are DREAM-bound and differentially expressed in cell cycle but lacking conserved CHR and E2F elements, the MEME program (39) in its version 4.6.1 was used with ‘-dna -mod anr -nmotifs 5 –minw 6 –maxw 9 -revcomp –maxsites 150’ options to search for motifs of length 6 to 9 on both strands. Comparison of motifs from MEME predictions with published transcription factor binding sites was performed with the motif comparison tool TOMTOM (40) from the MEME suite (website, version 4.9.0) using standard settings.
RESULTS
DREAM can bind to non-canonical CHR elements in vitro
Since only a few functional CHR sites that deviate from the canonical TTTGAA sequence had been identified previously, we performed a comprehensive search for alternative CHR elements. First, we established an assay system that allows for comparison of in vitro DREAM binding to CHR-derived elements independent of promoter context. For the mouse cyclin B2 promoter we had shown earlier that a single canonical CHR element is sufficient for cell cycle-dependent regulation and binding of DREAM, although binding is supported by an adjacent CDE element (2). We observed that the promoter context and additional elements were not required for DREAM binding since the isolated CDE/CHR tandem, placed in the irrelevant DNA context of the pGL4.10 vector backbone, was bound by representative DREAM components with similar affinity as the wild-type Ccnb2 promoter (Figure 1A). In order to identify CHR-like sequences that may be functional while differing from the wild-type CHR, we assayed all possible 18 single-nucleotide permutations of the canonical TTTGAA element for DREAM binding (Figure 1B). Probes were based on the mouse cyclin B2 CDE/CHR element since DREAM binding to the tandem element instead of the isolated CHR results in more robust western blot signals. We observed that the strongest binding occurs to the canonical CHR TTTGAA as well as to the sequence TTTAAA. Other related sites are able to bind DREAM with varying affinity. Especially binding to TTCGAA and TTTGAT elements is clearly above background. These results suggest that other non-canonical but functional CHR elements exist in mammalian genomes.
Figure 1.
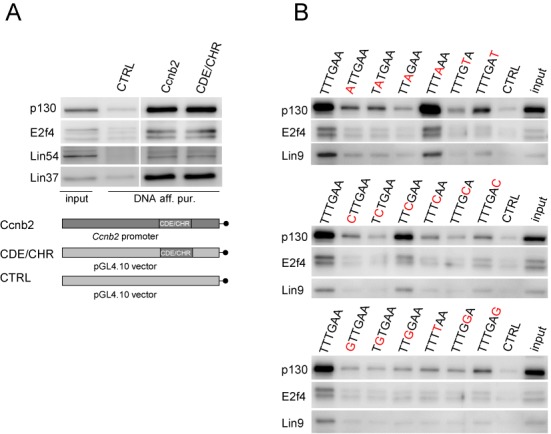
DREAM can bind to non-canonical CHR elements in DNA affinity purification assays. (A) Analysis of DREAM complex binding to the isolated cyclin B2 CDE/CHR element flanked by irrelevant DNA and to a 220-bp cyclin B2 promoter probe. Biotinylated DNA probes based on the mouse Ccnb2 promoter (Ccnb2) and on the CDE/CHR element from the same promoter (CDE/CHR) were cloned in the pGL4.10 vector backbone and subjected to DNA affinity purifications with density-arrested NIH3T3 cells. Binding of DREAM components (p130, E2f4, Lin37, Lin54) to both probes was analyzed with polyclonal antibodies. As a non-DREAM binding negative control, a DNA probe based on the empty pGL4.10 vector was utilized (CTRL). All samples are from the same blot. (B) DREAM complex binding to CHR elements differing from the canonical sequence TTTGAA in one nucleotide analyzed in an in vitro DNA affinity purification assay. Biotinylated probes based on the CDE/CHR probe with all possible 1 bp permutations were prepared and subjected to DNA affinity purifications with nuclear extracts from density-arrested NIH3T3 cells. A DNA probe based on the empty pGL4.10 vector was used as a non-DREAM binding negative control (CTRL). Eluates were probed in western blots for the DREAM components p130, E2f4 and Lin9. All samples were processed in parallel.
A comprehensive analysis of DREAM-bound late cell cycle genes identifies many candidates with non-canonical CHRs
For a genome-wide identification of genes regulated by DREAM through CHR elements in human, experimental data for DREAM binding and cell cycle regulation were incorporated in a computational screen. We utilized DREAM-binding data (4) and three data sets on cell cycle-dependent mRNA expression by Whitfield et al. (28), Bar-Joseph et al. (29), and Sadasivam et al. (8), referred to as WF, BJ and SV, respectively. After we had finished the initial identification of non-canonical CHR elements, a new data set on genes differentially expressed during the cell cycle became available (34). As this screen significantly increases the number of available cell cycle annotations, we decided to include these data for further computational analyses referring to it as GT. The four data sets had little overlap in the group of cell cycle regulated genes and in time points of peak expression (Figure 2A). This seems to be caused by the different methods used to classify the expression peaks rather than by differences in the measured expression profiles. While annotations of consecutive cell cycle phases (e.g. G1/S and S; G2, G2/M, and M/G1) are often inconsistent, classification into early and late cell cycle phases yielded similar data sets from the four analyses. In order to prevent loss of late cell cycle genes falsely annotated in S phase, we decided to define ‘late cell cycle genes’ to be expressed in S, G2 and M at this point of the analysis. Thus, peak expression was assigned to 2486 genes, of which 1408 have a maximum expression in the late cell cycle.
Figure 2.
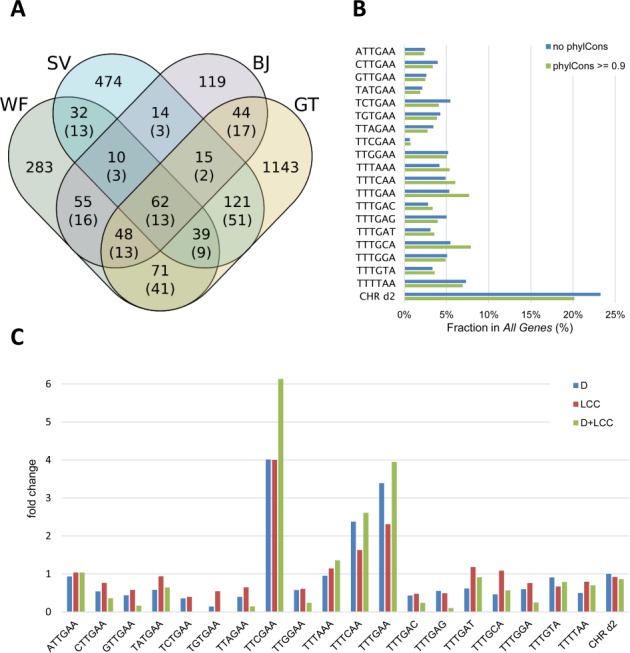
Identification of potential CHR elements in cell cycle genes. (A) Numbers of genes annotated as cell cycle-regulated by one or several of the data sets reported by Whitfield et al. (WF), Bar-Joseph et al. (BJ), Sadasivam et al. (SV) and Grant et al. (GT). The numbers of genes with identically annotated cell cycle phases of peak expression for each intersection are shown in parentheses. (B) Distribution of CHR-like elements in promoter regions of All Genes with (phylCons> = 0.9) or without (no phylCons) phylogenetical conservation. (C) Fold change of phylogenetically conserved CHR-like elements in the sets of DREAM-binding genes (D), late cell cycle-expressed genes (LCC), or late cell cycle genes binding DREAM (D+LCC). For all elements the fold change was calculated using their relative frequencies in the corresponding set divided by their relative frequencies in All Genes. A fold change above one indicates an enrichment of the element in the corresponding set.
The CHR-like sequences searched for in the genome-wide screen cover the canonical TTTGAA and all 18 single-nucleotide mutations, also comprising the already described functional CHR elements TTTAAA and CTTGAA. Additionally, elements differing in two positions from the canonical element TTTGAA were included in the search to systematically identify elements such as the TAGGAA CHR motif which had been described in the B-myb promoter (15,17). As swapping two bases in a sequence of six nucleotides results in 135 different permutations, many of which can represent target sites for transcription factors different from DREAM, we restricted our search to report CHR-like sites with two-nucleotide permutations only if a CDE with a 4-bp spacer upstream of the element was present.
The distribution of CHR-like elements in the promoter regions from all protein-coding genes (All Genes) was nearly uniform, independent from the evolutionary conservation (Figure 2B). Selection by phylogenetical conservation (phastCons score ≥ 0.9) reduced the number of potential CHRs from 47 444 to 7570. For all further analyses, the phastCons score was set to ≥ 0.9 for putative binding sites. Subsets of All Genes were defined containing genes binding DREAM (D), expressed during the late cell cycle (LCC), or the overlap of both groups (D+LCC). In contrast to All Genes, in these subsets a few CHR-like elements were enriched (Figure 2C and Supplementary Table S3): the canonical TTTGAA as well as the elements TTCGAA and TTTCAA. The element TTTCAA can resemble the inverse canonical CHR (TTCAAA) when shifted by one nucleotide which was observed in 85% of all cases. The same accounts for TTTTAA overlapping with a TTTAAA in 83% of the hits.
Since CHR-regulated genes are described to be bound by DREAM and to have peak expression in late cell cycle phases, candidate genes for experimental validation of CHR-like elements were selected from the D+LCC data set. Such candidates were not identified for all screened CHR-like sequences since either the putative element was not phylogenetically conserved or the corresponding gene harbored another previously described CHR that was located closer to the TSS. Furthermore, 82% of genes with the CHR-like element TTTGCA were identified as histone genes. This element resembles a canonical Oct1 binding site. Since it had been shown that histone genes are regulated through such Oct1 sites (41) and DREAM did not bind to this element with high affinity in the DNA affinity purification screen (Figure 1B), we did not further test these elements as potential CHRs.
Finally, 22 CHR-like elements were tested in promoters carrying such sites. (Supplementary Table S2). For more than half of these elements binding of DREAM to the promoter was not observed or mutation of the element did not affect promoter activity. Consequently, we do not regard these elements as functional CHRs. This also applies to the CHR-like elements in the promoters of Exo1 and Nek2, which will be presented in more detail below. However, we discovered six novel non-canonical and functional CHRs differing in one or two nucleotides from the canonical element: TTCGAA (Bub1), TTTGTA (Chek2), TTTGAT (Melk), TTTGAG (Pold1), TTCGAG (Rad18) and TTCGAT (Rad54l). Function and binding properties of these sites were tested in the listed genes serving as examples for the new CHR sites (Figures 3–5).
Figure 3.
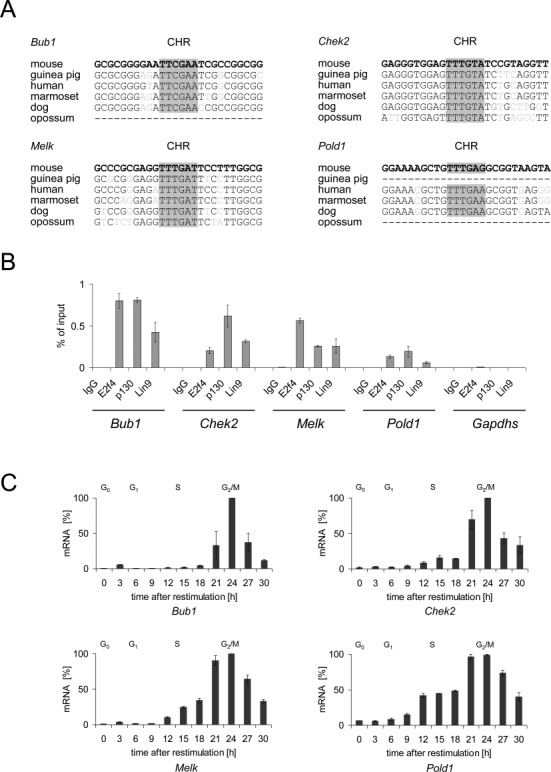
Genes with non-canonical CHR elements bind DREAM and are expressed in late cell cycle phases. (A) Alignments of putative CHR elements in Bub1, Chek2, Melk and Pold1 promoters from different mammals. CHRs are highlighted in gray. (B) Chromatin immunoprecipitations in density-arrested NIH3T3 cells to promoters with putative CHR elements. Antibodies targeting the representative DREAM components E2f4, p130 and Lin9 as well as a non-targeting rabbit antibody (IgG) were applied. Binding of DREAM components to the Gapdhs promoter was measured as a negative control. Precipitated DNA fragments were quantified by qPCR and normalized to the input. (C) mRNA expression of Bub1, Chek2, Melk and Pold1 in different cell cycle phases was measured in NIH3T3 cells synchronized by serum deprivation followed by serum re-stimulation. FACS analyses for the different time points are shown in Supplementary Figure S1.
Figure 4.
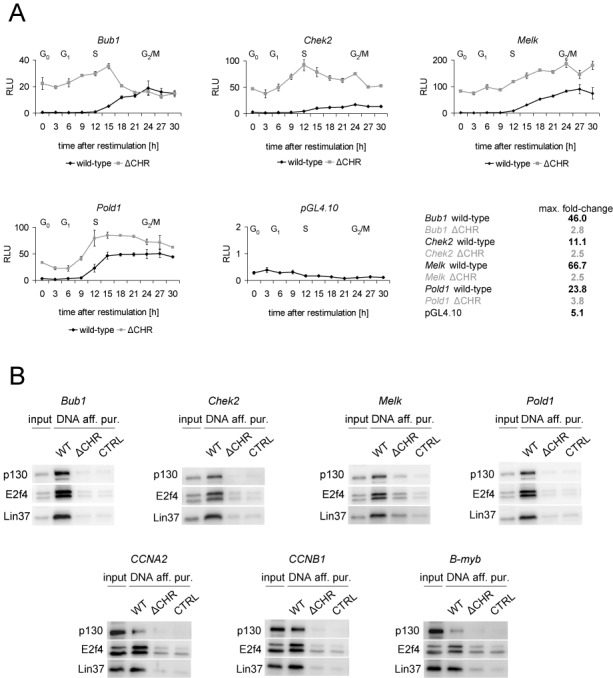
Non-canonical CHR elements are essential for cell cycle-dependent gene transcription and DREAM binding. (A) Promoter activity of Bub1, Chek2, Melk and Pold1 was analyzed with luciferase reporter assays in NIH3T3 cells synchronized by serum starvation followed by serum re-stimulation. Activities of wild-type promoters were measured in different cell cycle phases and compared to the activity of the corresponding CHR mutants (ΔCHR). pGL4.10 empty vector served as a negative control. FACS analyses for the different time points are shown in Supplementary Figure S1. (B) DREAM binding to promoters with non-canonical CHR elements was analyzed by DNA affinity purification followed by western blot. Proteins from nuclear extracts of density-arrested NIH3T3 cells binding to wild-type promoters (WT) and CHR mutants (ΔCHR) were probed with antibodies targeting p130, E2f4 and Lin37 as representative components of DREAM. A DNA probe of the Gapdhs promoter served as a negative control (CTRL).
Figure 5.
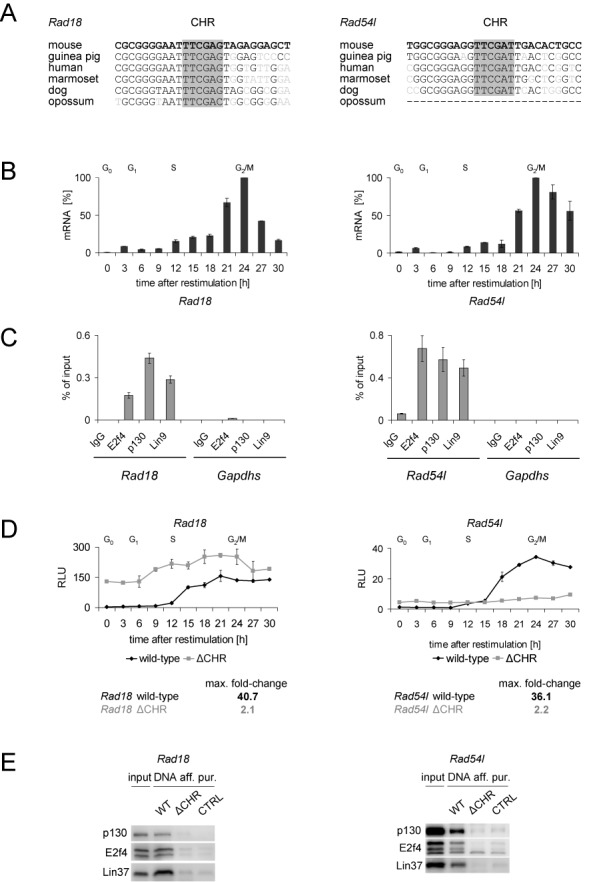
The Rad18 and Rad54l CHRs deviate in two nucleotides from the consensus TTTGAA element but are still functional. (A) Comparison of nucleotide sequences in Rad18 and Rad54l promoters from several mammals. (B)Rad18 and Rad54l mRNA expression measured in synchronized NIH3T3 cells at indicated time points after serum starvation and serum re-stimulation. (C)In vivo binding of DREAM components (E2f4, p130, Lin9) in density-arrested NIH3T3 cells to the Rad18 and Rad54l promoters measured by ChIP. A non-targeting rabbit antibody (IgG) served as a negative control. Binding of DREAM proteins to Rad18 and Rad54l was quantified by qPCR and compared to binding to the Gapdhs promoter, which is not targeted by DREAM. (D) Promoter activity of Rad18 and Rad54l measured in synchronized NIH3T3 cells with luciferase reporter assays. Activity of the wild-type promoters is compared to CHR mutants (ΔCHR). (E) DNA affinity purification from nuclear extracts of density-arrested NIH3T3 cells tested for representative DREAM components E2f4, p130 and Lin37 by western blot. A DNA probe of the Gapdhs promoter served as a negative control (CTRL).
Mouse ortholog genes with non-canonical CHRs are cell cycle regulated and bind DREAM in vivo
Having identified human candidate genes harboring potential non-canonical CHRs in their promoters which differ in one nucleotide from the canonical element (Figure 3A), we tested whether their mouse orthologs are expressed in late cell cycle phases and whether they bind DREAM in mouse NIH3T3 cells to confirm the observation we have made for many CDE/CHR-regulated genes that regulation for a given human or mouse promoter appears identical in mouse and human cells (2). We performed ChIPs in density-arrested NIH3T3 cells and showed that E2f4, p130 and Lin9 as representative DREAM components are enriched at the Bub1, Chek2, Melk and Pold1 promoters (Figure 3B). Additionally, we measured cell cycle-dependent mRNA expression of these genes in synchronized NIH3T3 cells. Expression is low in G0 and G1 phase and starts to increase in S phase. Maximal mRNA expression is reached in late S, G2 and M phases (Figure 3C, Supplementary Figure S1). mRNA expression of the non-cell cycle regulated U6 snRNA was measured as a control (Supplementary Figure S1). These findings strongly suggest that the mouse and human genes are regulated similarly and that the identified elements represent indeed functional novel non-canonical CHRs.
Non-canonical CHR elements mediate cell cycle-dependent regulation and bind DREAM
In order to test the potential elements differing in one nucleotide from the canonical sequence for function, wild-type and CHR mutant promoters of Bub1, Chek2, Melk and Pold1 were analyzed in reporter assays during the cell cycle (Figure 4A). To create promoters with non-functional CHRs, the sequence of the elements was changed to TGCATA (ΔCHR). In G0 and G1 phases, the activity of all wild-type promoters was very low and started to increase in S phase to reach their maxima in late S and G2/M phases (Figure 4A, Supplementary Figure S1). The expression from the promoters in the reporter assays was consistent with their mRNA expression patterns (Figure 3C). Importantly, all promoters with mutated CHRs were deregulated. As an example for this deregulation, the maximal fold-change from G1 to G2/M for the Melk gene shifts from 66.7 for the wild-type construct to 2.5 for the CHR mutant reporter (Figure 4A). A common feature of all tested mutants is de-repression in the early cell cycle phases.
The ability of DREAM to bind the non-canonical CHR elements in vitro was examined using DNA affinity purification with extracts of density-arrested NIH3T3 cells. Additionally, we analyzed the CHR elements in the promoters of CCNA2 (CTTGAA), CCNB1 (TTTAAA) and B-myb (TAGGAA) genes. These non-canonical CHRs had been shown to be functional, but interaction of the elements with DREAM had yet to be investigated (11,14–16). The representative components of the DREAM complex p130, E2f4 and Lin37 bound to all wild-type promoters, while binding was disrupted when the CHR elements were mutated (Figure 4B). Taken together, the results from functional and protein-binding assays provide evidence that the analyzed elements are non-canonical CHRs.
Functional CHRs can deviate from the canonical element by two nucleotides
In combination with an E2F site, the non-canonical CHR TAGGAA of the B-myb promoter was shown to control cell cycle-dependent transcription (15,17). This is the only element that had been described previously as a CHR which differs from the canonical sequence by two nucleotides. In the computational screen, we identified seven additional candidate genes harboring CHR-like elements with two nucleotide variations and tested them for function (Supplementary Table S2). Three putative CHR elements (TTCGGA, TTGGAT, TCTGTA) in the candidate genes ATAD2, Blm and BRCA1 could not be verified to be functional in the reporter assays (data not shown). The CHR-like element in NEK2 is not conserved between human (GTTAAA) and mouse (GTTGAC) genes (Supplementary Figure S2A). We cloned the promoters of both ortholog genes and showed that their cell cycle-dependent transcription was mediated by these elements. While the wild-type promoters showed a pattern of activity that is typical for late-expressed cell cycle genes, mutation of the CHR-like elements resulted in deregulation (Supplementary Figure S2B). However, although binding of DREAM components to the promoters in ChIP assays was detected, we did not observe binding of the proteins in DNA affinity purifications (Supplementary Figure S2C and D). Similar observations were made for the CHR-like element ATCGAA in the mouse Exo1 promoter (Supplementary Figure S3). Taken together, the CHR-like sequences exhibit features of functional CHRs and were shown to be central elements for cell cycle-dependent regulation in the DREAM-bound promoters of NEK2 and Exo1 genes. Yet, we do not designate them as CHRs since direct DREAM binding to the CHR-like elements could not be shown in vitro.
Nevertheless, we could identify two novel functional non-canonical CHR elements with two-nucleotide variations in the promoters of Rad18 (TTCGAG) and Rad54l (TTCGAT). Both CHRs are highly conserved and located close to the transcription start (Figure 5A). Human RAD18 and RAD54L mRNA were already shown to be differentially expressed during the cell cycle (42,43). We confirmed the cell cycle-dependent expression with a peak in G2/M phases for Rad18 in the mouse NIH3T3 cell system and observed a similar pattern for Rad54l (Figure 5B). More importantly, we detected binding of DREAM components to the region close to the TSS by ChIP (Figure 5C). Subsequently, we analyzed the regions 500 bp upstream from the translational starts of the both genes and found that these promoters were repressed in G0 and early G1 and became activated when the cells progressed to S phase. When the CHR-like elements were mutated, cell cycle-dependent regulation was completely abolished (Figure 5D) and DREAM binding was lost (Figure 5E). Thus, the cell cycle-dependent expression of Rad18 and Rad54l genes is regulated through non-canonical CHRs that deviate by two nucleotides from the canonical element.
CHR elements are enriched in promoters binding DREAM, MMB or FOXM1
The MuvB core complex participates in DNA binding of DREAM, MMB and FOXM1-MuvB by differentially associating with E2F4-DP1-p130, B-MYB or FOXM1. With E2F elements (binding E2F/DP dimers), CHRs (binding LIN54), MBS elements (MYB-binding sites) and FBS elements (forkhead binding sites for FOXM1), four different elements have been suggested as binding sites for MuvB-containing complexes (2–4,8,9). However, it appears that not all interactions of those transcription factors with their canonical DNA binding sites are necessary for the MuvB-containing complexes to function. For instance, although a significant overlap of genes bound by MMB and FOXM1 is observed, no enrichment of MYB-binding sites (MBS) or forkhead-binding sites (FBS) was detected in the data set representing FOXM1-binding genes (6).
Here, the presence of all 10 identified CHR sequences in combination with either E2F, MBS or FBS elements was analyzed and compared in sets of promoters bound by DREAM, MMB and FOXM1 (Figure 6). We observed a substantial enrichment of promoters containing single CHR (+CHR-E2F) or E2F (−CHR+E2F) elements or combinations of both (+CHR+E2F) in the data set representing genes bound by DREAM (D) compared to the All Genes data set. CHR elements are even more enriched in promoters bound by MMB or FOXM1 and accumulate to 82% in the set of genes bound by DREAM, MMB and FOXM1. In contrast, the fraction of promoters containing E2F elements only (−CHR+E2F) was found reduced in this comparison (Figure 6A) and was almost depleted of genes bound by DREAM and MMB as well as by FOXM1. This is in agreement with published data showing an enrichment of E2F sites in DREAM-bound, but not in MMB- and FOXM1-bound genes (4,6,8).
Figure 6.
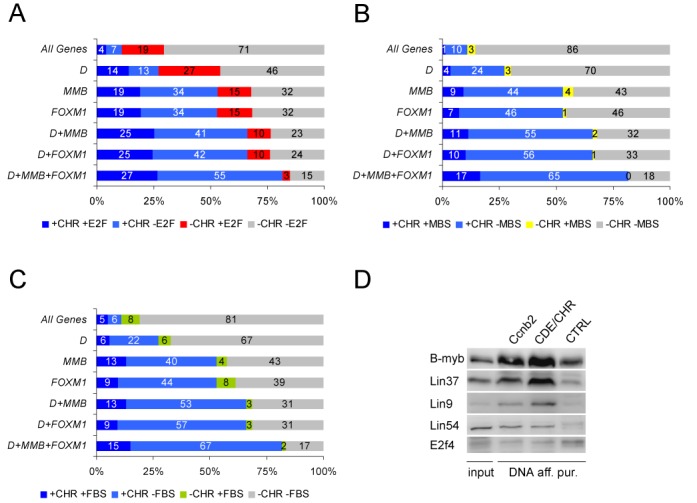
CHR elements are enriched in promoters of genes bound by DREAM, MMB and FOXM1. (A) Percentage of promoters with or without CHR elements (+CHR or –CHR) and with or without E2F sites (+E2F or –E2F) in different promoter subsets: all promoters (All Genes), DREAM-bound promoters (D), promoters bound by the MMB components B-MYB and LIN9 (MMB), promoters bound by FOXM1 (FOXM1) and promoters bound by two or all three of the complexes. (B) Fraction of promoters with or without Myb-binding sites (+MBS or –MBS) and with or without CHRs (+CHR, –CHR) in different promoter subsets. (C) Percentage of promoters with or without forkhead-binding sites (+FBS or –FBS) and with or without CHRs (+CHR, –CHR) in different promoter subsets. (D) Binding of MMB to the isolated cyclin B2 CDE/CHR site flanked by irrelevant DNA in comparison to a 220 bp cyclin B2 promoter probe. Biotinylated DNA probes derived from the mouse Ccnb2 promoter (Ccnb2) and from the cyclin B2 CDE/CHR element cloned into the pGL4.10 vector backbone (CDE/CHR) were subjected to DNA affinity purifications with nuclear extracts of proliferating F9 cells which do not form DREAM. MMB components B-Myb, Lin54, Lin37 and Lin9 were detected by western blot. To control the absence of DREAM binding to the probes, E2f4 as part of the DREAM complex was analyzed. As a non-MMB-binding negative control, a DNA probe based on the empty pGL4.10 vector was employed (CTRL).
MMB and FOXM1-MuvB appear to bind promoters through CHR sites
In genes bound by MMB or FOXM1, the co-occurrence of CHR and MBS elements (+CHR+MBS) was enriched about 10-fold compared to All Genes (Figure 6B). Nevertheless, the fraction of genes in the MMB and FOXM1 data sets containing CHR elements only (+CHR−MBS) was about four to five times larger than for those containing both sites. In contrast, single MBS elements (−CHR+MBS) were not enriched in MMB and FOXM1 compared to their fraction observed in All Genes. Interestingly, in the data set of genes bound by all three complexes, not a single promoter holding a conserved MBS was identified provided that no CHR was found in the promoter (Figure 6B). Comparable observations were made for combinations of FBS elements and CHR sites (Figure 6C). The finding that CHRs, but not FBS elements, are strongly enriched supports the conclusion that FOXM1 does not bind to FBS but is recruited via MuvB. This is consistent with recent observations (6).
As MBS elements were suggested to be necessary for recruiting MMB to promoters (8,9), we tested whether this complex can bind to an isolated CHR not surrounded by natural promoter DNA, specifically devoid of an MBS element. To this end, we assayed binding of MMB to the isolated CDE/CHR element of Ccnb2 flanked by irrelevant vector DNA or to the Ccnb2 core promoter which holds an MBS (Figure 6D). MMB bound to the probe carrying the isolated element with comparable affinity as to the Ccnb2 promoter probe. Thus, MMB can bind to isolated CDE/CHR elements without the necessity of additional elements such as an MBS.
Recently, four MBS elements in the Birc5 (survivin) promoter were implicated to cooperate in activating the promoter through binding of MMB (9). These elements are not phylogenetically conserved and thus were not identified in our screen. The impact of a functional CHR in the Birc5 promoter (44) was not analyzed in this report. We tested whether mutation of the MBS and the CHR changes promoter activity in different cell cycle phases. Birc5 promoter luciferase reporter constructs containing the reported four MBS elements as well as the CHR in the wild-type reporter and mutant reporter constructs lacking several or all MBS elements or the CHR were generated. While wild-type promoters and all MBS mutants showed nearly identical activities with low expression in G0/G1 and maximal expression in G2/M, cell cycle-dependent regulation was completely disrupted upon CHR mutation (Supplementary Figure S4). Thus, the CHR is the main element in the Birc5 promoter necessary for cell cycle-dependent regulation while MBS elements are not required.
Taken together, our data show that among the analyzed binding sites, only the CHR is strongly enriched in promoters bound by MMB and FOXM1. There is no indication that MBS or FBS elements are generally required to recruit MMB or FOXM1-MuvB.
CHR sites are predominantly found in genes expressed during G2/M
The presence of CHR and E2F elements was analyzed for DREAM-bound genes expressed in specific cell cycle phases (Figure 7A). Nearly all genes harboring a CHR without an E2F site in their promoters (+CHR−E2F) were expressed late in the cell cycle with maximal expression in G2 or M. In case of sole E2F elements in the promoter (−CHR+E2F), 68% of the genes showed peak expression in early cell cycle phases (G1 and S). Interestingly, genes that contain both elements in their promoters (+CHR+E2F) accumulated to 85% in fractions expressed in late cell cycle phases, a distribution found similarly for genes carrying only a CHR (Figure 7A). Furthermore, a significant difference in the time point of expression start could not be detected when comparing expression profiles in the SV data set for these genes (data not shown). These results suggest that time points of transcriptional activation and maximal expression are mainly regulated through the CHRs even if additional conserved E2F sites are present in these promoters.
Figure 7.
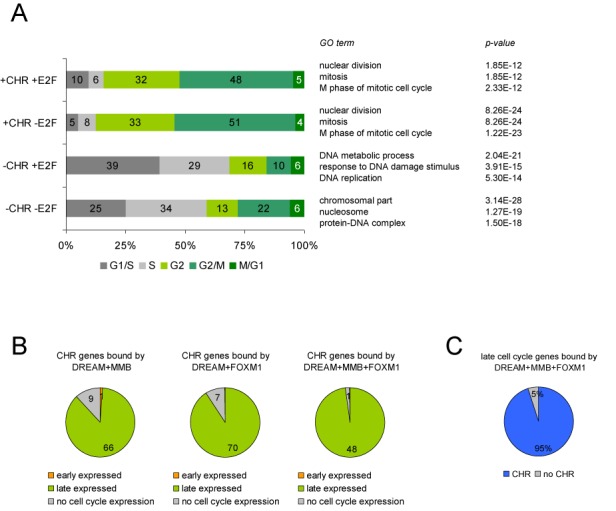
CHR genes bound by DREAM, MMB and FOXM1 are cell cycle regulated with peak expression in late cell cycle phases. (A) Correlation of the time point of maximal gene expression with the occurrence of CHR elements (+CHR or –CHR) and E2F sites (+E2F or –E2F) in promoters. Enriched functional clusters obtained using each of the four subsets along with their P-values are given. (B) Peak expression of CHR genes bound by DREAM, MMB and FOXM1 in early and late cell cycle phases. Numbers of genes in the different subsets are given. (C) Rate of CHR sites in genes bound by DREAM, MMB and FOXM1. Percentages of genes with (CHR) and without (no CHR) CHR element are given.
In general, the number of DREAM-bound genes expressed in early or late cell cycle phases was nearly equal. Genes with promoters carrying no conserved CHR or E2F elements (−CHR−E2F) were expressed in early cell cycle phases in nearly 60% (Figure 7A). Interestingly, 40% of those genes represent histones with a peak expression in S phase. For DREAM-bound and cell cycle regulated genes, functionally related groups based on GO terms were analyzed using the DAVID Functional Annotation Tool (45). Consistent with the enrichment of late and early peak expression for promoters harboring either CHRs or E2F elements, related GO terms were found enriched for those genes (Figure 7A). Namely, the GO terms nuclear division, mitosis and M phase of mitotic cell cycle were found highly enriched for genes carrying CHR elements in their promoters (+CHR−E2F). In contrast, genes with only E2F elements (−CHR+E2F) were annotated mainly in the categories DNA metabolic process, response to DNA damage stimulus and DNA replication. Again, the presence of E2F elements in addition to CHRs (+CHR+E2F) had no detectable effect compared to genes with single CHRs since for both sets the same GO terms were enriched. In promoters without identified CHRs or E2F sites, only the terms chromosomal part, nucleosome, protein–DNA complex were found enriched due to the high number of histone genes in this group (Figure 7A). A de novo motif prediction from the −CHR−E2F data set of genes followed by a comparison with known transcription factor binding sites identified CCAAT-boxes, Oct1 sites as well as GC-rich and AT-rich regions. While the CCAAT-boxes are common elements in many eukaryotic promoters that provide basal transcriptional activity, the Oct1-binding site mediates regulation of cell cycle dependent expression in histone genes (41). Thus, in promoters without CHR and E2F sites, evidence for an alternative mechanism by other transcription factor binding sites that could substitute for E2F elements or CHRs did not emerge by de novo motif prediction.
Most CHR genes bound by DREAM, MMB and FOXM1 are cell cycle regulated with peak expression in G2/M
After having identified CHR elements in most genes bound by DREAM, MMB and FOXM1 and having observed that most CHR genes bound by DREAM are expressed during the late cell cycle, we asked whether binding of the three complexes to a CHR gene generally results in a cell cycle-dependent regulation. We analyzed CHR-containing genes bound by DREAM and MMB or FOXM1 for cell cycle-dependent expression and found a late cell cycle annotation for 66 out of 76 genes in the DREAM+MMB gene set and for 70 out of 77 genes in DREAM+FOXM1 (Figure 7B). From the non-regulated genes, five (H2AFZ, KIF24, MXD3, NCAPG, PARPBP) were identified as cell cycle regulated with peak expression in G2/M by quantification of mRNA in synchronized HFF cells (Supplementary Figure S5). Interestingly, another six (METTL15, DCAF16, IFT80, PDZD11, PTRHD1, HSCB) are positioned head-to-head in close proximity to genes that were identified as cell cycle regulated (Supplementary Figure S5). Thus, in these examples it is likely that binding of DREAM, MMB and FOXM1-MuvB to the region shared by two genes is responsible only for the regulation of one gene. For the three remaining genes (HNRNPH2, RBMX, LSM5), no significant increase in mRNA expression during the late cell cycle was measured. In CHR genes bound by all three complexes, only one gene (METTL15) was not annotated as a late cell cycle gene (Figure 7B).
Taken together, binding of transcription factors and expression data is consistent with a model of sequential DREAM, MMB and FOXM1–MuvB binding to CHR genes resulting in repression of transcription in early cell cycle phases and activation of expression during the late cell cycle.
Regulation of late cell cycle genes by binding of DREAM, MMB and FOXM1-MuvB depends on CHR elements
Interestingly, in the group of late cell cycle genes bound by DREAM and MMB as well as by FOXM1, our computational analysis identified highly conserved and validated CHR sites in 48 out of 55 genes. In one of the seven remaining genes, NEK2, we had identified a CHR-like element (Supplementary Figure S2). In another four of those genes (TOP2A, CKAP2, DZIP3, KIAA1524), CHR elements are located close to the TSS, but were not detected in the screen because their conservation scores are below 0.9. However, these elements still show considerable conservation with scores between 0.36 and 0.71 (Supplementary Table S3). As these genes exhibit all characteristics of CHR-regulated genes, it is likely that their cell cycle-dependent expression is regulated through these elements. In only two genes (SKA2, SFPQ,) no CHR was located in the region of +/−200 bp to the TSS; however, highly conserved CHRs are found exactly on the TSS of an alternative SKA2 transcript and 400 bp upstream of the SFPQ gene.
Thus, we identified evolutionary conserved CHR sites close to the TSS in 95% of all late cell cycle genes which are bound by DREAM, MMB and FOXM1 (Figure 7C). All of these genes show peak expression in G2 and M phases. Taken together, the results show that a CHR is always present in promoters of late cell cycle genes if they are regulated by binding of DREAM, MMB and FOXM1.
CHR elements exhibit low sequence variability and can function in forward and reverse orientations
Based on the results of our computational screen and the analysis of potential CHR genes and their promoters, we identified 148 genes that show clear features of CHR-regulated genes (Table 1). This group of genes also includes CEP152, GSG2 (Haspin), NCAPG2, SASS6 and SMC2. These genes were identified as late cell cycle genes by quantification of mRNA in synchronized HFF cells although they are not annotated accordingly in the four genome-wide screens (Supplementary Figure S5). Additionally, GAS2L3 was reported as a DREAM target before (46) and identified here as a CHR gene.
Table 1.
Late cell cycle genes harboring evolutionary conserved CHR elements
| AKIRIN2 | CCDC99 | CEP152 | ESCO2 | KIF15 | NCAPD2 | PRC1 | SPAG5 |
| ANLN | CCNA2 | CHEK2 | ESPL1 | KIF18A | NCAPG | PRR11 | SPC25 |
| ANP32E | CCNB1 | CIT | FAM110A | KIF20B | NCAPG2 | PTTG1 | SPRY4 |
| ARHGAP11A | CCNB2 | CKAP2 | FAM64A | KIF22 | NCAPH | RACGAP1 | TMPO |
| ARL6IP1 | CDC20 | CKAP2L | FBXO5 | KIF23 | NDC80 | RAD18 | TOP1 |
| ASF1B | CDC25C | CKAP5 | FOXM1 | KIF24 | NEIL3 | RAD54L | TOP2A |
| ASPM | CDCA2 | CKS1B | GAS2L3 | KIF2C | NRD1 | RANGAP1 | TPX2 |
| AURKA | CDCA3 | CKS2 | GSG2 | KIF4A | NUCKS1 | REEP4 | TRAIP |
| AURKB | CDCA8 | CSE1L | GTSE1 | KPNA2 | NUF2 | RNF26 | TROAP |
| BIRC5 | CDK1 | CSTF1 | H2AFX | LMNB1 | NUP35 | RTKN2 | TTK |
| BORA | CDK2 | DBF4B | H2AFZ | LOC81691 | NUSAP1 | SASS6 | UACA |
| BUB1 | CDKN2D | DCP2 | HJURP | MAD2L1 | PARPBP | SCLT1 | UBE2C |
| BUB1B | CDKN3 | DEPDC1 | HMMR | MELK | PIF1 | SETD8 | UBE2S |
| BUB3 | CENPA | DEPDC1B | INCENP | METTL4 | PLK1 | SGOL1 | WEE1 |
| C11orf82 | CENPE | DLGAP5 | IQGAP3 | MIS18BP1 | PLK4 | SGOL2 | ZRANB3 |
| C12orf32 | CENPF | DNMT3B | KIAA1524 | MND1 | POC5 | SHCBP1 | |
| C2orf69 | CENPM | DZIP3 | KIAA1731 | MYBL2 | POLD1 | SKA1 | |
| C9orf100 | CENPO | EIF2AK3 | KIF11 | MXD3 | POLQ | SMC2 | |
| CASC5 | CEP55 | ENC1 | KIF14 | MZT1 | POU2F1 | SMC4 |
All genes are bound by DREAM. Genes also observed to bind MMB and FOXM1 are marked in bold.
Generally, the identified CHR elements showed little sequence variation. Highest variability was observed for the last nucleotide position of CHR sites (Figure 8A). We performed a quantitative analysis of sequence variation covering CHRs and their flanking regions from all 148 genes. Importantly, the canonical TTTGAA sequence was the predominant CHR element (Figure 8B). Nevertheless, also the sequence TTTAAA and the newly identified TTCGAA occurred frequently. Reverse orientation of non-palindromic CHRs was detected in about 45% of the genes. Another important feature of the identified CHR sites was their location on or very close to the annotated TSS (Figure 8C).
Figure 8.
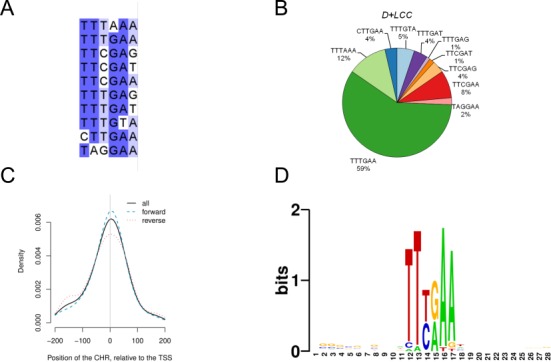
CHR sites predominantly have the sequence TTTGAA and are found close to the transcription start site. (A) Alignment of functional CHR elements. (B) Relative occurrence of the 10 validated CHR sequences in DREAM-bound genes expressed during the late cell cycle (D+LCC). (C) Positions of the identified CHR elements relative to the transcription start site (TSS) in DREAM-bound genes expressed during the late cell cycle (D+LCC). (D) Sequence logo derived from verified CHR sequences identified in promoters of the D+LCC data set with flanking nucleotides.
Although initial reports had suggested that CDE sites four nucleotides upstream from CHRs are often required for function (1), only the CHR elements themselves were found to be conserved (Figure 8D). Searching directly for CDE elements combined with validated CHRs, we found CDE/CHR tandem combinations are not standard. From the CHR-containing promoters bound by DREAM and expressed late in the cell cycle only 23% contained a CDE (Supplementary Table S3). Therefore, we conclude that the majority of CHRs can function without an upstream CDE.
DISCUSSION
CHR promoter elements have been identified as important transcriptional regulatory elements almost 20 years ago (11). These elements are usually found in TATA-less promoters of genes that are differentially expressed during the cell cycle with a maximal expression in G2 and M phases. Four different CHR sequences had been characterized in several late cell cycle genes, but a comprehensive approach with the aim to identify a broader spectrum of these important regulatory elements has been missing until now.
Identification of novel CHR sequences
A DNA affinity purification assay provided evidence that additional CHR-like sequences can be bound by the DREAM complex (Figure 1) and thus may constitute functional elements. We therefore searched for such non-canonical CHRs in the promoters of cell cycle genes. Screening DREAM-bound genes with peak expression in S, G2 and M phases, we identified and then experimentally validated four new sequences that deviate in one nucleotide from the most common element TTTGAA and two sequences that deviate in two nucleotides, resulting in six novel DNA sequences that can be designated as CHR elements. All six genes tested as examples for harboring these novel CHR elements (Bub1, Chek2, Melk, Pold1, Rad18 and Rad54l) play important roles in cell cycle regulation and DNA repair (44,47–58). Cell cycle-dependent expression had already been established for POLD1, MELK, RAD18 and RAD54L. Interestingly, cell cycle-dependent transcription of the human POLD1 gene had been shown to be regulated through a CHR (59). The CHR in the human promoter deviates from the sequence in mouse and resembles the canonical sequence TTTGAA (Figure 3A) in contrast to the TTTGAG element we have identified in mouse. Thus, POLD1 is a good example for a gene with evolutionary changes in the CHR sequence that do not disrupt functionality of the element. The Melk gene had been shown to be differentially regulated during the cell cycle depending on E2F4 and p130/p107, however, a CHR element had not been reported (60). This is also the case for human RAD18 and RAD54L, whose mRNAs had been shown to be cell cycle regulated without further investigation of the promoter (42,43).
Most of the previously reported genes with CHRs were detected in our computational screen showing that our search strategy was appropriate. Nevertheless, additional CHR sites may have escaped our search. As it is not feasible to examine all potential CHR elements by mutation analysis, we focused on example candidates for each identified sequence. In particular, we cannot rule out that a CHR-like sequence that was non-functional in one tested gene could be functional in another promoter context. We may also have missed functional CHRs due to incorrect classifications of cell cycle-regulated genes, erroneous annotation of the TSS or failure to detect representative FOXM1-MuvB, MMB or DREAM components at promoters. Given the small overlap of cell cycle-regulated genes in the screens used for classification of peak expression, particularly these assays seem to be prone to artifacts (Figure 2A). Moreover, when we searched for CHR elements with two deviations from TTTGAA, we had to include the CDE as an anchor sequence to reduce the number of hits for experimental validation. This dramatically reduced the number of candidate CHR elements in the search and hence may have led us to ignore CHRs unlinked to a CDE. Nevertheless, we think that we missed only a small number of sites. We reason that most potential CHR elements that differ in more than one nucleotide from the canonical sites are most likely not functional, because already most elements with single nucleotide exchanges are clearly underrepresented in comparison to the canonical elements. Also, the three sites that deviate in two nucleotides from TTTGAA can only be found in four promoters.
Furthermore, we have identified CHR-like sequences in the human and mouse NEK2 promoters and in the mouse Exo1 promoter that show clear features of CHR elements since mutation disrupts cell cycle-dependent transcription and results in a strong elevation of promoter activity already in G0. Even though DREAM binding to these promoters is observed in ChIP assays, we could not detect an interaction of DREAM with the potential CHRs in DNA affinity purifications. However, since the DNA probes used for these experiments were amplified on basis of the cloned promoters that contain all evolutionary conserved elements around the TSS and show proper regulation during the cell cycle (Supplementary Figures S2 and S3), it seems unlikely that DREAM binds to elements that are located outside the tested promoter region. It is more likely that DREAM has a low affinity to the CHR-like elements in vitro that prevents detection of the interaction. Also, we have screened the D+LCC data set for these elements and did not find additional candidates for testing. Therefore, we can rule out that there is a large group of genes regulated through these elements. Taken together, it appears that the three most common CHR sequences are present in 80% of all CHR promoters: TTTGAA, TTTAAA and TTCGAA (Figures 6 and 8). Interestingly, DREAM bound with the highest affinity to DNA probes carrying these sites (Figure 1B). The remaining seven elements account for only 20% of all CHRs and also bind DREAM with a lower affinity in vitro. Since we found at least one of the 10 CHR sequences in 95% of late cell cycle genes bound by DREAM and MMB as well as by FOXM1, it is likely that we identified most of the CHR sequences associated with late cell cycle genes.
Forty five percent of the identified CHR elements had a reverse orientation to the TSS, which is in agreement with the finding that CHRs can be functional in both orientations (2). Interestingly, when found close to the transcription start sites of head-to-head orientated genes, the CHR elements did not mediate co-expression of these genes. This was not expected since bidirectional genes sharing one promoter region are often functionally related, co-regulated and co-expressed (61). In all observed cases, only one of the head-to-head-orientated genes was identified as a late cell cycle gene although the CHRs were located in a window of 200 bp around the TSS of both genes (Supplementary Figure S5). As we generally find other regulatory elements like CCAAT-boxes to be located upstream of the CHRs (1), it is more likely that the specific localization of all elements determines the direction of transcription. Additionally, insulator elements may contribute to the different expression of these genes.
Analysis of other potential binding sites: E2F, CDE, MBS, FBS
The exact mechanism of DREAM, MMB and FOXM1-MuvB binding to promoter elements is still unclear. Five components of the MuvB core or its associated proteins are considered to be DNA-binding transcription factors: E2F4, DP1, LIN54, B-MYB and FOXM1. LIN54 is the only component that is part of both the repressing as well as the activating complexes and was found to be able to bind the CHR in the Cdk1 promoter in vitro (3). Recently, we have shown that DREAM, MMB and also FOXM1-MuvB can bind DNA through CHR sites (2,6,62). Consistently, the computational approach in our current study identified CHRs in 95% of late cell cycle genes bound by DREAM and MMB as well as FOXM1 which clearly indicates that CHR sites are required for the regulation of late cell cycle genes by these proteins. Moreover, we observed that 27% of DREAM-bound genes contain conserved E2F sites in their proximal promoters. It had been shown that DREAM can bind to promoters with E2F sites, while MMB and FOXM1 do not (4,6,8). Even though the interaction of DREAM with E2F binding sites has not been studied in detail, the available data suggest that DREAM, in addition to contacting the CHR and regulating G2/M genes without the requirement of E2F sites, also binds and represses early cell cycle genes through an interaction of E2F4/DP1 with E2F-binding sites. Thus, both types of DNA sites may complement each other in mediating repression of early and late cell cycle genes by DREAM. Interestingly, we identified a large group of genes with promoters bound by DREAM that harbor evolutionary conserved CHRs as well as E2F sites (Figure 6A). More than 80% of these promoters show peak expression in late cell cycle phases like promoters that solely harbor CHRs (Figure 7A). We also analyzed whether E2F/CHR promoters are activated significantly earlier in the cell cycle than CHR promoters, but did not observe obvious timing differences based on currently available data. The timing of gene expression during the cell cycle thus appears to be regulated through CHR elements in cases where both elements are located in the same promoter. However, as these E2F sites are phylogenetically conserved, it is highly likely that they also have a functional significance, for example for reaching the full promoter activity by binding activating E2F proteins.
Initial reports had identified CDE sites four nucleotides upstream from CHRs and suggested that in most genes a combination of CDE and CHR elements is necessary for cell cycle-dependent regulation (1). In contrast to this notion, we found the CHR elements but not CDEs to be highly conserved (Figure 8D). Moreover, in the group of CHR-containing promoters bound by DREAM and expressed late during the cell cycle only 23% contained a CDE (Supplementary Table S3). These findings are consistent with our observation that CDEs support binding of DREAM to CHR elements and increase the regulation of promoters, but CDE sites are not essential for function (2). DREAM can bind to CHR elements without support of a CDE (2). Additionally, MMB and FOXM1-MuvB do not interact with CDE elements (2,6,62). These observations lead us to conclude that evolutionary pressure on CHR elements is much higher than on CDEs. This also explains that CHR sites, in contrast to CDEs, are highly conserved.
In contrast, a general role of MBS and FBS in recruiting the MMB- and MuvB–FOXM1 complexes is unlikely. We did not find an enrichment of evolutionary conserved MBS elements in promoters without a CHR bound by MMB or FOXM1. Moreover, in promoters bound by DREAM, MMB and FOXM1, such elements are completely absent. However, 17% of the genes bound by all three complexes contain evolutionary conserved MBS co-occurring with CHR elements. In contrast, CHRs without MBS are found in 65% of such promoters (Figure 6B). ChIP experiments with transiently transfected promoter constructs suggested that MMB is recruited to the Birc5/survivin promoter through four non-conserved MBS elements (9). In another report from the same group, it was shown that binding of FOXM1 to the survivin promoter depends also on the four MBS (7). However, a canonical CHR site in the survivin promoter located close to the TSS was not investigated in both reports, although it had been reported to be regulating this promoter (44). We analyzed the CHR and MBS elements in the survivin promoter and found that the promoter is largely deregulated upon mutation of the CHR, while its activity remained essentially unchanged after mutation of the MBS elements in all phases of the cell cycle (Supplementary Figure S4). Thus, the results provide strong evidence that the CHR is the main element necessary for cell cycle-dependent transcriptional regulation also of the survivin promoter. Furthermore, since only 3% of all cell cycle promoters bound by MMB contain MBS elements without also carrying CHRs, it is unlikely that recruiting MMB through MBS elements to promoters of late cell cycle genes without participation of CHRs is a common mechanism. This model is in agreement with the observation that loss of Myb in Drosophila leads to reduced expression of late cell cycle genes and adult lethality, but these defects do not depend on the ability of Myb to bind to DNA since DNA-binding-deficient Myb mutants can rescue the Myb-null mutants (63). Additionally, MMB is still targeted to essential late cell cycle genes even in the absence of Myb, and Myb only contributes to targeting of MMB to a subset of non-cell-cycle-regulated genes (64). However, since we found evolutionary conserved MBS in 17% of CHR promoters binding DREAM, MMB and FOXM1, it may be possible that MBS are involved in the regulation of a subgroup of mammalian CHR promoters. An interaction of B-MYB with MBS may support binding of MMB to CHRs, similar to CDE elements supporting binding of DREAM to CHR sites. In contrast to CDE sites, the distance between the identified MBS and CHRs is not conserved. Thus, a possible participation of such elements co-occurring with CHRs in the regulation of late cell cycle genes and mechanistic details need to be investigated.
We also explored the possible involvement of FBS elements in binding FOXM1 in late cell cycle promoters. We had shown before that CHRs are highly enriched in a set of FOXM1-bound promoters using two motif finding tools, but FBS elements did not accumulate (6). In contrast, Sadasivam et al. observed in addition to CHRs also an enrichment of MBS and FBS elements in a set of MMB-bound promoters utilizing de novo motif discovery tools (8). In order to support one of the two binding models, we searched for conserved FBS elements in the data sets of MMB- or FOXM1-bound promoters. FBS elements were identified in 8% of promoters bound by FOXM1 and in only 4% of the promoters bound by MMB compared to 13% in All Genes. In the set of promoters bound by DREAM, MMB and FOXM1, the fraction of promoters displaying only FBS elements decreased to 2%, whereas CHR promoters accumulated to 67%. In this data set, 15% of promoters contain CHRs as well as FBS elements (Figure 6C). These results are clearly inconsistent with an enrichment of FBS elements in MMB- or FOXM1-bound cell cycle genes. Instead it favors a mechanism in which FOXM1 is recruited to CHRs via MMB (6). This model is further supported by the finding that FOXM1 in general has a very low affinity to FBS elements (65).
In general, it has been observed that mutation of CHR elements results in an activation of all promoters in G0 and G1 phases when tested in reporter assays which clearly shows that CHRs are the central repressive elements in the promoters of late cell cycle genes. The importance of the CHR for activation is not as clear-cut. For the cyclin B2 promoter, we have shown the DREAM, MMB and FOXM1 bind through the CHR and that mutation of the element results not only in an increased promoter activity in G0/G1, but also in a decreased activity in G2/M (2,6,62). Furthermore, knockdown of B-MYB leads to a reduced activity of the wild-type promoter, but not of the CHR mutant (2). Thus, the CHR mediates repression as well as activation of the cyclin B2 gene, most likely through an interaction with Lin54 (3). In contrast, the activity of promoters like Chek2, Melk and Pold1 was lower than the respective CHR mutants in all phases of the cell cycle (Figure 4). In addition, only for a subgroup of CHR promoters bound by DREAM an additional binding of MMB and FOXM1 (4,6,8) (Table 1) and a reduced expression after knockdown of MMB components (9,66) was reported. Thus, it remains open if activation by MMB and FOXM1-MuvB is a general feature of all CHR genes. However, in 95% of late cell cycle genes bound by DREAM and MMB as well as by FOXM1, we have identified evolutionary conserved CHR elements, which supports a general mechanism for this group of genes: If late cell cycle genes are regulated by sequential binding of DREAM, MMB and FOXM1-MuvB, then interaction of the complexes with the promoters is mediated through CHR elements (Figure 9).
Figure 9.
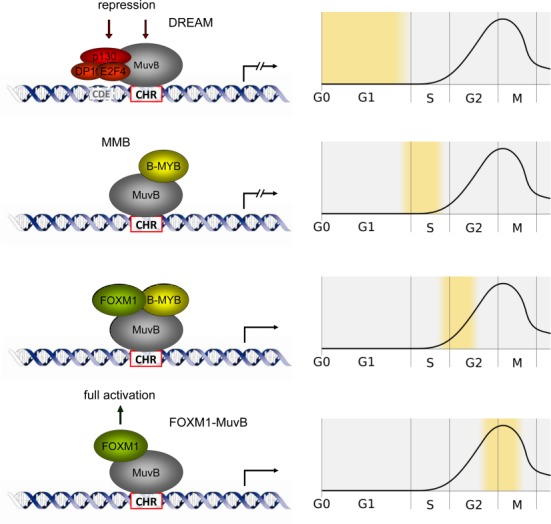
Regulation of late cell cycle genes through sequential binding of DREAM, MMB and FOXM1-MuvB to the CHR. In the derived model, all complexes are recruited to CHR elements via MuvB. Expression of late cell cycle genes is repressed in G0 and G1 by DREAM. CDE elements can support binding of DREAM to the CHR. In early S phase, MMB binds to the CHR. Later, MMB recruits FOXM1, which results in initiation of transcription. In G2 and M phases, B-MYB is degraded and expression of late cell cycle genes reaches its maximum through activation by FOXM1-MuvB.
Binding of DREAM to non-cell cycle genes
In ChIP-chip analyses, DREAM binds also to non-cell cycle-regulated genes (4). It is not clear whether these binding events contribute to transcription or if they represent just non-functional protein–DNA interactions. Moreover, bound transcription factors may become functional only in certain tissues or after triggering by specific stimuli. Interestingly, it was shown for the DREAM component E2F4 that protein binding to promoters is highly conserved between tissues of one species, while only 20% of bound genes overlap between human and mouse (67). Consistent with our observations, in the group of promoters bound in both species, genes with functions in cell cycle regulation and proliferation were highly overrepresented (67). Important functions of DREAM in cell cycle regulation are therefore conserved between species and tissues, while other interactions of DREAM with DNA may contribute to unknown species-specific regulation or, more likely, may be non-functional in most cases. This hypothesis is supported by the finding that only 25% of genes bound by MMB in their promoters vary in their expression after depletion of MMB by siRNA in Drosophila (64). In agreement with the concept that functional binding of transcription factors to their binding elements is often evolutionary conserved, we find CHR elements identified in human to be functional in mouse as well (Figures 3–5). Even in cases in which we observed differences in CHR nucleotide sequences of ortholog human and mouse genes, these variations did not impair function as shown for the CHRs in the mouse and human Pold1 promoters (Figures 3 and 4), (59).
In contrast to the group of genes only bound by DREAM, we found a massive enrichment of cell cycle genes in the group additionally bound by MMB or FOXM1. We identified only three genes holding an evolutionary conserved CHR and being bound by DREAM and MMB or FOXM1 that were not cell cycle-regulated in our analysis: HNRNPH2, LSM5 and RBMX. It remains to be clarified if binding of the complexes to these non-cell cycle genes has a physiological significance. In a recent publication, DREAM was implicated in the repression of genes during osteoblast differentiation (68). In undifferentiated proliferating MC3T3-E1 cells, DREAM was reported to bind to Bglap and Alpl promoters and to repress these osteoblast-specific genes. Since both promoters lack CHRs or E2F sites, we were interested in elucidating the binding of DREAM to these genes. However, we could not detect binding of DREAM to Bglap or Alpl, neither in ChIP assays nor in DNA affinity purifications using preparations of undifferentiated MC3T3-E1 cells, even when comparing the Bglap and Alpl results to many positive controls proving that the assays were able to detect DREAM binding to cell cycle promoters (data not shown). As our data stand in contrast to the findings by Flowers et al., it remains unresolved if mammalian DREAM, like the ortholog complexes in Drosophila and C. elegans, also participates in regulation of differentiation in mammals (20,69,70).
In summary, we define CHR motifs and provide a comprehensive overview of CHR elements with their distribution in the human genome. We have identified evolutionary conserved CHR elements in 95% of late cell cycle promoters bound by DREAM and MMB as well as FOXM1. Such genes are generally differentially expressed during the cell cycle with a maximum in G2 and M phases. We conclude that the CHR is the central promoter element in the transcriptional regulation of late cell cycle genes by DREAM, MMB and FOXM1-MuvB (Figure 9).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors are indebted to Carola Koschke and Andrea Rothe for expert technical assistance and Juliane Röthe, Marianne Quaas and Martin Fischer for experiments in the initial phase of this project. Andreas Lösche, Kathrin Jäger (FACS) and Birgit Oelzner (DNA sequencing) performed analyses at the IZKF Leipzig core unit.
We thank Jim DeCaprio and Roger Watson for kind gifts of antibodies and Christine Engeland for critically reading the manuscript.
Author contributions: G.A.M. and A.W. conceived the experiments. A.W. performed the computational analyses. G.A.M. and K.S. performed all other experiments. G.A.M., A.W., K.E., S.J.P. and P.F.S. discussed results and wrote the manuscript. All authors read and approved the final manuscript.
FUNDING
This work was funded in part by the Bundesministerium für Bildung und Forschung (BMBF) through grants by the Interdisciplinary Center for Clinical Research [IZKF project D02] at the University of Leipzig to K.E. and by the John Templeton Foundation through the grant ‘Origins and Evolution of Regulation in Biological Systems’ [24332] to S.J.P. and P.F.S. The opinions expressed in this publication are those of the authors and do not necessarily reflect the views of the John Templeton Foundation. K.S. and A.W. are recipients of graduate fellowships provided by the Freistaat Sachsen and by the European Social Fund. The Open Access Publication charge was partially waived by Oxford University Press. Funding for open access charge: German Research Foundation (DFG) and University of Leipzig within the program of Open Access Publishing.
Conflict of interest statement. None declared.
REFERENCES
- 1.Müller G.A., Engeland K. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J. 2010;277:877–893. doi: 10.1111/j.1742-4658.2009.07508.x. [DOI] [PubMed] [Google Scholar]
- 2.Müller G.A., Quaas M., Schumann M., Krause E., Padi M., Fischer M., Litovchick L., DeCaprio J.A., Engeland K. The CHR promoter element controls cell cycle-dependent gene transcription and binds the DREAM and MMB complexes. Nucleic Acids Res. 2012;40:1561–1578. doi: 10.1093/nar/gkr793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmit F., Cremer S., Gaubatz S. LIN54 is an essential core subunit of the DREAM/LINC complex that binds to the cdc2 promoter in a sequence-specific manner. FEBS J. 2009;276:5703–5716. doi: 10.1111/j.1742-4658.2009.07261.x. [DOI] [PubMed] [Google Scholar]
- 4.Litovchick L., Sadasivam S., Florens L., Zhu X., Swanson S.K., Velmurugan S., Chen R., Washburn M.P., Liu X.S., DeCaprio J.A. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 5.Schmit F., Korenjak M., Mannefeld M., Schmitt K., Franke C., von E.B., Gagrica S., Hanel F., Brehm A., Gaubatz S. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle. 2007;6:1903–1913. doi: 10.4161/cc.6.15.4512. [DOI] [PubMed] [Google Scholar]
- 6.Chen X., Muller G.A., Quaas M., Fischer M., Han N., Stutchbury B., Sharrocks A.D., Engeland K. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol. Cell Biol. 2013;33:227–236. doi: 10.1128/MCB.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Down C.F., Millour J., Lam E.W., Watson R.J. Binding of FoxM1 to G2/M gene promoters is dependent upon B-Myb. Biochim. Biophys. Acta. 2012;1819:855–862. doi: 10.1016/j.bbagrm.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Sadasivam S., Duan S., DeCaprio J.A. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26:474–489. doi: 10.1101/gad.181933.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knight A.S., Notaridou M., Watson R.J. A Lin-9 complex is recruited by B-Myb to activate transcription of G2/M genes in undifferentiated embryonal carcinoma cells. Oncogene. 2009;28:1737–1747. doi: 10.1038/onc.2009.22. [DOI] [PubMed] [Google Scholar]
- 10.Lange-zu Dohna C., Brandeis M., Berr F., Mössner J., Engeland K. A CDE/CHR-tandem element regulates cell cycle-dependent repression of cyclin B2 transcription. FEBS Lett. 2000;484:77–81. doi: 10.1016/s0014-5793(00)02133-5. [DOI] [PubMed] [Google Scholar]
- 11.Zwicker J., Lucibello F.C., Wolfraim L.A., Gross C., Truss M., Engeland K., Müller R. Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 1995;14:4514–4522. doi: 10.1002/j.1460-2075.1995.tb00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura M., Uchida C., Takano Y., Kitagawa M., Okano Y. Cell cycle-dependent regulation of the human aurora B promoter. Biochem. Biophys. Res. Commun. 2004;316:930–936. doi: 10.1016/j.bbrc.2004.01.178. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida K. Cell-cycle-dependent regulation of the human and mouse Tome-1 promoters. FEBS Lett. 2005;579:1488–1492. doi: 10.1016/j.febslet.2005.01.055. [DOI] [PubMed] [Google Scholar]
- 14.Wasner M., Tschöp K., Spiesbach K., Haugwitz U., Johne C., Mössner J., Mantovani R., Engeland K. Cyclin B1 transcription is enhanced by the p300 coactivator and regulated during the cell cycle by a CHR-dependent repression mechanism. FEBS Lett. 2003;536:66–70. doi: 10.1016/s0014-5793(03)00028-0. [DOI] [PubMed] [Google Scholar]
- 15.Bennett J.D., Farlie P.G., Watson R.J. E2F binding is required but not sufficient for repression of B- myb transcription in quiescent fibroblasts. Oncogene. 1996;13:1073–1082. [PubMed] [Google Scholar]
- 16.Catchpole S., Tavner F., Le C.L., Sardet C., Watson R.J. A B-myb promoter corepressor site facilitates in vivo occupation of the adjacent E2F site by p107 x E2F and p130 x E2F complexes. J. Biol. Chem. 2002;277:39015–39024. doi: 10.1074/jbc.M202960200. [DOI] [PubMed] [Google Scholar]
- 17.Liu N., Lucibello F.C., Zwicker J., Engeland K., Müller R. Cell cycle-regulated repression of B-myb transcription: cooperation of an E2F site with a contiguous corepressor element. Nucleic Acids Res. 1996;24:2905–2910. doi: 10.1093/nar/24.15.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer M., Quaas M., Wintsche A., Muller G.A., Engeland K. Polo-like kinase 4 transcription is activated via CRE and NRF1 elements, repressed by DREAM through CDE/CHR sites and deregulated by HPV E7 protein. Nucleic Acids Res. 2014;42:163–180. doi: 10.1093/nar/gkt849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korenjak M., Anderssen E., Ramaswamy S., Whetstine J.R., Dyson N.J. RBF binding to both canonical E2F targets and noncanonical targets depends on functional dE2F/dDP complexes. Mol. Cell Biol. 2012;32:4375–4387. doi: 10.1128/MCB.00536-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tabuchi T.M., Deplancke B., Osato N., Zhu L.J., Barrasa M.I., Harrison M.M., Horvitz H.R., Walhout A.J., Hagstrom K.A. Chromosome-biased binding and gene regulation by the Caenorhabditis elegans DRM complex. PLoS Genet. 2011;7:e1002074. doi: 10.1371/journal.pgen.1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirschner R.D., Sänger K., Müller G.A., Engeland K. Transcriptional activation of the tumor suppressor and differentiation gene S100A2 by a novel p63-binding site. Nucleic Acids Res. 2008;36:2969–2980. doi: 10.1093/nar/gkn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tavner F., Frampton J., Watson R.J. Targeting an E2F site in the mouse genome prevents promoter silencing in quiescent and post-mitotic cells. Oncogene. 2007;26:2727–2735. doi: 10.1038/sj.onc.1210087. [DOI] [PubMed] [Google Scholar]
- 23.Hsu F., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 24.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitfield M.L., Sherlock G., Saldanha A.J., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bar-Joseph Z., Siegfried Z., Brandeis M., Brors B., Lu Y., Eils R., Dynlacht B.D., Simon I. Genome-wide transcriptional analysis of the human cell cycle identifies genes differentially regulated in normal and cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2008;105:955–960. doi: 10.1073/pnas.0704723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alibes A., Yankilevich P., Canada A., Diaz-Uriarte R. IDconverter and IDClight: conversion and annotation of gene and protein IDs. BMC Bioinformatics. 2007;8:9. doi: 10.1186/1471-2105-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 32.Tai Y.C., Speed T.P. On gene ranking using replicated microarray time course data. Biometrics. 2009;65:40–51. doi: 10.1111/j.1541-0420.2008.01057.x. [DOI] [PubMed] [Google Scholar]
- 33.Durinck S., Moreau Y., Kasprzyk A., Davis S., De M.B., Brazma A., Huber W. BioMart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 34.Grant G.D., Brooks L., III, Zhang X., Mahoney J.M., Martyanov V., Wood T.A., Sherlock G., Cheng C., Whitfield M.L. Identification of cell cycle-regulated genes periodically expressed in U2OS cells and their regulation by FOXM1 and E2F transcription factors. Mol. Biol. Cell. 2013;24:3634–3650. doi: 10.1091/mbc.E13-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.Y., Chou A., Ienasescu H., et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biedenkapp H., Borgmeyer U., Sippel A.E., Klempnauer K.H. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988;335:835–837. doi: 10.1038/335835a0. [DOI] [PubMed] [Google Scholar]
- 37.Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 40.Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts S.B., Segil N., Heintz N. Differential phosphorylation of the transcription factor Oct1 during the cell cycle. Science. 1991;253:1022–1026. doi: 10.1126/science.1887216. [DOI] [PubMed] [Google Scholar]
- 42.Kanaar R., Troelstra C., Swagemakers S.M., Essers J., Smit B., Franssen J.H., Pastink A., Bezzubova O.Y., Buerstedde J.M., Clever B., et al. Human and mouse homologs of the Saccharomyces cerevisiae RAD54 DNA repair gene: evidence for functional conservation. Curr. Biol. 1996;6:828–838. doi: 10.1016/s0960-9822(02)00606-1. [DOI] [PubMed] [Google Scholar]
- 43.Masuyama S., Tateishi S., Yomogida K., Nishimune Y., Suzuki K., Sakuraba Y., Inoue H., Ogawa M., Yamaizumi M. Regulated expression and dynamic changes in subnuclear localization of mammalian Rad18 under normal and genotoxic conditions. Genes Cells. 2005;10:753–762. doi: 10.1111/j.1365-2443.2005.00874.x. [DOI] [PubMed] [Google Scholar]
- 44.Otaki M., Hatano M., Kobayashi K., Ogasawara T., Kuriyama T., Tokuhisa T. Cell cycle-dependent regulation of TIAP/m-survivin expression. Biochim. Biophys. Acta. 2000;1493:188–194. doi: 10.1016/s0167-4781(00)00142-1. [DOI] [PubMed] [Google Scholar]
- 45.Huang d.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolter P., Schmitt K., Fackler M., Kremling H., Probst L., Hauser S., Gruss O.J., Gaubatz S. GAS2L3, a target gene of the DREAM complex, is required for proper cytokinesis and genomic stability. J. Cell Sci. 2012;125:2393–2406. doi: 10.1242/jcs.097253. [DOI] [PubMed] [Google Scholar]
- 47.Bartek J., Falck J., Lukas J. CHK2 kinase–a busy messenger. Nat. Rev. Mol. Cell Biol. 2001;2:877–886. doi: 10.1038/35103059. [DOI] [PubMed] [Google Scholar]
- 48.Bi X., Barkley L.R., Slater D.M., Tateishi S., Yamaizumi M., Ohmori H., Vaziri C. Rad18 regulates DNA polymerase kappa and is required for recovery from S-phase checkpoint-mediated arrest. Mol. Cell Biol. 2006;26:3527–3540. doi: 10.1128/MCB.26.9.3527-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bugreev D.V., Mazina O.M., Mazin A.V. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 50.Huang J., Huen M.S., Kim H., Leung C.C., Glover J.N., Yu X., Chen J. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat. Cell Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang P., Zhang D. Maternal embryonic leucine zipper kinase (MELK): a novel regulator in cell cycle control, embryonic development, and cancer. Int. J. Mol. Sci. 2013;14:21551–21560. doi: 10.3390/ijms141121551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meraldi P., Sorger P.K. A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J. 2005;24:1621–1633. doi: 10.1038/sj.emboj.7600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishida C., Reinhard P., Linn S. DNA repair synthesis in human fibroblasts requires DNA polymerase delta. J. Biol. Chem. 1988;263:501–510. [PubMed] [Google Scholar]
- 54.Perera D., Tilston V., Hopwood J.A., Barchi M., Boot-Handford R.P., Taylor S.S. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev. Cell. 2007;13:566–579. doi: 10.1016/j.devcel.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 55.Tang Z., Sun Y., Harley S.E., Zou H., Yu H. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc. Natl. Acad. Sci. U.S.A. 2004;101:18012–18017. doi: 10.1073/pnas.0408600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uchimura A., Hidaka Y., Hirabayashi T., Hirabayashi M., Yagi T. DNA polymerase delta is required for early mammalian embryogenesis. PLoS One. 2009;4:e4184. doi: 10.1371/journal.pone.0004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoon J.H., Prakash S., Prakash L. Requirement of Rad18 protein for replication through DNA lesions in mouse and human cells. Proc. Natl. Acad. Sci. U.S.A. 2012;109:7799–7804. doi: 10.1073/pnas.1204105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Z., Fan H.Y., Goldman J.A., Kingston R.E. Homology-driven chromatin remodeling by human RAD54. Nat. Struct. Mol. Biol. 2007;14:397–405. doi: 10.1038/nsmb1223. [DOI] [PubMed] [Google Scholar]
- 59.Song N., Zhu X., Shi L., An J., Wu Y., Sang J. Identification and functional analysis of a CDE/CHR element in the POLD1 promoter. Sci. China C. Life Sci. 2009;52:551–559. doi: 10.1007/s11427-009-0077-5. [DOI] [PubMed] [Google Scholar]
- 60.Verlinden L., Eelen G., Beullens I., Van C.M., Van H.P., Engelen K., Van H.R., Marchal K., De M.B., Foijer F., et al. Characterization of the condensin component Cnap1 and protein kinase Melk as novel E2F target genes down-regulated by 1, 25-dihydroxyvitamin D3. J. Biol. Chem. 2005;280:37319–37330. doi: 10.1074/jbc.M503587200. [DOI] [PubMed] [Google Scholar]
- 61.Adachi N., Lieber M.R. Bidirectional gene organization: a common architectural feature of the human genome. Cell. 2002;109:807–809. doi: 10.1016/s0092-8674(02)00758-4. [DOI] [PubMed] [Google Scholar]
- 62.Quaas M., Muller G.A., Engeland K. p53 can repress transcription of cell cycle genes through a p21(WAF1/CIP1)-dependent switch from MMB to DREAM protein complex binding at CHR promoter elements. Cell Cycle. 2012;11:4661–4672. doi: 10.4161/cc.22917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wen H., Andrejka L., Ashton J., Karess R., Lipsick J.S. Epigenetic regulation of gene expression by Drosophila Myb and E2F2-RBF via the Myb-MuvB/dREAM complex. Genes Dev. 2008;22:601–614. doi: 10.1101/gad.1626308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Georlette D., Ahn S., MacAlpine D.M., Cheung E., Lewis P.W., Beall E.L., Bell S.P., Speed T., Manak J.R., Botchan M.R. Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes Dev. 2007;21:2880–2896. doi: 10.1101/gad.1600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Littler D.R., Alvarez-Fernandez M., Stein A., Hibbert R.G., Heidebrecht T., Aloy P., Medema R.H., Perrakis A. Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence. Nucleic Acids Res. 2010;38:4527–4538. doi: 10.1093/nar/gkq194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Osterloh L., von E.B., Schmit F., Rein L., Hubner D., Samans B., Hauser S., Gaubatz S. The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. EMBO J. 2007;26:144–157. doi: 10.1038/sj.emboj.7601478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Conboy C.M., Spyrou C., Thorne N.P., Wade E.J., Barbosa-Morais N.L., Wilson M.D., Bhattacharjee A., Young R.A., Tavare S., Lees J.A., et al. Cell cycle genes are the evolutionarily conserved targets of the E2F4 transcription factor. PLoS One. 2007;2:e1061. doi: 10.1371/journal.pone.0001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flowers S., Beck G.R., Jr, Moran E. Tissue-specific gene targeting by the multiprotein mammalian DREAM complex. J. Biol. Chem. 2011;286:27867–27871. doi: 10.1074/jbc.C111.255091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee H., Ohno K., Voskoboynik Y., Ragusano L., Martinez A., Dimova D.K. Drosophila RB proteins repress differentiation-specific genes via two different mechanisms. Mol. Cell Biol. 2010;30:2563–2577. doi: 10.1128/MCB.01075-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee H., Ragusano L., Martinez A., Gill J., Dimova D.K. A dual role for the dREAM/MMB complex in the regulation of differentiation-specific E2F/RB target genes. Mol. Cell Biol. 2012;32:2110–2120. doi: 10.1128/MCB.06314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


