This article describes mapping and molecular analysis of the differential transcriptional activity of a MULE1 DNA transposon in two commonly used lab strains. The study is expanded to over 200 accessions to describe the overall polymorphism of this transposon in the Arabidopsis lineage. This study provides insight into natural variation of transposon silencing.
Abstract
Transposons are massively abundant in all eukaryotic genomes and are suppressed by epigenetic silencing. Transposon activity contributes to the evolution of species; however, it is unclear how much transposition-induced variation exists at a smaller scale and how transposons are targeted for silencing. Here, we exploited differential silencing of the AtMu1c transposon in the Arabidopsis thaliana accessions Columbia (Col) and Landsberg erecta (Ler). The difference persisted in hybrids and recombinant inbred lines and was mapped to a single expression quantitative trait locus within a 20-kb interval. In Ler only, this interval contained a previously unidentified copy of AtMu1c, which was inserted at the 3′ end of a protein-coding gene and showed features of expressed genes. By contrast, AtMu1c(Col) was intergenic and associated with heterochromatic features. Furthermore, we identified widespread natural AtMu1c transposition from the analysis of over 200 accessions, which was not evident from alignments to the reference genome. AtMu1c expression was highest for insertions within 3′ untranslated regions, suggesting that this location provides protection from silencing. Taken together, our results provide a species-wide view of the activity of one transposable element at unprecedented resolution, showing that AtMu1c transposed in the Arabidopsis lineage and that transposons can escape epigenetic silencing by inserting into specific genomic locations, such as the 3′ end of genes.
INTRODUCTION
Up to 90% of eukaryotic genomes consist of sequences derived from transposable elements (TEs), which were originally described as selfish DNA (Schnable et al., 2009; Tenaillon et al., 2010; Fedoroff, 2012). However, more recent findings indicate that they also benefit the host organisms through their function in genome organization and gene regulation (Levin and Moran, 2011; Tsuchiya and Eulgem, 2013). Importantly, they form a source of genetic variation that can be utilized by natural selection (Lisch, 2013a). The success of TEs is based on their ability to mobilize and transpose in the host genome, and this may be activated by stress or hybridization (McClintock, 1984; Biémont and Vieira, 2006; Lisch, 2013a). TEs are also an invaluable tool to generate insertional mutants for research and breeding (Lisch, 2012).
The class II TE Robertson’s Mutator element was originally isolated from maize (Zea mays), where it transposes frequently (Lisch, 2012). The autonomous element contains two homologous terminal inverted repeats (TIRs) and the mudrA and mudrB genes (Lisch, 2013b). mudrA encodes a highly conserved transposase; mudrB is much less conserved and contains a protein of unknown function, which may be important for autonomous transposition. Mutator elements from Arabidopsis (AtMu) always lack the mudrB gene (Singer et al., 2001). AtMu1 is targeted by several epigenetic silencing pathways, which converge to stably silence transcription (Singer et al., 2001; Lippman et al., 2003; Bäurle et al., 2007). They comprise mainly DNA methylation (requiring DECREASED DNA METHYLATION1 [DDM1] and METHYLTRANSFERASE1 [MET1]) and RNA-directed DNA methylation (RdDM); in RdDM, the generation of small RNA (sRNA) triggers the deposition of chromatin-silencing marks such as DNA methylation and histone H3K9 dimethylation at target loci (Rigal and Mathieu, 2011; Castel and Martienssen, 2013). While the current model accounts well for the maintenance of epigenetic silencing, we are only beginning to understand how de novo silencing is established (Panda and Slotkin, 2013).
Although possibly intertwined topics, the regulation of TE transposition receives much less attention than TE transcriptional suppression (Bucher et al., 2012). In Arabidopsis thaliana, transposition has been described for several class I and class II TEs, including AtMu1 in sensitized backgrounds with globally defective DNA methylation, such as ddm1 or met1 (Miura et al., 2001; Singer et al., 2001; Mirouze et al., 2009; Tsukahara et al., 2009). Natural transposition was also detected in the vegetative nucleus of pollen, where global reactivation of TEs occurs (Slotkin et al., 2009). However, as the vegetative nucleus does not contribute to the gametes, this transposition is not transmitted to the next generation (Slotkin et al., 2009). Thus, the extent of (natural) transposition that is transmitted to the next generation currently remains elusive.
Species-wide transposition activity in natural Arabidopsis populations has not been addressed so far. Compared with the outcrossing Arabidopsis lyrata, A. thaliana is relatively poor in TEs, with only 10% of genomic sequences derived from TEs and retroelements (Tenaillon et al., 2010; Hollister et al., 2011). This may be connected with the selfing life strategy and a higher activity of silencing pathways. Methylated, but not unmethylated, TEs have a negative effect on nearby gene expression (Hollister and Gaut, 2009), but it remains unclear how methylation levels are determined.
Using expression quantitative trait locus (eQTL) mapping, here we identify transposition as the cause for natural variation of AtMu1c silencing. We show that differential activity of the alleles is epigenetically stable through many generations and correlates with stable chromatin properties. While the silent AtMu1c(Col) displays characteristics of heterochromatin, the active AtMu1c(Ler) displays euchromatic features. Thus, AtMu1c(Ler) escapes epigenetic silencing, likely by inserting into a protective chromosomal environment. Our extensive analysis of the transposition of AtMu1c in the Arabidopsis lineage, based on genome sequences and transcriptome data of over 200 accessions, uncovers the extent of AtMu1c transposition and suggests that insertion location is a key determinant of AtMu1 silencing.
RESULTS
Characterization of Expressed AtMu1 Copies in Columbia and Landsberg erecta
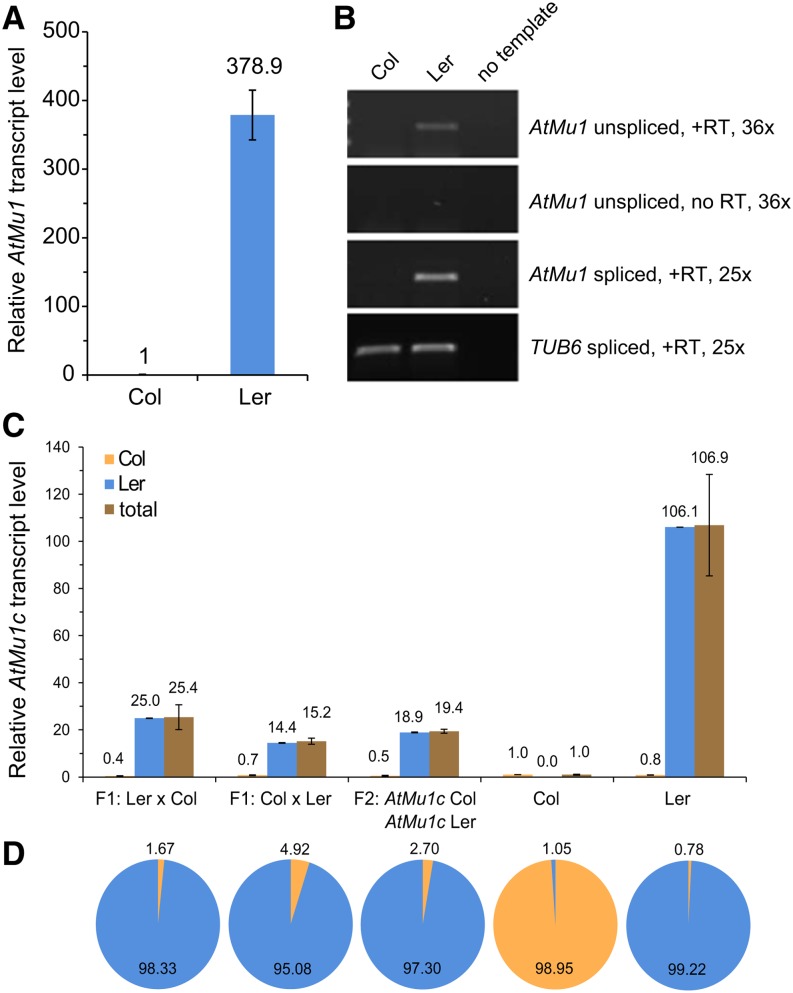
In agreement with previous reports (Singer et al., 2001; Slotkin et al., 2009), transcript levels of AtMu1 in Landsberg erecta (Ler) were two orders of magnitude higher than in Columbia (Col) (Figure 1A), suggesting a loss of silencing in Ler. Of note, Col transcript levels were still robustly detected by quantitative RT-PCR (qRT-PCR). To estimate whether silencing in Ler was reduced at the transcriptional or posttranscriptional level, we analyzed unspliced transcripts as a proxy for transcriptional activity (Bäurle et al., 2007; Le Masson et al., 2012; Stief et al., 2014). We detected unspliced AtMu1 transcripts in Ler but not in Col (Figure 1B), suggesting that a differential rate of transcription contributed at least in part to the observed difference in steady state transcript levels.
Figure 1.
AtMu1c Escapes Epigenetic Silencing in Ler but Not in Col.
(A) Relative AtMu1 transcript levels in Col and Ler as determined by qRT-PCR. Expression values (primers 44 and 45) were normalized to TUB6 and Col. Error bars are se of six biological replicates.
(B) RT-PCR of spliced and unspliced AtMu1 transcript levels. TUB6 served as a loading control. Representative gels of three biological replicates are shown.
(C) Total AtMu1c transcript levels were determined by qRT-PCR, and relative allele-specific transcript levels were calculated using the frequency data from (D). Error bars are se of three replicates. Total expression values were normalized to TUB6 and Col.
(D) Allele frequencies of AtMu1c(Col) and AtMu1c(Ler) in cDNA from parents and F1 and F2 hybrid progeny were determined by pyrosequencing (Col, orange; Ler, blue). The ∼1% false signal in the parents is a systemic technical error of the pyrosequencing assay. Data represent averages of three biological replicates.
Besides the commonly studied AtMu1a copy (At4g08680), two highly similar copies of AtMu1 are present in the Col genome (AtMu1b [At1g78095] and AtMu1c [At5g27345]). AtMu1a and AtMu1b show 99% sequence identity across the complete region (including TIRs), while AtMu1c is 87% identical to the two other copies (Supplemental Figure 1A) (Singer et al., 2001). All three copies can be detected by the primers used in Figures 1A and 1B. In Ler, AtMu1b is not present (Singer et al., 2001). The TIRs in each copy show 91 to 98% identity (Supplemental Table 1). To determine the origin of the AtMu1 transcripts in Col and Ler, we established an assay that distinguished the three copies based on the detection of two single-nucleotide polymorphisms (SNPs) (Supplemental Figure 1B). All 26 clones analyzed for each Col and Ler were derived from AtMu1c. Thus, in both accessions, the major expressed copy is AtMu1c, with AtMu1a and AtMu1b not or only weakly expressed.
AtMu1c(Col) Remains Epigenetically Silenced in Hybrids
Epigenetic silencing is stably inherited across generations, and thus AtMu1c(Col) could be expected to remain silent in Col × Ler hybrids. Conversely, AtMu1c(Ler) may become silenced if combined in a cell with AtMu1c(Col). Alternatively, it is possible that the silencing state of both alleles is determined by one or more trans-acting factor(s) whose activity differs between the two accessions; hence, silencing states may be equalized in hybrids. To distinguish between these possibilities, we measured AtMu1c transcript levels (using from here on AtMu1c-specific primers) in pools of F1 plants from reciprocal crosses between Ler and Col (Figure 1C). Transcript levels in F1 plants were intermediate (25.4 and 15.2) compared with the parental strains (Col, 1; Ler, 106.9). On the same material, allele frequencies were determined by pyrosequencing (Figure 1D), and the relative transcript level for each allele was calculated (Figure 1C). Pyrosequencing derives allele frequencies by the detection of photons emitted upon nucleotide incorporation. In F1 hybrids, the frequency of the Col allele–derived transcripts was 0.017 and 0.049, respectively. We also studied an F2 pool of plants genotyped for the presence of both AtMu1c haplotypes. We obtained similar results compared with the F1, with intermediate overall transcript level and a very low frequency of the Col allele–derived transcripts (19.4 relative transcript level; Col frequency of 0.027). Thus, the Col allele remained silenced in the hybrid F1 and F2 generations. We noted that the overall transcript levels in the F1 and F2 were consistently less than 50% of the Ler transcript levels, suggesting that there may either be an activating interaction between the two Ler alleles or a weak trans-silencing effect from the Col allele.
Mapping of eQTL-Mu1 and Interaction with AtMu1c
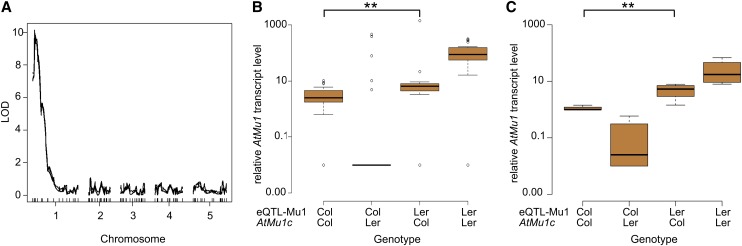
To determine the molecular basis for the differential silencing of AtMu1c in Col and Ler, we identified quantitative trait loci (QTLs) that affected AtMu1c expression. To this end, we quantified AtMu1 transcript levels in a Col × Ler recombinant inbred line (RIL) population (Lister and Dean, 1993) and performed eQTL analysis. A single eQTL was identified on chromosome 1 and named eQTL-Mu1 (Figure 2A). eQTL-Mu1 did not overlap with AtMu1b, and there was no eQTL associated with AtMu1c, suggesting that a single trans-acting eQTL was responsible for differential AtMu1c transcript levels. We mapped eQTL-Mu1 to a 20-kb interval between 2.86 and 2.88 Mb (Figure 3A).
Figure 2.
eQTL Mapping Identifies a Single eQTL for AtMu1 Expression, and the eQTL-Mu1(Ler) Promotes AtMu1 Expression.
(A) eQTL mapping identified one major QTL in Col × Ler RILs based on the analysis of 98 RILs. LOD, likelihood of difference score. The LOD significance threshold (5%, 1000 permutations) was 3.22.
(B) AtMu1 transcript levels in 98 RILs with the indicated genotypes at AtMu1c and eQTL-Mu1. **P < 0.005, by Wilcoxon rank-sum test.
(C) AtMu1c transcript levels in HIF lines with indicated genotypes at AtMu1c and eQTL-Mu1. F8 HIF individuals were genotyped and pooled for qRT-PCR analysis. Data were averaged over four different HIFs with three technical replicates each. **P < 0.005, by Student’s t test.
[See online article for color version of this figure.]
Figure 3.
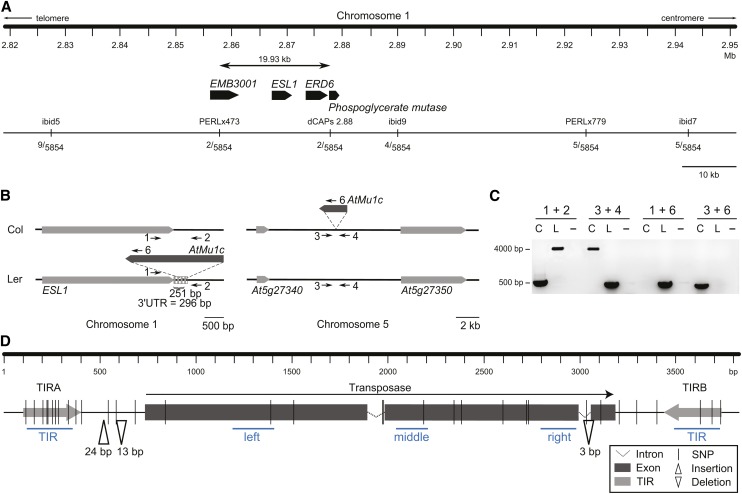
Fine-Mapping of eQTL-Mu1 Identifies a Novel Insertion of AtMu1c Underlying the eQTL.
(A) Fine-mapping of eQTL-Mu1. The number of recombinants for each marker from 5854 chromosomes is shown. The mapping interval contained four protein-coding genes (At1g08910 [EMB3001], At1g08920 [ESL1], At1g08930 [ERD6], and At1g08940). None of them had a nonsynonymous SNP between Col and Ler.
(B) Schematic representation of AtMu1c insertion positions in Col and Ler. Neighboring genes, light gray; 3′ UTR, dashed box; numbered arrows indicate the primers used in (C).
(C) Products of AtMu1c amplification with flanking primers (empty site, ∼450 bp; with AtMu1c insertion, ∼4000 bp). Primer positions are indicated in (B). C, Col; L, Ler.
(D) Schematic representation of genomic sequence differences between AtMu1c(Col) and AtMu1c(Ler). Sequences were determined by sequencing multiple independent PCR products. Blue lines represent amplicons assayed in Figure 4. Primer sequences can be found in Supplemental Table 3.
[See online article for color version of this figure.]
We next tested the interaction between AtMu1c and eQTL-Mu1 in the RILs (Figure 2B). Transcript levels were highest when AtMu1c and eQTL-Mu1 were both derived from Ler. Transcript levels were slightly lower for AtMu1c(Col) and eQTL-Mu1(Ler). In plants fixed for the Col allele at eQTL-Mu1, the direction of the effect of the AtMu1c genotype was reversed; AtMu1c transcript levels were hardly detectable when AtMu1c was Ler and slightly higher when it was Col. The effect could be recapitulated in a heterogeneous inbred family (HIF) that was homozygous for most of the genome and segregated for eQTL-Mu1 and for AtMu1c (Figure 2C). AtMu1c transcript levels were highest when both regions were homozygous Ler and lowest when AtMu1c was Ler and eQTL-Mu1 was Col. For both sets of lines and homozygous AtMu1c(Col), AtMu1c expression was significantly higher when eQTL-Mu1 was Ler compared with when it was Col. Thus, eQTL-Mu1 from Ler had a positive effect on AtMu1c transcription that persisted for many generations.
Sequence Analysis of eQTL-Mu1
For the four genes within the eQTL-Mu1 interval (Figure 3A), no changes in protein sequence or transcript levels were found between Col and Ler (Lempe et al., 2005; Schmitz et al., 2013). Thus, we explored the possibility of a new insertion of AtMu1c within this interval by screening Ler Illumina data for reads matching AtMu1c TIR sequences. Several reads suggested an insertion of AtMu1c within eQTL-Mu1 very close to the 3′ end of ERD (EARLY RESPONSE TO DEHYDRATION) SIX-LIKE1 (ESL1) at 2.87 Mb. We confirmed the insertion of a full-length copy of AtMu1c next to ESL1 in Ler but not in Col by PCR and sequencing (Figures 3B and 3C). The insertion was flanked by a typical 9-bp target site duplication (TSD). Furthermore, in Ler, we did not find a copy of AtMu1c at the Col site on chromosome 5. Together with the allele-specific transcript analysis (Figures 2B and 2C), these results demonstrate that this new copy caused the high AtMu1c transcription in Ler.
AtMu1c(Ler) was inserted within the annotated 3′ untranslated region (UTR) of the ESL1 gene (Figure 3B; after 251 bp of 296 bp), with the orientation of the transposase gene in the opposite direction to ESL1. We did not find evidence for altered ESL1 transcript levels in Ler compared with Col (Supplemental Figure 2; Lempe et al., 2005; Schmitz et al., 2013). Neither did we find evidence for read-through transcription using a primer in the coding region of ESL1 and one in the TIR of AtMu1c (primers 1 and 6; Figure 3B; Supplemental Figure 2).
To compare the sequences of AtMu1c(Col) and AtMu1c(Ler), both were amplified with flanking primers (primers 1/2 and 3/4, respectively) and sequenced. Thirty-three SNPs and three insertions/deletions were identified (Figure 3D). The 3- to 24-bp-long insertions/deletions were between the transposase and the TIR and within the intron, respectively. The SNP frequency was highest in the TIRs (13 of 581 bp, 2.24%) and lower in the transposase region (13 of 2413 bp, 0.54%). Eight of the 13 SNPs in the transposase region were nonsynonymous. In summary, AtMu1c is located at different positions in Col and Ler, suggesting that genomic location is responsible for differential silencing. Moreover, our results suggest that AtMu1c actively transposes in Arabidopsis.
AtMu1c(Col) and AtMu1c(Ler) Differ in Chromatin State and DNA Methylation
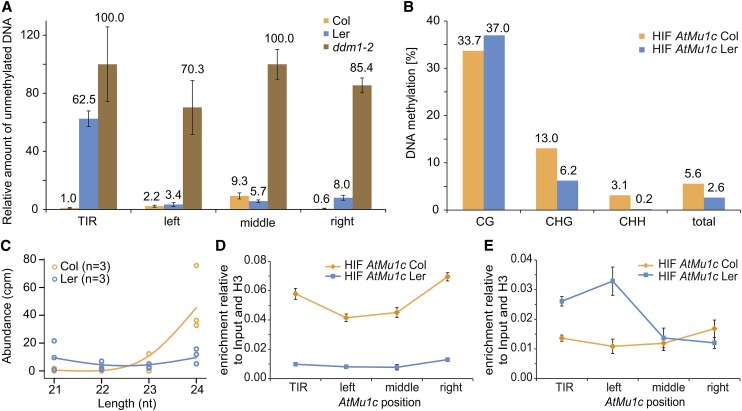
To test whether differential expression of AtMu1c(Col) and AtMu1c(Ler) correlated with different chromatin states, we analyzed DNA methylation by McrBC quantitative PCR (qPCR) across AtMu1c (for amplicon positions, see Figure 3D). Methylated DNA is cleaved by McrBC irrespective of the sequence context, and thus the amplification value is inversely correlated with DNA methylation. TIR methylation in Col was higher than in Ler, whereas the transposase region was highly methylated in both Col and Ler (Figure 4A). Methylation levels were very low in the hypomethylated ddm1-2, which was used as a control (Vongs et al., 1993). Previously, TIR methylation was correlated with Mutator element silencing (Raizada et al., 2001). Bisulfite sequencing of the TIR from HIFs containing AtMu1c from Col or Ler indicated that methylation in Ler was reduced in the CHG and CHH sequence contexts (Figure 4B; Supplemental Table 2). Strongly reduced CHG methylation in the TIR of Ler compared with Col was also found in a BS-seq data set (Supplemental Figure 3A) (Schmitz et al., 2013). CHG and CHH methylation is targeted by RdDM and requires sRNAs (Law and Jacobsen, 2010; Stroud et al., 2014). AtMu1 was previously described as a target of RdDM, and complementary sRNAs were observed (Lippman et al., 2003; Bäurle et al., 2007). Analyzing a next-generation sequencing data set (Li et al., 2012) from Col and Ler for sRNA of 21 to 24 nucleotides, we found high levels of AtMu1c-specific 24-nucleotide sRNA in Col but not in Ler (Figure 4C). RdDM also involves dimethylation of histone H3K9. Indeed, H3K9 dimethylation was enriched throughout AtMu1c(Col) but not AtMu1c(Ler) (Figure 4D).
Figure 4.
AtMu1c from Col, but Not Ler, Displays Hallmarks of Epigenetic Silencing.
(A) DNA methylation analysis of AtMu1c from Col and Ler by McrBC qPCR. Amplification values were normalized to mock-treated DNA. Four different regions of AtMu1c were investigated (cf. Figure 3D). Error bars are se of three replicates.
(B) DNA methylation analysis by bisulfite sequencing of AtMu1c TIR in an HIF segregating for eQTL-Mu1 and AtMu1c(Col). Percentages of DNA methylation were calculated from 23 independent clones each (cf. Supplemental Table 2).
(C) Abundance of 21- to 24-nucleotide sRNAs (Li et al., 2012) perfectly matching AtMu1c (including TIRs) from Ler and Col. Dots represent individual data points of three replicates.
(D) and (E) Histone H3K9 dimethylation (D) and H3K4 trimethylation (E) levels at AtMu1c(Col) and AtMu1c(Ler) in the HIF lines determined by chromatin immunoprecipitation. Data normalized to input and H3 occupancy were averaged over three biological replicates. Error bars represent se.
As a mark for active chromatin, we next assessed histone H3K4 trimethylation (H3K4me3), which correlates with transcription and peaks at the 5′ end of active genes (Shilatifard, 2012). AtMu1c(Ler) displayed a peak in H3K4me3 enrichment at the 5′ end of the transposase region (Figure 4E). No corresponding peak was observed in AtMu1c(Col). Thus, AtMu1c(Col) displayed hallmarks of transcriptional silencing, while AtMu1c(Ler) chromatin resembled actively transcribed genes.
Identification of 32 AtMu1c Insertion Sites from 217 Accessions
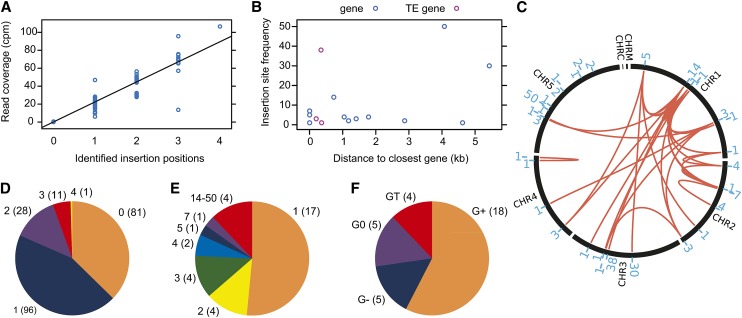
The analysis of eQTL-Mu1 indicated that AtMu1c has transposed in the Arabidopsis lineage. Therefore, we analyzed AtMu1c transposition at a species-wide level. In total, 32 novel AtMu1c insertion sites were identified from genomic next-generation sequencing data from 217 accessions (Schmitz et al., 2013; Figure 5; Supplemental Data Set 1). Three additional insertion sites were selected for experimental validation by PCR, and all were successfully confirmed, corroborating the accuracy of the computational predictions (Supplemental Data Set 1 and Supplemental Figure 4). The new insertion sites were distributed across the whole genome with no obvious insertion site preferences (Figure 5C). With the exception of Cal-0, no accession had more than three distinct insertions of AtMu1c (with 81 accessions lacking any detectable insertion), and there was a high correlation between the number of identified insertion positions and read coverage (Figures 5A and 5D). Each insertion site was present in up to 50 accessions (Figure 5C; Supplemental Figure 5 and Supplemental Data Set 1). The three most frequent insertions were present in 14 to 23% of accessions, and 17 insertion sites were represented only by a single accession (Figures 5C and 5E). In total, 82% of insertions had a perfect 9-bp TSD (Supplemental Data Set 1). We next characterized the genomic environment of the insertion sites relative to the neighboring gene. Four insertions were within 2 kb of a TE gene (Figure 5F, GT). Five insertions were within an annotated gene (G0), with four being in the 3′ UTR and one in an intron. None of the G0 insertions disrupted the coding sequence of the “host” gene, which may be a result of natural purifying selection or insertion preference. Eighteen insertions were outside an annotated gene but within 2 kb of it (G+). Five insertions were more than 2 kb away from the next annotated gene (G−). The frequent insertions tended to be farther away from genes or close to TE genes (Figure 5B). A phylogenetic tree of AtMu1c sequences from single-insertion accessions clustered AtMu1c copies according to their insertion sites, indicating clonal origin (Supplemental Figure 6). Thus, AtMu1c actively transposed during the natural history of Arabidopsis. The observed mix-and-match pattern suggests that transposition preceded substantial hybridization (Figure 5C; Supplemental Figure 5). We next tested how the expression of neighboring genes was affected with the distance of the AtMu1c insertion by determining the transcript level ratio of this gene in accessions with a given insertion and accessions lacking it. For G0 and G+, there was no obvious effect (Figure 6A). Insertions farther than 2 kb from a gene appeared to have a negative effect on its transcript levels.
Figure 5.
Evidence for AtMu1c Transposition in the Arabidopsis Lineage and Characterization of Novel Insertion Sites from the Analysis of 217 Accessions.
(A) Correlation of Illumina read coverage and number of identified AtMu1c insertions.
(B) Insertion site frequency (number of accessions where this insertion was present) depending on the distance to the neighboring gene/TE gene.
(C) Number and distribution of AtMu1c insertion sites across chromosomes. The red lines connect insertions that occurred together in individual accessions. The blue numbers indicate the number of accessions where this particular insertion was found. For details, see Supplemental Data Set 1.
(D) Distribution of AtMu1c copy numbers in the accessions analyzed; numbers in parentheses are number of accessions.
(E) Classification of insertion sites according to their frequency; numbers in parentheses are number of insertions.
(F) Classification of insertion sites according to their distance from the next neighboring gene/TE; numbers in parentheses are number of insertions. G0, insertion site within an annotated protein-coding gene; G+, insertion site outside but within 2.0 kb of a protein-coding gene; G−, insertion site more than 2.0 kb away from a protein-coding gene; GT, insertion site within or next to a TE gene.
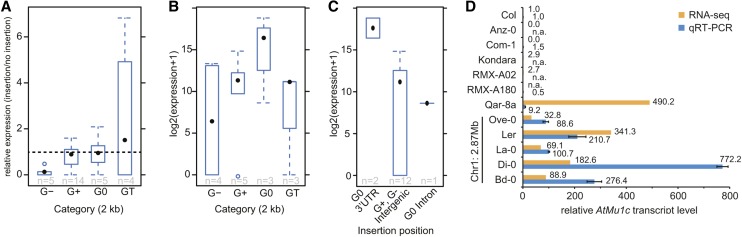
Figure 6.
Novel AtMu1c Insertions Indicate a Correlation between Insertion Site and Transcript Level.
(A) Box plot illustrating neighboring gene transcript levels. The ratio of neighboring gene transcript levels of accessions with the insertion divided by all other accessions is displayed. For GT, transcript levels of the neighboring TE gene were calculated. G0, insertion site within an annotated protein-coding gene; G+, insertion site outside but within 2.0 kb of a protein-coding gene; G−, insertion site more than 2.0 kb away from a protein-coding gene; GT, insertion site within or next to a TE gene.
(B) and (C) Box plots for AtMu1c transcript levels for different categories of insertion sites. Expression data from accessions with only one AtMu1c insertion were used. Expression values for accessions with the same insertion site were averaged to avoid weighting of insertion frequencies.
(D) AtMu1c transcript levels in selected accessions determined by qRT-PCR and RNA-seq (Schmitz et al., 2013). All accessions analyzed have only one AtMu1c insertion. Data were normalized to Col. Error bars are se of three biological replicates. n.a., not available. Values represent numbers of AtMu1c insertions analyzed.
AtMu1c Insertion in 3′ UTRs Correlates with High Expression
If the difference in transcript levels between AtMu1c(Col) and AtMu1c(Ler) was caused by the proximity of a protein-coding gene, there might be a negative correlation between AtMu1c transcription and distance to the neighboring gene. We tested this by analyzing RNA sequencing (RNA-seq) data from those accessions with a single AtMu1c insertion (Schmitz et al., 2013). Average AtMu1c transcript levels for each insertion site were grouped according to the distance from the neighboring gene/TE (Figure 6B). G0 accessions showed a tendency toward higher average AtMu1c transcript levels compared with the other groups. We classified G0 accessions further according to the position of the insertion and found that the highest expressing AtMu1c copies were located within 3′ UTRs (Figure 6C). Thus, it appears that genomic location is an important determinant for AtMu1c activity. We did not detect evidence for read-through transcription in the Ler group or in Qar-8a (Supplemental Figure 2). To test the possibility of sequence effects, we analyzed AtMu1c transcript levels in several accessions with only one insertion that represented either the highest expressing insertion sites (Ler group and Qar-8a) or insertion sites that cluster together with Ler and Qar-8a based on their sequence (Supplemental Figure 6) but that are G+ or G− accessions (RMX-A180 and Com-1). We found high transcript levels only in the Ler group and Qar-8a but not in RMX-A180 or Com-1 (Figure 6D). Three additional accessions (Anz-0, Kondara, and RMX-A02) were not tested experimentally but had low expression based on the RNA-seq data (Schmitz et al., 2013). Together, these results indicate that insertion position and not sequence variation is a major determinant for the escape from silencing.
We next asked whether there was a global correlation between AtMu1c expression and DNA methylation. To this end, we performed a global methylome analysis based on available BS-seq data (Schmitz et al., 2013). Average AtMu1c methylation levels for each insertion site were grouped according to the distance from the neighboring gene/TE as was done for the global expression analysis (Figure 6B; Supplemental Figure 3B). This revealed a tendency for lower CG and CHG methylation for TIRA but not the transposase region in G0 accessions, which showed overall higher AtMu1c expression. This trend was also observed when CG and CHG methylation levels from all accessions with a single insertion and detectable AtMu1c expression were plotted against AtMu1c expression (Supplemental Figure 3C). Thus, our analysis detects a trend that AtMu1c expression is negatively correlated with CG and CHG methylation in TIRA, while there is no such correlation with methylation levels in the transposase region.
DISCUSSION
The strong difference in transcription of AtMu1 in Col and Ler was reported previously and was suggested to result in differential transposition (Singer et al., 2001; Slotkin et al., 2009). However, the cause of this difference remained unknown. Natural variation in trans-acting factors was previously identified as a determinant of silencing state (Woo et al., 2007). Here, we mapped eQTL-Mu1 to a 20-kb interval containing a novel insertion of AtMu1c in Ler but not in Col. Our findings indicate that AtMu1c(Ler) has high transcript levels through escape from silencing. The most straightforward explanation for this is the location of AtMu1c(Ler) immediately downstream of the 3′ UTR of the ESL1 gene. This is in contrast to the location of AtMu1c(Col) within a 10-kb region without any protein-coding gene and containing heterochromatic features (Roudier et al., 2011). To corroborate our hypothesis, we performed a population-wide analysis of AtMu1c transposition. The other AtMu1c copy with very high transcript levels was also inserted into the 3′ UTR of a protein-coding gene. Further AtMu1c copies, which had very high sequence similarity with those two copies but were located in intergenic regions, did not show high transcript levels.
In Col, there are three closely related copies of AtMu1. While the silencing of AtMu1a through DNA methylation and RdDM pathways was studied extensively (Singer et al., 2001; Lippman et al., 2003), we identified AtMu1c as the only copy with detectable transcriptional activity in the wild type. This suggests that, despite the high sequence identity (87%), the degree of silencing differs between individual AtMu1 copies. Interestingly, there was no trans-silencing between AtMu1a and AtMu1c. As AtMu1c transcript levels in the F1 hybrids were less than 50% of the Ler levels, some trans-silencing between AtMu1c(Col) and AtMu1c(Ler) may occur, which, however, is not sufficient to fully silence AtMu1c(Ler) or trigger stable silencing. This suggests that the action of AtMu1c sRNAs is largely restricted to their site of production or that an antisilencing effect caused by transcription of the ESL1 locus can stably overcome RdDM. Conversely, AtMu1c(Col) remains epigenetically silenced even after many generations in the presence of AtMu1c(Ler).
We identified 32 new AtMu1c insertions from the analysis of 217 accessions. Interestingly, no accession contained more than four insertions, and one-third of the accessions had no detectable copy. The insertion sites were distributed in a mix-and-match pattern that suggested either that they were generated in one or very few individuals and then dispersed by outcrossing or that they were all generated in separate individuals that were later subject to substantial hybridization. The two alternatives have different implications for the likely cause of AtMu1c activation: the first explanation favors an event limited to one individual (such as hybridization or spontaneous mutation), while the second favors an event that affects many plants within a population similarly (such as environmental stress). At any rate, most of the insertions presumably are relatively old. Their distribution pattern is consistent with a transient phase of active transposition caused by a “genomic shock” that was potentially triggered by hybridization or extreme environmental conditions (McClintock, 1984). Of note, none of the identified AtMu1c-like sequences from A. lyrata were syntenic orthologs with the known Arabidopsis copies.
The transcript levels of AtMu1c from single-insertion copies were highest for the two insertion sites within a 3′ UTR (Ler group and Qar-8a) that could be analyzed. Another two insertion sites within a 3′ UTR could not be analyzed because the corresponding accessions had additional copies of AtMu1c. Beyond the 3′ UTR, position effects decayed rapidly. It remains an interesting question for future study to examine whether AtMu1c itself requires a certain genomic environment in order to attract silencing or whether AtMu1c induces silencing by default and is actively protected from silencing by certain locations (such as 3′ UTRs). We favor this latter hypothesis, as none of the other insertion sites that could be analyzed in single-copy accessions had consistently similarly high AtMu1c transcript levels as Qar-8a and the Ler group. Also, it is unlikely that all of them were inserted by chance into a genomic environment that promotes silencing. In addition, TE insertions in promoter regions have been implicated in the silencing of nearby genes, while no such example is known for insertions into 3′ regions (Kinoshita et al., 2007; Martin et al., 2009; Lisch, 2013a).
The distribution of AtMu1c insertion sites did not indicate any obvious preference for certain genomic regions. However, there was a striking absence of insertions within 5′ UTRs and exons. This may suggest that 5′ UTR insertions have a more deleterious effect on gene expression (and thus may have been eradicated through natural selection). Maize Mu inserts preferentially into the 5′ end of genes with open chromatin conformation (Liu et al., 2009). It remains to be seen whether AtMu1c displays a similar preference of inserting into the 5′ end of genes with open chromatin conformation. Consistent with our findings, a negative effect of methylated TEs on nearby gene expression, but not of unmethylated TEs, was reported (Hollister and Gaut, 2009). It is unclear how de novo silencing of a TE is triggered; however, most models invoke the production of long double-stranded RNAs (Marí-Ordóñez et al., 2013; Nuthikattu et al., 2013), which may be more frequent in gene-poor and TE-rich regions, caused by spurious transcription from neighboring TEs.
What is the significance of high AtMu1c transcription in Ler? One new germinal transposition event of AtMu1 was reported from Ler but none from Col (Singer et al., 2001), suggesting that higher AtMu1c transcription in Ler may result in increased transposition. AtMu1 transcript levels are highest in the vegetative nucleus of pollen, and transposition has been detected in Col pollen (Slotkin et al., 2009). An attractive hypothesis is that a nonsilenced TE next to a protein-coding gene could act as a latent source of phenotypic variation that becomes effective only after some internal or external cue triggers silencing of the TE and, subsequently, the neighboring gene.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana plants were grown in long-day conditions on soil in a greenhouse or on GM plates (1% [w/v] Glc) at 23°C day/21°C night cycles for 10 to 14 d before analysis. The Col × Ler RILs (Lister and Dean, 1993) and accession stocks were obtained from the European Arabidopsis Stock Centre. ddm1-2 in Col has been described (Vongs et al., 1993).
eQTL-Mu1 Mapping
eQTL mapping was performed using expectation-maximization, multiple imputation, and Haley-Knott regression interval mapping algorithms as implemented in R (Broman and Sen, 2009). For fine-mapping, we screened 5854 chromosomes from segregating populations for recombination between flanking markers and phenotyped the progeny of recombinants. Pools of progeny plants that were genotyped as homozygous Col or Ler at the segregating marker were generated and their AtMu1c transcript levels determined. The HIF was generated by selecting a plant with two recombination break points flanking either side of eQTL-Mu1 and repeated selfing. The segregating interval was flanked by ibid5 (2.827 Mb) (Figure 3A) and AC003114-0979 (2.965 Mb) and segregates for the eQTL region in between and for the AtMu1c region on chromosome 5. F8 plants of the desired genotypes were used for analyses.
Gene Expression Analysis
RNA was extracted from 10- to 14-d-old seedlings using a hot-phenol RNA extraction protocol, and DNase-treated RNA was reverse-transcribed and subjected to qRT-PCR as described (Stief et al., 2014). Primer sequences are listed in Supplemental Table 3. The pyrosequencing assay was designed with PyroMark Assay Design 2.0 (Qiagen) and performed on a PyroMark Q24 instrument, with manually optimized dispensation order (5′-ATTCTGATATCGTAGCACT-3′). Data were analyzed using the PyroMark Q24 software. For the cleaved-amplified polymorphic sequence marker–based assay, AtMu1 transcripts were amplified from oligo(dT)-primed cDNA using primer 496 and 497, gel-purified, and cloned into pGEM-T Easy (Promega). Inserts were reamplified from 26 individual clones and digested with MspI or DdeI.
DNA Methylation and Chromatin Immunoprecipitation Analysis
To determine DNA methylation levels through qPCR of McrBC-digested DNA, 100 ng of CTAB-extracted DNA was incubated for 1 h with 15 units of McrBC (New England Biolabs). After inactivation, qPCR was performed using 2 ng of digested DNA per reaction. Bisulfite sequencing was performed as described (Bäurle et al., 2007), and the success of conversion was confirmed (Foerster and Mittelsten Scheid, 2010). Twenty-six individual clones per genotype were analyzed with CyMATE (Hetzl et al., 2007).
Chromatin was extracted from 7-d-old seedlings as described (Bastow et al., 2004) and sheared with a Bioruptor (Diagenode). Chromatin immunoprecipitation was performed as described (Kaufmann et al., 2010) using anti-histone H3K4 trimethyl (Abcam; ab8580), anti-histone H3 (ab1791), or anti–histone H3K9 dimethyl (Wako; 302-32369) antibodies.
Computational Analyses: Insertion Site Identification
Illumina 100-bp paired-end genome sequencing data for 217 accessions (Schmitz et al., 2013) were downloaded from the National Center for Biotechnology Information (NCBI). Additional Ler sequencing data were obtained from M. Lenhard (Universität Potsdam). Reads were trimmed to 51 bp to avoid overlapping ends using Trimmomatic (Bolger et al., 2014) and mapped to the Arabidopsis TAIR10 reference genome using bwa mem (Li, 2013). Discordant read pairs with at least one pair mapping to the TAIR10 AtMu1c region were filtered using samtools (Li and Durbin, 2009). New insertion sites were considered if they were characterized by (1) multiple read pairs coming from both ends of the reference region and (2) multiple read pairs pointing toward an insertion position from both sides. Spanning read pairs over the reference region identified deletions at this position. Identified insertion positions were checked visually using IGV (Thorvaldsdóttir et al., 2013). Junction sequences were manually assembled after combining untrimmed read pairs using FLASH (Magoč and Salzberg, 2011). Exact insertion positions were read out after assembly and corresponded to the start and end of the TSD. In the case of one degenerated end, the TAIR10 reference was used to identify the TSD. Insertion sites were classified based on the distance to the closest gene (G−, >2 kb; G+, <2 kb; G0, within the gene; GT, within or close to another transposon) or the region type (3′ UTR, intergenic, or intron) using TAIR10 annotations. Insertion sites were illustrated in a Circos plot (Krzywinski et al., 2009). Statistics and illustrations were done using R.
Phylogenetic Analyses
Consensus sequences of the whole AtMu1c region (chromosome 5, 9,640,955 to 9,644,581) for accessions with a single insertion site were called using GATK after samtools mpileup variant calling (McKenna et al., 2010). Sequences were multiple sequence aligned using MUSCLE (Edgar, 2004), and alignments are provided in Supplemental Data Sets 2 and 3. A maximum likelihood tree was generated using PHYLIP 3.6 dnaml (Felsenstein, 1989) and visualized using the R package ape (Paradis et al., 2004).
Gene Expression and Methylation Analyses
Processed RNA-seq expression data for 144 accessions (Schmitz et al., 2013) were downloaded from the NCBI. Processed BS-seq data (Schmitz et al., 2013) were also downloaded. Methylations were calculated as the coverage count indicating methylated sites divided by the total coverage at these sites. Statistics and illustrations were done using R.
sRNA Analysis
sRNA sequencing reads (Li et al., 2012) were downloadad from the NCBI. Reads were aligned using bwa aln (Li and Durbin, 2009) after adaptor removal using Trimmomatic (Bolger et al., 2014). All reads between 21 and 24 nucleotides in length and mapping to the whole AtMu1c region were counted and normalized to the total number of reads of the same size range. Statistics and illustrations were done using R.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AtMu1a (At4g08680), AtMu1b (At1g78095), AtMu1c (At5g27345), and ESL1 (At1g08920).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. AtMu1 Phylogeny in Col and Transcript Discrimination Assay.
Supplemental Figure 2. Absence of Evidence for Read-Through Transcription from the Adjacent Gene into AtMu1c Inserted into Annotated 3′ UTRs.
Supplemental Figure 3. Global DNA Methylation Analysis of AtMu1c Based on BS-seq Methylome Data.
Supplemental Figure 4. Experimental Validation of Novel AtMu1c Insertion Sites.
Supplemental Figure 5. Mosaic Table of AtMu1c Insertion Sites in Accessions.
Supplemental Figure 6. Phylogenetic Clustering of AtMu1c from Accessions with Single AtMu1c Copies Indicates Clustering of Accessions with Insertion Position.
Supplemental Table 1. Comparison of AtMu1 TIR Sequences.
Supplemental Table 2. Details of Bisulfite Sequencing of AtMu1c Presented in Figure 4B.
Supplemental Table 3. List of Primers Used in This Study.
Supplemental Data Set 1. AtMu1c Insertion Sites and Their Characterization.
Supplemental Data Set 2. Text File of Alignment Corresponding to the Phylogenetic Analysis in Supplemental Figure 1A.
Supplemental Data Set 3. Text File of Alignment Corresponding to the Phylogenetic Analysis in Supplemental Figure 6.
Supplementary Material
Acknowledgments
We thank the European Arabidopsis Stock Centre for seeds. We thank M. Lenhard for sharing Ler sequencing data and M. Lenhard and A. Sicard for help with QTL analysis. We thank V. Ketmaier, M. Lenhard, and members of our laboratory for helpful comments. I.B. was supported by a Royal Society University Research Fellowship, a Sofja-Kovalevskaja Award from the Alexander-von-Humboldt Foundation, the Deutsche Forschungsgemeinschaft (Grant SFB 973, Project A02), and the John Innes Centre.
AUTHOR CONTRIBUTIONS
I.B. conceived research. T.K., C.K., C.N., and I.B. designed and analyzed research. All authors performed research. T.K. and I.B. wrote the article with input from all authors.
Glossary
- TE
transposable element
- TIR
terminal inverted repeat
- RdDM
RNA-directed DNA methylation
- sRNA
small RNA
- eQTL
expression quantitative trait locus
- Ler
Landsberg erecta
- Col
Columbia
- qRT-PCR
quantitative RT-PCR
- SNP
single-nucleotide polymorphism
- RIL
recombinant inbred line
- HIF
heterogeneous inbred family
- TSD
target site duplication
- UTR
untranslated region
- qPCR
quantitative PCR
- H3K4me3
histone H3K4 trimethylation
- RNA-seq
RNA sequencing
- NCBI
National Center for Biotechnology Information
- QTL
quantitative trait locus
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Bastow R., Mylne J.S., Lister C., Lippman Z., Martienssen R.A., Dean C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167. [DOI] [PubMed] [Google Scholar]
- Bäurle I., Smith L., Baulcombe D.C., Dean C. (2007). Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science 318: 109–112. [DOI] [PubMed] [Google Scholar]
- Biémont C., Vieira C. (2006). Genetics: Junk DNA as an evolutionary force. Nature 443: 521–524. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman, K.W., and Sen, S. (2009). A Guide to QTL Mapping with R/qtl. (New York: Springer), pp. 75–236. [Google Scholar]
- Bucher E., Reinders J., Mirouze M. (2012). Epigenetic control of transposon transcription and mobility in Arabidopsis. Curr. Opin. Plant Biol. 15: 503–510. [DOI] [PubMed] [Google Scholar]
- Castel S.E., Martienssen R.A. (2013). RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14: 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N.V. (2012). Presidential address. Transposable elements, epigenetics, and genome evolution. Science 338: 758–767. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1989). PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5: 164–166. [Google Scholar]
- Foerster A.M., Mittelsten Scheid O. (2010). Analysis of DNA methylation in plants by bisulfite sequencing. Methods Mol. Biol. 631: 1–11. [DOI] [PubMed] [Google Scholar]
- Hetzl J., Foerster A.M., Raidl G., Mittelsten Scheid O. (2007). CyMATE: A new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 51: 526–536. [DOI] [PubMed] [Google Scholar]
- Hollister J.D., Gaut B.S. (2009). Epigenetic silencing of transposable elements: A trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 19: 1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister J.D., Smith L.M., Guo Y.L., Ott F., Weigel D., Gaut B.S. (2011). Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc. Natl. Acad. Sci. USA 108: 2322–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Østerås M., Farinelli L., Krajewski P., Angenent G.C. (2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5: 457–472. [DOI] [PubMed] [Google Scholar]
- Kinoshita Y., Saze H., Kinoshita T., Miura A., Soppe W.J., Koornneef M., Kakutani T. (2007). Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J. 49: 38–45. [DOI] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Masson I., Jauvion V., Bouteiller N., Rivard M., Elmayan T., Vaucheret H. (2012). Mutations in the Arabidopsis H3K4me2/3 demethylase JMJ14 suppress posttranscriptional gene silencing by decreasing transgene transcription. Plant Cell 24: 3603–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempe J., Balasubramanian S., Sureshkumar S., Singh A., Schmid M., Weigel D. (2005). Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 1: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin H.L., Moran J.V. (2011). Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 12: 615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. (May 26, 2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Arvix (online), /arXiv/1303.3997v2.
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Varala K., Moose S.P., Hudson M.E. (2012). The inheritance pattern of 24 nt siRNA clusters in Arabidopsis hybrids is influenced by proximity to transposable elements. PLoS ONE 7: e47043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z., May B., Yordan C., Singer T., Martienssen R. (2003). Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 1: E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. (2012). Regulation of transposable elements in maize. Curr. Opin. Plant Biol. 15: 511–516. [DOI] [PubMed] [Google Scholar]
- Lisch D. (2013a). How important are transposons for plant evolution? Nat. Rev. Genet. 14: 49–61. [DOI] [PubMed] [Google Scholar]
- Lisch D. (2013b). Regulation of the Mutator system of transposons in maize. Methods Mol. Biol. 1057: 123–142. [DOI] [PubMed] [Google Scholar]
- Lister C., Dean C. (1993). Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 4: 745–750. [DOI] [PubMed] [Google Scholar]
- Liu S., Yeh C.T., Ji T., Ying K., Wu H., Tang H.M., Fu Y., Nettleton D., Schnable P.S. (2009). Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet. 5: e1000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T., Salzberg S.L. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí-Ordóñez A., Marchais A., Etcheverry M., Martin A., Colot V., Voinnet O. (2013). Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 45: 1029–1039. [DOI] [PubMed] [Google Scholar]
- Martin A., Troadec C., Boualem A., Rajab M., Fernandez R., Morin H., Pitrat M., Dogimont C., Bendahmane A. (2009). A transposon-induced epigenetic change leads to sex determination in melon. Nature 461: 1135–1138. [DOI] [PubMed] [Google Scholar]
- McClintock B. (1984). The significance of responses of the genome to challenge. Science 226: 792–801. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. (2010). The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M., Reinders J., Bucher E., Nishimura T., Schneeberger K., Ossowski S., Cao J., Weigel D., Paszkowski J., Mathieu O. (2009). Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461: 427–430. [DOI] [PubMed] [Google Scholar]
- Miura A., Yonebayashi S., Watanabe K., Toyama T., Shimada H., Kakutani T. (2001). Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411: 212–214. [DOI] [PubMed] [Google Scholar]
- Nuthikattu S., McCue A.D., Panda K., Fultz D., DeFraia C., Thomas E.N., Slotkin R.K. (2013). The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162: 116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda K., Slotkin R.K. (2013). Proposed mechanism for the initiation of transposable element silencing by the RDR6-directed DNA methylation pathway. Plant Signal. Behav. 8: e25206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K. (2004). APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Raizada M.N., Benito M.I., Walbot V. (2001). The MuDR transposon terminal inverted repeat contains a complex plant promoter directing distinct somatic and germinal programs. Plant J. 25: 79–91. [DOI] [PubMed] [Google Scholar]
- Rigal M., Mathieu O. (2011). A “mille-feuille” of silencing: Epigenetic control of transposable elements. Biochim. Biophys. Acta 1809: 452–458. [DOI] [PubMed] [Google Scholar]
- Roudier F., et al. (2011). Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30: 1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R.J., Schultz M.D., Urich M.A., Nery J.R., Pelizzola M., Libiger O., Alix A., McCosh R.B., Chen H., Schork N.J., Ecker J.R. (2013). Patterns of population epigenomic diversity. Nature 495: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. (2012). The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81: 65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T., Yordan C., Martienssen R.A. (2001). Robertson’s Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev. 15: 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzić M., Becker J.D., Feijó J.A., Martienssen R.A. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stief A., Altmann S., Hoffmann K., Pant B.D., Scheible W.-R., Bäurle I. (2014). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26: 1792–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Do T., Du J., Zhong X., Feng S., Johnson L., Patel D.J., Jacobsen S.E. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon M.I., Hollister J.D., Gaut B.S. (2010). A triptych of the evolution of plant transposable elements. Trends Plant Sci. 15: 471–478. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. (2013). Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T., Eulgem T. (2013). An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc. Natl. Acad. Sci. USA 110: E3535–E3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara S., Kobayashi A., Kawabe A., Mathieu O., Miura A., Kakutani T. (2009). Bursts of retrotransposition reproduced in Arabidopsis. Nature 461: 423–426. [DOI] [PubMed] [Google Scholar]
- Vongs A., Kakutani T., Martienssen R.A., Richards E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928. [DOI] [PubMed] [Google Scholar]
- Woo H.R., Pontes O., Pikaard C.S., Richards E.J. (2007). VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 21: 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.